Genomic Disorders on 22q11 (original) (raw)
Abstract
The 22q11 region is involved in chromosomal rearrangements that lead to altered gene dosage, resulting in genomic disorders that are characterized by mental retardation and/or congenital malformations. Three such disorders—cat-eye syndrome (CES), der(22) syndrome, and velocardiofacial syndrome/DiGeorge syndrome (VCFS/DGS)—are associated with four, three, and one dose, respectively, of parts of 22q11. The critical region for CES lies centromeric to the deletion region of VCFS/DGS, although, in some cases, the extra material in CES extends across the VCFS/DGS region. The der(22) syndrome region overlaps both the CES region and the VCFS/DGS region. Molecular approaches have revealed a set of common chromosome breakpoints that are shared between the three disorders, implicating specific mechanisms that cause these rearrangements. Most VCFS/DGS and CES rearrangements are likely to occur by homologous recombination events between blocks of low-copy repeats (e.g., LCR22), whereas nonhomologous recombination mechanisms lead to the constitutional t(11;22) translocation. Meiotic nondisjunction events in carriers of the t(11;22) translocation can then lead to offspring with der(22) syndrome. The molecular basis of the clinical phenotype of these genomic disorders has also begun to be addressed. Analysis of both the genomic sequence for the 22q11 interval and the orthologous regions in the mouse has identified >24 genes that are shared between VCFS/DGS and der(22) syndrome and has identified 14 putative genes that are shared between CES and der(22) syndrome. The ability to manipulate the mouse genome aids in the identification of candidate genes in these three syndromes. Research on genomic disorders on 22q11 will continue to expand our knowledge of the mechanisms of chromosomal rearrangements and the molecular basis of their phenotypic consequences.
Introduction
The term “genomic disorders” refers to those diseases that are caused by chromosomal rearrangements involving large regions of one to several megabase pairs (reviewed in Lupski 1998). Chromosomal rearrangements can result in interstitial or terminal deletions or duplications, as well as unbalanced translocations, and all of these consequences may subsequently result in imbalanced gene dosage. Meiotic nondisjunction events in normal carriers of balanced translocations may also lead to a disturbance of gene dosage in offspring. Each of these rearrangements occurs sporadically in the population and therefore represents the product of de novo mutations. Many of the rearrangements occur in specific regions of the genome, suggesting specific mechanisms. Although each individual disorder is rare in the population, when combined, they are responsible for a substantial proportion (0.7% of live births) of birth defects (Borgaonkar 1989). Most of the rearrangements are associated with both congenital malformations and mental retardation. Therefore, genomic disorders have a large impact on human health. The 22q11 region serves as a model for genomic disorders because it is particularly susceptible to chromosomal rearrangements, leading to three different congenital malformation syndromes. In the present review, we focus on efforts to determine both the mechanism for the rearrangements and the molecular basis of these disorders.
Clinical and Cytogenetic Features of Genomic Disorders on 22q11
The most common genomic disorder on 22q11 is velocardiofacial syndrome/DiGeorge syndrome (VCFS/DGS [MIM 192430; MIM 601362]). With a frequency of 1/4,000 live births (Burn and Goodship 1996), VCFS/DGS is the most common genomic disorder in humans. The main clinical findings include learning disabilities, characteristic facial appearance, velopharyngeal insufficiency, hypernasal speech, occult cleft palate, and conotruncal heart defects (outflow-tract defects) (Shprintzen et al. 1978). A more severely affected subset of patients have a reduced or absent thymus gland and hypocalcemia (DiGeorge 1965). There are many additional clinical findings that are associated with this disorder. Most features show variable expressivity and penetrance. In addition to having physical malformations, most affected children have learning disabilities and behavioral disorders (Swillen et al. 1999). Adults with this syndrome develop major psychiatric illnesses, including schizophrenia and bipolar disorder (Chow et al. 1994; Pulver et al. 1994; Papolos et al. 1996; Murphy et al. 1999; Shprintzen 2001; reviewed in Murphy and Owen 2001). Most patients with VCFS/DGS have a deletion in 22q11, making this region hemizygous (fig. 1) (de la Chapelle et al. 1981; Driscoll et al. 1992; Scambler et al. 1992; Lindsay et al. 1993). The deletion was first identified by cytogenetic methods and was later confirmed by molecular approaches, including FISH (Driscoll et al. 1993) and haplotype analysis with genetic markers (Morrow et al. 1995).
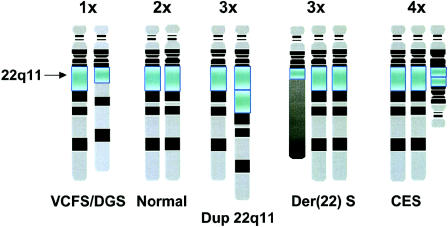
Figure 1.
Rearrangements on 22q11. A multitude of germline rearrangements on 22q11 (boxed) are associated with congenital anomaly disorders. VCFS/DGS is associated with interstitial hemizygous deletions; a family has been reported with an interstitial duplication of the same interval that is deleted in patients with VCFS/DGS (Edelmann et al. 1999_b_). Der(22) syndrome occurs in offspring of unaffected carriers of the constitutional t(11;22) translocation; patients with der(22) syndrome have a partial trisomy for 22pter-q11 and 11q23-qter. Patients with CES harbor a bisatellited supernumerary chromosome 22 that results in a partial tetrasomy.
In contrast to VCFS/DGS, cat-eye syndrome (CES [MIM 115470]) and der(22) syndrome are rare disorders. The clinical findings for CES are distinct from those in patients with VCFS/DGS, and features of CES include ocular colobomata, anal atresia, congenital heart defects (typically, total anomalous pulmonary venous return [TAPVR]), renal malformations, craniofacial anomalies (e.g., preauricular skin tags and pits), male genital anomalies, skeletal defects, and borderline-to-moderate mental retardation (Schinzel et al. 1981; reviewed in Berends et al. 2001). As with VCFS/DGS, most features show variable expressivity and penetrance. Patients with CES have a partial tetrasomy (i.e., four copies) of the region that spans the p-arm and part of 22q11 (fig. 1) (Schinzel et al. 1981; McDermid et al. 1986). The extra copies of this region are usually in the form of a chromosome that is supernumerary, bisatellited, and dicentric—that is, an inv dup(22), or the CES chromosome. Features of CES (including colobomata and TAPVR) have also been associated with interstitial duplications (i.e., three copies) of part of the 22q11 region (Reiss et al. 1985; Knoll et al. 1995).
The second disorder associated with increased gene dosage of 22q11 is der(22) syndrome. Many of the clinical findings for der(22) syndrome are the same as those for CES, although there are also distinct differences (Fraccaro et al. 1980; Van Hove et al. 1992). Features of der(22) syndrome that are similar to CES include heart defects (primarily, atrial septal defects; conotruncal heart defects are not a finding in this syndrome; Lin et al. 1986), renal malformations, craniofacial anomalies (e.g., preauricular tags and pits and cleft palate), male genital anomalies, anal stenosis or atresia, skeletal defects, and mental retardation (reviewed in Knoll et al. 1995). Interestingly, colobomata, microphthalmia, and TAPVR, which are common in CES, are not seen in these patients. Der(22) syndrome occurs by nondisjunction events from normal carriers of the constitutional t(11;22) translocation (fig. 1) (Zakai and Emanuel 1980). This is the only known constitutional germline translocation that recurs in humans. Patients with der(22) syndrome have a partial trisomy of both the 11q23-qter region and the 22q11-qter region (Zakai and Emanuel 1980; Schinzel et al. 1981).
