Sturge–Weber Syndrome and Port-Wine Stains Caused by Somatic Mutation in GNAQ (original) (raw)
. Author manuscript; available in PMC: 2013 Nov 23.
Published in final edited form as: N Engl J Med. 2013 May 8;368(21):1971–1979. doi: 10.1056/NEJMoa1213507
Abstract
BACKGROUND
The Sturge–Weber syndrome is a sporadic congenital neurocutaneous disorder characterized by a port-wine stain affecting the skin in the distribution of the ophthalmic branch of the trigeminal nerve, abnormal capillary venous vessels in the leptomeninges of the brain and choroid, glaucoma, seizures, stroke, and intellectual disability. It has been hypothesized that somatic mosaic mutations disrupting vascular development cause both the Sturge–Weber syndrome and port-wine stains, and the severity and extent of presentation are determined by the developmental time point at which the mutations occurred. To date, no such mutation has been identified.
METHODS
We performed whole-genome sequencing of DNA from paired samples of visibly affected and normal tissue from 3 persons with the Sturge–Weber syndrome. We tested for the presence of a somatic mosaic mutation in 97 samples from 50 persons with the Sturge–Weber syndrome, a port-wine stain, or neither (controls), using amplicon sequencing and SNaPshot assays, and investigated the effects of the mutation on downstream signaling, using phosphorylation-specific antibodies for relevant effectors and a luciferase reporter assay.
RESULTS
We identified a nonsynonymous single-nucleotide variant (c.548G→A, p.Arg183Gln) in GNAQ in samples of affected tissue from 88% of the participants (23 of 26) with the Sturge–Weber syndrome and from 92% of the participants (12 of 13) with apparently nonsyndromic port-wine stains, but not in any of the samples of affected tissue from 4 participants with an unrelated cerebrovascular malformation or in any of the samples from the 6 controls. The prevalence of the mutant allele in affected tissues ranged from 1.0 to 18.1%. Extracellular signal-regulated kinase activity was modestly increased during transgenic expression of mutant G_α_q.
CONCLUSIONS
The Sturge–Weber syndrome and port-wine stains are caused by a somatic activating mutation in GNAQ. This finding confirms a long-standing hypothesis. (Funded by the National Institutes of Health and Hunter’s Dream for a Cure Foundation.)
A port-wine stain is a cutaneous capillary malformation (Fig. 1A, 1B, and 1C) that occurs in approximately 3 of every 1000 newborns1,2 and usually involves the head and neck.3 The Sturge–Weber syndrome, also known as encephalofacial angiomatosis, is a neurocutaneous disorder that occurs as a sporadic congenital condition; it is characterized by a port-wine stain that affects the skin in the distribution of the ophthalmic branch of the trigeminal nerve (Fig. 1A and 1B) and is associated with venous-capillary abnormalities of the leptomeninges (Fig. 1D, 1E, and 1F) and the eye. It occurs in both male and female newborns, in approximately 1 in 20,000 to 50,000 live births.1 A child born with a port-wine stain on the face has approximately a 6% chance of having the Sturge–Weber syndrome,2 and this risk increases to 26% when the port-wine stain is located in the distribution of the ophthalmic branch of the trigeminal nerve.3 Port-wine stains usually have underlying soft-tissue and bony-tissue overgrowth that may be mild or massive.4 A long-standing but unproven hypothesis is that the Sturge–Weber syndrome and port-wine stains are caused by the same underlying somatic mutations,5,6 with the precise clinical manifestations dependent on where and when in the developing fetus the somatic mutation occurs. We tested this hypothesis through whole-genome sequencing of affected and unaffected tissue to identify the causative somatic mutation.
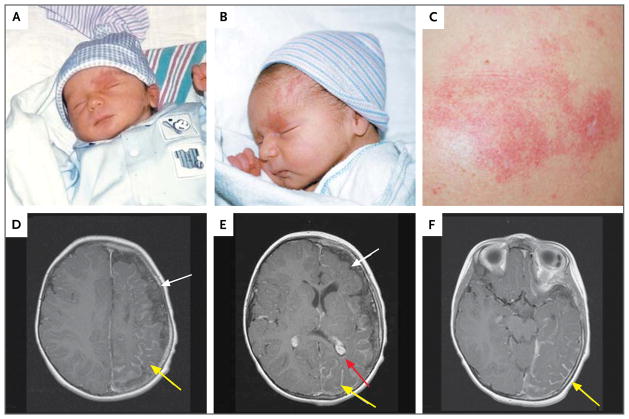
Figure 1. Representative Photographs and Magnetic Resonance Imaging (MRI) Scans from Study Participants with the Sturge–Weber Syndrome or Isolated Port-Wine Stains.
