Common polygenic variation contributes to risk of schizophrenia that overlaps with bipolar disorder (original) (raw)
. Author manuscript; available in PMC: 2014 Feb 4.
Published in final edited form as: Nature. 2009 Jul 1;460(7256):748–752. doi: 10.1038/nature08185
Abstract
Schizophrenia (SCZ) is a severe mental disorder with a lifetime risk of about 1%, characterized by hallucinations, delusions and cognitive deficits with heritability estimated at up to 80%1,2. We adopted two analytic approaches to determine the extent to which common genetic variation underlies risk of SCZ using genome-wide association study (GWAS) data from 3,322 European individuals with SCZ and 3,587 controls. First, we implicate the major histocompatibility complex (MHC). Second, we provide molecular genetic evidence for a substantial polygenic component to risk of SCZ involving thousands of common alleles of very small effect. We show that this component also contributes to risk of bipolar disorder (BPD), but not to multiple non-psychiatric diseases.
We genotyped the International Schizophrenia Consortium (ISC) case-control sample for up to ~1 million single nucleotide polymorphisms (SNPs) augmented by imputed common HapMap SNPs. In the GWAS (λGC=1.09; Table S1, Figure S1–S3), the most associated genotyped SNP (P = 3.4×10−7) was located in the first intron of myosin XVIIIB (MYO18B) on chromosome 22. The second strongest association comprised over 450 SNPs on chromosome 6p spanning the MHC (Figure 1). There is some evidence for between-site heterogeneity in both allele frequencies and odds ratios (Table 1). We observed associations consistent with previous reports in the 22q11.2 deletion region and ZNF804A3 (Table S2, Figure S2 and Section S5).
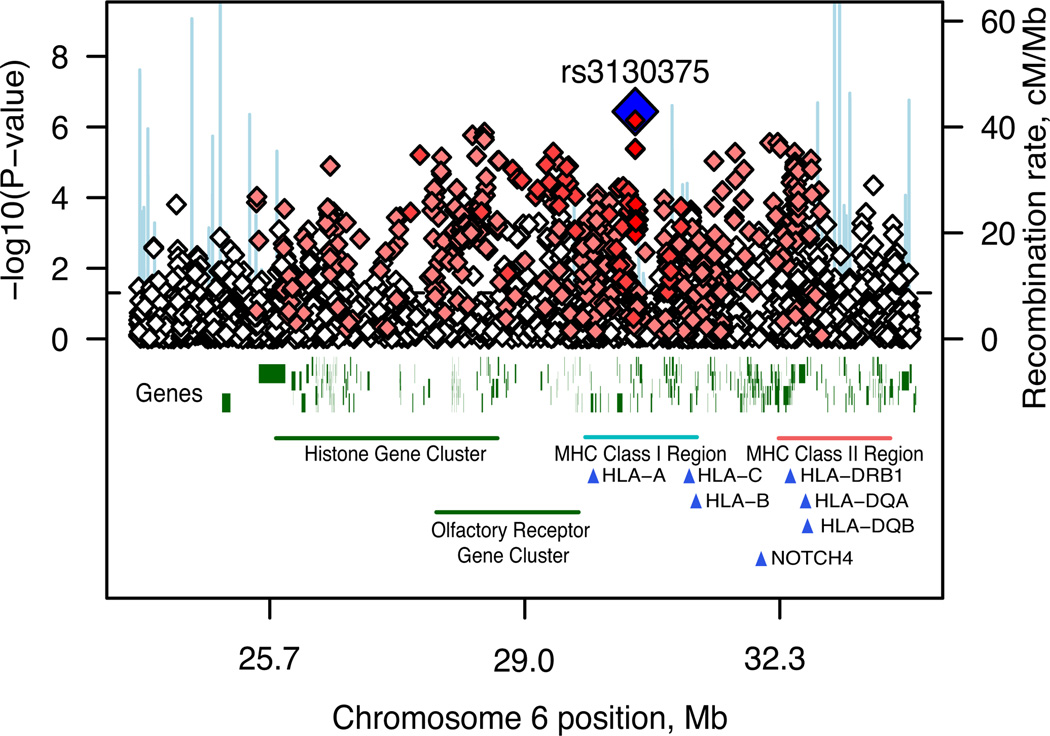
Figure 1. Association results across the MHC region.
Results are shown as –log10 (P-value) for genotyped SNPs. The most associated SNP is shown as a blue diamond. The colour of the remaining markers reflects r2 with rs3130375, light pink,r2 > 0.1, red,r2 > 0.8. The recombination rate from the CEU HapMap (second Y-axis) is plotted in light blue.
Table 1.