Definition of the Regions Harboring Chromosomal Breakpoints
Definition of the intervals that are rearranged in genomic disorders on 22q11 has led to insight into the overlap between the syndromes, as well as into how the rearrangements occur. High-density genetic markers were used to define the extent of the deletion in patients with VCFS/DGS. As many as 15 consecutive genetic markers in the 22q11 region were used to genotype >150 patients and their unaffected parents (Carlson et al. 1997). Haplotypes were then deduced on the basis of the genotypes. Of the 83% of patients with a detectable deletion, 90% had a similar 3-Mb deletion. Another 7% of deleted patients had the same proximal breakpoint as those with the 3-Mb deletion but had a nested distal deletion endpoint resulting in a 1.5-Mb deletion (fig. 2) (Carlson et al. 1997). Also, there was no sex-based bias (in terms of whether the deletions occurred on the maternal or paternal chromosome), indicating a lack of imprinting in the region (Morrow et al. 1995). Deletions of other sizes have also been identified in the interval, in a small subset of patients (Edelmann et al. 1999_b;_ Shaikh et al. 2000).
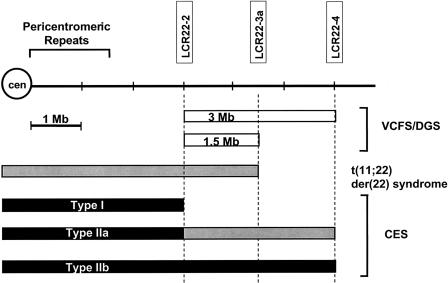
Figure 2.
LCR22s in mediation of chromosomal rearrangements. LCR22 designations have been described elsewhere (Dunham et al. 1999). The proximal endpoints for the 1.5- and 3-Mb VCFS/DGS deletions occur in LCR22-2, and the distal endpoints occur in LCR22-3a and LCR22-4, respectively. The constitutional t(11;22) translocation disrupts LCR22-3a. Three CES-duplication endpoints—those for types I, IIa, and IIb—are shown. Deletions (white boxes), partial trisomies (i.e., three copies; gray boxes), and partial tetrasomies (i.e., four copies; black boxes) are depicted.
A combination of FISH and quantitative dosage analysis identified two recurrent regions for the CES breakpoints (fig. 2) (Mears et al. 1994; McTaggart et al. 1998). Each inv dup(22) CES chromosome contains two 22q11 breakpoints (fig. 1), but the location of the individual breakpoints can differ, thereby resulting in CES chromosomes with duplications of different sizes. The two recurrent CES breakpoint regions occur in the same intervals as do the proximal and distal breakpoints of the 3-Mb VCFS/DGS deletion (McTaggart et al. 1998; Edelmann et al. 1999_b_). Thus, type I CES chromosomes are symmetrical, with two proximal breakpoints (fig. 2). Type II CES chromosomes contain either one proximal and one distal breakpoint (type IIa, asymmetrical) or two distal breakpoints (type IIb, symmetrical), resulting in an additional one or two copies of the VCFS/DGS region. There is no obvious difference between the phenotypes of individuals with type I or type II CES chromosomes, although the syndrome is highly variable and although relatively few patients have been studied (McTaggart et al. 1998). That the CES critical region is centromeric to the 3-Mb region that is deleted in VCFS/DGS suggests that duplication of the 3-Mb VCFS/DGS region alone would have a subtle phenotype.
For der(22) syndrome, the t(11;22) breakpoint occurs in the same interval as does the nested distal 1.5-Mb VCFS/DGS breakpoint (fig. 2) (Funke et al. 1999; Shaikh et al. 1999). Thus, this syndrome overlaps the regions for both CES and VCFS/DGS, suggesting that 22q11 is exquisitely prone to a multitude of rearrangements.
Together, the coincident locations of the breakpoints in most patients with the three genomic disorders on 22q11 that we herein review provide strong evidence for site-specific mechanisms for the sporadic rearrangements on 22q11. This evidence was further elucidated using BAC and cosmid libraries to generate physical maps of 22q11 (Collins et al. 1995). Analysis of the breakpoint regions demonstrated that sequences that are present in the common 3-Mb proximal breakpoint interval were repeated in the 1.5- and 3-Mb distal breakpoint intervals (e.g., LCR22; see fig. 2) (Edelmann et al. 1999_a;_ Shaikh 2000), suggesting that recombination between the intervals results in the chromosomal rearrangements.
Mechanisms for Rearrangements on 22q11
All three breakpoint regions, each of which are 1.5 Mb apart, harbor a similar low-copy repeat (LCR) that is known as an “LCR22” (fig. 2) (Edelmann et al. 1999_a_). Each LCR22 is larger than the inserts in the genomic clones that were used to construct the map. Therefore, the strategy to isolate overlapping clones that span each LCR22 was to identify distinguishing characteristics in each. Unique PCR-based landmarks that flanked each LCR22 were used to anchor genomic clones to individual LCR22s (Edelmann et al. 1999_a_). Clones that spanned the LCR22s have been sequenced and analyzed (Dunham et al. 1999). The two LCR22s that are 3 Mb apart are ∼200 kb and contain direct and inverted segments or modules (Dunham et al. 1999; Bailey et al. 2002). These regions contain genes, pseudogenes, and other genomic sequences that are 94%–99% identical within each component—both in and between each LCR22. The central LCR22, LCR22-3a (fig. 2) contains a gap in the physical map that has not been fully cloned, despite efforts to do so.
Therefore, these three LCR22s are present in the regions of chromosome breakage in three different genomic disorders. The VCFS/DGS common proximal endpoint and the CES type I endpoint map to LCR22-2, whereas the 3-Mb distal deletion endpoint in patients with VCFS/DGS and the CES type II endpoint map to LCR22-4. The 1.5-Mb distal deletion endpoint in patients with VCFS/DGS and the t(11;22) breakpoint map to LCR22-3a (fig. 2) (Funke et al. 1999). We also predict that there are CES chromosomes with one or both breakpoints at the central LCR22-3a. A probable example was described by Crolla et al. (1997), in which a supernumerary inv dic(22) in a child and her father was shown by FISH to contain the HIRA gene (between LCR22-2 and LCR22-3a) but not the genetic marker D22S264 (between LCR22-3a and LCR22-4). That the child displayed only developmental delay, severe hypotonia, and seizures and that the father was unaffected indicate the wide phenotypic variation seen for partial tetrasomy of 22q11.
Homologous recombination events between the LCR22s during meiosis have been implicated in VCFS/DGS and CES rearrangements on the basis of haplotype analysis of three generations of individuals with VCFS/DGS (Baumer et al. 1998; Edelmann et al. 1999_b;_ reviewed in Emanuel and Shaikh 2001). The reason for such a multitude of rearrangements lies in the complicated inverted and direct orientation of sequences in the LCR22s. For VCFS/DGS, both intrachromosomal and interchromosomal mechanisms occur (Baumer et al. 1998; Edelmann et al. 1999_b_). Homologous recombination between inverted and direct repeats in LCRs of sister chromatids and homologous chromosomes could also lead to _U_-type exchange (rather than the classic crossover _X_-type exchange), which would result in the formation of the inv dic(22) of CES, as well as an acentric fragment that would be lost (Schreck et al. 1977; Van Dyke et al. 1977).