Photographs of a participant with the Sturge–Weber syndrome (Patient 36), obtained at birth, show a facial port-wine stain with a left-sided V1 distribution (Panels A and B). The child began having seizures at 7 months of age. An isolated port-wine stain birthmark on the left shoulder from a participant without the Sturge–Weber syndrome (Patient 10) shows a birthmark that is flat and red without evidence of hypertrophy or cobblestoning or any other associated vascular or lymphatic anomaly (Panel C). Axial contrast-enhanced MRI scans of the brain in a participant with the Sturge–Weber syndrome (Panels D, E, and F; Patient 36 at 17 months of age) show left-sided hemispheric leptomeningeal enhancement (yellow arrows), an enlarged and enhancing left-sided choroid plexus (red arrow), and left hemispheric brain atrophy (white arrows).
METHODS
STUDY OVERSIGHT
All the sequencing was performed with approval from the institutional review board at the Johns Hopkins University or Duke University. Deidentified samples (see Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org) were obtained with written informed consent from persons with the Sturge–Weber syndrome or from the Brain and Tissue Bank for Developmental Disorders of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD). The age, sex, race or ethnic group, and syndrome status (apparently non-syndromic port-wine stain or the Sturge–Weber syndrome) of the participants were confirmed and recorded from the source, along with information regarding the surgery, autopsy, or other procedure that was used for obtaining the tissue.
TISSUE SAMPLES
We obtained tissue samples from research participants and from the NICHD Brain and Tissue Bank for Developmental Disorders (Table S1 in the Supplementary Appendix). Included were samples of skin with port-wine stains and of visibly normal skin from participants with the Sturge–Weber syndrome (23 samples from 9 participants), samples of skin with port-wine stains and of visibly normal skin from participants without the Sturge–Weber syndrome (14 samples from 13 participants), samples of brain tissue from persons with the Sturge–Weber syndrome (50 samples from 18 persons), samples of brain tissue from presumably normal persons (controls; 6 samples from 6 persons), and samples of brain tissue from persons with the unrelated cerebral cavernous vascular malformation (4 samples from 4 persons). Amplicon sequencing and single-base extension interrogation (SNaPshot analysis) were used to investigate each tissue sample for the c.548G→A mutation. We assayed SNaPshot specificity by testing brain tissue from 5 normal controls (data not shown). When multiple samplings of biopsied tissue or multiple sequencing assays were performed, we considered the participant to be positive for the mutation if at least 1 tissue sample tested positive (≥1% mutant allele) and to be negative if every tissue sample tested negative for the mutation (<1% mutant allele).
WHOLE-GENOME SEQUENCING
Genomic DNA was purified from paired samples of affected tissue and unaffected tissue or blood from three participants (for a total of six samples). Whole-genome sequencing was performed on an Illumina HiSeq 2000 sequencer to a mean per-sample depth of coverage between 33X and 51X. Details regarding the preparation of the samples and bioinformatic methods are provided in the Supplementary Appendix.
TARGETED AMPLICON SEQUENCING
GNAQ exon 4 and the adjacent intronic sequence were amplified by means of polymerase chain reaction in a two-stage reaction. Paired-end reads were generated with the use of an Illumina MiSeq sequencer and were evaluated at the c.548 position for base calls supporting the c.548G→A mutation. Samples were considered to have a mutation if the percentage of reads supporting a mutation exceeded 1% (10 times the expected base miscall rate of 0.1%). Details regarding sequencing adapters, barcodes, and primer sequences are provided in the Supplementary Appendix.
PLASMIDS
Two specific mutations, c.548G→A (encoding p.Arg183Gln) and c.626A→T (encoding p.Gln209 Leu), were introduced separately into GNAQ with the use of primers for site-directed mutagenesis (Table S4 in the Supplementary Appendix). Serum response element plasmid (pSRE)–Luc (Agilent Technologies) and pSV40-RL (Roche) were used as reporter plasmids for the luciferase assay.
CELL CULTURE AND WESTERN BLOTTING
Human embryonic kidney (HEK) 293T cell lysates were analyzed by means of Western blotting with the use of standard methods. Details of culture conditions, antibodies, and methods of Western blotting are provided in the Supplementary Appendix.
LUCIFERASE ASSAY
GNAQ, GNAQ p.Arg183Gln or GNAQ encoding p.Gln209Leu, pSRE-Luc, and pSV40-RL were transfected into HEK293T cells, which were lysed after 20 to 24 hours of incubation. At the end of the incubation period, luciferase activity was measured.
PRIMER EXTENSION-BASED ASSAY
DNA was extracted from formalin-fixed, paraffin-embedded samples from participants with portwine stains. Primers designed to amplify exon 4 of GNAQ (Table S5 in the Supplementary Appendix) were used to amplify genomic DNA from each of the samples. Amplicons were interrogated for their sequence at position c.548 with the use of the SNaPshot Multiplex Kit (Life Technologies) and analyzed on an ABI Prism 3130 genetic analyzer (Life Technologies). Reference and mutant allele frequencies were calculated on the basis of the area of the resulting peaks.
