Interferon-Stimulated Genes: A Complex Web of Host Defenses (original) (raw)
. Author manuscript; available in PMC: 2015 Feb 6.
Abstract
Interferon-stimulated gene (ISG) products take on a number of diverse roles. Collectively, they are highly effective at resisting and controlling pathogens. In this review, we begin by introducing interferon (IFN) and the JAK-STAT signaling pathway to highlight features that impact ISG production. Next, we describe ways in which ISGs both enhance innate pathogen-sensing capabilities and negatively regulate signaling through the JAK-STAT pathway. Several ISGs that directly inhibit virus infection are described with an emphasis on those that impact early and late stages of the virus life cycle. Finally, we describe ongoing efforts to identify and characterize antiviral ISGs, and we provide a forward-looking perspective on the ISG landscape.
Keywords: innate immunity, pathogen recognition, desensitization, antiviral effectors
INTRODUCTION
The interferon (IFN)-mediated innate immune response, selected by evolution, is hardwired within genomes and provides a robust first line of defense against invading pathogens. Following pathogen detection and subsequent IFN production, IFN molecules bind to cell surface receptors and initiate a signaling cascade through the Janus kinase signal transducer and activator of transcription (JAK-STAT) pathway, leading to the transcriptional regulation of hundreds of IFN-regulated genes (IRGs) (reviewed in 1). This leads to a remarkable antiviral state, effective against positive-, negative-, and double-stranded RNA viruses, DNA viruses, and intracellular bacteria and parasites. IFN signaling also plays an important role in shaping the adaptive immune response (reviewed in 2, 3).
Through years of dedicated effort, investigators have described the mediators of signal transduction and the DNA response elements involved in JAK-STAT signaling, yet the functions of only a handful of IRGs have been studied in detail. Investigation into the mechanisms of IRGs has begun to elucidate how the IFN-induced state reprograms cellular biology to prime cells for enhanced pathogen detection, enables effective pathogen defense, and allows cells to recover to normal function. In addition, aberrant IFN signatures have been found in a variety of autoimmune disorders, thereby highlighting the importance of maintaining strict control over IFN signaling (reviewed in 4).
In this review, we begin by providing a brief historical perspective on the discovery of IFN and describe critical features of the JAK-STAT signaling pathway. In describing these features, we highlight ways in which complexity can arise from differences in the endogenous levels of signaling pathway components and IFN properties. These differences may ultimately lead to variation in the nature and number of genes that are transcriptionally up- and downregulated and, as a result, lead to distinct biological outcomes. In this review, we focus primarily on the upregulated, IFN-stimulated genes (ISGs). We describe ISGs that positively and negatively regulate type I and III IFN signaling along with those that directly inhibit virus infection. A number of reviews have been written describing individual ISGs. Here, we describe several ISGs, emphasizing those that impact the early and late stages of the virus life cycle. Furthermore, we touch upon screening efforts to identify ISGs that are important for antiviral activity and upon the impact that a better understanding of ISG biology will have on future antiviral therapies. We conclude by discussing how contemporary technologies will influence our knowledge of the ISG landscape and how new insights are challenging the classical definitions of ISGs.
A BRIEF HISTORY OF INTERFERON
By the early 1950s, it was well established that under certain conditions virus-infected cells are resistant to a second virus infection. Therefore, by some mechanism, viruses interfere with each other. Similarly, it had been demonstrated that inactivated influenza virus is capable of interfering with live influenza virus, but at the time the agent or substance responsible was a mystery (reviewed in 5). In 1957, the term “interferon” was coined by Isaacs & Lindenmann (6, 7) to describe a substance, likely produced by cells, that interferes with influenza infection. IFN was later shown to be a small protein, produced and secreted by cells (reviewed in 8) following cellular detection of pathogen-associated molecular patterns, commonly known as PAMPs, by pattern-recognition receptors (PRRs; reviewed in this volume; see Reference 9).
In the years following these initial discoveries, crude protein fractions obtained from the medium of stimulated human (or nonhuman primate) white blood cells served as a source of IFN (10, 11). It was nearly two decades after the initial description of IFN before methods were developed to allow sufficient purification and more rigorous characterization of IFN’s properties (reviewed in 12). As purification schemes were being refined, it became apparent that IFN was not one, but a family of distinct proteins that fall into three discrete classes differentiated by their receptor complexes (13–15). Signaling triggered by all three IFN types—as well as subtypes—gives rise to important and distinct outcomes that vary with respect to ISG profiles, induction kinetics, antiviral and antiproliferative activity, and immunomodulatory potential. Type I IFNs (α and β) are currently approved for treating a variety of diseases including chronic hepatitis B and C virus infection, multiple sclerosis, and melanoma.
INTERFERONS AND THEIR RECEPTORS
IFN signaling complexes contain two unique receptor chains, one with low affinity and one with high affinity for IFN binding. Several reviews have been dedicated to IFNs and their receptors (16–18). Here, we provide a brief overview.
Type I IFNs and IFNARs
Type I IFNs constitute the largest IFN class. In humans, this class comprises IFN-α, IFN-β, IFN-ε, IFN-κ, and IFN-ω, all of which are clustered on chromosome 9 and signal through the type I IFN heterodimeric receptor complex comprising IFN-α receptor 1 (IFNAR1) and IFNAR2 subunits (Figure 1). Each type I IFN is encoded by a single gene with the exception of IFN-α, which comprises 13 subtypes in human. Nearly every cell is capable of producing IFN-α/β; however, during the course of an infection, specialized immune cells known as plasmacytoid dendritic cells produce the vast majority of IFN-α (19; reviewed in 20). The various type I IFNs display differential tissue expression and binding affinities for the IFNAR1/2 receptor complex (reviewed in 12, 17), and consequently, the distinct subtypes give rise to various outcomes with respect to antiviral, antiproliferative, and immunomodulatory activity (21–23).
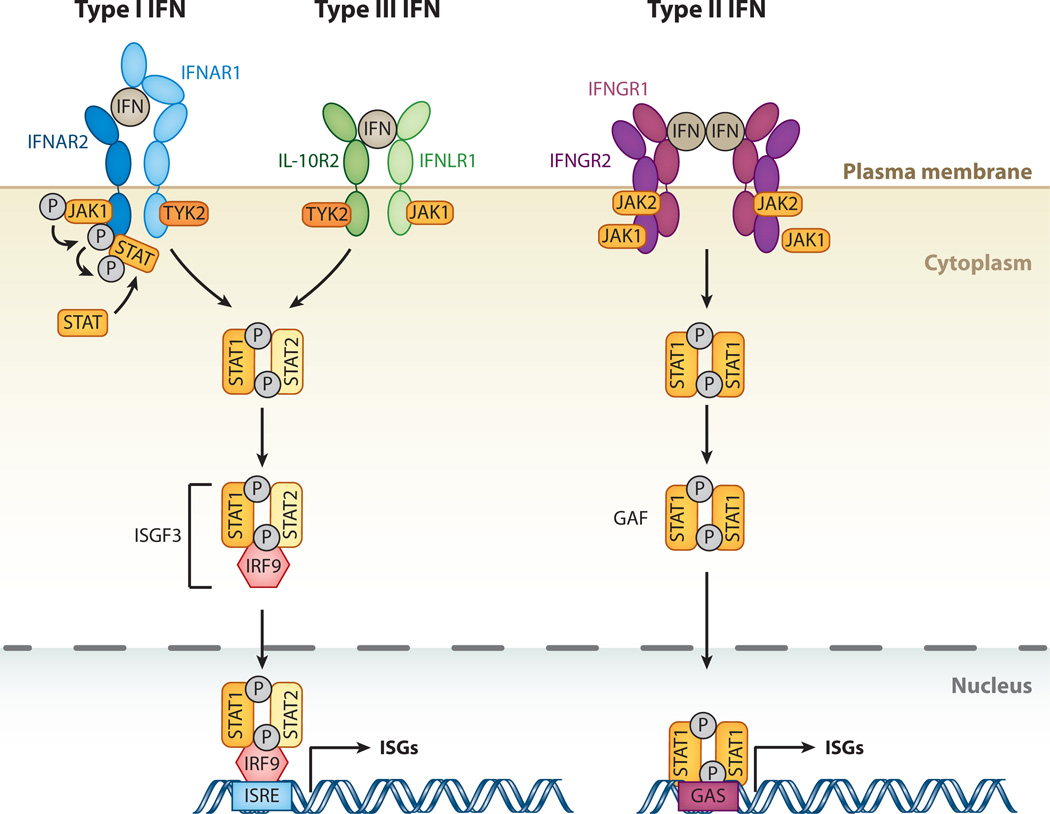
Figure 1.
The interferon (IFN)-signaling cascade. The three different classes of IFNs signal through distinct receptor complexes on the cell surface: type I IFNs act through IFN-α receptor 1 (IFNAR1) and 2 (IFNAR2) heterodimers; type III IFN through interleukin-10 receptor 2 (IL-10R2) and IFN-λ receptor 1 (IFNLR1) heterodimers; and type II IFN through dimers of heterodimers consisting of IFN-γ receptors 1 (IFNGR1) and 2 (IFNGR2). Binding of both type I and type III IFNs to their IFNAR1/2 or IL-10R2/IFNLR1 complexes, respectively, triggers phosphorylation of preassociated Janus kinase 1 (JAK1) and tyrosine kinase 2 (TYK2), which in turn phosphorylate the receptors at specific intracellular tyrosine residues. This leads to the recruitment and phosphorylation of signal transducers and activators of transcription 1 and 2 (STAT1 and 2). STAT1 and 2 associate to form a heterodimer, which in turn recruits the IFN-regulatory factor 9 (IRF9) to form the IFN-stimulated gene factor 3 (ISGF3). Binding of type II IFN dimers to the IFNGR1/2 complex leads to phosphorylation of preassociated JAK1 and JAK2 tyrosine kinases, and transphosphorylation of the receptor chains leads to recruitment and phosphorylation of STAT1. Phosphorylated STAT1 homodimers form the IFN-γ activation factor (GAF). Both ISGF3 and GAF translocate to the nucleus to induce genes regulated by IFN-stimulated response elements (ISRE) and gamma-activated sequence (GAS) promoter elements, respectively, resulting in expression of antiviral genes.
Type II IFN and IFNGR
IFN-γ is the sole type II IFN. It forms a homodimer and signals through the IFN-γ receptor complex (IFNGR; Figure 1), initiated by interaction with two IFNGR1 subunits. This leads to binding of two additional IFNGR2 subunits and results in receptor activation (24). IFN-γ production is largely restricted to cells of the immune system (25); however, the IFNGR1/2 proteins are widely expressed, and therefore nearly all cell types are capable of responding to IFN-γ (25). The signaling that results plays a major role in establishing cellular immunity, and it also induces gene products that prime the type I IFN response (26–28). Likewise, type I IFN signaling primes IFN-γ signaling as well (29, 30). Overall, IFN-γ plays a pivotal role in regulating immune function and bridging the innate and adaptive responses, and therefore it deserves much more attention than can be accommodated within the scope of this review. For a more thorough description of IFN-γ function, the reader is referred to recent reviews (31–33).
Type III IFN, IL-10, and IFNLR
The type III IFNs—IFNL1, IFNL2, and IFNL3 [also known as IFN-λ1, IFN-λ2, and IFN-λ3, or interleukin (IL)-29, IL-28A, and IL-28B, respectively]—were described independently by two research groups in 2003, making them the most recently discovered members of the IFN family (34, 35). A fourth member of the family, IFNL4, has also been recently described (36, 37). These IFNs share structural features with members of the IL-10 cytokine family and utilize the same broadly distributed low-affinity receptor subunit (IL-10R2) as the cytokines IL-10, IL-22, and IL-26 (Figure 1). In contrast, the high-affinity type III IFN receptor subunit (IFN-λ receptor 1, IFNLR1) is uniquely utilized by the type III IFNs, and its expression is restricted to epithelial cells (38). This receptor complex signals through a similar JAK-STAT pathway as the type I IFN receptor complex and induces many of the same ISGs (39, 40).
INTERFERON SIGNALING: THE JAK-STAT PATHWAY
Upon IFN binding to cell surface receptors, a signal is transmitted through the membrane and into the cell, leading to dramatic changes in cellular properties. One of the most striking features of IFN signaling is the speed at which it occurs, made possible because the synthesis of new proteins is not required—the necessary components are present at baseline (41, 42).
All IFNs signal through the JAK-STAT signaling pathway (Figure 1). A remarkable combination of perspicacity and hard work led to the elucidation of this prototypic pathway, laying the groundwork for our current understanding of how extracellular events lead to transcriptional activation. Here we outline key events that drive ISG transcription. For a more thorough account of the events that occur during JAK-STAT signaling and the important discoveries that led to elucidation of the pathway, the reader is referred to a recent review (1).
Janus Kinases (JAKs)
Three of the four known JAKs [JAK1, JAK2, and tyrosine kinase 2 (TYK2)] are ubiquitously expressed, and all three function in IFN signaling (43). They bind to receptor chains on the inner side of the membrane and provide receptors with stability, facilitate their cell surface localization, and serve as key components of signaling complexes (44–46). In any given cell, various distinct receptor chains compete for the same JAK protein, and as a result limited JAK concentration is an important determinant of cell surface receptor levels and signaling potential (reviewed in 47).
In the absence of a stimulus, the cytoplasmic domain of each IFN receptor chain is bound by a specific JAK protein in an inactive conformation. Upon IFN binding, receptor chains are brought into close proximity, and the two JAK kinase domains juxtapose and undergo transphosphorylation and sustained activation. Juxtaposition, however, is not sufficient—ligand-induced structural changes are required for activation, and information in the form of biochemical and biophysical differences among the various IFNs is likely transmitted through the membrane as structural changes in the receptor chains. This mechanism for transmitting information may at least partly explain how various type I IFNs can give rise to qualitatively distinct biological outcomes (reviewed in 48).
Once activated, JAKs phosphorylate IFN receptor chains on highly conserved tyrosine residues, which leads to the repositioning or binding of STAT proteins via Src homology 2(SH2) domain interactions (49). As a result, STATs are phosphorylated on conserved tyrosine residues and released from the receptor, where conformational changes lead to homo- (type II IFN) or heterodimerization (type I and III IFN). This change exposes a nuclear localization signal that facilitates nuclear translocation (Figure 1) (50–56). Once in the nucleus, STATs serve as transcriptional activators that drive ISG expression.
Signal Transducers and Activators of Transcription (STATs)
There are seven STAT proteins in mammals, STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, and STAT6 (reviewed in 57), all of which play some role in innate immune signaling. However, when it comes to signaling through any of the IFN receptor complexes, STAT1 and STAT2 are the most important.
Type II IFN signaling entails phosphorylation of STAT1 on tyrosine 701 (50, 58), followed by homodimerization, nuclear translocation, and DNA binding at gamma-activated sequence (GAS) elements upstream of IFN-γ-induced genes (Figure 1) (59). These events result in transcriptional activation of IFN-γ-induced genes (reviewed in 60). Type I and III IFN signaling leads to phosphorylation of both STAT1 and STAT2, which leads to heterodimerization and interaction with IFN regulatory factor (IRF) 9 (formerly termed p48), forming the ISG factor 3 (ISGF3) complex (Figure 1) (61–63). ISGF3 then translocates to the nucleus, where it binds IFN-stimulated regulatory elements (ISREs) in the DNA upstream of ISGs, resulting in the transcription of hundreds of type I and III ISGs (64–66).