Der(22) syndrome occurs in offspring of carriers of the constitutional t(11;22) translocation. The balanced t(11;22) translocation, although a rare event, is recurrent in the population (Zackai and Emanuel 1980). The intervals on 22q11 (Funke et al. 1999) and 11q23 (Edelmann et al. 1999_c_) that contain the region of chromosome breakage have been mapped and cloned (Kurahashi et al. 2000_a,_ 2000_b;_ Edelmann et al. 2001). The sites of chromosome breakage on 11q23 and 22q11 occur in AT-rich palindromic sequences. Such sequences are known to be unstable in eukaryotic genomes. The sites of chromosome breakage occur in the center of the palindromes of both parental chromosomes 11q23 and 22q11 (Edelmann et al. 2001; Kurahashi and Emanuel 2001). We hypothesize that double-strand breaks at the tip of a putative hairpin, the center of the palindrome, lead to nonhomologous end-joining mechanisms that result in the formation of a stable, nonpalindromic sequence. Such rearrangements have been described in mammalian cell-culture systems (Akgun et al. 1997).
Since interchromosomal recombination events occur sporadically in the population, individuals that carry a reciprocal duplication of the same region that is deleted in VCFS/DGS should exist, if the condition is not lethal. This region's presence in one or two extra copies in some cases of CES without obvious additional clinical features indicates that the phenotype of the VCFS/DGS-region duplication alone would likely show subtle or no effect. One such family was identified, in which the proband, her mother, and her grandmother carried a VCFS/DGS-region duplication (fig. 1) (Edelmann et al. 1999_b_). The proband was mildly affected, with developmental delay and hypotonia, but the mother and grandmother appeared to be unaffected (Edelmann et al. 1999_b_). Because of the mild nature of the anomalies and the difficulty of detecting submicroscopic duplications, it is likely that there are many individuals in the population with a similar rearrangement but that they remain undiagnosed. This would represent a fourth genomic disorder on 22q11.
Both homologous and nonhomologous recombination mechanisms that involve LCR22s lead to germline rearrangements on 22q11. Therefore, the genomic disorders on the 22q11 region serve as models to understand the molecular basis of chromosomal rearrangements in humans.
Animal Models for Genomic Disorders on 22q11
Since genomic disorders on 22q11 involve large regions of the genome, it is possible that the altered dosage of several contiguous genes is responsible for their etiology. Alternatively, only a single gene may be responsible. To determine the molecular basis of genomic disorders, it is possible to take mouse-genetics approaches. The generation of the complete sequence of chromosome 22 (Dunham et al. 1999) and orthologous regions in the mouse have provided the tools for such efforts (Dunham et al. 1999; Lund et al. 1999).
Although candidate genes can be chosen for study on the basis of both their presence in a critical region and their expression patterns, ultimate proof of their involvement in a genomic disorder can be difficult. The discovery of patients who harbor balanced translocations in 22q11 and have VCFS/DGS-related malformations was a disappointment, because there were no obvious candidates that were interrupted (Budarf et al. 1995; Levy et al. 1995; Sutherland et al. 1996; Holmes et al. 1997), thereby implying that the translocations created positional effects on neighboring genes. Another more successful strategy is the generation of mouse models that contain large nested deletions or duplications (Ramirez-Solis et al. 1995; Zheng et al. 2000; reviewed in Yu and Bradley 2001) This is possible as long as the genes of the human critical region are conserved as a unit on an individual mouse chromosome. Once the critical region for the syndrome is narrowed in the mouse model that harbors large rearrangements, each individual gene within the region can be inactivated using conventional gene-targeting approaches. The gene-targeted mutants can be compared with those mice that harbor chromosomal rearrangements to ascertain whether the disorder is a single or contiguous gene syndrome.
The 1.5-Mb region shared between VCFS/DGS and der(22) syndrome (and sometimes duplicated or triplicated in CES) lies on mouse chromosome 16 (MMU16) (Puech et al. 1997; Lund et al. 1999). It was possible to generate nested deletions within chromosome 16 to generate mouse models of VCFS/DGS (see below), since patients with VCFS/DGS who have the 1.5-Mb deletion have the same phenotypic spectrum as patients with the larger, 3-Mb deletion (Morrow et al. 1995). The distal 1.5-Mb VCFS/DGS region lies on mouse chromosome 10 (MMU10; see Human-Mouse Homology Map). For confirmation that altered dosage of this region does not affect the overall phenotype of VCFS/DGS, a deletion of this interval on MMU10 could also be generated.
Making CES mouse models may be more problematic. Most of the CES critical region (from IL-17R to ATP6E and MIL1; see fig. 3) maps to mouse chromosome 6 (MMU6; see Human-Mouse Homology Map) (Puech et al. 1997; Footz et al. 1998). Although a duplication of this region could be created, one of the human candidate genes (CECR1; see next section) is missing from this region of MMU6 and probably does not exist in the mouse genome (Footz et al. 2001). In addition, the CES critical region includes the 22q11 pericentromeric DNA, which contains a primate-specific patchwork of fragments that are duplicated from other human chromosomes (Footz et al. 2001; Bailey et al. 2002). Although this region is unlikely to contain active genes, any that do exist would not be present in the mouse.
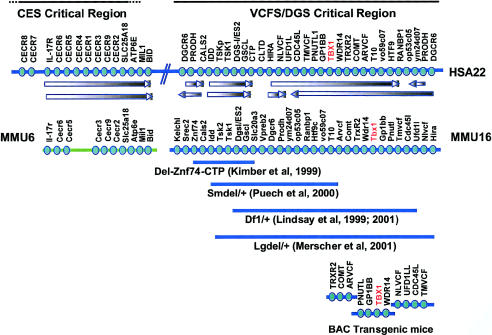
Figure 3.
Map of human 22q11 region and orthologous regions in mouse. The relative order of genes (circles) on the 22q11 region (HSA22) and orthologous genes on MMU6 (for CES) and MMU16 (for VCFS/DGS) are shown. The arrows between the human and mouse maps indicate orientation. Deletions that have been generated in MMU16, to generate mouse models of VCFS/DGS, are indicated by a solid blue line below MMU16. BAC transgenic lines that harbor human genes in the distal half of the 1.5-Mb region that is deleted in patients with VCFS/DGS have also been generated, to overexpress sets of genes for genetic complementation studies; the genes included in these lines are indicated at the bottom of the figure.
Tbx1 as a Candidate Gene for VCFS/DGS in Mouse Models
Many of the tissues and structures affected in patients with VCFS/DGS derive, during embryonic development, from the pharyngeal arches (reviewed in Kirby and Waldo 1995), which are conserved among all vertebrate organisms. Neural-crest cells migrate from a position adjacent to the neural tube and participate in the formation of both the pharyngeal arches and their derivatives. It has been hypothesized that defects in neural-crest cells are responsible for the characteristic features of VCFS/DGS (Kirby et al. 1983, 1985; Bockman and Kirby 1984; Phillips et al. 1987; Couly and Le Douarin 1987; reviewed in Kirby and Waldo 1995). Neural-crest ablation generates mice with malformations that are similar to those in patients with VCFS/DGS (Kirby et al. 1983; Bockman and Kirby 1984; Bockman et al. 1987). Therefore, a gene that is important for neural-crest cell function would be a candidate for VCFS/DGS.
