TOGp, the Human Homolog of XMAP215/Dis1, Is Required for Centrosome Integrity, Spindle Pole Organization, and Bipolar Spindle Assembly (original) (raw)
Abstract
The XMAP215/Dis1 MAP family is thought to regulate microtubule plus-end assembly in part by antagonizing the catastrophe-promoting function of kin I kinesins, yet XMAP215/Dis1 proteins localize to centrosomes. We probed the mitotic function of TOGp (human homolog of XMAP215/Dis1) using siRNA. Cells lacking TOGp assembled multipolar spindles, confirming results of Gergely et al. (2003. Genes Dev. 17, 336–341). Eg5 motor activity was necessary to maintain the multipolar morphology. Depletion of TOGp decreased microtubule length and density in the spindle by ∼20%. Depletion of MCAK, a kin I kinesin, increased MT lengths and density by ∼20%, but did not disrupt spindle morphology. Mitotic cells lacking both TOGp and MCAK formed bipolar and monopolar spindles, indicating that TOGp and MCAK contribute to spindle bipolarity, without major effects on MT stability. TOGp localized to centrosomes in the absence of MTs and depletion of TOGp resulted in centrosome fragmentation. TOGp depletion also disrupted MT minus-end focus at the spindle poles, detected by localizations of NuMA and the p150 component of dynactin. The major functions of TOGp during mitosis are to focus MT minus ends at spindle poles, maintain centrosome integrity, and contribute to spindle bipolarity.
INTRODUCTION
Rapid microtubule (MT) turnover is required for many cellular processes, including assembly of the mitotic spindle. The assembly and dynamic turnover of purified tubulin is too slow to generate the rates of MT growth and catastrophe measured in living cells, indicating that associated proteins are necessary to speed MT turnover in vivo (Desai and Mitchison, 1997). It is generally thought that a delicate balance between MT stabilizing and destabilizing proteins generates the MT dynamics observed in cells (Kinoshita et al., 2002).
One of the key structural proteins regulating the dynamic turnover of MTs is XMAP215/Dis1. The founding member of the XMAP215/Dis1 protein family, XMAP215, stimulates rapid assembly of MT plus ends and protects plus ends from the catastrophe-promoting activity of the kin I kinesin, XKCM1 (Gard and Kirschner, 1987; Vasquez et al., 1994; Wilde and Zheng, 1999; Tournebize et al., 2000; Kinoshita et al., 2001). Consistent with an MT assembly stimulating function, depletion of XMAP215 from Xenopus egg extracts (Wilde and Zheng, 1999; Tournebize et al., 2000) or inactivation of the protein in Xenopus oocytes (Becker et al., 2003) results in assembly of short MTs. XMAP215 also increases the dynamicity of MTs in vitro by promoting both rapid polymerization and rapid depolymerization (Vasquez et al., 1994), perhaps by inhibiting MTs from entering a pause state between growth and shortening (Shirasu-Hiza et al., 2003).
In other organisms, XMAP215 homologues can either stabilize or destabilize MTs. TOGp (Homo sapiens) stimulates MT assembly (Charrassee et al., 1998) and is required for MT aster formation in HeLa mitotic extracts (Dionne et al., 2000), whereas mutations in Zyg-9 (Caenorhabditis elegans; Matthews et al., 1998; Srayko et al., 2003; Bellanger and Gonczy, 2003) or MOR1 (Arabidopsis; Whittington et al., 2001) result in shorter MTs. In contrast, Stu2p (Saccharomyces cerevisiae) destabilizes MTs (Kosco et al., 2001; van Breugel et al., 2003). It is not yet known whether Dis1 and Alp14 (Schizosaccharomyces pombe homologues) destabilize MTs, but these two XMAP215/Dis1 homologues function synergistically, not antagonistically, with the Kin I kinesins klp5 and 6 during mitosis (Garcia et al., 2002). The synergistic function of these two protein families raises the possibility that S. pombe XMAP215 homologues also destabilize MTs.
The functions of XMAP215/Dis1 proteins within the cell may not solely reflect their ability to modulate MT plus-end assembly. All members of the XMAP215/Dis1 family are localized to centrosomes or spindle pole bodies (Nabeshima et al., 1995; Wang and Huffaker, 1997; Charrasse et al., 1998; Matthews et al., 1998; Cullen et al., 1999; Graf et al., 2000), where they may contribute to microtubule nucleation (Lee et al., 2001; Popov et al., 2002) or microtubule anchorage to the centrosome (Usui et al., 2003).
TOGp, the human homolog of XMAP215/Dis1, is required for proper assembly of bipolar spindles (Gergely et al., 2003), but it is not yet known whether TOGp contributes to MT stability during mitosis or whether TOGp antagonizes MCAK, the human homolog of XKCM1. Here we use siRNA to deplete TOGp and/or MCAK to examine their roles in spindle assembly. Because TOGp localizes to centrosomes during mitosis, we also used siRNA to examine how TOGp contributes to centrosome and spindle pole organization. Our results indicate that the major functions of TOGp during mitosis are to tightly focus the spindle poles and maintain centrosome integrity.
MATERIALS AND METHODS
Cell Culture
HeLa cells were grown in MEM (Life Technologies, Rockville, MD) supplemented with 10% FBS, l-glutamine, and antibiotics/antimycotic. In some experiments monastrol (100 μM; a gift from Tarun Kapoor) was added to the culture medium for the times indicated.
siRNA
The 21-nucleotide siRNA duplexes were designed (Elbashir et al., 2001) based on a 19-bp ch-TOG cDNA region (GAGCCCAGAGUGGUCCAAA, position 123–142), MCAK cDNA region (GAUCCAACGCAGUAAUGGU, position 54–72; sequence suggested by C. Walczak), or hTPX (Garrett et al., 2002). siRNA duplexes were synthesized with symmetric 3′ dTdT overhangs (RNAs synthesized by Dharmacon Research [Boulder, CO] and double-stranded RNA (dsRNA) annealed according to the manufacturer's instructions). A BLAST search of the NCBI database ensured specific targeting to ch-TOG or MCAK mRNA. HeLa cells were plated on coverslips at 0.5 × 105 cells/cm2 in 35-mm dishes the day before transfection. Cells were incubated overnight in MEM lacking antibiotics and then transfected with 1 μg of dsRNA using GeneSilencer siRNA Transfection Reagent (Gene Therapy Systems, San Diego, CA). Cotransfection of 1 μg each dsRNA was used to simultaneously deplete TOG and MCAK. Single-stranded sense RNA sequences were used as controls. Typically, cells were analyzed for the loss of TOGp or MCAK 48 h after transfection using either immunoblotting or immunofluorescence. Depletion of hTPX2 was analyzed by immunofluorescence 36–39 h after transfection (Garrett et al., 2002).
Immunoblotting
Forty-eight hours after RNA transfection, each 35-mm dish was washed two times with PBS. Lysis buffer (50 mM HEPES, pH 7.3, 250 mM NaCl, 0.1% NP-40, 1 mM DTT, 1 mM Pefabloc, 1 mM EDTA, and a protease inhibitor cocktail; Boerhinger Mannheim, Indianapolis, IN) was added to each dish to yield a cell concentration of ∼1 × 107 cells/ml. Cells were scraped from the dish and lysed for 30 min on ice. The lysate was clarified by centrifugation (10 min at top speed in a microfuge) A small aliquot of the supernatant was used for protein determination (Bio-Rad, Richmond, CA) using IgG as the standard. 5× SDS sample buffer was added to the remaining supernatant. Typically, each gel lane was loaded with 5 μg total soluble protein.