RESULTS
IDENTIFICATION OF GNAQ SOMATIC VARIANT
To test the hypothesis that the Sturge–Weber syndrome is associated with a somatic mosaic mutation, we sequenced the whole genomes of paired DNA samples from affected regions (biopsied tissue with port-wine stain or hemispherectomized brain tissue) and matched, presumably normal regions (blood or unaffected skin or brain tissue) from three persons with the Sturge–Weber syndrome. This sequencing resulted in the identification of 1294 somatic single-nucleotide variants found in at least one of three affected samples. We calculated the prevalence of the variant allele at each of these 1294 sites in all affected and normal samples and identified 658 single-nucleotide variants that were present in two or three affected samples and were not present in any normal samples (Fig. S1 in the Supplementary Appendix). We functionally annotated and ranked the 1294 somatic single-nucleotide variants (Table S6 in the Supplementary Appendix) using the Variant Annotation, Analysis and Search Tool (VAAST).7 This resulted in the identification of one nonsynonymous somatic single-nucleotide variant that was present in all three affected samples and was not present in the samples that were presumed to be normal — a c.548G→A nucleotide transition in GNAQ on chromosome 9q21, encoding guanine nucleotide binding protein (G protein), q polypeptide (G_α_q). The variant is predicted to result in the amino acid substitution p.Arg183Gln. The affected arginine residue, at position 183, is conserved in 24 human proteins paralogous to G_α_q (Fig. S2 in the Supplementary Appendix).
DETECTION OF GNAQ SOMATIC VARIANT IN SAMPLES FROM PATIENTS WITH THE STURGE–WEBER SYNDROME
The results of our studies of skin samples were as follows: 100% of participants (9 of 9) with the Sturge–Weber syndrome were positive for the c.548G→A mutation in port-wine–stained skin, 86% of participants (6 of 7) with the syndrome were negative for the mutation in visibly normal skin, and 92% of participants (12 of 13) with apparently nonsyndromic port-wine stains were positive for the mutation (Table 1). The mutation was also detected in brain samples from 83% of participants (15 of 18) with the Sturge–Weber syndrome, whereas 100% of brain samples (6 of 6 samples) from normal controls were negative. The results were negative in 100% of formalin-fixed, paraffin-embedded brain samples (4 of 4 samples) from persons with cerebral cavernous malformation (an unrelated cerebrovascular malformation) (Table 2), as well as in 99.3% of exomes from the 1000 Genomes database (664 of 669 exomes) (Table S7 in the Supplementary Appendix). In total, 88% of the participants (23 of 26) with the Sturge–Weber syndrome were positive for the c.548G→A mutation in either port-wine–stained skin or brain tissue.
Table 1.
Somatic Mutation of GNAQ in Skin Samples.*
| Patient No. | Mutation Present† | PWS | SWS | Mutant Allele Frequency‡ | Total No. of Samples Assayed |
|---|---|---|---|---|---|
| percent | |||||
| 1 | Yes | Yes | Yes | 3.60 | 1 |
| 1 | No | No | Yes | 0.11 | 1 |
| 2 | Yes | Yes | Yes | 3.17 | 1 |
| 2 | No | No | Yes | 0.13 | 1 |
| 3 | Yes | Yes | Yes | 6.06–6.46 | 2 |
| 3 | No | No | Yes | 0.62–0.93 | 2 |
| 4 | Yes | Yes | Yes | 3.50–4.51 | 2 |
| 4 | No | No | Yes | 0.13–0.90 | 2 |
| 5 | Yes | Yes | Yes | 3.38 | 1 |
| 5 | No | No | Yes | 0.11 | 1 |
| 6 | Yes | Yes | Yes | 3.99 | 1 |
| 7 | Yes | Yes | Yes | 2.05–2.16 | 2 |
| 7 | Yes | No | Yes | 0.09–2.00 | 2 |
| 8 | Yes | Yes | Yes | 4.08 | 1 |
| 8 | No | No | Yes | 0.06 | 1 |
| 9 | Yes | Yes | No | 5.58 | 1 |
| 10 | Yes | Yes | No | 2.76 | 1 |
| 10 | Yes | No | No | 1.14 | 1 |
| 11 | Yes | Yes | No | 6.70 | 1 |
| 12 | No | Yes | No | 0.00 | 1 |
| 13 | Yes | Yes | No | 5.90 | 1 |
| 14 | Yes | Yes | No | 6.20 | 1 |
| 15 | Yes | Yes | No | 14.20 | 1 |
| 16 | Yes | Yes | No | 1.70 | 1 |
| 17 | Yes | Yes | No | 4.50 | 1 |
| 18 | Yes | Yes | No | 5.30 | 1 |
| 19 | Yes | Yes | No | 4.70 | 1 |
| 20 | Yes | Yes | No | 4.30 | 1 |
| 21 | Yes | Yes | No | 18.10 | 1 |
| 22 | Yes | Yes | Yes | 5.00 | 1 |
Table 2.