The above description of STAT activation pathways is simplified for the purposes of this review but provides a framework in which one can begin to glimpse the complexity of the IFN response. Many cell types respond to IFN with varying transcriptional responses, and of course in vivo IFN signaling does not occur in isolation. Furthermore, unphosphorylated STATs can prolong the induction of a subset of ISGs (67, 68), and an increasing number of reports suggest a role for additional STAT proteins in the IFN response. Clearly, the simplified model of JAK-STAT signaling presented here is only part of a much larger sequence of events leading to ISG induction.
INTERFERON-STIMULATED GENES
ISGs take on a wide range of activities. PRRs, IRFs, and several signal transducing proteins described above such as JAK2, STAT1/2, and IRF9 are present at baseline but are also ISGs and reinforce the IFN response. Many ISGs control viral, bacterial, and parasite infection by directly targeting pathways and functions required during pathogen life cycles. Upregulation of chemokines and chemokine receptors enables cell-to-cell communication, whereas negative regulators of signaling help resolve the IFN-induced state and facilitate the return to cellular homeostasis. Additional ISGs encode for proapoptotic proteins, leading to cell death under certain conditions. To provide an overview of ISG function, here we highlight a number of ISG activities using well-known and recently discovered examples.
IFN-Induced Pathogen-Sensing Sensitization
PRRs and many IRFs are present in cells at baseline, but their gene expression is enhanced by IFN. Upon induction, this set of ISGs acts to reinforce IFN signaling and prime cells for enhanced pathogen detection. Subsets of ISGs are induced directly by IRF activation in a pathway that is independent of the JAK-STAT pathway, and this alternative pathway of ISG induction likely evolved to counteract pathogen-mediated innate immune evasion strategies (Figure 2) (reviewed in Reference 69 and, in this volume, in Reference 9).
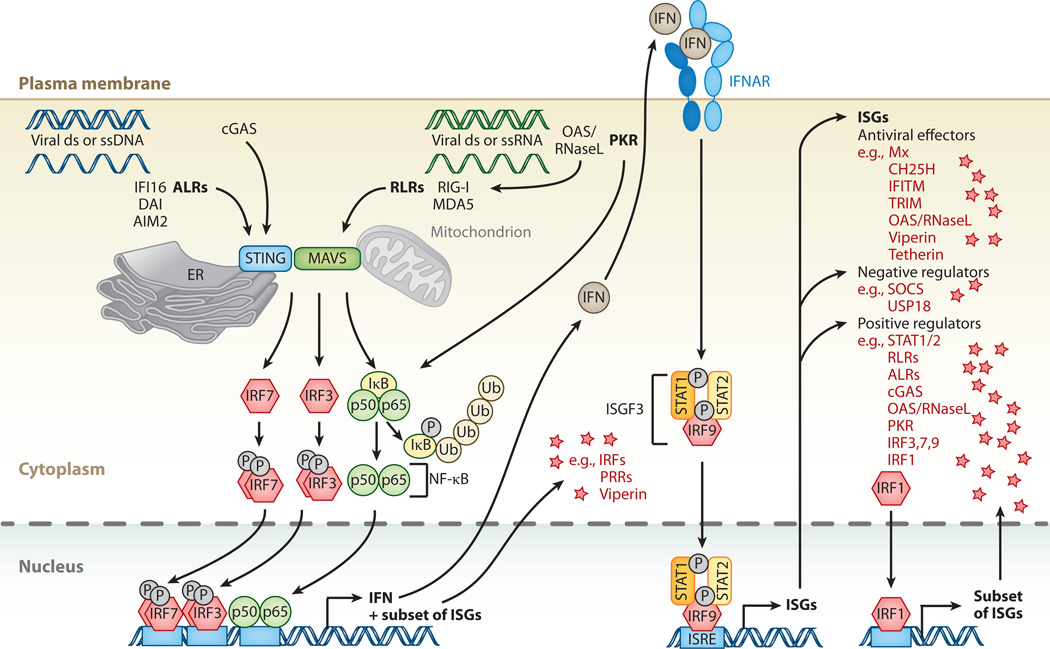
Figure 2.
Cytosolic nucleic acid pattern recognition and activation of ISGs. Cytosolic pattern-recognition receptors (PRRs) recognize viral double-stranded (ds) or single-stranded (ss) DNA or RNA. AIM2-like receptors (ALRs), such as IFI16, DAI, or AIM2 itself, specialize in DNA detection, whereas RIG-I-like receptors (RLR)—RIG-I and MDA5—specialize in RNA detection. Cyclic GMP-AMP synthase acts as an additional DNA sensor. 2′-5′-oligoadenylate synthetase (OAS) senses foreign RNA and produces 2′-5′ adenylic acid, which activates latent RNase (RNaseL). Degradation products produced by RNaseL further stimulate RLRs. Protein kinase R (PKR) is an additional sensor for foreign RNA. PRR signals are transduced to transcription factor activity by stimulator of IFN genes (STING) and mitochondrial antiviral-signaling protein (MAVS) at the ER/mitochondrion-associated membrane. Activation of STING/MAVS leads to phosphorylation of interferon (IFN) response factors 3 or 7 (IRF3/7), or to phosphorylation and ubiquitin-mediated degradation of IκB. Phosphorylated dimers of IRF3/7 or NF-κB translocate to the nucleus, where they bind to and activate specific promoters, triggering expression of IFN as well as a subset of ISGs. These ISGs include IRFs and PRRs but also antiviral effectors such as viperin. IFN induces gene expression via the JAK-STAT pathway, resulting in expression of a large spectrum of ISGs that can be divided into antiviral effectors and negative or positive regulators of IFN signaling. A special case of positive regulators is IRF1, which upon expression directly translocates to the nucleus to enhance expression of a subset of ISGs.
There are a number of PRRs, each specializing in the detection of distinct PAMPs (reviewed in 69, 70). Retinoic acid-inducible gene 1 (RIG-I)-like receptors (RLRs) recognize cytosolic double-stranded RNA and 5'pppRNA (71, 72), whereas AIM2-like receptors (ALRs) recognize cytosolic DNA (73; reviewed in 74). Nucleotide-binding oligomerization domain-like receptors (NLRs) recognize a number of cytosolic PAMPs produced largely by bacteria (reviewed in 75). For example, NLRs recognize flagellin, lipopolysaccharide (LPS), peptidoglycans, bacterial toxins, and bacterial and viral nucleic acids. Toll-like receptors (TLRs) 1, 2, and 4 are transmembrane proteins that localize to the cell surface, where they detect viral glycoproteins and a variety of bacterial PAMPs, similar to those described for the NLRs above. Similarly, TLR3, 7, and 9 are also transmembrane proteins, but this group of receptors localizes in endosomes, where they specialize in the detection of virus and bacterial nucleic acids, similar to the cytosolic RLRs (reviewed in 76).
Foreign RNA is also detected by oligoadenylate synthetase (OAS) and latent endoribonuclease (RNaseL). Cytosolic PAMP detection by OAS leads to the synthesis of 2′-5′-oligoadenylates, which then act as intracellular second messengers to activate latent RNaseL. This leads to indiscriminate cleavage of both host and viral RNA and the production of additional PAMPs, which reinforce the innate immune response (Figure 2) (reviewed in 77). In addition to the OAS/RNaseL system, activation of protein kinase R (PKR) by cytosolic PAMPs leads to a dramatic reduction in host cell translation as well as to the degradation of the inhibitor of κB (IκB)—the latter results in activation of the NF-κB signaling pathway (reviewed in 78).
Adding to the family of PRRs, cyclic GMP-AMP (cGAMP) synthase (cGAS) was recently identified as a cytosolic PAMP sensor (79–82). By a mechanism that remains unclear, cytosolic detection of a PAMP—likely DNA—activates cGAS nucleotidyltransferase activity and leads to the production of cGAMP; this molecule binds to and activates the stimulator of IFN genes (STING) (83). STING is located on the ER, specifically at regions known as the mitochondrial-associated membrane, a critical hub for innate immune activation (reviewed in 84, 85). In addition to STING, the mitochondrial antiviral-signaling protein (MAVS, also known as VISA, IPS-1, or Cardif) (86–88) is also found in this membrane-rich region, where it interacts with the RLRs RIG-I and MDA5 to further propagate innate immune signaling (Figure 2).
Upon activation of STING and/or MAVS, IRF3 and IRF7 are phosphorylated. This results in their homodimerization and translocation to the nucleus. Similarly, activation of STING and/or MAVS triggers activation of the NF-κB pathway by inducing the phosphorylation and ubiquitination of IκB, leading to IκB degradation and exposure of nuclear localization signal sequences on NF-κB. Once in the nucleus, IRF3, IRF7, and NF-κB bind to specific binding sites in the IFN-β promoter as well as to the promoters of a subset of ISGs, including IRFs and PRRs. DNA binding then leads to transcriptional activation of IFN-β mRNA along with a subset of ISGs.
IFN stimulation enhances production of many IRFs and PRRs and sensitizes cells to pathogen detection; however, most positive regulators require additional activation to fulfill their signaling roles. For example, PRRs are inactive until PAMPs are detected, and IRF3 and IRF7 require phosphorylation for nuclear translocation and transcription-enhancing activity; canonical signaling by the STAT proteins requires phosphorylation as well. A notable exception in this class of ISGs is IRF1, which is capable of gene activation with no additional post-translational modification or signals (89).
Overall, a number of ISGs act to enhance pathogen detection and innate immune signaling. The likely reason that many of these proteins are present at low levels before IFN stimulation is to minimize the risk of aberrant signaling that may lead to inflammation, while at the same time allowing for PAMP detection. In addition to heightened pathogen sensing following IFN signaling, alternative pathways of ISG induction exist—at least for subsets of ISGs—and this mechanism of ISG induction likely evolved to control infection in situations in which canonical ISG induction is inhibited.
IFN Desensitization
The IFN response is tightly controlled, and shortly after IFN exposure, cultured cells enter an IFN-desensitized state that can last up to several days (41). This desensitized state allows cells to recover from IFN signaling, whereas dysregulation of IFN production and signaling manifests in autoimmune disorders such as systemic lupus erythematosus and Sjögren’s syndrome, among others (reviewed in 4). These are complex diseases with multiple etiologies; they nonetheless highlight the importance of negatively regulating the IFN response. The IFN-desensitized state is established in cells by multiple mechanisms; some are cell intrinsic and others are mediated by the actions of ISGs.
Cell-intrinsic desensitization
Receptor endocytosis and turnover play important roles in rapidly reducing the level of JAK-STAT signaling (Figure 3_a_) (reviewed in 90). Signaling is further decreased by the action of phosphatases that inactivate the JAKs and STATs (91–96). In addition, STAT activity can be modulated by the protein inhibitors of activated STAT (PIAS) family of proteins.
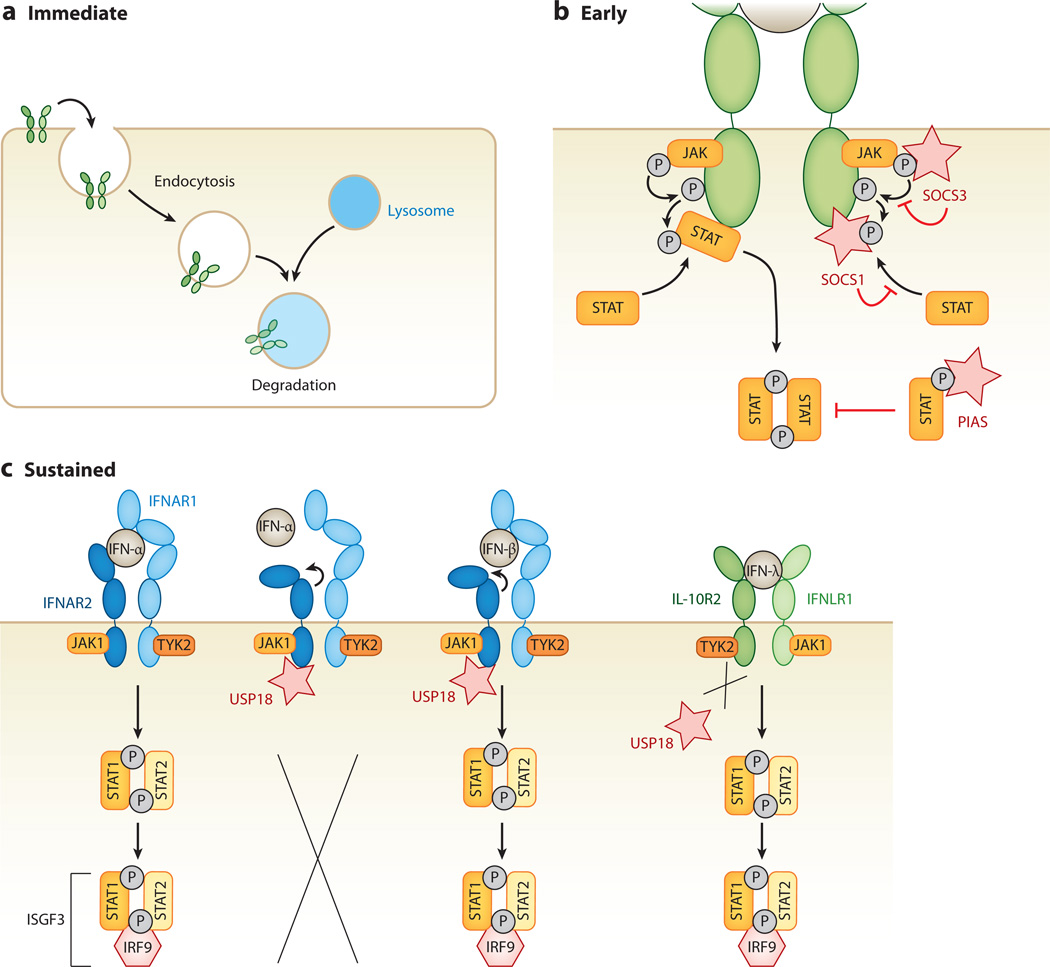
Figure 3.
Interferon (IFN) desensitization pathways. IFN signaling is negatively regulated by various mechanisms. (a) An immediate mechanism of IFN desensitization is endocytosis and turnover of IFN receptors, which rapidly reduces the level of JAK-STAT signaling within the cell. (b) Another early mechanism of IFN desensitization requires de novo synthesis of inhibitory proteins. IFN-stimulated suppressor of cytokine signaling (SOCS) proteins act as kinase inhibitors within the JAK-STAT phosphorylation cascade. Both SOCS1 and SOCS3 act as pseudosubstrates for receptor-associated JAKs. Protein inhibitors of activated STAT (PIAS) proteins bind to and inhibit phosphorylated STATs, thereby interrupting the signaling cascade. (c) The ubiquitin-specific peptidase 18 (USP18), expressed from an IFN-stimulated gene, leads to a more sustained shutdown of JAK-STAT signaling. USP18 binds to the intracellular side of the IFN-α receptor 2 (IFNAR2), resulting in conformational changes in the extracellular domains of IFNAR2, which keeps low-affinity IFNs such as IFN-α from binding, and hence from inducing, the JAK-STAT signaling cascade. In contrast, IFNs with higher receptor affinity, such as IFN-β, are still able to bind and initiate the signaling cascade. USP18 binding is specific to IFNAR2, and thus it does not interfere with type II or type III IFN signaling.