Gel electrophoresis and immunoblotting were performed as described previously (Howell et al., 1999) except that 5% PAGE gels were used to allow >95% transfer of the 215-kDa TOGp. Primary antibodies used included mouse antitubulin (Sigma Chemical Co., St. Louis, MO; T5168), rabbit antiactin (Sigma A5060), rabbit anti-TOGp (Cassimeris et al., 2001), and sheep anti-MCAK (provided by Linda Wordeman). Appropriate anti-mouse or anti-sheep antibodies, conjugated to HRP (Sigma), were used in conjunction with enhanced chemiluminescence (Amersham Pharmacia Biotech, Piscataway, NJ) to detect protein bands.
Immunofluorescence
Cells were fixed in -20°C methanol/EDTA and stained as described previously (Piehl and Cassimeris, 2003). Antibodies used included: rabbit anti-TOGp, mouse antitubulin (Sigma T5168), rabbit anti γ-tubulin (Sigma T3320), mouse anti γ-tubulin (Sigma T6557), rabbit antipericentrin (Covance Laboratories, Madison, WI; RRB-432), rabbit anti-p150 (Vaughan and Vallee, 1995, provided by Kevin Vaughan), human CREST serum (provided by Bill Brinkley), rabbit anti-NuMA (whole serum, provided by Duane Compton), rabbit anti-hTPX2 (Garrett et al., 2002; serum provided by Duane Compton), rabbit anti-Eg5 (provided by Tarun Kapoor), and rabbit anti-XKCM1 (provided by Clare Walczak). Secondary antibodies included goat anti-mouse FITC (Jackson ImmunoResearch Laboratories, West Grove, PA), goat anti-rabbit Alexa 568 (Molecular Probes, Eugene, OR) and goat anti-human Alexa 594. Coverslips were mounted in Vectashield (Vector Laboratories, Inc., Burlingame, CA) and examined by wide-field or confocal microscopy (described below).
Anti-tubulin–stained cells were used to determine mitotic index. At least 200 cells were counted per coverslip in each of four separate experiments.
Our TOGp antibodies were raised in rabbits, as were many of the antibodies used for these localization studies. Therefore, it was not always possible to demonstrate TOGp depletion and localization of another spindle protein in the same cell. In these cases, we confined our observations to cells with highly disrupted mitotic spindles (see Figure 1B). These disrupted spindle structures were never observed in untreated or sense RNA-transfected cells and were always depleted of TOGp.
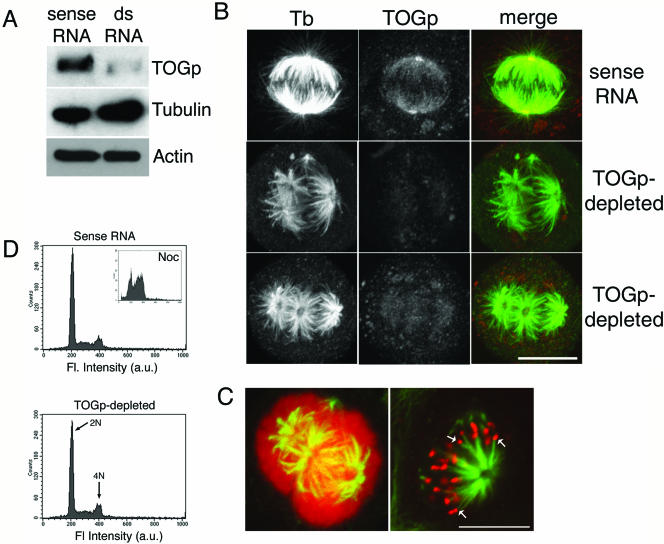
Figure 1.
TOGp depletion results in multipolar spindle formation. (A) Immunoblot demonstrating decreased TOGp levels 48 h after transfection with dsRNA. Tubulin and actin levels were not changed by TOGp depletion. (B) Immunofluorescence micrographs of spindle morphology in TOGp-depleted cells 48 h after transfection. Images were collected as Z-stacks using confocal microscopy and converted to maximium intensity projections. Images were acquired and printed with identical settings. TOGp-depleted cells have multipolar spindles with prominent holes in the center of the spindle poles. (C) Chromosomes associate with all MT asters in TOGp-depleted cells. Left image, maximium intensity projection of TOGp-depleted cell double-labeled for tubulin (green) and DNA (red). Right image, single confocal optical section from TOGp-depleted spindle showing tubulin (green) and kinetochores (CREST staining, red). Arrows mark kinetochores associated with the ends of MT bundles. Bars (B and C), 10 μm. (D) FACS analysis of DNA content after TOGp depletion. Cells depleted of TOGp have normal DNA content, indicating that the multipolar spindles did not arise from a previous failed division. Inset, DNA content of cells incubated overnight with nocodazole. The peaks were used to assign the 2N and 4N peaks. In all cases, control cells were transfected with the sense RNA sequence alone.
FACs Analysis
Cells were fixed and stained for FACs analysis 48 h after transfection. Cells were trypsinized and combined with any floating cells, pelleted, and resuspended in 0.5 ml PBS to a concentration of ∼1 × 107 cells/ml. Ice cold ethanol (70%) was added, and samples were incubated on ice for ≥2 h. Cells were rehydrated by pelleting and resuspension in PBS. Finally, the cells were pelleted again and resuspended in 500 μl staining solution (2 mg/100 μl propidium iodine, 0.2 mg/ml RNAse A, 0.1% NP-40). DNA content was measured using the 488 line from the argon laser of a Becton Dickinson FACScan (Mountain View, CA; Kenna and Skibbens, 2003). To assign the 4N peak, cells were blocked in mitosis by overnight incubation in 33 μM nocodazole.
Wide Field Microscopy
Fixed and stained cells were examined using a 60×/1.4 NA planapo objective on an inverted microscope (Nikon TE300; Garden City, NY) equipped for epi-illumination (Piehl and Cassimeris, 2003). Images were projected to a CCD camera (Hamamatsu C4742–95; Bridgewater, NJ) and acquired and stored as 12-bit files using MetaMorph software (Universal Imaging, West Chester, PA).
Confocal Microscopy
Coverslips were also examined using a 63×/1.4 NA plan apo objective on an inverted microscope (Zeiss Axiovert 200M) equipped with a Zeiss LSM510 META scan head. Argon ion and 543 HeNe lasers were used to generate the 488 and 543 nm wavelengths used for excitation. Pinholes were typically set to 1–1.5 airy units. Images, usually 1024 × 1024 or 512 × 512 pixels, were acquired at ∼4× zoom using four-line mean averaging. Each Z-series typically contained 20–40 slices ∼0.3-μm-thick for a total stack depth of ∼5 μm. For presentation, some image stacks were converted to maximum intensity projections using the Zeiss LSM510 META 3.0 software. Images were exported as JPEG files and printed using Photoshop 7.0 (Adobe, San Jose, CA).