Somatic Mutation of GNAQ in Brain-Tissue Samples.*
| Patient No. | Mutation Present | SWS | Mutant Allele Frequency | Total No. of Samples Assayed |
|---|---|---|---|---|
| percent | ||||
| 7 | Yes | Yes | 5.57–5.63 | 2 |
| 23 | Yes | Yes | 5.56–5.78 | 2 |
| 24 | Yes | Yes | 2.67–3.51 | 2 |
| 25 | No | Yes | 0.02–0.10 | 2 |
| 26 | Yes | Yes | 0.13–3.06 | 4 |
| 27 | Yes | Yes | 2.19–5.12 | 2 |
| 28 | Yes | Yes | 6.95–8.13 | 4 |
| 29 | Yes | Yes | 6.04–11.15 | 5 |
| 30 | Yes | Yes | 4.14 | 1 |
| 31 | Yes | Yes | 4.78 | 1 |
| 32 | Yes | Yes | 0.22–1.48 | 4 |
| 33 | Yes | Yes | 4.04–5.74 | 2 |
| 34 | No | Yes | 0.05–0.12 | 2 |
| 35 | Yes | Yes | 0.05–1.51 | 7 |
| 36 | Yes | Yes | 0.35–6.03 | 5 |
| 37 | Yes | Yes | 5.74–6.49 | 2 |
| 38 | No | Yes | 0.03–0.05 | 2 |
| 39 | Yes | Yes | 1.83 | 1 |
| 40 | No | No | 0.11 | 1 |
| 41 | No | No | 0.05 | 1 |
| 42 | No | No | 0.08 | 1 |
| 43 | No | No | 0.09 | 1 |
| 44 | No | No | 0.04 | 1 |
| 45 | No | No | 0.04 | 1 |
| 46 | No | No, CCM | 0.00 | 1 |
| 47 | No | No, CCM | 0.00 | 1 |
| 48 | No | No, CCM | 0.00 | 1 |
| 49 | No | No, CCM | 0.00 | 1 |
Amplicon sequencing showed that mutant allele frequencies ranged from 1.0 to 18.1% and read depth ranged from 2446 to 93,008 (median, 12,947). Mutant allele frequencies in exomes in the 1000 Genomes database ranged from 1.0 to 1.5%, and read depth ranged from 100 to 453 (median, 271). GNA11 mutations have also been found in patients with uveal melanoma.8 We tested GNAQ Arg183Gln mutation–negative samples from participants with the Sturge–Weber syndrome and those with nonsyndromic port-wine stains for the presence of previously identified GNA11 mutations (p.Arg183Cys, c.547C→T and c.546C→T; p.Arg183His, c.548G→A; p.Gln209Leu, c.626A→T and c.627G→A; and p.Gln209Pro, c.626A→C) using SNaPshot analysis. We did not detect any of these mutations (data not shown).
EFFECT OF MUTATION ON MAPK SIGNALING PATHWAY
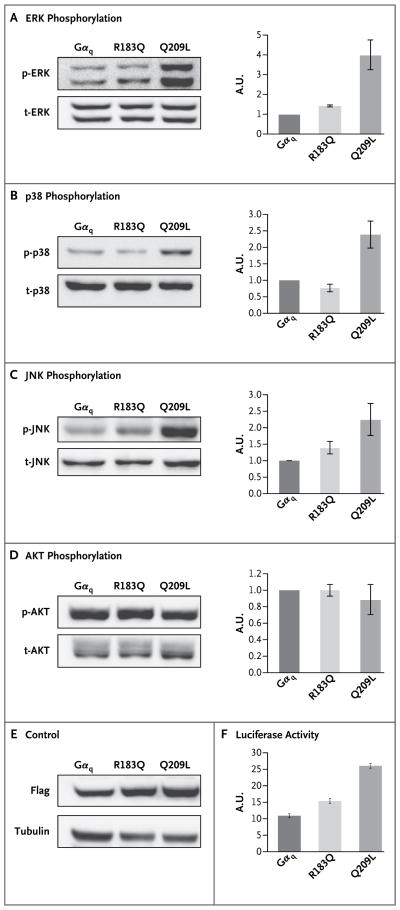
The somatic substitutions in GNAQ encoding p.Gln209Leu and p.Arg183Gln are found in patients with uveal melanoma. The more common p.Gln209Leu has been shown to overactivate the mitogen-activated protein kinase (MAPK) pathway. 9 We examined whether p.Arg183Gln would likewise overactivate the MAPK pathway. As shown in Figure 2A, cells transfected with GNAQ p.Gln209Leu or GNAQ p.Arg183Gln, as compared with cells transfected with nonmutant GNAQ, showed significant activation of extracellular signal-regulated kinase (ERK) (P<0.05). However, the activation induced by p.Arg183Gln was modest as compared with the activation induced by p.Gln209Leu. We also examined the effect of these substitutions on additional downstream signaling pathways. G_α_q p.Gln209Leu strongly activated p38 and Jun N-terminal kinase (JNK), other MAPK pathway members, whereas p.Arg183Gln did not (Fig. 2B and 2C). Neither substitution had an effect on the AKT signaling pathway (Fig. 1D). These data show that p.Arg183Gln has a gain-of-function effect that activates downstream signaling pathways. However, the effect of p.Arg183Gln in MAPK signal transduction appeared to be both weaker and less promiscuous with respect to the activation of downstream effectors than the effect of the substitution p.Gln209Leu that is found more commonly in uveal melanoma tissue.