For the identification of candidates for VCFS/DGS, nested deletions and duplications of the orthologous region on MMU16 were generated (fig. 3) (Puech et al. 1997; Kimber et al. 1999; Lindsay et al. 1999, 2001; Merscher et al. 2001). Mice that harbor a large, 1.5-Mb deletion (fig. 3), containing >24 genes and mimicking the nested 1.5-Mb deletion in humans, had reduced viability, conotruncal heart defects, and hypoparathyroidism. A critical region for these cardiovascular defects was defined through genetic complementation between different-sized deletions and BAC-containing transgenic mice (fig. 3) (Lindsay et al. 2001; Merscher et al. 2001). A BAC that harbored four human genes—GP1Bβ, PNUTL1, TBX1, and _WDR14_—provided complete rescue in most mice (Merscher et al. 2001). A PAC that contained the mouse homologues of the four genes provided complete rescue of the cardiovascular findings in embryos (Lindsay et al. 2001), suggesting that one of these four genes is responsible for the defects. It was noteworthy that the human transgene could not provide complete rescue, although this may have been due to either the insufficient expression of the transgenes or the inability of the human genes to completely replace the endogenous mouse genes.
One of the four genes in the BAC—TBX1, a member of the T-box–containing family of transcription-factor genes—is highly expressed in the pharyngeal arches during mouse embryonic development (Chapman et al. 1996). This gene was targeted for inactivation by three independent research groups (Jerome and Papaioannou 2001; Lindsay et al. 2001; Merscher et al. 2001). Tbx1 hemizygotes had mild cardiovascular defects (Jerome and Papaioannou 2001; Lindsay et al. 2001; Merscher et al. 2001) but did not show reduced viability, whereas homozygotes had more severe defects (Jerome and Papaioannou 2001). Tbx1 homozygosity was perinatally lethal, with thymus and parathyroid gland aplasia and major ear malformations. Homozygotes also showed cleft palate and truncus arteriosus, a more severe conotruncal heart defect than that shown in the heterozygotes (Jerome and Papaoiannou 2001). Many of these anomalies occur in patients with VCFS/DGS but are milder. On the basis of the three studies, Tbx1 is responsible for VCFS/DGS-related malformations in mouse models.
Overexpression of Tbx1 may also shed light on other genomic syndromes on 22q11. BAC-containing transgenic mice with overexpression of the four human genes that are listed above had a similar VCFS/DGS phenotype of their own. This phenotype included cleft palate, conotruncal heart defects, thymus gland hypoplasia, and ear defects (including chronic otitis media, a common finding in VCFS/DGS), indicating that one of these four genes is dosage sensitive for this aspect of development (Funke et al. 2001; Merscher et al. 2001). These results for the transgenic mouse support the proposal that one of the four genes on the human BAC is dosage sensitive for the development of relevant structures and is therefore a candidate gene for VCFS/DGS. It is also possible that overexpression of one of these genes may contribute to the overall phenotype in der(22) syndrome or in some cases of CES.
Other genes in the interval have been considered candidate genes for VCFS/DGS. They include HIRA (similar to yeast Hir1p and Hir2p) (Lamour et al. 1995; Roberts et al. 1997, 2002), UFD1L (Pizzuti et al. 1997; Yamagishi et al. 1999), and goosecoid-like (GSCL) (Lindsay et al. 1998; Saint-Jore et al. 1998; Wakamiya et al. 1998), but each has waned from interest because they did not fulfill the criteria for a candidate gene—that is, the gene was not expressed in the precursors or the affected structures (GSCL) or neither heterozygotes nor homozygotes had a relevant phenotype (HIRA, UFD1L, and GSCL). Another candidate gene of interest is the CRKL gene, a member of the family of protein-tyrosine kinases with SH2 and SH3 (src-homology) domains, which maps to the 22q11 region. Although heterozygotes are normal, mice that harbor null mutations of Crkl have craniofacial anomalies, outflow-tract heart defects, abnormal thymus and parathyroid glands, and abnormalities in the cranial nerves (Guris et al. 2001). The CRKL gene maps in the 3-Mb deleted interval, which is distal to the 1.5-Mb region of overlap between the three disorders. Therefore, it is unlikely that this gene provides a major contribution to VCFS/DGS. On the other hand, the deletion or mutation of this gene may contribute to the overall phenotype in some patients. It addition, CRKL may be responsible for the defects in rare patients with deletions that are distal to the 1.5-Mb region on 22q11 who have congenital anomalies that are distinct but related to those in patients with VCFS/DGS (Kurahashi et al. 1996; O'Donnell et al. 1997; Amati et al. 1999; McQuade et al. 1999; Rauch et al. 1999).
Candidate Genes for Overexpression Genomic Syndromes on 22q11
The CES critical region was narrowed to the most proximal 2–2.5 Mb of 22q11 by characterization of an unusual supernumerary r(22) chromosome in a child with all the features typical of CES (Mears et al. 1995). Fourteen genes have been identified in this region, two of which are present in the pericentromeric repeats and therefore may be aberrant transcripts without function (fig. 3) (Footz et al. 2001). Of the remaining 12 transcripts, 2 genes stand out as excellent candidates for involvement in the duplication phenotype, on the basis of their putative functions. One of the genes, CECR1, encodes a homologue of a family of secreted growth factors that are best characterized in invertebrates (fig. 3) (Riazi et al. 2000). The presence of an adenosine deaminase (ADA) domain in these family members suggests that these genes function by regulating the concentration of extracellular adenosine. Several insect homologues have been shown to have ADA activity (Li and Aksoy 2000; Charlab et al. 2001; Zurovec et al., in press). In a 35-d human embryo, CECR1 was expressed in the outflow tract and atrium of the heart and in the VII/VIII cranial nerve ganglion, suggesting potential involvement in CES heart and facial defects (Riazi et al. 2000). A second gene, CECR2, encodes a putative transcriptional coactivator, which may be sensitive to dosage changes (Footz et al. 1998; G. S. Banting, unpublished data).
Der(22) syndrome and CES share a similar region of extra dosage on 22q11. CES is usually associated with four copies of this region; however, patients with interstitial duplications who have features of CES have been reported (Reiss et al. 1985; Knoll et al. 1995), which suggests that CES candidate genes are relevant to der(22) syndrome. Although the phenotype of der(22) syndrome shows many similarities to that of CES, it also shows some significant differences. For instance, mental retardation, cleft palate, and hypotonia are all much more common in der(22) syndrome than in CES (Fraccaro et al. 1980; Berends et al. 2001). This presumably results from the additional presence of three copies of 11q23-qter in der(22) syndrome. However, genes in this region of chromosome 11 may also counteract some of the effect of the 22q11 duplication, since colobomata, microphthalmia, and TAPVR are common features of CES, yet are rarely seen in the der(22) syndrome.