Despite years of research, the precise mechanisms by which PIAS proteins inhibit IFN signaling remain unclear. As a family, PIAS proteins have been found to regulate a wide range of transcription factors (both positively and negatively) through various molecular mechanisms (reviewed in 97, 98). One function is to provide specificity for the covalent attachment of small ubiquitin-like modifier (SUMO), in a process called SUMOylation, to protein targets in a process that mirrors the ubiquitination pathway. In addition, they can negatively regulate gene expression in a SUMO-independent manner by preventing the binding of transcription factors to DNA. As such, PIAS1 has been shown to inhibit STAT1 independently of SUMOylation (Figure 3_b_) (99); however, STAT1 proteins can be SUMOylated, and this modification impacts ISG induction as well (100, 101).
ISG-mediated desensitization
Inhibition of protein translation with cycloheximide extends the length of time that ISGs are transcribed following IFN treatment and decreases the interval of time necessary for reactivation, thereby demonstrating that new protein synthesis is required to establish and maintain an IFN-desensitized state (41). Here we describe two well-known ISGs that negatively regulate IFN signaling by inhibiting the JAK-STAT signaling pathway.
SOCS proteins
SOCS proteins inhibit JAK-STAT signaling by binding to phosphorylated tyrosine residues, on either the IFN receptors or the JAK proteins, resulting in inhibition of STAT binding as well as JAK activity (Figure 3_b_). Signaling is further decreased through the SOCS box domain present in the C terminus of all SOCS proteins. This domain recruits proteins involved in receptor ubiquitination and proteasome degradation.
SOCS proteins are induced early in the IFN response and play an important role in early IFN desensitization. Similarly, for some cells, in the absence of IFN signaling an elevated baseline level of SOCS1 protein expression renders cells less responsive to IFN (102). This reduced responsiveness is believed to be partly responsible for the weak response to type I IFN that results from IFN stimulation of human pluripotent stem cells (102). Similarly, it is possible that additional naturally occurring cell populations exist with elevated SOCS protein levels and reduced sensitivity to IFN signaling.
USP18
USP18 is arguably the ISG with the most important role in establishing and maintaining long-term desensitization to type I IFN signaling. For this reason, here we provide a detailed account of USP18 activity.
In a process known as ISGylation, a small IFN-induced ubiquitin-like protein (ISG15) is covalently attached to proteins through a series of steps that mirror the ubiquitination pathway; the removal of ISG15 conjugates (deISGylation) is performed by the isopeptidase activity of the ISG USP18 (103). Accordingly, _Usp18_−/− mice display a dramatic increase in the level of ISGylation (104, 105).
_Usp18_−/− mice are hypersensitive to type I IFN and more resistant to virus infection (106). In 2003, studies with _Usp18_−/− cells in culture revealed that JAK-STAT signaling is prolonged and that a dramatic increase in apoptosis occurs upon IFN-β treatment, suggesting that USP18 is important in negatively regulating the IFN response. It was unclear at the time whether this effect was at all related to protein ISGylation (105). However, it was later demonstrated that a catalytically inactive form of USP18 confers the same level of IFN desensitization as the wild-type protein (107). This isopeptidase-independent activity is mediated by the binding of USP18 to the intracellular domain of IFNAR2, which prevents the binding of JAK1. Mutation of arginine residues in USP18 that disrupt binding to IFNAR2 impairs its ability to inhibit IFN signaling (107). These results clearly demonstrated that the major role of USP18 in the inhibition of JAK-STAT signaling is independent of ISGylation (108–110).
The discovery that USP18 maintains long-term IFN desensitization through an interaction with IFNAR2 (Figure 3) suggests that USP18-mediated inhibition may be restricted to type I IFN signaling. Indeed, studies have shown that type III IFN signaling remains intact in the presence of USP18, but cells that are prestimulated with type I or III IFN are refractory to type I IFN signaling due to USP18 upregulation (111, 112). These studies have also found that USP18-mediated desensitization to type I IFN is differential—IFN-α signaling is blocked, whereas IFN-β continues to activate the JAK-STAT pathway (111, 112). These findings seem to conflict with the initial studies of USP18-mediated IFN densensitization in which IFN-β was employed (103, 105, 107).
In the initial experiments demonstrating IFN desensitization, ISGF3 gel shift assays and STAT1 phosphorylation levels indicated that the presence of IFN-β failed to result in prolonged JAK-STAT signaling in USP18-expressing cells; in contrast, signaling was maintained in _Usp18_−/− cells (105). These data suggest that although IFN-β may be less sensitive to USP18-mediated desensitization, its ability to signal through the JAK-STAT pathway is nonetheless affected. More recent data also show that in some cell types, the second round of IFN-β stimulation is less robust than the first (111). Thus, cell type differences may be important factors in determining the degree to which a cell is refractory to IFN-β.
The differential sensitivity of different type I IFNs to USP18-mediated inhibition raises important mechanistic questions and invokes a long-standing puzzle in the field: How do different IFN proteins that signal through the same receptor give rise to varied outcomes? As highlighted above in the JAK section, differential binding affinities by the IFNs themselves lead to altered receptor conformations that are propagated through the membrane and may alter ISG induction profiles (18, 21, 23, 113). In the case of USP18 binding, the converse would be true: Changes in receptor conformations on the cytoplasmic face of the membrane may affect IFN binding on the surface. This model is supported by recent data documenting the differential sensitivity of IFN-α and IFN-β to USP18-mediated desensitization; however, the precise molecular basis for this difference remains uncertain.
Moving forward, it will be important to determine whether a low (even undetectable) level of signaling by IFN-α is sufficient to maintain USP18 expression and sustained IFN desensitization. This question is of clinical importance because elevated levels of USP18 mRNA predict a poor treatment response to IFN-α therapy for hepatitis C virus (HCV) infection (114, 115). A more basic understanding of the ISG-based mechanisms involved in desensitization is necessary to better understand what occurs in cells under conditions of prolonged IFN stimulus, such as autoimmune disorders, chronic viral infection, and IFN-based therapy.
Antiviral Effectors
For many years, IFN-based therapies have been used to treat chronic hepatitis B and C virus infection, and a better understanding of ISG function will shed light on the underlying biology influencing treatment outcomes. Additionally, developing an arsenal of tools to combat acute and emerging viral infections is of high interest. In this regard, identifying and characterizing direct-acting antiviral effector ISGs have the potential to uncover evolutionarily selected mechanisms of pathogen defense that can be mimicked or manipulated to create novel therapies.
To complete their life cycle, viruses must enter cells, translate and replicate their genomes, and exit in order to infect new cells. Every stage of the virus life cycle is a potential target for ISG intervention, and indeed, there are examples of ISGs targeting each one. Here, we focus on the antiviral mechanism of several recently characterized ISGs, highlighting those affecting early and late stages of infection (Figure 4).
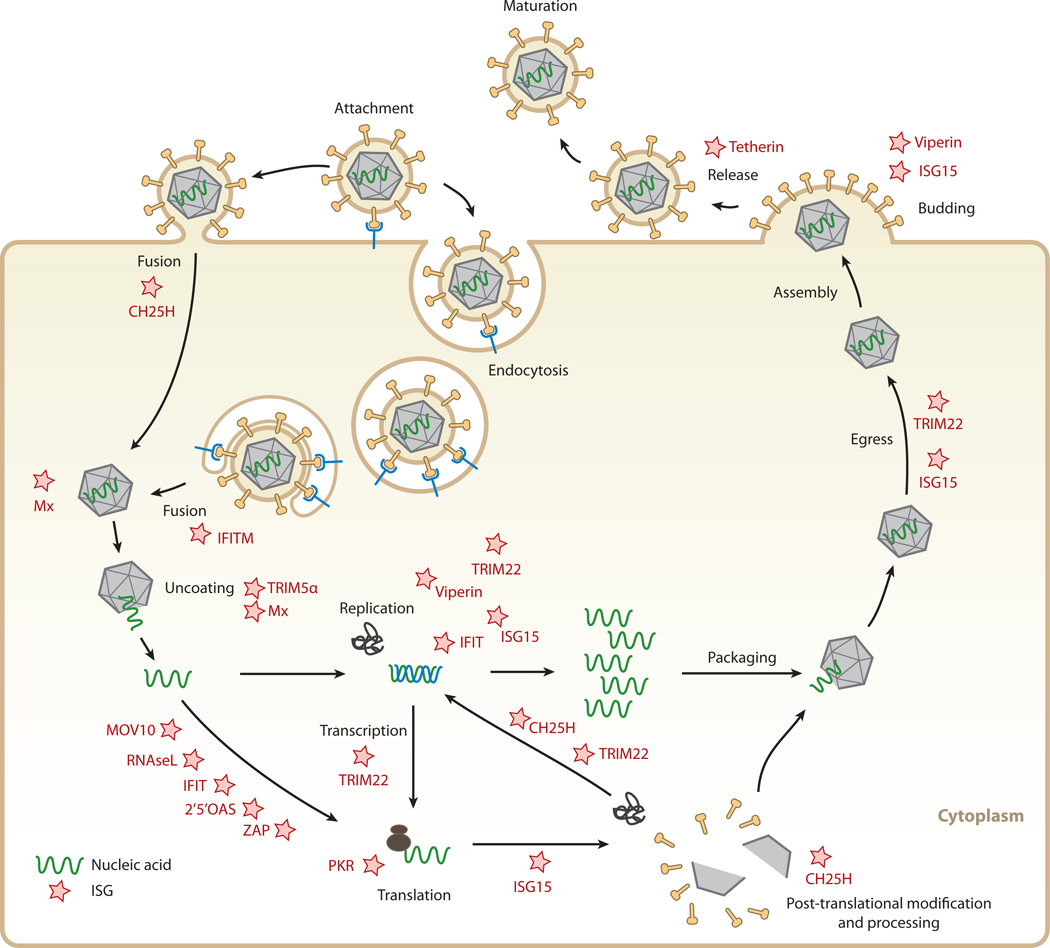
Figure 4.
Targets for interferon (IFN)-stimulated proteins within viral life cycles. IFN-stimulated gene (ISG) products (stars) interfere with different stages of different viral life cycles. Cholesterol-25-hydroxylase (CH25H) affects viruses early, presumably at the host-membrane fusion event; at protein maturation of viral structural proteins by prenylation; and at protein maturation of viral replication enzymes. IFN-induced transmembrane (IFITM) protein members inhibit endocytic-fusion events of a broad spectrum of viruses. Tripartite motif protein 5 α (TRIM5 α) inhibits human immunodeficiency virus 1 (HIV-1) uncoating of the viral RNA. The myxoma resistance protein 1 (Mx1) inhibits a wide range of viruses by blocking endocytic traffic of incoming virus particles and uncoating of ribonucleocapsids. Some ISGs inhibit viruses by degrading viral RNA and/or blocking translation of viral mRNAs, such as 2′,5′-oligoadenylate synthetase (OAS) and latent ribonuclease L (RNase L), protein kinase R (PKR), Moloney leukemia virus 10 homolog (MOV10), and zinc-finger antiviral protein (ZAP). IFN-induced proteins with tetratricopeptide repeats (IFIT) inhibit protein translation and have been implicated in viral RNA degradation as well. TRIM22 inhibits viral transcription, replication, or trafficking of viral proteins to the plasma membrane. ISG15 can inhibit viral translation, replication, or egress. Viperin has been shown to inhibit viral replication or virus budding at the plasma membrane. Finally, tetherin traps otherwise mature virus particles on the plasma membrane and thus inhibits viral release, exerting its effect broadly on many enveloped viruses.
Inhibition of virus entry
Here, we describe several mechanisms by which ISGs affect virus entry into cells.
Myxovirus resistance (Mx)
The murine myxovirus resistance 1 (Mx1) gene product was one of the first described inhibitors of virus entry. Human cells express two Mx proteins, Mx1 and Mx2 (also known as human MxA and MxB, respectively). These two IFN-induced proteins belong to a small family of dynamin-like large guanosine triphosphatases (GTPases), which is closely related to the dynamin GTPase family.
Mx1 is broadly inhibitory and acts prior to genome replication at an early postentry step of the virus life cycle. Evidence suggests that Mx1 traps incoming viral components, such as nucleocapsids, and prevents them from reaching their cellular destination. The Mx1 structure was recently solved, providing new insight into its mode of action (116). Like dynamin GTPases, Mx1 contains a middle stalk domain and a GTPase effector domain, which are both essential for self-oligomerization and formation of ring-like structures that bind to and impose conformational changes on interaction partners (117). For Mx1, formation of these structures is important for its antiviral activity, as mutations that abolish self-oligomerization result in the loss of antiviral activity against LaCrosse and influenza A viruses (116). In the current model of Mx1-mediated inhibition, viral nucleocapsids are surrounded by Mx1 oligomer rings, and the resulting ring-ring interactions stimulate GTPase activity, which possibly directs them to sites of degradation. The viral structures targeted by Mx1 and the precise mechanism of antiviral activity have yet to be fully defined (reviewed in 118, 119).
Mx2 has been recently characterized as an antiretroviral effector protein (120–122). Overexpression of Mx2 potently inhibits HIV-1 and HIV-2 (120–122) but has less potent or no antiviral activity against other retrovirus family members (120, 121) or the orthomyxovirus influenza A (122). Indeed, the presence of Mx2 is necessary for full antiviral potency of IFN-α against HIV-1 (120–122). Mx2 acts at the level of nuclear entry and keeps the reverse-transcribed genome from reaching its nuclear destination, thereby ultimately inhibiting chromosomal integration, which is a key event of the HIV-1 replication cycle (120–122). Intriguingly, mutations in the HIV-1 capsid protein render the virus resistant to Mx2-mediated inhibition, suggesting that Mx2 specifically inhibits the capsid’s function in nuclear entry (120–122).
Cholesterol-25-hydroxylase (CH25H)
Expression of the CH25H gene is upregulated by both type I and type II IFNs (123–125). The protein product CH25H is an enzyme that converts cholesterol into 25-hydroxycholesterol (25HC). Treating cells directly with 25HC or transferring supernatants from CH25H-expressing cells protects against infection by a diverse set of enveloped viruses yet has no impact on infection by adenovirus, a nonenveloped virus (126, 127). These findings suggest that CH25H-mediated protection occurs at an early step in the infectious cycle, possibly at the step of virus-host membrane fusion. However, 25HC may impact virus infection by additional mechanisms as well.
Oxysterols, including 25HC produced by CH25H, have long been implicated in innate immunity, but the mechanisms by which they act are unclear (128–131). Recently, investigators have proposed that changes in the physical properties of membranes resulting from high concentrations of 25HC preclude virus-host membrane fusion (126). Alternatively, the antiviral activity of 25HC may partly result from its involvement in regulation of the sterol biosynthesis pathway.
Genes involved in sterol biosynthesis contain sterol regulatory elements (SRE)in their promoters that are recognized by transcription factors, designated as SRE-binding proteins (SREBPs). SREBP levels are tightly controlled by products of the sterol biosynthesis pathway through a negative feedback mechanism; sterol-replete conditions lead to an accumulation of 25HC, inhibiting sterol biosynthesis. Owing to its ability to permeate membranes, 25HC can inhibit sterol biosynthesis in both an autocrine and paracrine manner (reviewed in 132). In addition to generating cholesterol and 25HC, the sterol biosynthesis pathway also generates isoprenoids, such as farnesol and geranylgeraniol, critical for protein prenylation—a modification known to affect numerous viral and cellular proteins (including ISGs) (133–135). Indeed, protein prenylation plays a critical role in the life cycle of several viruses. For example, hepatitis delta virus large antigen is modified by prenylation, and preventing this modification abolishes infectious particle production (136–138). Similarly, for HCV infection, geranylgeranylation of a host protein (Fox-box and leucine-rich repeat protein 2, known as FBL2) is required for replication (139). 25HC also inhibits the replication of HCV subgenomic viral RNA (virus replicon), thereby displaying antiviral activity even in the absence of membrane fusion (139–142). Thus, an increased production of 25HC may affect virus infection by multiple mechanisms, including, but not exclusively restricted, to viral entry inhibition.