Figure 2. Downstream Effectors of G_α_q.
Plasmid encoding nonmutant G_α_q, p.Arg183Gln, and p.Gln209Leu were transfected into human embryonic kidney (HEK) 293T cells. Strongly increased phosphorylation of extracellular signal-regulated kinase (ERK) is seen with G_α_q p.Gln209Leu, and weaker but marked activation with G_α_q p.Arg183Gln (P<0.05 for both comparisons) (Panel A). Increased phosphorylation of p38 is seen with G_α_q p.Gln209Leu (P<0.05) but not with G_α_q p.Arg183Gln (Panel B). Increased phosphorylation of Jun N-terminal kinase (JNK) is seen with G_α_q p.Gln209Leu (P<0.05), and weaker activation with G_α_q p.Arg183Gln (P = 0.052) (Panel C). No change in phosphorylation of AKT is seen with either the G_α_q p.Arg183Gln or the p.Gln209Leu construct (Panel D). A control for transfection efficiency, transfected into HEK 293T cells, shows similar amounts of the three transfected, Flag-tagged proteins (Panel E). A serum-response- element (SRE) luciferase assay (Panel F) shows the relative luciferase activity expressed under the control of the SRE promoter, coexpressed with GNAQ encoding p.Arg183Gln and p.Gln209Leu, as compared with nonmutant G_α_q (P<0.05 for both comparisons). AU denotes arbitrary units, the prefix p antibody recognizing phosphorylated antigen, and the prefix t antibody recognizing total antigen.
EFFECT OF GNAQ MUTATION ON SRE PROMOTER ACTIVITY
A different substitution in GNAQ encoding a variant at the same amino acid residue, p.Arg183Cys, was previously shown to overstimulate the serum response element (SRE) in a promoter reporter assay. 10 We investigated whether the p.Arg183Gln substitution had the same stimulatory effect on SRE promoter activity. We transfected HEK 293T cells with pSRE-Luc, pSV40-RL (reporter constructs), and GNAQ, GNAQ p.Arg183Gln, or GNAQ p.Gln209Leu plasmids and measured luciferase activity after 24 hours. Both p.Gln209Leu and p.Arg183Gln showed significantly increased reporter activity as compared with nonmutant GNAQ (P<0.05), confirming that the p.Arg183Gln mutation is a gain-of-function or activating mutation (Fig. 2F). In this assay, the p.Gln209Leu substitution again showed a stronger effect than did p.Arg183Gln.
