Axon Regeneration in C. elegans (original) (raw)
. Author manuscript; available in PMC: 2015 Aug 1.
Published in final edited form as: Curr Opin Neurobiol. 2014 May 4;0:199–207. doi: 10.1016/j.conb.2014.04.001
Abstract
Single axon transection by laser surgery has made C. elegans a new model for axon regeneration. Multiple conserved molecular signaling modules have been discovered through powerful genetic screening. in vivo imaging with single cell and axon resolution has revealed unprecedented cellular dynamics in regenerating axons. Information from C. elegans has greatly expanded our knowledge of the molecular and cellular mechanisms of axon regeneration.
Introduction
To maintain nervous system function throughout the lifetime of an animal, animals have evolved the ability to repair damaged axons by regeneration. Axon regeneration requires the neuron to recognize that it has been damaged, to initiate regrowth (usually by forming a growth cone), to sustain long-distance extension, to navigate to an appropriate target, and to form a functional connection by building a synapse. Recent work in multiple species—including mouse, zebrafish, Drosophila, and _C. elegans—_has identified common biological pathways that utilize conserved molecules to regulate regeneration. One striking finding is that in general the intrinsic mechanisms that function in neurons to mediate regeneration are conserved across species. Thus, axon regeneration, like axon guidance and synaptic transmission, is a fundamental and ancient property of neurons that predates the evolution of vertebrates.
The model organism C. elegans is a particularly useful species for discovering gene function and for studying the cellular mechanisms of axon regeneration. The transparent body and the simple nervous system of C. elegans make it possible to observe individual neurons in vivo throughout the life of the animal. The development of optical and genetic techniques to injure axons enables the study of regeneration without disruptive surgery. Powerful genetic and cell biological tools facilitate the in-depth analysis of regeneration mechanisms. In this review, we focus on how regeneration is studied in C. elegans. We summarize genetic screens in C. elegans that have identified a large number of new genes that display novel functions in axon regeneration. We discuss key findings from C. elegans research on the cell biology of axon regeneration, as studied by live imaging at single axon resolution. Finally, we offer our views on the promises of, and challenges for, C. elegans axon regeneration research.
Overview of axon regeneration in C. elegans
Since the demonstration that GABAergic motor neurons in adult C. elegans can regenerate after ultrafast laser microsurgery [1], multiple types of neurons have been shown to have the potential to regenerate [2–4]. Among these, the mechanosensory neurons (ALM, PLM, AVM) and the GABAergic motor neurons (DD, VD) are the most extensively used to address the mechanisms of regeneration, and we discuss these neurons below. A common theme among all regenerating axons is that their response to injury is quite variable—that is, identical neurons do not always respond in the same way to similar injury, even under tightly controlled conditions. The source of this neuron-to-neuron variability is not completely clear, but one contributing factor is initial neuronal calcium conditions at the time of injury [5]. Another common observation is that axon regrowth in the mature nervous system is error-prone, unlike the precise growth that occurs during normal nervous system development [3,4].
Despite the variability of axon regeneration at the level of single neurons, the average regenerative potential of a given neuron is remarkably reproducible across multiple independent studies, labs, and investigators. Using this stable background to investigate factors that operate in different types of neurons to mediate axon regeneration has proven to be very powerful, enabling gene discovery as well as the analysis of other factors—such as age—that affect regeneration. These findings are discussed below.
Large-scale screening reveals the genetic landscape of axon regeneration
To date, two large-scale genetic screens for regeneration have been conducted in C. elegans. Together, these screens represent the most complete genetic analysis of regeneration in any organism, and have identified large numbers of new genes with clear functional roles in axon regeneration. Both screens used restricted lists of target genes rather than random mutagenesis, and target genes were chosen in order to focus on genes most likely to have conserved functions in neurons.
PLM mechanosensory neurons: screening by genetic mutations
The PLM (Posterior Lateral Microtubule) mechanosensory neurons are a pair of bilaterally symmetric cells whose cell bodies lie in the lumbar ganglion [6]. Each neuron extends a long anterior axon of average length 300 μm in young adults and a short posterior neurite that is presumed as dendrite. Along the anterior axon, a collateral branch forms and extends ventrally into the nerve cord to form synapses to other neurites. The function of PLM ensures sensory response to gentle touch for the posterior half of the worm [7].
