SPAR1/RTEL1 maintains genomic stability by suppressing homologous recombination (original) (raw)
. Author manuscript; available in PMC: 2013 Jul 29.
SUMMARY
Inappropriate homologous recombination (HR) can cause gross chromosomal rearrangements that in mammalian cells may lead to tumorigenesis. In yeast, the Srs2 protein is an anti-recombinase that eliminates inappropriate recombination events, but the functional equivalent of Srs2 in higher eukaryotes has proven to be elusive. In this work, we identify C. elegans SPAR-1 as a functional analogue of Srs2 and describe its vertebrate counterpart, SPAR1/RTEL1, which is required for genome stability and tumour avoidance. We find that spar-1 mutant worms and SPAR1 knockdown human cells share characteristic phenotypes with yeast srs2 mutants, including inviability upon deletion of the sgs1/BLM homologue, hyper-recombination, and DNA damage sensitivity. In vitro, purified human SPAR1 antagonises HR by promoting the disassembly of D loop recombination intermediates in a reaction dependent upon ATP hydrolysis. We propose that loss of HR control following deregulation of SPAR1/RTEL1 may be a critical event that drives genome instability and cancer.
INTRODUCTION
Homologous recombination (HR) is an essential conserved process for dividing cells. In mitosis HR is required not only for the accurate repair of DNA double-strand breaks (DSBs), but also for the restart of stalled replication forks. Furthermore HR is crucial for meiotic DSB repair, which is required for accurate chromosome segregation at the first meiotic division. However inappropriate HR can give rise to genome instability and cancer as a result of erroneous chromosomal rearrangements and the persistence of intermediate recombination structures that cannot be resolved. Hence HR must be tightly regulated and temporally coordinated with cell cycle progression and replication.
Current models of eukaryotic HR (Krogh and Symington, 2004) propose that a DSB is resected to produce 3′-single stranded DNA tails that are bound by the DNA strand exchange protein RAD51 to form a nucleoprotein filament. These filaments are the catalyst for strand invasion into homologous duplex DNA, resulting in the formation of a D loop structure. The invading 3′ end provides a primer for DNA synthesis and D loop extension, which can be resolved either through displacement of the invading strand from the D loop and annealing to the other DSB end (synthesis-dependent strand annealing), or by the capture of the other resected end by the extruded strand of the D loop to form a double Holliday junction (dHJ). HR can be completed by endonucleolytic cleavage of the two HJs, which may result in a crossover.
In yeast, the initiation of strand invasion is antagonised by Srs2 to ensure that HR occurs at the appropriate time and place. Srs2, first identified 30 years ago (Lawrence and Christensen, 1979), is a 3′-5′ SF1 helicase related both by sequence and function to bacterial UvrD (Aboussekhra et al., 1989). S. cerevisiae srs2 and E. coli uvrD mutants exhibit elevated rates of spontaneous recombination (Aguilera and Klein, 1988; Arthur and Lloyd, 1980; Bierne et al., 1997; Zieg et al., 1978). Yeast srs2 mutants are synthetic lethal with deletion of the yeast RecQ helicase, sgs1 (Lee et al., 1999; Wang et al., 2001). It was subsequently found that the inviability of srs2 sgs1 mutants results from the accumulation of toxic HR intermediates, as viability can be restored by loss of RAD51 or RAD54, which are essential for the formation of the nucleoprotein filament and extension of the invading strand (Gangloff et al., 2000; Klein, 2001). Loss of srs2 also results in significant sensitivity to a range of DNA damaging agents, including IR and bleomycin that directly cause DSBs (Bennett et al., 2001), and ultraviolet radiation (UV), camptothecin and DNA interstrand cross-links (ICLs) that lead to replication-blocking lesions (Aboussekhra et al., 1992; Birrell et al., 2002). Biochemical studies have shown that both UvrD and Srs2 act to inhibit strand exchange by disrupting RecA/Rad51 filaments (Krejci et al., 2003; Morel et al., 1993; Veaute et al., 2003). This has led to the model that UvrD and Srs2 negatively regulate HR by disassembling the nucleoprotein filament.
Sequence homologues of SRS2 are not apparent in the genomes of higher eukaryotes. It has therefore been proposed that other helicases act in combination to substitute for Srs2 in order to negatively regulate HR and ensure genome stability. The function of an Srs2-related DNA helicase, Fbh1, overlaps with Srs2 in the processing of recombination intermediates in Schizosaccharomyces pombe (Morishita et al., 2005; Osman et al., 2005), and expression of human FBH1 is able to rescue some recombination defects in yeast srs2 mutants (Chiolo et al., 2007). Fbh1 is not conserved in budding yeast, C. elegans, Drosophila, or Arabidopsis. Although orthologues are found in humans, mice, and chicken (Kim et al., 2002; Kohzaki et al., 2007), Fbh1 deletion mutants are viable and exhibit only a mild phenotype in DT40 cells (Kohzaki et al., 2007). Furthermore, the RecQ family helicases BLM and RECQL5 are able to disrupt RAD51 filaments in vitro and inhibit the initiation of HR (Bachrati et al., 2006; Bugreev et al., 2007; Hu et al., 2007).
In metazoans there is a distinct family of helicases, defined by the discovery and characterization of dog-1 in C. elegans (_d_eletion o_f g_uanine rich DNA), which is essential for the maintenance of polyG/C-tracts (Cheung et al., 2002). In mouse, another member of this family, Rtel, is essential for telomere maintenance (Ding et al., 2004). Cells derived from the Rtel−/− null mouse exhibit reduced proliferative capacity and chromosomal abnormalities (Ding et al., 2004), which may be phenotypes attributable to a more general role in the maintenance of genome stability. The human homolog of Rtel is amplified in gastric tumours (Bai et al., 2000), but how Rtel functions and how its deregulated expression promotes tumorigenesis remains unclear.
Here we have utilized the genetic tractability of the nematode Caenorhabditis elegans to screen for a functional equivalent of Srs2 in a metazoan. We have identified a previously uncharacterized RAD3-like helicase, SPAR-1 (SuPpression of Aberrant Recombination defective), which is the C. elegans homologue of murine Rtel. We show that spar-1 mutant worms and SPAR1/RTEL1 knockdown human cells share a number of characteristic phenotypes with yeast srs2 mutants, and demonstrate that recombinant human SPAR1 is a potent antagonist of HR that acts specifically to disrupt D loop recombination intermediates. Our results imply a role for SPAR1 as an anti-recombinase, and suggest that SPAR1 is a functional analogue of Srs2 in metazoans.
RESULTS
Identification of spar-1, the C. elegans homologue of Rtel
One of the best characterised antagonists of HR is the yeast helicase Srs2 (Krejci et al., 2003; Veaute et al., 2003), yet sequence analysis has failed to identify putative homologues of SRS2 in higher eukaryotes. As it would be detrimental for a cell to undergo inappropriate recombination, it is expected that mechanisms to restrain HR must exist in higher eukaryotes. We used a genetic approach to identify potential antagonists of recombination in the nematode C. elegans. One of the characteristic phenotypes of both budding and fission yeast srs2 mutants is that growth is severely impaired by the additional loss of sgs1/rqh1, a homologue of the human Bloom’s Syndrome helicase (BLM) (Lee et al., 1999; Wang et al., 2001). We therefore conducted a candidate based synthetic lethal screen to look for helicase genes that result in significantly impaired viability when mutated in combination with the C. elegans BLM homolog, him-6 (Wicky et al., 2004); Supplementary Information). This screen identified F25H2.13, which we have named spar-1. This gene is the C. elegans homolog of Rtel (Ding et al., 2004) (Fig. S1A), which is essential for embryonic development, genome stability and telomere maintenance in mice (Ding et al., 2004).
