A PH domain in ACAP1 possesses key features of the BAR domain in promoting membrane curvature (original) (raw)
. Author manuscript; available in PMC: 2015 Oct 13.
SUMMARY
The BAR (Bin-Amphiphysin-Rvs) domain undergoes dimerization to produce a curved protein structure, which superimposes onto membrane through electrostatic interactions to sense and impart membrane curvature. In some cases, a BAR domain also possesses an amphipathic helix that inserts into the membrane to induce curvature. ACAP1 (Arfgap with Coil coil, Ankyrin repeat and PH domain protein 1) contains a BAR domain. Here, we show that this BAR domain can neither bind membrane nor impart curvature, but instead, requires a neighboring PH (Pleckstrin Homology) domain to achieve these functions. Specific residues within the PH domain are responsible for both membrane binding and curvature generation. The BAR domain adjacent to the PH domain instead interacts with the BAR domains of neighboring ACAP1 proteins to enable clustering at the membrane. Thus, we have uncovered the molecular basis for an unexpected and unconventional collaboration between PH and BAR domains in membrane bending.
Keywords: BAR domain, PH domain, membrane remodeling, crystal structure, electron microscopy, molecular simulation
INTRODUCTION
Membrane remodeling is critical for many aspects of cellular function, including the biogenesis of organelles and transport carriers, cell motility, and cytokinesis. One of the best characterized protein families that participate in membrane remodeling are those that contain the BAR domain. The BAR domain can be further sub-classified into four categories, F-BAR, BAR/N-BAR, I-BAR (inverse-BAR) and PinkBAR (Mim and Unger, 2012). F-BAR and BAR/N-BAR promote positive membrane curvatures through shallow or steep concave surfaces, respectively. I-BAR has a negative curvature and induces membrane extrusion. PinkBAR does not possess intrinsic curvature and acts on planar membrane surface (Pykäläinen et al., 2011).
Currently, two general mechanisms are known for how the BAR domains induce membrane curvature. One mechanism is based on scaffolding, which is initiated through dimerization that results in a banana-like structure. This structure imposes curvature on membrane through positively charged patches on the concave surface of BAR proteins that interact with the negatively charged surface of membrane through electrostatic interactions. Further cooperative membrane binding and curvature induction is achieved through tip-to-tip and lateral interactions among the dimeric BAR structures coating the membrane (Frost et al., 2008; Mizuno et al., 2010; Yu and Schulten, 2013). The other general mechanism involves membrane insertion, which typically involves an amphipathic helix, such as that seen for the N-BAR-containing endophilin (Gallop et al., 2006; Mim et al., 2012). Curiously, some BAR domains have been found to require an additional neighboring PH domain to bind membrane and induce curvature (Peter et al., 2004; Pylypenko et al., 2007), but why the PH domain is required has been unclear.
ACAP1 was initially characterized as a GTPase-activating protein (GAP) for ADP-ribosylation factor 6 (ARF6) (Jackson et al., 2000). Subsequently, it was found to act also as an ARF6 effector, which involves its role as a coat component to promote endocytic recycling (Dai et al., 2004; Li et al., 2007b). ACAP1 contains four domains, with the BAR and PH domains comprising its amino portion, and ArfGAP and ANK (ankryin repeat) domains comprising its carboxy portion. We recently provided a molecular explanation for how the tandem ArfGAP-ANK domains act in cargo binding by ACAP1 (Bai et al., 2012). Besides cargo binding, the other major function of coat components involves membrane bending to generate transport carriers. Here, we have sought to elucidate this role of ACAP1 through studies on its tandem BAR-PH domains.
RESULTS
ACAP1BAR-PH is sufficient for membrane binding and tubulation
ACAP1 has similar domain structures as multiple other members of the ArfGAP family (Figure S1A). Within this subfamily of ArfGAPs, ACAP2 (also known as centaurin β) is the most similar to ACAP1. ACAP2 has been characterized previously to require its neighboring PH domain to achieve membrane binding and curvature generation (Peter et al., 2004). Thus, we focused on the corresponding tandem BAR-PH domains in ACAP1 (referred hereon as ACAP1BAR-PH) for detailed analysis.
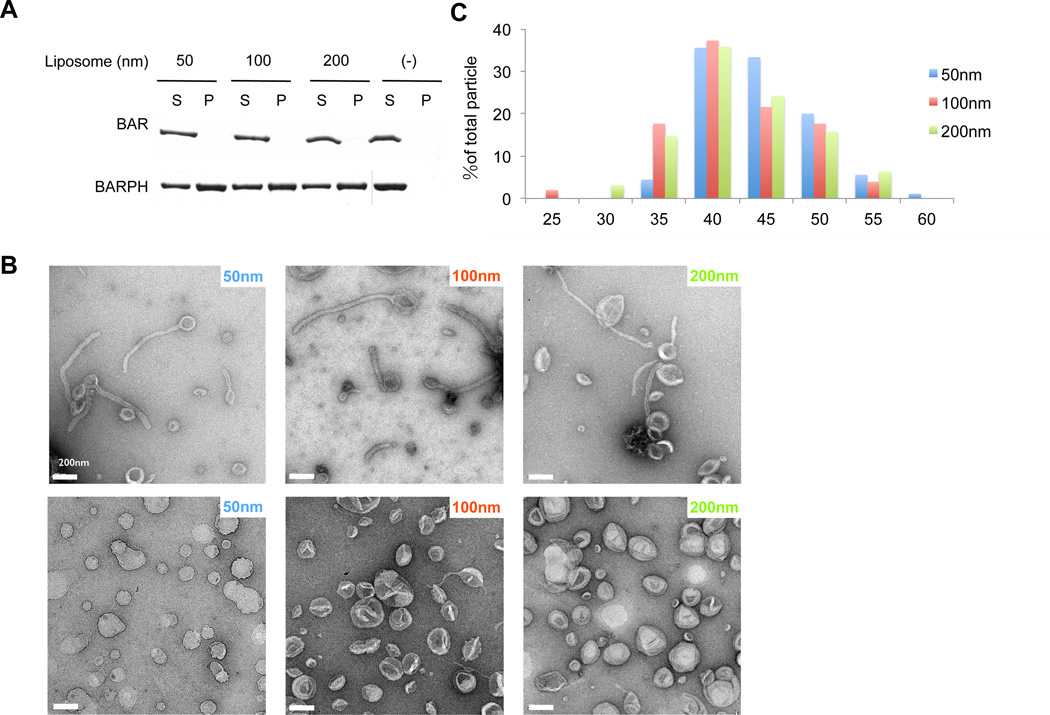
We initially generated liposomes that contained the major phospholipids of organellar membrane. To this mixture, we also added phosphatidylinositol 4,5-bisphosphate, PI(4,5)P2, as this phosphoinositide has been found previously to promote membrane tubulation by ACAP1 (Shinozaki-Narikawa et al., 2006). The generated liposomes were then incubated with ACAP1BAR-PH and analyzed for binding by centrifugation. Consistent with the previous finding on the BAR domain of ACAP2 (Peter et al., 2004), the BAR domain of ACAP1 also showed little affinity to the generated liposomes, regardless of their size (Figure 1A). In contrast, ACAP1BAR-PH bound to these liposomes, which was independent of their size that ranged from 50 nm to 200 nm in diameter (Figure 1A).
Figure 1. BAR-PH domain of ACAP1 is sufficient for membrane binding and tubulation.
(A) Binding of different forms of ACAP1 (BAR or BAR-PH) to liposomes of varying sizes (as indicated) is assessed by centrifugation; S, supernatant; P, pellet. (B) Negative-stain electron microscopy visualizing liposomes incubated either with (first row) or without (second row) ACAP1BAR-PH; bar, 200 nm. The diameters of the liposomes are also indicated. (C) Statistical histogram of the diameters of tubules generated by ACAP1BAR-PH, plotted as percentage of all tubules, n = 90 for 50 nm-liposomes, n = 51 for 100 nm-liposomes and n = 95 for 200 nm-liposomes.
[See also Figure S1.]
We next found that ACAP1BAR-PH also induces membrane curvature, as reflected by the tubulation of liposomes, which was visualized by negative-stain electron microscopy (EM) (Figure 1B). The diameters of membrane tubules were similar regardless of the liposome size added to the incubation, with quantitation revealing 44.7±5.0nm for liposomes of 50 nm diameter (n = 90), 42.4±5.8 nm for liposomes of 100 nm diameter (n = 51) and 42.7±5.8 nm for liposomes of 200 nm diameter (n = 95) (Figure 1C). Thus, different from the properties of prototypic F-BAR and N-BAR proteins, whose ability to induce membrane tubulation is dependent on liposome size (Shimada et al., 2007), ACAP1BAR-PH can act on a range of liposome sizes to induce membrane tubules that have relatively constant diameters.