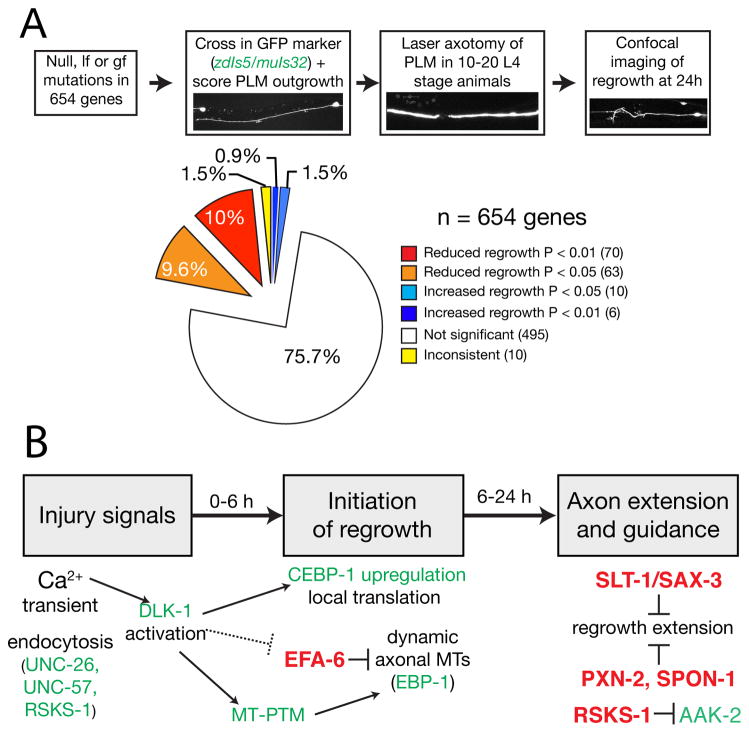
After laser-induced injury is delivered to the anterior axon >40–50um away from soma, the severed axon stumps exhibit morphological growth signs, such as filopodia or growth cones, within a few hours, and then extend with a misguided trajectory over a day or more [3]. A systematic screen using genetic null or strong loss of function mutations was carried out for > 650 conserved genes [8]. These genes fall into all functional categories, and the homozygous mutants generally do not exhibit discernable morphological defects in PLM axon development. PLM axon regeneration was assessed based on quantitative measurement of regrowth length, as well as growth-cone like structures and axon extension trajectory. This screen uncovered ~ 100 genes, loss of function in which resulted in significant reduced regrowth, and about ten genes, loss of function in which caused enhanced regrowth (Figure 1). Further investigation of select genes included genetic epistasis and temporal requirement, and led to the establishment of a regeneration pathway depicting three distinct phases in PLM axon regeneration: immediate response, regrowth initiation, regrowth extension and guidance (Figure 1, and see later).
Figure 1.
Overview of axon regeneration screen in PLM neurons (modified from [8]). A) Illustration of the screen (top panels) and summary of the outcome. B). Model of the regeneration pathway based on multiple lines of evidences described in the text.
GABA motor neurons: screening by RNAi
The GABA motor neurons are a set of 19 neurons with cell bodies distributed along the ventral side of the animal. Each GABA neuron extends a process in the ventral nerve cord that eventually defasciculates and extends a commissure to the dorsal nerve cord, where it again extends and fasciculates [6]. The GABA motor neurons receive excitatory inputs from the cholinergic motor neurons, and transmit inhibitory GABA signals onto body wall muscles on the opposite side of the animal. There are two types of GABA motor neurons: the DD neurons, which receive ventral inputs and inhibit dorsal muscles, and the VD neurons, which receive dorsal inputs and inhibit ventral muscles.
The GABA neurons were the first neurons analyzed by laser axotomy [1]. Axons are typically severed at the dorsal-ventral midline, and regenerating axons grow back toward and often reach the dorsal nerve cord. Multiple independent studies demonstrate that after laser axotomy in L4-stage animals, approximately 70% of injured axons can initiate regrowth [1,3,9–12]. Except for two neurons (DD4 and VD8) in which regeneration is impeded by vulval structures, each neuron has a similar chance of regenerating, and the average regeneration rate is 2–3 um/h [3]. This growth rate is comparable to the growth rate reported during initial development of the GABAergic VD neurons [45], and also to that reported for mouse CNS axon sprouting following a pre-conditioning injury [46]. If nearly all (~15/19) GABA motor neurons are severed, animals exhibit a ‘shrinker’ movement phenotype--a specific and characteristic behavioral deficit associated with loss of GABA function [13]. 24 hours later (when many severed axons have regenerated to the dorsal cord) animals largely recover wild type movement, and this recovery does not occur when regeneration is reduced [1,10]. It remains to be determined whether there is a precise correlation between movement improvement and synaptic activity. Nonetheless, these results demonstrate that regenerated GABA neurons can rewire into proper circuits to mediate normal behavior.
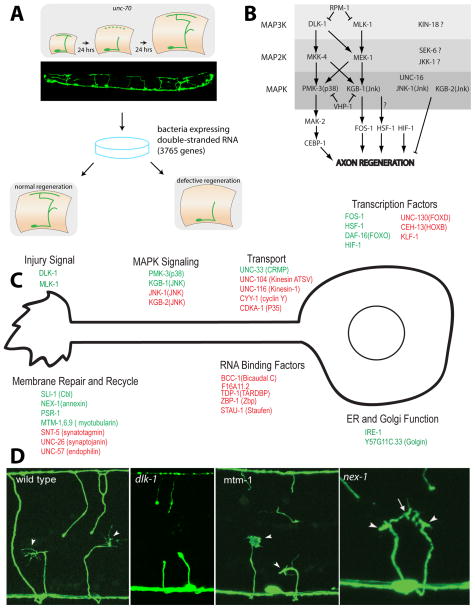
Regeneration of the GABA neurons can also be initiated without laser surgery, by use of the unc-70 mutation. unc-70 encodes β-spectrin, and GABA neurons in animals that lack β-spectrin break spontaneously and regenerate [14]. Breakage is due to mechanical weakness and is likely independent of the functional properties of the neuron. This phenotype was exploited for an RNAi screen for genes that altered regeneration [15]. Gene targets were selected with clear human orthologs, and 3765 independent experiments were done in a genetic background that contained the unc-70 mutation (to initiate breakage and regeneration) and the eri-1; lin-15b mutations (to sensitize neurons to RNAi). 70 genes showed a strong regeneration phenotype in this background. Each of these genes was then tested by axotomy. In the end, the screen identified 15 genes that are required for normal regeneration, and 16 genes that inhibit regeneration. One group of genes identified by this screen controls growth cone behavior during regeneration, including microvesicle shedding, growth cone morphology, and filopodia. A second group of genes includes multiple MAP kinase pathway genes that control growth cone initiation (Figure 2).