We obtained a nematode mutant allele of spar-1, tm1866, in which a 1346 bp region comprising exons 2–5 is deleted. The deletion truncates the predicted protein and results in a premature stop codon downstream of the conserved IA helicase motif. spar-1 (tm1866) animals are viable, although the brood size is reduced relative to the wild-type N2 strain (Table 1), and the life cycle is retarded (at 20°C, spar-1 worms take 24 hours longer to reach the gravid adult stage). The smaller brood size of spar-1 mutants may result from replicative stress as we observe a three-fold increase in germ line apoptosis (data not shown), and a high incidence (21 %) of the protruding vulva phenotype, which is often associated with a persistence of unrepaired DNA damage during development (Weidhaas et al., 2006).
Table 1. Genetic interactions of C. elegans spar-1.
Analysis of brood size, embryonic lethality, and viable progeny reaching adulthood in single and double gene deletion mutants. Yeast and human homologues are indicated and “−” denotes genes not found in yeast.
| Equivalent Genotype | C. elegans Genotype | Total Brood Size | Percent Embryonic Lethality | Percent Viable Progeny | Number scored | |
|---|---|---|---|---|---|---|
| Yeast | Human | |||||
| wild- type | wild-type | N2 (wild-type) | 257 ± 20 | 0.6 ± 0.05 | 99.4 ± 0.05 | n=5 |
| - | SPAR1 | spar-1 | 68 ± 12 | 3.0 ± 0.9 | 86.0 ± 9.0 | n=20 |
| sgs1 | BLM | him-6 | 214 ± 14 | 49.7 ± 1.4 | 48.5 ± 1.6 | n=20 |
| - | FANCJ | dog-1 | 229 ± 11 | 3.9 ± 1.7 | 96.1 ± 1.7 | n=15 |
| mus81 | MUS81 | mus-81 | 153 ± 13 | 15.2 ± 6.0 | 84.8 ± 6.0 | n=20 |
| - | RECQ5 | rcq-5 | 209 ± 20 | 0.4 ± 0.1 | 99.6 ± 0.1 | n=20 |
| - | WRN1 | wrn-1 | 279 ± 13 | 0.3 ± 0.2 | 99.7 ± 0.1 | n=20 |
| - | SPAR1 BLM | spar-1; him-6 | 54 ± 11 | 72.1 ± 2.5 | 7.1 ± 1.6 | n=20 |
| - | SPAR1 FANCJ | spar-1 dog-1 | 0 | 0 | 0 | n=30 |
| - | SPAR1 MUS81 | spar-1 mus-81 | 22 ± 7 | 100 | 0 | n=30 |
| - | SPAR1 RECQ5 | spar-1; rcq-5 | 168 ± 19 | 100 | 0 | n=17 |
| - | SPAR1 WRN1 | spar-1; wrn-1 | 47 ± 13 | 4.0 ± 6.3 | 84.5 ± 7.8 | n=20 |
| - | FANCJ BLM | dog-1; him-6* | 185 ± 13 | 72.5 ± 1.8 | 27.6 ±1.8 | n=41 |
| - | RECQ5 BLM | rcq-5; him-6 | 111 ± 20 | 49.4 ± 4.2 | 50.6 ± 4.2 | n=10 |
| - | WRN1 BLM | wrn-1; him-6 | 170 ± 23 | 56.4 ± 1.6 | 43.6 ± 1.6 | n=10 |
| - | FANCJ RECQ5 | dog-1; rcq-5 | 201 ± 19 | 1.3 ± 0.7 | 98.7 ± 0.7 | n=20 |
| - | FANCJ WRN1 | dog-1; wrn-1 | 247 ± 12 | 4.1 ± 2.4 | 95.9 ± 2.4 | n=15 |
| - | RECQ5 WRN1 | rcq-5; wrn-1 | 243 ± 16 | ND | 97.3 ± 1.4 | n=10 |
We observed that the viability of spar-1; him-6 double mutant is severely compromised compared with either the him-6 or spar-1 single mutants, with only 7% of progeny surviving to adulthood (Table 1). The synthetic lethality of spar-1; him-6 is in stark contrast to the 49% progeny viability observed for the him-6 mutant, consistent with a previous report (Wicky et al., 2004). DOG-1 (Deletion of Guanine rich DNA) is the C. elegans homolog of FANCJ and is the helicase most closely related to SPAR-1 (Cheung et al., 2002; Youds et al., 2008). An earlier study by Youds et al. has shown that dog-1; him-6 double mutants have fewer viable progeny compared to either single mutant (Youds et al., 2006); Table 1). However, the reduced viability of later larval stages is far more severe in spar-1; him-6 animals (Table 1).
In addition to him-6/BLM, there are three other genes encoding RecQ family helicases in the C. elegans genome: wrn-1/WRN, rcq-5/RECQL5, and K02F3.12/RECQL1 (Jeong et al., 2003; Lee et al., 2004). We found that spar-1 is also synthetic lethal with rcq-5 (Table 1), with 100% lethality at the embryonic stage. Interestingly, although the eggs are inviable, the total number of eggs laid by spar-1; rcq-5 double mutant worms is similar to wild-type, suggesting that the rcq-5 mutation is able to rescue the reduced brood size in spar-1 mutants (Table 1). In contrast to spar-1; him-6 and spar-1; rcq-5, we did not observe synthetic lethality in the spar-1; wrn-1 double mutants, and the K02F3.12 helicase is essential, which precludes analysis. Therefore, the genetic relationship of spar-1 with RecQ helicases appears restricted to him-6 and rcq-5.
It has been shown that the human homologue of rcq-5 (RECQL5) is able to disrupt RAD51 nucleoprotein filaments in vitro (Hu et al., 2007), hence we also considered the RecQ family genes in our primary screen for synthetic lethality with him-6. We observed that wrn-1 and rcq-5 single mutant worms had brood sizes and viability similar to wild-type animals, while double mutants of each of these genes with him-6 displayed viability similar to him-6 single mutants (Table 1). Thus, wrn-1 and rcq-5 do not exhibit the characteristic synthetic lethality with him-6, which is predicted of a potential SRS2 analogue. In contrast, the synthetic lethality of spar-1 in combination with him-6 or rcq-5 raised the possibility that SPAR-1 could be a candidate antagonist of homologous recombination.
spar-1 mutants are synthetic lethal with other factors required for DNA metabolism
S. cerevisiae srs2 has been found to cause synthetic growth defects in a range of mutant backgrounds that impact on DNA repair and genome stability, in addition to sgs1 (Chiolo et al., 2005; Klein, 2001; Palladino and Klein, 1992; Tong et al., 2004). These include mutants in genes encoding: double strand break repair factors such as Mre11, Rad50, Xrs2, and Rad54; the helicases Chl1 and Mph1; and nucleases such as the flap endonuclease Rad27 and the structure-specific heterodimeric nuclease Mus81_-_Mms4. Therefore we investigated the genetic relationship between spar-1 and other factors involved in DNA metabolism. In the absence of viable C. elegans deletion mutants in mre-11, rad-50, rad-54, rad-27, chl-1, mph-1, or a putative homologue of mms4, we examined the interaction between spar-1 and mus-81. In S. cerevisiae, mus81 shows a reduced growth rate in the srs2 background (Fabre et al., 2002; Pan et al., 2006), and mms4 displays a synthetic sick phenotype with srs2 (Tong et al., 2004). The C. elegans mus-81 strain is largely viable, whereas the mus-81 spar-1 double mutant worms lay eggs that are completely inviable (Table 1). Furthermore the brood size is severely reduced, beyond that of the spar-1 single mutant.