Perspectives
In the past few years, there has been much progress toward understanding the molecular basis of chromosomal rearrangements that lead to genomic disorders. Several other well-characterized genomic disorders are associated with their own region-specific blocks of low-copy repeats. These include Charcot-Marie-Tooth disease type 1A/hereditary neuropathy with pressure palsies (CMT1A/HNPP), Smith-Magenis syndrome, Williams-Beuren syndrome, and Prader-Willi syndrome/Angelman syndrome, among others (reviewed in Lupski 1998; Emanuel and Shaikh 2001). For CMT1A/HNPP, the region of chromosome breakage and strand exchange has been narrowed to a single 557-bp hotspot within 27-kb CMT1A repeats on 17p11-12. This interval lies near a dysfunctional Mariner transposase sequence, and it has been hypothesized that this might create a hotspot for rearrangement (Reiter et al. 1996, 1998). For the other genomic disorders, including VCFS/DGS and CES, the precise sites of chromosome breakage and strand exchange are not known. One essential question is whether similar hotspots exist within the larger 200-kb LCR22s or whether they occur randomly in regions of sequence similarity. The presence of hotspots would suggest precise mechanisms for recombination events. Alterations in the sequence of these hotspots might alter susceptibility to rearrangements, and this could be used to predict the chances for occurrence of a particular chromosomal rearrangement in an individual. Unlike VCFS/DGS and CES, which are mediated by homologous recombination events between two LCRs, the recurrent constitutional t(11;22) translocation has been defined to occur in AT-rich palindromes. Many nonrecurrent translocations disrupt the 22q11 region. It is not known whether these other translocations preferentially occur in the same LCR, different LCRs, or unique sequences. This frequency will give us an indication of the relative importance that this AT-rich sequence has in human translocations. Of interest, a family with a t(17;22) translocation that was associated with neurofibromatosis type 1 had a disruption of the same region on 22q11 as had a family with the t(11;22) translocation (Kehrer-Sawatzki et al. 1997). As does 11q23, the interval on chromosome 17 that is disrupted in the family contains AT-rich palindromic sequence. This supports the potential mutation mechanism discussed in previous sections.
With respect to the molecular basis of rearrangement disorders on 22q11, much progress has been made by use of genetic approaches with mouse models. For VCFS/DGS, a single candidate gene, TBX1, has been implicated as causing the main clinical findings for the disorder. However, proof that haploinsufficiency of this gene is responsible for the syndrome in humans is lacking, since mutations have not been identified in this gene in nondeleted cases (Chieffo et al. 1997; Gong et al. 2001). Since the regulatory regions have not been defined, the possibility of an inactivating mutation cannot be excluded. However, one problem in these screens was that none of the nondeleted cases were familial, which makes a clear genetic basis for the phenotype questionable in these cases. It is still surprising that not a single patient has been identified who has a de novo inactivating mutation in the coding region of this gene. This suggests that, perhaps, in humans, haploinsufficiency of additional genes may contribute to the etiology of the disorder. Another important question is what the basis for psychiatric disorders associated with VCFS/DGS is and whether haploinsufficiency of TBX1, a gene not expressed in the brain, could be responsible for its etiology.
In contrast to VCFS/DGS, for which a clear candidate gene has been identified through mouse-genetics approaches, efforts are still under way to identify candidate genes for CES and der(22) syndrome in mouse mutants. Generating large duplications of the interval and then rescuing mice by use of nested deletions, in the opposite manner as for Tbx1 and VCFS/DGS, would be an obvious approach. Der(22) syndrome is particularly challenging because two different human chromosomes are involved. Despite the challenges ahead, we feel that the uncovering of the molecular basis of these relatively rare human disorders can provide insight into the molecular basis of mental retardation and congenital malformations.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Human-Mouse Homology Map, http://www.ncbi.nlm.nih.gov/Homology/ (for MMU6 and MMU10)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for VCFS/DGS [MIM 192430; MIM 601362] and CES [MIM 115470])
References
- Akgun E, Zahn J, Baumes S, Brown G, Liang F, Romanienko PJ, Lewis S, Jasin M (1997) Palindrome resolution and recombination in the mammalian germ line. Mol Cell Biol 17:5559–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati F, Conti E, Novelli A, Bengala M, Diglio MC, Marino B, Giannotti A, Gabrielli O, Novelli G, Dallapiccola B (1999) Atypical deletions suggest five 22q11.2 critical regions related to the DiGeorge/velo-cardio-facial syndrome. Eur J Hum Genet 7:903–909 [DOI] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Viggiano L, Misceo D, Horvath JE, Archidiacono N, Schwartz S, Rocchi M, Eichler EE (2002) Human-specific duplication and mosaic transcripts: the recent paralogous structure of chromosome 22. Am J Hum Genet 70:83–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer A, Dutly F, Balmer D, Riegel M, Tukel T, Krajewska-Walasek M, Schinzel AA (1998) High level of unequal meiotic crossovers at the origin of the 22q11.2 and 7q11.23 deletions. Hum Mol Genet 7:887–894 [DOI] [PubMed] [Google Scholar]
- Berends MJ, Tan-Sindhunata G, Leegte B, van Essen AJ (2001) Phenotypic variability of cat-eye syndrome. Genet Couns 12:23–34 [PubMed] [Google Scholar]
- Bockman DE, Kirby ML (1984) Dependence of thymus development on derivatives of the neural crest. Science 223:498–500 [DOI] [PubMed] [Google Scholar]
- Bockman DE, Redmond ME, Waldo K, Davis H, Kirby ML (1987) Effect of neural crest ablation on development of the heart and arch arteries in the chick. Am J Anat 180:332–341 [DOI] [PubMed] [Google Scholar]
- Borgaonkar DS (1989) Chromosomal variation in man: a catalog of chromosomal variants and anomalies, 5th ed. Alan R Liss, New York [Google Scholar]
- Budarf ML, Collins J, Gong W, Roe B, Wang Z, Bailey LC, Sellinger B, Michaud D, Driscoll DA, Emanuel BS (1995) Cloning a balanced translocation associated with DiGeorge syndrome and identification of a disrupted candidate gene. Nat Genet 10:269–278 [DOI] [PubMed] [Google Scholar]
- Burn J, Goodship J (1996) Congenital heart disease. In: Rimoin DL, Connor JM, Pyeritz RE (eds) Emery and Rimoin's principles and practice of medical genetics, 3d ed. Vol 1, pp 767–828 [Google Scholar]