Figure 2.
Overview of axon regeneration screen in GABA motor neurons [15]. A) Overview of the screen. B) MAP kinase networks that regulate regeneration. C) Summary of genes that affect axon regeneration. D) Growth cone phenotypes in regenerating axons. A adapted from [14]; B and C adapted from [15], D adapted from [9,15].
Summary
Although the two screens were done in different neurons using different injury paradigms, they identified a substantial number of overlapping genes [8,15]. This finding suggests that the mechanism of regeneration is broadly similar in different cell types. However, there are also many genes that are unique to each screen, and other studies have shown that even for the same gene, different neurons exhibit different dependencies for regeneration. For example, loss of dlk-1 function blocks regeneration in PLM and GABAergic motor neurons by > 90%, but only reduces ALM axon regeneration by less than 50% [9,16,17]. One possible explanation for these neuron-specific differences is that multiple partially-redundant processes can mediate regeneration, but only a subset of these processes are present in a given neuron type. Similar observations are known for vertebrate axon regeneration; for example, removing PTEN greatly enhances optic and peripheral nerve regeneration, but has a modest effect on spinal cord axons [18–20]. These findings underscore the important notion that the study of diverse neuronal types is necessary for generating a deeper understanding of regeneration.
Novel pathways discovered from C. elegans
Building on information from the genetic screens as well as candidate gene approaches, recent studies have made rapid progress towards in-depth understanding of the molecular and cellular mechanisms that mediate axon regeneration. One major thread has been the analysis of the conserved DLK-1 MAP kinase pathway and other MAP kinase pathways in regeneration [9,11,12,15,16,21,22]. Additional studies have also elucidated mechanisms for several new regeneration pathways, including Notch signaling [10], microtubule regulators [8,23,24], insulin signaling [12], and caspase [17]. Further, careful analysis has yielded novel insights into known regeneration factors, including calcium [5], axon guidance signaling [4], and the extracellular matrix [25]. The specific effects and the signaling components of these pathways have been thoroughly reviewed recently [26–31]. Here, we refer readers to the general information summarized in Table 1. Suffice it to say that the findings originated from C. elegans have rapidly expanded the molecular landscape of axon regeneration.
Table 1.
Summary of major pathways in C. elegans axon regeneration
| Neuron/Axon type | Effects on regeneration | Signaling pathway | homologs | references | |
|---|---|---|---|---|---|
| dlk-1 | GABAergic motor neuron; Touch neurons ASJ | Lf: eliminate or reduceGof: enhance | PMK-3CEBP-1/bZip | DLKLZK | [9,16,22] |
| lin-12 | GABAergic motor neuron | Lf: enhanceGof: reduce | ADAM presenilin | Notch | [10] |
| ced-3 ced-4 | ALMTouch neuron | Lf: reduce | Caspase Apaf-4 | [17] | |
| pde-4 | PLM | Lf: enhance | cAMP | phosphodi esterase | [5] |
| efa-6 | ALM, PLM, Touch neuron | Lf: enhanceGof: reduce | Microtubule | Efa6(A–D) | [8] |
| mlk-1 | GABAergic motor neuron | Lf:Gof? | KGB-1JNKFos? | MLK | [11] |
| svh-1/svh-2 | GABAergic motor neurons | LfGof? | MLK-KGB/JNK? | HGF-MSP | [21] |
| rsks-1 | ALM, PLMTouch neurons | Lf: enhance | AAK-2 | S6 kinase/AM P kinase | [36] |
| daf-18 | GABAergic motor neuron | Lf: enhance | mTOR | PTEN | [12] |
| Tubulin & MT PTM | PLM Touch neuron | Lf: variable effects depending on the genes in the types of MT-PTM | Tubulin | Tubulin CCPP Kinesin-13 MCAK | [8,23,24] |
| Guidance signaling sax-3/slt: vab-1 unc-129 unc-40 | PLMAVM | Lf: extension and projection | Robo/slitEphTGF-bDCC | [8][4] | |
| miRNA_alg-1_ | AVM | alg-1(lf): enhance_let-7_(lf): enhance | lin-41 | Argonaut Let7 | [38] |
| ECM_: pxn-2_ | PLM | Lf: enhance | [25] | ||
| daf-2 | GABAergic motor neurons | Lf: enhance in aged animals | FOXO/daf-16 | insulin/IG FR | [12] |
Advent of in vivo imaging of regenerating axon uncovers cellular dynamics
The transparent body of C. elegans enables the in vivo, non-invasive study of cells and cellular components labeled with fluorescent proteins. The invention of genetically encoded cellular activity sensors (such as calcium sensors) has further aided the investigation of cellular dynamics at single-cell, single-axon resolution with temporal and spatial precision. Recently, studies that combine these sensors with time-lapse imaging during regeneration have begun to reveal the complex subcellular dynamics that accompany regeneration, and to identify molecular mechanisms that regulate them.