We also investigated how loss of the dog-1 helicase might affect progeny viability in the spar-1 background. Strikingly, the spar-1 dog-1 double mutants produce no embryos, whereas both of the single mutant strains produce progeny with 80% viability (Table 1). Cytological analysis of the parental spar-1 dog-1 animals revealed that the germ line fails to develop appropriately. The mitotic tip contains only a few enlarged mitotic nuclei, which are indicative of replication stress (Ahmed et al., 2001), and these nuclei have fragmented chromosomes (Fig. S2). Clearly the mitotic nuclei do not progress into meiosis, resulting in sterility. Thus loss of both spar-1 and the related helicase dog-1 results in profound proliferation defects in the germ line and synthetic sterility. Together, spar-1 exhibits synthetic sterility or lethality in combination with dog-1, him-6, rcq-5, or mus-81, reminiscent of the genetic interactions observed between srs2 and factors required for DNA metabolism in yeast. These data reinforce the possibility that SPAR-1 may function to negatively regulate HR in metazoans in a similar manner to Srs2 in yeast.
Synthetic lethality correlates with a massive accumulation of recombination intermediates
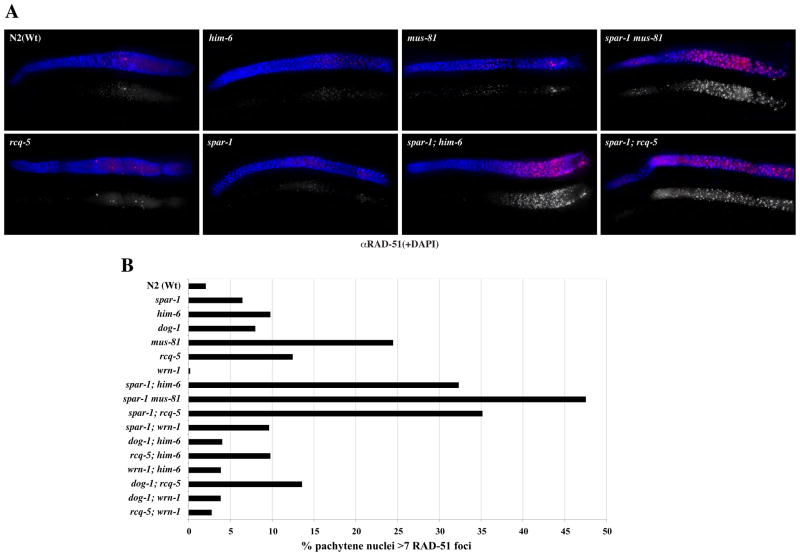
It has been proposed that the inviability of yeast srs2 sgs1 is caused by the accumulation of toxic recombination intermediates as viability can be partially restored by the additional loss of RAD51 or RAD54 (Gangloff et al., 2000; Klein, 2001). Unfortunately, C. elegans RAD-51 and RAD-54 are essential for meiotic DSB repair and subsequent progeny viability (Alpi et al., 2003; Martin et al., 2005) (data not shown) and this precludes similar analyses of the synthetic lethal interactions observed with spar-1. In view of this, we directly measured the occurrence of RAD-51 foci in the worm germ line to determine if the terminal phenotype associated with the synthetic lethality of spar-1 with him-6, dog-1, rcq-5 and mus-81 correlates with persistent HR intermediates. Nuclei in the C. elegans germ line are spatially ordered, progressing proximally from a region of mitosis in the distal tip through the different stages of meiotic prophase I, enabling the separate analysis of mitotic and meiotic RAD-51 foci (Alpi et al., 2003) (Fig. S3).
In spar-1 mutants, normal levels of RAD-51 foci are observed in early meiotic prophase nuclei, corresponding to the initiation and repair of SPO-11-induced DSBs (Fig. 1B and Fig. S3). A normal incidence of RAD-51 foci is also seen in him-6, dog-1, wrn-1 and rcq-5 worms, whereas mus-81 germ lines exhibit a slightly elevated level of RAD-51 foci suggestive of a reduced capacity to resolve mitotic and/or meiotic DSBs, or an increased incidence of spontaneous DNA damage (Boddy et al., 2001) (Fig. 1A and Fig. S3). In contrast, a massive accumulation of RAD-51 foci (up to 10-fold higher incidence of nuclei with more than six RAD-51 foci, relative to wild-type) is observed in the pachytene region of all of the double mutant germ lines that give rise to progeny with severely impaired viability, namely i) spar-1; him-6, ii) spar-1; rcq-5, and iii) mus-81 spar-1 (Table 1, Fig. 1A, B). Elevated levels of RAD-51 foci were also observed in the spar-1 dog-1 double mutant. However, we were unable to quantify the occurrence of RAD-51 foci in the same manner as for the other strains due to the mitotic catastrophe and lack of an intact germ line in these animals (Fig. S2). Importantly, elevated levels of RAD-51 foci are not observed for double mutant combinations that have been shown to be viable, including spar-1; wrn-1 and all possible double mutant combinations of the RecQ family of genes and dog-1 (Table 1, Fig. 1B). We conclude that the terminal phenotype of the synthetic lethality of spar-1 with dog-1, him-6, rcq-5, and mus-81 corresponds to a massive accumulation of recombination intermediates that persist and fail to be appropriately repaired.
Figure 1. Synthetic lethality correlates with elevated levels of RAD-51 foci.
A. Representative images of germ lines (genotype as indicated) stained with α-RAD-51 (Red) and DNA is counter-strained with DAPI (blue). Top RAD-51 + DAPI merge; bottom RAD-51 in greyscale. The distal end of the germline is on the left.
B. Quantification of nuclei containing >6 RAD-51 foci in the pachytene region of the germline. At least 5 animals were scored for each genotype. A detailed quantification of total germline RAD-51 staining is shown in Fig. S3.
Meiotic recombination frequencies are elevated in spar-1 mutants
In addition to the growth defects of yeast srs2 mutants in combination with sgs1/rqh1, loss of srs2 has also been shown to influence the frequency of recombination events (Ira et al., 2003; Rong et al., 1991). Therefore, we examined meiotic exchange in the spar-1 deletion strain. Using the visible mutant phenotypes “Dpy” (short, fat worms) and “Unc” (uncoordinated movement), we measured the frequency of crossing over for two intervals on two separate chromosomes. On chromosome V, crossing over in the interval between dpy-11 and unc-42 was increased more than 2.5-fold in the spar-1 background compared to the wild-type (Table 2). Loss of either dog-1 or rcq-5 did not significantly alter the meiotic recombination frequency (Table 2). A similar result was observed between dpy-17 and unc-36 on chromosome III (Table 2). Crossover frequency was elevated 4-fold in spar-1 progeny relative to wild-type, whereas deletion of dog-1 did not significantly affect the recombination frequency (Table 2). We were unable to analyse recombination frequencies for rcq-5 due to the close proximity of the rcq-5 gene to the dpy-17 unc-36 interval. Thus, SPAR-1 is the first example of a helicase that increases meiotic crossover frequencies when absent from C. elegans.
Table 2. spar-1 mutants display elevated meiotic recombination frequencies.