Liu et al. (126) recently proposed that 25HC can directly block membrane fusion by altering cellular membranes, and they speculate that this may be due to membrane expansion or aggregation. Several enveloped viruses were tested in this study, and in all cases the cellular protection by 25HC occurred at an early step. Overexpression of individual SREBPs or the addition of intermediates in the sterol biosynthesis pathway such as mevalonate did not rescue 25HC-mediated virus inhibition. These data are consistent with the idea that 25HC blocks membrane fusion; however, results from an additional study by Blanc et al. (127) suggest that 25HC may inhibit viruses by alternative mechanisms.
In these studies, both groups tested 25HC effects on herpes simplex virus 1 (HSV-1) and found that inhibition occurs at an early step in the virus life cycle (126, 127). However, Blanc et al. (127) also tested an additional herpesvirus—murine cytomegalovirus (MCMV)—and found that inhibition occurs at a postentry step (and thus post-membrane fusion), at or prior to viral DNA replication. Using the enantiomer of 25HC (ent-25HC), which is expected to affect membrane properties similar to 25HC but to lose the conformation-specific interaction with proteins necessary for sterol biosynthesis inhibition, Blanc et al. (127) found that higher concentrations of ent-25HC are required to achieve the same level of MCMV inhibition seen with 25HC. This provides evidence that negative feedback of the sterol biosynthesis pathway may be involved in the inhibition of some viruses by 25HC.
It is likely that 25HC exerts its antiviral effects by multiple mechanisms—altering membrane properties directly, inhibiting sterol biosynthesis through negative feedback, and affecting preny-lation of both virus and host proteins. Teasing these apart will require careful comparisons of different 25HC concentrations under various conditions as well as targeted strategies to disrupt sterol biosynthesis. Different viruses will likely vary in their susceptibility to each of these mechanisms.
IFITM proteins
With the possible exception of CH25H, the only ISGs shown to have a bona fide role in blocking virus entry are members of the IFN-inducible transmembrane (IFITM) family. In humans, the IFITM family of proteins is composed of four members, IFITM1, IFITM2, IFITM3, and IFITM5. These proteins were recently shown to be potent inhibitors of influenza A virus infection in a loss-of-function screen (143). Since then, they have been shown to inhibit a diverse range of viruses, and this broad antiviral activity strongly suggests that inhibition is not at the level of receptor engagement (89, 144–148).
IFITM proteins are enriched in late endosomes and lysosomes. Consistent with this, viruses most affected by IFITM expression are those that require transit to these compartments for productive entry (144, 149). Although this appears to be a common feature of virus restriction, IFITM family members display selectivity in the viruses they inhibit. IFITM1 inhibits SARS-coronavirus (CoV) and the filoviruses, Ebola and Marburg, with greater efficiency than does IFITM3 (144). IFITM3, on the other hand, has higher potency against influenza A virus. In contrast, HCV infection is inhibited by IFITM1, but not by IFITM3 (145). IFITM-mediated inhibition of human immunodeficiency virus 1 (HIV-1) is slightly more complex. IFITM1 has no effect on HIV-1 entry, but it effectively inhibits virus production (147). IFITM2 and IFITM3 also inhibit HIV-1 virus production, but they affect entry as well (147).
The varying profile of virus restriction among the IFITM proteins may result in part from differences in cellular localization. IFITM3 is modified by S-palmitoylation and ubiquitination, possibly influencing membrane topology and localization (150, 151). Furthermore, the membrane topology of IFITM proteins may be dynamic (i.e., influenced by binding partners) and may differ among family members. Relative to IFITM3, the IFITM1 protein has a shorter N-terminal region and displays a different pattern of localization. When 21 amino acids are deleted from the N terminus of IFITM3, its association with endosomes is lost and it localizes to the periphery of the cell (152). The biological significance of these findings is highlighted by results from a genome-wide association study that compared a control population to patients hospitalized for severe symptoms associated with influenza infection. This study uncovered a statistically significant enrichment for a polymorphism in the IFITM3 gene (153). Interestingly, this polymorphism generates an altered splice acceptor site resulting in an IFITM3 protein lacking 21 amino acids from the N terminus. The resulting mislocalization of IFITM3 and the correspondingly compromised antiviral activity provide a plausible biological mechanism for the association of this polymorphism with severe disease.
Although the emerging theme of IFITM inhibition is that restriction occurs at late endosomes and lysosomes, the actual mechanism is unclear. IFITM3 has recently been shown to restrict infection with reovirus, a nonenveloped virus (146). This finding is surprising and may help refine or expand current models of antiviral action. One model suggests that IFITM proteins alter the kinetics of endosome acidification, potentially increasing nonspecific protease activity or otherwise perturbing the normal sequence of events required for productive viral entry. This could explain the inhibition of both enveloped and nonenveloped viruses. Alternatively, IFITM proteins may affect virus entry by inhibiting steps prior to membrane hemifusion, potentially through physical changes in membrane properties such as curvature and fluidity (154). Neither reovirus nor other nonenveloped viruses undergo membrane fusion; however, the membrane must be traversed, and physical changes in membrane properties could impact this process. More work is required to elucidate the precise mechanism(s) by which IFITM proteins function. For additional reading on IFITM proteins, the reader is referred to a recent review (155).
TRIM proteins
The tripartite motif (TRIM) family of proteins is large—composed of more than 60 members in humans and mice—and exhibits a wide range of activities including E3 ubiquitin ligase activity (reviewed in 156). A defining feature of this family is the presence of an N-terminal RING domain, followed by one or two B-box domains and a C-terminal coiled-coil domain. The RING domain and B-box domains serve as a platform for protein-protein interactions and are important for TRIM protein roles as E3 ubiquitin ligases. However, TRIM protein functions are not limited to protein ubiquitination, and like many E3 ligases they can be involved in SUMOylation (157) and ISGylation as well (158). The C-terminal coiled-coil domain is involved in self-association and can lead to the formation of large complexes that occupy many discrete and uncharacterized subcellular compartments (159). Individual TRIM proteins are classified based on additional subdomains present in the C terminus, including the PRY subdomain, the SPRY subdomain, and a fusion of the two, PRYSPRY, also known as the B30.2 subdomain. Together, these subdomains are responsible for conferring additional mechanisms of target specificity for protein-protein interaction (156).
One of the best-studied members of the TRIM family, TRIM5α, was originally identified as a potent inhibitor of early stages in HIV-1 infection (160). Upon entry into cells, TRIM5α binds directly to viral capsid proteins, leading to accelerated disassembly of the capsid shell (uncoating) and premature exposure of the nucleoprotein complex known as the retroviral preintegration complex. TRIM5α is autoubiquitinated and rapidly turned over, and although this appears to be important to virus restriction, components of the preintegration complex do not appear to be modified (161). Evolutionary signatures of positive selection on the PRYSPRY indicate that this domain is under strong selective pressure (162). Accordingly, the capsid-TRIM5α interface maps to the hypervariable region of the retroviral capsid. These and similar studies underlie a burgeoning field of virus-host coevolution known as paleovirology that encompasses the study of positive evolutionary selection of host factors imposed by pathogens, within populations and across species (reviewed in 163). This evolutionary perspective of intrinsic cellular immunity has proven very useful in characterizing and identifying ISGs that directly interact with pathogens.
TRIM5α is but one of many members of the TRIM family with antiviral activity (164). Another TRIM family member, TRIM22, has also been shown to play a role in virus restriction. TRIM22 was originally identified based on its ability to inhibit a reporter under control of the HIV-1 long terminal repeat (165); additional work demonstrated that TRIM22 also inhibits trafficking of the HIV-1 Gag protein to the host cell plasma membrane, resulting in decreased particle production (166). The importance of TRIM22 as an HIV-1 restriction factor in vivo is highlighted by the finding that higher patient TRIM22 levels correlate with lower viral loads and higher CD4+ T cells counts (167). Since its initial characterization as an HIV-1 inhibitor, TRIM22 has also been shown to inhibit multiple viruses by various mechanisms (reviewed in 168). For example, it inhibits transcription from the core promoter of hepatitis B virus (169), blocks encephalomyocarditis virus replication by promoting ubiquitination of the viral 3C protease (170), and targets influenza A virus nucleoprotein for proteasomal degradation (171).
Other notable TRIM family members include TRIM19, also known as PML, which is present in multiple isoforms and localizes predominantly to nuclear bodies (reviewed in 172); TRIM25, which leads to ubiquitination and enhanced activity of RIG-I (173); and TRIM56, involved in ubiquitination and activation of STING (174). For additional review of TRIM proteins, the reader is referred to References 156, 175, 176.
Inhibition of virus translation and replication
Viruses rely on host ribosomes for protein synthesis, and translation is a common target for ISG intervention (Figure 4). A number of ISGs that inhibit translation, including zinc-finger antiviral protein (ZAP), the IFN-induced protein with tetratricopeptide repeats (IFIT) family, the OAS-RNaseL pathway, and PKR have been reviewed recently elsewhere (78, 155, 177, 178). In addition to translation, post-translational modification of viral or host proteins is also important for virus replication and infectivity. Here, we highlight ISG15 as an exemplar of ISG complexity and of a novel role in coupling translation with antiviral post-translational modification.
ISG15 is one of the most highly induced ISGs (179), and as mentioned above, it is a ubiquitin-like protein that can be covalently attached to target proteins in a process known as ISGylation (reviewed in 180). Many of the proteins involved in ISGylation and deISGylation—UBE2L6, HERC5, HERC6, UBE1LA, TRIM25, and _USP18_—are also induced by IFN (reviewed in 181). After decades of study, investigators have uncovered a rich and tangled history for ISG15. Studies have cataloged numerous proteins—both pathogen and host derived—targeted for ISGylation, and modification gives rise to pleiotropic effects. For example, ISGylation of IRF3 increases its stability by preventing polyubiquitination, which leads to sustained transcription factor activity (182). Similarly, ISGylation of the host protein 4EHP—a negative regulator of translation—increases its affinity for binding to the 5′ cap structure of mRNAs, thereby enhancing its ability to block translation initiation (183). ISGylation can also negatively affect targeted proteins. For example, ISGylation of cyclin D1 leads to protein destabilization, reduced activity, and cell cycle inhibition (184).
These are only a few examples of the possible consequences of protein ISGylation. The staggering number of proteins that are ISGylated upon ISG induction raises the question of whether all proteins that are targeted for ISGylation are targeted with specificity. More recently, ISG15 was found to be preferentially conjugated to newly synthesized proteins cotranslationally, which suggests that ISGylation may be a general, nonspecific mechanism of host defense (185). If this is true, then in addition to numerous host proteins, ISGylation may potentially impact all viral proteins translated in IFN-stimulated cells (185).
Despite the many reports describing ISG15 function (and ISG15 targets) in vitro, its in vivo role remains a subject of debate. One study found that wild-type and _ISG15_−/− mice are equally susceptible to vesicular stomatitis virus and lymphocytic choriomeningitis virus (109); in contrast, _ISG15_−/− mice are more susceptible to infection with influenza A and B, HSV-1, and Sindbis virus (186). Furthermore, ISG15 plays a protective role in vaccinia virus infection, but only when the virus lacks the E3 viral antagonist (187).
In addition to being involved in the diverse effects of protein ISGylation, ISG15 is also secreted from immune cells, thus taking on the role of a cytokine and stimulating the production of IFN-γ (reviewed in 188). Recently, a study performed on patients with inborn ISG15 deficiencies suggested that secreted ISG15 may be clinically important because these individuals suffer from increased risk of severe mycobacterial infections yet have no history of increased viral susceptibility (189). Hence, the complete repertoire of ISG15 activities and possible host species– and pathogen-specific differences have yet to be fully defined.
Inhibitors of viral egress
During late stages of the virus life cycle, viral nucleic acids are packaged into capsids, and the particles exit cells, by either cell lysis, exocytosis, or direct budding from the plasma membrane. Viral envelopes are acquired in this process, and lipid bilayer compositions vary depending on the site of budding. Relative to other stages in the virus life cycle, few ISGs are known to inhibit viral assembly and viral egress. This may be partly due to the greater challenges associated with performing large-scale virus screens to selectively identify late-stage inhibitors (Figure 4).
Viperin: virus inhibitory protein, endoplasmic reticulum–associated, IFN-inducible
Viperin, also known as RSAD2, is one of the better-studied, most highly induced antiviral effectors. It can be induced by at least two different innate immune pathways: via JAK-STAT signaling (115, 190–192) or via direct activation by IRF1/3 (Figure 2) (193–195). Interestingly, viperin expression is stimulated directly by human cytomegalovirus (HCMV) glycoprotein B to the benefit of the virus, and therefore in at least one known case, viperin has been usurped to act as a proviral factor (196).
Viperin normally resides in the ER and in ER-derived lipid droplets—organelles important for lipid metabolism. Upon HCMV infection, viperin is relocalized to mitochondria by the HCMV-encoded mitochondrial inhibitor of apoptosis (vMIA) protein where it interacts with the mitochondrial trifunctional protein, leading to the inhibition of ATP generation (196). Decreased ATP levels lead to disruption of the cellular cytoskeleton and facilitate HCMV infection by an unknown mechanism.
Viperin inhibits many enveloped viruses, and various modes of antiviral action—possibly influenced by different viral life cycles—have been described (reviewed in 197). Viperin inhibits HIV-1 and influenza A virus budding at the host cell membrane by inhibiting farnesyl diphosphate synthase (FPPS), an enzyme involved in isoprenoid biosynthesis (198, 199). Influenza A virus buds from lipid rafts—lipid microdomains with specific membrane fluidity—and decreased FPPS activity alters membrane fluidity, thereby interfering with virus budding (198). Inhibition of HIV-1 particle release is believed to occur by a similar mechanism (199).
Viperin can also affect earlier steps in the virus life cycle. For example, viperin inhibits RNA replication of HCV subgenomic replicons (200). HCV replicates in altered subcellular membrane structures that are referred to as the membranous web (201), which are intimately associated with lipid droplets. Within these droplets viperin interacts with both the host [vesicle-associated membrane protein–associated protein A (VAP-A, also known as VAP-33)] and the HCV non-structural protein 5A (NS5A), a key protein required for viral replication, assembly, and egress (202). Binding of viperin to both of these factors might disrupt the VAP-A/NS5A interaction, resulting in inhibition of HCV genome replication (200, 203). A similar mode of action has been proposed for the inhibition of dengue virus, where viperin also interacts with components of the viral replication complex (204).
Tetherin
Tetherin is encoded by the ISG BST2 and inhibits virus budding by using two membrane anchors to trap HIV-1 virions on the plasma membrane (Figure 4) (205). Initially described in 2009, tetherin has since been shown to have antiviral activity against many enveloped viruses (listed in Reference 206).
Tetherin’s importance as a direct-acting antiviral effector is highlighted by the evolution of viral evasion strategies. A number of viral proteins—namely glycoproteins and accessory proteins— are charged with the task of subverting tetherin function. These include HIV-1 Vpu, HIV-2 Env, simian immunodeficiency virus (SIV) Nef, Ebola virus surface glycoprotein VP40, Kaposi-sarcoma-associated herpes virus (KSHV) K5 ubiquitin ligase, and influenza A virus neuraminidase (NA) proteins. Both HIV-1 Vpu and KSHV K5 are involved in tetherin ubiquitination—Vpu by recruiting the E3 ubiquitin ligase complex and K5 by directly catalyzing the reaction—resulting in tetherin degradation (206).