Calcium transients immediately after injury promote regeneration
PLM axon severing by laser is accompanied by an immediate calcium transient at the injury site (detected by GCaMP) that propagates bidirectionally. The amplitude of the initial transient is positively correlated to the extent of axon regrowth, and depends on the voltage-gated calcium channel EGL-19 as well as internal calcium stores and the IP3 receptor [5]. Similarly, ALM axon severing by laser initiates an immediate increase of cellular calcium levels in the soma (detected by cameleon-based FRET) that depends on the ER calcium-binding chaperone calreticulin, CRT-1 [17]. The axonal calcium transients induced by laser injury are reminiscent of those described in Aplysia axons severed mechanically [32] as well as a recent finding in mouse DRG neurons [33]. This calcium transient promotes axon regeneration likely through multiple pathways, including activation of the DLK-1 kinase [22], and caspase CED-3 and its activator CED-4 [17].
Membrane and cytoskeleton dynamics during regeneration
Similar to mechanical severing of axons in larger animals, laser microsurgery of C. elegans axons causes traumatic damage to axons, including cytoskeleton disruption, membrane breakage and resealing. Initiation of regrowth from injured axon stumps generally has been assessed as morphological changes visualized by fluorescent protein-labeled axons. Such changes include rapid filopodia protrusions and formation of growth-cone like structures [2,3,9,34,35]. To some extent, it has been possible to mechanistically separate these processes—for example, transient filopodia after GABA neuron injury do not require the DLK-1 signaling pathway, while subsequent growth cone formation does [9]. Interestingly, in the ALM neuron regeneration-like morphological changes are also observed after axotomy in the distal axon fragment, which is no longer attached to the cell body [17]. These experiments suggest that the initial steps of regeneration do not require the nucleus.
A thorough analysis of microtubule (MT) dynamics was recently reported using the MT plus end binding protein EBP-GFP reporter [23]. In mature and uninjured PLM axons, MTs are relatively stable, and axons contain comparatively few comets that grow for short periods. An early response to injury is local upregulation of growing MTs close to the severed axon stump. Three hours after axotomy the number of EBP-GFP tracks increases near the cut site, although the track length and duration remain similar to uninjured axons. Within the next three hours, EBP-GFP tracks decrease in catastrophe frequency and double in length, indicating an increase in persistent MT growth. The transition to more persistent MT growth correlates with reformation of a growth cone and the beginning of axon extension. Injury-induced MT dynamics are in part regulated by a set of genes known to affect MT post-translational modification, such as the MT depolymerizing kinesin and carboxyl peptidases, and are also under the control of the DLK-1 MAP kinase cascade [23]. In addition, the conserved EFA-6 protein acts as an intrinsic inhibitor of MT dynamics (Table 1) [8]. Overexpression of efa-6 inhibits PLM axon regrowth, which can be partially ameliorated by exposure to exogenous taxol. These studies show remarkable parallels to those undertaken in the field of mammalian spinal cord injury [47, 48].
Axon extension, synapse formation, and function recovery
The error-prone regrowth of injured axons during extension is likely due to multiple causes, such as loss of intrinsic ability to detect guidance cues, absence or insufficient guidance information, as well as inhibitory factors in the environment. Several studies have addressed the roles of classical developing axon guidance pathways, including EphR/ephrin, Slit/Robo, Netrin/DCC, and TGF-b [3,4,8]. These data indicate that guidance pathways remain active in the mature nervous system and can influence regrowing axons. Despite the apparent lack of myelin-producing glia and microglia in C. elegans, several extracellular matrix proteins, such as pxn-2 and spondin, are inhibitory to regeneration [8,25].
The rate of regrowth affects the eventual outcome of regeneration, and likely relies on synergistic and coordinated activities of multiple pathways. One important pathway functions downstream of the growth inhibitor PTEN, which inhibits regeneration in vertebrates [18–20]. In C. elegans, loss of PTEN improves regeneration, and this increase is mediated by TOR [12]. A complementary study shows that the S6 kinase homolog, RSKS-1, acts as an intrinsic regeneration inhibitor in part through controlling the AMP kinase, AAK-2, and has a prominent influence on regrowth extension [36]. These data identify a conserved pathway for regulating regeneration, and suggest that modulating the intrinsic growth potential of injured neurons can sustain vitality and increase regeneration.
Increasing cAMP promotes growing regrowth of axons towards targets, delivery of synaptic cargos and formation of synapses that have rudimentary morphological organization [5]. As mentioned earlier, functional improvement of movement after regrowth of motor neurons is also observed [1,10]. In addition to conventional axon regeneration, the mechanosensory neurons can regenerate via fusion, a phenomenon also observed in leech and crayfish. Both ALM and PLM exhibit axonal fusion between regrowing axons and severed distal axon segments, as demonstrated by ultrastructural analyses as well as using a photoactivable fluorescent reporter [5,37]. Such fusion appears to be in part dependent on fusogens, such as EFF-1 [5]. Increasing cAMP in the cell can increase the incidence of axonal fusion [5], possibly leading to preservation of some function. By contrast, functional recovery by conventional regeneration via precise synapse formation is less robust than observed for fusion, and genetic mechanisms that might permit full functional recovery by conventional regeneration have not yet been identified. The failure of full functional recovery by injured C. elegans axons despite the lack of glial scar (which is a major obstacle to functional recovery in vertebrate axon regeneration injury models) raises interesting questions for future research.