Meiotic recombination frequencies within the intervals defined by dpy-11 to unc-42, and dpy-17 to unc-36, were determined in different genetic backgrounds, as indicated.
| Genetic Interval Tested | Genotype | Total Progeny | Number of Recombinants | Map Distance in cM (95% CI) |
|---|---|---|---|---|
| dpy-11 to unc-42 | wild-type | 3135 | 114 | 3.71 (3.05–4.44) |
| dog-1 | 3273 | 84 | 2.60 (2.06–3.21) | |
| rcq-5 | 3824 | 94 | 2.48 (1.99–3.05) | |
| spar-1 | 723 | 66 | 9.59 (7.37–12.25) | |
| dpy-17 to unc-36 | wild-type | 3299 | 32 | 0.97 (0.65–1.35) |
| dog-1 | 3495 | 42 | 1.21 (0.86–1.60) | |
| spar-1 | 459 | 18 | 4.00 (2.47–6.24) |
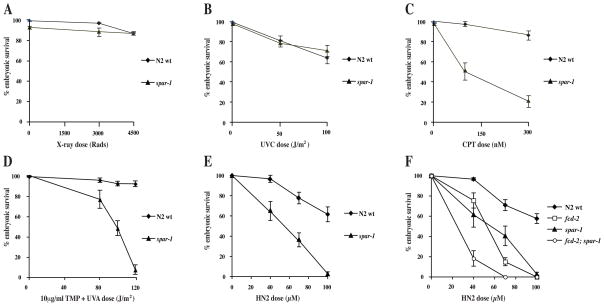
Loss of spar-1 confers sensitivity to a range of DNA damaging agents
Another characteristic phenotype of yeast srs2 mutants is sensitivity to DNA damaging agents (Aboussekhra et al., 1992; Bennett et al., 2001; Birrell et al., 2002). We investigated whether loss of C. elegans spar-1 had any effect on the capacity to tolerate different types of DNA damage, using progeny viability as a measure of sensitivity. Embryonic survival after exposure to X-rays or UVC was not significantly affected in either the wild-type or spar-1 mutant worms (Fig 2A and 2B). However, the response to DNA interstrand cross-links (ICLs) was affected in the absence of spar-1. Treatment with either UVA-activated trimethylpsoralen or nitrogen mustard led to a marked reduction in spar-1 progeny survival compared to the wild-type (Fig. 2D, E). Furthermore spar-1 worms were sensitive to treatment with the topoisomerase I inhibitor, camptothecin (Fig. 2C). Hence spar-1 is required for the C. elegans response to DNA damage, although this appears to be specific to lesions that affect replication fork progression.
Figure 2. spar-1 mutants are sensitive to specific types of DNA damage.
(A–E) Percentage progeny survival of worms treated with the indicated doses of (A) X-rays, (B) UVC (245 nm), (C) camptothecin (D) trimethylpsoralen activated with increasing doses of UVA (365 nm), and (E) nitrogen mustard (HN2). (F) Epistasis analysis of spar-1, fcd-2 and fcd-2; spar-1 mutants for sensitivity to HN2.
SPAR-1 function is distinct from that of the related helicase DOG-1
In humans, sensitivity to ICL-inducing agents is a characteristic of cells deficient for any of the genes associated with the inherited cancer-associated disorder Fanconi anaemia (FA) (Kennedy and D’Andrea, 2005). C. elegans dog-1 is the homologue of human FANCJ, and is epistatic to the FANCD2 homologue, fcd-2 (Youds et al., 2008). Given the considerable sensitivity of the spar-1 mutant to ICLs, we investigated whether spar-1 genetically interacts with the FA pathway in DNA repair. In contrast to dog-1; fcd-2 double mutants (Youds et al., 2008), the progeny of spar-1; fcd-2 double mutants were significantly more sensitive to nitrogen mustard than either single mutant alone (Fig. 2F). Hence spar-1 is not epistatic with fcd-2, one of the key factors in the FA pathway. In response to replication stress, FCD-2 is recruited to nuclear foci (Collis et al., 2006). We found that loss of spar-1/SPAR1 has no effect on the relocation of FCD-2/FANCD2 to repair foci in either C. elegans or human cells (data not shown).
In C. elegans, dog-1 mutants accumulate deletions at polyG/C-tracts; hence DOG-1 is proposed to unwind secondary structures in these tracts during replication (Cheung et al., 2002). We investigated whether SPAR-1 is also required to maintain polyG/C-tracts, or alternative repetitive DNA sequences (polyA/T, (CAG)8). However no deletions were identified in spar-1 mutants at any of the tracts investigated (data not shown). These results indicate that SPAR-1 has a role distinct from DOG-1 in DNA replication and repair in C. elegans.
Human SPAR1 suppresses homologous recombination and is required for DNA repair
To investigate whether or not the function of C. elegans SPAR-1 is conserved in humans, we depleted human SPAR1 (RTEL1, NHL) by siRNA in HeLa cells and tested for phenotypes in common with the C. elegans spar-1 mutant (Fig. S4). Delivery of human SPAR1 siRNA to cells decreased the level of mRNA expression detected by quantitative real time PCR by 90% relative to cells treated with control or FANCJ siRNA (Fig. S4A). We were unable to directly assess protein levels using the limited number of cells targeted in siRNA experiments, as SPAR1 is not sufficiently abundant to be detected in whole cell or nuclear extracts. The low abundance of SPAR1 is consistent with previous reports in mouse (Ding et al., 2004). However, we were able to show that these _SPAR1_-specific siRNAs, but not the non-targeting control siRNA, can effectively repress the expression of SPAR1 from a tetracycline-inducible promoter in a stable integrated HEK293 cell line (Fig. S4B).
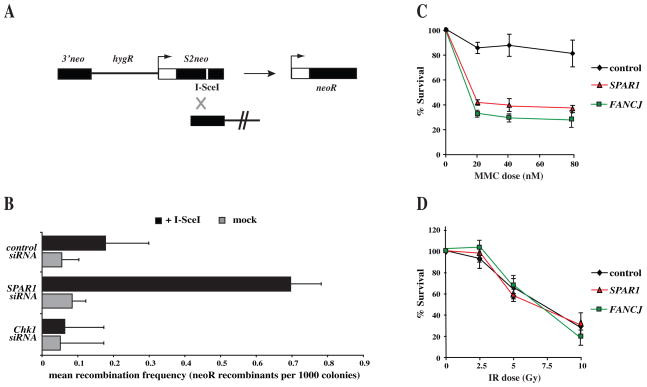
To investigate whether the human SPAR1 homologue is involved in the regulation of HR repair, we employed an I-SceI inducible DSB assay to quantify the frequencies of HR repair within an integrated SCneo substrate (Fig. 3A) (Johnson and Jasin, 2001; Mohindra et al., 2002). As has been shown previously (Collis et al., 2007; Sorensen et al., 2005), depletion of CHK1 leads to a reduction in the frequency of HR relative to the control (Fig. 3B). In contrast, siRNA depletion of SPAR1 consistently resulted in a 4-fold increase in the frequency of HR repair when compared to control (Fig. 3B). Hence, as in C. elegans, human SPAR1 suppresses the incidence of HR.
Figure 3. Human cells depleted for SPAR1 exhibit similar hyper-recombination and DNA damage sensitivity phenotypes to C. elegans spar-1 mutants.
A. Schematic of the integrated SCneo substrate used to measure recombination frequencies, comprising two non-functional alleles of the neomycin resistance gene. Initiation of a DSB at the I-SceI restriction site (white line) induces HR, restoring a functional neoR cassette through gene conversion.
B. Analysis of I-SceI induced HR at the SCneo construct after non-targeting control, SPAR1, or CHK1 siRNA depletion, and subsequent transfection of I-SceI expression vector in SW480/SN3 cells. Neomycin resistant colonies were scored at 10 days after transfection. Error bars indicate s.e.m. from 3 independent experiments.
C, D. Sensitivity of siRNA depleted cells to the indicated doses of (C) mitomycin C and (D) IR. Error bars indicate s.e.m. from 3 independent experiments.
We next examined human SPAR1 depleted cells for sensitivity to different DNA damaging agents. Knockdown of SPAR1 in HeLa cells had no significant effect on survival after treatment with IR (Fig. 3C). In contrast cells depleted for human SPAR1 exhibit a 50% reduction in cell survival after exposure to the ICL-inducing drug mitomycin C, relative to a non-targeting control siRNA treatment (Fig. 3D). These data are consistent with the DNA damage sensitivity of C. elegans spar-1 mutants (Fig. 2), and suggest that SPAR-1/SPAR1 function is conserved.