Image Quantitation
MT staining intensity was measured using either 12-bit images obtained by wide-field microscopy or 8-bit images obtained by confocal microscopy. For wide-field microscopy, the exposure time and neutral density filters were kept constant. For confocal microscopy, the detector gain and offset were kept constant. Under the imaging conditions used, no pixels were saturated. To measure the average MT staining intensity of half spindles, the outline of each half spindle was traced using MetaMorph software. The mean pixel intensity within this region was then calculated by the software. Mean background intensity, measured in a region devoid of cells was subtracted from each half-spindle measurement. The results from three experiments (15–20 spindles for each condition per experiment) were pooled by setting the mean value for the control (untransfected or transfected with sense RNA) intensity to 100%.
The microtubule staining intensity of monopolar spindles assembled in monastrol were measured from single sections obtained by confocal microscopy. The section at the center of the Z-dimension was used and mean pixel intensity measured within a circle of 4-μm radius positioned near the brightly stained central region. Lengths of half spindles or diameters of monopolar spindles were measured from confocal images using the measure tool in the Zeiss LSM 510 META software.
The areas occupied by centrosomes were measured for anti–γ-tubulin–stained cells. The outline of γ-tubulin spots were traced and the area calculated using the overlay tools in the Zeiss LSM 510 software. Individual images from Z-series were used for measurements, using the section where each γ-tubulin spot appeared largest. The diameters of the holes at the centers of asters were measured from tubulin and NuMA stained spindles. For cells transfected with the sense RNA sequence (controls), the image stacks were converted to 180° 3D projections and rotated to view the spindle poles from their ends (see Figure 3). These projections were used to measure any small holes present at the spindle poles of control cells.
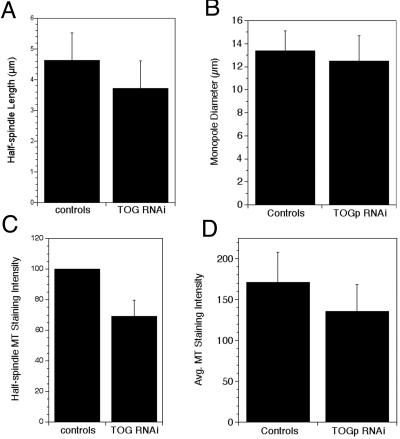
Figure 3.
TOGp does not make major contributions to MT length and density in the spindle. (A) Half spindle lengths measured from control (untreated or sense RNA transfected) and TOGp-depleted cells. Measured from 62 (control) and 90 (TOGp depleted) antitubulin-stained cells. Data from nine experiments, images collected by confocal microscopy. (B) Widths of monasters assembled in the presence of monastrol (100 μM). Data from three separate experiments, images collected by confocal microscopy (49 control and 58 TOGp-depleted cells). (C) MT staining intensity of half-spindles in control and TOGp-depleted cells. Data were normalized and pooled from three experiments. For each experiment the average intensity in controls (untreated and sense RNA transfected cells) was set to 100. Data from three experiments (102 control half spindles, 142 TOGp half spindles), images collected by wide-field microscopy. (D) MT staining intensity of monopolar spindles. Intensity was measured in a circle placed at the brightest region of the monoaster, adjacent to the central hole. Image stacks were collected by confocal microscopy, and a single optical section at the center of the monaster was used for quantitation. Data shown are from a representative experiment (15 control cells and 22 TOGp-depleted cells). All data are means ± SD.
Means were compared at the 95% confidence level using the ANOVA function in Microsoft Excel (Redmond, WA).
RESULTS
TOGp Depletion Delays Mitosis and Results in Multipolar Spindle Formation
To investigate the role of TOGp in mitosis, we reduced TOG protein level using siRNA. Based on immunoblotting, the level of TOGp was significantly reduced 48 h after transfection with dsRNA, but was unchanged by transfection with the sense RNA sequence alone (Figure 1A). Tubulin and actin levels were also unchanged by TOG siRNA, suggesting that RNA transfection did not simply cause a general decline in cell health or mRNA stability. By immunofluorescence, our antibodies only recognize TOGp in mitotic cells. Based on immunofluorescent staining of these mitotic cells, TOGp was greatly reduced in the majority of cells (∼80% of mitotic cells in a typical experiment contained undetectable levels of TOGp), whereas a small number of cells expressed TOGp at approximately normal levels. MTs in interphase cells were of normal length and density (our unpublished results).
Spindle morphology was severely disrupted in cells depleted of TOGp. As shown in Figure 1B, TOGp depletion resulted in multipolar spindle formation in ∼50% of mitotic cells, confirming recent results of Gergely et al. (2003). In TOGp-depleted cells, mitotic asters often contained a large central hole that did not stain positively for tubulin (Figure 1B). In contrast, a small percentage of untreated HeLa cells (or those transfected with the sense RNA sequence) assembled multipolar spindles (∼10%; 5 experiments), but these spindles always had tightly focused spindle poles (our unpublished results). In the TOGp-depleted spindles, one aster was often observed to form two half spindles (e.g., Figure 1B, bottom panel), similar to the phenotype observed in Drosophila embryos having mutations in the XMAP215/Dis1 homolog, msps (Cullen et al., 1999). Chromosomes were associated with all the asters, and kinetochores (visualized by CREST localization) were attached to MTs (Figure 1C). Multipolar spindles often lack astral MTs because MTs that would normally extend away from the chromosomes are recruited into other half-spindles. Consistent with the defects in bipolar spindle assembly, TOGp depletion increased the mitotic index from 5% in sense RNA–transfected cells to 13% in TOGp-depleted cells (average of 4 experiments). Both the siRNA sequence used here (targeted to the 5′ end) and one overlapping the stop codon (Gergely et al., 2003) yielded nearly identical TOGp depletion and multipolar spindle phenotypes (our unpublished results).
Multipolar spindles were observed 48 h after transfection with dsRNA. Because cells should have divided twice after transfection, it was possible that multipolar spindles resulted from a previous failed division during the 48-h incubation time. It is unlikely that this was the case because examination of cells fixed 24 h after transfection showed that only 16% of mitotic cells assembled multipolar spindles (our unpublished results). At this time most cells also had TOGp staining intensities similar to untreated cells (our unpublished results). A large percentage of multipolar spindles was not observed until ∼40 h after transfection. By 48 h after transfection, the DNA content of dsRNA transfected cells showed only 2N and 4N peaks (Figure 1D). Taken together, these results indicate that the multipolar spindles assembled in TOGp-depleted cells did not arise from previous failed divisions, confirming previous results by Gergely et al. (2003).
Eg5 Is Required for Multipolar Spindle Formation
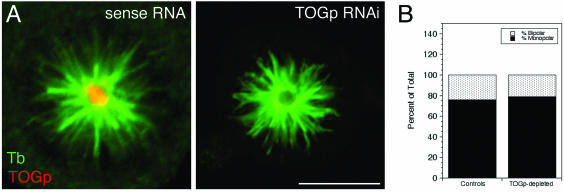
Bipolar spindle formation requires the kinesin, Eg5, for proper separation of microtubule asters (Gaglio et al., 1996; Kapoor et al., 2000). In TOGp-depleted cells, Eg5 was localized at spindle poles and along spindle MTs, similar to its distribution in untreated cells (our unpublished results; Kapoor et al., 2000), indicating that TOGp was not required for Eg5 localization. To determine whether Eg5 motor activity was necessary to maintain the multipolar spindle morphology of TOGp-depleted cells, cells were incubated in 100 μM monastrol for 4 h before fixation (44–48 h posttransfection with RNAs). Under these conditions, the majority of mitotic cells contained monopolar spindles (Figure 2). Most remaining mitotic cells had bipolar spindles, similar to that observed in untreated or sense RNA–transfected cells (Figure 2).