HIV-2 and most SIV strains do not encode Vpu; rather, their antitetherin function is executed by HIV-2 Env and SIV Nef, respectively. Both effectively keep tetherin from sites of viral assembly, but only for HIV-2 Env have the mechanisms been identified. HIV-2 Env not only sequesters tetherin in the trans-Golgi network, disrupting proper trafficking, but it also triggers clathrin-dependent endocytosis. Unlike HIV-1 Vpu, the latter process does not seem to involve tetherin degradation (207–209). Ebola VP40 and influenza A virus NA both sequester tetherin on the cell surface, but how this accomplishes virus inhibition is uncertain (210, 211). For a more detailed review on tetherin, the reader is referred to two recent reviews (206, 212).
Screening for ISG antiviral effectors
Many of the well-characterized classic ISGs are, not surprisingly, those that display the most potent antiviral activity against commonly studied viruses. These ISGs represent only a handful of the total number of ISGs produced during infection. What, then, of the hundreds of others? To address this question, a number of gene knockdown and overexpression screens have been performed to identify ISGs that act as direct antiviral effectors.
Considering the large number of ISGs activated upon IFN signaling, considerable redundancy in the system is likely. Redundancy is a major challenge when attempting to identify antiviral ISGs using gene expression knockdown approaches, but in spite of this, several groups have been successful. With the use of both genome-wide (213) and ISG-targeted (214) siRNA-based approaches, genes have been identified for which, when their expression is reduced, IFN is less effective at HCV replicon inhibition. Similarly, the discovery that IFITM proteins potently inhibit influenza A virus infection was also made by performing a genome-wide siRNA screen (143). Use of a lentiviral-based shRNA approach was also successful; a recent report identified a number of ISGs that are important for the ability of IFN-β to inhibit infection by West Nile virus. This screen also identified the ISG activating signal cointegrator complex 3 (ASCC3), and further confirmatory experiments showed it to be involved in negative regulation of IFN signaling (215). The identification of several hits in each of these studies validates the feasibility of this approach for antiviral effector discovery. From these studies, and additional screens yet to be performed, we may be able to classify ISGs into groups according to their mechanism of inhibition and virus specificity.
A number of groups have performed ectopic overexpression screens to identify direct antiviral effectors. In a small-scale study, overexpression of 7 ISGs in murine fibroblasts identified genes with the ability to inhibit alphavirus infection (216). Similarly, the individual overexpression of 18 (217) and 29 (218) ISGs identified inhibitors of HCV replicons, and overexpression of 36 ISGs identified inhibitors of dengue virus and West Nile virus replicons (219). More recently, 39 genes were screened for their ability to inhibit West Nile virus in primary neurons (220). In all cases, ISGs with direct antiviral activity were described, but from these studies the breadth and specificity of the antiviral factors are unclear.
Recently, a comprehensive large-scale screen for antiviral ISGs was performed with a library of over 380 ISGs against a diverse panel of viruses (89, 221). Each virus tested was susceptible to inhibition by several ISGs, some that are broadly inhibitory and others that display virus-specific activity. The transcription factor IRF1 and cGAS (formerly c6orf150) were among the few ISGs that displayed broad, potent antiviral activity. Upon further study, IRF1 overexpression was found to transcriptionally activate a set of genes that overlaps with those induced by type I IFN (89), and cGAS was shown to play an important role in activating the innate pathogen sensor STING (79, 81). Subsequently, experiments performed using _cGAS_−/− mice suggested that cGAS may protect cells from infection by RNA viruses, and therefore it may play a broader role in innate immunity than was previously expected (221). Unlike IRF1 and cGAS, most ISGs do not inhibit virus infection when expressed individually, and those that do display moderate effects. Net antiviral activity does, however, increase when various ISGs are expressed in combination, as has been shown by measuring the inhibition of virus infection following the expression of ISG pairs (89). Similarly, performing an ISG overexpression screen in a ZAP-inducible cell line identified ISGs that enhance ZAP’s antiviral activity (89, 222). These results are consistent with the notion that the emergent power of type I IFN derives from many ISGs acting in concert.
When further exploring the mechanisms of virus inhibition for ISGs identified by an overexpression screen, Schoggins et al. (89) reported that protein translation is a key target for many ISGs. In the Schoggins study, inhibition was monitored after a single round of infection, and therefore ISGs that inhibit viral egress or decrease the specific infectivity of the virus particle would not have been identified. Additional screens geared toward identifying ISGs affecting late stages of the virus life cycle would be of interest to pursue. Also, performing screens that utilize viruses bearing defects in viral antagonist proteins or screens with gene libraries from related, but distinct, species may uncover ISGs that viruses evolved to inactivate or evade.
Harnessing the power of ISGs
It is clear that the IFN system did not evolve to have one potent virus-specific ISG per virus, but rather to work in a combinatorial fashion. The benefit of this system manifests when considering decades of clinical data describing viral resistance to drugs and success rates of combination versus single-agent therapies. Similarly, the modest reductions in virus infection resulting from overexpression of individual ISGs contrast starkly with the extraordinary potency of IFN. A better mechanistic understanding of individual ISGs may lead to the development of novel therapeutics, but from a practical standpoint the most powerful ISG-based therapeutics in the near future may be those that harness the collective power of ISGs—similar to IFN itself.
Developing and utilizing novel chimeric IFNs and IFN-like molecules have the potential to activate subsets of ISGs, minimizing side effects and at the same time providing potent virus restriction. This approach is promising but has been met with challenges—viruses have developed innovative strategies to evade innate immunity, and therefore an effective therapy may need to overcome several viral mechanisms of immune evasion. Another approach is to identify small molecule compounds capable of activating innate immunity at distal nodes in the host response pathway. For example, direct activation of the ISGF3 complex or downstream transcription factors, such as IRF1 or IRF3, may stimulate a number of ISGs directly under the radar of viral antagonism mechanisms that evolved to target upstream signaling pathways.
BEYOND CONVENTIONAL ISGs: ADDITIONAL CONSEQUENCES OF INTERFERON STIMULATION
Recent studies utilizing contemporary sequencing and proteomics technologies are changing our understanding of the IFN biology landscape. This section discusses additional consequences of IFN stimulation (other than classical ISG induction) (Figure 5).
Figure 5.
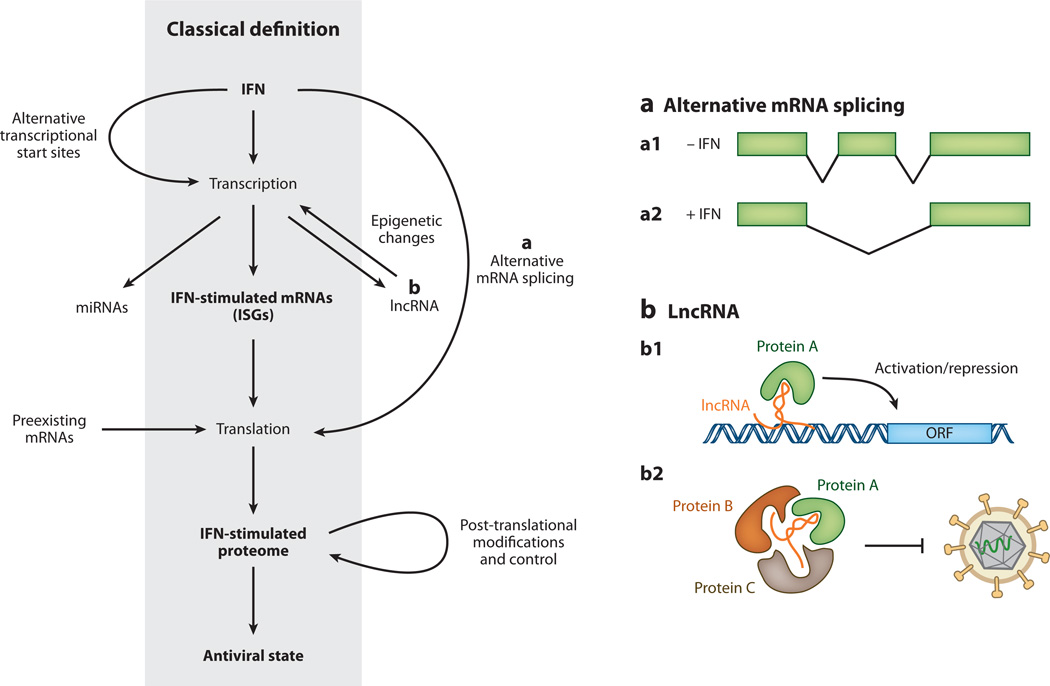
Canonical and noncanonical definition of interferon (IFN)-stimulated antiviral effectors. For years the canonical understanding of the IFN-mediated antiviral response has been that IFN triggers the transcription of IFN-stimulated genes (ISGs), which leads to changes in the cellular proteome and establishment of an antiviral state within cells. Recent studies suggest that the situation is likely more complex. (a) IFN treatment of cells promotes alternative transcriptional start site usage, and IFN-induced alternative mRNA splicing may give rise to transcript isoforms that encode for different protein products or transcripts with altered stability or translational efficiency (a1/a2). (b) In addition, IFN stimulation may alter the expression of microRNAs (miRNAs) or long noncoding RNAs (lncRNAs). LncRNAs may influence gene expression through interaction with chromatin remodeling complexes (b1) or may serve as a scaffold for the formation of RNA-protein complexes that confer antiviral activity (b2). IFN stimulation or pathogen recognition may promote translation of preexisting mRNAs. Finally, IFN stimulation influences the proteome directly by promoting proteins’ post-translational modification, altering protein stability, and increasing protein secretion.
IFN-Stimulated Transcripts
Strictly speaking, ISGs are genes in which transcriptional output increases in response to IFN, largely due to the presence of GAS and/or ISRE sequences in promoter and enhancer regions. Initial genome-wide surveys of transcription following IFN stimulation used microarray technology (115, 190, 223–228); these studies utilized mRNA enriched by polyA selection and arrays that probe for protein-coding transcripts. With this approach, only RNAs with corresponding probes are measured, and as a result these studies are blind to changes in a variety of RNA species including, but not limited to, transcript isoforms, long noncoding RNAs (lncRNAs), microRNAs (miRNAs), and several classes of small RNAs.
miRNAs
The impact of small RNAs and miRNAs on mRNA translation and stability is well documented. Numerous studies have begun to address the impact of miRNAs in virus infection, and indeed, multiple viruses interact with miRNAs in various ways; some viruses encode miRNAs in their genomes (reviewed in 229), whereas others require specific miRNAs for replication (230). Modifying miRNA transcription or activity may have important consequences in the context of the IFN response, and recent reports have suggested that IFN stimulation can impact the expression of cellular miRNAs, leading to suppression of type I IFN–induced apoptosis (231) and type II IFN–induced cell cycle arrest (232). Other data suggest that the greatest impact of miRNA expression may occur at the interface between the innate and adaptive responses (reviewed in 233).
lncRNAs
Advances in RNA sequencing technology in recent years have led to an explosion in the number of lncRNAs identified, and researchers have begun to seek potential roles for lncRNAs in innate immunity (234). Most lncRNAs have unknown function; however, most studies support a role in modulation of gene transcription by guiding epigenetic chromatin modification (Figure 5) (reviewed in 235). Recently, and for the first time, a lncRNA was shown to play a critical role in conferring resistance to persistent virus infection (236). This lncRNA, NeST, is short for _ne_ttoie Salmonella pas Theiler’s (which translates into “cleanup Salmonella not Theiler’s”), and it received its name for the phenotype associated with its chromosomal location. In B10.S mice, this locus—which also contains the gene encoding IFN-γ—is associated with an increased susceptibility to Salmonella infection and is also associated with reduced susceptibility to persistent infection by Theiler’s virus (237).
While attempting to reveal the underlying mechanisms responsible for this locus-associated phenotype, Gomez et al. (236) found that NeST expression had a positive impact on the production of IFN-γ mRNA and protein. The authors then went on to show that NeST was expressed at very low levels in CD8+ T cells under conditions of immune activation and proposed a model in which NeST RNA acts by recruiting a methyltransferase complex to the IFN-γ locus, resulting in epigenetic chromatin modification and altered gene expression (236). The results from this landmark study are supported by prior work identifying polymorphisms in regions surrounding the IFN-γ gene that were previously defined as distal regulatory elements; these polymorphisms lie within NeST (237). This discovery opens the possibility that additional genes may be affected by NeST and raises the question of whether other lncRNAs will have roles in the type I or III IFN response.
Alternative Splicing
It is well established that the human genome contains approximately 23,000 genes, many fewer than original estimates, but we now know this low number is more than compensated for by rampant alternative splicing (reviewed in 238). More than 95% of genes yield multiple transcripts, yet the consequences of this are largely unstudied in the context of IFN stimulation, making it another frontier in IFN biology.
Several ISGs are present in multiple isoforms and give rise to proteins with differences in cellular localization and activity. The PML gene, a member of the TRIM family and mentioned previously, is one such example (reviewed in 172). Similarly, IFN-γ stimulation leads to the production of alternative isoforms of tryptophanyl-tRNA synthetase, resulting in antiproliferative and antiangiogenic effects (239). The use of alternative transcriptional start sites can also impact protein function. The adenosine deaminase acting on RNA-1 (ADAR1) gene is constitutively expressed and shuttles between the nucleus and cytoplasm. Following IFN signaling, transcription initiates from an upstream alternative transcriptional start site, giving rise to an extended protein that is predominantly localized to the cytoplasm (240).
On average, IFN treatment leads to transcriptional stimulation of up to 5% of the total number of cellular genes; however, this statistic only captures those genes with changes in mRNA abundance. Thus, IFN stimulation may lead to changes in isoform abundance that are undetected by conventional methods (Figure 5). Furthermore, many transcript isoforms differ not only in coding exons but also (some exclusively) in their 5′ and 3′ untranslated regions. These changes can have dramatic effects on mRNA stability and translation efficiency (241, 242).
IFN-Stimulated Translation
Viruses employ myriad strategies to counteract pathogen sensing and JAK-STAT signaling, making it clear that the need for speed is under strict evolutionary pressure.
Type I IFN signaling activates phosphatidylinositol-3 kinase and the mammalian target of rapamycin pathway, leading to rapid changes in translational regulation (reviewed in 243). The ability to promote efficient translation of select mRNAs even prior to ISG transcription would provide a mechanism to rapidly enhance pathogen detection and/or amplify IFN signaling. It has been reported that IFN-β mRNA preexists in cells, but the translational competence and stability of the message are impaired (244). By a mechanism that remains unclear, PKR is believed to influence mRNA stability, thereby enhancing translation and IFN-β production (244). Several additional mechanisms have been described for controlling the translation of cytokine mRNAs (reviewed in 245), and one can envision that similar regulation may exist for ISG or non-ISG mRNAs that may be present at low basal levels before IFN receptor engagement but are rapidly translated upon IFN or PRR signaling.