DISCUSSION
Rudolf Happle first suggested that sporadic asymmetric or scattered birth defects involving the skin are caused by somatic mosaic mutations that would be lethal if they occurred in very early embryonic development.11 Somatic mosaic activating mutations have been identified in several disorders, including the McCune–Albright syndrome12 and the Proteus syndrome.13 In the current study, we found that a specific somatic mosaic activating mutation in GNAQ is associated with both the Sturge–Weber syndrome, a neurocutaneous disorder, and apparently nonsyndromic port-wine stains. GNAQ encodes G_α_q, a member of the q class of G-protein alpha subunits that mediates signals between G-protein–coupled receptors and downstream effectors. We have identified somatic mosaic GNAQ encoding p.Arg183Gln amino acid substitutions in skin and brain tissue from patients with the Sturge–Weber syndrome and in skin tissue with nonsyndromic port-wine stains and have shown that this mutation, much like the GNAQ variant encoding p.Gln209Leu, activates downstream MAPK signaling. G_α_q Arg183 is conserved in the guanosine triphosphate (GTP) binding pocket of all human G_α_ subunits, where it plays a critical role in the hydrolysis of GTP, the key step required for inactivation of the protein. Substitution of cysteine at this position results in a reduction in the intrinsic GTPase activity, leading to increased signaling activity.10,14–18
Activating mutations in genes encoding G_α_ subunits have previously been shown to be associated with relevant phenotypes, including the McCune–Albright syndrome, which is characterized by skeletal abnormalities and abnormal skin pigmentation.12 Activating somatic GNAQ mutations have been identified in blue nevi and the more extensive nevi of Ota.9 When these melanocytic nevi are colocalized with port-wine stains, the disorder is termed phakomatosis pigmentovascularis, which is occasionally found in association with the Sturge–Weber syndrome.19 Mutations in GNAQ were also identified in a chemical mutagenesis screen for a dark-skin phenotype in laboratory mice.20 Two of the dark-skin mutant alleles were identified at positions corresponding to human G_α_q p.Val179Met and p.Phe335Leu. These germline-derived amino acid substitutions cause an increase in the number of neural-crest cells that differentiate into melanoblasts. The abnormal early melanocytic development resulting from these mutations in the neural-crest cells is mediated through endothelin, a G-protein– coupled receptor.20 Since endothelin also has important roles in vasculogenesis,21 dysregulation of this G-protein–coupled receptor as a result of the G_α_q p.Arg183Gln mutation in persons with the Sturge–Weber syndrome and those with nonsyndromic port-wine stains may also bring about vascular malformation.
A somatic activating mutation may have oncogenic potential. In fact, somatic mutations of GNAQ in melanocytes are associated with uveal melanoma. The most common mutation, causing G_α_q p.Gln209Leu, is an activating mutation that leads to increased downstream signaling through the MAPK pathway. The activation of this pathway increases cell proliferation and inhibits apoptosis.9 A few uveal melanomas have been reported to harbor a somatic mutation in GNAQ encoding p.Arg183Gln, although the functional consequence of this substitution has not been reported.8 The pathogenesis of uveal melanoma is likely to be very different from the pathogenesis of nonsyndromic port-wine stains and the Sturge–Weber syndrome. Melanomas frequently have several somatic mutations.22 We found no evidence of accumulating mutations on whole-genome sequencing of our three paired samples (affected and unaffected tissue) from participants with the Sturge–Weber syndrome. In addition, the Sturge–Weber syndrome, nonsyndromic port-wine stains, and melanocytic nevi are thought to originate during fetal development; therefore, the effects of the same GNAQ somatic mutation may be quite different, depending on the cell type and the point in development at which they arise. There are reported cases of uveal melanoma associated with phakomatosis pigmentovascularis,23 and the coincidence of the blue nevus and port-wine stain phenotypes in a patient with the Sturge–Weber syndrome may indicate an increased risk of uveal melanoma, although such coincidences are rare.