Human SPAR1 inhibits the formation of recombination intermediates in vitro
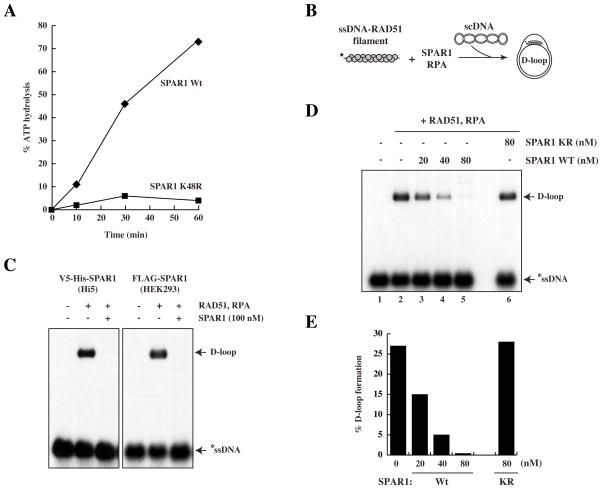
Yeast Srs2 has been shown to suppress HR by disrupting RAD51 nucleoprotein filaments and preventing strand invasion (Krejci et al., 2003; Veaute et al., 2003). Our genetic data from both C. elegans and human cells suggest that SPAR1 may act to restrict the occurrence of HR, which we next sought to test biochemically. We utilised the baculovirus system to express and purify both wild-type human SPAR1 and a K48R mutant to near homogeneity (Fig. S4C). As expected from comparison with mutations in the Walker A ATP-hydrolysing motif of other helicases, including Srs2 (Krejci et al., 2004), the K48R mutant was ATPase-dead (Fig. 4A).
Figure 4. Human SPAR1 inhibits D loop formation in an ATP-dependent manner.
A. ATP hydrolysis assay of human wild-type and mutant (K48R) SPAR1 performed in the presence of single stranded DNA.
B. Schematic of the D loop assay.
C. D loop assay with 100nM of Wt SPAR1 purified from Hi5 insect cells or HEK293 cells, as indicated. The D loop species migrates slower than the ssDNA probe.
D. D loop assay with Wt or K48R SPAR1, as indicated.
E. Quantification of D loop formation.
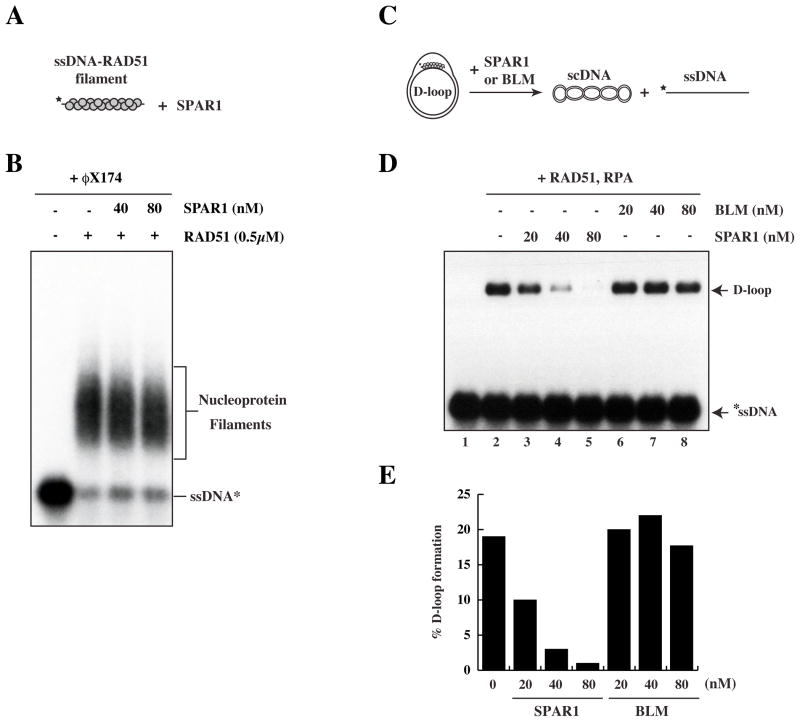
An in vitro HR assay was used to assess whether SPAR1 is able to modulate recombination reactions catalyzed by RAD51 (McIlwraith et al., 2000). In this assay, a nucleoprotein filament is first formed between RAD51 and 32P-labelled ssDNA. Upon addition of supercoiled (sc)DNA template, the nucleoprotein filament invades the scDNA duplex to form a joint molecule known as a D loop (Fig. 4B, 4C). The heterotrimeric ssDNA-binding protein RPA is added to the reaction to stabilise the displaced strand of the D loop, and also to prevent any re-association of RAD51 with ssDNA if the nucleoprotein filament is disrupted (Krejci et al., 2003). It is clear that addition of wild-type SPAR1 purified from insect or human cells prior to scDNA severely impairs the formation of a D loop (Fig. 4C, 4D, lanes 3–5). SPAR1 activity was dependent upon ATP-hydrolysis, as the K48R mutant protein was found to be incapable of blocking D loop formation (Fig. 4D lane 6, 4E).
SPAR1 does not disrupt the RAD51-ssDNA filament, but can disrupt pre-formed D loops
A number of proteins, including Srs2, BLM, and RECQL5, have been shown to disrupt the RAD51 nucleoprotein filament (Bugreev et al., 2007; Hu et al., 2007; Krejci et al., 2003; Veaute et al., 2003). To further investigate how SPAR1 antagonises D loop formation, we performed gel shifts to examine whether SPAR1 is able to dissociate the RAD51 nucleoprotein filament (Fig. 5A). Surprisingly, we were unable to detect any measurable effect of SPAR1 on the stability of the RAD51 nucleoprotein filament, either in the presence or absence of RPA or following addition of a 200-fold excess of unlabelled φX174 ssDNA competitor (Fig. 5B and data not shown). This result suggested that SPAR1-dependent D loop inhibition occurs by a different mechanism to that exhibited by BLM and RECQL5. We next employed the D loop assay with a modified order of addition (Fig. 5C) to determine whether SPAR1 instead acts on the D loop structure to reverse strand invasion. Figure 5D (lanes 3–5) shows that SPAR1 disrupts pre-formed D loops to reverse the HR process in vitro. D loop disruption by SPAR1 is concentration-dependent, and is almost entirely inhibited at 80 nM SPAR1 (Fig. 5D lane 5, E). Furthermore, SPAR1 disrupts D loops in the presence of calcium (Fig. 5D, E), which stabilises RAD51 filaments (Bugreev and Mazin, 2004). In contrast, BLM cannot dissociate pre-formed D loops under these conditions (Fig. 5D lanes 6–8, E), consistent with previous findings (Bugreev et al., 2007). These results demonstrate that SPAR1 antagonises recombination by disrupting D loop intermediates.
Figure 5. SPAR1 specifically disrupts performed D loops.
A. Schematic of the ssDNA–RAD51 filament disruption assay.
B. Pre-formed ssDNA-RAD51 filaments were incubated with the indicated concentrations of SPAR1 followed by addition of a 200-fold excess of cold φX174 ssDNA competitor.