Age-dependency of axon regeneration
The short life cycle and tractable genetics of C. elegans have made it a classic model for developmental and aging studies. Recently, these same advantages have been applied to studying the effect of age on axon regeneration. These studies have begun to identify age-dependent mechanisms that regulate regenerative potential.
Regeneration of the ALM and AVM mechanosensory axons exhibits increased guidance errors and decreased overall growth as animals age to and beyond young adulthood [3,4]. In AVM, regeneration declines during larval development to relatively stable levels maintained during adulthood. This larval decline is regulated by a regulatory loop consisting of the microRNA let-7 and its target gene lin-41 [38]. During the initial development of AVM in early larvae, lin-41 inhibits let-7 expression to allow growth. Shortly after AVM outgrowth is complete, when lin-41 levels are still high, regeneration can be observed at high levels. As animals age into later larval stages, increased let-7 expression reduces lin-41 expression, and this in turns reduces regeneration. Thus, the let-7/lin-41 pathway regulates a switch from the high growth potential of active neuronal development to the lower growth potential of the mature neuron.
By contrast, the GABA motor neurons (which develop around the same time as ALM and AVM) retain most of their regenerative ability into adulthood, but regeneration declines steeply during adult life [9,12]. The insulin/IGF receptor daf-2 regulates this decline in regeneration by inhibiting the FOXO homolog daf-16. While the daf-2/daf-16 pathway also regulates lifespan, its effect on axon regeneration can be decoupled from lifespan, and acts specifically in neurons to regulate regeneration. One output of daf-2/daf-16 signaling is transcriptional regulation of genes in the DLK MAP kinase pathway, including dlk-1 itself [12].
Together, these studies demonstrate that C. elegans animals regulate the regenerative potential of neurons in response to age. The axon regeneration process is autonomous to the nervous system and contributes to healthspan, rather than lifespan. However, much work remains to be done to understand the effect of age on neuronal regeneration. Neither disrupting let-7 nor daf-2 can entirely eliminate age-related decline of regeneration, suggesting that other pathways are also functioning to limit the regenerative potential of neurons as animals age.
Outlook: Promises and challenges
Within less than ten years, research on axon regeneration using the C. elegans model has greatly expanded the molecular and genetic landscape, and provided deep mechanistic insights into cellular dynamics triggered by axonal injury. It is worth noting that the discovery of DLK-1 kinase as a hub for axon regeneration in C. elegans has spurred the efforts to unravel the multifaceted roles and downstream signaling pathways of the vertebrate DLK kinase in both axon regeneration and degeneration in different neurons [39], underscoring the relevance of the axon regeneration investigation from C. elegans to translational research in higher mammals.
Looking to the future, we envision that more sophisticated genetic screening in C. elegans, in conjunction with chemical screening and finer tools for in vivo observation and perturbation of cellular function, will offer even broader perspectives. Future screens could analyze regeneration in different neurons, could use different techniques for gene ablation, and could analyze parameters of regeneration other than axon morphology. The application of microfluidics to speed animal handling, and the automation of imaging and even of laser surgery, can increase screening throughput [40–42] making it suitable for drug discovery [43,44]. Finally, a theme emerging from genetic studies so far is the identification of molecules and signaling pathways (e.g. MAP kinases, Notch, microRNAs, FOXO) that can coordinately regulate large numbers of genes. A challenge will be to describe the functional outputs of this genetic regulation, either by identifying specific relevant targets or by adopting a systems biology-oriented approach. Together, such genetic approaches will help to clarify the complete genetic requirements for axon regeneration and could identify additional targets for therapies aimed at improving regeneration.
The ability in the worm to study the cell biology of regeneration in vivo and with single neuron resolution has aided precise description of calcium and microtubule dynamics and growth cone morphology during regeneration. But many cell-biological questions remain. For example, the actin cytoskeleton has not been studied in detail. Identification of genes that affect growth cone morphology during regeneration may provide a genetic entry point into additional factors that directly drive growth. Further, for regeneration to matter, morphological reconstruction is not sufficient: regenerated neurons must form functional synapses onto relevant targets and rebuild damaged circuits. Powerful tools exist in C. elegans to study the cell biology of synaptogenesis, and even to analyze circuit function at the level of individual neurons. The application of these tools to the study of axon regeneration should yield information about the cellular mechanisms that govern functional regeneration.
Highlights.