- Carlson C, Sirotkin H, Pandita R, Goldberg R, McKie J, Wadey R, Patanjali SR, Weissman SM, Anyane-Yeboa K, Warburton D, Scambler P, Shprintzen R, Kucherlapati R, Morrow EB (1997) Molecular definition of 22q11 deletions in 151 velo-cardio-facial syndrome patients. Am J Hum Genet 61:620–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson-Brown JJ, Cebra-Thomas J, Bollag RJ, Silver LM, Papaioannou VE (1996) Expression of the T-box family genes, Tbx1_–_Tbx5, during early mouse development. Dev Dyn 206:379–390 [DOI] [PubMed] [Google Scholar]
- Charlab R, Valenzuela JG, Andersen J, Ribeiro JM (2001) The invertebrate growth factor/CECR1 subfamily of adenosine deaminase proteins. Gene 267:13–22 [DOI] [PubMed] [Google Scholar]
- Chieffo C, Garvey N, Gong W, Roe B, Zhang G, Silver L, Emanuel BS, Budarf ML (1997) Isolation and characterization of a gene from the DiGeorge chromosomal region homologous to the mouse Tbx1 gene. Genomics 43:267–277 [DOI] [PubMed] [Google Scholar]
- Chow EW, Bassett AS, Weksberg R (1994) Velo-cardio-facial syndrome and psychotic disorders: implications for psychiatric genetics. Am J Med Genet 54:107–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J, Cole C, Smink L, Garret C, Leversham M, Soderlund C, Maslen G, et al (1995) A high-density YAC contig map of human chromosome 22. Nature Suppl 377:367–379 [DOI] [PubMed] [Google Scholar]
- Couly GF, Le Douarin NM (1987) Mapping of the early neural primordium in quail-chick chimeras. II. The prosencephalic neural plate and neural folds: implications for the genesis of cephalic human congenital abnormalities. Dev Biol 120:198–214 [DOI] [PubMed] [Google Scholar]
- Crolla JA, Howard P, Mitchell C, Long FL, Dennis NR (1997) A molecular and FISH approach to determining karyotype and phenotype correlations in six patients with supernumerary marker(22) chromosomes. Am J Med Genet 72:440–447 [DOI] [PubMed] [Google Scholar]
- de la Chapelle A, Herva R, Koivisto M, Aula P (1981) A deletion in chromosome 22 can cause DiGeorge syndrome. Hum Genet 57:253–256 [DOI] [PubMed] [Google Scholar]
- DiGeorge A (1965) A new concept of the cellular basis of immunity. J Pediatr 67:907 [Google Scholar]
- Driscoll DA, Budarf ML, Emanuel BS (1992) A genetic etiology for DiGeorge syndrome: consistent deletions and microdeletions of 22q11. Am J Hum Genet 50:924–933 [PMC free article] [PubMed] [Google Scholar]
- Driscoll DA, Salvin J, Sellinger B, Budarf ML, McDonald-McGinn DM, Zackai EH, Emanuel BS (1993) Prevalence of 22q11 microdeletions in DiGeorge and velocardiofacial syndromes: implications for genetic counselling and prenatal diagnosis. J Med Genet 30:813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, et al (1999) The DNA sequence of human chromosome 22. Nature 402:489–495 [DOI] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Morrow BE (1999_a_) Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am J Hum Genet 64:1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, Magenis E, Shprintzen RJ, Morrow BE (1999_b_) A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet 8:1157–1167 [DOI] [PubMed] [Google Scholar]
- Edelmann L, Spiteri E, Koren K, Pulijaal V, Bialer MG, Shanske A, Goldberg R, Morrow BE (2001) AT-rich palindromes mediate the constitutional t(11;22) translocation. Am J Hum Genet 68:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Spiteri E, McCain N, Goldberg R, Pandita RK, Duong S, Fox J, Blumenthal D, Lalani SR, Shaffer LG, Morrow BE (1999_c_) A common breakpoint on 11q23 in carriers of the constitutional t(11;22) translocation. Am J Hum Genet 65:1608–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel BS, Shaikh TH (2001) Segmental duplications: an “expanding” role in genomic instability and disease. Nat Rev Genet 2:791–800 [DOI] [PubMed] [Google Scholar]
- Footz TK, Birren B, Minoshima S, Asakawa S, Shimizu N, Riazi MA, McDermid HE (1998) The gene for death agonist BID maps to the region of human 22q11.2 duplicated in cat eye syndrome chromosomes and to mouse chromosome 6. Genomics 51:472–475 [DOI] [PubMed] [Google Scholar]
- Footz TK, Brinkman-Mills P, Banting GS, Maier SA, Riazi MA, Bridgland L, Hu S, et al (2001) Analysis of the cat eye syndrome critical region in humans and the region of conserved synteny in mice: a search for candidate genes at or near the human chromosome 22 pericentromere. Genome Res 11:1053–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraccaro M, Lindsten J, Ford CE, Iselius L (1980) The 11q;22q translocation: a European collaborative analysis of 43 cases. Hum Genet 56:21–51 [DOI] [PubMed] [Google Scholar]
- Funke B, Edelmann L, McCain N, Pandita R, Ferreira J, Merscher S, Zohouri M, Cannizzaro L, Shanske A, Morrow BE (1999) Der(22) syndrome and velo-cardio-facial syndrome/DiGeorge syndrome share a 1.5-Mb region of overlap on chromosome 22q11. Am J Hum Genet 64:747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke B, Epstein JA, Kochilas LK, Lu MM, Pandita RK, Liao J, Bauerndistel R, Schuler T, Schorle H, Brown MC, Adams J, Morrow BE (2001) Mice overexpressing genes from the 22q11 region deleted in velo-cardio-facial syndrome/DiGeorge syndrome have middle and inner ear defects. Hum Mol Genet 10:2549–2556 [DOI] [PubMed] [Google Scholar]
- Gong W, Gottlieb S, Collins J, Blescia A, Dietz H, Goldmuntz E, McDonald-McGinn DM, Zackai EH, Emanuel BS, Driscoll DA, Budarf ML (2001) Mutation analysis of TBX1 in non-deleted patients with features of DGS/VCFS or isolated cardiovascular defects. J Med Genet 38:E1–E5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guris DL, Fantes J, Tara D, Druker BJ, Imamoto A (2001) Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat Genet 27:293–298 [DOI] [PubMed] [Google Scholar]
- Holmes SE, Riazi MA, Gong W, McDermid HE, Sellinger BT, Hua A, Chen F, Wang Z, Zhang G, Roe B, Gonzalez I, McDonald-McGinn DM, Zackai E, Emanuel BS, Budarf ML (1997) Disruption of the clathrin heavy chain-like gene (CLTCL) associated with features of DGS/VCFS: a balanced (21;22)(p12;q11) translocation. Hum Mol Genet 6:357–367 [DOI] [PubMed] [Google Scholar]
- Jerome LA, Papaioannou VE (2001) DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet 27:286–291 [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Haussler J, Krone W, Bode H, Jenne DE, Mehnert KU, Tummers U, Assum G (1997) The second case of a t(17;22) in a family with neurofibromatosis type 1: sequence analysis of the breakpoint regions. Hum Genet 99:237–247 [DOI] [PubMed] [Google Scholar]
- Kimber WL, Hsieh P, Hirotsune S, Yuva-Paylor L, Sutherland HF, Chen A, Ruiz-Lozano P, Hoogstraten-Miller SL, Chien KR, Paylor R, Scambler PJ, Wynshaw-Boris A (1999) Deletion of 150 kb in the minimal DiGeorge/velocardiofacial syndrome critical region in mouse. Hum Mol Genet 8:2229–2237 [DOI] [PubMed] [Google Scholar]