C. Schematic of the modified D loop disruption assay.
D. Preformed D loops were incubated with the indicated concentrations of SPAR1 and BLM.
E. Quantification of D loop disruption.
DISCUSSION
HR repair is an essential cellular process that must be tightly regulated to prevent genome instability through inappropriate recombination events. In this study, we have identified a helicase SPAR-1/SPAR1, conserved from C. elegans to humans, which demonstrates functional similarity to yeast Srs2. Although SPAR1 shows no sequence similarity to Srs2, loss of SPAR-1/SPAR1 in either the nematode or in human cells gives rise to a range of phenotypes analogous to srs2 mutant yeast, including: synthetic lethality with the sgs1/BLM homologue (Lee et al., 1999; Wang et al., 2001) that is associated with an accumulation of persistent recombination intermediates (Gangloff et al., 2000; Klein, 2001); hyper-recombination in human cell culture (Ira et al., 2003; Rong et al., 1991) and during C. elegans meiosis; and sensitivity to complex DNA damage such as ICLs (Birrell et al., 2002). Collectively, the phenotypes observed in the absence of SPAR-1/SPAR1 strongly suggest that it has a conserved function as an anti-recombinase. Indeed, we demonstrate that human SPAR1 has potent D loop dissociation activity in vitro, which is dependent upon ATP hydrolysis. D loop unwinding by SPAR1, unlike BLM, occurs in the presence of calcium, which is known to stabilise RAD51 filaments (Bachrati et al., 2006; Bugreev et al., 2007; van Brabant et al., 2000). This implies that SPAR1 can antagonise HR at an early stage, post strand invasion, and could promote synthesis-dependent strand annealing.
It should be noted that SPAR1 and Srs2 also differ in some respects, perhaps unsurprisingly given the lack of sequence homology. Loss of SPAR-1/SPAR1 confers sensitivity to a more select range of DNA damaging agents than for Srs2, and this could be indicative of a broader functional scope for Srs2, perhaps with different factors accountable for distinct roles in higher eukaryotes. Certainly our candidate screen was not exhaustive, and additional anti-recombinases may subsequently be identified. Furthermore, the exact mechanisms by which SPAR1 and Srs2 antagonise recombination may not be identical. Srs2 has previously been shown to disrupt the Rad51 nucleoprotein filament (Krejci et al., 2003; Veaute et al., 2003), yet this was not observed for SPAR1. Also, whilst both SPAR1 and Srs2 can antagonise the formation of a D loop, it is not clear whether Srs2 can act on a preformed D loop structure. Previous work had suggested that Srs2 is unable to unwind D loop intermediates (Krejci et al., 2003; Veaute et al., 2003), although a recent study by Dupaigne and colleagues has demonstrated Srs2 activity on the related PX junction (Dupaigne et al., 2008). Nevertheless, SPAR-1/SPAR1 is clearly a novel anti-recombinase that shares some features with Srs2, and further study of SPAR-1/SPAR1 will facilitate the understanding of HR regulation in metazoans.
A role for SPAR-1/SPAR1 in the negative regulation of HR by disrupting D loop intermediates provides molecular insight into the phenotypes observed in this study and for the Rtel knockout mice. It is predicted that an inability to antagonise HR would result in elevated levels of HR, as we have demonstrated in human cell culture and during C. elegans meiosis. The specific sensitivity to complex DNA damage such as ICLs but not IR or UV lesions may result from a failure to temporally regulate HR during the repair process. HR has been shown to be required as part of the composite pathway for the efficient repair of ICLs, most likely downstream of incision and processing of the lesion (Li and Heyer, 2008). However, if unrestricted, invoking HR on an inappropriate intermediate of ICL repair could lead to persistent damage from irreparable intermediates. Furthermore, an inability to reverse non-productive HR intermediates provides a plausible explanation for the synthetic lethality of spar-1 in combination with him-6, rcq-5, mus-81, and dog-1. It is possible that the synthetic sterility observed in spar-1 dog-1 animals is due to a requirement for SPAR-1 function at replication forks stalled by polyG/C-tract secondary structure in dog-1 mutants. However, it is equally possible that the phenotype of the spar-1 dog-1 double mutant is due to overlapping roles for SPAR-1 and DOG-1 at telomeres or other DNA structures in C. elegans.
Rtel is the murine homologue of C. elegans SPAR-1 and was originally identified by genomic mapping of loci that control telomere length differences between M. musculus and M. spretus (Ding et al., 2004). Rtel plays a critical role in genome stability as knockout mice are embryonic lethal and cells derived from these mice exhibit a rapid reduction in proliferative capacity upon differentiation, accompanied with an increased incidence of chromosomal abnormalities and telomere loss (Ding et al., 2004). Based on homology to C. elegans DOG-1, it was proposed that Rtel might function to unwind G-rich DNA secondary structures formed during DNA replication and at the telomere (Ding et al., 2004). However, the human homologue of dog-1 has now been identified as FANCJ (Youds et al., 2008), and no mechanistic evidence has yet been provided to support this model. While a role for Rtel in unwinding DNA secondary structures remains a possibility, C. elegans spar-1 mutants differ from dog-1 mutants in that they do not exhibit instability at G-rich sequences. Our finding that SPAR-1/SPAR1 is an anti-recombinase leads us to propose an alternative possibility: the inviability and severe genomic instability exhibited by the _Rtel_−/− null mouse results from an inability to correctly regulate HR. HR has also been shown to cause deletion of the protective T loop structure formed by the sequestration of the 3′ telomeric end into a sub-telomeric duplex TTAGGG repeat (Wang et al., 2004). The T loop structure requires both HR and telomere specific proteins for its assembly and has therefore been proposed to resemble a D loop HR intermediate (de Lange, 2004). It is therefore possible that in the absence of SPAR1/Rtel the T loop structure may be erroneously resolved as a substrate by the HR machinery. Hence the telomere deficiency in _Rtel_−/− mouse cells may result from the inability to antagonise inappropriate HR at the T loop. Alternatively (or additionally), Rtel may function to unwind/disengage the T loop structure to allow telomerase access to complete chromosome end replication during each cell cycle.
The importance of correctly restraining HR is also highlighted by the observation that human SPAR1 is located in a four gene cluster that is over-expressed in some tumours of the gastrointestinal tract (Bai et al., 2000). It is likely that excessive SPAR1 activity could repress productive HR events, and in this way mimic the loss of an essential HR factor, such as BRCA2, which is associated with familial breast and ovarian cancers (Venkitaraman, 2002). Amplification of SPAR1 may also drive genome instability and tumourigenesis by inappropriately disengaging the T loop structure, leading to telomere de-protection. It will now be imperative to screen human cancers for mutations, amplified expression and copy number changes in SPAR1, especially given the association of other helicase regulators of HR with tumorigenesis (BLM (German, 1995), RECQL5 (Hu et al., 2007)).
In summary, we have identified SPAR-1/SPAR1 as a novel _s_u_p_pressor of _a_berrant _r_ecombination that is conserved in metazoans. Our genetic and biochemical analysis reveal SPAR-1/SPAR1 as an anti-recombinase with some functional similarities to yeast Srs2. Our observations provide a possible mechanistic explanation for the genomic instability seen in Rtel−/− mice (Ding et al., 2004). Finally, the overexpression of human SPAR1 in gastrointestinal tract tumours (Bai et al., 2000) raises the possibility that SPAR1 may prove to be a valuable target for new anti-cancer therapeutics.
EXPERIMENTAL PROCEDURES
C. elegans assays
Nematode strains were maintained as previously described (Brenner, 1974). C. elegans immunofluorescence microscopy was performed as previously described (Colaiacovo et al., 2003; Youds et al., 2008). The frequency of meiotic recombination was measured by scoring the number of recombinant progeny of a cis_-heterozygote, as previously described_ (Rose and Baillie, 1979). C. elegans DNA damage sensitivity assays were performed as previously described (Collis et al., 2006; Youds et al., 2008).