- genetic screening reveals relevant molecules and expands molecular landscape of axon regeneration
- in-depth mechanistic dissection uncovers signaling pathways
- in vivo imaging analyzes subcellular processes in regenerating axons
- functional conservation underscores the impact of the C. elegans model for axon regeneration
Acknowledgments
Work in M.H.’s lab is supported by grants from the NIH and the Ellison Medical Foundation. Work in Y.J.’s lab is supported by Howard Hughes Medical Institute and a grant from NIH. We are grateful for our lab members for their dedication, and our colleagues for valuable discussions and their contributions, to the axon regeneration research in C. elegans.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References (annotations follow)
** Of outstanding interest:
* Of interest:
- 1.Yanik MF, Cinar H, Cinar HN, Chisholm AD, Jin Y, Ben-Yakar A. Neurosurgery: functional regeneration after laser axotomy. Nature. 2004;432:822. doi: 10.1038/432822a. [DOI] [PubMed] [Google Scholar]
- 2.Yanik MF, Cinar H, Cinar HN, Gibby A, Chisholm AD, Jin Y, Ben-Yakar A. Nerve Regeneration in Caenorhabditis elegans After Femtosecond Laser Axotomy. IEEE J Select Topics Quantum Electron. 2006;12:1283–1291. [Google Scholar]
- 3.Wu Z, Ghosh-Roy A, Yanik MF, Zhang JZ, Jin Y, Chisholm AD. Caenorhabditis elegans neuronal regeneration is influenced by life stage, ephrin signaling, and synaptic branching. 2007:15132–15137. doi: 10.1073/pnas.0707001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabel CV, Antoine F, Chuang C-F, Samuel ADT, Chang C. Distinct cellular and molecular mechanisms mediate initial axon development and adult-stage axon regeneration in C. elegans. Development. 2008;135:1129–1136. doi: 10.1242/dev.013995. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh-Roy A, Wu Z, Goncharov A. Calcium and cyclic AMP promote axonal regeneration in Caenorhabditis elegans and require DLK-1 kinase. The Journal of …. 2010 doi: 10.1523/JNEUROSCI.5464-09.2010. no volume. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White JG, Southgate E, Thomson JN, Brenner S, White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond, B, Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 7.Chalfie M, SULSTON J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol. 1981;82:358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- 8**.Chen L, Wang Z, Ghosh-Roy A, Hubert T, Yan D, O’Rourke S, Bowerman B, Wu Z, Jin Y, Chisholm AD. Axon Regeneration Pathways Identified by Systematic Genetic Screening in C. elegans. Neuron. 2011;71:1043–1057. doi: 10.1016/j.neuron.2011.07.009. This paper reports first functional screening of 654 genes using genetic null or strong loss of function mutations. It reveals several functional clusters that act at distinct phases of PLM axon regeneration, such as synaptic vesicle endocytosis genes at early phase of regrowth and Slit/Robo axon guidance signaling at regrowth extension. Furthermore, the studies uncover an inhibitory function of the conserved gene EFA-6 in axon regeneration via regulation of the microtubule cytoskeleton. Together, their data establish a genetic framework for PLM axon regeneration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammarlund M, Nix P, Hauth L, Jorgensen EM, Bastiani M. Axon regeneration requires a conserved MAP kinase pathway. Science. 2009;323:802–806. doi: 10.1126/science.1165527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10**.Bejjani El R, Hammarlund M. Notch signaling inhibits axon regeneration. Neuron. 2012;73:268–278. doi: 10.1016/j.neuron.2011.11.017. This paper reports a novel role for Notch signaling in limiting axon regeneration. Notch signaling acts in injured GABA neurons, at the time of injury, to suppress growth cone formation. Notch signials via its canonical mechanism involving ADAM and gamma-secretase cleavage, and the Notch intracellular domain (NICD) alone can function in the nucleus to inhibit regeneration. Blocking Notch signaling after injury is sufficient to increase regeneration, and Notch signaling regulates both morphological and functional regeneration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nix P, Hisamoto N, Matsumoto K, Bastiani M. Axon regeneration requires coordinate activation of p38 and JNK MAPK pathways. Proc Natl Acad Sci USA. 2011;108:10738–10743. doi: 10.1073/pnas.1104830108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12**.Byrne A, Walradt T, Gardner KE, Hubbert A, Reinke V, Hammarlund M. Insulin/IGF1 Signaling Inhibits Age-Dependent Axon Regeneration. Neuron. 2014 doi: 10.1016/j.neuron.2013.11.019. [no volume]. This paper characterizes adult-age-related decline in axon regeneration. C. elegans GABA motor neurons lose the ability to regenerate with advanced adult age. The study shows that loss of regenerative ability in aged animals is regulated by the insulin signaling pathway (DAF-2/INSR and DAF-16/FOXO). The insulin pathway regulates regeneration via the DLK-1 pathway and independently of PTEN and TOR signaling. Using tissue-specific mosaics, the authors found that insulin signaling regulates regeneration cell autonomously and independently of its role in lifespan determination. Together, these findings demonstrate that loss of regenerative ability with increased adult age is a genetically regulated process and is not a secondary consequence of a decrepit animal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McIntire SL, Jorgensen E, Horvitz HR. Genes required for GABA function in Caenorhabditis elegans. Nature. 1993;364:334–337. doi: 10.1038/364334a0. [DOI] [PubMed] [Google Scholar]