Crystal structure of ACAP1BAR-PH
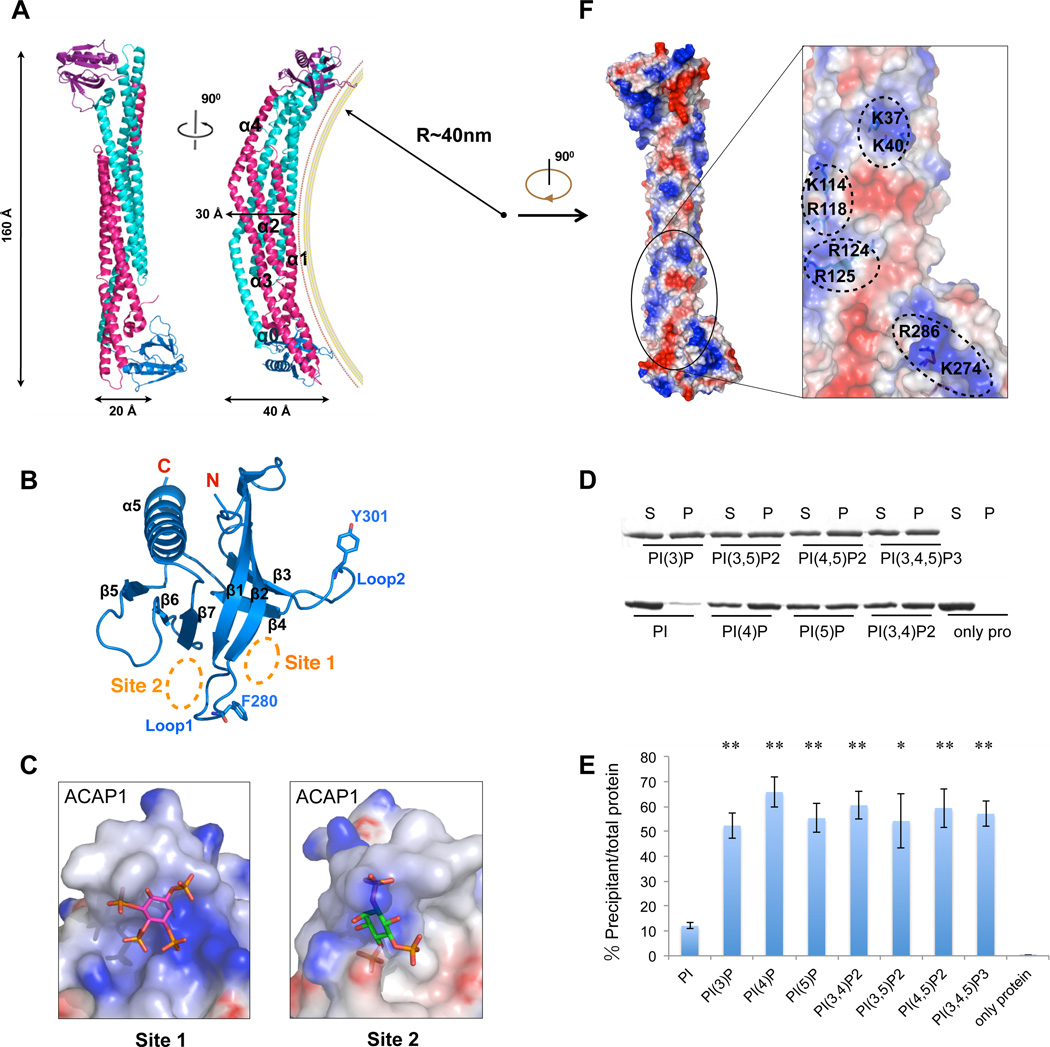
To understand how ACAP1BAR-PH imparts membrane curvature, we next solved its crystal structure to 2.2 Å resolution by single wavelength anomalous diffraction method (Table 1). As would be predicted for a BAR-containing protein, the structure appears as a banana-shaped dimer within one asymmetric unit (Figure 2A). The final refined model contains 364 residues (a.a. 1–364) for one monomer and 361 residues (a.a. 1–361) for the other. The BAR domains within the dimer form a bundle that contains eight-helices, which is similar to the BAR domains of APPL1 and APPL2 (King et al., 2012; Li et al., 2007a; Zhu et al., 2007), but different from other reported BAR domains (Gallop et al., 2006; Peter et al., 2004; Pykäläinen et al., 2011; Shimada et al., 2007). The dimer interface has a large area (4100 Å2), and contains mainly hydrophobic residues that likely mediate dimerization. Dimerization of ACAP1BAR-PH was confirmed from size exclusion chromatography and light scattering (Figure S2A). The in vitro disassociation constant of the dimerization was calculated to be below 5 µM, according to the quantitative gel-filtration experiment (Figure S2B), which is lower than the reported constant for the N-BAR protein endophilin A2 (Ross et al., 2011). This higher affinity could be attributed to the larger dimerization interface (4100 Å2) of ACAP1BAR-PH.
Table 1. Crystallographic data collection and refinement statistics for ACAP1BAR-PH.
Corresponding parameters for the highest-resolution shell are shown in parentheses.
| ACAP1BAR-PH | Se- ACAP1BAR-PH | |
|---|---|---|
| Data collection and processing | ||
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 42.5, 59.7,168.9 | 42.4, 59.8, 169.2 |
| α, β, γ (°) | 90.0, 91.4, 90.0 | 90.0, 91.1, 90.0 |
| Wavelength (Å) | 1.0000 | 0.9790 |
| Resolution (Å) | 50.0-2.20 (2.24-2.20) | 50.0-2.90 (2.95-2.90) |
| Rmerge(%) | 8.2 (41.8) | 12.3(55.2) |
| <I/σ(I)> | 21.3 (3.9) | 14.7(1.34) |
| Completeness (%) | 96.9 (95.6) | 98.4(81.9) |
| Redundancy | 3.6 (3.2) | 6.9 (4.4) |
| Refinement | ||
| Resolution (Å) | 30.0-2.20 | |
| No. reflections | 41114 | |
| Rwork/Rfree | 0.211/0.266 | |
| No. atoms | ||
| Protein | 5839 | |
| Water | 146 | |
| _B_-factors(Å2) | ||
| Protein | 24.22 | |
| Water | 23.22 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.013 | |
| Bond angle (°) | 1.458 |
Figure 2. Crystal structure ofACAP1BAR-PH.
(A) Overall structure of dimeric ACAP1BAR-PH. The two BAR domains are shown in cyan and pink, while the corresponding PH domains are colored in magenta and blue, respectively. Secondary structural elements of BAR domain are labeled as indicated. A hypothetic membrane is depicted according to the curvature (40 nm−1) of the BAR domains. (B) Structure of PH domain in cartoon representation. Secondary structural elements are labeled as indicated. Relevant residues (F280 and Y301) in Loop1 and Loop2 are shown as stick structures. The potential phosphoinositides binding sites (Site 1 and Site 2) are indicated via the orange dashed circles. (C) Site1 and Site2 in the PH domain of ACAP1. The electrostatic potential is mapped onto the surface of the structures, with blue coloring positive charge and red coloring negative charge. The docked inositol ligands for the binding pockets are shown as stick models. For Site 1, the docked PI(3,4,5)P3 is based on the crystal structure of DAPP1 (PDB code, 1FAO). For Site 2, the docked PI(4,5)P2 is based on the crystal structure of ArhGAP9 (PDB code, 2P0D). (D) Binding of ACAP1BAR-PH to liposome that contains various phospholipids (as indicated) is assessed by centrifugation; S, supernatant; P, pellet. (E) Quantitation of results in (D). The level of pelleted protein is expressed as the percentage of the total protein input. All error bars represent standard deviation (s.d.) from three independent experiments. The significances of the differences in comparison to PI containing liposome are expressed as ** for p < 0.001 and * for p < 0.01. (F) Electrostatic surface representation on the concave surface of ACAP1BAR-PH, and a close-up view of the positively charged patches, with blue coloring positive charges and red coloring negative charges.
[See also Figure S2.]
The BAR domain in each monomer contains five helices, α0, α1, α2, α3 and α4 (Figures 2A and S1B). Helix α1, α2 and α3 form an antiparallel coiled-coil and helix α4 forms a helical bundle with the antiparallel coiled-coil of another monomer. This is followed by the PH domain (Figures 2A and 2B), which contains a seven-stranded β barrel (β1–β7) followed a helix (α5) that is located at the extreme carboxy terminus (Figures 2B and S1B).
The two PH domains are positioned at the opposite ends of the dimeric structure. Moreover, the relative positions between the PH and BAR domains are not identical when the two monomers are superimposed by aligning the BAR domains (Figure S2C). A linker region (from residue 249 to 267) between the BAR and PH domains (Figure S1B) suggests the possibility of flexibility in the movement of the PH domain. Indeed, this feature turned out to be important for membrane binding and bending by ACAP1BAR-PH (to be detailed further below).
In comparison to other BAR structures, such as those from APPL1 (Li et al., 2007a; Zhu et al., 2007), the N-BAR Amphiphysin (Peter et al., 2004), and the F-BAR FBP17 (Shimada et al., 2007) (see also Figure S2D), the positive curvature of ACAP1BAR-PH, which is ~1/40nm−1 (Figure 2A), is smaller than that of APPL1 (~1/17 nm−1) and amphiphysin (~1/22 nm−1), but larger than that of FBP17 (~1/60 nm−1). Although the overall fold of ACAP1BAR-PH is similar to that of the tandem BAR-PH domains in APPL1 (Li et al., 2007a; Zhu et al., 2007), the structural r.m.s.d. (root mean square deviation) of the BAR domains between ACAP1 and APPL1 is 2.9 Å for 190 aligned Cα atoms. Furthermore, there is ~ 20° outward rotation of helix α4 in ACAP1BAR-PH when compared to that of APPL1 (Figure S2E). This feature explains the more shallow curvature formed by ACAP1BAR-PH as compared to the corresponding counterpart in APPL1.
Studies on multiple PH domains have revealed a common β1/β2 loop that plays a key role in binding to phosphoinositides (Ceccarelli et al., 2007; Cronin et al., 2004; Ferguson et al., 2000). However, when compared to these PH domains, such as that of DAPP1 and Grp1 (Figure S2F), the corresponding region in the PH domain of ACAP1 (Site 1, Figures 2B and 2C) exhibits less positive electrostatic potential and also forms a less significant cavity for binding to an inositol group. The PH domain of ArhGAP9 has been shown previously to bind inositol trisphosphate at an opposite site to the β1/β2 loop (Ceccarelli et al., 2007) (see also Figure S2G). The corresponding site in ACAP1 (Site 2, Figures 2B and 2C) shows similar electrostatic potential but different surface contour. Collectively, these structural comparisons suggest that the PH domain of ACAP1 may form binding pockets that would have less specificity toward a particular phosphoinositides species. Indeed, revisiting the liposome binding assay, we found that ACAP1BAR-PH can bind to a range of phosphoinositides, including PI(3)P, PI(4)P, PI(5)P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2 and PI(3,4,5)P3 (Figure 2D). We also found that ACAP1BAR-PH cannot bind liposomes that contain PI (Figure 2D), suggesting that the binding pocket in the PH domain only recognizes inositol ligands when they are phosphorylated. These binding results were also quantified (Figure 2E).
A positively charged patch in the PH domain contributes importantly to membrane binding
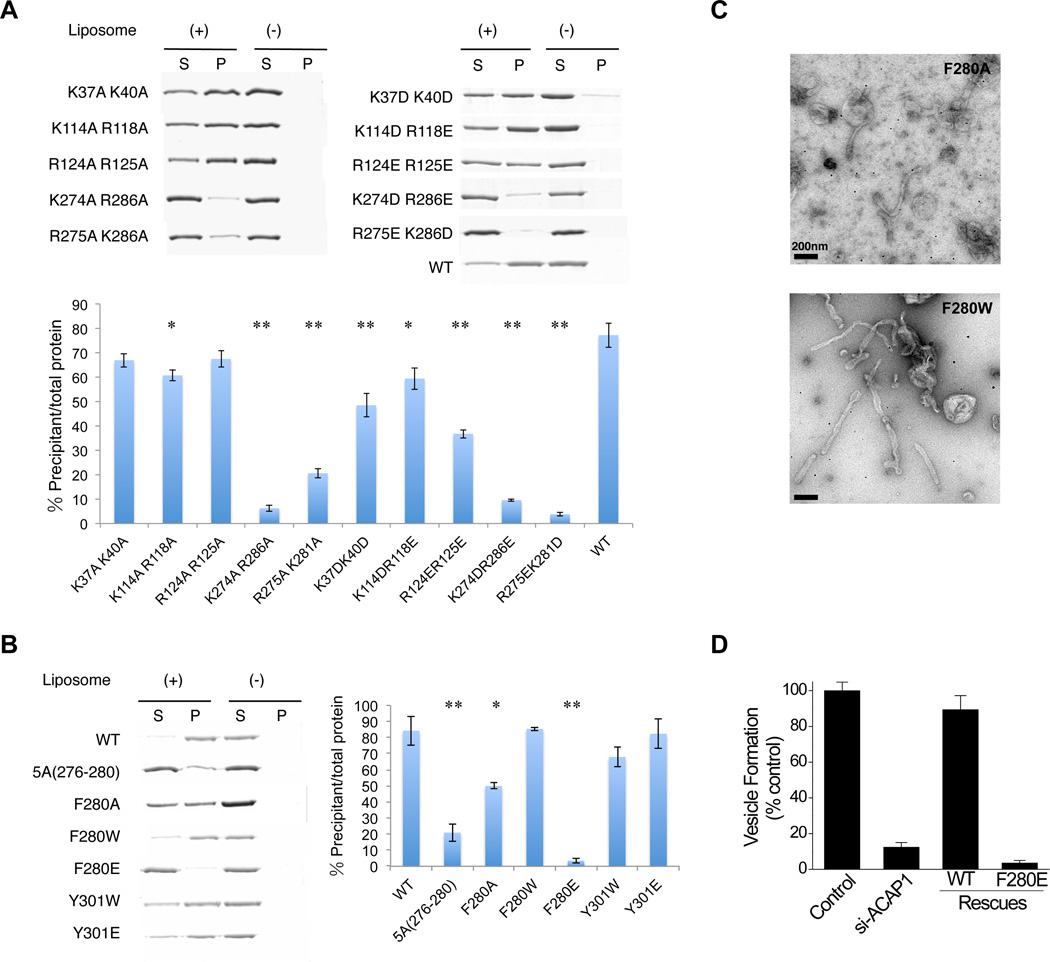
Previous elucidations have revealed that the banana-like structure formed through the dimerization of the BAR domain binds to the negatively charged membrane surface through positively charged patches on the concave surface of the BAR structure (Itoh and De Camilli, 2006; Peter et al., 2004; Shimada et al., 2007). On the concave surface of the BAR domain in ACAP1, we also noted multiple positively charged patches, specifically K37/K40, K114/R118 and R124/R125 (Figures 2F and S1B). Thus, we targeted these residues for mutagenesis, which involves substitutions with alanines, and also by substituting lysines with aspartates and arginines with glutamates. These mutations resulted in mild to modest decreases in liposome binding by ACAP1BAR-PH, as confirmed by quantitation (Figure 3A), suggesting that the positively charged patches in the BAR domain do not constitute the main determinant for membrane binding by ACAP1BAR-PH.
Figure 3. Mutagenesis studies to assess membrane binding and tubulation.
(A) Binding of mutant forms of ACAP1BAR-PH (as indicated) to liposomes is assessed by centrifugation; S, supernatant; P, pellet. The top panel shows a representative result, while the bottom panel shows quantitation from three independent experiments. All error bars represent the s.d. from three independent experiments. The degree of significance involves comparison between wild-type and different mutants, with ** for p < 0.001 and * for p < 0.01. (B) Binding of mutant forms of ACAP1BAR-PH (as indicated) to liposomes is assessed by centrifugation; S, supernatant; P, pellet. The left panel shows a representative result, while the right panel shows quantitation from three independent experiments. All error bars represent the s.d. from three independent experiments. The degree of significance involves comparison between wild-type and different mutants, with ** for p < 0.001 and * for p < 0.01. (C) Negative-stain electron microscopy visualizing liposome tubulation induced by two mutant forms of ACAP1BAR-PH (as indicated); scale bar, 200nm. (D) The reconstitution of recycling vesicles from endosomal membrane was performed by incubating membrane and cytosol fractions derived from HeLa cells, and then tracking the redistribution of a recycling cargo, TfR, from the compartmental membrane fraction to the vesicular membrane fraction. Cytosol was derived from cells with different treatment as indicated. The level of vesicle formation after the incubation was then quantified.
[See also Figure S3.]
We next considered that the concave surface of the banana-like structure of ACAP1BAR-PH extends to the PH domain (Figure 2A). Intriguingly, there exists a positively charged patch on this side of the PH domain, K274/R286 (Figure 2F). When this patch was mutated, again by substituting with alanines, or by substituting lysines with aspartates and arginines with glutamates, we detected a more dramatic effect on liposome binding by ACAP1BAR-PH (Figure 3A). Thus, unlike the conventional BAR domains, the BAR domain in ACAP1 requires cooperation from the PH domain for membrane binding.
A loop in the PH domain contributes importantly to both membrane binding and curvature induction
We then noted another intriguing feature of the PH domain in ACAP1. Near the vicinity of the positively charged patch characterized above is a predicted loop. This structure, Loop1 (a.a. 276–282, ASNAFKT), occurs between the β1 and β2 folds of the PH domain (Figure 2B), which could potentially insert into the membrane, when the overall orientation of the ACAP1BAR-PH structure is considered with respect to the membrane (Figure 2A). In addition, by aligning the structures of PH domains between ACAP1 and Grp1, a corresponding loop is predicted to exist in Grp1, which has also been suggested previously to penetrate into lipid bilayer (Lumb et al., 2011) (see also Figure S2H). Thus, to test whether Loop1 can be involved in membrane insertion, we mutated the residues (276–280) in Loop1 to alanines. We found that the resulting mutant form of ACAP1BAR-PH, 5A (276–280), exhibits substantially decreased binding to the membrane (Figure 3B).
We next noted that an aromatic residue (F280) resides near the distal region of Loop1, which could potentially be important for membrane insertion. Focusing on this residue, we first mutated it to alanine, which weakened the membrane binding by ACAP1BAR-PH (Figure 3B). When F280 was mutated more dramatically to glutamate, we found that membrane binding by ACAP1BAR-PH was virtually eliminated (Figure 3B). Consistent with the bulky hydrophobic nature of F280 being important, we found that a more conservative mutation of F280 to tryptophan (F280W) allows membrane binding by ACAP1BAR-PH to be preserved (Figure 3B).
We next examined mutations in the F280 residue with respect to the ability of ACAP1BAR-PH to induce membrane curvature. Point mutant forms were incubated with liposomes and then examined by EM. We found that F280 is also critical for the induction of membrane curvature (Figure 3C). In particular, mutation of this residue to alanine (F280A) reduced the ability of ACAP1BAR-PH to induce liposome tubulation, while a more conservative mutation (F280W) preserved the ability to tubulate liposomes.
We also sought confirmation that a critical role for the F280 residue in membrane insertion underlies the ability of ACAP1 to act as a coat component in generating recycling vesicles from endosomal membrane. We had previously established a reconstitution system to show that ACAP1 acts in vesicle formation from endosomal membrane, which involved the incubation of membrane and cytosolic fractions and then tracking the redistribution of a recycling cargo from compartmental membrane to vesicular membrane (Li et al., 2012). Taking a similar approach, which in this case involves the incubation of membrane and cytosolic fractions derived from HeLa cells and then tracking the redistribution of the recycling transferrin receptor (TfR) between these two fractions, we found that, whereas cytosol that expressed the wild-type ACAP1 promotes vesicle formation, cytosol that expressed the mutant ACAP1 (F280E) shows markedly impaired ability (Figure 3D). Thus, the result confirmed that the F280 residue in ACAP1 is also critical for its ability to act as a coat component in promoting vesicle formation.