Human cell culture assays
Sub-confluent cultures were transfected with 100 nM siRNA (Dharmacon ON-TARGET_plus SPAR1_ and FANCJ; SMARTPool Chk1) using Dharmafect #1 reagent in antibiotic-free media. FLP-In T-Rex-293 pDEST-Flag/FRT/TO-SPAR1 cells were selected with 200 μg/ml Hygromycin (Invitrogen) and 15 μg/ml Blasticidin (Autogen Bioclear). Tetracycline (Sigma) was used at 1 μg/ml. Quantitative PCR analysis of mRNA levels was performed as previously described (Collis et al., 2007) using Superarray RT2 primer sets for SPAR1, FANCJ, and ACTG1. Homologous recombination frequencies were measured in SW480/SN3 cells that contain a single integrated copy of an SCneo substrate, as previously described (Collis et al., 2007; Johnson and Jasin, 2001; Mohindra et al., 2002). MMC (Sigma, Poole, UK) sensitivity was assayed 48 h after siRNA treatment, as previously described (Collis et al., 2007).
Protein purification and antibodies
Wild-type and K48R mutant SPAR1 fused to a V5 epitope at the N-terminus and MYC-6HIS at the C-terminus were expressed in Hi5 insect cells using the baculovirus system. Five litres of Hi5 cells were infected at a concentration of 1×106 cells/ml, and collected 72 h post infection. The cell pellet was lysed in 50 mM Tris HCl pH8, 0.5 M NaCl, 4 mM MgCl2, 1 mM DTT, EDTA-free complete protease inhibitor cocktail, 30 units/ml benzonase and sonicated three times on ice (30 s, max amplitude, 2 min rest interval). The soluble fraction was enriched for SPAR1 protein by 1.5 M ammonium sulphate precipitation, resuspended and dialysed in 20 mM Tris HCl pH7.5, 0.5 M NaCl. The protein was purified to near homogeneity using anti-V5 agarose, and cleaved from the affinity matrix by AcTEV protease digestion, 3 h 16°C. Purified protein was concentrated by serial passage over a Biomax ultrafree 5k NMWL spin column. Protein concentration was estimated by Bradford assay, and purity was confirmed by SDS-PAGE followed by Coomassie staining. Human RAD51 and RPA proteins were purified as published previously (Baumann et al., 1997; Henricksen et al., 1994). Human BLM protein was a kind gift from Ian Hickson (Oxford, UK). Primary antibodies used were FLAG-HRP (Sigma) and human Actin (Abcam).
ATPase assay
60 μl reactions contained 15 nM Wt or K48R SPAR1 with 40 μM φX174 ssDNA in 70 mM Tris HCl pH7.6, 10 mM MgCl2, 3 μCi of [γ-32P]ATP (3000 Ci/mmol), 5 mM DTT, 0.1 mg/ml BSA, at 37°C. At the times indicated, 10 μl aliquots were removed, and the reaction terminated by the addition of 5 μl 0.5 M EDTA. Samples were analyzed by thin-layer chromatography on CEL 300 PEI/UV254 plates in 1 M formic acid, 0.5 M LiCl. ATP hydrolysis was quantified as the percentage of [γ-32P]ATP hydrolysed to [γ-32P]ADP, using a Storm 860 Phosphorimager.
D loop Recombination assay
10 μl reactions contained 1 μM 5′-32P-end-labeled 100mer ssDNA in recombination buffer (25 mM Tris-acetate pH 7.5, 5 mM CaCl2, 2 mM MgCl2, 2 mM ATP, 1 mM DTT, 100 μg/ml BSA). The following were then added sequentially, separated by 5 min incubations at 37°C: 0.5 μM RAD51; SPAR1 or BLM; 0.2 μM RPA; 0.3 mM supercoiled pPB4.3 DNA. After a further 10 minute incubation the products were deproteinized by the addition of one-fifth volume stop buffer (0.1 M Tris-HCl pH 7.5, 0.1 M MgCl2, 3% SDS and 10 mg/ml proteinase K) with 20 min incubation at 37°C. DNA products were analyzed by 1% agarose gel electrophoresis, dried onto filter paper, visualized by autoradiography and quantified using a Phosphorimager.
Nucleoprotein filament disruption assay
10 μl reactions contained1 μM 5′-32P-end-labeled 100mer ssDNA in recombination buffer. 0.5 μM RAD51 was added then SPAR1, then 216 μM single-stranded φX174 virion DNA, each separated by 5 min incubations at 37°C. After a further 5 min, DNA products were analyzed by 1% agarose gel electrophoresis, dried onto filter paper and visualized by autoradiography.
Supplementary Material
01
Acknowledgments
We wish to thank: Shohei Mitani (National Bioresource Project, Japan) and the C. elegans Knockout Consortium for providing nematode strains; Cancer Research UK Cell Services, Helen Bryant, and Thomas Helleday for cells; Ian Hickson for BLM protein; and Helen Bryant, Thomas Helleday, and Janet Cronshaw for plasmids. H.A.T. is funded by the William Randolph Hearst Foundation and a grant from the NIH (AG25891 to HAT); A.M.R. is funded by the Natural Sciences and Engineering Research Council; J.L.Y. was funded by National Cancer Institute of Canada and Michael Smith Foundation for Health Research; S.C.W. is supported by the Louis-Jeantet Foundation and is part of the EU DNA Repair consortium; the labs of S.C.W and S.J.B. receive funding from Cancer Research UK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol Cell Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboussekhra A, Chanet R, Zgaga Z, Cassier-Chauvat C, Heude M, Fabre F. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 1989;17:7211–7219. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, Klein HL. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, Alpi A, Hengartner MO, Gartner A. C. elegans RAD-5/CLK-2 defines a new DNA damage checkpoint protein. Curr Biol. 2001;11:1934–1944. doi: 10.1016/s0960-9822(01)00604-2. [DOI] [PubMed] [Google Scholar]
- Alpi A, Pasierbek P, Gartner A, Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma. 2003;112:6–16. doi: 10.1007/s00412-003-0237-5. [DOI] [PubMed] [Google Scholar]
- Arthur HM, Lloyd RG. Hyper-recombination in uvrD mutants of Escherichia coli K-12. Mol Gen Genet. 1980;180:185–191. doi: 10.1007/BF00267368. [DOI] [PubMed] [Google Scholar]
- Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Connolly B, Metzker ML, Hilliard CA, Liu X, Sandig V, Soderman A, Galloway SM, Liu Q, Austin CP, et al. Overexpression of M68/DcR3 in human gastrointestinal tract tumors independent of gene amplification and its location in a four-gene cluster. Proc Natl Acad Sci U S A. 2000;97:1230–1235. doi: 10.1073/pnas.97.3.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Benson FE, Hajibagheri N, West SC. Purification of human Rad51 protein by selective spermidine precipitation. Mutat Res. 1997;384:65–72. doi: 10.1016/s0921-8777(97)00028-1. [DOI] [PubMed] [Google Scholar]
- Bennett CB, Lewis LK, Karthikeyan G, Lobachev KS, Jin YH, Sterling JF, Snipe JR, Resnick MA. Genes required for ionizing radiation resistance in yeast. Nat Genet. 2001;29:426–434. doi: 10.1038/ng778. [DOI] [PubMed] [Google Scholar]
- Bierne H, Seigneur M, Ehrlich SD, Michel B. uvrD mutations enhance tandem repeat deletion in the Escherichia coli chromosome via SOS induction of the RecF recombination pathway. Mol Microbiol. 1997;26:557–567. doi: 10.1046/j.1365-2958.1997.6011973.x. [DOI] [PubMed] [Google Scholar]
- Birrell GW, Brown JA, Wu HI, Giaever G, Chu AM, Davis RW, Brown JM. Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc Natl Acad Sci U S A. 2002;99:8778–8783. doi: 10.1073/pnas.132275199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, 3rd, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci U S A. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung I, Schertzer M, Rose A, Lansdorp PM. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat Genet. 2002;31:405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- Chiolo I, Carotenuto W, Maffioletti G, Petrini JH, Foiani M, Liberi G. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol Cell Biol. 2005;25:5738–5751. doi: 10.1128/MCB.25.13.5738-5751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Saponaro M, Baryshnikova A, Kim JH, Seo YS, Liberi G. The human F-Box DNA helicase FBH1 faces Saccharomyces cerevisiae Srs2 and postreplication repair pathway roles. Mol Cell Biol. 2007;27:7439–7450. doi: 10.1128/MCB.00963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Collis SJ, Barber LJ, Clark AJ, Martin JS, Ward JD, Boulton SJ. HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat Cell Biol. 2007;9:391–401. doi: 10.1038/ncb1555. [DOI] [PubMed] [Google Scholar]