- Kirby ML, Gale TF, Stewart DE (1983) Neural crest cells contribute to normal aorticopulmonary septation. Science 220:1059–1061 [DOI] [PubMed] [Google Scholar]
- Kirby ML, Turnage KL 3d, Hays BM (1985) Characterization of conotruncal malformations following ablation of “cardiac” neural crest. Anat Rec 213:87–93 [DOI] [PubMed] [Google Scholar]
- Kirby ML, Waldo KL (1995) Neural crest and cardiovascular patterning. Circ Res 77:211–215 [DOI] [PubMed] [Google Scholar]
- Knoll JH, Asamoah A, Pletcher BA, Wagstaff J (1995) Interstitial duplication of proximal 22q: phenotypic overlap with cat eye syndrome. Am J Med Genet 55:221–224 [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Emanuel BS (2001) Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum Mol Genet 10:2605–2617 [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Nakayama T, Osugi Y, Tsuda E, Masuno M, Imaizumi K, Kamiya T, Sano T, Okada S, Nishisho I (1996) Deletion mapping of 22q11 in CATCH22 syndrome: identification of a second critical region. Am J Hum Genet 58:1377–1381 [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H, Shaikh TH, Hu P, Roe BA, Emanuel BS, Budarf ML (2000_a_) Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22). Hum Mol Genet 9:1665–1670 [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Shaikh TH, Zackai EH, Celle L, Driscoll DA, Budarf ML, Emanuel BS (2000_b_) Tightly clustered 11q23 and 22q11 breakpoints permit PCR-based detection of the recurrent constitutional t(11;22). Am J Hum Genet 67:763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamour V, Lecluse Y, Desmaze C, Spector M, Bodescot M, Aurias A, Osley MA, et al (1995) A human homolog of the S. cerevisiae HIR1 and HIR2 transcriptional repressors cloned from the DiGeorge syndrome critical region. Hum Mol Genet 4:791–799 [DOI] [PubMed] [Google Scholar]
- Levy A, Demczuk S, Aurias A, Depetris D, Mattei MG, Philip N (1995) Interstitial 22q11 microdeletion excluding the ADU breakpoint in a patient with DiGeorge syndrome. Hum Mol Genet 4:2417–2419 [DOI] [PubMed] [Google Scholar]
- Li S, Aksoy S (2000) A family of genes with growth factor and adenosine deaminase similarity are preferentially expressed in the salivary glands of Glossina m. morsitans. Gene 252:83–93 [DOI] [PubMed] [Google Scholar]
- Lin AE, Bernar J, Chin AJ, Sparkes RS, Emanuel BS, Zackai EH (1986) Congenital heart disease in supernumerary der(22),t(11;22) syndrome. Clin Genet 29:269–275 [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Botta A, Jurecic V, Carattini-Rivera S, Cheah YC, Rosenblatt HM, Bradley A, Baldini A (1999) Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature 401:379–383 [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Halford S, Wadey R, Scambler PJ, Baldini A (1993) Molecular cytogenetic characterization of the DiGeorge syndrome region using fluorescence in situ hybridization. Genomics 17:403–407 [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Harvey EL, Scambler PJ, Baldini A (1998) ES2, a gene deleted in DiGeorge syndrome, encodes a nuclear protein and is expressed during early mouse development, where it shares an expression domain with a goosecoid-like gene. Hum Mol Genet 7:629–635 [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A (2001) Tbx1 haploinsufficiency in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 410:97–101 [DOI] [PubMed] [Google Scholar]
- Lund J, Roe B, Chen F, Budarf M, Galili N, Riblet R, Miller RD, Emanuel BS, Reeves RH (1999) Sequence-ready physical map of the mouse chromosome 16 region with conserved synteny to the human velocardiofacial syndrome region on 22q11.2. Mamm Genome 10:438–443 [DOI] [PubMed] [Google Scholar]
- Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422 [DOI] [PubMed] [Google Scholar]
- McDermid HE, Duncan AM, Brasch KR, Holden JJ, Magenis E, Sheehy R, Burn J, Kardon N, Noel B, Schninzel A, Teshima I, White BN (1986) Characterization of the supernumerary chromosome in cat eye syndrome. Science 232:646–648 [DOI] [PubMed] [Google Scholar]
- McQuade L, Christodoulou J, Budarf M, Sachdev R, Wilson M, Emanuel B, Colley A (1999) Patient with a 22q11.2 deletion with no overlap of the minimal DiGeorge syndrome critical region (MDGCR). Am J Med Genet 86:27–33 [PubMed] [Google Scholar]
- McTaggart KE, Budarf ML, Driscoll DA, Emanuel BS, Ferreira P, McDermid HE (1998) Cat eye syndrome chromosome breakpoint clustering: identification of two intervals also associated with 22q11 deletion syndrome breakpoints. Cytogenet Cell Genet 81:222–228 [DOI] [PubMed] [Google Scholar]
- Mears AJ, Duncan AM, Budarf ML, Emanuel BS, Sellinger B, Siegel-Bartelt J, Greenberg CR, McDermid HE (1994) Molecular characterization of the marker chromosome associated with cat eye syndrome. Am J Hum Genet 55:134–142 [PMC free article] [PubMed] [Google Scholar]
- Mears AJ, el-Shanti H, Murray JC, McDermid HE, Patil SR (1995) Minute supernumerary ring chromosome 22 associated with cat eye syndrome: further delineation of the critical region. Am J Hum Genet 57:667–673 [PMC free article] [PubMed] [Google Scholar]
- Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, Tokooya K, Jore BS, Lopez M, Pandita RK, Lia M, Carrion D, Xu H, Schorle H, Kobler JB, Scambler P, Wynshaw-Boris A, Skoultchi AI, Morrow BE, Kucherlapati R (2001) TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell 104:619–629 [DOI] [PubMed] [Google Scholar]
- Morrow B, Goldberg R, Carlson C, Das Gupta R, Sirotkin H, Collins J, Dunham I, et al (1995) Molecular definition of the 22q11 deletions in velo-cardio-facial syndrome. Am J Hum Genet 56:1391–1403 [PMC free article] [PubMed] [Google Scholar]
- Murphy KC, Jones LA, Owen MJ (1999) High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry 56:940–945 [DOI] [PubMed] [Google Scholar]
- Murphy KC, Owen MJ (2001) Velo-cardio-facial syndrome: a model for understanding the genetics and pathogenesis of schizophrenia. Br J Psychiatry 179:397–402 [DOI] [PubMed] [Google Scholar]
- O'Donnell H, McKeown C, Gould C, Morrow B, Scambler P (1997) Detection of an atypical 22q11 deletion that has no overlap with the DiGeorge syndrome critical region. Am J Hum Genet 60:1544–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papolos DF, Faedda GL, Veit S, Goldberg R, Morrow B, Kucherlapati R, Shprintzen RJ (1996) Bipolar spectrum disorders in patients diagnosed with velo-cardio-facial syndrome: does a hemizygous deletion of chromosome 22q11 result in bipolar affective disorder? Am J Psychiatry 153:1541–1547 [DOI] [PubMed] [Google Scholar]
- Phillips MT, Kirby ML, Forbes G (1987) Analysis of cranial neural crest distribution in the developing heart using quail-chick chimeras. Circ Res 60:27–30 [DOI] [PubMed] [Google Scholar]