We also noted that, besides Loop1, the PH domain is predicted to possess another loop structure, Loop2 (a.a. 299–305), which resides between β3 and β4 (Figure 2B). Although this loop also contains a bulky hydrophobic residue (Y301), it is predicted to be oriented less favorably with respect to the possibility of inserting into the membrane. As confirmation, we mutated Y301 in Loop2 to either tryptophan (Y301W) or glutamate (Y301E), and found that either mutation has minimal effects on membrane binding by ACAP1BAR-PH (Figure 3B). Thus, the result suggested that Loop2 is less likely to be involved in membrane insertion by ACAP1.
N-terminal helix α0 of ACAP1BAR-PH is not involved in membrane interaction
In some BAR domains, an N-terminal amphipathic helix, such as that seen in endophilin, has been elucidated to be important for membrane insertion and curvature induction (Bai et al., 2010; Boucrot et al., 2012; Gallop et al., 2006). The N-terminal short helix α0 (residues 6–12, DFEECLK) of ACAP1BAR-PH exhibits a semi-amphipathic pattern according to helical wheel analysis (http://rzlab.ucr.edu/scripts/wheel/wheel.cgi), having hydrophobic surface on one side and charged surface on the other (Figure S3A). However, the crystal structure of the dimer suggested that helix α0 forms close contacts with helices α1, α2 and α3 of the same monomer, and also additional contacts with the other monomer, via both hydrophobic and electrostatic interactions (Figure S3A). These interactions suggested that helix α0 might not be sufficiently flexible to participate in membrane insertion. As confirmation, we deleted the first twelve residues of ACAP1BAR-PH, and found that it had no significant effect on membrane binding (Figure S3B) or membrane tubulation (Figure S3C) of liposomes. Thus, the N-terminal helix α0 is unlikely to act in membrane insertion.
Cryo-electron microscopy of liposomes coated with ACAP1BAR-PH
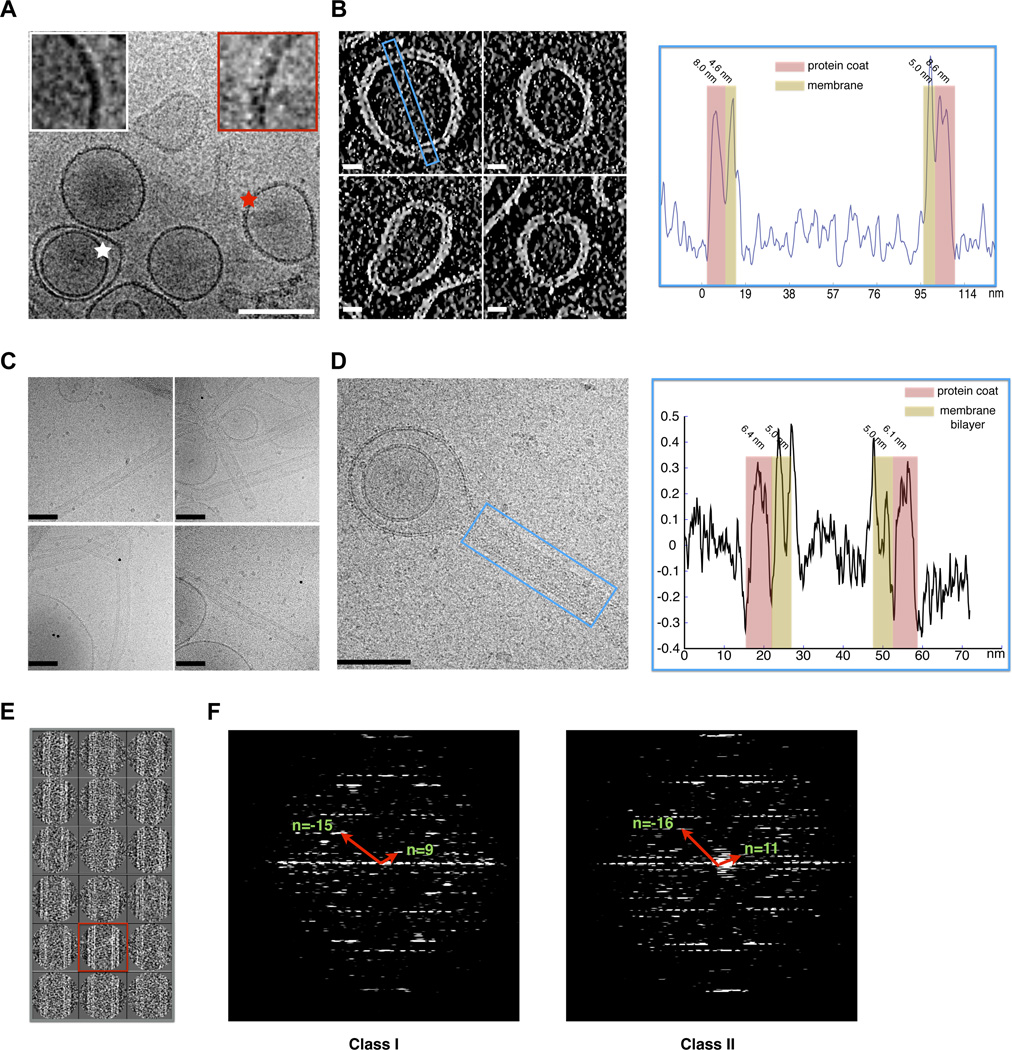
Having defined the key features of the PH domain that enable the ACAP1BAR-PH structure to bind and bend membrane, we next sought further insight into how it achieves this role by pursuing cryo-electron microscopy (cryoEM). After incubation of ACAP1BAR-PH with liposome, the sample was flash frozen and subjected to cryoEM observation. From the cryoEM micrographs, we observed that some liposomes are multilamellar and only the outer layer can be detected to have coating by the ACAP1BAR-PH protein while the inner layer appears as smooth uncoated membrane (Figure 4A). This observation suggested that the coating observed on liposomal membrane is unlikely due to artifacts, such as cryoEM imaging noise or defocusing effects. Pursuing cryo-electron tomography, we measured the thickness of the coated layer to be about 8 nm (Figure 4B and Movie S1), which is thicker than the width (~4 nm) but not the length (~16 nm) of the banana-like structure formed by the dimerization of ACAP1BAR-PH (Figure 2A), suggesting that ACAP1BAR-PH binds to the liposome membrane in a tilted orientation.
Figure 4. CryoEM of ACAP1BAR-PH coating liposomal membrane.
(A) Raw cryoEM micrograph of ACAP1BAR-PH coating liposomes. The coated outer lamellar and the uncoated inner lamellar are indicated by red and white stars, respectively; bar, 100 nm. Insets show magnified views (B) Cryo-electron tomograms of liposomes coated with ACAP1BAR-PH; bar, 20 nm (left panel). A region was selected for line-scanning analysis, as highlighted by the blue rectangle. The integrated density across the liposome is plotted, with the regions for protein coat and membrane indicated and thickness labeled (right panel). (C) Raw cryoEM micrographs of tubules coated with ACAP1BAR-PH; bar, 100 nm. (D) CryoEM micrograph of liposome with tubule coated with ACAP1BAR-PH; bar, 100 nm (left panel). A region was selected for line-scanning analysis, as highlighted by the blue rectangle. The integrated density across the tubule is plotted, with the regions for protein coat and membrane indicated and thickness labeled (right panel). (E) Two-dimensional classification of images showing dynamics of ACAP1BAR-PH coating liposomal tubules. (F) Helical diffraction patterns of tubules exemplifying Class I and II. The Bessel functions for basis vectors are shown and labeled accordingly.
[See also Movie S1.]
We then focused on the tubulated portions of liposomes coated by ACAP1BAR-PH (Figure 4C). The diameters of these tubules were in the range of 40 nm to 50 nm (Figure S4A), which is similar to our previous measurement of these tubules based on negative-stain EM (Figure 1C). Portions of the tubules were selected to generate a density profile across the diameter of the tubule (Figure 4D). The membrane bilayer could be easily discerned, and the thickness of the protein coat was measured to be about 6 nm (Figure 4D). The thickness of this coating varied depending on whether the measurement was taken on the tubulated or the non-tubulated portion of the liposome, suggesting a different assembly of ACAP1BAR-PH on the membrane upon tubulation.
We next performed three-dimensional (3D) reconstructions of the linear portions of the tubules (boxed region in Figure 4D), which were segmented for image analysis and classification. The spiral feature of some class averages (Figure 4E) suggested helical symmetry in the packing of ACAP1BAR-PH on the tubules, which was further confirmed based on the layer-line diffraction patterns of selected well-ordered long tubules (Figure 4F). Two types of helical diffraction patterns were observed and indexed, Class I and II (Figure 4F). We utilized an iterative helical real space reconstruction (IHRSR) approach (Egelman, 2000) to determine the 3D structures of these two classes. The layer-line diffraction patterns of the final reconstructed cryoEM maps (Figure S4B) matched the ones of the original micrographs (Figure 4F) and further validations of the reconstructions were provided via the match of maps with the atomic models (below). We achieved ~14 Å resolution for Class I and ~17 Å resolution for Class II, as assessed by visual comparison between the cryoEM maps and the fitted crystallography structural models. These two resolutions are between the ones based on the 0.5-cutoff and 0.143-cutoff criterions on the Fourier shell correlation (FSC) curves (Figures S4C and S4D).