- 14.Hammarlund M, Jorgensen EM, Bastiani MJ. Axons break in animals lacking beta-spectrin. The Journal of Cell Biology. 2007;176:269–275. doi: 10.1083/jcb.200611117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15**.Nix P, Hammarlund M, Hauth L, Lachnit M, Jorgensen EM, Bastiani MJ. Axon regeneration genes identified by RNAi screening in C. elegans. Journal of Neuroscience. 2014 doi: 10.1523/JNEUROSCI.3859-13.2014. [no volume]. This paper reports on a large RNAi screen for axon regeneration using the unc-70 genetic background to trigger axon injury in the GABA motor neurons. Laser axotomy was used to confirm phenotypes and test additional candidates. The screen identified 31 novel regeneration genes, which define a large number of molecular pathways that mediate regeneration. One group of genes affects growth cone morphology during regeneration, suggesting that multiple pathways act to support normal growth cone behavior in regenerating axons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan D, Wu Z, Chisholm AD, Jin Y. The DLK-1 Kinase Promotes mRNA Stability and Local Translation in C. elegans Synapses and Axon Regeneration. Cell. 2009;138:1005–1018. doi: 10.1016/j.cell.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Pinan-Lucarre B, Gabel CV, Reina CP, Hulme SE, Shevkoplyas SS, Slone RD, Xue J, Qiao Y, Weisberg S, Roodhouse K, et al. The Core Apoptotic Executioner Proteins CED-3 and CED-4 Promote Initiation of Neuronal Regeneration in Caenorhabditis elegans. PLoS Biol. 2012;10:e1001331. doi: 10.1371/journal.pbio.1001331. This study systematically examined ALM axon regeneration. They describe an injury triggered calcium transient that is dependent on the calcium internal store chaperone CRT-1/calreticulin. They show that CED-3 caspase and its activator CED-4/Apaf-1 are strongly required for ALM axon regeneration, likely acting upstream of DLK-1. Interestingly, other upstream regulators of classic apototic pathway are not required for ALM axon regeneation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christie KJ, Webber CA, Martinez JA, Singh B, Zochodne DW. PTEN inhibition to facilitate intrinsic regenerative outgrowth of adult peripheral axons. J Neurosci. 2010;30:9306–9315. doi: 10.1523/JNEUROSCI.6271-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu K, Lu Y, Lee JK, Samara R, Willenberg R, Sears-Kraxberger I, Tedeschi A, Park KK, Jin D, Cai B. PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat Neurosci. 2010;13:1075–1081. doi: 10.1038/nn.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Hisamoto N, Nix P, Kanao S, Mizuno T, Bastiani M, Matsumoto K. The growth factor SVH-1 regulates axon regeneration in C. elegans via the JNK MAPK cascade. Nat Neurosci. 2012 doi: 10.1038/nn.3052. [DOI] [PubMed] [Google Scholar]
- 22**.Yan D, Jin Y. Regulation of DLK-1 kinase activity by calcium-mediated dissociation from an inhibitory isoform. Neuron. 2012;76:534–548. doi: 10.1016/j.neuron.2012.08.043. Through systematic analysis of genetic mutations of dlk-1, this study reveals an auto-regulatory mechanism controlling DLK-1 kinase activation. They show that a short isoform of DLK-1 acts as an endogenous inhibitor to the active long isoform. The The heteromeric interaction can be modulated by calcium and also depends on a short protein motif in the active form of DLK-1. Remarkably, this motif is completely conserved in MAP3K13, the vertebrate homologs of DLK-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23**.Ghosh-Roy A, Goncharov A, Jin Y, Chisholm AD. Kinesin-13 and Tubulin Posttranslational Modifications Regulate Microtubule Growth in Axon Regeneration. Dev Cell. 2012 doi: 10.1016/j.devcel.2012.08.010. This study provides first in vivo analysis of microtubule (MT) dynamics triggered by axonal injury. Using EBP::GFP reporter, they observed two phases of local up-regulation of MT dynamics in the injured axon. The activity of the MT depolymerizing Kinesin-13, or KLP-7, is important for the increase of growing MTs in early phase, and the cytosolic carboxypeptidase is required for persistent MT growth. DLK-1 kinase cascade coordinates both phases of MT growth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Kirszenblat L, Neumann B, Coakley S, Hilliard MA. A dominant mutation in mec-7/β-tubulin affects axon development and regeneration in Caenorhabditis elegans neurons. Mol Biol Cell. 2013;24:285–296. doi: 10.1091/mbc.E12-06-0441. This paper describes a point mutation in mec-7/β-tubulin that causes defects in initiation of regeneration but not subsequent axon growth, suggesting that these two processes are mediated by different functions of microtubules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gotenstein JR, Swale RE, Fukuda T, Wu Z, Giurumescu CA, Goncharov A, Jin Y, Chisholm AD. The C. elegans peroxidasin PXN-2 is essential for embryonic morphogenesis and inhibits adult axon regeneration. Development. 2010 doi: 10.1242/dev.049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hilliard MA. Axonal degeneration and regeneration: a mechanistic tug-of-war. J Neurochem. 2009;108:23–32. doi: 10.1111/j.1471-4159.2008.05754.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Jin Y. Genetic dissection of axon regeneration. Curr Opin Neurobiol. 2010 doi: 10.1016/j.conb.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bejjani El R, Hammarlund M. Neural regeneration in Caenorhabditis elegans. Annu Rev Genet. 2012;46:499–513. doi: 10.1146/annurev-genet-110711-155550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh-Roy A, Chisholm AD. Caenorhabditis elegans: a new model organism for studies of axon regeneration. Dev Dyn. 2010;239:1460–1464. doi: 10.1002/dvdy.22253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Chisholm AD. Axon regeneration mechanisms: insights from C. elegans. Trends in Cell Biology. 2011;21:577–584. doi: 10.1016/j.tcb.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chisholm AD. Cytoskeletal dynamics in Caenorhabditis elegans axon regeneration. Annu Rev Cell Dev Biol. 2013;29:271–297. doi: 10.1146/annurev-cellbio-101512-122311. [DOI] [PubMed] [Google Scholar]