Post-Translational Modification, Relocalization, and Secretion
Post-translational modification has the ability to alter protein stability, cellular localization, and activity with exceptional speed. Obvious examples of post-translational modification in the IFN response are the phosphorylation of JAKs and IFN receptor chains and the STAT proteins, which are phosphorylated and also relocalized to the nucleus. A number of additional examples exist, such as acetylation of IFNAR2 (246), ISGylation of IRF3 (182), and ubiquitination of RIG-I (247); however, few attempts have been made to perform a global survey of post-translational changes that occur rapidly upon IFN signaling. State-of-the-art mass spectrometry technologies are beginning to make such studies feasible.
Recently, Meissner et al. (248) employed a high-sensitivity quantitative mass spectrometry approach to characterize the secretome of macrophages activated with LPS. A variety of pro-and anti-inflammatory cytokines, protease inhibitors, and complement components were found to be secreted in a time-resolved fashion. Many of these proteins showed neither increased transcription nor increased intracellular protein levels, suggesting that the innate pathogen-sensing response regulates protein secretion as a defense tactic. Along these same lines, Li et al. (249) recently described IFN-α-mediated cell-to-cell transfer of exosomes containing antiviral factors. These exosome “care packages” facilitate pathogen defense in cells that have been rendered incapable of IFN signaling by viral antagonism. Further exploration in these areas will improve our understanding of novel innate immune strategies for pathogen resistance.
CONCLUSION
Despite decades of IFN biology, we are still striving to understand IFN-regulated antiviral mechanisms. In light of many recent findings at the transcriptome and proteome level, the classical definition of ISGs is dated. Numerous diverse and tightly regulated events contribute to the antiviral state such as changes in transcriptomes (beyond mRNA upregulation), proteomes, and secretomes. A substantial number of genes are transcriptionally downregulated as a result of IFN signaling, yet the consequence this has on virus infection is underexplored. Understanding this enormous complexity will require novel mechanistic insight into the actions of individual components coupled with cutting-edge systems biology approaches.
ACKNOWLEDGMENTS
W.M.S. was supported by National Institute of Diabetes and Digestive and Kidney Diseases National Research Service Award DK095666. M.D.C. was supported by a research grant from the German Research Foundation (DFG) and the Rockefeller Women and Science award. This work was funded in part by National Institutes of Health Grant AI091707 (to C.M.R.). Additional funding was provided by the Greenberg Medical Research Institute, the Starr Foundation, and the Ronald A. Shellow Memorial Fund (to C.M.R.). We give tremendous thanks to M. Li, M. MacDonald, J. Sable, and M. Saeed for critical reading of the manuscript and express our sincerest apologies to the many authors whose outstanding work we could not cite because of space limitations.
Glossary
JAK
Janus kinase
STAT
signal transducer and activator of transcription
IRG
IFN-regulated gene
ISG
IFN-stimulated gene
PAMP
pathogen-associated molecular pattern
PRR
pattern-recognition receptor, such as PKR, MDA5, RIG-I, AIM2
IRF
IFN-regulatory factor
PIAS
protein inhibitor of activated STAT
SOCS
suppressor of cytokine signaling
USP18
ubiquitin-specific peptidase 18 (also known as Ubp43 in mice, but herein referred to as USP18 for both species)
ISGylation
covalent linkage of proteins to ISG15, a ubiquitin-like protein
Virus replicon
viral nucleic acid capable of autonomous replication used to study viral replication events uncoupled from entry and egress
Viral entry
earliest step of the virus life cycle, where viral material is introduced into the host cell
Uncoating
removal of the protein capsid or envelope from a virus to release its genome
Gag
group-specific antigen, a polyprotein containing retrovirus matrix-, capsid-, and nucleoprotein
Viral assembly
assembly of viral structural proteins and nucleic acids to form a virus particle
Viral egress
virus leaving its host cell after assembly (by host cell lysis, exocytosis, or budding)
vMIA
viral mitochondrial inhibitor of apoptosis
lncRNA
long noncoding RNA
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might affect the objectivity of this review.
Contributor Information
William M. Schneider, Email: wschneider@rockefeller.edu.
Meike Dittmann Chevillotte, Email: mchevillot@rockefeller.edu.
Charles M. Rice, Email: ricec@rockefeller.edu.
LITERATURE CITED
- 1.Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonzalez-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 2012;12:125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoenborn JR, Wilson CB. Regulation of interferon-gamma during innate and adaptive immune responses. Adv. Immunol. 2007;96:41–101. doi: 10.1016/S0065-2776(07)96002-2. [DOI] [PubMed] [Google Scholar]
- 4.Rönnblom L. The type I interferon system in etiopathogenesis of autoimmune diseases. Upsala J. Med. Sci. 2011;116:227–237. doi: 10.3109/03009734.2011.624649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henle W. Interference phenomena between animal viruses; a review. J. Immunol. 1950;64:203–236. [PubMed] [Google Scholar]
- 6.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc. R. Soc. B. 1957;147:258–267. [PubMed] [Google Scholar]
- 7.Isaacs A, Lindenmann J, Valentine RC. Virus interference. II. Some properties of interferon. Proc. R. Soc. B. 1957;147:268–273. [PubMed] [Google Scholar]
- 8.Lengyel P. Biochemistry of interferons and their actions. Annu. Rev. Biochem. 1982;51:251–282. doi: 10.1146/annurev.bi.51.070182.001343. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 10.Cantell K, Hirvonen S, Kauppinen HL, Myllyla G. Production of interferon in human leukocytes from normal donors with the use of Sendai virus. Methods Enzymol. 1981;78:29–38. doi: 10.1016/0076-6879(81)78094-7. [DOI] [PubMed] [Google Scholar]
- 11.Cantell K, Hirvonen S, Koistinen V. Partial purification of human leukocyte interferon on a large scale. Methods Enzymol. 1981;78:499–505. doi: 10.1016/0076-6879(81)78161-8. [DOI] [PubMed] [Google Scholar]
- 12.Pestka S. The interferons: 50 years after their discovery, there is much more to learn. J. Biol. Chem. 2007;282:20047–20051. doi: 10.1074/jbc.R700004200. [DOI] [PubMed] [Google Scholar]
- 13.Knight E., Jr Heterogeneity of purified mouse interferons. J. Biol. Chem. 1975;250:4139–4144. [PubMed] [Google Scholar]
- 14.Rubinstein M, Levy WP, Moschera JA, Lai CY, Hershberg RD, et al. Human leukocyte interferon: isolation and characterization of several molecular forms. Arch. Biochem. Biophys. 1981;210:307–318. doi: 10.1016/0003-9861(81)90194-6. [DOI] [PubMed] [Google Scholar]
- 15.Maeda S, McCandliss R, Gross M, Sloma A, Familletti PC, et al. Construction and identification of bacterial plasmids containing nucleotide sequence for human leukocyte interferon. Proc. Natl. Acad. Sci. USA. 1980;77:7010–7013. doi: 10.1073/pnas.77.12.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Weerd NA, Samarajiwa SA, Hertzog PJ. Type I interferon receptors: biochemistry and biological functions. J. Biol. Chem. 2007;282:20053–20057. doi: 10.1074/jbc.R700006200. [DOI] [PubMed] [Google Scholar]
- 17.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 18.Uze G, Schreiber G, Piehler J, Pellegrini S. The receptor of the type I interferon family. Curr. Top. Microbiol. Immunol. 2007;316:71–95. doi: 10.1007/978-3-540-71329-6_5. [DOI] [PubMed] [Google Scholar]
- 19.Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, et al. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y-J. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 21.Moraga I, Harari D, Schreiber G, Uze G, Pellegrini S. Receptor density is key to the alpha2/beta interferon differential activities. Mol. Cell. Biol. 2009;29:4778–4787. doi: 10.1128/MCB.01808-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaitin DA, Roisman LC, Jaks E, Gavutis M, Piehler J, et al. Inquiring into the differential action of interferons (IFNs): an IFN-α mutant with enhanced affinity to IFNAR1 is functionally similar to IFN-β. Mol. Cell. Biol. 2006;26:1888–1897. doi: 10.1128/MCB.26.5.1888-1897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalie E, Jaitin DA, Podoplelova Y, Piehler J, Schreiber G. The stability of the ternary interferon-receptor complex rather than the affinity to the individual subunits dictates differential biological activities. J. Biol. Chem. 2008;283:32925–32936. doi: 10.1074/jbc.M806019200. [DOI] [PubMed] [Google Scholar]
- 24.Walter MR, Windsor WT, Nagabhushan TL, Lundell DJ, Lunn CA, et al. Crystal structure of a complex between interferon-γ and its soluble high-affinity receptor. Nature. 1995;376:230–235. doi: 10.1038/376230a0. [DOI] [PubMed] [Google Scholar]
- 25.Valente G, Ozmen L, Novelli F, Geuna M, Palestro G, et al. Distribution of interferon-y receptor in human tissues. Eur. J. Immunol. 1992;22:2403–2412. doi: 10.1002/eji.1830220933. [DOI] [PubMed] [Google Scholar]
- 26.Decker T, Lew DJ, Cheng YS, Levy DE, Darnell JE., Jr Interactions of α- and γ-interferon in the transcriptional regulation of the gene encoding a guanylate-binding protein. EMBO J. 1989;8:2009–2014. doi: 10.1002/j.1460-2075.1989.tb03608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lew DJ, Decker T, Darnell JE., Jr Alpha interferon and gamma interferon stimulate transcription of a single gene through different signal transduction pathways. Mol. Cell. Biol. 1989;9:5404–5411. doi: 10.1128/mcb.9.12.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy DE, Lew DJ, Decker T, Kessler DS, Darnell JE., Jr Synergistic interaction between interferon-α and interferon-γ through induced synthesis of one subunit of the transcription factor ISGF3. EMBO J. 1990;9:1105–1111. doi: 10.1002/j.1460-2075.1990.tb08216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujimoto M, Naka T. SOCS1, a negative regulator of cytokine signals and TLR responses, in human liver diseases. Gastroenterol. Res. Pract. 2010 doi: 10.1155/2010/470468. http://dx.doi.org/10.1155/2010/470468. [DOI] [PMC free article] [PubMed]
- 30.Fenner JE, Starr R, Cornish AL, Zhang JG, Metcalf D, et al. Suppressor of cytokine signaling 1 regulates the immune response to infection by a unique inhibition of type I interferon activity. Nat. Immunol. 2006;7:33–39. doi: 10.1038/ni1287. [DOI] [PubMed] [Google Scholar]
- 31.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 32.McLaren JE, Ramji DP. Interferon gamma: a master regulator of atherosclerosis. Cytokine Growth Factor Rev. 2009;20:125–135. doi: 10.1016/j.cytogfr.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Zaidi MR, Merlino G. The two faces of interferon-gamma in cancer. Clin. Cancer Res. 2011;17:6118–6124. doi: 10.1158/1078-0432.CCR-11-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, et al. IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 35.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 36.Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fox BA, Sheppard PO, O’Hara PJ. The role of genomic data in the discovery, annotation and evolutionary interpretation of the interferon-λ family. PLoS ONE. 2009;4:e4933. doi: 10.1371/journal.pone.0004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-λ) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, et al. Interferons α and λ inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 40.Bolen CR, Ding S, Robek MD, Kleinstein SH. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology. 2014 doi: 10.1002/hep.26657. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larner AC, Chaudhuri A, Darnell JE., Jr Transcriptional induction by interferon. New protein(s) determine the extent and length of the induction. J. Biol. Chem. 1986;261:453–459. [PubMed] [Google Scholar]
- 42.Larner AC, Jonak G, Cheng YS, Korant B, Knight E, Darnell JE., Jr Transcriptional induction of two genes in human cells by β interferon. Proc. Natl. Acad. Sci. USA. 1984;81:6733–6737. doi: 10.1073/pnas.81.21.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawamura M, McVicar DW, Johnston JA, Blake TB, Chen YQ, et al. Molecular cloning of L-JAK, a Janus family protein-tyrosine kinase expressed in natural killer cells and activated leukocytes. Proc. Natl. Acad. Sci. USA. 1994;91:6374–6378. doi: 10.1073/pnas.91.14.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pellegrini S, John J, Shearer M, Kerr IM, Stark GR. Use of a selectable marker regulated by α interferon to obtain mutations in the signaling pathway. Mol. Cell. Biol. 1989;9:4605–4612. doi: 10.1128/mcb.9.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ragimbeau J, Dondi E, Alcover A, Eid P, Uze G, Pellegrini S. The tyrosine kinase Tyk2 controls IFNAR1 cell surface expression. EMBO J. 2003;22:537–547. doi: 10.1093/emboj/cdg038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gauzzi MC, Barbieri G, Richter MF, Uze G, Ling L, et al. The amino-terminal region of Tyk2 sustains the level of interferon α receptor 1, a component of the interferon α/β receptor. Proc. Natl. Acad. Sci. USA. 1997;94:11839–11844. doi: 10.1073/pnas.94.22.11839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haan C, Kreis S, Margue C, Behrmann I. Jaks and cytokine receptors—an intimate relationship. Biochem. Pharmacol. 2006;72:1538–1546. doi: 10.1016/j.bcp.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 48.van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 49.Heim MH, Kerr IM, Stark GR, Darnell JE., Jr Contribution of STAT SH2 groups to specific interferon signaling by the Jak-STAT pathway. Science. 1995;267:1347–1349. doi: 10.1126/science.7871432. [DOI] [PubMed] [Google Scholar]
- 50.Shuai K, Schindler C, Prezioso VR, Darnell JE., Jr Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science. 1992;258:1808–1812. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- 51.Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 52.Greenlund AC, Morales MO, Viviano BL, Yan H, Krolewski J, Schreiber RD. Stat recruitment by tyrosine-phosphorylated cytokine receptors: an ordered reversible affinity-driven process. Immunity. 1995;2:677–687. doi: 10.1016/1074-7613(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 53.Sekimoto T, Imamoto N, Nakajima K, Hirano T, Yoneda Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 1997;16:7067–7077. doi: 10.1093/emboj/16.23.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McBride KM, Banninger G, McDonald C, Reich NC. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-α. EMBO J. 2002;21:1754–1763. doi: 10.1093/emboj/21.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fagerlund R, Melen K, Kinnunen R, Julkunen I. Arginine/lysine-rich nuclear localization signals mediate interactions between dimeric STATs and importin α5. J. Biol. Chem. 2002;277:30072–30078. doi: 10.1074/jbc.M202943200. [DOI] [PubMed] [Google Scholar]
- 56.Melen K, Kinnunen L, Julkunen I. Arginine/lysine-rich structural element is involved in interferon-induced nuclear import of STATs. J. Biol. Chem. 2001;276:16447–16455. doi: 10.1074/jbc.M008821200. [DOI] [PubMed] [Google Scholar]
- 57.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 58.Shuai K, Stark GR, Kerr IM, Darnell JE., Jr A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science. 1993;261:1744–1746. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- 59.Decker T, Lew DJ, Mirkovitch J, Darnell JE., Jr Cytoplasmic activation of GAF, an IFN-gamma-regulated DNA-binding factor. EMBO J. 1991;10:927–932. doi: 10.1002/j.1460-2075.1991.tb08026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Decker T, Kovarik P, Meinke A. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J. Interferon Cytokine Res. 1997;17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- 61.Fu XY, Kessler DS, Veals SA, Levy DE, Darnell JE., Jr ISGF3, the transcriptional activator induced by interferon α, consists of multiple interacting polypeptide chains. Proc. Natl. Acad. Sci. USA. 1990;87:8555–8559. doi: 10.1073/pnas.87.21.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levy DE, Kessler DS, Pine R, Darnell JE., Jr Cytoplasmic activation of ISGF3, the positive regulator of interferon-α-stimulated transcription, reconstituted in vitro. Genes Dev. 1989;3:1362–1371. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- 63.Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon α. Proc. Natl. Acad. Sci. USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levy DE, Kessler DS, Pine R, Reich N, Darnell JE., Jr Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 1988;2:383–393. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- 65.Levy D, Larner A, Chaudhuri A, Babiss LE, Darnell JE., Jr Interferon-stimulated transcription: isolation of an inducible gene and identification of its regulatory region. Proc. Natl. Acad. Sci. USA. 1986;83:8929–8933. doi: 10.1073/pnas.83.23.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reich N, Evans B, Levy D, Fahey D, Knight E, Jr, Darnell JE., Jr Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc. Natl. Acad. Sci. USA. 1987;84:6394–6398. doi: 10.1073/pnas.84.18.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheon H, Stark GR. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc. Natl. Acad. Sci. USA. 2009;106:9373–9378. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheon H, Yang J, Stark GR. The functions of signal transducers and activators of transcriptions 1 and 3 as cytokine-inducible proteins. J. Interferon Cytokine Res. 2011;31:33–40. doi: 10.1089/jir.2010.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iwasaki A. A virological view of innate immune recognition. Annu. Rev. Microbiol. 2012;66:177–96. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barber GN. Cytoplasmic DNA innate immune pathways. Immunol. Rev. 2011;243:99–108. doi: 10.1111/j.1600-065X.2011.01051.x. [DOI] [PubMed] [Google Scholar]
- 71.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 72.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 73.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Di Virgilio F. The therapeutic potential of modifying inflammasomes and NOD-like receptors. Pharmacol. Rev. 2013;65:872–905. doi: 10.1124/pr.112.006171. [DOI] [PubMed] [Google Scholar]
- 76.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 77.Chakrabarti A, Jha BK, Silverman RH. New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Munir M, Berg M. The multiple faces of proteinkinase Rin antiviral defense. Virulence. 2013;4:85–89. doi: 10.4161/viru.23134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao D, Wu J, Wu YT, Du F, Aroh C, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu J, Sun L, Chen X, Du F, Shi H, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang X, Shi H, Wu J, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wallach D, Kovalenko A. How do cells sense foreign DNA? A new outlook on the function of STING. Mol. Cell. 2013;50:1–2. doi: 10.1016/j.molcel.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 85.Zemirli N, Arnoult D. Mitochondrial anti-viral immunity. Int. J. Biochem. Cell Biol. 2012;44:1473–1476. doi: 10.1016/j.biocel.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 86.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 87.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, et al. IPS-1, an adaptor triggering RIG-I-and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 88.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 89.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coccia EM, Uze G, Pellegrini S. Negative regulation of type I interferon signaling: facts and mechanisms. Cell Mol. Biol. 2006;52:77–87. [PubMed] [Google Scholar]
- 91.ten Hoeve J, de Jesus Ibarra-Sanchez M, Fu Y, Zhu W, Tremblay M, et al. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol. Cell. Biol. 2002;22:5662–5668. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.David M, Chen HE, Goelz S, Larner AC, Neel BG. Differential regulation of the α/β interferon-stimulated Jak/Stat pathway by the SH2 domain-containing tyrosine phosphatase SHPTP1. Mol. Cell. Biol. 1995;15:7050–7058. doi: 10.1128/mcb.15.12.7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.You M, Yu DH, Feng GS. Shp-2 tyrosine phosphatase functions as a negative regulator of the interferon-stimulated Jak/STAT pathway. Mol. Cell. Biol. 1999;19:2416–2424. doi: 10.1128/mcb.19.3.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Myers MP, Andersen JN, Cheng A, Tremblay ML, Horvath CM, et al. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J. Biol. Chem. 2001;276:47771–47774. doi: 10.1074/jbc.C100583200. [DOI] [PubMed] [Google Scholar]
- 95.Irie-Sasaki J, Sasaki T, Matsumoto W, Opavsky A, Cheng M, et al. CD45 is a JAK phosphatase and negatively regulates cytokine receptor signalling. Nature. 2001;409:349–354. doi: 10.1038/35053086. [DOI] [PubMed] [Google Scholar]
- 96.Simoncic PD, Lee-Loy A, Barber DL, Tremblay ML, McGlade CJ. The T cell protein tyrosine phosphatase is a negative regulator of Janus family kinases 1 and 3. Curr. Biol. 2002;12:446–453. doi: 10.1016/s0960-9822(02)00697-8. [DOI] [PubMed] [Google Scholar]
- 97.Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat. Rev. Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 98.Palvimo JJ. PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem. Soc. Trans. 2007;35:1405–1408. doi: 10.1042/BST0351405. [DOI] [PubMed] [Google Scholar]
- 99.Rogers RS, Horvath CM, Matunis MJ. SUMO modification of STAT1 and its role in PIAS-mediated inhibition of gene activation. J. Biol. Chem. 2003;278:30091–30097. doi: 10.1074/jbc.M301344200. [DOI] [PubMed] [Google Scholar]
- 100.Ungureanu D, Vanhatupa S, Kotaja N, Yang J, Aittomaki S, et al. PIAS proteins promote SUMO-1 conjugation to STAT1. Blood. 2003;102:3311–3313. doi: 10.1182/blood-2002-12-3816. [DOI] [PubMed] [Google Scholar]
- 101.Ungureanu D, Vanhatupa S, Gronholm J, Palvimo JJ, Silvennoinen O. SUMO-1 conjugation selectively modulates STAT1-mediated gene responses. Blood. 2005;106:224–226. doi: 10.1182/blood-2004-11-4514. [DOI] [PubMed] [Google Scholar]
- 102.Hong XX, Carmichael GG. Innate immunity in pluripotent human cells: attenuated response to interferon-β. J. Biol. Chem. 2013;288:16196–16205. doi: 10.1074/jbc.M112.435461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang DE. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 2002;277:9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 104.Ritchie KJ, Malakhov MP, Hetherington CJ, Zhou L, Little MT, et al. Dysregulation of protein modification by ISG15 results in brain cell injury. Genes Dev. 2002;16:2207–2212. doi: 10.1101/gad.1010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Malakhova OA, Yan M, Malakhov MP, Yuan Y, Ritchie KJ, et al. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 2003;17:455–460. doi: 10.1101/gad.1056303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ritchie KJ, Hahn CS, Kim KI, Yan M, Rosario D, et al. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat. Med. 2004;10:1374–1378. doi: 10.1038/nm1133. [DOI] [PubMed] [Google Scholar]
- 107.Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KG, et al. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25:2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Knobeloch KP, Utermohlen O, Kisser A, Prinz M, Horak I. Reexaminationofthe role of ubiquitin-like modifier ISG15 in the phenotype of UBP43-deficient mice. Mol. Cell. Biol. 2005;25:11030–11034. doi: 10.1128/MCB.25.24.11030-11034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Osiak A, Utermöhlen O, Niendorf S, Horak I, Knobeloch K-P. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol. Cell. Biol. 2005;25:6338–6345. doi: 10.1128/MCB.25.15.6338-6345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim KI, Yan M, Malakhova O, Luo JK, Shen MF, et al. Ube1L and protein ISGylation are not essential for α/β interferon signaling. Mol. Cell. Biol. 2006;26:472–479. doi: 10.1128/MCB.26.2.472-479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Francois-Newton V, Magno de Freitas Almeida G, Payelle-Brogard B, Monneron D, Pichard-Garcia L, et al. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon α response. PLoS ONE. 2011;6:e22200. doi: 10.1371/journal.pone.0022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Makowska Z, Duong FH, Trincucci G, Tough DF, Heim MH. Interferon-β and interferon-λ signaling is not affected by interferon-induced refractoriness to interferon-α in vivo. Hepatology. 2011;53:1154–1163. doi: 10.1002/hep.24189. [DOI] [PubMed] [Google Scholar]
- 113.Coelho LF, Magno de Freitas Almeida G, Mennechet FJ, Blangy A, Uze G. Interferon-α and -β differentially regulate osteoclastogenesis: role of differential induction of chemokine CXCL11 expression. Proc. Natl. Acad. Sci. USA. 2005;102:11917–11922. doi: 10.1073/pnas.0502188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen L, Borozan I, Feld J, Sun J, Tannis LL, et al. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128:1437–1444. doi: 10.1053/j.gastro.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 115.Sarasin-Filipowicz M, Oakeley EJ, Duong FHT, Christen V, Terracciano L, et al. Interferon signaling and treatment outcome in chronic hepatitis C. Proc. Natl. Acad. Sci. USA. 2008;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, et al. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature. 2010;465:502–506. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- 117.Klockow B, Tichelaar W, Madden DR, Niemann HH, Akiba T, et al. ThedynaminAring complex: molecular organization and nucleotide-dependent conformational changes. EMBO J. 2002;21:240–250. doi: 10.1093/emboj/21.3.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Haller O, Kochs G. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J. Interferon Cytokine Res. 2011;31:79–87. doi: 10.1089/jir.2010.0076. [DOI] [PubMed] [Google Scholar]
- 119.Sadler AJ, Williams BRG. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, et al. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature. 2013;502:563–566. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Goujon C, Moncorgé O, Bauby H, Doyle T, Ward CC, et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu Z, Pan Q, Ding S, Qian J, Xu F, et al. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe. 2013;14:398–410. doi: 10.1016/j.chom.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 123.Bauman DR, Bitmansour AD, McDonald JG, Thompson BM, Liang G, Russell DW. 25-Hydroxycholesterol secreted by macrophages in response to Toll-like receptor activation suppresses immunoglobulin A production. Proc. Natl. Acad. Sci. USA. 2009;106:16764–16769. doi: 10.1073/pnas.0909142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Diczfalusy U, Olofsson KE, Carlsson AM, Gong M, Golenbock DT, et al. Marked upregulation of cholesterol 25-hydroxylase expression by lipopolysaccharide. J. Lipid Res. 2009;50:2258–2264. doi: 10.1194/jlr.M900107-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Park K, Scott AL. Cholesterol 25-hydroxylase production by dendritic cells and macrophages is regulated by type I interferons. J. Leukoc. Biol. 2010;88:1081–1087. doi: 10.1189/jlb.0610318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, et al. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity. 2013;38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Blanc M, Hsieh WY, Robertson KA, Kropp KA, Forster T, et al. The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response. Immunity. 2013;38:106–118. doi: 10.1016/j.immuni.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Blanc M, Hsieh WY, Robertson KA, Watterson S, Shui G, et al. Host defense against viral infection involves interferon mediated down-regulation of sterol biosynthesis. PLoS Biol. 2011;9:e1000598. doi: 10.1371/journal.pbio.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Moog C, Aubertin AM, Kirn A, Luu B. Oxysterols, but not cholesterol, inhibit human immunodeficiency virus replication in vitro. Antivir. Chem. Chemother. 1998;9:491–496. doi: 10.1177/095632029800900605. [DOI] [PubMed] [Google Scholar]
- 130.Pezacki JP, Sagan SM, Tonary AM, Rouleau Y, Belanger S, et al. Transcriptional profiling of the effects of 25-hydroxycholesterol on human hepatocyte metabolism and the antiviral state it conveys against the hepatitis C virus. BMC Chem. Biol. 2009;9:2. doi: 10.1186/1472-6769-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, et al. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. USA. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Espenshade PJ, Hughes AL. Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 2007;41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 133.Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 134.Einav S, Glenn JS. Prenylation inhibitors: a novel class of antiviral agents. J. Antimicrob. Chemother. 2003;52:883–886. doi: 10.1093/jac/dkg490. [DOI] [PubMed] [Google Scholar]
- 135.Wilson SJ, Schoggins JW, Zang T, Kutluay SB, Jouvenet N, et al. Inhibition of HIV-1 particle assembly by 2′,3′-cyclic-nucleotide 3′-phosphodiesterase. Cell Host Microbe. 2012;12:585–597. doi: 10.1016/j.chom.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bordier BB, Ohkanda J, Liu P, Lee SY, Salazar FH, et al. In vivo antiviral efficacy of prenylation inhibitors against hepatitis delta virus. J. Clin. Investig. 2003;112:407–414. doi: 10.1172/JCI17704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bordier BB, Marion PL, Ohashi K, Kay MA, Greenberg HB, et al. A prenylation inhibitor prevents production of infectious hepatitis delta virus particles. J. Virol. 2002;76:10465–10472. doi: 10.1128/JVI.76.20.10465-10472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Glenn JS, Watson JA, Havel CM, White JM. Identification of a prenylation site in delta virus large antigen. Science. 1992;256:1331–1333. doi: 10.1126/science.1598578. [DOI] [PubMed] [Google Scholar]
- 139.Wang C, Gale M, Jr, Keller BC, Huang H, Brown MS, et al. Identification of FBL2 as a geranyl-geranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell. 2005;18:425–434. doi: 10.1016/j.molcel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 140.Gower TL, Graham BS. Antiviral activity of lovastatin against respiratory syncytial virus in vivo and in vitro. Antimicrob. Agents Chemother. 2001;45:1231–1237. doi: 10.1128/AAC.45.4.1231-1237.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ye J, Wang C, Sumpter R, Jr, Brown MS, Goldstein JL, Gale M., Jr Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci. USA. 2003;100:15865–15870. doi: 10.1073/pnas.2237238100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kapadia SB, Chisari FV. Hepatitis C virus RNAreplication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. USA. 2005;102:2561–2566. doi: 10.1073/pnas.0409834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, et al. The IFITM proteins mediate cellular resistance to influenza AH1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wilkins C, Woodward J, Lau DT, Barnes A, Joyce M, et al. IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology. 2013;57:461–469. doi: 10.1002/hep.26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Anafu AA, Bowen CH, Chin CR, Brass AL, Holm GH. Interferon-inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry. J. Biol. Chem. 2013;288:17261–17271. doi: 10.1074/jbc.M112.438515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lu J, Pan Q, Rong L, He W, Liu SL, Liang C. The IFITM proteins inhibit HIV-1 infection. J. Virol. 2011;85:2126–2137. doi: 10.1128/JVI.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chan YK, Huang IC, Farzan M. IFITM proteins restrict antibody-dependent enhancement of dengue virus infection. PLoS ONE. 2012;7:e34508. doi: 10.1371/journal.pone.0034508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Feeley EM, Sims JS, John SP, Chin CR, Pertel T, et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7:e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yount JS, Karssemeijer RA, Hang HC. S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J. Biol. Chem. 2012;287:19631–19641. doi: 10.1074/jbc.M112.362095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yount JS, Moltedo B, Yang YY, Charron G, Moran TM, et al. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 2010;6:610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jia R, Pan Q, Ding S, Rong L, Liu SL, et al. The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J. Virol. 2012;86:13697–13707. doi: 10.1128/JVI.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Everitt AR, Clare S, Pertel T, John SP, Wash RS, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li K, Markosyan RM, Zheng YM, Golfetto O, Bungart B, et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9:e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chu Y, Yang X. SUMO E3 ligase activity of TRIM proteins. Oncogene. 2011;30:1108–1116. doi: 10.1038/onc.2010.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zou W, Zhang DE. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J. Biol. Chem. 2006;281:3989–3994. doi: 10.1074/jbc.M510787200. [DOI] [PubMed] [Google Scholar]
- 159.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5 α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 161.Campbell EM, Perez O, Anderson JL, Hope TJ. Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5 α. J. Cell Biol. 2008;180:549–561. doi: 10.1083/jcb.200706154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sawyer SL, Emerman M, Malik HS. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 2007;3:e197. doi: 10.1371/journal.ppat.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Duggal NK, Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat. Rev. Immunol. 2012;12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, et al. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tissot C, Mechti N. Molecular cloning of a new interferon-induced factor that represses human immunodeficiency virus type 1 long terminal repeat expression. J. Biol. Chem. 1995;270:14891–14898. doi: 10.1074/jbc.270.25.14891. [DOI] [PubMed] [Google Scholar]
- 166.Barr SD, Smiley JR, Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 2008;4:e1000007. doi: 10.1371/journal.ppat.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Singh R, Gaiha G, Werner L, McKim K, Mlisana K, Team CAIS, et al. Association of TRIM22 with the type 1 interferon response and viral control during primary HIV-1 infection. J. Virol. 2011;85:208–216. doi: 10.1128/JVI.01810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hattlmann CJ, Kelly JN, Barr SD. TRIM22: a diverse and dynamic antiviral protein. Mol. Biol. Int. 2012;2012:153415. doi: 10.1155/2012/153415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gao B, Duan Z, Xu W, Xiong S. Tripartite motif-containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear-located RING domain. Hepatology. 2009;50:424–433. doi: 10.1002/hep.23011. [DOI] [PubMed] [Google Scholar]
- 170.Eldin P, Papon L, Oteiza A, Brocchi E, Lawson TG, Mechti N. TRIM22 E3 ubiquitin ligase activity is required to mediate antiviral activity against encephalomyocarditis virus. J. Gen. Virol. 2009;90:536–545. doi: 10.1099/vir.0.006288-0. [DOI] [PubMed] [Google Scholar]
- 171.Di Pietro A, Kajaste-Rudnitski A, Oteiza A, Nicora L, Towers GJ, et al. TRIM22 inhibits influenza A virus infection by targeting the viral nucleoprotein for degradation. J. Virol. 2013;87:4523–4533. doi: 10.1128/JVI.02548-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Geoffroy MC, Chelbi-Alix MK. Role of promyelocytic leukemia protein in host antiviral defense. J. Interferon Cytokine Res. 2011;31:145–58. doi: 10.1089/jir.2010.0111. [DOI] [PubMed] [Google Scholar]
- 173.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, et al. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 175.Munir M. TRIM proteins: another class of viral victims. Sci. Signal. 2010;3 doi: 10.1126/scisignal.3118jc2. jc2. [DOI] [PubMed] [Google Scholar]
- 176.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 177.Goff SP. Retrovirus restriction factors. Mol. Cell. 2004;16:849–859. doi: 10.1016/j.molcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 178.Kok K-H, Jin D-Y. Balance of power in host-virus arms races. Cell Host Microbe. 2013;14:5–6. doi: 10.1016/j.chom.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 179.Farrell PJ, Broeze RJ, Lengyel P. Accumulation of an mRNA and protein in interferon-treated Ehrlich ascites tumour cells. Nature. 1979;279:523–525. doi: 10.1038/279523a0. [DOI] [PubMed] [Google Scholar]
- 180.Zhao C, Collins MN, Hsiang T-Y, Krug RM. Interferon-induced ISG15 pathway: an ongoing virus-host battle. Trends Microbiol. 2013;21:181–186. doi: 10.1016/j.tim.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sgorbissa A, Brancolini C. IFNs, ISGylation and cancer: Cui prodest? Cytokine Growth Factor Rev. 2012;23:307–314. doi: 10.1016/j.cytogfr.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 182.Shi H-X, Yang K, Liu X, Liu X-Y, Wei B, et al. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol. Cell. Biol. 2010;30:2424–2436. doi: 10.1128/MCB.01466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Okumura F, Zou W, Zhang DE. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 2007;21:255–260. doi: 10.1101/gad.1521607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Feng Q, Sekula D, Guo Y, Liu X, Black CC, et al. UBE1L causes lung cancer growth suppression by targeting cyclin D1. Mol. Cancer Ther. 2008;7:3780–3788. doi: 10.1158/1535-7163.MCT-08-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Durfee LA, Lyon N, Seo K, Huibregtse JM. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol. Cell. 2010;38:722–732. doi: 10.1016/j.molcel.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, et al. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. USA. 2007;104:1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Guerra S, Ca´ceres A, Knobeloch K-P, Horak I, Esteban M. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog. 2008;4:e1000096. doi: 10.1371/journal.ppat.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bogunovic D, Boisson-Dupuis S, Casanova J-L. ISG15: leading a double life as a secreted molecule. Exp. Mol. Med. 2013;45:e18. doi: 10.1038/emm.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bogunovic D, Byun M, Durfee LA, Abhyankar A, Sanal O, et al. Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science. 2012;337:1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Indraccolo S, Pfeffer U, Minuzzo S, Esposito G, Roni V, et al. Identification of genes selectively regulated by IFNs in endothelial cells. J. Immunol. 2007;178:1122–1135. doi: 10.4049/jimmunol.178.2.1122. [DOI] [PubMed] [Google Scholar]
- 191.Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, Hartmann R. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J. Virol. 2007;81:7749–7758. doi: 10.1128/JVI.02438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Severa M, Coccia EM, Fitzgerald KA. Toll-like receptor-dependent and -independent viperin gene expression and counter-regulation by PRDI-binding factor-1/BLIMP1. J. Biol. Chem. 2006;281:26188–26195. doi: 10.1074/jbc.M604516200. [DOI] [PubMed] [Google Scholar]
- 193.Chin KC, Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA. 2001;98:15125–15130. doi: 10.1073/pnas.011593298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Stirnweiss A, Ksienzyk A, Klages K, Rand U, Grashoff M, et al. IFN regulatory factor-1 bypasses IFN-mediated antiviral effects through viperin gene induction. J. Immunol. 2010;184:5179–5185. doi: 10.4049/jimmunol.0902264. [DOI] [PubMed] [Google Scholar]
- 195.DeFilippis VR, Robinson B, Keck TM, Hansen SG, Nelson JA, Fru¨h KJ. Interferon regulatory factor 3 is necessary for induction of antiviral genes during human cytomegalovirus infection. J. Virol. 2006;80:1032–1037. doi: 10.1128/JVI.80.2.1032-1037.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Seo JY, Yaneva R, Hinson ER, Cresswell P. Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science. 2011;332:1093–1097. doi: 10.1126/science.1202007. [DOI] [PubMed] [Google Scholar]
- 197.Szretter KJ, Brien JD, Thackray LB, Virgin HW, Cresswell P, Diamond MS. The interferon-inducible gene viperin restricts West Nile virus pathogenesis. J. Virol. 2011;85:11557–11566. doi: 10.1128/JVI.05519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang X, Hinson ER, Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007;2:96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 199.Nasr N, Maddocks S, Turville SG, Harman AN, Woolger N, et al. HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood. 2012;120:778–788. doi: 10.1182/blood-2012-01-407395. [DOI] [PubMed] [Google Scholar]
- 200.Helbig KJ, Eyre NS, Yip E, Narayana S, Li K, et al. The antiviral protein viperin inhibits hepatitis C virus replication via interaction with nonstructural protein 5A. Hepatology. 2011;54:1506–1517. doi: 10.1002/hep.24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bartenschlager R, Penin F, Lohmann V, Andre P. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 2011;19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 202.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat. Rev. Microbiol. 2007;5:453–63. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 203.Wang S, Wu X, Pan T, Song W, Wang Y, et al. Viperin inhibits hepatitis C virus replication by interfering with binding of NS5A to host protein hVAP-33. J. Gen. Virol. 2012;93:83–92. doi: 10.1099/vir.0.033860-0. [DOI] [PubMed] [Google Scholar]
- 204.Helbig KJ, Carr JM, Calvert JK, Wati S, Clarke JN, et al. Viperin is induced following dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the C-terminal end of viperin. PLoS Neglected Trop. Dis. 2013;7:e2178. doi: 10.1371/journal.pntd.0002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, et al. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009;139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Swiecki M, Omattage NS, Brett TJ. BST-2/tetherin: structural biology, viral antagonism, and immunobiology of a potent host antiviral factor. Mol. Immunol. 2013;54:132–139. doi: 10.1016/j.molimm.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 207.Le Tortorec A, Neil SJD. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J. Virol. 2009;83:11966–11978. doi: 10.1128/JVI.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, et al. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 2009;5:e1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, et al. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe. 2009;6:54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc. Natl. Acad. Sci. USA. 2009;106:2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Mangeat B, Cavagliotti L, Lehmann M, Gers-Huber G, Kaur I, et al. Influenza virus partially counteracts restriction imposed by tetherin/BST-2. J. Biol. Chem. 2012;287:22015–22029. doi: 10.1074/jbc.M111.319996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Blanco-Melo D, Venkatesh S, Bieniasz PD. Intrinsic cellular defenses against human immunodeficiency viruses. Immunity. 2012;37:399–411. doi: 10.1016/j.immuni.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zhao H, Lin W, Kumthip K, Cheng D, Fusco DN, et al. A functional genomic screen reveals novel host genes that mediate interferon-α’s effects against hepatitis C virus. J. Hepatol. 2012;56:326–333. doi: 10.1016/j.jhep.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Metz P, Dazert E, Ruggieri A, Mazur J, Kaderali L, et al. Identification of type I and type II interferon-induced effectors controlling hepatitis C virus replication. Hepatology. 2012;56:2082–2093. doi: 10.1002/hep.25908. [DOI] [PubMed] [Google Scholar]
- 215.Li J, Ding SC, Cho H, Chung BC, Gale M, Jr, et al. A short hairpin RNA screen of interferon-stimulated genes identifies a novel negative regulator of the cellular antiviral response. MBio. 2013;4:e00385–13. doi: 10.1128/mBio.00385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhang Y, Burke CW, Ryman KD, Klimstra WB. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 2007;81:11246–11255. doi: 10.1128/JVI.01282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Itsui Y, Sakamoto N, Kurosaki M, Kanazawa N, Tanabe Y, et al. Expressional screening of interferon-stimulated genes for antiviral activity against hepatitis C virus replication. J. Viral. Hepat. 2006;13:690–700. doi: 10.1111/j.1365-2893.2006.00732.x. [DOI] [PubMed] [Google Scholar]
- 218.Jiang D, Guo H, Xu C, Chang J, Gu B, et al. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J. Virol. 2008;82:1665–1678. doi: 10.1128/JVI.02113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Jiang D, Weidner JM, Qing M, Pan XB, Guo H, et al. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J. Virol. 2010;84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Cho H, Proll SC, Szretter KJ, Katze MG, Gale M, Jr, Diamond MS. Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses. Nat. Med. 2013;19:458–464. doi: 10.1038/nm.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Schoggins JW, Macduff DA, Imanaka N, Gainey MD, Shrestha B, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Karki S, Li MM, Schoggins JW, Tian S, Rice CM, MacDonald MR. Multiple interferon stimulated genes synergize with the zinc finger antiviral protein to mediate anti-alphavirus activity. PLoS ONE. 2012;7:e37398. doi: 10.1371/journal.pone.0037398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.de Veer MJ, Holko M, Frevel M, Walker E, Der S, et al. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- 224.Hilkens CM, Schlaak JF, Kerr IM. Differential responses to IFN-α subtypes in human T cells and dendritic cells. J. Immunol. 2003;171:5255–5263. doi: 10.4049/jimmunol.171.10.5255. [DOI] [PubMed] [Google Scholar]
- 225.Hultcrantz M, Huhn MH, Wolf M, Olsson A, Jacobson S, et al. Interferons induce an antiviral state in human pancreatic islet cells. Virology. 2007;367:92–101. doi: 10.1016/j.virol.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 226.Lanford RE, Guerra B, Lee H, Chavez D, Brasky KM, Bigger CB. Genomic response to interferon-α in chimpanzees: implications of rapid downregulation for hepatitis C kinetics. Hepatology. 2006;43:961–972. doi: 10.1002/hep.21167. [DOI] [PubMed] [Google Scholar]
- 227.Leaman DW, Chawla-Sarkar M, Jacobs B, Vyas K, Sun Y, et al. Novel growth and death related interferon-stimulated genes (ISGs) in melanoma: greater potency of IFN-β compared with IFN-α2. J. Interferon Cytokine Res. 2003;23:745–756. doi: 10.1089/107999003772084860. [DOI] [PubMed] [Google Scholar]
- 228.Rani MR, Shrock J, Appachi S, Rudick RA, Williams BR, Ransohoff RM. Novel interferon-β-induced gene expression in peripheral blood cells. J. Leukoc. Biol. 2007;82:1353–1360. doi: 10.1189/jlb.0507273. [DOI] [PubMed] [Google Scholar]
- 229.Kincaid RP, Sullivan CS. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog. 2012;8:e1003018. doi: 10.1371/journal.ppat.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 231.Yang CH, Yue J, Fan M, Pfeffer LM. IFNinducesmiR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res. 2010;70:8108–8116. doi: 10.1158/0008-5472.CAN-10-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Schmitt MJ, Philippidou D, Reinsbach SE, Margue C, Wienecke-Baldacchino A, et al. Interferon-gamma-induced activation of signal transducer and activator of transcription 1 (STAT1) up-regulates the tumor suppressing microRNA-29 family in melanoma cells. Cell Commun. Signal. 2012;10:41. doi: 10.1186/1478-811X-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 234.Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio. 2010;1:e00206–e00210. doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, et al. TheNeSTlongncRNAcontrols microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Vigneau S, Rohrlich PS, Brahic M, Bureau JF. Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J. Virol. 2003;77:5632–5638. doi: 10.1128/JVI.77.10.5632-5638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Liu J, Shue E, Ewalt KL, Schimmel P. A new gamma-interferon-inducible promoter and splice variants of an anti-angiogenic human tRNA synthetase. Nucleic Acids Res. 2004;32:719–727. doi: 10.1093/nar/gkh240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Keegan LP, Leroy A, Sproul D, O’Connell MA. Adenosine deaminases acting on RNA (ADARs): RNA-editing enzymes. Genome Biol. 2004;5:209. doi: 10.1186/gb-2004-5-2-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Araujo PR, Yoon K, Ko D, Smith AD, Qiao M, et al. Before it gets started: regulating translation at the 5’ UTR. Comp. Funct. Genomics. 2012;2012:475731. doi: 10.1155/2012/475731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Zhao W, Blagev D, Pollack JL, Erle DJ. Toward a systematic understanding of mRNA 3 untranslated regions. Proc. Am. Thorac. Soc. 2011;8:163–166. doi: 10.1513/pats.201007-054MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 244.Schulz O, Pichlmair A, Rehwinkel J, Rogers NC, Scheuner D, et al. Protein kinase R contributes to immunity against specific viruses by regulating interferon mRNA integrity. Cell Host Microbe. 2010;7:354–361. doi: 10.1016/j.chom.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Anderson P. Post-transcriptional control of cytokine production. Nat. Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 246.Tang X, Gao JS, Guan YJ, McLane KE, Yuan ZL, et al. Acetylation-dependent signal transduction for type I interferon receptor. Cell. 2007;131:93–105. doi: 10.1016/j.cell.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 247.Gack MU, Shin YC, Joo CH, Urano T, Liang C, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 248.Meissner F, Scheltema RA, Mollenkopf HJ, Mann M. Direct proteomic quantification of the secretome of activated immune cells. Science. 2013;340:475–478. doi: 10.1126/science.1232578. [DOI] [PubMed] [Google Scholar]
- 249.Li J, Liu K, Liu Y, Xu Y, Zhang F, et al. Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat. Immunol. 2013;14:793–803. doi: 10.1038/ni.2647. [DOI] [PubMed] [Google Scholar]