MHC association for a) rs3130375, the most significant genotyped SNP, stratified by sample b) imputed classical HLA alleles
| a) | |||
|---|---|---|---|
| Frequency (rs3130375, A allele) | |||
| Sample | Cases | Controls | P value |
| Scottish (Aberdeen) | 0.132 | 0.168 | 0.0060 |
| Scottish (Edinburgh) | 0.137 | 0.135 | 0.8930 |
| British (UCL) * | 0.132 | 0.143 | 0.4836 |
| Irish (TCD) | 0.110 | 0.170 | 0.0012 |
| Bulgarian (Cardiff) | 0.077 | 0.084 | 0.5602 |
| Portuguese (PIC) | 0.048 | 0.061 | 0.3510 |
| Swedish (KI, 5.0) | 0.043 | 0.119 | 0.0004 |
| Swedish (KI, 6.0) | 0.089 | 0.142 | 0.0040 |
| b) | |||
|---|---|---|---|
| HLA Allele | Frequency | Odds ratio | P value |
| HLA-A* 0101 | 0.10 | 0.79 | 4×10−5 |
| HLA-C* 0701 | 0.11 | 0.78 | 5×10−5 |
| HLA-B* 0801 | 0.07 | 0.76 | 3×10−5 |
| HLA-DRB* 0301 | 0.12 | 0.77 | 3×10−6 |
| HLA-DQB* 0201 | 0.21 | 0.86 | 4×10−4 |
| HLA-DQA* 0501 | 0.21 | 0.80 | 6×10−7 |
The best imputed SNP, which reached genome-wide significance (rs3130297,P = 4.79×10−8, T allele, OR=0.747, MAF=0.114, 32.3Mb), was also in the MHC, 7kb from NOTCH4, a gene with previously reported associations with SCZ4. We imputed classical human leukocyte antigen (HLA) alleles; 6 were significant at P < 10−3, found on the ancestral European haplotype5 (Table 1, Table S3, Section S3). However, it was not possible to ascribe the association to a specific HLA allele, haplotype or region (Table S3, Figure S4).
We exchanged GWAS summary results with the Molecular Genetics of Schizophrenia (MGS) and SGENE consortia for genotyped SNPs with P < 10−3. There were 8,014 cases and 19,080 controls of European descent in the combined sample (see companion manuscripts, Section S7). Our top genotyped MHC SNP (rs3130375) had P = 0.086 and P = 0.14 in MGS and SGENE. Considering combined results for genotyped and imputed SNPs across the MHC region more broadly, rs13194053 had a genome-wide significant combined_P_ = 9.5×10−9 (ISC, MGS and SGENE_P_ = 3×10−4, 1×10−2 and 1×10−4respectively; C allele OR = 0.82, 0.88 and 0.78) and was in LD with rs3130375 (_r2_=0.35 in HapMap). Across the region 11 other SNPs had P < 10−7 at 27.1 – 27.3Mb and 32.7Mb (Table S5).
Our second approach was to evaluate whether common variants play an important role en masse, directly testing the classic theory of polygenic inheritance6, applied to SCZ by Gottesman & Shields7. While our GWAS analysis did not identify a large number of strongly associated loci, there could still be many – potentially thousands – of very small individual effects that collectively account for a substantial proportion of variation in risk. We summarized variation across many nominally-associated loci into quantitative scores and related the scores to disease state in independent samples8. Although variants of small effect (e.g. genotypic relative risk, GRR=1.05) are unlikely to achieve even nominally significant p-values, increasing proportions will be detected at increasingly liberal significance thresholds (pT), for example pT < 0.1 or pT < 0.5. Using such thresholds, we defined large sets of “score alleles” in a discovery sample, in order to generate aggregate risk scores for individuals in independent target samples. We use the term score, instead of_risk_, as we cannot differentiate the minority of true risk alleles from unassociated variants.
We performed the score analyses on a reduced set of SNPs to facilitate analysis and interpretation. After filtering on MAF, genotyping rate and LD (independent of association with SCZ) we obtained a subset of 74,062 autosomal SNPs in approximate linkage equilibrium (Table S5, S6). In each discovery sample, we selected sets of score alleles at different association test pT thresholds. For each individual in the target sample, we calculated the number of score alleles they possessed, each weighted by the log odds ratios from the discovery sample. To assess whether the aggregate scores reflect SCZ risk, we tested for a higher mean score in target cases compared to controls (Sections S9–S11, Table S7).
We selected males (2176 cases, 1642 controls) and females (1146 cases, 1945 controls) to form arbitrary discovery and target samples (Table S8). Score alleles designated in the discovery sample were significantly enriched among target cases and the effect was larger for increasingly liberal pT thresholds. The score based on all SNPs with male discovery pT < 0.5 (N=37,655 SNPs) was highly correlated with SCZ in target females (P_=9.4×10−19), explaining ~3% of the variance (Nagelkerke’s pseudo_R2 from logistic regression), with higher scores in cases. The results were not driven by only a few highly associated regions (Section S12).