- Collis SJ, Barber LJ, Ward JD, Martin JS, Boulton SJ. C. elegans FANCD2 responds to replication stress and functions in interstrand cross-link repair. DNA Repair (Amst) 2006;5:1398–1406. doi: 10.1016/j.dnarep.2006.06.010. [DOI] [PubMed] [Google Scholar]
- de Lange T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, Pourvali R, Poon S, Vulto I, Chavez E, et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Dupaigne P, Le Breton C, Fabre F, Gangloff S, Le Cam E, Veaute X. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: implications for crossover incidence during mitotic recombination. Mol Cell. 2008;29:243–254. doi: 10.1016/j.molcel.2007.11.033. [DOI] [PubMed] [Google Scholar]
- Fabre F, Chan A, Heyer WD, Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci U S A. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- German J. Bloom’s syndrome. Dermatol Clin. 1995;13:7–18. [PubMed] [Google Scholar]
- Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YS, Kang Y, Lim KH, Lee MH, Lee J, Koo HS. Deficiency of Caenorhabditis elegans RecQ5 homologue reduces life span and increases sensitivity to ionizing radiation. DNA Repair (Amst) 2003;2:1309–1319. doi: 10.1016/j.dnarep.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Johnson RD, Jasin M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem Soc Trans. 2001;29:196–201. doi: 10.1042/0300-5127:0290196. [DOI] [PubMed] [Google Scholar]
- Kennedy RD, D’Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim JH, Lee SH, Kim DH, Kang HY, Bae SH, Pan ZQ, Seo YS. The novel human DNA helicase hFBH1 is an F-box protein. J Biol Chem. 2002;277:24530–24537. doi: 10.1074/jbc.M201612200. [DOI] [PubMed] [Google Scholar]
- Klein HL. Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Delta with other DNA repair genes in Saccharomyces cerevisiae. Genetics. 2001;157:557–565. doi: 10.1093/genetics/157.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohzaki M, Hatanaka A, Sonoda E, Yamazoe M, Kikuchi K, Vu Trung N, Szuts D, Sale JE, Shinagawa H, Watanabe M, et al. Cooperative roles of vertebrate Fbh1 and Blm DNA helicases in avoidance of crossovers during recombination initiated by replication fork collapse. Mol Cell Biol. 2007;27:2812–2820. doi: 10.1128/MCB.02043-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, Macris M, Li Y, Van Komen S, Villemain J, Ellenberger T, Klein H, Sung P. Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J Biol Chem. 2004;279:23193–23199. doi: 10.1074/jbc.M402586200. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Lawrence CW, Christensen RB. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J Bacteriol. 1979;139:866–876. doi: 10.1128/jb.139.3.866-876.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Yook JS, Han SM, Koo HS. A Werner syndrome protein homolog affects C. elegans development, growth rate, life span and sensitivity to DNA damage by acting at a DNA damage checkpoint. Development. 2004;131:2565–2575. doi: 10.1242/dev.01136. [DOI] [PubMed] [Google Scholar]
- Lee SK, Johnson RE, Yu SL, Prakash L, Prakash S. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science. 1999;286:2339–2342. doi: 10.1126/science.286.5448.2339. [DOI] [PubMed] [Google Scholar]
- Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JS, Winkelmann N, Petalcorin MI, McIlwraith MJ, Boulton SJ. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol Cell Biol. 2005;25:3127–3139. doi: 10.1128/MCB.25.8.3127-3139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwraith MJ, Van Dyck E, Masson JY, Stasiak AZ, Stasiak A, West SC. Reconstitution of the strand invasion step of double-strand break repair using human Rad51 Rad52 and RPA proteins. J Mol Biol. 2000;304:151–164. doi: 10.1006/jmbi.2000.4180. [DOI] [PubMed] [Google Scholar]
- Mohindra A, Hays LE, Phillips EN, Preston BD, Helleday T, Meuth M. Defects in homologous recombination repair in mismatch-repair-deficient tumour cell lines. Hum Mol Genet. 2002;11:2189–2200. doi: 10.1093/hmg/11.18.2189. [DOI] [PubMed] [Google Scholar]
- Morel P, Hejna JA, Ehrlich SD, Cassuto E. Antipairing and strand transferase activities of E. coli helicase II (UvrD) Nucleic Acids Res. 1993;21:3205–3209. doi: 10.1093/nar/21.14.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T, Furukawa F, Sakaguchi C, Toda T, Carr AM, Iwasaki H, Shinagawa H. Role of the Schizosaccharomyces pombe F-Box DNA helicase in processing recombination intermediates. Mol Cell Biol. 2005;25:8074–8083. doi: 10.1128/MCB.25.18.8074-8083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Dixon J, Barr AR, Whitby MC. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol Cell Biol. 2005;25:8084–8096. doi: 10.1128/MCB.25.18.8084-8096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F, Klein HL. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics. 1992;132:23–37. doi: 10.1093/genetics/132.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Rong L, Palladino F, Aguilera A, Klein HL. The hyper-gene conversion hpr5–1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics. 1991;127:75–85. doi: 10.1093/genetics/127.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AM, Baillie DL. The Effect of Temperature and Parental Age on Recombination and Nondisjunction in CAENORHABDITIS ELEGANS. Genetics. 1979;92:409–418. doi: 10.1093/genetics/92.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- van Brabant AJ, Ye T, Sanz M, German IJ, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Wang SW, Goodwin A, Hickson ID, Norbury CJ. Involvement of Schizosaccharomyces pombe Srs2 in cellular responses to DNA damage. Nucleic Acids Res. 2001;29:2963–2972. doi: 10.1093/nar/29.14.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas JB, Eisenmann DM, Holub JM, Nallur SV. A Caenorhabditis elegans tissue model of radiation-induced reproductive cell death. Proc Natl Acad Sci U S A. 2006;103:9946–9951. doi: 10.1073/pnas.0603791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicky C, Alpi A, Passannante M, Rose A, Gartner A, Muller F. Multiple genetic pathways involving the Caenorhabditis elegans Bloom’s syndrome genes him-6, rad-51, and top-3 are needed to maintain genome stability in the germ line. Mol Cell Biol. 2004;24:5016–5027. doi: 10.1128/MCB.24.11.5016-5027.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, Barber LJ, Ward JD, Collis SJ, O’Neil NJ, Boulton SJ, Rose AM. DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol Cell Biol. 2008;28:1470–1479. doi: 10.1128/MCB.01641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, O’Neil NJ, Rose AM. Homologous recombination is required for genome stability in the absence of DOG-1 in Caenorhabditis elegans. Genetics. 2006;173:697–708. doi: 10.1534/genetics.106.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieg J, Maples VF, Kushner SR. Recombinant levels of Escherichia coli K-12 mutants deficient in various replication, recombination, or repair genes. J Bacteriol. 1978;134:958–966. doi: 10.1128/jb.134.3.958-966.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01