- 32.Ziv NE, Spira ME. Localized and transient elevations of intracellular Ca2+ induce the dedifferentiation of axonal segments into growth cones. J Neurosci. 1997;17:3568–3579. doi: 10.1523/JNEUROSCI.17-10-03568.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho Y, Cavalli V. HDAC5 is a novel injury-regulated tubulin deacetylase controlling axon regeneration. The EMBO Journal. 2012 doi: 10.1038/emboj.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bourgeois F, Ben-Yakar A. Femtosecond laser nanoaxotomy properties and their effect on axonal recovery in C. elegans. Opt Express. 2007;15:8521–8531. doi: 10.1364/oe.15.008521. [DOI] [PubMed] [Google Scholar]
- 35.Rao GN, Kulkarni SS, Koushika SP, Rau KR. In vivo nanosecond laser axotomy: cavitation dynamics and vesicle transport. Opt Express. 2008;16:9884–9894. doi: 10.1364/oe.16.009884. [DOI] [PubMed] [Google Scholar]
- 36*.Hubert T, Wu Z, Chisholm AD, Jin Y. S6 Kinase Inhibits Intrinsic Axon Regeneration Capacity via AMP Kinase in Caenorhabditis elegans. Journal of Neuroscience. 2014;34(3):758–63. doi: 10.1523/JNEUROSCI.2886-13.2014. This study reveals an unexpected inhibitory function of the ribosomal S6 kinase, RSKS-1, in mechanosensory axon regeneration. They show that the function of RSKS-1 is likely parallel to DLK-1, and involves the AMP kinase AAK-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neumann B, Nguyen KCQ, Hall DH, Ben-Yakar A, Hilliard MA. Axonal regeneration proceeds through specific axonal fusion in transected C. elegans neurons. Dev Dyn. 2011;240:1365–1372. doi: 10.1002/dvdy.22606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38**.Zou Y, Chiu H, Zinovyeva A, Ambros V, Chuang C-F, Chang C. Developmental decline in neuronal regeneration by the progressive change of two intrinsic timers. Science. 2013;340:372–376. doi: 10.1126/science.1231321. This study identified a dramatic effect of the microRNA let-7 on axon regeneration in the ALM neuron. let-7 is found to act in a regulatory loop with its target lin-41, and this loop also requires the argonaute homolog alg-1. The effect of this signaling loop is to mediate the developmental transformation from axons in young larvae with high growth potential to axons in older larvae and adults with lower growth potential. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tedeschi A, Bradke F. The DLK signalling pathway—a double-edged sword in neural development and regeneration. EMBO Rep. 2013;14:605–614. doi: 10.1038/embor.2013.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng F, Rohde CB, Yanik MF. Sub-cellular precision on-chip small-animal immobilization, multi-photon imaging and femtosecond-laser manipulation. Lab on a Chip. 2008;8:653–656. doi: 10.1039/b804808h. [DOI] [PubMed] [Google Scholar]
- 41.Chung K, Crane MM, Lu H. Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nat Meth. 2008;5:637–643. doi: 10.1038/nmeth.1227. [DOI] [PubMed] [Google Scholar]
- 42.Guo SX, Bourgeois F, Chokshi T, Durr NJ, Hilliard MA, Chronis N, Ben-Yakar A. Femtosecond laser nanoaxotomy lab-on-a-chip for in vivo nerve regeneration studies. Nat Meth. 2008;5:531–533. doi: 10.1038/nmeth.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ben-Yakar A, Bourgeois F. Ultrafast laser nanosurgery in microfluidics for genome-wide screenings. Curr Opin Biotechnol. 2009;20:100–105. doi: 10.1016/j.copbio.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samara C, Rohde CB, Gilleland CL, Norton S, Haggarty SJ, Yanik MF. Large-scale in vivo femtosecond laser neurosurgery screen reveals small-molecule enhancer of regeneration. Proc Natl Acad Sci USA. 2010;107:18342–18347. doi: 10.1073/pnas.1005372107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knobel KM, Davis WS, Jorgensen EM, Bastiani MJ. UNC-119 suppresses axon branching in C. elegans. Development. 2001;128:4079–4092. doi: 10.1242/dev.128.20.4079. [DOI] [PubMed] [Google Scholar]
- 46.Ylera B, Erturk A, Hellal F, Nadrigny F, Hurtado A, Tahirovic S, Oudega M, Kirchhoff F, Bradke F. Chronically CNS-injured adult sensory neurons gain regenerative competence upon a lesion of their peripheral axon. Curr Biol. 2009;19:930–936. doi: 10.1016/j.cub.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 47.Hellal F, Hurtado A, Ruschel J, Flynn K, Laskowski CJ, Umlauf M, et al. Microtubule stabilization reduces scarring and causes axon regeneration after spinal cord injury. Science. 331:928–931. doi: 10.1126/science.1201148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sengottuvel V, Leibinger M, Pfreimer M, Andreadaki A, Fischer D. Taxol facilitates axon regeneration in the mature CNS. J Neurosci. 2011;31:2688–2699. doi: 10.1523/JNEUROSCI.4885-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]