- Pizzuti A, Novelli G, Ratti A, Amati F, Mari A, Calabrese G, Nicolis S, Silani V, Marino B, Scarlato G, Ottolenghi S, Dallapiccola B (1997) UFD1L, a developmentally expressed ubiquitination gene, is deleted in CATCH 22 syndrome. Hum Mol Genet 6:259–265 [DOI] [PubMed] [Google Scholar]
- Puech A, Saint-Jore B, Funke B, Gilbert DJ, Sirotkin H, Copeland NG, Jenkins NA, Kucherlapati R, Morrow B, Skoultchi AI (1997) Comparative mapping of the human 22q11 chromosomal region and the orthologous region in mice reveals complex changes in gene organization. Proc Natl Acad Sci USA 94:14608–14613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver AE, Nestadt G, Goldberg R, Shprintzen RJ, Lamacz M, Wolyniec PS, Morrow B, et al (1994) Psychotic illness in patients diagnosed with velo-cardio-facial syndrome and their relatives. J Nerv Ment Dis 182:476–478 [DOI] [PubMed] [Google Scholar]
- Ramirez-Solis R, Liu P, Bradley A (1995) Chromosome engineering in mice. Nature 378:720–724 [DOI] [PubMed] [Google Scholar]
- Rauch A, Pfeiffer RA, Leipold G, Singer H, Tigges M, Hofbeck M (1999) A novel 22q11.2 microdeletion in DiGeorge syndrome. Am J Hum Genet 64:659–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss JA, Weleber RG, Brown MG, Bangs CD, Lovrien EW, Magenis RE (1985) Tandem duplication of proximal 22q: a cause of cat-eye syndrome. Am J Med Genet 20:165–171 [DOI] [PubMed] [Google Scholar]
- Reiter LT, Hastings PJ, Nelis E, De Jonghe P, Van Broeckhoven C, Lupski JR (1998) Human meiotic recombination products revealed by sequencing a hotspot for homologous strand exchange in multiple HNPP deletion patients. Am J Hum Genet 62:1023–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Pentao L, Muzny DM, Gibbs RA, Lupski JR (1996) A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet 12:288–297 [DOI] [PubMed] [Google Scholar]
- Riazi MA, Brinkman-Mills P, Nguyen T, Pan H, Phan S, Ying F, Roe BA, Tochigi J, Shimizu Y, Minoshima S, Shimizu N, Buchwald M, McDermid HE (2000) The human homolog of insect-derived growth factor, CECR1, is a candidate gene for features of cat eye syndrome. Genomics 64:277–285 [DOI] [PubMed] [Google Scholar]
- Roberts C, Daw SC, Halford S, Scambler PJ (1997) Cloning and developmental expression analysis of chick Hira (Chira), a candidate gene for DiGeorge syndrome. Hum Mol Genet 6:237–245 [DOI] [PubMed] [Google Scholar]
- Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, Brickman JM, Wynshaw-Boris A, Scambler PJ (2002) Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol Cell Biol 22:2318–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore B, Puech A, Heyer J, Lin Q, Raine C, Kucherlapati R, Skoultchi AI (1998) Goosecoid-like (Gscl), a candidate gene for velocardiofacial syndrome, is not essential for normal mouse development. Hum Mol Genet 7:1841–1849 [DOI] [PubMed] [Google Scholar]
- Scambler PJ, Kelly D, Lindsay E, Williamson R, Goldberg R, Shprintzen R, Wilson DI, Goodship JA, Cross IE, Burn J (1992) Velo-cardio-facial syndrome associated with chromosome 22 deletions encompassing the DiGeorge locus. Lancet 339:1138–1139 [DOI] [PubMed] [Google Scholar]
- Schinzel A, Schmid W, Auf der Maur P, Moser H, Degenhardt KH, Geisler M, Grubisic A (1981) Incomplete trisomy 22. I. Familial 11/22 translocation with 3:1 meiotic disjunction: delineation of a common clinical picture and report of nine new cases from six families. Hum Genet 56:249–262 [DOI] [PubMed] [Google Scholar]
- Schreck RR, Breg WR, Erlanger BF, Miller OJ (1977) Preferential derivation of abnormal human G-group-like chromosomes from chromosome 15. Hum Genet 36:1–12 [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Budarf ML, Celle L, Zackai EH, Emanuel BS (1999) Clustered 11q23 and 22q11 breakpoints and 3:1 meiotic malsegregation in multiple unrelated t(11;22) families. Am J Hum Genet 65:1595–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TH, Kurahashi H, Saitta SC, O'Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, Emanuel BS (2000) Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet 9:489–501 [DOI] [PubMed] [Google Scholar]
- Shprintzen RJ (2001) Velo-cardio-facial syndrome: a distinctive behavioral phenotype. Ment Retard Dev Disabil Res Rev 6:142–147 [DOI] [PubMed] [Google Scholar]
- Shprintzen RJ, Goldberg RB, Lewin ML, Sidoti EJ, Berkman MD, Argamaso RV, Young D (1978) A new syndrome involving cleft palate, cardiac anomalies, typical facies, and learning disabilities: velo-cardio-facial syndrome. Cleft Palate J 15:56–62 [PubMed] [Google Scholar]
- Sutherland HF, Wadey R, McKie JM, Taylor C, Atif U, Johnstone KA, Halford S, Kim UJ, Goodship J, Baldini A, Scambler PJ (1996) Identification of a novel transcript disrupted by a balanced translocation associated with DiGeorge syndrome. Am J Hum Genet 59:23–31 [PMC free article] [PubMed] [Google Scholar]
- Swillen A, Devriendt K, Legius E, Prinzie P, Vogels A, Ghesquiere P, Fryns JP (1999) The behavioural phenotype in velo-cardio-facial syndrome (VCFS): from infancy to adolescence. Genet Couns 10:79–88 [PubMed] [Google Scholar]
- Van Dyke DL, Weiss L, Logan M, Pai GS (1977) The origin and behavior of two isodicentric bisatellited chromosomes. Am J Hum Genet 29:294–300 [PMC free article] [PubMed] [Google Scholar]
- Van Hove JL, McConkie-Rosell A, Chen YT, Iafolla AK, Lanman JT Jr, Hennessy MD, Kahler SG (1992) Unbalanced translocation 46,XY,−15,+der(22)t(15;22)(q13;q11)pat: case report and review of the literature. Am J Med Genet 44:24–30 [DOI] [PubMed] [Google Scholar]
- Wakamiya M, Lindsay EA, Rivera-Pérez JA, Baldini A, Behringer RR (1998) Functional analysis of Gscl in the pathogenesis of the DiGeorge and velocardiofacial syndromes. Hum Mol Genet 7:1835–1840 [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Garg V, Matsuoka R, Thomas T, Srivastava D (1999) A molecular pathway revealing a genetic basis for human cardiac and craniofacial defects. Science 283:1158–1161 [DOI] [PubMed] [Google Scholar]
- Yu Y, Bradley A (2001) Engineering chromosomal rearrangements in mice. Nat Rev Genet 2:780–790 [DOI] [PubMed] [Google Scholar]
- Zackai EH, Emanuel BS (1980) Site-specific reciprocal translocation t(11;22)(q23;q11), in several unrelated families with 3:1 meiotic disjunction. Am J Med Genet 7:507–521 [DOI] [PubMed] [Google Scholar]
- Zheng B, Sage M, Sheppeard EA, Jurecic V, Bradley A (2000) Engineering mouse chromosomes with Cre-loxP: range, efficiency, and somatic applications. Mol Cell Biol 20:648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurovec M, Dolezal T, Gazi M, Pavlova E, Bryant PJ. Adenosine deaminase-related growth factors stimulate cell proliferation in Drosophila by depleting extracellular adenosine. Proc Natl Acad Sci USA (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]