We have shown that the G_α_q p.Arg183Gln substitution can activate ERK and does not activate p38 or JNK in the same way that p.Gln209Leu does. We propose that the moderate activation of ERK, the differential effect on p38 and JNK pathways, or both may contribute to the port-wine stains and syndromic characteristics of the Sturge–Weber syndrome. This may occur through either upstream regulation of G_α_q or downstream modulation of the G-protein–coupled receptor signaling cascade. To provide insight into possible mechanisms underlying the partial activation of G_α_ downstream signaling, we considered an interesting corollary in other G_α_ proteins. The regulator of G-protein signaling (RGS) proteins serve as GTPase-activating proteins for G_α_ proteins, inhibiting downstream activation. Of these, RGS4 regulates G_α_q and G_α_i, whereas RGS2 is selective for G_α_q.24 On examination of the ability of RGS4 to regulate G_α_i1 with activating mutations in positions p.Arg178Cys and p.Gln204Leu, homologous to G_α_q p.Arg183Gln and p.Gln209Leu, it was found that all regulatory ability was lost for p.Gln204Leu, whereas GTPase activity was partially maintained for p.Arg178Cys.18 Thus, the weaker and less promiscuously activating effects of G_α_q p.Arg183Gln, as compared with G_α_q p.Gln209Leu, may be a result of partial regulation by a member of the RGS family. G_α_q is also able to initiate sustained RhoA and Rac1 activation independently of PLC-β, through direct interaction with Trio, a guanine nucleotide exchange factor. It has been shown that G_α_q-mediated oncogenic proliferation, mediated through p38 and JNK, is significantly reduced after Trio knockdown without affecting PLC or ERK activation levels.25 This provides a possible mechanism, related to altered affinity of protein–protein interactions with both regulators (RGS family) and cascade activators (Trio), to explain the non-oncogenic proliferation seen in the Sturge–Weber syndrome and nonsyndromic port-wine stains. We hypothesize that only the weaker effect of somatic G_α_q p.Arg183Gln would be compatible with the abnormal but nonlethal development of the cerebrovascular system seen in the Sturge–Weber syndrome. We also hypothesize that during vulnerable periods in embryonic development, moderately increased baseline signaling downstream of G_α_q, or dysregulated signaling through G-protein–coupled receptors such as that for endothelin,21 may result in the malformed, progressively dilated, and abnormally innervated blood vessels underlying port-wine stains. There is some evidence in the literature to support this hypothesis. Shirazi et al. reported the localization of phosphorylated ribosomal protein S6 (RPS6), which is downstream of MAPK signaling, to endothelial cells lining the luminal wall of abnormal blood vessels in port-wine stain tissue from patients with the Sturge–Weber syndrome.26
The nonsyndromic port-wine stains may represent a late origin of the somatic GNAQ mutation in vascular endothelial cells, whereas the Sturge–Weber syndrome mutation may occur earlier in development, in progenitor cells that are precursors to a larger variety of cell types and tissues, leading to the syndromic phenotype. We found that 0.7% of samples of blood from the 1000 Genomes database (5 of 669 samples) that were tested for the presence of the GNAQ mutation encoding p.Arg183Gln were positive. The reported prevalence of port-wine stains1,2 is 0.3 to 0.5%. We therefore hypothesize that the 0.7% prevalence in this database represents the occurrence of port-wine stains in this population.
Our data indicate that there is a single underlying mechanism for the Sturge–Weber syndrome and nonsyndromic port-wine stains and add a molecular basis for a decades-old hypothesis regarding the cause of these malformations. The scientific and translational novelty of this discovery lies in the association of both apparently nonsyndromic port-wine stains and the Sturge-Weber syndrome with a mutation in a specific gene, a specific genetic mechanism, and a set of potential pathways, which provides a foundation for further scientific and clinical research.
Supplementary Material
Supplement1
Acknowledgments
Supported by grants from the National Institute of Neurological Disorders and Stroke (NINDS) (National Institutes of Health [NIH] U54NS065705, to Drs. Marchuk, Comi, and Pevsner) and from Hunter’s Dream for a Cure Foundation (to Dr. Comi). The Brain Vascular Malformation Consortium (U54NS065705) is a part of the NIH Rare Disease Clinical Research Network, supported through a collaboration between the NIH Office of Rare Diseases Research at the National Center for Advancing Translational Science and the NINDS. Human tissue was obtained from the Brain and Tissue Bank for Developmental Disorders of the Eunice Kennedy Shriver National Institute of Child Health and Human Development at the University of Maryland, Baltimore (funded by NIH Contract No. #HHSN275200900011C, Ref. No. NO1-HD-9-0011).
We thank the participants with the Sturge–Weber syndrome and their families for generously agreeing to participate in this study; Kira Lanier and Cathy Bachur for their excellent assistance with coordinating this research; the Sturge-Weber Foundation for its encouragement of this work and assistance in obtaining samples; Drs. Harry Chugani and Diane Chugani as well as Peter Black for providing brain and skin tissue; and Drs. Sarah Wheelan and Srinivasan Yegnasubramanian and the Next Generation Sequencing Center at the Johns Hopkins Sidney Kimmel Comprehensive Cancer Center for their support with the high-throughput sequencing experiments.