We eliminated several possible confounders, with emphasis on subtle population stratification (Table S9–S15). Defining score alleles in British Isles samples and testing in samples from Sweden, Portugal and Bulgaria, and vice versa, we observed a similar pattern of results. It is unlikely that the same substructure is overrepresented in the corresponding phenotype class when discovery and target samples are from distinct populations. The effect is also stronger for SNPs within annotated genes (Table S16).
We used independent GWAS samples a) to replicate the polygenic component, b) to examine whether this component is shared with BPD9 and c) to demonstrate specificity by considering non-psychiatric diseases. We used the entire ISC for the discovery sample, considering the five most informative pT thresholds from the intra-ISC analyses. The independent target samples were the MGS European-American (MGS-EA), the MGS African-American (MGS-AA) and the UK sample described by O’Donovan et al3. The ISC-derived score was highly associated with disease in both European SCZ samples (Figure 2, Figure S6 and Table S17). The MGS-EA had a significantly higher mean pT < 0.5 score in cases (P = 2×10−28;R2 = 3.2%), as did the smaller O’Donovan sample (P = 5×10−11;R2 = 2.3%). Aggregate differences in allele frequencies and patterns of LD between Europeans and African-Americans are expected to lead to an attenuated effect. Still, MGS-AA cases carried more of the European-derived score alleles than MGS-AA controls (P = 0.008;R2 = 0.4%).
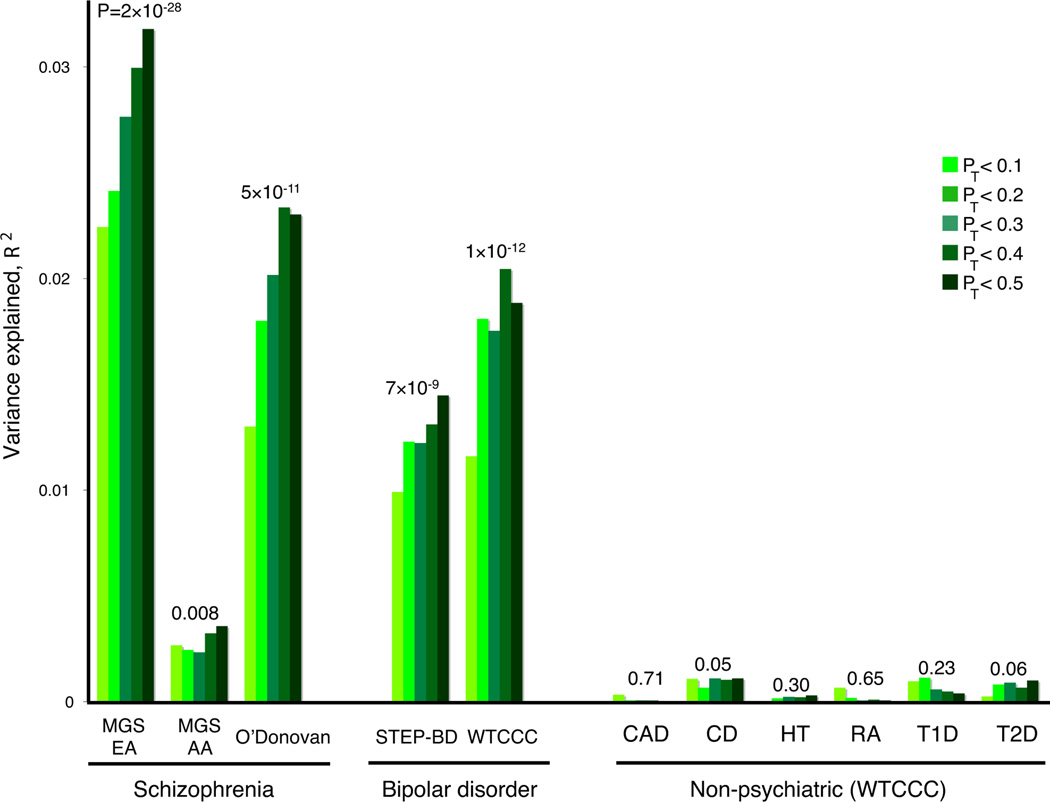
Figure 2. Replication of the ISC-derived polygenic component in independent SCZ and BPD samples.
Variance explained in the target samples based on scores derived in the entire ISC for five significance thresholds (pT < 0.1, 0.2, 0.3, 0.4 and 0.5, plotted left to right in each study). The y-axis indicates Nagelkerke’s pseudo-R2; the number above each set of bars is the P-value for the pT < 0.5 target sample analysis. Numbers for cases/controls: MGS-EA 2687 / 2656; MGS-AA 1287 / 973; O’Donovan 479 / 2938; STEP-BD 955 / 1498; WTCCC 1829 / 2935; CAD 1926 / 2935; CD 1748 / 2935; HT 1952 / 2935; RA 1860 / 2935 ; T1D 1963 / 2935 ; T2D 1924 / 2935.
The ISC-derived score alleles were also associated with BPD in two independent samples. Both STEP-BD10 and WTCCC11 had higher mean pT < 0.5 scores in cases (P = 7×10−9, R2 = 1.9%; and P = 1×10−12,R2 = 1.4% respectively) indicating a substantial, shared genetic component.
To test disease specificity, we selected all six non-psychiatric WTCCC samples (coronary artery disease, Crohn’s disease, hypertension, rheumatoid arthritis, type I diabetes and type II diabetes). Controls are shared among the WTCCC case samples, including BPD. In contrast to SCZ and BPD, there was no association (p>0.05) between the ISC-derived SCZ scores and these non-psychiatric diseases, for any pT threshold.
We next investigated the genetic models consistent with our data. The total additive genetic variance (VA) reflects the number of causal alleles as well as their frequency and effect size distributions. However, the variance explained by the markers that tag these causal alleles (VM) will be attenuated, reflecting the average extent of LD between marker and causal allele. In our target samples, the variance explained by the observed score alleles, VS, will be further attenuated by sampling variation and pT threshold, such that VS≤ VM ≤ VA.
We used simulation to estimate possible values for VM and VA, by identifying models that produced profiles of VS across pT threshold that were similar to those observed in ISC data, as indexed by the target sample R2. Under a variety of genetic models, we simulated discovery and target datasets of comparable sample size to the ISC. Based on the empirical allele frequency distribution, we simulated marker SNPs, varying the proportion that were in LD with causal variants, for which we varied allele frequency (uniform, U-shaped) and effect size distributions (fixed GRRs, exponential GRRs or fixed variance explained) as well as the extent of LD (Section S16).
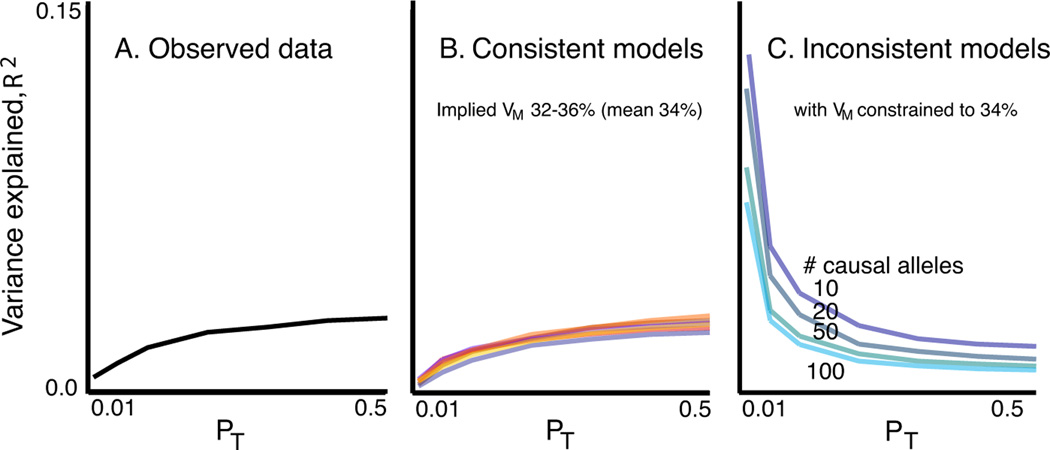
From a broad range of models, a subset produced results consistent with the ISC data (Figures 3 and S7). Among these, all led to similar estimates of VM (mean 34%, range 32 to 36%). In models in which the causal alleles were imperfectly tagged (r2<1) estimates of VA can be considerably larger. Therefore, our estimate that common polygenic variation accounts for one third of the total variation in SCZ risk is a lower bound for the true value, which could be much higher. Figure 3b shows seven examples from the range of consistent models, detailed in Table S18.
Figure 3. Observed and simulated profiles of target sample variance explained.
Panel A shows the observed variance explained (R2, black line). Panel B shows a subset of models that produced results consistent with the observed data. All yielded similar estimates of the total variance explained by the SNPs that tag the causal variants, VM, with a mean value of 34%. The seven models were: (% SNPs, Mean GRR/variance explained (V) per causal allele, LD, frequency model) M1: 6.25%, GRR=1.05,r2 =1, empirical; M2: 25%, GRR=1.025, r2 =1, empirical; M3: 12%, GRR=1.05, r2<1, uniform; M4: 32%, GRR=1.04,r2 <1, U-shaped; M5: 11%, V=0.00006, r2 =1, empirical; M6: 25%, GRR(Exponential)=1.025,r2 <1, uniform; M7: 100%, GRR(Exponential)=1.012, r2 <1, uniform. Panel C shows four inconsistent models with fewer variants of larger effect.
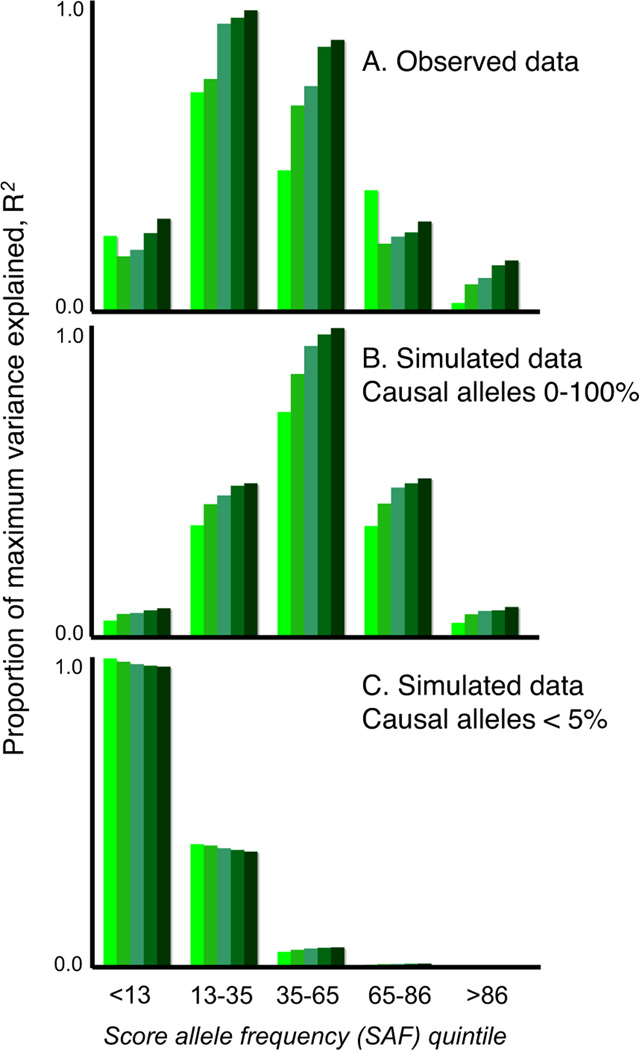
The simulated models consistent with our observed results all implied a substantial number of common variants, whereas models that invoked only a few common variants of large effect or only rare variants were not able to account for our findings. For example, if VM ≈ 34% arose from only 100 common causal alleles, with GRRs at the tagging marker between ~1.2–1.5, the majority would be detected at pT<0.01, and so the variance explained would decline, not increase, as more SNPs were added (Figure 3c, Table S19). It is possible that an observed GRR of ~1.05 could represent a large effect of a weakly tagged rare variant, e.g. a 10-fold effect of a 1/10,000 variant in complete LD (D’ = 1, but low_r2_) with a genotyped SNP. However, as this would only hold for low frequency markers (MAF < ~0.1), we stratified our analysis by score allele frequency (Figure 4a). For simulated models in which all causal variants were of low frequency (<0.05), a stratified analysis revealed the expected, skewed distribution (Figure 4c, Section S17), which was more pronounced for rarer causal alleles, e.g. 1/1,000 (data not shown). In contrast, models in which causal alleles followed a uniform frequency distribution provided a closer fit to our data (Figure 4b; although note some enrichment in the 2nd quintile, of ~13–30% score alleles). Moreover, rare variants are likely to be population specific and if recurrent, in LD with different common alleles within and between populations. As such, they could not account for the observation of disease variation that is largely shared across our different populations.
Figure 4. Analysis stratified by score allele frequency.
Panel A shows the observed data for the ISC/MGS-EA comparison. The y-axis is the target sample pseudo R2, scaled within each figure as a proportion of the maximum value observed. The two other plots show results for simulated data: the common variant model, with a uniform frequency distribution for causal risk-increasing alleles (panel B) and a multiple rare variant (MRV) model, in which the collective frequency of rare variants at a locus that all reside on the same haplotypic background with respect to the genotyped SNP was bounded at a maximum of 5% (panel C).
Decreased reproductive fitness in SCZ12 suggests that risk alleles of large to moderate effect will be under negative selection and therefore very rare13,14. This is not inconsistent with our results, since the common variants indexed by our polygenic score will not be subjected to strong selection, by virtue of their very small individual effect sizes. Our results do not exclude important contributions of rare variants for SCZ13, since rare variants are expected as part of the allele frequency/effect size spectrum of a polygenic model. We and others recently reported higher genome-wide rates of rare copy number variants in SCZ15,16,17. However, our results imply that medical sequencing and studies of structural variation to identify rare, highly penetrant variants will not alone fully characterize the genetic risk factors.
In conclusion, our molecular genetic data strongly support a polygenic basis to SCZ that a) involves common SNPs, b) explains at least a third of the total variation in liability, c) is substantially shared with BPD, and d) is largely not shared with several non-psychiatric diseases. We also identified variants in the MHC region that received support in two independent studies, although the population specificity and extensive LD will make follow-up challenging.
A highly polygenic model suggests that genetically influenced individual differences across domains of brain development and function may form a diathesis for major psychiatric illness, perhaps as multiple growth and metabolic pathways influence human height18. Our results may also reflect heterogeneity, such that some patients have aetiologically distinct diseases. The shared genetic liability between SCZ and BPD, previously suggested by clinical and genetic epidemiology9,19, opens up the possibility of genetically-based refinements in diagnosis. However, the scores derived here have little value for individual risk prediction, meaning that application to clinical genetic testing for SCZ would be unwarranted. In the future, measures of polygenic burden, along with known risk loci and non-genetic factors such as season of birth, life stress, obstetrical complications, viral infections and epigenetics, could open new avenues for studying gene-gene and gene-environment interplay.
Increasing the discovery sample size should substantially refine the polygenic scores derived here. The variance explained by the observed score increases from ~3% to over 20% in extended simulations of 20,000 case/control pair, as will soon be available via international meta-analytic efforts such as the Psychiatric GWAS Consortium20–22 (Section S18, Figure S8). In addition, analyses that focus on gene pathways, clinical features and non-additivity may increase the variance captured by the score and identify genes or biological systems, that are either shared by, or unique to, SCZ and BPD.
We identified fewer unambiguously associated variants than studies of some non-psychiatric diseases of comparable size23. Nonetheless, for other diseases replicated variants typically account for only a modest fraction of risk. The nature of this “missing heritability” is a general problem now faced by complex disease geneticists24. For SCZ, our data point to a genetic architecture that includes many common variants of small effect. The extent to which similar models characterize genetic variation within and across other complex diseases remains to be investigated.
METHODS SUMMARY
Cases satisfied criteria for SCZ. Clinical characteristics and copy number variation have been described previously15. DNA was extracted from whole blood, with approval from institutional review boards. Genotypes were called using the Birdseed/Birdsuite algorithm25 and analyses were performed with PLINK v1.0526. Association analyses used a Cochran-Mantel-Haenszel test and logistic regression with covariates for sample site and ancestry. In the simulations, we generated datasets with pairs of unobserved variants and marker SNPs in varying degrees of within-pair LD, based on the effective number of independent SNPs in the ISC and assuming Hardy-Weinberg equilibrium and linkage equilibrium between different pairs of SNPs. We considered a large grid of possible values for allele frequency and effect size distributions, also varying the proportion of non-null variants and the LD between causal allele and observed marker. We retained models that produced similar profiles of target sample R2 compared to the original ISC analysis, for the same range of pT thresholds, and calculated the implied total genetic variance under these models, assuming additivity within and across loci. See Supplementary Information for details.
Supplementary Material
Supplementary Data
Acknowledgements
We thank the patients and families who contributed to these studies. We also thank Eric Lander, Nick Patterson and members of the Medical and Population Genetics group at the Broad Institute of Harvard and Massachusetts Institute of Technology for valuable discussion and members of the Broad Biological Samples and Genetic Analysis Platforms for sample management and genotyping. We particularly thank Douglas Levinson and Pablo Gejman for allowing access to the MGS samples, and Jianxin Shi for analytic support with the MGS samples. The group at the Stanley Center for Psychiatric Research at the Broad Institute was supported by the Stanley Medical Research Institute (E.M.S.), the Sylvan C. Herman Foundation (E.M.S.), and MH071681 (P.S.). The Cardiff University group was supported by a Medical Research Council (UK) Programme grant and the National Institutes of Mental Health (USA) (CONTE: 2 P50 MH066392-05A1). The group at Karolinska Institutet was supported by the Swedish Council for Working Life and Social Research (FO 184/2000; 2001-2368). The Massachusetts General Hospital group was supported by the Stanley Medical Research Institute (P.S.), MH071681 (P.S.) and a Narsad Young Investigator Award (S.P.). The group at the Queensland Institute of Medical Research was supported by the Australian National Health and Medical Research Council (grants 389892, 442915, 496688, 496674) and thanks Scott Gordon for data preparation. The Trinity College Dublin group was supported by Science Foundation Ireland, the Health Research Board (Ireland), the Stanley Medical Research Institute and the Wellcome Trust; Irish controls were supplied by J. McPartlin from the Trinity College Biobank. The work at the University of Aberdeen was partly funded by GlaxoSmithKline and Generation Scotland, Genetics Health Initiative. University College London clinical and control samples were collected with support from the Neuroscience Research Charitable Trust, the Camden and Islington Mental Health and Social Care Trust, East London and City Mental Heath Trust, the West Berkshire NHS Trust, the West London Mental Health Trust, Oxfordshire and Buckinghamshire Mental Health Partnership NHS Trust, South Essex Partnership NHS Foundation Trust, Gloucestershire Partnership NHS Foundation Trust, Mersey Care NHS Trust, Hampshire Partnership NHS Trust and the North East London Mental Health Trust. The collection of the University of Edinburgh cohort was supported by the Wellcome Trust Clinical Research Facility (Edinburgh) and grants from The Wellcome Trust, London and the Chief Scientist Office of the Scottish Government. The group at the University of North Carolina, Chapel Hill, was supported by MH074027, MH077139 and MH080403, the Sylvan C. Herman Foundation (P.F.S.) and the Stanley Medical Research Institute (P.F.S.) The group at the University of Southern California thanks the patients and their families for their collaboration, and acknowledges the support of the National Institutes of Mental Health and the Department of Veterans Affairs.
The International Schizophrenia Consortium
Manuscript Preparation Shaun M. Purcell1–4, Naomi R. Wray5, Jennifer L. Stone1–4, Peter M. Visscher5, Michael C. O’Donovan6, Patrick F. Sullivan7, Pamela Sklar1–4; Data Analysis Shaun M. Purcell1–4 (leader), Jennifer L. Stone1–4 GWAS analysis subgroup Patrick F. Sullivan7, Douglas M. Ruderfer1–4, Andrew McQuillin8, Derek W. Morris9, Aiden Corvin9, Colm T. O'Dushlaine9, Peter A. Holmans6, Michael C. O’Donovan6, Pamela Sklar1–4,Polygenic analysis subgroup Naomi R. Wray5, Stuart MacGregor5, Pamela Sklar1–4, Patrick F. Sullivan7, Michael C. O’Donovan6, Peter M. Visscher5; Management Committee Hugh Gurling8, Aiden Corvin9, Douglas H.R. Blackwood10, Nick J. Craddock6, Michael Gill9, Christina M. Hultman11,12, George K. Kirov6, Paul Lichtenstein11, Andrew McQuillin8, Michael C. O’Donovan6, Michael J. Owen6, Carlos N. Pato13, Shaun M. Purcell1–4, Edward M. Scolnick2,3, David St. Clair14, Jennifer L. Stone1–4, Patrick F. Sullivan7, Pamela Sklar1–4 (leader); Cardiff University Michael C. O’Donovan6, George K. Kirov6, Nick J. Craddock6, Peter A. Holmans6, Nigel M. Williams6, Lucy Georgieva6, Ivan Nikolov6, N. Norton, H. Williams6, Draga Toncheva15, Vihra Milanova16, Michael J. Owen6; Karolinska Institutet/University of North Carolina at Chapel Hill Christina M. Hultman11,12, Paul Lichtenstein11, Emma F. Thelander11, Patrick Sullivan7; Trinity College DublinDerek W. Morris9, Colm T. O’Dushlaine9, Elaine Kenny9, Emma M. Quinn9, Michael Gill9, Aiden Corvin9;University College London Andrew McQuillin8, Khalid Choudhury8, Susmita Datta8, Jonathan Pimm8, Srinivasa Thirumalai18, Vinay Puri8, Robert Krasucki8, Jacob Lawrence8, Digby Quested19, Nicholas Bass8, Hugh Gurling8; University of Aberdeen Caroline Crombie21, Gillian Fraser21, Soh Leh Kuan14, Nicholas Walker22, David St Clair14;University of Edinburgh Douglas H. R. Blackwood10, Walter J. Muir10, Kevin A. McGhee10, Ben Pickard10, Pat Malloy10, Alan W. Maclean10, Margaret Van Beck10; Queensland Institute of Medical Research Naomi R. Wray5, Peter M. Visscher5, Stuart Macgregor5; University of Southern California Michele T. Pato13, Helena Medeiros13, Frank Middleton23, Celia Carvalho13, Christopher Morley23, Ayman Fanous13,24–26, David Conti13, James A. Knowles13, Carlos Paz Ferreira27, Antonio Macedo28, M. Helena Azevedo28, Carlos N. Pato13; Massachusetts General Hospital Jennifer L. Stone1–4, Douglas M. Ruderfer1–4, Manuel A. R. Ferreira1–4, Mark J. Daly2–4, Shaun M. Purcell1–4, Pamela Sklar1–4 Stanley Center for Psychiatric Research and Broad Institute of MIT and Harvard Shaun M. Purcell1–4, Jennifer L. Stone1–4, Kimberly Chambert2,3, Douglas M. Ruderfer1–4, Finny Kuruvilla3, Stacey B. Gabriel3, Kristin Ardlie3, Mark J. Daly2–4, Edward M. Scolnick2,3, Pamela Sklar1–4
1Psychiatric and Neurodevelopmental Genetics Unit, Department of Psychiatry, Massachusetts General Hospital and Harvard Medical School, 185 Cambridge Street, Boston, Massachusetts 02114, USA.
2Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, Massachusetts 02142, USA.
3Broad Institute of Harvard and Massachusetts Institute of Technology, Cambridge, Massachusetts 02142, USA.
4Center for Human Genetic Research, Massachusetts General Hospital, 185 Cambridge Street, Boston, Massachusetts 02114, USA.
5Queensland Institute of Medical Research, 300 Herston Road, Brisbane, Queensland 4006 Australia.
6School of Medicine, Department of Psychological Medicine, Cardiff University, Cardiff C14 4XN, UK.
7Departments of Genetics, Psychiatry, and Epidemiology, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.
8Molecular Psychiatry Laboratory, Department of Mental Health Sciences, University College London Medical School, Windeyer Institute of Medical Sciences, 46 Cleveland Street, London W1T 4JF, UK.
9Neuropsychiatric Genetics Research Group, Department of Psychiatry and Institute of Molecular Medicine, Trinity College Dublin, Dublin 2, Ireland.
10Division of Psychiatry, University of Edinburgh, Royal Edinburgh Hospital, Edinburgh, EH10 5HF, UK.
11Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Se-171 77 Stockholm, Sweden.
12Department of Neuroscience, Psychiatry, Ulleråker, Uppsala University, Se- 750 17 Uppsala, Sweden.
13Center for Genomic Psychiatry, University of Southern California, Los Angeles, California 90033, USA.
14Institute of Medical Sciences, University of Aberdeen, Foresterhill, Aberdeen AB25 2ZD, UK.
15Department of Medical Genetics, University Hospital Maichin Dom, Sofia 1431, Bulgaria.
16Department of Psychiatry, First Psychiatric Clinic, Alexander University Hospital, Sofia 1431, Bulgaria.
18West Berkshire NHS Trust, 25 Erleigh Road, Reading RG3 5LR, UK.
19Department of Psychiatry, University of Oxford, Warneford Hospital, Headington, Oxford OX3 7JX, UK.
21Department of Mental Health, University of Aberdeen, Aberdeen, AB25 2ZD, UK.
22Ravenscraig Hospital, Inverkip Road, Greenock PA16 9HA, UK.
23State University of New York – Upstate Medical University, Syracuse, New York, 13210, USA.
24Washington VA Medical Center, Washington DC 20422, USA.
25Department of Psychiatry, Georgetown University School of Medicine, Washington DC 20057, USA.
26Department of Psychiatry, Virginia Commonwealth University, Richmond, VA 23298, USA.
27Department of Psychiatry, Sao Miguel, 9500-310 Azores, Portugal.
28Department of Psychiatry University of Coimbra, 3004-504 Coimbra, Portugal
REFERENCES
- 1.Cardno AG, Gottesman II. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–17. [PubMed] [Google Scholar]
- 2.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 3.O'Donovan MC, et al. Identification of loci associated with schizophrenia by genomewide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 4.Wei J, Hemmings GP. The NOTCH4 locus is associated with susceptibility to schizophrenia. Nat Genet. 2000;25:376–377. doi: 10.1038/78044. [DOI] [PubMed] [Google Scholar]
- 5.Horton R, et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher R. The correlation between relatives on the supposition of Mendelian inheritance. Trans Roy Soc Edinb. 1918;52:399–433. [Google Scholar]
- 7.Gottesman II, Shields J. A polygenic theory of schizophrenia. Proc Natl Acad Sci U S A. 1967;58:199–205. doi: 10.1073/pnas.58.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17:1520–1528. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craddock N, O'Donovan MC, Owen MJ. Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr Bull. 2006;32:9–16. doi: 10.1093/schbul/sbj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sklar P, et al. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13:558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svensson AC, Lichtenstein P, Sandin S, Hultman CM. Fertility of first-degree relatives of patients with schizophrenia: a three generation perspective. Schizophr Res. 2007;91:238–245. doi: 10.1016/j.schres.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 13.McClellan JM, Susser E, King MC. Schizophrenia: a common disease caused by multiple rare alleles. Br J Psychiatry. 2007;190:194–199. doi: 10.1192/bjp.bp.106.025585. [DOI] [PubMed] [Google Scholar]
- 14.Craddock N, O'Donovan MC, Owen MJ. Phenotypic and genetic complexity of psychosis. Invited commentary on ... Schizophrenia: a common disease caused by multiple rare alleles. Br J Psychiatry. 2007;190:200–203. doi: 10.1192/bjp.bp.106.033761. [DOI] [PubMed] [Google Scholar]
- 15.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 17.Xu B, et al. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 18.Weedon MN, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lichtenstein P, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Psychiatric GWAS Consortium Steering Committee. A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry. 2009;14:10–17. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
- 21.The Psychiatric GWAS Consortium. Genome-wide association studies: History, rationale and prospects for psychiatric disorders. Am J Psychiatry. 2009 doi: 10.1176/appi.ajp.2008.08091354. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cross Disorder Phenotype Group of the Psychiatric GWAS Consortium. Dissecting the phenotype in genome-wide association studies of psychiatric illness. Br J Psychiatry. 2009 doi: 10.1192/bjp.bp.108.063156. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maher B. The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 25.Korn JM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, et al. PLINK: A Tool Set for Whole-Genome Association and Population- Based Linkage Analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data