Figure 2.
Eg5 activity is required for multipolar spindle formation in TOGp-depleted cells. (A) Immunofluorescence micrographs (tubulin green, TOGp red) of monastrol-treated cells. Images are maximium intensity projections generated from confocal stacks. Scale bar, 10 μm. (B) Quantitation of the numbers of monopolar and bipolar spindles in monastrol-treated cells. No multipolar spindles were observed in TOGp-depleted cells and the percents of monopolar and bipolar spindles were identical to control cells (untreated or sense RNA-transfected cells).
TOGp Depletion Results in Small Changes to MT Polymer
To determine whether TOGp regulates MT stability in mitosis, we measured the length and density of spindle MTs in antitubulin-stained cells. TOGp depletion resulted in a small (20%) decrease in the length of MT half-spindles compared with sense RNA or untransfected cells at metaphase (Figure 3). This decrease in average length could reflect a reduced MT stability in TOGp-depleted cells or differences in MT length at prometaphase (the depleted cells resemble prometaphase because all chromosomes are not aligned on metaphase plates) or metaphase. Because factors other than MT stability could contribute to MT lengths in the TOGp-depleted multipolar spindles, we also measured the diameter of monastrol-induced monopolar spindles. In TOGp-depleted cells treated with monastrol, the diameter of the single aster was not significantly different from that in untreated or sense RNA–treated cells (Figure 3B). TOGp depletion also had only a small effect on spindle MT density. The mean antitubulin fluorescence intensity of half-spindles in TOGp-depleted cells was ∼70% of the value in sense RNA–treated or untreated cells (Figure 3C). In monastrol-treated cells, TOGp depletion also resulted in a drop in MT density (to ∼80% of the level in untreated cells or cells transfected with the sense RNA sequence).
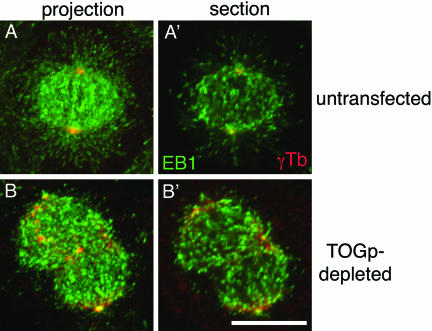
The length and density of MTs in the TOGp-depleted spindles suggested that TOGp did not make major contributions to MT stability during mitosis. We have not been able to express GFP-tubulin in TOGp-depleted cells to measure MT dynamics directly. To estimate MT dynamics in the spindle, we stained cells with antibodies to EB1 as a marker for growing plus ends (Mimori-Kiyosue et al., 2000; Tirnauer et al., 2002). It was difficult to interpret the EB1 staining patterns without double-labeling cells with antibodies to γ-tubulin to mark the centrosomes. As shown in Figure 4, TOGp depletion did not significantly change the density of growing MT plus ends within the spindle. Taken together, the length and density of spindle MTs and the density of growing MT plus ends indicate that TOGp makes small contributions to MT stability within the spindle.
Figure 4.
Growing MT plus ends are abundant in TOGp-depleted cells. Growing MT plus ends were identified using an antibody to EB1 (green) and costained with antibodies against γ-tubulin to identify centrosomes (red). Images shown are maximium intensity projections and single optical sections from the center of the spindle. Growing MT plus ends were abundant in TOGp-depleted cells are were qualitatively of a density similar to that in untransfected spindles. Bar, 10 μm.
TOGp and MCAK Contribute to Spindle Bipolarity without Causing Major Changes in Spindle MT Polymer Levels
To test whether TOGp and MCAK function in opposition to each other during mitosis, we examined cells depleted of TOGp, MCAK, or both. Cells transfected with siRNA to deplete MCAK had significantly reduced levels of MCAK protein 48 h after transfection (Figure 5). In mitotic cells, nearly 100% of cells had levels of MCAK that were undetectable by immunofluorescence (our unpublished results). Depletion of MCAK did not alter TOGp levels or localization (Figure 5 and our unpublished results). Likewise, depletion of TOGp did not alter MCAK protein level or localization (Figure 5 and our unpublished results).
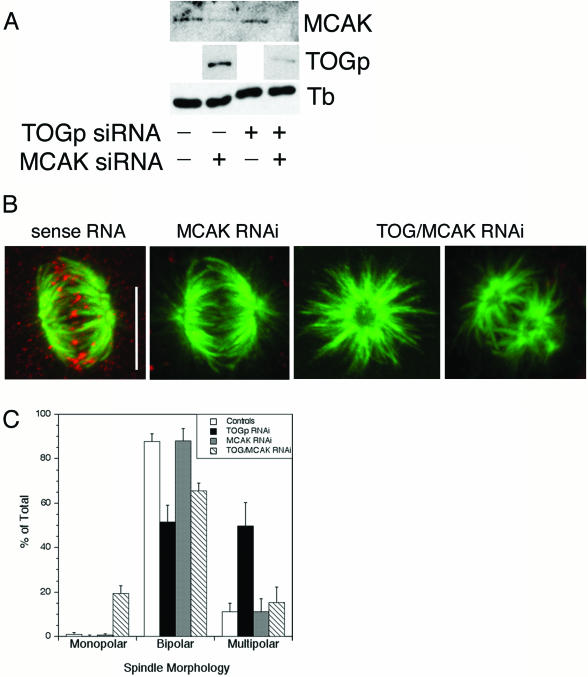
Figure 5.
Simultaneous depletion of TOGp and MCAK results in bipolar and monopolar spindle formation. (A) Cell lysates were prepared 48 h after transfection with siRNA for TOGp, MCAK or both. Depletion of MCAK does not affect TOGp level and vice versa. Transfection of both TOG and MCAK siRNAs depletes cells of both proteins. Tubulin levels were unchanged by siRNA transfection. (B) Immunofluorescence micrographs of spindles assembled in MCAK- or TOGp/MCAK-depleted cells. MCAK localizes to kinetochores in cells transfected with sense RNA (red). MCAK depletion did not alter spindle morphology. Depletion of both TOGp and MCAK results in mono- and bipolar spindles. The TOG/MCAK-depleted cells were stained with rabbit antibodies to both TOGp and XKCM1 (MCAK). Bar, 10 μm. (C) Quantitation of spindle morphologies. Simultaneous depletion of TOGp and MCAK reverses the multipolar phenotype observed after TOGp depletion and results in a large increase in the number of monopolar spindles. Means ± SD from four experiments.
Cells depleted of MCAK assembled bipolar spindles with normal morphology and a normal MT focus at spindle poles (Figure 5B). A normal spindle morphology was also observed by Walczak et al. (2002) after displacement of full-length XKCM1 from centromeres by an N-terminal fragment. Spindle MT staining intensity in HeLa cells depleted of MCAK were 1.2 times higher than that measured in untreated cells or cells transfected with the sense RNA sequence. When treated with monastrol, MCAK-depleted cells formed monopolar spindles (our unpublished results). These monasters had an average diameter of 15.9 ± 1.7 μm (mean of three experiments), wider than that measured in cells treated with monastrol alone (14.3 ± 1.5 μm). Our results are consistent with loss of a microtubule destabilizing activity, but the magnitude of change associated with MCAK depletion is considerably less than that observed after XKCM1 depletion from Xenopus egg extracts (Tournebize et al., 2000). Interphase MT arrays were qualitatively similar to those in untransfected cells (our unpublished results).
Transfection of cells with siRNAs for both TOGp and MCAK resulted in reduced levels of both proteins after 48 h (Figure 5A). Cells depleted of both TOGp and MCAK were still able to assemble mitotic spindles, and chromosomes formed attachments to these spindles (based on double-labeling with antitubulin and CREST antibodies; our unpublished results). A large percentage of spindles were bipolar, and the percentage of multipolar spindles was reduced to that seen in untreated cells (Figure 5C). Spindles in the TOGp- and MCAK-depleted cells retained the disrupted spindle pole structure observed in cells depleted of TOGp alone (Figure 5B). Surprisingly, ∼20% of mitotic cells depleted of both TOGp and MCAK contained monopolar spindles (4/4 experiments). In contrast, <1% of spindles were monopolar in untreated cells or cells depleted of either TOGp or MCAK alone (Figure 5C).
We used the monopolar spindles assembled in TOGp/MCAK-depleted cells to quantify changes in MT length and polymer density. These monopolar spindles had a diameter similar to that in untreated cells (14.3 ± 1.5 μm in untreated cells vs. 14.6 ± 2.1 μm in TOGp/MCAK-depleted cells), indicating that removal of both proteins reversed the longer MT phenotype observed in cells lacking MCAK. The antitubulin staining intensity measured in the monopolar spindles was 1.27 times that in untransfected, monastrol-treated cells (mean of 3 experiments). This small, but significant increase in MT density was nearly identical to that measured in cells depleted of MCAK alone.
TOGp Depletion Results in Centrosome Fragmentation
Because TOGp did not make major contributions to MT stability in the spindle, we next examined TOGp function at centrosomes. In untreated HeLa cells, we find that TOGp colocalized with γ-tubulin at the centrosome and within the spindle (Figure 6). TOGp remained localized to the centrosome after a 4-h incubation in 30 μM nocodazole, demonstrating that TOGp binding to the centrosome is independent of MTs (Figure 6).
Figure 6.
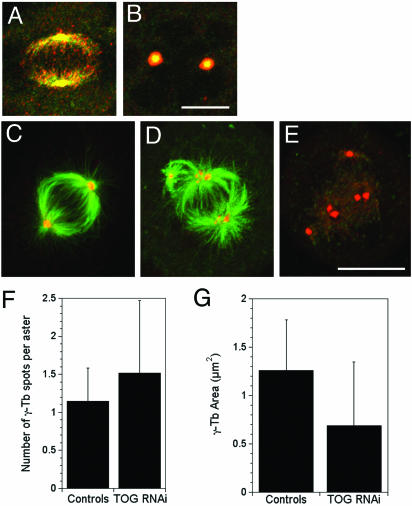
TOGp localizes to centrosomes in the absence of MTs and is required to maintain centrosome cohesion. (A and B) TOGp colocalizes with γ-tubulin in the presence (A) and absence (B) of MTs. (A and B) γ-tubulin (green) and TOGp (red). Cells in B were incubated in 30 μM nocodazole for 4 h before fixation. (C–F) Centrosomes fragment in the absence of TOGp. (C) Sense RNA–transfected cell; (D and E) cell fixed 48 h after transfection with siRNA to deplete TOGp. Cells are stained with antibodies to tubulin (C and D, green), TOGp (E, green), and γ-tubulin (C–E, red). In the TOGp-depleted cell, many asters contain multiple γ-tubulin foci. Bars, 10 μm. All images shown are maximium intensity projections. (E) The number of γ-tubulin foci per aster increased after TOGp depletion. The area of each γ-tubulin focus decreased after TOGp depletion. Data pooled from three experiments (34 asters in control cells [untransfected and sense RNA transfected] and 55 in TOGp-depleted cells).
To examine the function of TOGp at the centrosome, we examined the distributions of the centrosome components γ-tubulin and pericentrin in TOGp-depleted cells. Untransfected cells or cells transfected with the sense RNA sequence had normal distributions of γ-tubulin (Figure 6) or pericentrin (our unpublished results). In contrast, asters in TOGp-depleted cells often had two or more γ-tubulin foci at the center of the aster (Figure 6) and in rare cases the γ-tubulin staining was present as numerous small speckles (Figure 6). Pericentrin was also often observed as two or more foci per aster (our unpublished results). TOGp-depleted cells have a greater number of γ-tubulin foci per aster (p < 0.05), and the average area of each γ-tubulin foci is reduced (p < 0.05; Figure 6), indicating that centrosomes fragment in the absence of TOGp.
Spindle Pole Organization in TOGp-depleted Cells
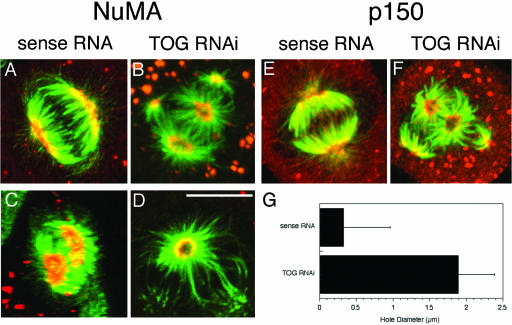
TOGp-depleted spindles have holes, lacking MTs, at the center of the asters, indicating that these spindles have aberrant spindle poles. To characterize the structure of the spindle poles, we examined the distribution of the spindle pole proteins NuMA and the p150 component of dynactin. In cells transfected with the sense RNA sequence, both NuMA and p150 were localized to a crescent-shaped structure at the spindle poles (Figure 7; Gaglio et al., 1996, 1997). In TOGp-depleted cells, NuMA was localized to the edges of the holes in the asters and appeared as a donut-like shape ∼2 μm across (Figure 7). Likewise, p150, a component of the dynactin complex, also localized to a donut-shaped structure at spindle poles in these cells. Because dynein/dynactin transports NuMA to the MT minus ends (Merdes et al., 2000) and both NuMA and p150 colocalized with MT minus ends in TOGp-depleted cells, the holes in the center of MT asters cannot result from mislocalization of NuMA or p150. Instead, TOGp may function to stabilize MT minus ends at the spindle poles or anchor these minus ends.
Figure 7.
TOGp depletion displaces MT minus ends from their tight focus at spindle poles. NuMA (A and C) and p150 (E) localized to a crescent-shaped structure at the spindle poles in sense RNA–transfected cells. In TOGp-depleted cells, NuMA (B and D) and p150 (F) localized to the edges of the holes in the center of each aster. (A, B, E, and F) are maximium intensity projections; (C and D) are single confocal sections showing end-on views of spindle poles. Panel C was generated from a 180° 3D projection tilted to view the spindle poles in a sense RNA–transfected cell. The NuMA crescent completely covers the front spindle pole. In other untransfected cells or cells transfected with sense RNA, small holes were sometimes observed in NuMA staining. Tubulin shown in green; NuMA or p150 shown in red. Bar, 10 μm. (G) The diameter of the central hole in the aster was ∼5 times wider after TOGp depletion.
The Phenotype of TOGp-depleted Spindles Does Not Result from hTPX2 Mislocalization
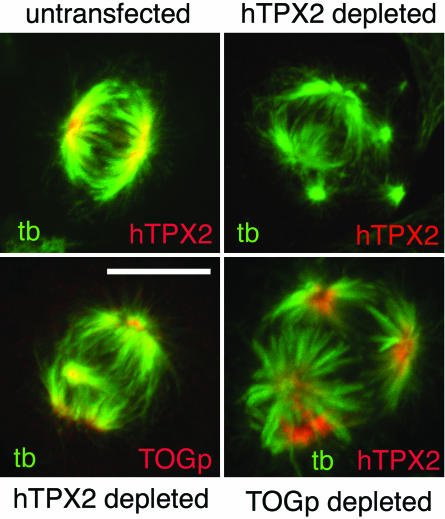
The phenotype of the TOGp-depleted spindles is similar to that described recently in cells depleted of hTPX2 (Garrett et al., 2002). In each case, multipolar spindles result from depletion of either protein, Eg5 activity is required to maintain the multipolar morphology and the centrosome fragments. The similar phenotypes of cells depleted of TOGp or hTPX2 raised the possibility that depletion of one protein could cause mis-localization of the other, generating a similar phenotype. We examined this possiblity by localizing TOGp in hTPX2-depleted spindles and hTPX2 localization in TOGp-depleted spindles. Depletion of hTPX2 is shown by immunofluorescence in Figure 8. TOGp localizes to the centrosomes in hTPX2-depleted spindles, similar to its localization in untransfected cells (Figure 6). In TOGp-depleted spindles, hTPX2 is localized to the spindle poles (Figure 8), and its localization mirrors that of NuMA or p150 (Figure 7). Although both TOGp and hTPX2 form multipolar spindles, these spindles differ in the morphology of the spindle poles. The spindle pole remains tightly focused in the hTPX2 depleted spindles, whereas it is present as an expanded ring in the TOGp-depleted spindles.
Figure 8.
hTPX2 and TOGp localize to the spindle independently of each other. Cells were transfected with siRNAs to deplete hTPX2 or TOGp, as marked. hTPX2 is undetectable in a large number of cells 36–40 h after siRNA transfection. Depletion of hTPX2 resulted in multipolar spindles, but TOGp remained localized to the centrosomes of these cells. hTPX2 retained spindle pole localization in the absence of TOGp. Bar, 10 μm.
DISCUSSION
TOGp and Spindle MT Assembly
Depletion of TOGp reduced the length and density of spindle microtubules by ∼20%, indicating that TOGp does not play a major role in regulating MT assembly in mitotic tissue cells. Because we find that TOGp depletion varies from cell to cell and we must rely on immunofluorescence to identify cells with reduced TOGp, it is possible that some cells contained low amounts of TOGp that are not detected by immunofluorescence. This low concentration is unlikely to have influenced our results. In Xenopus egg extracts, antibody depletion removed only ∼70% of XMAP215, yet this partial depletion was sufficient to generate significant changes in MT lengths and catastrophe frequencies (Tournebize et al., 2000).
Inactivation of TOGp homologues in Xenopus and C. elegans embryonic systems results in a significant decrease in MT lengths during mitosis (Tournebize et al., 2000; Bellanger and Gonczy, 2003; Srayko et al., 2003). The difference between our results and those in Xenopus and C. elegans embryos may reflect a difference in activity for TOGp and its homologues or may reflect differences between embryonic and nonembryonic cells. For example, spindles in embryonic cells may be more sensitive to changes in XMAP215/TOGp levels. Although spindle MTs were not drastically changed by TOGp depletion in mammalian cells, the spindle does change MT length and density in response to other perturbations. In cells overexpressing active oncoprotein 18/stathmin, the length and density of spindle MTs are significantly reduced (Larsson et al., 1997).
Although TOGp makes a small contribution to mitotic MT assembly, depletion of TOGp results in multipolar spindle formation (Figure 1; Gergely et al., 2003), indicating that TOGp is necessary for bipolar organization of the spindle. The multipolar phenotype is strikingly different from the very short MT asters seen after XMAP215 inactivation/depletion from Xenopus eggs or extracts (Tournebize et al., 2000; Becker et al., 2003) and inactivation of Zyg9 in C. elegans (Matthews et al., 1998; Bellanger and Gonczy, 2003; Srayko et al., 2003). Instead, the phenotype is similar to that observed in mutants of the Drosophila homolog, msps (Cullen et al., 1999; Cullen and Ohkura, 2001; Lee et al., 2001).
TOGp and MCAK Act Antagonistically To Regulate Spindle Morphology
Depletion of MCAK increased the amount of MT polymer assembled into spindles, but the magnitude of the increase was relatively small (1.2×) compared with that observed after XKCM1 inactivation in Xenopus egg extracts (Walczak et al., 1996; Tournebize et al., 2000) or oocytes (Becker et al., 2003). Spindle length and bipolar structure were also not affected by MCAK depletion. These results suggest that MCAK is not a major MT destabilizer in mitotic mammalian cells. Similar results were seen after antisense (Maney et al., 1998) or siRNA (Holmfeldt and Gullberg, personal communication) depletion of MCAK. In contrast, microinjection of anti-XKCM1 antibodies into PtK1 cells resulted in increased MT density and length (Smith-Kline and Walczak, 2002). The different results from RNA knockdown and antibody injection experiments may reflect differences in the time required to deplete/inactivate MCAK or differences in the cell types (HeLa vs. PtK cells) under study.
Depletion of TOGp and MCAK resulted in several phenotypes consistent with general antagonistic functions for these proteins during bipolar spindle assembly. The individual depletions of TOGp and MCAK are consistent with experiments in Xenopus extracts demonstrating antagonistic MT regulatory functions for these two proteins, but in mammalian cells each protein makes relatively smaller contributions to MT length and density within the spindle. The most striking phenotype suggesting antagonistic functions for TOGp and MCAK is the formation of bipolar and monopolar spindles in cells lacking both proteins, compared with the multipolar spindles present in TOGp-depleted cells. Overexpression of XKCM1 in PtK cells results in monopolar spindle formation (Smith-Kline and Walczak, 2002), again indicating a role for the kin I kinesins in spindle morphology. Depletion of TOGp or MCAK may generate small changes in MT dynamics that are sufficient to disrupt bipolar spindle organization. Alternatively, TOGp and MCAK may have antagonistic functions at centrosomes and centromeres, respectively, that contribute to bipolar spindle morphology by mechanisms independent of plus-end MT assembly dynamics. In support of this model, depletion of TACC3 results in loss of TOGp from the spindle, but TOGp remains at the centrosomes (Gergely et al., 2003). TACC3-depleted cells assemble a normal bipolar spindle (Gergely et al., 2003), indicating that loss of TOGp from the centrosome, and not the spindle MTs, is necessary for disruption of spindle morphology.
TOGp Contributes to Centrosome Cohesion and Spindle Pole Organization
TOGp was localized to centrosomes in the absence of MTs, as are several other members of the XMAP215/Dis1 family (Figure 6; Graf et al., 2000; Popov et al., 2001). TOGp-depleted cells often contained more than one γ-tubulin focus per MT aster (Figure 6). These γ-tubulin foci likely represent fragmented centrosomes because the average area of the γ-tubulin foci also decreased. Thus, TOGp contributes to centrosome integrity and in this way could regulate centrosome number. Recently, the Dictyostelium homolog, DdCP224, was also demonstrated to control centrosome number (Graf et al., 2003). Several other factors lead to centrosome fragmentation including inactivation of astrin, hTPX2, or Nek2B kinase (Uto and Sagata, 2000; Garrett et al., 2002; Gruber et al., 2002), DNA damage (Hut et al., 2003), cocaine treatment (Combelles et al., 2000), and RanBP1 overexpression (Di Fiore et al., 2003).
Although TOGp is associated with the centrosome, its depletion also causes disruption of the spindle poles. Cells lacking TOGp assemble MT asters with prominent holes in their centers (Figures 1 and 3). These holes are lined with NuMA and the p150 component of dynactin. Both NuMA and p150 are normally found at MT minus ends within the spindle (Merdes et al., 2000), where they are necessary gather MT minus ends into a spindle pole (Gordon et al., 2001). Previous experiments using HeLa mitotic extracts demonstrated that Eg5 depletion also caused a widening of the spindle poles, with NuMA localized at the edge of the ring (Gaglio et al., 1996). We find that TOGp depletion did not remove Eg5 from the spindle poles (our unpublished results), indicating roles for both TOGp and Eg5 in organizing the spindle pole. Inactivation of NuMA or dynein cause a different phenotype: splaying of MT minus ends (Gordon et al., 2001). In TOGp-depleted spindles, the minus ends of MTs are gathered into a broad ring and are no longer as tightly focused.
hTPX2 and TOGp Depletions Result in Similar Phenotypes
Depletion of hTPX2 or TOGp results in multipolar spindle formation and centrosome fragmentation (Figures 6 and 8; Garrett et al., 2002). Multipolar spindles formed in either TOGp-depleted or hTPX2-depleted cells collapse to a monopolar morphology when Eg5 motor activity is inhibited by monastrol (Figure 2; Garrett et al., 2002). Direct comparison of hTPX2- and TOGp-depleted spindles demonstrated that the localization of each protein was not dependent on the presence of the other (Figure 8), indicating that the loss of either protein alone was sufficient to disrupt spindle morphology. These results suggest that both TOGp and hTPX2 must be present to counteract Eg5 motor activity and limit spindles to a bipolar morphology.
CONCLUSIONS
During mitosis, TOGp functions primarily to maintain the integrity of the centrosomes and the spindle poles and to limit spindles to a bipolar morphology. TOGp likely exerts these functions at the centrosome and not by regulating MT plus-end dynamics. At the centrosome, TOGp could anchor MTs after their release from nucleation sites (Bornens, 2002). Similar functions have been proposed for homologues in S. cerevisiae (Stu2p, Usui et al., 2003) and Drosophila (msps, Lee et al., 2001). The loss of MT anchorage in TOGp-depleted cells could result in widening of the spindle pole and could also reduce the stability of MT minus ends. Alternatively, the anchoring function of TOGp may be to keep nucleation sites attached to the pericentriolar matrix. Loss of nucleation site anchoring could generate the fragmented foci of γ-tubulin and pericentrin observed in TOGp-depleted cells.
Acknowledgments
We thank Duane Compton, Tarun Kapoor, Claire Walczak, Linda Wordeman, and Bill Brinkley for providing reagents and Claire Walczak, Martin Gullberg, Fanni Gergely, and Tarun Kapoor for sharing data before publication. We also thank Bob Skibbens for help with FACs and Bob Skibbens, Michelle Piehl, and Mimi Shirasu-Hiza for critically reading the manuscript. This work was supported by National Institutes of Health Grant GM58025 and by a National Science Foundation equipment grant (DBI-0200322) supporting the confocal microscope facility.
References
- Becker, B.E., Romney, S.J., and Gard, D.L. (2003). XMAP215, XKCM1, NuMA and cytoplasmic dynein are required for the assembly and organization of the transient microtubule array during maturation of Xenopus oocytes. Dev. Biol. 261, 488-505. [DOI] [PubMed] [Google Scholar]
- Bellanger, J.M., and Gonczy, P. (2003). TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Curr Biol. 13, 1488-1498. [DOI] [PubMed] [Google Scholar]
- Bornens, M. (2002). Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14, 25-34. [DOI] [PubMed] [Google Scholar]
- Cassimeris, L., Gard, D., Tran, P.T., and Erickson, H.P. (2001). XMAP215 is a long thin molecule which does not increase microtubule stiffness. J. Cell Sci. 114, 3025-3033. [DOI] [PubMed] [Google Scholar]
- Charrasse, S., Schroeder, M., Gauthier-Rouviere, C., Ango, F., Cassimeris, L., Gard, D.L., and Larroque, C. (1998). The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J. Cell Sci. 111, 1371-1383. [DOI] [PubMed] [Google Scholar]
- Combelles, C.M.H., Carabatsos, M.J., London, S.N., Mailhes, J.B., and Albertini, D.F. (2000). Centrosome-specific perturbations during in vitro maturation of mouse oocytes exposed to cocaine. Exp. Cell Res. 260, 116-126. [DOI] [PubMed] [Google Scholar]
- Cullin, C.F., and Ohkura, H. (2001). Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat. Cell Biol. 3, 637-642. [DOI] [PubMed] [Google Scholar]
- Cullen, C.F., Deak, P., Glover, D.M., and Ohkura, H. (1999). mini spindles: A gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J. Cell Biol. 146, 1005-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., and Mitchison, T.J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Biol. 13, 83-117. [DOI] [PubMed] [Google Scholar]
- Di Fiore, B., Ciciarello, M., R. Mangiacasale, R., Palena, A., Tassin, A.M., Cundari, E., and Lavia, P. (2003). Mammalian RanBP1 regulates centrosome cohesion during mitosis. J. Cell Sci. 116, 3399-3411. [DOI] [PubMed] [Google Scholar]
- Dionne, M.A., Sanchez, A., and Compton, D.A. (2000). ch-TOGp is required for microtubule aster formation in a mammalian mitotic extract. J. Biol. Chem. 275, 12346-12352. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- Gaglio, T., Saredi, A., Bingham, J.B., Hasbani, M.J., Gill, S.R., Schroer, T.A., and Compton, D.A. (1996). Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 135, 399-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio, T., Dionne, M.A., and Compton, D.A. (1997). Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J. Cell Biol. 138, 1055-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, M.A., Koonrugsa, N., and Toda, T. (2002). Spindle-kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 21, 6015-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard, D.L., and Kirschner, M.W. (1987). A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J. Cell Biol. 105, 2203-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett, S., Auer, K., Compton, D.A., and Kapoor, T.M. (2002). hTP2 is required for normal spindle morphology and centrosome integrity during vertebrate cell division. Curr. Biol. 12, 2055-2059. [DOI] [PubMed] [Google Scholar]
- Gergely, F., Daviam, V.M., and Raff, J.W. (2003). The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 17, 336-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, M.B., Howard, L., and Compton, D.A. (2001). Chromosome movement in mitosis requires microtubule anchorage at spindle poles. J. Cell Biol. 152, 425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf, R., Daunderer, C., and Schliwa, M. (2000). Dictyostelium DdCP224 is a microtubule-associated protein and a permanent centrosomal resident involved in centrosome duplication. J. Cell Sci. 113, 1747-1758. [DOI] [PubMed] [Google Scholar]
- Graf, R., Euteneuer, U., Ho, T.H., and Rehberg, M. (2003). Regulated expression of the centrosomal protein DdCP224 affects microtubule dynamics and reveals mechanisms for the control of supernumerary centrosome number. Mol. Biol. Cell 14, 4067-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber, J., Harborth, J. Schnabel, J., Weber, K., and Hatzfeld, M. (2002). The mitotic-spindle-associated protein astrin is essential for progression through mitosis. J. Cell Sci. 115, 4053-4059. [DOI] [PubMed] [Google Scholar]
- Howell, B.J., Deacon, H., and Cassimeris, L. (1999). Decreasing oncoprotein 18 levels reduces microtubule catastrophes and increases microtubule polymer in vivo. J. Cell Sci. 112, 3713-3722. [DOI] [PubMed] [Google Scholar]
- Hut, H.M.J., Lemstra, W., Blaauw, E.H., van Cappellen, G.W.A., Kampinga, H.H., and Sibon, O.C.M. (2003). Centrosomes split in the presence of impaired DNA integrity during mitosis. Mol. Biol. Cell 14, 1993-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, T.M., Mayer, T.U., Coughlin, M.L., and Mitchison, T.J. (2000). Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150, 975-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna, M., and Skibbens, R.V. (2003). A mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different RFC Complexes. Mol. Cell. Biol. 8, 2999-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, K., Arnal, I., Desai, A., Drechsel, D.N., and Hyman, A.A. (2001). Reconstitution of physiological microtubule dynamics using purified components. Science 294, 1340-1343. [DOI] [PubMed] [Google Scholar]
- Kinoshita, K., Habermann, B., and Hyman, A.A. (2002). XMAP 215, A key component of the dynamic microtubule cytoskeleton. Trends Cell Biol. 12, 267-273. [DOI] [PubMed] [Google Scholar]
- Kosco, K.A., Pearson, C.G., Maddox, P.S., Wang, P.J., Adams, I.R., Salmon, E.D., Bloom, K., and Huffaker, T.C. (2001). Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol. Biol. Cell 12, 2870-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, N., Marklund, U., Melander Gradin, H., Brattsand, G., and Gullberg, M. (1997). Control of microtubule dynamics by oncoprotein 18: dissection of the regulatory role of multisite phosphorylation during mitosis. Mol. Cell. Biol. 17, 5530-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.J., Gergely, F., Jeffers, K., Peak-Chew, S., and Raff, J.W. (2001). Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behavior. Nat. Cell Biol. 3, 643-649. [DOI] [PubMed] [Google Scholar]
- Maney, T., Hunter, A.W., Wagenbach, M., and Wordeman, L. (1998). Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 142, 787-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, L.R., Carter, P., Thierry-Mieg, D., and Kemphues, K. (1998). ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J. Cell Biol. 141, 1159-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes, A., Heald, R., Samejima, K., Earnshaw, W.C., and Cleveland, D.W. (2000). Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149, 851-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue, Y., Shiina, N., and Tsukita, S. (2000). The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 10, 865-868. [DOI] [PubMed] [Google Scholar]
- Nabeshima, K., Kurooka, H., Takeuchi, M., Kinoshita, K., Nakaseko, Y., and Yanagida, M. (1995). p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the cdc2 target sites. Genes Dev. 9, 1572-1585. [DOI] [PubMed] [Google Scholar]
- Piehl, M., and Cassimeris, L. (2003). Organization and dynamics of growing microtubule plus ends during early mitosis. Mol. Biol. Cell 14, 916-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov, A.V., Pozniakovsky, A., Arnal, I., Antony, C., Ashford, A.J., Kinoshita, K., Tournebize, R., Hyman, A.A., and Karsenti, E. (2001). XMAP215 regulates microtubule dynamics through two distinct domains. EMBO J. 20, 397-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov, A.V., Severin, F., and Karsenti, E. (2002). XMAP215 is required for the microtubule-nucleating activity of centrosomes. Cur. Biol. 12, 1326-1330. [DOI] [PubMed] [Google Scholar]
- Shirasu-Hiza, M., Coughlin, P., and Mitchison, T. (2003). Identification of XMAP215 as a microtubule-destabilizing factor in Xenopus egg extract by biochemical purification. J. Cell Biol. 161, 349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Kline, S.L., and Walczak, C.E. (2002). The microtubule-destabilizing kinesin XKCM1 regulates microtubule dynamic instability in cells. Mol. Biol. Cell 13, 2718-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko, M., Quintin, S., Schwager, A., and Hyman, A.A. (2003). Caenorhabditis elegans TAC-1 and ZYG-9 form a complex that is essential for long astral and spindle microtubules. Curr. Biol. 13, 1506-1511. [DOI] [PubMed] [Google Scholar]
- Tirnauer, J.S., Canman, J.C., Salmon, E.D., and Mitchison, T.J. (2002). EB1 targets to kinetochores with attached, polymerizing microtubules. Mol. Biol. Cell 13, 4308-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize, R., Popov, A., Kinoshita, K., Ashford, A.J., Rybina, S., Pozniakovsky, A., Mayer, T.U., Walczak, C.E., Karsenti, E., and Hyman, A.A. (2000). Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2, 13-19. [DOI] [PubMed] [Google Scholar]
- Usui, T., Maekawa, H., Pereira, G., and Schiebel, E. (2003). The XMAP215 homologue Stu2 at yeast spindle pole bodies regulates microtubule dynamics and anchorage. EMBO J. 22, 4779-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uto, K., and Sagata, N. (2000). Nek2B, a novel maternal form of Nek2 kinase, is essential for the assembly or maintenance of centrosomes in early Xenopus embryos. EMBO J. 19, 1816-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bruegel, M., Drechsel, D., and Hyman, A. (2003). Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus end-binding microtubule destabilizer. J. Cell Biol. 161, 359-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez, R.J., Gard, D.L., and Cassimeris, L. (1994). XMAP from Xenopus Eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J. Cell Biol. 127, 985-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan, K.T., and Vallee, R.B. (1995). Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J. Cell Biol. 131, 1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak, C.E., Mitchison, T.J., and Desai, A. (1996). XKCM 1, A Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell 84, 37-47. [DOI] [PubMed] [Google Scholar]
- Walczak, C.E., Gan, E.C., Desai, A., Mitchison, T.J., and Kline-Smith, S.L. (2002). The microtubule-destabilizing kinesin XKCM1 is required for chromosome positioning during spindle assembly. Curr. Biol. 12, 1885-1889. [DOI] [PubMed] [Google Scholar]
- Wang, P.J., and Huffaker, T.C. (1997). Stu2p: a microtubule-binding protein that is an essential component of the yeast spindle pole body. J. Cell Biol. 139, 1271-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington, A.T., Vugrek, O., Wei, K.J., Hasenbein, N.G., Sugimoto, K., Rashbrooke, M.C., and Wasteneys, G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411, 610-613. [DOI] [PubMed] [Google Scholar]
- Wilde, A., and Zheng, Y. (1999). Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science 284, 1359-1362. [DOI] [PubMed] [Google Scholar]