Footnotes
The views expressed in this article are those of the authors and do not necessarily represent the official views of the National Institutes of Health.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Comi AM. Update on Sturge-Weber syndrome: diagnosis, treatment, quantitative measures, and controversies. Lymphat Res Biol. 2007;5:257–64. doi: 10.1089/lrb.2007.1016. [DOI] [PubMed] [Google Scholar]
- 2.Piram M, Lorette G, Sirinelli D, Herbreteau D, Giraudeau B, Maruani A. Sturge-Weber syndrome in patients with facial port-wine stain. Pediatr Dermatol. 2012;29:32–7. doi: 10.1111/j.1525-1470.2011.01485.x. [DOI] [PubMed] [Google Scholar]
- 3.Ch’ng S, Tan ST. Facial port-wine stains — clinical stratification and risks of neuro-ocular involvement. J Plast Reconstr Aesthet Surg. 2008;61:889–93. doi: 10.1016/j.bjps.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Greene AK, Taber SF, Ball KL, Padwa BL, Mulliken JB. Sturge-Weber syndrome: soft-tissue and skeletal overgrowth. J Craniofac Surg. 2009;20(Suppl 1):617–21. doi: 10.1097/SCS.0b013e318192988e. [Erratum, J Craniofac Surg 2009;20:1629–30.] [DOI] [PubMed] [Google Scholar]
- 5.Gasparini G, Perugini M, Vetrano S, Cassoni A, Fini G. Angiodysplasia with osteohypertrophy affecting the oromaxillofacial area: clinical findings. J Craniofac Surg. 2001;12:485–9. doi: 10.1097/00001665-200109000-00016. [DOI] [PubMed] [Google Scholar]
- 6.Vissers W, Van Steensel M, Steijlen P, Renier W, Van De Kerkhof P, Van Der Vleuten C. Klippel-Trenaunay syndrome and Sturge-Weber syndrome: variations on a theme? Eur J Dermatol. 2003;13:238–41. [PubMed] [Google Scholar]
- 7.Yandell M, Huff C, Hu H, et al. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011;21:1529–42. doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takasaki J. A novel Gα(q/11)-selective inhibitor. J Biol Chem. 2004;279:47438–45. doi: 10.1074/jbc.M408846200. [DOI] [PubMed] [Google Scholar]
- 11.Happle R. Lethal genes surviving by mosaicism: a possible explanation for sporadic birth defects involving the skin. J Am Acad Dermatol. 1987;16:899–906. doi: 10.1016/s0190-9622(87)80249-9. [DOI] [PubMed] [Google Scholar]
- 12.Weinstein LS, Shenker A, Gejman PV, Merino MJ, Friedman E, Spiegel AM. Activating mutations of the stimulatory G protein in the McCune–Albright syndrome. N Engl J Med. 1991;325:1688–95. doi: 10.1056/NEJM199112123252403. [DOI] [PubMed] [Google Scholar]
- 13.Lindhurst MJ, Sapp JC, Teer JK, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med. 2011;365:611–9. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conklin BR, Chabre O, Wong YH, Federman AD, Bourne HR. Recombinant Gq alpha: mutational activation and coupling to receptors and phospholipase C. J Biol Chem. 1992;267:31–4. [PubMed] [Google Scholar]
- 15.Orth JHC, Preuss I, Fester I, Schlosser A, Wilson BA, Aktories K. Pasteurella multocida toxin activation of heterotrimeric G proteins by deamidation. Proc Natl Acad Sci U S A. 2009;106:7179–84. doi: 10.1073/pnas.0900160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coleman DE, Berghuis AM, Lee E, Linder ME, Gilman AG, Sprang SR. Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis. Science. 1994;265:1405–12. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 17.Kimple AJ, Bosch DE, Giguère PM, Siderovski DP. Regulators of G-protein signaling and their Gα substrates: promises and challenges in their use as drug discovery targets. Pharmacol Rev. 2011;63:728–49. doi: 10.1124/pr.110.003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berman DM, Wilkie TM, Gilman AG. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell. 1996;86:445–52. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 19.Al Robaee A, Banka N, Alfadley A. Phakomatosis pigmentovascularis type IIb associated with Sturge-Weber syndrome. Pediatr Dermatol. 2004;21:642–5. doi: 10.1111/j.0736-8046.2004.21605.x. [DOI] [PubMed] [Google Scholar]
- 20.Van Raamsdonk CD, Fitch KR, Fuchs H, de Angelis MH, Barsh GS. Effects of G-protein mutations on skin color. Nat Genet. 2004;36:961–8. doi: 10.1038/ng1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kajio F, Nakazawa M. Vascular effects of endothelin-1 in stage 21 chick embryos. Heart Vessels. 1997;12:300–5. doi: 10.1007/BF02766807. [DOI] [PubMed] [Google Scholar]
- 22.Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shields CL, Kligman BE, Suriano M, et al. Phacomatosis pigmentovascularis of cesioflammea type in 7 patients: combination of ocular pigmentation (melanocytosis or melanosis) and nevus flammeus with risk for melanoma. Arch Ophthalmol. 2011;129:746–50. doi: 10.1001/archophthalmol.2011.135. [DOI] [PubMed] [Google Scholar]
- 24.Hepler JR, Berman DM, Gilman AG, Kozasa T. RGS4 and GAIP are GTPase-activating proteins for Gq alpha and block activation of phospholipase C beta by gamma-thio-GTP-Gq alpha. Proc Natl Acad Sci U S A. 1997;94:428–32. doi: 10.1073/pnas.94.2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaqué JP, Dorsam RT, Feng X, et al. A genome-wide RNAi screen reveals a trio-regulated Rho GTPase circuitry transducing mitogenic signals initiated by G protein- coupled receptors. Mol Cell. 2013;49:94–108. doi: 10.1016/j.molcel.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirazi F, Cohen C, Fried L, Arbiser JL. Mammalian target of rapamycin (mTOR) is activated in cutaneous vascular malformations in vivo. Lymphat Res Biol. 2007;5:233–6. doi: 10.1089/lrb.2007.1012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement1