Optimization of the analogue-sensitive Cdc2/Cdk1 mutant by in vivo selection eliminates physiological limitations to its use in cell cycle analysis (original) (raw)
Abstract
Analogue-sensitive (as) mutants of kinases are widely used to selectively inhibit a single kinase with few off-target effects. The analogue-sensitive mutant cdc2-as of fission yeast (Schizosaccharomyces pombe) is a powerful tool to study the cell cycle, but the strain displays meiotic defects, and is sensitive to high and low temperature even in the absence of ATP-analogue inhibitors. This has limited the use of the strain for use in these settings. Here, we used in vivo selection for intragenic suppressor mutations of cdc2-as that restore full function in the absence of ATP-analogues. The cdc2-asM17 underwent meiosis and produced viable spores to a similar degree to the wild-type strain. The suppressor mutation also rescued the sensitivity of the cdc2-as strain to high and low temperature, genotoxins and an anti-microtubule drug. We have used cdc2-asM17 to show that Cdc2 activity is required to maintain the activity of the spindle assembly checkpoint. Furthermore, we also demonstrate that maintenance of the Shugoshin Sgo1 at meiotic centromeres does not require Cdc2 activity, whereas localization of the kinase aurora does. The modified cdc2-asM17 allele can be thus used to analyse many aspects of cell-cycle-related events in fission yeast.
Keywords: cell cycle, cyclin-dependent kinase, chemical genetics, analogue-sensitive mutant, fission yeast
2. Introduction
Phosphorylation is involved in many cellular events, often serving as a molecular switch to regulate signalling pathways. The fission yeast genome contains 96 protein kinases (www.pombase.org [1]). A variety of genetic materials and methods have been developed to investigate the function of each kinase in Schizosaccharomyces pombe. For kinases that are indispensable for cell growth, it is common to use conditional mutants to knockdown the gene function. Those include temperature-sensitive (ts) mutants, which lose functions at the restrictive temperature. Although this method provides a powerful genetic tool, it poses practical problems for cell biology. For instance, it is difficult to reduce kinase activity rapidly during live-cell imaging because of the technical difficulties involved in changing the temperature. Furthermore, most mutants are not well characterized with regard to how fast the activity is lost following shift to the restrictive temperature. Small molecules are frequently used as kinase inhibitors, particularly for the analysis of cultured mammalian cells; however, many of these work poorly on yeast cells (for example, fig. 1 of [2]).
These technical difficulties were solved by a so-called chemical genetics approach [3]. Substitution of a single amino acid in the adenosine triphosphate (ATP)-binding pocket of a kinase (the so-called gatekeeper residue) renders the mutant kinase sensitive to ATP-analogue molecules that cannot fit into the active site of an unmodified kinase. This confers specificity to the inhibitor, as genetically unmodified kinases are unaffected by ATP-analogues. They also inhibit the kinase function rapidly (approximately minutes after a drug addition to the medium [4]), permitting time-lapse observation over short time scales. In principle, the gatekeeper residue of any kinase can be predicted from its amino acid sequence [5,6], which has prompted the construction of an as-mutant collection of fission yeast essential kinases [7]. Analogue-sensitive mutants are now widely used for analyses of cell cycle regulation.
Mitotic progression is controlled by protein kinases that have been conserved from yeast to human [8]. The main mitotic kinases include cyclin-dependent kinase 1 (Cdk1), known as Cdc2 in fission yeast and Cdc28 in budding yeast [9–11]; the polo kinase, known as Plo1 in fission yeast and Cdc5 in budding yeast [12–14]; and the single aurora kinase (equivalent to aurora B), which is known as Ark1 in fission yeast and Ipl1 in budding yeast [15,16]. Analogue-sensitive mutants of these mitotic kinases have been described: fission yeast cdc2-as [17] and budding yeast cdc28-as1 [3] for Cdk1; cdc5-as [18] and plo1-as [7] for polo kinase; and ark1-as2/as3 [19] and ipl1-as [5] for aurora B kinase.
Cdc2/Cdc28 regulates both the G1/S and G2/M transitions in S. pombe and Saccharomyces cerevisiae. Comprehensive proteomics analyses using cdc28-as1 in budding yeast have identified more than 300 Cdk1 substrates [4,20]. In S. pombe, Cdc2 is required both for the G1/S and G2/M transitions (for review, see [21]). Many mitotic substrates have been identified; for example, Cdc2 is required for activation of Plo1 [22], for faithful chromosome segregation through controlling localization of the chromosomal passenger complex (CPC) [23] and Nsk1 [24], for chromosome condensation through nuclear accumulation of the condensin Cut3/SMC4 [25,26], and for localization of the microtubule-associated protein (MAP) Dis1/tumour overexpressed gene (TOG) at kinetochores [27,28]. Cdc2 is also required for activation of the anaphase-promoting complex (APC/cyclosome) [29–31].
Combining analogue-sensitive mutants of mitotic kinases with microscopy of living cells provides a way to investigate kinase function during short periods of the cell cycle (e.g. metaphase or anaphase): addition of the inhibitory analogue decreases the activity of the kinase rapidly, to the extent desired [17,32]. This approach has revealed that Cdc2 is required for the nuclear accumulation of the MAP complex Alp7/transforming acidic coiled-coil-Alp14/TOG in early stages of mitosis, which facilitates bipolar spindle assembly [33–35], and in late mitosis, downregulation of Cdc2 promotes the asymmetric localization of septum initiation network proteins at spindle pole bodies (SPBs, the centrosome equivalent in yeast) [17,36].
Although analogue-sensitive mutants can be easily made by substitution of the gatekeeper amino acid, this mutation occasionally has deleterious effects. The fission yeast cdc2-as mutant is generated by the F84G mutation. However, in addition to analogue sensitivity, the cells are elongated at 25°C, indicating a delay in mitotic commitment, and they are also heat-sensitive, particularly at 36°C, even in the absence of the ATP-analogue molecule [17]. In addition, the cdc2-as mutant is defective in sexual differentiation (mating and meiosis) and the mutant zygotes produce two-spore asci instead of four-spore ones of the wild-type zygotes [17]. Such phenotypes have been observed in other hypomorphic alleles of Cdc2 [37–39] and the meiotic mutant cdc2-N22/tws1 [40,41], indicating that the gatekeeper mutation reduces Cdc2–Cdc13 activity per se. This limits the usefulness of the cdc2-as allele in certain circumstances. For example, it is difficult to combine cdc2-as with many heat-sensitive mutants that require incubation at 36°C to arrest efficiently (e.g. cdc25–22 [42]); cdc2-as also shows a negative interaction with mutants that arrest in mitosis, such as the β-tubulin mutants nda3-311 (cs) and nda3/alp12-1828 (ts) [43–45]. This incompatibility of the cdc2-as mutation with key mutants used to impose cell cycle arrests limits its utility for the analysis of some mitotic functions of Cdc2–Cdc13.
Meiosis in fission yeast consists of pre-meiotic DNA replication, meiotic recombination during meiotic prophase, and two consecutive rounds of chromosome segregation (meiosis I and meiosis II) without an intervening S phase, prior to sporulation. To achieve this meiosis-specific cell cycle progression, the Cdc2 activity is regulated in a special manner during meiosis. A fraction of Cdc13/cyclin B is protected from degradation even after anaphase onset of meiosis I to provide CDK activity for the onset of meiosis II [46]; this contrasts with the situation in mitosis, where Cdc13 is entirely degraded at anaphase onset. Degradation of Cdc13 by the APC/cyclosome is inhibited by Mes1 after anaphase onset of meiosis I [46], whereas CDK must be fully inactivated after meiosis II to avoid ‘meiosis III’ [47]. The unique modulation of Cdc2–Cdc13 implies that the function of Cdc2 in meiosis may differ from that in mitosis. The multiple roles of Cdc2 in meiosis have limited the use of conditional mutants to analyse its function. The existing cdc2-as mutant is also limited in its suitability for studies in meiosis, owing to production of dyads, in contrast to tetrads that wild-type zygotes produce [17].
Thus, although the previously described cdc2-as mutant is a powerful tool, it also has technical limitations in some experimental settings. We therefore decided to use natural selection to modify the cdc2-as allele to eliminate the undesirable hypomorphic phenotypes by additional mutations. We have used this improved cdc2-as allele to examine various functions of cdc2 during mitosis and meiosis.
3. Results and discussion
3.1. Isolation of intragenic suppressor mutants of cdc2-as
The original cdc2-as mutant gene contains a single amino acid substitution (F84G) at the gatekeeper residue [17] (figure 1a). We performed an error-prone PCR to introduce additional mutations to the cdc2-as gene containing the open reading frame and 500 bp upstream and downstream flanking regions (the strategy is summarized in the electronic supplementary material, figure S1_a_). The amplified 2.2 kb fragment was used for transformation of the original cdc2-as mutant, selecting for colony formation at 36°C (electronic supplementary material, figure S1_a,b_). We expected that these survivors should include intragenic suppressor mutations. We chose 17 colonies that survived at 36°C, and restreaked onto the YE5S plate (rich medium) containing phloxine B, which stains dead cells. Eight of 17 colonies (named M1, M2, M6, M8, M10, M11, M12 and M17) grew well at 36°C as well as at 20°C (electronic supplementary material, figure S1_b_), therefore those candidates were neither ts nor cs. Importantly, all of these survivors remained sensitive to the ATP-analogue molecule 1NM-PP1 (electronic supplementary material, figure S1_b_), indicating that none of the survivors were revertants of the as mutation.
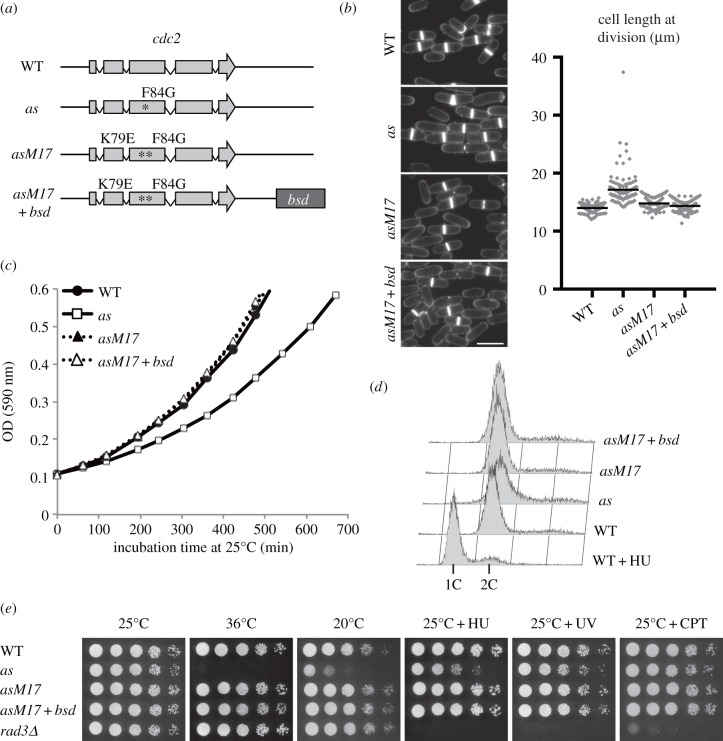
Figure 1.
Characterization of the cdc2-asM17 mutant in mitotic cell cycle. (a) Schematic of wild-type cdc2 (WT), cdc2-as (as), cdc2-asM17 (asM17) and cdc2-asM17 + bsd (asM17 + bsd) mutant genes. The bsd marker was inserted in the downstream of the cdc2 coding sequence. (b) Calcofluor staining of vegetative cells at 25°C. Scale bar, 10 µm. The scatter-dot plot indicates distribution of cell length at cell division (µm; n ≥ 100). Black bars indicate mean values (mean ± s.e.: WT = 14.0 ± 0.1, as = 17.2 ± 0.3, asM17 = 14.8 ± 0.1, asM17 + bsd = 14.3 ± 0.1). (c) OD (590 nm) measurement of log-phase cultures at 25°C. (d) FACS results showing the DNA content of vegetative cells at 25°C. For control of 1C DNA content, 12 mM HU was added to the WT culture (WT + HU). (e) Fivefold dilutions of the indicated strains were spotted onto the following media: YE containing 5 mM HU or 5 µM CPT, YE irradiated with 100 J m−2 UV. Plates were incubated at the indicated temperature for 3–6 days.
Based upon these assays, we retained the M17 mutant, in which ts and cs phenotypes were significantly suppressed, for further analyses (the mutant allele is called cdc2-asM17 hereafter; figure 1a). We next inserted the bsd gene (conferring the blasticidin S resistance [48]) as the selection marker for the modified cdc2-asM17 gene. The bsd gene was inserted at the approximately 0.5 kb downstream of the termination codon of the cdc2-asM17 gene (the allele is called cdc2-asM17 + bsd hereafter; figure 1a), and this did not affect the function of Cdc2 (electronic supplementary material, figure S1_b,c_).
To validate cdc2-asM17 and cdc2-asM17 + bsd mutants as new tools for general purposes, we evaluated whether they behave normally in the absence of ATP-analogues, because the original cdc2-as mutant is slightly deficient in cell cycle [17]. First, we measured the cell size of cdc2-asM17 (±bsd) mutants at cell division. As shown in figure 1b, cdc2-as cells were slightly longer than wild-type (WT) cells [17], indicating compromised CDK activity. The elongation was not observed in the cdc2-asM17 and cdc2-asM17 + bsd strains. This was confirmed by a growth curve assay of four strains (WT, cdc2-as, -asM17, -asM17 + bsd; figure 1c): the growth of the cdc2-as strain was slightly slower than WT, whereas the cdc2-asM17 and -asM17 + bsd strains grew at the same rate as WT. Next, we performed FACS analysis to examine the DNA content of cdc2-asM17 mutants. As shown in figure 1d, cdc2-asM17 and -asM17 + bsd mutants displayed similar DNA content profiles compared with WT, indicating that cell cycle progression of those mutants is similar to WT in the absence of the inhibitory drug. Because the DNA structure check point depends upon Cdc2 for activity, we examined whether cdc2-asM17 mutants were sensitive to genotoxins. Although the original cdc2-as strain was slightly sensitive to hydroxyurea (HU), UV and camptothecin (CPT), the cdc2-asM17 strains were not (figure 1e). Finally, we investigated whether cdc2-asM17 remains associated with Suc1 (p13suc1), which is known to interact with the Cdc2-cyclin B complex [49]. A pulldown assay using p13 Suc1-beads indicated that the cdc2-asM17 mutation did not affect interaction between CDK and Suc1, in the absence of ATP-analogues (electronic supplementary material, figure S2).
Together the data described above, we demonstrate that the cdc2-asM17 mutant (±bsd) behaves similarly to WT in assays where the original cdc2-as allele shows clear defects, thereby validating its use to study the role of Cdc2 activity during mitotic cell cycle in several conditions where the original cdc2-as was not functional. Insertion of the bsd marker gene at the downstream of the cdc2 gene did not cause abnormality as far as we have tested (figure 1b–e).
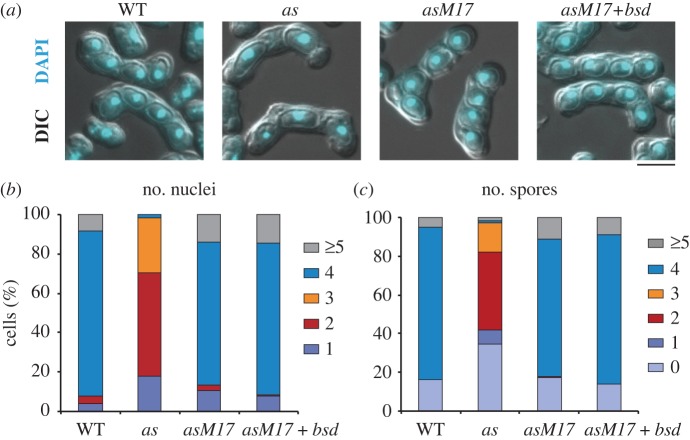
Next, we investigated if the cdc2-asM17 mutant suppressed the meiotic defects seen in the original cdc2-as mutant. Homothallic h90 cdc2-asM17 cells underwent mating and meiosis, and mostly generated four nuclei and four spores per cell, as in WT, whereas the original cdc2-as cells generated abnormal two or three nuclei and two or three spores [17] (figure 2a–c). cdc2-asM17 + bsd also generated four nuclei and four spores per cell (figure 2a–c). Spore viabilities of cdc2-asM17 (91%, n = 108) and cdc2-asM17 + bsd (94%, n = 108) are comparable with that of WT (100%, n = 104), indicating that cdc2-asM17 and cdc2-asM17 + bsd mutants undergo meiosis and produce viable spores to a similar extent to WT. Thus, these mutants are suitable for analysis of Cdc2 functions in meiosis, which could not be addressed using the original cdc2-as mutant. Sporulation of five other suppressor mutants (M1–M11) was also examined, and sporulation defects of the original cdc2-as mutant were in general suppressed to some extent (electronic supplementary material, figure S3).
Figure 2.
Characterization of the cdc2-asM17 mutant in meiosis. (a) DAPI staining of cells that underwent mating, meiosis and sporulation. DAPI and differential interference contrast (DIC) images are shown merged. Scale bar, 5 µm. (b,c) The numbers of (b) nuclei and (c) spores in each ascus shown in (a) were counted for the indicated strains and the percentages are shown. (n > 200).
3.2. Characterization of the suppressor mutations
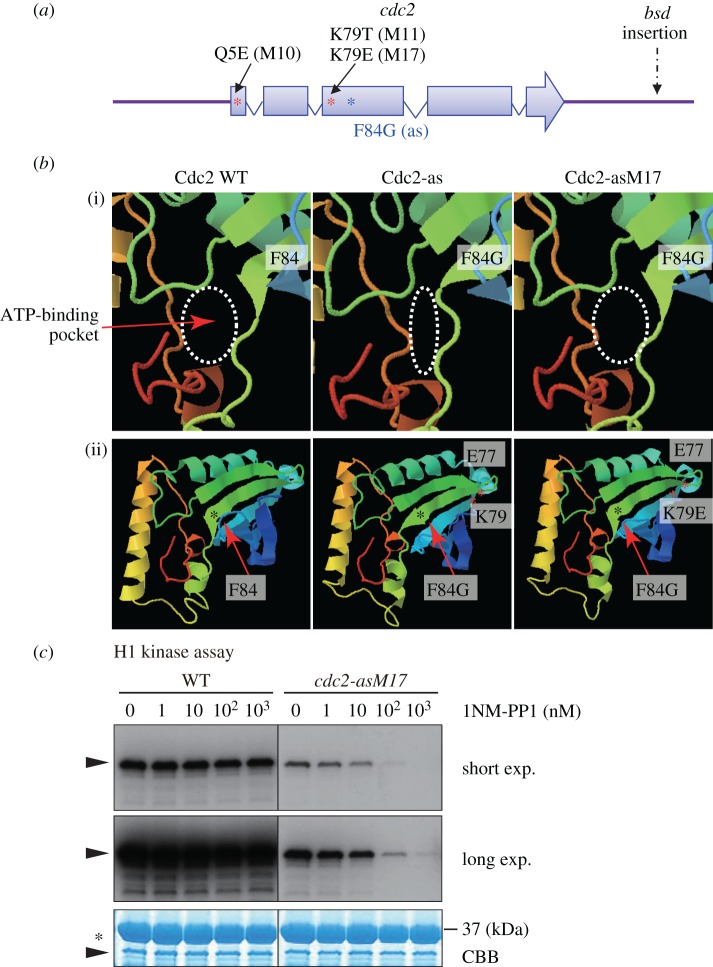
We then sequenced the cdc2 gene of the suppressor mutants. Interestingly, mutants M11 and M17 possessed mutations at the same residue, lysine 79 (figure 3a). M11 had a substitution of K79 to T (threonine), whereas M17 had a substitution of K79 to E (glutamic acid). The mutation site was close to the as mutation site F84G in the primary structure. M10 had a substitution of glutamine at five to glutamic acid (Q5E). To explore how the suppressor mutation in the mutant M17 (K79E) suppressed the as mutation (F84G), the secondary and tertiary structure of the mutant protein was subjected to the protein folding prediction program Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/) [50]. For the sake of simplicity, we used amino acid residues 1–149 (from the N-terminus to the β7 sheet), which form the N-terminal lobe of the kinase [51]. Introduction of the analogue-sensitive mutation F84G was predicted to generate a structural alteration around the ATP binding pocket (Cdc2-as; figure 3b). The deformation of the ATP binding pocket was suppressed by the additional introduction of K79E (Cdc2-asM17; figure 3b). Consistent with this result, mutations within a β sheet in the N-terminal lobe have been reported to suppress the gatekeeper mutations that are not tolerated in several kinases [52]. It is possible that the introduction of K79E, which locates at the edge of the β sheet, may twist the sheet, so that the opposite edge of the sheet harbouring F84G alters the angle at the ATP-binding pocket. Kinase assays of WT Cdc2 and Cdc2-asM17 revealed that the activity of asM17, but not of WT Cdc2, was inhibited by addition of 102–103 nM of the ATP-analogue 1NM-PP1 (figure 3c). Although the suppressor mutation rescues all the phenotypic defects of cdc2-as, the in vitro kinase assay nonetheless reveals that Cdc2-asM17 is less active than WT Cdc2 in the absence of 1NM-PP1 (figure 3c).
Figure 3.
Intragenic suppressor mutations of the isolated cdc2 mutants. (a) The schematic of the cdc2 gene. The analogue-sensitive mutation is F84G [17]. The suppressor mutant M10 contained the Q5E substitution. The M11 and M17 mutants shared the mutation site K79 to T (M11) and to E (M17) in the open reading frame of the cdc2 gene, in addition to the F84G mutation. (b) The prediction of secondary and tertiary structure of Cdc2 WT (wild-type), Cdc2-as and Cdc2-asM17 proteins made by the Phyre program. (i) Magnified view around the ATP-binding pocket and the gatekeeper residue F84 (or F84G). (ii) The overview for (i). The mutation site K79E and a reference site E77 are shown with red asterisks, and F84G is shown with black asterisks. (c) In vitro kinase assay using Cdc2 WT and Cdc2-asM17 proteins purified from WT and the mutant strains, respectively. The Cdk1 substrate histone H1 was incubated with purified Cdc2 proteins and [32P]-α-ATP in the indicated concentration of 1NM-PP1. Autoradiograph images with a short and long exposure are shown. CBB; Coomassie brilliant blue straining for the loading control. Arrowheads indicate the position of histone H1, and the asterisk band corresponds to GST-tagged p13Suc1 derived from Suc1-beads used in affinity purification.
3.3. Cdc2 and the SAC: the use of cdc2-asM17 with MBC or the β-tubulin ts mutant
Elevation of the Cdc2 kinase activity induces assembly of the bipolar spindle at mitotic onset. If microtubule formation is perturbed by internal mutations or drugs, then the attachment of kinetochores to microtubules may be inhibited. The spindle assembly checkpoint (SAC) monitors kinetochore–microtubule attachment to ensure faithful chromosome segregation in mitosis. The SAC components Mad1–Mad2 recognize unattached kinetochores and arrest cell cycle progression at metaphase until correct attachment has been accomplished. Recent studies in yeast, flies, frogs and humans have suggested that Cdk1 can serve as an upstream activator of the SAC [53–58]. We therefore used the cdc2-asM17 mutant to examine whether CDK activity is required to maintain an SAC arrest in S. pombe.
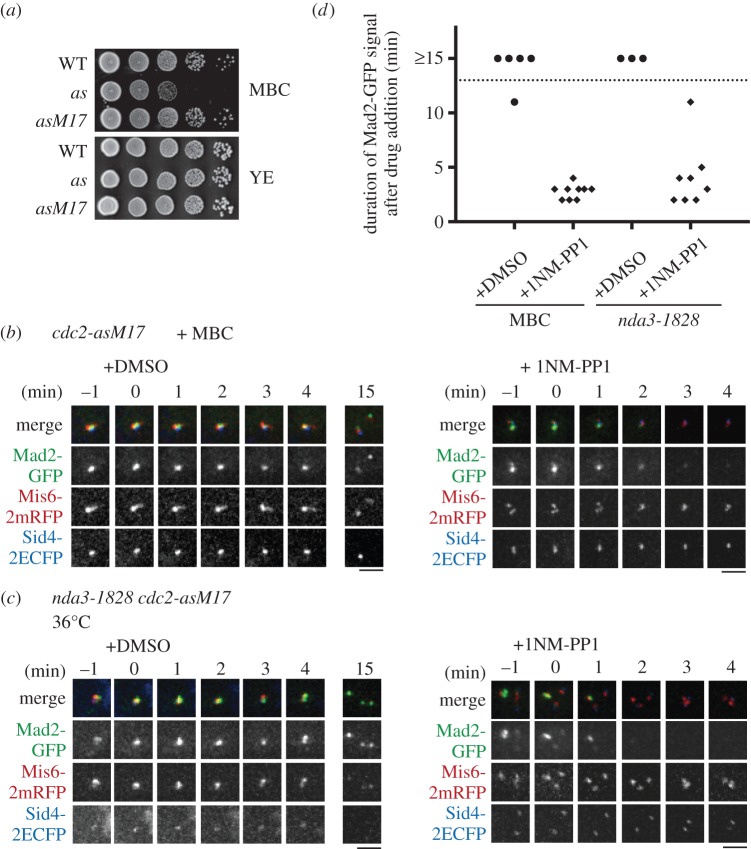
First, we tested whether the microtubule drug methyl benzimidazol-2-yl-carbamate (MBC) induces cdc2-as cells to arrest at metaphase; the original cdc2-as mutant was sensitive to a low concentration of MBC (10 µg ml−1), which did not prevent colony formation in WT cells (figure 4a). By contrast, the cdc2-asM17 mutant was not sensitive to MBC (figure 4a). We therefore constructed the cdc2-asM17 strain expressing Mad2-GFP, the kinetochore marker Mis6-2mRFP and the SPB marker Sid4-2ECFP. Cells were treated with MBC to induce metaphase arrest; bright Mad2-GFP signal was observed at kinetochores, which is a hallmark of SAC activation (−1 min, figure 4b). Cells were then filmed and 1NM-PP1 or DMSO was added to the medium (0 min, figure 4b). In the DMSO treatment, Mad2-GFP dots remained at kinetochores for longer than 15 min (+DMSO, figure 4b,d). By contrast, Mad2-GFP dots mostly disappeared within 4 min after 1NM-PP1 addition (+1NM-PP1, figure 4b,d). This demonstrates that Cdc2 kinase activity is required for maintenance of SAC activation in the presence of microtubule perturbation, through Mad2 recruitment to kinetochores. A very recent study in human cells has shown that Mad2 recruitment to kinetochore requires Cdk1 activity [59], suggesting that this mechanism has been conserved through evolution.
Figure 4.
The requirement of Cdc2 in SAC maintenance was revealed by use of the cdc2-asM17 mutant. (a) The original cdc2-as mutant (as) was sensitive to a low dose of the microtubule drug MBC (10 µg ml−1), whereas the revived mutant cdc2-asM17 (asM17) was not. This enabled use of the analogue-sensitive cdc2 mutant in the presence of MBC. (b) The c_dc2-asM17_ mutant was used in combination with MBC treatment. cdc2-asM17 cells were treated with MBC to arrest cells at metaphase without spindles. Cells with Mad2-GFP dots at kinetochores were chosen for filming. After 1NM-PP1 addition (t = 0 min), Mad2-GFP dots disappeared within 4 min. DMSO was added as negative control. Mis6-2mRFP, a kinetochore marker; Sid4-2ECFP, an SPB marker. (c) The cdc2-asM17 mutation was combined with the β-tubulin ts mutation nda3/alp12–1828. Cells were arrested at metaphase at 36°C. Filming was done similarly to (b). After 1NM-PP1 addition (t = 0 min), Mad2-GFP dots disappeared within 3 min. DMSO was added as negative control. Scale bars, 2 µm. (d) Duration of Mad2-GFP dots residence at kinetochores after addition of DMSO or 1NM-PP1 in (b,c) was measured.
We examined this further using the ts β-tubulin mutant nda3/alp12-1828, which was impossible to combine with the original cdc2-as mutant, because the long G2-phase of S. pombe results in a predominant G2-arrest owing to temperature sensitivity of the cdc2-as allele. After shift to 36°C, the nda3-1828 cdc2-asM17 mutant efficiently arrested at prometaphase without a spindle, and Mad2-GFP dots were associated with kinetochores (−1 min, figure 4c). In the presence of 1NM-PP1, Mad2-GFP dots mostly disappeared within 4 min (+1NM-PP1, figure 4c,d), confirming that SAC maintenance requires Cdc2 activity during mitosis. These experiments also demonstrate the utility of the analogue-sensitive mutant cdc2-asM17 to investigate the involvement of Cdc2 in mitotic events such as the SAC.
3.4. SAC silencing and PP1: the use of cdc2-asM17 with the β-tubulin cs mutant
Recently, it was reported that SAC silencing (inactivation) during mitosis is achieved by the protein phosphatase PP1 [60–63]. The fission yeast has two PP1 phosphatases, Dis2 and Sds21 [64,65], though only Dis2 has a function in SAC silencing [61]. To test whether Dis2/PP1 is required for the SAC silencing following Cdc2 inactivation (as shown in figure 4), we created the double mutant cdc2-asM17 nda3-KM311. The double mutant of the original cdc2-as and nda3-KM311 did not enter mitosis efficiently, owing to the cold-sensitivity of cdc2-as [17]. By contrast, the cdc2-asM17 nda3-KM311 cells arrested efficiently in mitosis at 18°C with a strong nuclear signal of cyclin B/Cdc13-YFP, (0 min, figure 5a,c). When 1NM-PP1 was added to the medium, the frequency of cells harbouring Cdc13-YFP signal decreased (60 min, figure 5a,c). In contrast, when the dis2+ gene was disrupted, the triple mutant _dis2_Δ nda3-KM311 cdc2-asM17 retained a Cdc13-YFP signal even after addition of 1NM-PP1 (figure 5b,c). This indicates that Cdc2 and Dis2/PP1 are required to maintain and silence the checkpoint machinery, respectively. Ark1/aurora B kinase, a component of the CPC, is also required for SAC maintenance [61]. As centromere targeting of CPC depends on Cdk1 [23], maintenance of the SAC by Cdc2 might be achieved through the control of CPC localization to centromeres. Alternatively, Cdc2 might regulate the SAC independently of CPC localization, because centromeric retention of CPC in anaphase does not result in APC/C inhibition (SAC activation) in human cells [66]. How Dis2 /PP1 counteracts Cdc2 in SAC silencing is an important question and will be addressed in future studies. The _dis2_Δ nda3-KM311 cdc2-asM17 mutant frequently resulted in unequal chromosome segregation with persistent nuclear Cdc13-YFP (figure 5b), indicating that checkpoint adaptation (slippage into anaphase) may occur even without complete cyclin destruction, when the Cdc2 activity is inhibited.
Figure 5.
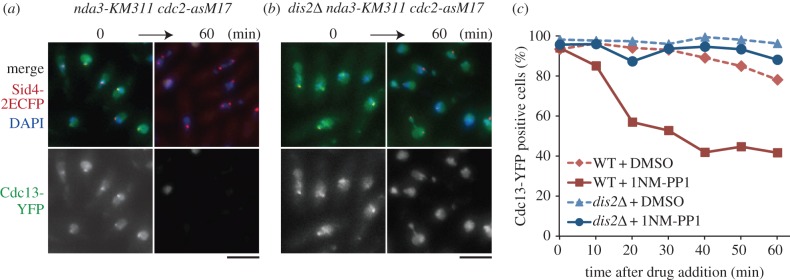
Dis2/PP1 is required for checkpoint inactivation triggered by Cdc2 inhibition. (a) The cdc2-asM17 mutation was combined with the cs β-tubulin mutation nda3-KM311. The mutant was arrested at metaphase at 18°C with high concentration of cyclin B1/Cdc13-YFP in the nucleus and at SPBs (0 min). Then, 1NM-PP1 was added to the culture. After 60 min, the percentage of cells with Cdc13-YFP decreased. Sid4–2ECFP (SPB) and DAPI are also shown. (b) Similar experiments were done with the triple mutant _dis2_Δ nda3-KM311 cdc2-asM17. Cells were cultured at 18°C (0 min) and then 1NM-PP1 was added. In contrast to dis2+ cells (a), Cdc13-YFP remained in the nucleus. Scale bars, 5 µm. (c) Frequencies of cells with nuclear Cdc13-YFP signal in dis2+ (WT) and _dis2_Δ strains in the nda3-KM311 cdc2-asM17 background were plotted in response to 1NM-PP1 (or DMSO for negative control) addition (n ≥ 100).
3.5. Shugoshin and Cdc2: the use of cdc2-asM17 in meiosis
Genetic perturbation of Cdc2 prevents entry into meiosis I [67], making it difficult to examine the function of Cdc2 in spindle organization and chromosome segregation during meiosis I. Because the cdc2-asM17 strain does not show meiotic defects in the absence of ATP-analogues we used it to address this question.
We have previously used cdc2-asM17 strain to show that reorganization of SPBs at the onset of meiosis I requires Cdc2 activity [68]. We have now used it to examine the role of Cdc2 activity at the metaphase–anaphase transition in meiosis I. To ensure faithful segregation of chromosomes during meiosis I, proteins such as aurora B and protein phosphatase 2A (PP2A) are localized transiently to centromeres, until the onset of anaphase I [69,70]. It is unclear whether delocalization of aurora B and PP2A at the onset of anaphase I depends on reduction of the Cdc2 activity or proteasome-dependent protein degradation.
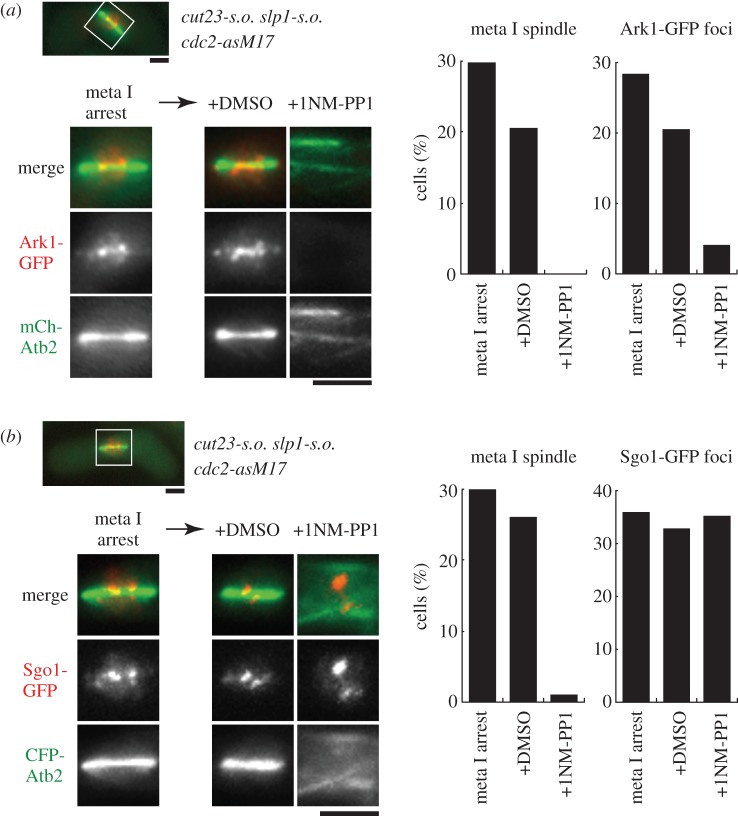
To distinguish these two possibilities, we used slp1-s.o. cut23-s.o. mutations, in which meiotic expression of the APC/C component Cut23 and its activator Slp1 is repressed [71], thereby preventing proteasome-dependent protein degradation. Then, 1NM-PP1 was added to inhibit Cdc2 activity of cdc2-asM17 cells. First, we examined localization of Ark1 (aurora B). Cdc2 phosphorylates the CPC component Bir1/survivin to recruit Ark1 to centromeres in fission yeast mitosis [23], but it has not been tested whether this mechanism is conserved during meiosis. Ark1 localized to centromeres in cells arrested in metaphase I, and delocalized from centromeres when Cdc2 activity was reduced by addition of 1NM-PP1 (figure 6a). This indicates that delocalization of Ark1 from centromeres depends on reduction of the Cdc2 activity, but not on degradation of CPC components. Second, we examined the localization of Shugoshin (Sgo1), which forms a complex with PP2A and is essential for its localization at centromeres [70]. Consistent with previous studies [69], Sgo1 localized to centromeres in metaphase I-arrested cells. Interestingly, Sgo1 did not alter its localization at centromeres even when Cdc2 activity was decreased by 1NM-PP1 (figure 6b). Thus, maintenance of the Sgo1 localization at centromeres does not require Cdc2 activity, raising the possibility that APC-mediated Sgo1 degradation might be a trigger of Sgo1 delocalization from centromeres.
Figure 6.
Cdc2 activity is required for aurora-B localization but not for the Shugoshin Sgo1 during meiosis I. (a) The cdc2-asM17 mutation was combined with slp1-s.o. and cut23-s.o. mutations, in which meiotic transcription of these APC/C factors is shut off and cells are arrested in metaphase I. The rectangular region was magnified and is shown below. The aurora B/Ark1-GFP localized to centromeres on the metaphase spindle, which was visualized by CFP-Atb2 (α2-tubulin). Ark1-GFP foci dispersed after 1NM-PP1 treatment, but not after DMSO treatment (negative control). Images corresponding to the nuclear region were taken 1 h after addition of 1NM-PP1 or DMSO. The frequencies of cells with the metaphase I spindle (the left graph) and with Ark1-GFP foci (the right graph) are also shown (n > 100). (b) Similar experiments to (a) were done for Sgo1-GFP. Sgo1-GFP foci at centromeres did not disperse even 1 h after 1NM-PP1 addition (n > 100). Scale bars, 2 µm.
4. Conclusion
The generation of an analogue-sensitive mutant is a powerful tool that enables chemical inhibition of any kinase of interest. In general, analogue sensitivity is conferred by introducing a single amino acid substitution of the ‘gatekeeper’ residue in the active site. This sometimes causes a partial loss of kinase function, as in the case of the cdc2-as mutant [17]. The original cdc2-as mutant (F84G) showed defects in growth at high and low temperature and in meiosis. We have used natural selection to suppress these defects, which occurred through an additional mutation at K79E, to generate the cdc2-asM17 mutant.
A systematic generation of analogue-sensitive mutants of all essential kinases in fission yeast found that three (of 16) essential kinases could not be generated (Cdc7, Hsk1 and Sid1), because an introduction of the analogue-sensitive mutation caused a significant loss of function [7]. For instance, it was impossible to create the sid1-as mutant, because replacement of the wild-type sid1+ with the sid1-as mutant gene caused lethality (Y.A., S.A.K., V.S., M.Y. & M.S. 2011, unpublished data). Those issues, however, might be solved by applying the method described in this study. Specifically, if the gatekeeper mutation resides at the end of a β sheet, then the suppressor mutation could be introduced at the opposite end of the sheet (figure 3a,b). Alternatively, it might be more judicious to allow in vivo selection to generate the required mutant, as we have done here (electronic supplementary material, figure S1_a_). The library of mutagenized DNA fragments could be used for transformation of the existing ts (or other) mutant of the kinase. Transformants viable at the restrictive temperature should contain the suppressor mutation in addition to the gatekeeper mutation.
The in vivo selected cdc2-asM17 mutant permitted us to undertake experiments that were not possible previously, such as using microtubule-depolymerizing drugs, high and low temperatures, and during meiosis. This mutant has revealed an important role of Cdc2/Cdk1 in SAC maintenance, and the dispensability of Cdc2/Cdk1 for localization of Shugoshin in meiosis. Although mitotic kinases represented by Cdk1, Polo and aurora regulate many aspects of mitosis and meiosis, the availability of this chemical genetic tool will allow us to increase our understanding of the role of Cdc2/CDK1.
5. Material and methods
5.1. Yeast genetics and general manipulations
The strains used in this study are listed in table 1. We used standard methods for fission yeast genetics, as described previously [72]. Tagging of a single copy of a fluorescent protein (GFP) at the C-terminus of genes, and insertion of the bsd marker gene, was performed using standard PCR-based methods [73,74]. Tagging of multiple tandem copies of fluorescent proteins (2mRFP and 2ECFP) at the C-terminus of genes was performed as previously described [34]. All the fluorescent protein-fused genes are expressed by the native promoter and the adh terminator [73], except for cdc13-YFP, mCherry-atb2 and CFP-atb2 fusion genes. The cdc13-YFP fusion gene is expressed by the cdc13+ promoter and terminator. The mCherry-atb2 gene is expressed by the P_adh15_ promoter and the adh terminator, and the CFP-atb2 gene is expressed by the P_adh13_ promoter and the adh terminator [75]. 1NM-PP1 (Calbiochem, CA) was added to media at the concentration of 2 µM. For figure 1b, living cells were stained with Calcofluor (Sigma, MO), and the cell length was measured on the ImageJ software (NIH). For the FACS analysis in figure 1d, the DNA content was measured using BD FACSCalibur (BD, NJ). For figure 2a–c, homothallic _h_90 cells were induced to mating, meiosis and sporulation on sporulation agar plates. After incubation for 24 h at 30°C, cells were fixed with methanol and stained with 4′,6-diamidino-2-phenylindole (DAPI; Wako Pure Chemicals, Japan), and the number of nuclei and spores in each ascus were counted. For the spore viability assay, spores were dissected by the micromanipulator (Singer Instruments, Somerset, UK), and the percentage of spores that formed colonies was calculated. The checkpoint silencing assay in figure 5 was performed as previously described [61]. The prometaphase-arrested nda3-KM311 cells at 18°C were treated with 1 µM 1NM-PP1 or DMSO. Cells were collected at the indicated time point, fixed with methanol and stained with DAPI (Wako Pure Chemicals).
Table 1.
Strains used in this study. The origin of the strains is this study, except for JY878 and SP5959 (our stock) and PY328 (a gift from Y. Watanabe).
| strain | genotype | figures |
|---|---|---|
| MJ1172 | _h_90 sfi1-CFP-nat leu1–32 ura4-D18 ade6-M216 | 1b–e, 2a–c and electronic supplementary material, figure S2 |
| MJ1254 | _h_90 cdc2-as sfi1-CFP-nat leu1–32 ura4-D18 ade6-M216 | 1b–e, 2a–c and electronic supplementary material, figures S1_a–c_, S2 and S3_a–c_ |
| MJ1353 | _h_90 cdc2-asM17 sfi1-CFP leu1–32 ura4-D18 ade6-M216 | 1b–e, 2a–c and electronic supplementary material, figures S1_b,c_, S2 and S3_a–c_ |
| MJ1358 | _h_90 cdc2-asM17-bsd sfi1-CFP-nat leu1–32 ura4-D18 ade6-M216 | 1b–e, 2a–c, 3c, 4a and electronic supplementary material, figures S1_c_ and S2 |
| PY328 | _h_90 rad3 ::LEU2+ leu1–32 ura4-D18 ade6-M210 | 1e |
| SAK1 | h+ leu1–32 ura4-D18 ade6-M216 | 3c |
| MJ1360 | _h_90 cdc2-as-bsd sfi1-CFP-nat leu1-32 ura4-D18 ade6-M216 | 4a and electronic supplementary material, figure S1_c_ |
| YA1843 | h90 cdc2asM17bsd mad2GFPkan mis62mRFPhph sid42ECFPnat leu132 ura4D18 | 4b,d |
| YA1829 | h90 cdc2asM17bsd nda3(alp12)1828 mad2GFPkan mis62mRFPhph sid42ECFPnat leu132 ura4D18 ade6M216 | 4c,d |
| YA1900 | h90 cdc2asM17bsd nda3KM311 cdc13YFPTcdc13kan sid42ECFPnat leu132 ura4D18 ade6 | 5a,c |
| YA1893 | h90 cdc2asM17bsd nda3KM311 dis2::ura4+ cdc13YFPTcdc13kan sid42ECFPnat leu132 ura4D18 ade6 | 5b,c |
| SAK422 | _h_90 cdc2-asM17-bsd ark1-GFPFH-kan Prad21-slp1-kan Prad21-cut23-kan z::Padh15-mcherry-atb2-nat leu1-32 ade6-M216 | 6a |
| SAK420 | _h_90 cdc2-asM17-bsd sgo1+-flag-GFP par1--mCherry-hyg Prad21-slp1-kan Prad21-cut23-kan z::Padh13-CFP-atb2-nat leu1-32 ade6-M216 | 6b |
| JY878 | _h_90 leu1-32 ura4-D18 ade6-M216 | electronic supplementary material, figures S1_b,c_ and S3_a_ |
| SP5959 | _h_– cdc2-as ura4-D18 | electronic supplementary material, figure S1_a_ |
| MJ1346 | _h_90 cdc2-asM1 sfi1-CFP-nat leu1-32 ura4-D18 ade6-M216 | electronic supplementary material, figure S1_b_ |
| MJ1347 | _h_90 cdc2-asM2 sfi1-CFP-nat leu1-32 ura4-D18 ade6-M216 | electronic supplementary material, figure S1_b_ |
| MJ1348 | _h_90 cdc2-asM6 sfi1-CFP-nat leu1-32 ura4-D18 ade6-M216 | electronic supplementary material, figures S1_b_ and S3_a_ |
| MJ1349 | _h_90 cdc2-asM8 sfi1-CFP-nat leu1-32 ura4-D18 ade6-M216 | electronic supplementary material, figures S1_b_ and S3_a–c_ |
| MJ1350 | _h_90 cdc2-asM10 sfi1-CFP-nat leu1-32 ura4-D18 ade6-M216 | electronic supplementary material, figures S1_b,c_ and S3_a–c_ |
| MJ1351 | _h_90 cdc2-asM11 sfi1-CFP-nat leu1-32 ura4-D18 ade6-M216 | electronic supplementary material, figures S1_b,c_ and S3_a–c_ |
| MJ1352 | _h_90 cdc2-asM12 sfi1-CFP-nat leu1-32 ura4-D18 ade6-M216 | electronic supplementary material, figure S1_b_ |
| MJ1356 | _h_90 cdc2-asM10-bsd sfi1-CFP-nat leu1-32 ura4-D18 ade6-M216 | electronic supplementary material, figure S1_c_ |
| MJ1357 | _h_90 cdc2-asM11-bsd sfi1-CFP-nat leu1-32 ura4-D18 ade6-M216 | electronic supplementary material, figure S1_c_ |
| MJ1359 | _h_90 cdc2+-bsd leu1-32 ura4-D18 ade6-M216 | electronic supplementary material, figure S1_c_ |
| SAK163 | _h_– cdc2-33 | electronic supplementary material, figure S2 |
| MO1893 | _h_90 cdc2-asM17-bsd leu1-32 ura4-D18 ade6-M216 | Methods |
5.2. Creation of the cdc2-asM17 mutant
The creation of the original cdc2-as mutant has been described previously [17]. A flow chart for the method to generate the cdc2-asM17 mutant is depicted in the electronic supplementary material, figure S1_a_. Briefly, the genomic cdc2-as mutant gene was amplified from the original mutant [17]. The coding region of cdc2-as with flanking 0.5 kb up/downstream regions at both ends (in total 2.2 kb; electronic supplementary material, figure S1_a_) was amplified by a standard PCR method with PrimeSTAR HS DNA polymerase (Takara-Bio, Japan). The amplified fragment was gel-purified, and used as the template for the following error-prone PCR. To induce errors, thermal cycling was performed for 40 cycles using the Ex Taq polymerase (Takara-Bio) and the same pair of oligomers used for the first PCR. The amplified fragments were then used for transformation of the cdc2-as sfi1-CFP-nat strain MJ1254 (Sfi1-CFP is an SPB marker). Colonies that grew at 36°C were restreaked onto YE5S plates containing Phloxin B, to visualize suppression of temperature sensitivity. Cold sensitivity was also tested at 20°C. Confirmation of the analogue sensitivity was done with YE5S plates containing 10 µM 1NM-PP1. The bsd marker gene was inserted 528 bp downstream of the termination codon of the cdc2 gene. For figures 3–6, the cdc2-asM17 mutant with bsd insertion was used and denoted as cdc2-asM17 for simplicity. The sfi1-CFP SPB marker was removed through backcrossing of the cdc2-asM17-bsd sfi1-CFP-nat strain (MJ1358) with a wild-type strain without markers, to yield MO1893.
5.3. Microscopy
Images in figures 1b and 2a were acquired using an Axio Imager.M2 fluorescence microscope and AxioVision software (Zeiss, Germany). Live-cell imaging methods for figure 4 were described previously [34]. Briefly, live-cell imaging was performed with the DeltaVision-SoftWoRx system (GE Healthcare, UK). Cultured cells were mounted on a glass-bottom dish (Matsunami, Japan) coated with lectin and filled with minimal medium. Serial section images along the z_-axis were acquired and stacked using the ‘quick projection’ protocol in SoftWoRx. Temperature-sensitive strains were observed in a temperature-controlled chamber to maintain 36°C during observation. MBC (carbendazim; Sigma, MO) was added to liquid culture at the final concentration of 50 µg ml−1 [76]. 2 µM 1NM-PP1 or DMSO was added to liquid media during observation. Images in figure 5 and electronic supplementary material, figure S3_a were acquired by an Axioplan 2 fluorescence microscope (Zeiss) and SlideBook software (Leeds Precision, UK). Images in figure 6 were taken as described previously [77].
5.4. In vitro kinase assay and Western blotting
Schizosaccharomyces pombe protein extract from wild-type and cdc2-asM17 cells was prepared, and the Cdc2–Cdc13 complex was purified using Suc1-beads (Millipore, MA). The pulldowns containing Cdc2–Cdc13 were mixed with histone H1 (New England BioLabs, UK) as a substrate, in the absence or presence of 1NM-PP1 (0–103 nM). Samples were subjected to SDS–PAGE, and the gel was stained with Coomassie brilliant blue followed by autoradiography. For Western blotting in electronic supplementary material, figure S2, extracts or pulldowns by Suc1-beads were subjected to SDS–PAGE. The following antibodies were used: anti-Cdc2 monoclonal (1 : 1000; a gift from Y. Watanabe) and anti-tubulin monoclonal TAT-1 (1 : 5000; a gift from K. Gull).
Supplementary Material
Adobe PDF - rsob-14-0063-File008.pdf
Acknowledgements
We thank T. Toda for strains, Y. Watanabe for materials and support for the FACS system and K. Gull for an antibody.
Funding statement
Y.A. is a research fellow of Japan Society for the Promotion of Science (JSPS). This work was supported by grants-in-aid for Young Scientists (A; 26711001) from JSPS (to S.A.K), grant-in-aid for Scientific Research on Priority Areas ‘Cell Proliferation Control’ from MEXT, the Ministry of Education, Culture, Sports, Science and Technology of Japan and grants-in-aid for Young Scientists (A) (21687015) and for Scientific Research (B; 25291041) from JSPS (to M.S.), and grants-in-aid for Specially Promoted Research and Scientific Research (S; 21227007) from JSPS (to M.Y.). This work was also supported by the Naito Foundation, the Senri Life Science Foundation, the Sumitomo Foundation and Kato Life Science Foundation; Waseda University Grant for Special Research Projects (2013A-911, 2013B-178 and 2013A-6313; to M.S.), and in part by Global COE Programme (Integrative Life Science Based on the Study of Biosignaling Mechanisms), MEXT, Japan. Research in V.S. laboratory is supported by the Swiss National Science Foundation and EPFL.
References
- 1.Wood V, et al. 2012. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 40, D695–D699. (doi:10.1093/nar/gkr853) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawashima SA, Takemoto A, Nurse P, Kapoor TM. 2013. A chemical biology strategy to analyze rheostat-like protein kinase-dependent regulation. Chem. Biol. 20, 262–271. (doi:10.1016/j.chembiol.2013.01.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop AC, et al. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407, 395–401. (doi:10.1038/35030148) [DOI] [PubMed] [Google Scholar]
- 4.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425, 859–864. (doi:10.1038/nature02062) [DOI] [PubMed] [Google Scholar]
- 5.Kung C, Kenski DM, Dickerson SH, Howson RW, Kuyper LF, Madhani HD, Shokat KM. 2005. Chemical genomic profiling to identify intracellular targets of a multiplex kinase inhibitor. Proc. Natl Acad. Sci. USA 102, 3587–3592. (doi:10.1073/pnas.0407170102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregan J, Zhang C, Rumpf C, Cipak L, Li Z, Uluocak P, Nasmyth K, Shokat KM. 2007. Construction of conditional analog-sensitive kinase alleles in the fission yeast Schizosaccharomyces pombe. Nat. Protoc. 2, 2996–3000. (doi:10.1038/nprot.2007.447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cipak L, Zhang C, Kovacikova I, Rumpf C, Miadokova E, Shokat KM, Gregan J. 2011. Generation of a set of conditional analog-sensitive alleles of essential protein kinases in the fission yeast Schizosaccharomyces pombe. Cell Cycle 10, 3527–3532. (doi:10.4161/cc.10.20.17792) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nigg EA. 2001. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2, 21–32. (doi:10.1038/35048096) [DOI] [PubMed] [Google Scholar]
- 9.Hindley J, Phear GA. 1984. Sequence of the cell division gene CDC2 from Schizosaccharomyces pombe; patterns of splicing and homology to protein kinases. Gene 31, 129–134. (doi:10.1016/0378-1119(84)90203-8) [DOI] [PubMed] [Google Scholar]
- 10.Simanis V, Nurse P. 1986. The cell cycle control gene cdc2+ of fission yeast encodes a protein kinase potentially regulated by phosphorylation. Cell 45, 261–268. (doi:10.1016/0092-8674(86)90390-9) [DOI] [PubMed] [Google Scholar]
- 11.Reed SI, Hadwiger JA, Lörincz AT. 1985. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc. Natl Acad. Sci. USA 82, 4055–4059. (doi:10.1073/pnas.82.12.4055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bähler J, Steever AB, Wheatley S, Wang Y, Pringle JR, Gould KL, McCollum D. 1998. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J. Cell Biol. 143, 1603–1616. (doi:10.1083/jcb.143.6.1603) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulvihill DP, Petersen J, Ohkura H, Glover DM, Hagan IM. 1999. Plo1 kinase recruitment to the spindle pole body and its role in cell division in Schizosaccharomyces pombe. Mol. Biol. Cell 10, 2771–2785. (doi:10.1091/mbc.10.8.2771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golsteyn RM, Schultz SJ, Bartek J, Ziemiecki A, Ried T, Nigg EA. 1994. Cell cycle analysis and chromosomal localization of human Plk1, a putative homologue of the mitotic kinases Drosophila polo and Saccharomyces cerevisiae Cdc5. J. Cell Sci. 107, 1509–1517. [DOI] [PubMed] [Google Scholar]
- 15.Petersen J, Paris J, Willer M, Philippe M, Hagan IM. 2001. The S. pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J. Cell Sci. 114, 4371–4384. [DOI] [PubMed] [Google Scholar]
- 16.Francisco L, Chan CS. 1994. Regulation of yeast chromosome segregation by Ipl1 protein kinase and type 1 protein phosphatase. Cell Mol. Biol. Res. 40, 207–213. [PubMed] [Google Scholar]
- 17.Dischinger S, Krapp A, Xie L, Paulson JR, Simanis V. 2008. Chemical genetic analysis of the regulatory role of Cdc2p in the S. pombe septation initiation network. J. Cell Sci. 121, 843–853. (doi:10.1242/jcs.021584) [DOI] [PubMed] [Google Scholar]
- 18.Snead JL, et al. 2007. A coupled chemical-genetic and bioinformatic approach to Polo-like kinase pathway exploration. Chem. Biol. 14, 1261–1272. (doi:10.1016/j.chembiol.2007.09.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauf S, Biswas A, Langegger M, Kawashima SA, Tsukahara T, Watanabe Y. 2007. Aurora controls sister kinetochore mono-orientation and homolog bi-orientation in meiosis-I. EMBO J. 26, 4475–4486. (doi:10.1038/sj.emboj.7601880) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686. (doi:10.1126/science.1172867) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nurse P. 1990. Universal control mechanism regulating onset of M-phase. Nature 344, 503–508. (doi:10.1038/344503a0) [DOI] [PubMed] [Google Scholar]
- 22.Tanaka K, Petersen J, MacIver F, Mulvihill DP, Glover DM, Hagan IM. 2001. The role of Plo1 kinase in mitotic commitment and septation in Schizosaccharomyces pombe. EMBO J. 20, 1259–1270. (doi:10.1093/emboj/20.6.1259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsukahara T, Tanno Y, Watanabe Y. 2010. Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature 467, 719–723. (doi:10.1038/nature09390) [DOI] [PubMed] [Google Scholar]
- 24.Chen JS, Lu LX, Ohi MD, Creamer KM, English C, Partridge JF, Ohi R, Gould KL. 2011. Cdk1 phosphorylation of the kinetochore protein Nsk1 prevents error-prone chromosome segregation. J. Cell Biol. 195, 583–593. (doi:10.1083/jcb.201105074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M. 1999. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 13, 2271–2283. (doi:10.1101/gad.13.17.2271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakazawa N, Nakamura T, Kokubu A, Ebe M, Nagao K, Yanagida M. 2008. Dissection of the essential steps for condensin accumulation at kinetochores and rDNAs during fission yeast mitosis. J. Cell Biol. 180, 1115–1131. (doi:10.1083/jcb.200708170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nabeshima K, Kurooka H, Takeuchi M, Kinoshita K, Nakaseko Y, Yanagida M. 1995. p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev. 9, 1572–1585. (doi:10.1101/gad.9.13.1572) [DOI] [PubMed] [Google Scholar]
- 28.Aoki K, Nakaseko Y, Kinoshita K, Goshima G, Yanagida M. 2006. Cdc2 phosphorylation of the fission yeast Dis1 ensures accurate chromosome segregation. Curr. Biol. 16, 1627–1635. (doi:10.1016/j.cub.2006.06.065) [DOI] [PubMed] [Google Scholar]
- 29.Listovsky T, Zor A, Laronne A, Brandeis M. 2000. Cdk1 is essential for mammalian cyclosome/APC regulation. Exp. Cell Res. 255, 184–191. (doi:10.1006/excr.1999.4788) [DOI] [PubMed] [Google Scholar]
- 30.Golan A, Yudkovsky Y, Hershko A. 2002. The cyclin-ubiquitin ligase activity of cyclosome/APC is jointly activated by protein kinases Cdk1-cyclin B and Plk. J. Biol. Chem. 277, 15 552–15 557. (doi:10.1074/jbc.M111476200) [DOI] [PubMed] [Google Scholar]
- 31.Yoon HJ, Feoktistova A, Chen JS, Jennings JL, Link AJ, Gould KL. 2006. Role of Hcn1 and its phosphorylation in fission yeast anaphase-promoting complex/cyclosome function. J. Biol. Chem. 281, 32 284–32 293. (doi:10.1074/jbc.M603867200) [DOI] [PubMed] [Google Scholar]
- 32.Coudreuse D, Nurse P. 2010. Driving the cell cycle with a minimal CDK control network. Nature 468, 1074–1079. (doi:10.1038/nature09543) [DOI] [PubMed] [Google Scholar]
- 33.Sato M, Toda T. 2007. Alp7/TACC is a crucial target in Ran-GTPase-dependent spindle formation in fission yeast. Nature 447, 334–337. (doi:10.1038/nature05773) [DOI] [PubMed] [Google Scholar]
- 34.Sato M, Toya M, Toda T. 2009. Visualization of fluorescence-tagged proteins in fission yeast: the analysis of mitotic spindle dynamics using GFP-tubulin under the native promoter. Methods Mol. Biol. 545, 185–203. (doi:10.1007/978-1-60327-993-2_11) [DOI] [PubMed] [Google Scholar]
- 35.Okada N, Toda T, Yamamoto M, Sato M. In press. CDK-dependent phosphorylation of Alp7-Alp14 (TACC-TOG) promotes its nuclear accumulation and spindle microtubule assembly. Mol. Biol. Cell. (doi:10.1091/mbc.E13-11-0679) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krapp A, Simanis V. 2008. An overview of the fission yeast septation initiation network (SIN). Biochem. Soc. Trans. 36, 411–415. (doi:10.1042/BST0360411) [DOI] [PubMed] [Google Scholar]
- 37.MacNeill SA, Creanor J, Nurse P. 1991. Isolation, characterisation and molecular cloning of new mutant alleles of the fission yeast p34_cdc2_+ protein kinase gene: identification of temperature-sensitive G2-arresting alleles. Mol. Gen. Genet. 229, 109–118. (doi:10.1007/BF00264219) [DOI] [PubMed] [Google Scholar]
- 38.MacNeill SA, Nurse P. 1993. Genetic analysis of human p34CDC2 function in fission yeast. Mol. Gen. Genet. 240, 315–322. (doi:10.1007/BF00280381) [DOI] [PubMed] [Google Scholar]
- 39.Nurse P, Bissett Y. 1981. Gene required in G1 for commitment to cell cycle and in G2 for control of mitosis in fission yeast. Nature 292, 558–560. (doi:10.1038/292558a0) [DOI] [PubMed] [Google Scholar]
- 40.Grallert B, Sipiczki M. 1990. Dissociation of meiotic and mitotic roles of the fission yeast cdc2 gene. Mol. Gen. Genet. 222, 473–475. (doi:10.1007/BF00633860) [DOI] [PubMed] [Google Scholar]
- 41.Nakaseko Y, Niwa O, Yanagida M. 1984. A meiotic mutant of the fission yeast Schizosaccharomyces pombe that produces mature asci containing two diploid spores. J. Bacteriol. 157, 334–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell P, Nurse P. 1986. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell 45, 145–153. (doi:10.1016/0092-8674(86)90546-5) [DOI] [PubMed] [Google Scholar]
- 43.Radcliffe P, Hirata D, Childs D, Vardy L, Toda T. 1998. Identification of novel temperature-sensitive lethal alleles in essential β-tubulin and nonessential α2-tubulin genes as fission yeast polarity mutants. Mol. Biol. Cell 9, 1757–1771. (doi:10.1091/mbc.9.7.1757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toda T, Umesono K, Hirata A, Yanagida M. 1983. Cold-sensitive nuclear division arrest mutants of the fission yeast Schizosaccharomyces pombe. J. Mol. Biol. 168, 251–270. (doi:10.1016/S0022-2836(83)80017-5) [DOI] [PubMed] [Google Scholar]
- 45.Umesono K, Toda T, Hayashi S, Yanagida M. 1983. Cell division cycle genes NDA2 and NDA3 of the fission yeast Schizosaccharomyces pombe control microtubular organization and sensitivity to anti-mitotic benzimidazole compounds. J. Mol. Biol. 168, 271–284. (doi:10.1016/S0022-2836(83)80018-7) [DOI] [PubMed] [Google Scholar]
- 46.Izawa D, Goto M, Yamashita A, Yamano H, Yamamoto M. 2005. Fission yeast Mes1p ensures the onset of meiosis II by blocking degradation of cyclin Cdc13p. Nature 434, 529–533. (doi:10.1038/nature03406) [DOI] [PubMed] [Google Scholar]
- 47.Aoi Y, Arai K, Miyamoto M, Katsuta Y, Yamashita A, Sato M, Yamamoto M. 2013. Cuf2 boosts the transcription of APC/C activator Fzr1 to terminate the meiotic division cycle. EMBO Rep. 14, 553–560. (doi:10.1038/embor.2013.52) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kimura M, Kamakura T, Tao QZ, Kaneko I, Yamaguchi I. 1994. Cloning of the blasticidin S deaminase gene (BSD) from Aspergillus terreus and its use as a selectable marker for Schizosaccharomyces pombe and Pyricularia oryzae. Mol. Gen. Genet. 242, 121–129. (doi:10.1007/BF00391004) [DOI] [PubMed] [Google Scholar]
- 49.Brizuela L, Draetta G, Beach D. 1987. p13suc1 acts in the fission yeast cell division cycle as a component of the p34cdc2 protein kinase. EMBO J. 6, 3507–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371. (doi:10.1038/nprot.2009.2) [DOI] [PubMed] [Google Scholar]
- 51.De Bondt HL, Rosenblatt J, Jancarik J, Jones HD, Morgan DO, Kim SH. 1993. Crystal structure of cyclin-dependent kinase 2. Nature 363, 595–602. (doi:10.1038/363595a0) [DOI] [PubMed] [Google Scholar]
- 52.Zhang C, Kenski DM, Paulson JL, Bonshtien A, Sessa G, Cross JV, Templeton DJ, Shokat KM. 2005. A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat. Methods 2, 435–441. (doi:10.1038/nmeth764) [DOI] [PubMed] [Google Scholar]
- 53.D'Angiolella V, Mari C, Nocera D, Rametti L, Grieco D. 2003. The spindle checkpoint requires cyclin-dependent kinase activity. Genes Dev. 17, 2520–2525. (doi:10.1101/gad.267603) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mirchenko L, Uhlmann F. 2010. Sli15(INCENP) dephosphorylation prevents mitotic checkpoint reengagement due to loss of tension at anaphase onset. Curr. Biol. 20, 1396–1401. (doi:10.1016/j.cub.2010.06.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oliveira RA, Hamilton RS, Pauli A, Davis I, Nasmyth K. 2010. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat. Cell Biol. 12, 185–192. (doi:10.1038/ncb2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parry DH, Hickson GR, O'Farrell PH. 2003. Cyclin B destruction triggers changes in kinetochore behavior essential for successful anaphase. Curr. Biol. 13, 647–653. (doi:10.1016/S0960-9822(03)00242-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamaguchi S, Decottignies A, Nurse P. 2003. Function of Cdc2p-dependent Bub1p phosphorylation and Bub1p kinase activity in the mitotic and meiotic spindle checkpoint. EMBO J. 22, 1075–1087. (doi:10.1093/emboj/cdg100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morin V, Prieto S, Melines S, Hem S, Rossignol M, Lorca T, Espeut J, Morin N, Abrieu A. 2012. CDK-dependent potentiation of MPS1 kinase activity is essential to the mitotic checkpoint. Curr. Biol. 22, 289–295. (doi:10.1016/j.cub.2011.12.048) [DOI] [PubMed] [Google Scholar]
- 59.Vázquez-Novelle MD, Sansregret L, Dick AE, Smith CA, McAinsh AD, Gerlich DW, Petronczki M. 2014. Cdk1 inactivation terminates mitotic checkpoint surveillance and stabilizes kinetochore attachments in anaphase. Curr. Biol. 24, 638–645. (doi:10.1016/j.cub.2014.01.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pinsky BA, Nelson CR, Biggins S. 2009. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr. Biol. 19, 1182–1187. (doi:10.1016/j.cub.2009.06.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanoosthuyse V, Hardwick KG. 2009. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr. Biol. 19, 1176–1181. (doi:10.1016/j.cub.2009.05.060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.London N, Ceto S, Ranish JA, Biggins S. 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22, 900–906. (doi:10.1016/j.cub.2012.03.052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shepperd LA, Meadows JC, Sochaj AM, Lancaster TC, Zou J, Buttrick GJ, Rappsilber J, Hardwick KG, Millar JB. 2012. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22, 891–899. (doi:10.1016/j.cub.2012.03.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ohkura H, Kinoshita N, Miyatani S, Toda T, Yanagida M. 1989. The fission yeast dis2+ gene required for chromosome disjoining encodes one of two putative type 1 protein phosphatases. Cell 57, 997–1007. (doi:10.1016/0092-8674(89)90338-3) [DOI] [PubMed] [Google Scholar]
- 65.Kinoshita N, Ohkura H, Yanagida M. 1990. Distinct, essential roles of type 1 and 2A protein phosphatases in the control of the fission yeast cell division cycle. Cell 63, 405–415. (doi:10.1016/0092-8674(90)90173-C) [DOI] [PubMed] [Google Scholar]
- 66.Vázquez-Novelle MD, Petronczki M. 2010. Relocation of the chromosomal passenger complex prevents mitotic checkpoint engagement at anaphase. Curr. Biol. 20, 1402–1407. (doi:10.1016/j.cub.2010.06.036) [DOI] [PubMed] [Google Scholar]
- 67.Sato M, Okada N, Kakui Y, Yamamoto M, Yoshida M, Toda T. 2009. Nucleocytoplasmic transport of Alp7/TACC organizes spatiotemporal microtubule formation in fission yeast. EMBO Rep. 10, 1161–1167. (doi:10.1038/embor.2009.158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohta M, Sato M, Yamamoto M. 2012. Spindle pole body components are reorganized during fission yeast meiosis. Mol. Biol. Cell 23, 1799–1811. (doi:10.1091/mbc.E11-11-0951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kitajima TS, Kawashima SA, Watanabe Y. 2004. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427, 510–517. (doi:10.1038/nature02312) [DOI] [PubMed] [Google Scholar]
- 70.Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. 2006. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441, 46–52. (doi:10.1038/nature04663) [DOI] [PubMed] [Google Scholar]
- 71.Kawashima SA, Tsukahara T, Langegger M, Hauf S, Kitajima TS, Watanabe Y. 2007. Shugoshin enables tension-generating attachment of kinetochores by loading aurora to centromeres. Genes Dev. 21, 420–435. (doi:10.1101/gad.1497307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823. (doi:10.1016/0076-6879(91)94059-L) [DOI] [PubMed] [Google Scholar]
- 73.Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951. (doi:10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y) [DOI] [PubMed] [Google Scholar]
- 74.Sato M, Dhut S, Toda T. 2005. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast 22, 583–591. (doi:10.1002/yea.1233) [DOI] [PubMed] [Google Scholar]
- 75.Tada K, Susumu H, Sakuno T, Watanabe Y. 2011. Condensin association with histone H2A shapes mitotic chromosomes. Nature 474, 477–483. (doi:10.1038/nature10179) [DOI] [PubMed] [Google Scholar]
- 76.Akera T, Sato M, Yamamoto M. 2012. Interpolar microtubules are dispensable in fission yeast meiosis II. Nat. Commun. 3, 695 (doi:10.1038/ncomms1725) [DOI] [PubMed] [Google Scholar]
- 77.Kawashima SA, Takemoto A, Nurse P, Kapoor TM. 2012. Analyzing fission yeast multidrug resistance mechanisms to develop a genetically tractable model system for chemical biology. Chem. Biol. 19, 893–901. (doi:10.1016/j.chembiol.2012.06.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Adobe PDF - rsob-14-0063-File008.pdf