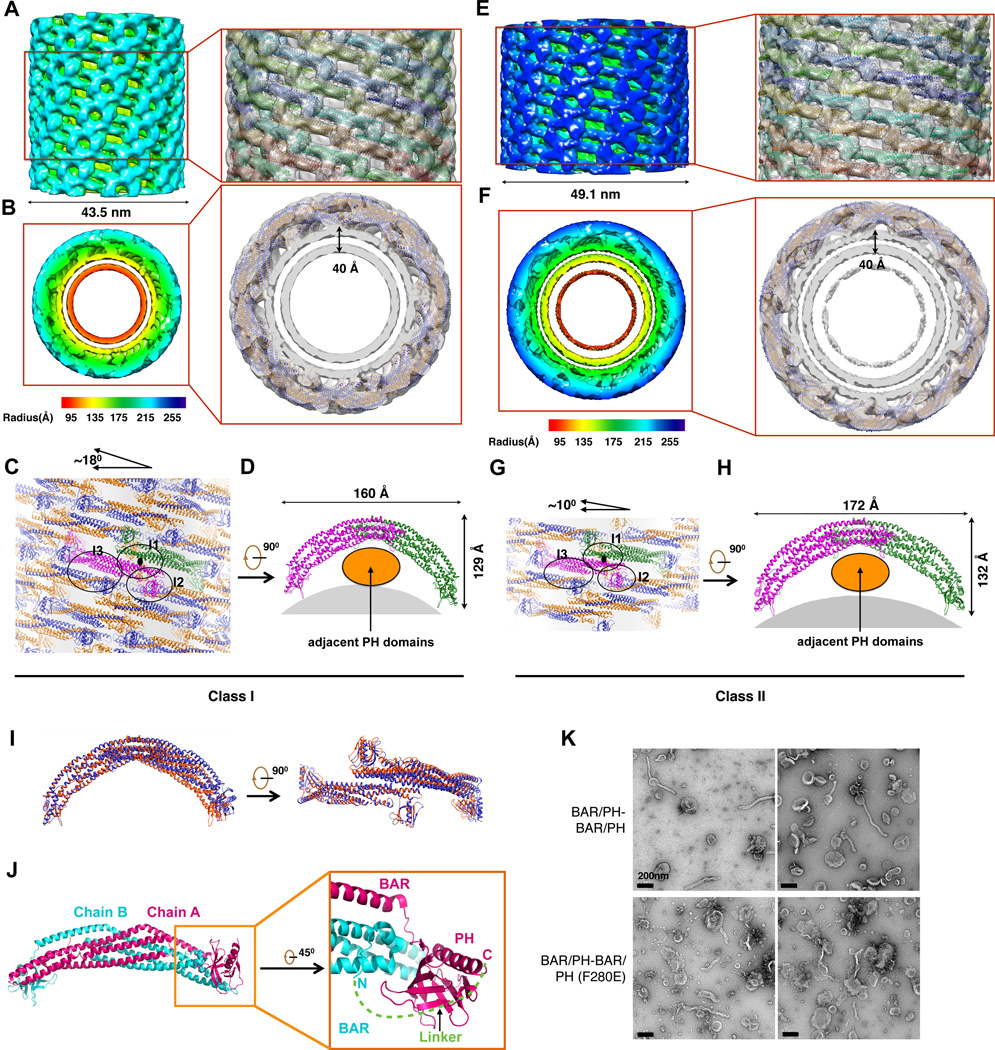
The cryoEM map revealed Class I tubule to have a diameter of 43.5 nm, with the double membrane thickness of 4.0 nm, and the ACAP1BAR-PH coating thickness of 6.0 nm (Figures 5A and 5B). We then sought to fit the structural model of ACAP1BAR-PH into the cryoEM map in determining how the ACAP1BAR-PH molecules could assemble on the tubulated liposome (Figures 5A and 5B; Movies S2 and S3). The analysis suggested that two ACAP1BAR-PH molecules pack laterally with each other via the interactions between BAR domains and thereby assembling into an ACAP1BAR-PH tetramer with a two-fold symmetry (Figures 5C and 5D). The resulting tetramer exhibited an elongated banana-like shape with two PH domains interacting with the membrane and two other PH domains not interacting with membrane. The tetramer formed an asymmetric unit of the ACAP1BAR-PH helical packing on the tubule, and the tilting angle with respect to the tubular cross session was ~18°. In this configuration, the tetramers were observed to pack onto the membrane via lateral interactions and “end-to-arch” interactions in which one tetramer inserts its end underneath the adjacent tetramer arch. Consequently, the convex part of the cryoEM map is composed of the arches and ends of tetramers, while the concave part is composed of lateral interacting BAR domains (Figure 5A). From the cross section of the cryoEM map (Figure 5B), the connection densities between the protein layer and the membrane could be clearly observed, which fitted well with Loop1 of the PH domain interacting with the membrane. Furthermore, only one monomer of the ACAP1BAR-PH dimer was observed to insert its Loop1 into the membrane.
Figure 5. 3D reconstructions of ACAP1BAR-PH coating tubules.
(A) and (B) CryoEM map of a liposomal tubule coated with ACAP1BAR-PH in Class I that is shown in iso-surface representation with the side view in (A) and top view in (B). The map is colored according to the cylinder radius from red to blue. The structural models of ACAP1BAR-PH in cartoon representation are fitted into the map and shown on the right. In (B), the two chains of ACAP1BAR-PH dimer are colored in gold and blue, respectively, and the thickness of the double-layer membrane is labeled. (C) and (D) Structural model of ACAP1BAR-PH helical assembly on the Class I liposomal tubule. The ACAP1BAR-PH tetramer in one asymmetric unit, colored in magenta for one dimer and green for another, is shown with the top view in (C) and side view in (D). The interaction interfaces (I1, I2 and I3) for ACAP1BAR-PH helical assembly are indicated by black circles and labeled in (C). The titling angle of ACAP1BAR-PH tetramer with respect to the cross section of the tubule is depicted on the top. In (D), the membrane region is shown in grey. (E) and (F) CryoEM map of a liposomal tubule coated with ACAP1BAR-PH in Class II that is shown in the side view (E) and the top view (F). The structural models of ACAP1BAR-PH in cartoon representation are fitted into the map and shown on the right. The color and labeling schemes are the same as that described in (A) and (B). (ACAP1 (BAR or BAR-PH) to liposomes of) and (H) Structural model of ACAP1BAR-PH helical assembly on the Class II liposomal tubule. The color and labeling schemes are the same as that described in (C) and (D). (I) Superposition between the ACAP1BAR-PH tetramers of Class I (red) and Class II (blue). (J) Structural basis for designing the fusion version of the ACAP1BAR-PH dimer (BAR/PH-BAR/PH). The overall crystal structure of ACAP1BAR-PH dimer is shown left and labeled with Chain A and Chain B for the two monomers. One distal region is zoomed in and shown (on right). The N-terminus of Chain B and the C-terminus of Chain A are labeled, respectively. The distance between the N-terminus of Chain B and the C-terminus of Chain A is ~37.8 Å. Thus, a linker (green) with the length of 14 amino acids (GGGSGGRLGSSNSG) is predicted to be sufficiently long to covalently link Chain A and Chain B via their termini. (K) Negative-stain electron microscopy visualizing liposome tubulation; bar, 200nm.
[See also Figures S4; Movies S2–5.]
Inspection of the ACAP1BAR-PH helical packing suggested three main interfaces (Figure 5C). One interface (labeled I1 in Figure 5C) would occur through the formation of tetramers of ACAP1BAR-PH, which involves their BAR domains. A second interface (labeled I2 in Figure 5C) occurs through “end-to-arch” interactions between tetramers, which are achieved by a PH domain that does not contact the underlying membrane interacting with an adjacent BAR domain. A third interface (labeled I3 in Figure 5C) occurs through lateral interactions among tetramers. Moreover, based on the residues likely to be involved in these interactions, they are predicted to be mainly electrostatic.
In contrast to Class I tubules, Class II tubules exhibit different helical symmetries (Figure 4F) and a longer diameter of 49.1 nm (Figure 5E). Nevertheless, the cryoEM reconstruction of Class II tubules suggested a similar helical packing of ACAP1BAR-PH on membrane (Figures 5E–H; Movies S4 and S5). The asymmetric unit of Class II tubules was still the tetramer of ACAP1BAR-PH, although with a smaller tilting angle of ~10°. Moreover, the tetramer assembly occurs via similar lateral and “end-to-arch” interactions (Figure 5G). The extended length of the banana-like tetramer in Class II tube was 172 Å, while the extended length was 160 Å for Class I tubules (Figures 5D, 5H and 5I). No other significant differences were observed between the tetramers in Class I and II (Figure 5I).
Overall, by inspecting the cryoEM reconstructions of the tubulated liposomes coated with ACAP1BAR-PH, we uncovered multiple remarkable findings. First, the interaction between the coating protein and the membrane is mediated by one PH domain of the ACAP1BAR-PH dimer. Second, there is no significant interaction between the BAR domain and the underlying membrane. Instead, the BAR domain contributes to the packing interfaces among ACAP1BAR-PH protein in assembling the coating on liposome membrane. Third, the PH domain in the ACAP1BAR-PH dimer that does not contact the membrane, interacts with the BAR domain of the adjacent dimer. As such, these collective findings reveal a distinctly different mechanism by which the BAR-containing ACAP1 achieves tubule formation, as compared to those previously elucidated for a prototypic F-BAR protein (Frost et al., 2008) and an N-BAR protein (Mim et al., 2012). Whereas those BAR-containing proteins achieve membrane binding and bending through the BAR domain playing the principal role, which involves either electrostatic interactions with membrane or insertion of a hydrophobic structure into membrane, we have found that the neighboring PH domain possesses these key features in explaining how ACAP1 achieves membrane binding and bending.
We also sought further confirmation of this overall conclusion. Based on the dimeric structure elucidated from crystallography, which predicts that the N-terminus of one monomer extends closely to the C-terminus of the other one (Figure 5J), we generated a fusion version of the ACAP1BAR-PH dimer (BAR/PH-BAR/PH), using a sequence (GGGSGGRLGSSNSG) that is predicted to be able to extend across the ends of the two monomers to covalently link them (Figure 5J). The resulting fusion construct is predicted to retain the overall orientations of the monomers within the banana-like structure, with the BAR domains residing at the central portion and the PH domains residing at the distal portions. Gel filtration experiments suggested that the fusion protein folds and assembles correctly, as it has a similar profile with the non-fusion dimer (Figure S4G). As functional assessment, we found that the fusion dimer (BAR/PH-BAR/PH) retains the ability to tubulate liposomes (Figure 5K). We then generated a mutant version, a point mutation that targets only one PH domain [BAR/PH-BAR/PH (F280E)], and found that it could still induce liposome tubulation (Figure 5K). Notably, this result is in contrast to the effect of the F280E mutation in the conventional (non-fusion) version, which would have this point mutation in both PH domains, as we had found previously that it could no longer bind liposomes (see F280E in Figure 3B). Thus, this additional finding suggests that one PH domain is sufficient to confer the ability of the ACAP1BAR-PH dimer to insert into membrane and impart curvature, which is in a good agreement with our cryoEM observations.
Molecular simulation of ACAP1BAR-PH binding to membrane
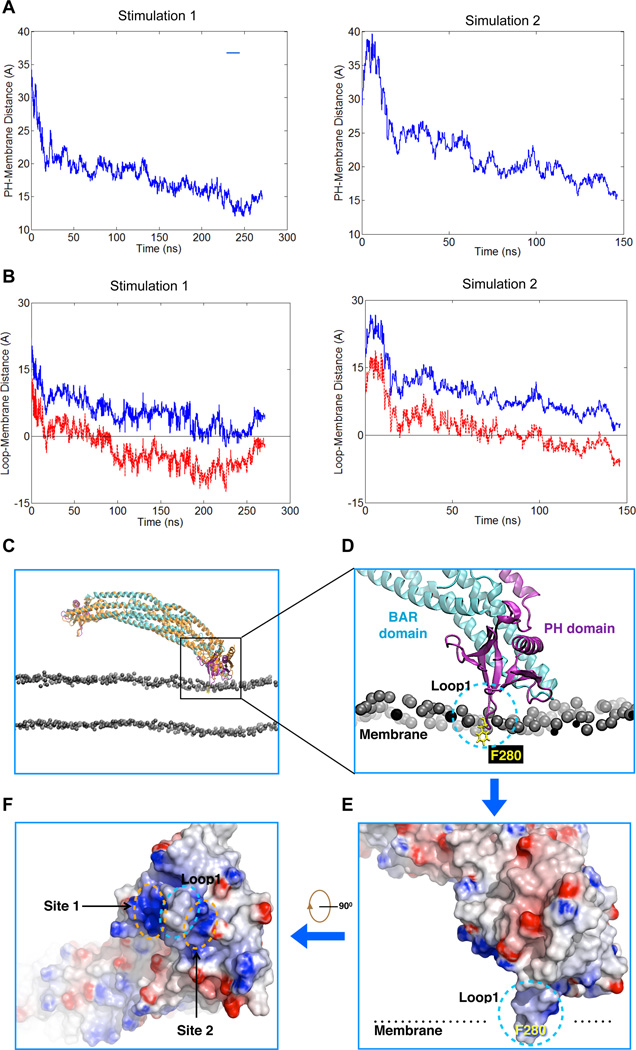
We next pursued molecular dynamics simulation to gain further insight into how these key features of the PH domain could be mediating interaction with the membrane. Two independent simulations (Simulation 1 and 2) were performed with the time scale of 270 nanoseconds (ns) and 150 ns. Notably, both simulations revealed that ACAP1BAR-PH dimer behaves asymmetrically, with only one PH domain (chain A in Simulation 1 and chain B in Simulation 2) having a significant interaction with the membrane. This is reflected from the distance between the center mass of PH domain and the membrane phosphate plane, which decreased monotonically. In contrast, the other PH domain (chain B in Simulation 1 and chain A in Simulation 2) remained away from the membrane (Figure 6A, Movies S6 and S7). Loop1 in the PH domain was suggested to insert into the membrane at ~80 ns for Simulation 1, and at ~100 ns for Simulation 2. These insertions remained until the end of the simulation (Figures 6B, 6C and Movie S8).
Figure 6. Molecular simulation of ACAP1BAR-PH binding to membrane.
(A) The time evolution of the distance between the center of mass of one PH domain and the surface membrane phosphate plane. (B) The time evolution of the distance between Loop 1 in the PH domain and the surface membrane phosphate plane. The blue tracing tracks the distance between the center of mass of Loop1 and the membrane phosphate plane, while the red tracing tracks the distance between the F280 residue in Loop1 and the surface membrane phosphate plane. Negative values indicate an insertion of a particular protein structure into the membrane. (C) Comparison of the crystal structure (shown in orange) and the final simulated model (shown in cyan and purple) of dimeric ACAP1BAR-PH interacting with the membrane bilayer. Only the phosphate atoms of phospholipids in the membrane are shown for simplification. (D) A zoomed-in view of (C), focusing on the PH domain engaging the membrane. The BAR and PH domains are shown as cartoon representation, and colored in cyan and purple respectively. The critical residue (F280) in Loop1 is shown embedded in membrane and represented in yellow. The phosphate atoms of the membrane phospholipids are shown as grey spheres. (E) Electrostatic surface representation of the PH domain shown in the configuration illustrated in (D). Blue coloring highlights positive charges, while red coloring highlights negative charges. (F) 90° rotation of image shown in (E) to better visualize the membrane-binding surface of the PH domain. The positively charged patches (Site 1 and 2) and the hydrophobic insertion site (Loop1) are indicated.
[See also Figure S5 and Movies S6–9.]
Notably, the simulations revealed another mechanistic insight regarding how membrane insertion is achieved via the PH domain of ACAP1. In comparison to the crystal structure of ACAP1BAR-PH, the membrane bound ACAP1BAR-PH from both simulations suggested a conformational rotation of the PH domain with respect to the BAR domain (Figure 6C and Movie S8), and then Loop1 adapts its orientation to facilitate the insertion of F280 into membrane (Figures 6D and 6E). As such, the insertion of F280 into one leaf of the bilayer could reduce membrane surface tension asymmetrically and thereby induce membrane deformation when the dimeric ACAP1BAR-PH becomes organized into higher-ordered helical packing onto membrane.
Both simulations also suggested that there are two potential phosphoinositide binding sites (Site 1 and Site 2) along the sides of Loop1 (Figure 6F and Movie S8). These two sites are equivalent to the previous ones predicted from the crystal structure (Figures 2B and 2C) and comprise of residues K274/R286 for Site 1 and residues R275/K281 for Site 2. The importance of K274/R286 has already been suggested (see Figure 3A). We performed further mutation analysis on Site 2 (R275/K281), by substituting with alanines (R275A/K281A), or by substituting lysines with aspartates and arginines with glutamates (R275E/K281D). These mutations revealed that alteration of Site2 also significantly eliminates the liposome binding of ACAP1BAR-PH (Figure 3A).
We also noted that the observed asymmetric binding of ACAP1BAR-PH dimer in both simulations further support the cryoEM model that predicts only one PH domain of one ACAP1BAR-PH dimer inserts into the membrane (Figures 5D and 5H). This observation was not due to the mild asymmetric position of ACAP1BAR-PH dimer at the start of the simulation because both PH domains of ACAP1BAR-PH dimer were found capable of interacting with the membrane during the corresponding simulations (Movies S6 and S7). We then compared the final membrane bound ACAP1BAR-PH with its starting conformation, i.e., the crystal structure. In both simulations, we found that, besides the rotation of the PH domain for membrane insertion, the BAR domains also exhibit significant distortion (Movie S9). The angle between the BAR domains and the membrane, which is defined as the angle between the 2-fold symmetric axis of the BAR domains and the membrane normal-plane that is across the BAR domains (Figure S5A), increased from 2.6 to 21 degree in Simulation 1 and decreased from 2.6 to −15 degree in Simulation 2 (Figure S5B). Thus, the repulsive distortion of the BAR domains could explain why the PH domains of the ACAP1BAR-PH dimer interact asymmetrically with the membrane.
DISCUSSION
We have elucidated in molecular detail how ACAP1 achieves membrane remodeling for its role as a coat component that promotes the generation of transport carrier in endocytic recycling. Notably, this elucidation has uncovered unexpected features in both the BAR and PH domains in explaining their relative contributions to the ability of ACAP1 to bind and bend membrane.
Although the BAR domain in ACAP1 undergoes dimerization to form a banana-like structure, the electrostatic interaction between this curved protein structure and membrane is relatively weak. Thus, ACAP1 has an unconventional BAR domain, as it can achieve neither membrane binding nor bending by itself. Instead, we have identified key features in the neighboring PH domain, which are typically attributed to a conventional BAR domain in promoting membrane binding and curvature induction. The initial insight came from the crystal structural of ACAP1BAR-PH. First, we identified a positively charged patch in the PH domain that could undergo electrostatic interaction with the negatively charged membrane surface to promote membrane binding. Second, we identified a loop that contains a bulky hydrophobic residue, which likely inserts into membrane to promote the generation of membrane curvature.
We then achieved significant new insights by examining how the ACAP1BAR-PH dimer is organized into a higher-order structure on tubulated membrane. Pursuing cryoEM studies, we find that the ACAP1BAR-PH dimer shows lateral interactions with additional ‘end-to-arch’ interactions. Notably however, whereas the lateral interactions are mediated mainly by the BAR domains, the ‘end-to-arch’ interactions are mediated mainly by the PH domains interacting with the BAR domains. Moreover, the ACAP1BAR-PH dimer uses only one of its PH domains to contact the underlying membrane, while the other PH domain acts primary to interact with adjacent ACAP1BAR-PH dimers. As such, rather than playing the primary role in membrane binding and curvature induction, the BAR domain in ACAP1 plays more of a supportive role in assisting the PH domain to bind and bend membrane.
Further insights into how the ACAP1BAR-PH dimer initiates membrane binding for curvature induction have come from molecular simulation studies. Besides accounting for the key features elucidated for the PH domain, namely its positively-charged patches and a key loop structure, in explaining how this domain acts in membrane binding and sculpting, the simulation studies revealed a further notable feature. Independently, both simulations suggested that only one PH domain of the ACAP1BAR-PH dimer undergoes a rotational shift with respect to the BAR domain, so that the positively charged patches in the PH domain can engage phosphoinositides of the membrane bilayer, and thereby allowing the key loop structure in this domain (loop 1) to be oriented properly for membrane insertion through a critical bulky hydrophobic residue (F280).
It is notable that, besides ACAP1, multiple other proteins are known to possess tandem BAR-PH domains. These include a number of ArfGAPs, as well as APPLs, and also sorting nexins that possess a PX domain that is highly similar to the PH domain (Cullen, 2008; Frost et al., 2009; Inoue and Randazzo, 2007). As such, our insights regarding the PH domain in ACAP1 are likely relevant to a better understanding of how they act in membrane remodeling. On a broader note, the PH/PX domain is a prevalent structure, as it exists in many proteins (Lemmon, 2008; Rebecchi and Scarlata, 1998). Thus, our findings have the intriguing prospect of being relevant to a greater set of proteins than those currently known to participate in membrane remodeling.
EXPERIMENTAL PROCEDURES
Protein crystallization
The ACAP1BAR-PH protein was dialyzed against 20mM Tris pH8.5 and concentrated to 3 mg/ml by ultrafiltration. The crystals were obtained using the hanging-drop vapor diffusion method at 16°C with the reservoir solution containing 0.2 M ammonium citrate, pH7.0 and 10% PEG 3350. The addition of 6.0mM Octyl beta-thioglucopyranoside yielded better single crystals that grew over a period of 3 days. The SeMet derivative was crystallized under the same condition as the native protein.
Data collection and structure determination
The ACAP1BAR-PH crystals were flash frozen in the reservoir solution with additional 20% glycerol. The diffraction data of native crystals and single-wavelength (λ=0.979Å) anomalous diffraction data of SeMet derivatives were collected at beamline BL5A and 17A (Photon Factory, Japan) and beamline BL17U (SSRF, China). All data were processed with the HKL2000 program suite (Table 1).
The crystal structure of ACAP1BAR-PH was determined by a hybrid method combining single wavelength anomalous dispersion and molecular replacement. The initial Selenium positions were determined using SHELXD (Uson and Sheldrick, 1999). The Selenium atoms were refined and initial phases were generated using SOLVE (Terwilliger and Berendzen, 1999). Phase improvement and density modification were performed using the program RESOLVE (Terwilliger, 2000). Initial model was built automatically using the Quick-Fold of ARP/WARP (Langer et al., 2008). Almost all BAR domain backbones were automatically traced, while the PH domain exhibited only several non-continuous fragments. MR of two PH domains were then performed using MOLREP (Vagin and Teplyakov, 2010) based on the SAD experimental density map. An iterative method (Yao et al., 2006; Zhang et al., 2010) combining OASIS (Fan and Gu, 1985), DM (Bailey, 1994) and REOLVE (Terwilliger, 2000)was then used to further improve the phases and thereby complete model building. Model building was further improved manually using COOT (Emsley and Cowtan, 2004). Structure refinement was done using Refmac5 (Murshudov et al., 1997) and CNS (Brunger et al., 1998). The statistics of the data processing and structure refinement is summarized in Table 1. All the Figures for structural illustration were generated with Pymol (http://www.pymol.org) or UCSF Chimera (Pettersen et al., 2004).
Cryo-electron microscopy and helical reconstruction
The ACAP1BAR-PH protein (4mg/ml) was incubated with 200 nm-diameter liposomes (2 mg/ml) at room temperature for 60minutes. A drop (3.5 µl) of the mixture was then applied onto a 300-mesh GiG holy carbon grid (Jiangsu LifeTrust, China) that was pre-treated in plasma cleaner (PDC-32G, HARRICK PLASMA). The grid was then blotted for 3.0 seconds with a blot force 2 at 100% humidity, using FEI Vitrobot (Mark IV), before it was quickly frozen in liquid ethane that was cooled by liquid nitrogen.
The ACAP1BAR-PH coating membrane tubules were imaged with a FEI Titan Krios cryo-electron microscope that was operated under 300kV and equipped with a Gatan UltraScan4000 CCD camera. Low-dose images (20 e−/Å2) were collected manually. The nominal magnification was set to 75,000×, which corresponds to a pixel size 1.196 Å. The defocus range was set to 2.5~3.5 µm. A total of 259 cryoEM micrographs were collected.
The defocus value of each micrograph was determined by CTFFIND3 (Mindell and Grigorieff, 2003). About 160 ACAP1BAR-PH tubules were selected using the helical option “helixboxer” in EMAN1 (Ludtke et al., 1999). All the selected tubules were padded to 4096×4096 pixels to obtain diffraction pattern in Fourier space. These tubules were then classified into two main classes (Class I and II) according to their different diffraction pattern. For each of class, further reference-free classification was performed using e2refine2d.py in EMAN2 (Tang et al., 2007). All the class averages were rotated to normalize the orientation of all the tubules in the vertical direction. The diameters of each class of ACAP1BAR-PH tubules were calculated by projecting the images in the horizontal direction to further sort the tubules. 304 segments were selected from 10 tubules with 95.5% overlap for Class I, while 352 segments were selected from 6 tubules with 88.3% overlap for Class II.
Initial helical parameters (Δφ and ΔZ), which denote the rise and the rotation angle between the two neighboring subunits, respectively, were estimated by calculating the order of Fourier-Bessel function from the layer lines of diffraction patterns (see Figure 4F). For Class I, the initial parameters (Δφ and ΔZ) are 42.7° and 23.2 Å, respectively, which yielded two starting numbers (n1 = −15 and n2 =9), implying the existence of a C3 symmetry in this class. For Class II, the initial parameters (Δφ and ΔZ) are 65.9° and 6.7 Å, respectively, which yielded two starting numbers (n1 = −16 and n2 = 11). These initial parameters were input into the IHRSR program (Egelman, 2000) by using a cylinder as a starting model. All the images of raw particles were binned 3 times before subjecting into the refinement by IHRSR. The out-of-plane tilt angles were considered during the refinement for both classes. The helical parameters finally converged to Δφ = 42.72°and ΔΖ = 23.20 Å for Class I, and to Δφ = 65.90°and ΔΖ = 6.67 Å for Class II. Nearly 82% of Class I filaments show a significant out-of-plane tilt angle and ~54% of Class II filaments with the significant out-of-plane tilt angles (see Figures S4E and S4F), which increases the difficulty of helical reconstructions.
The nominal resolutions of the reconstructed volumes were assessed by computing the Fourier shell correlation (FSC) between the cryoEM map and the fitted structural models and by using the threshold cutoff 0.5 and 0.143. The handedness of the cryoEM map was determined by judging the fit of the structural model of ACAP1BAR-PH dimer into the map. CryoEM maps were segmented, displayed and fitted with atomic models using UCSF Chimera (Pettersen et al., 2004). All structural figures were generated by UCSF Chimera (DeLano, 2002).
Molecular dynamics simulation
The molecular dynamics simulations were performed using NAMD 2.9 (Phillips et al., 2005) and the CHARM 22/27 force field with CMAP correction (Mackerell et al., 2004). Simulations were performed under constant NPT conditions and periodic boundary conditions. The temperature was maintained throughout at 310K using a Langevin thermostat with 5ps−1 damping coefficient. The system pressure was maintained at 1 atm using a Langevin piston barostat. Electrostatic interactions were calculated using the particle mesh Ewald sum method (Darden et al., 1993) with a cutoff of 12Å. An integration time step of 2 fs was used while constraining all hydrogen-containing covalent bonds with the SHAKE algorithm (Ryckaert et al., 1977).
The CHARMM-GUI (Jo et al., 2008) was employed to generate a lipid bilayer of 40% DOPC / 30%DOPE / 20%POPS / 10% DAPC. DAPC was then replaced by PI(4,5)P2. The crystal structure of ACAP1BAR-PH was placed to the top of the membrane. The closest point of the protein was 9.5 Å from the membrane surface phosphate plane. The system was solvated with TIP3P water and neutralized by K+ and Cl- counter ions. The membrane-protein system was equilibrated using a stepwise relaxation procedure. After all restraints on C-alpha of the protein atoms were removed, two independent regular NPT simulations were executed. A total of 270 and 150 ns of data were generated for Simulation 1 and Simulation 2, respectively.
Other methods are detailed in the supplementary materials section
ACCESSION CODES
The coordinate of ACAP1BAR-PH crystal structure has been deposited in the Protein Data Bank (PDB) with the accession code 4NSW. The EM maps of helical tubules have been deposited in Electron Microscopy Data Bank (EMDB) with the accession codes EMD-2546 for Class I and EMD-2547 for Class II, respectively. The corresponding fitted coordinates of ACAP1BAR-PH helical array on the tubule have been deposited in Protein Data Bank (PDB) with the accession codes 4CKG and 4CKH, respectively.
Supplementary Material
01
ACKNOWLEDGEMENTS
This work was supported with grants to F.S. by Chinese Ministry of Science and Technology (2011CB910301 and 2011CB910901), grants to X.P and Y.Z. by National Natural Science Foundation of China (31000635 and 31100617), grants to J.F. by City University of Hong Kong (Projects no. 7200350 and 9610291), grants to V.H. by the National Institutes of Health (GM073016 and GM058615), and grant to E.H.E. by the National Institutes of Health (EB001567). All the EM data were collected and most molecular dynamics simulations were performed at Center for Biological Imaging (CBI, http://cbi.ibp.ac.cn), Institute of Biophysics, Chinese Academy of Sciences. We are grateful to Dr. Gang (CBI) for his assistance in electron microscopy data collection.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTION
F.S. and V.H. initiated and supervised the project. X.P., V.H. and F.S. designed all experiments. X.P., K.Z., J.M. and Q.Z. performed crystallographic work. X.P., Y.Z., B.G. E.H.E. and Y.D. performed electron microscopy work. X.P. and B.G. performed mutagenesis studies. J.F. performed MD simulations. J.L. performed vesicle reconstitution studies. X.P., J.F., Y.Z., J.L., V.H. and F.S. analyzed the data. X.P., J.F. and Y.Z. contributed to manuscript preparation. V.H. and F.S. wrote the manuscript.
REFERENCES
- Bai J, Hu Z, Dittman JS, Pym EC, Kaplan JM. Endophilin functions as a membrane-bending molecule and is delivered to endocytic zones by exocytosis. Cell. 2010;143:430–441. doi: 10.1016/j.cell.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M, Pang X, Lou J, Zhou Q, Zhang K, Ma J, Li J, Sun F, Hsu VW. Mechanistic insights into regulated cargo binding by ACAP1 protein. The Journal of biological chemistry. 2012;287:28675–28685. doi: 10.1074/jbc.M112.378810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Boucrot E, Pick A, Camdere G, Liska N, Evergren E, McMahon HT, Kozlov MM. Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell. 2012;149:124–136. doi: 10.1016/j.cell.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Ceccarelli DF, Blasutig IM, Goudreault M, Li Z, Ruston J, Pawson T, Sicheri F. Non-canonical interaction of phosphoinositides with pleckstrin homology domains of Tiam1 and ArhGAP9. THE JOURNAL OF BIOLOGICAL CHEMISTRY. 2007;282:13864–13874. doi: 10.1074/jbc.M700505200. [DOI] [PubMed] [Google Scholar]
- Cronin TC, DiNitto JP, Czech MP, Lambright DG. Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains. The EMBO Journal. 2004;23:3711–3720. doi: 10.1038/sj.emboj.7600388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ. Endosomal sorting and signalling: an emerging role for sorting nexins. Nat Rev Mol Cell Biol. 2008;9:574–582. doi: 10.1038/nrm2427. [DOI] [PubMed] [Google Scholar]
- Dai J, Li J, Bos E, Porcionatto M, Premont RT, Bourgoin S, Peters PJ, Hsu VW. ACAP1 Promotes Endocytic Recycling by Recognizing Recycling Sorting Signals. Developmental Cell. 2004;7:771–776. doi: 10.1016/j.devcel.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Darden T, York D, Pedersen L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- DeLano WL. PyMOL Molecular Viewer. 2002 http://www.pymol.org. [Google Scholar]
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fan HF, Gu YX. Combining Direct Methods with Isomorphous Replacement or Anomalous Scattering Data. 3. The Incorporation of Partial Structure Information. Acta Crystallogr A. 1985;41:280–284. doi: 10.1107/s0907444998017703. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Kavran JM, Sankaran VG, Fournier E, Isakoff SJ, Skolnik EY, Lemmon MA. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol Cell. 2000;6:373–384. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT. Mechanism of endophilin N-BAR domain-mediated membrane curvature. The EMBO Journal. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Randazzo PA. Arf GAPs and their interacting proteins. Traffic. 2007;8:1465–1475. doi: 10.1111/j.1600-0854.2007.00624.x. [DOI] [PubMed] [Google Scholar]
- Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta. 2006;1761:897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Jackson TR, Brown FD, Nie ZZ, Miura K, Foroni L, Sun JL, Hsu VW, Donaldson JG, Randazzo PA. ACAPs are Arf6 GTPase-activating proteins that function in the cell periphery. Journal of Cell Biology. 2000;151:627–638. doi: 10.1083/jcb.151.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. Journal of computational chemistry. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- King GJ, Stockli J, Hu SH, Winnen B, Duprez WG, Meoli CC, Junutula JR, Jarrott RJ, James DE, Whitten AE, et al. Membrane curvature protein exhibits interdomain flexibility and binds a small GTPase. J Biol Chem. 2012;287:40996–41006. doi: 10.1074/jbc.M112.349803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nature Reviews Molecular Cell Biology. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Li J, Malaby AW, Famulok M, Sabe H, Lambright DG, Hsu VW. Grp1 plays a key role in linking insulin signaling to glut4 recycling. Developmental Cell. 2012;22:1286–1298. doi: 10.1016/j.devcel.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Mao X, Dong LQ, Liu F, Tong L. Crystal Structures of the BAR- PH and PTB Domains of Human APPL1. Structure. 2007a;15:525–533. doi: 10.1016/j.str.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Li J, Peters PJ, Bai M, Dai J, Bos E, Kirchhausen T, Kandror KV, Hsu VW. An ACAP1-containing clathrin coat complex for endocytic recycling. The Journal of Cell Biology. 2007b;178:453–464. doi: 10.1083/jcb.200608033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Lumb CN, He J, Xue Y, Stansfeld PJ, Stahelin RV, Kutateladze TG, Sansom MS. Biophysical and computational studies of membrane penetration by the GRP1 pleckstrin homology domain. Structure. 2011;19:1338–1346. doi: 10.1016/j.str.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- Mim C, Cui H, Gawronski-Salerno, Joseph A, Frost A, Lyman E, Voth, Gregory A, Unger, Vinzenz M. Structural Basis of Membrane Bending by the N-BAR Protein Endophilin. Cell. 2012;149:137–145. doi: 10.1016/j.cell.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mim C, Unger VM. Membrane curvature and its generation by BAR proteins. Trends Biochem Sci. 2012;37:526–533. doi: 10.1016/j.tibs.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Mizuno N, Jao CC, Langen R, Steven AC. Multiple modes of endophilin-mediated conversion of lipid vesicles into coated tubes: implications for synaptic endocytosis. J Biol Chem. 2010;285:23351–23358. doi: 10.1074/jbc.M110.143776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. Journal of computational chemistry. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pykäläinen A, Boczkowska M, Zhao H, Saarikangas J, Rebowski G, Jansen M, Hakanen J, Koskela EV, Peränen J, Vihinen H, et al. Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures. Nature Structural & Molecular Biology. 2011 doi: 10.1038/nsmb.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylypenko O, Lundmark R, Rasmuson E, Carlsson SR, Rak A. The PX-BAR membrane-remodeling unit of sorting nexin 9. The EMBO Journal. 2007;26:4788–4800. doi: 10.1038/sj.emboj.7601889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebecchi MJ, Scarlata S. Pleckstrin homology domains: a common fold with diverse functions. Annu Rev Biophys Biomol Struct. 1998;27:503–528. doi: 10.1146/annurev.biophys.27.1.503. [DOI] [PubMed] [Google Scholar]
- Ross JA, Chen Y, Muller J, Barylko B, Wang L, Banks HB, Albanesi JP, Jameson DM. Dimeric endophilin A2 stimulates assembly and GTPase activity of dynamin 2. Biophysical journal. 2011;100:729–737. doi: 10.1016/j.bpj.2010.12.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints - Molecular-Dynamics of N-Alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen LR, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Shinozaki-Narikawa N, Kodama T, Shibasaki Y. Cooperation of Phosphoinositides and BAR Domain Proteins in Endosomal Tubulation. Traffic. 2006;7:1539–1550. doi: 10.1111/j.1600-0854.2006.00480.x. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. Journal of structural biology. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uson I, Sheldrick GM. Advances in direct methods for protein crystallography. Curr Opin Struct Biol. 1999;9:643–648. doi: 10.1016/s0959-440x(99)00020-2. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- Yao DQ, Huang S, Wang JW, Gu YX, Zheng CD, Fan HF, Watanabe N, Tanaka I. SAD phasing by OASIS-2004: case studies of dual-space fragment extension. Acta Crystallogr D. 2006;62:883–890. doi: 10.1107/S0907444906006445. [DOI] [PubMed] [Google Scholar]
- Yu H, Schulten K. Membrane sculpting by F-BAR domains studied by molecular dynamics simulations. PLoS computational biology. 2013;9:e1002892. doi: 10.1371/journal.pcbi.1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Wu LJ, Gu YX, Zheng CD, Fan HF. Combining SAD/SIR iteration and MR iteration in partial-model extension of proteins. Chinese Phys B. 2010;19 [Google Scholar]
- Zhu G, Chen J, Liu J, Brunzelle JS, Huang B, Wakeham N, Terzyan S, Li X, Rao Z, Li G, et al. Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5. The EMBO Journal. 2007;26:3484–3493. doi: 10.1038/sj.emboj.7601771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01