A microRNA polycistron as a potential human oncogene (original) (raw)
. Author manuscript; available in PMC: 2015 Oct 9.
Published in final edited form as: Nature. 2005 Jun 9;435(7043):828–833. doi: 10.1038/nature03552
Abstract
To date, more than 200 microRNAs have been described in humans; however, the precise functions of these regulatory, non-coding RNAs remains largely obscure. One cluster of microRNAs, the _mir_-17_–_92 polycistron, is located in a region of DNA that is amplified in human B-cell lymphomas1. Here we compared B-cell lymphoma samples and cell lines to normal tissues, and found that the levels of the primary or mature microRNAs derived from the _mir_-17_–_92 locus are often substantially increased in these cancers. Enforced expression of the _mir_-17_–_92 cluster acted with _c_-myc expression to accelerate tumour development in a mouse B-cell lymphoma model. Tumours derived from haematopoietic stem cells expressing a subset of the _mir_-17_–_92 cluster and _c_-myc could be distinguished by an absence of apoptosis that was otherwise prevalent in _c_-_myc_-induced lymphomas. Together, these studies indicate that non-coding RNAs, specifically microRNAs, can modulate tumour formation, and implicate the _mir_-17_–_92 cluster as a potential human oncogene.
MicroRNAs (miRNAs) have emerged relatively recently as a new class of small, non-coding RNAs that regulate gene expression. Nascent primary miRNA transcripts (pri-miRNAs) are processed sequentially by two RNase III enzymes, Drosha and Dicer2,3, to yield mature miRNAs, ranging from 18 to 24 nucleotides (nt) in length. miRNAs are incorporated into the RNA interference (RNAi) effector complex, RISC, and target specific messenger RNAs for translational repression or mRNA cleavage4–6. Although hundreds of miRNAs have been cloned and/or predicted, only a handful have been functionally characterized. For example, lin-4 and let-7 regulate the timing of larval development in C. elegans7,8. Left/right asymmetric gene expression in C. elegans chemosensory neurons is controlled by lsy-6 and miR-273 (refs 9, 10). Bantam stimulates cell growth and prevents apoptosis in Drosophila11, and miR-181 potentiates B-cell differentiation in mammals12. These findings, in combination with computational target predictions, are consistent with miRNAs regulating a broad spectrum of physiological and developmental processes.
Microarray-based expression studies have indicated specific alterations in human miRNA expression profiles that correlate with particular tumour phenotypes (J.M.T. and S.M.H., unpublished data). Among those that show altered expression, the _mir_-17_–_92 cistron is located at 13q31, a genomic locus that is amplified in cases of diffuse large B-cell lymphoma, follicular lymphoma, mantle cell lymphoma, primary cutaneous B-cell lymphoma and several other tumour types1,13. There are only two annotated genes in the epicentre of this amplicon, c13orf25 and GPC5. Previous studies have shown that c13orf25 is the only one of the two genes for which increased expression correlates with the presence of the amplicon1. Therefore, c13orf25 had been implicated as a target of the 13q31 amplicon1. It is unlikely that c13orf25 actually encodes a protein, as predicted open reading frames (ORFs) encode only short peptides (<70 amino acids), which are not conserved in closely related species. Instead, the c13orf25 transcript appears to be the functional precursor of a series of seven microRNAs: miR-17-5p, miR-17-3p, miR-18, miR-19a, miR-20, miR-19b-1 and miR-92-1 (Fig. 1a). Additionally, this cluster is related to the homologous _mir_-106a_–_92 cluster on chromosome X and the _mir_-106b_–_25 cluster on chromosome 7 (ref. 14; Fig. 1a). Alignment of the human c13orf25 locus and its murine orthologue revealed extensive sequence conservation only within the _mir_-17_–_92 polycistron and its immediate flanking sequence. Several of the expressed-sequence-tags (ESTs) derived from c13orf25 and its mouse orthologue terminate at the 3′ end of _mir_-17_–_92 cluster, consistent with the presence of a Drosha processing site at this location (Fig. 1b).
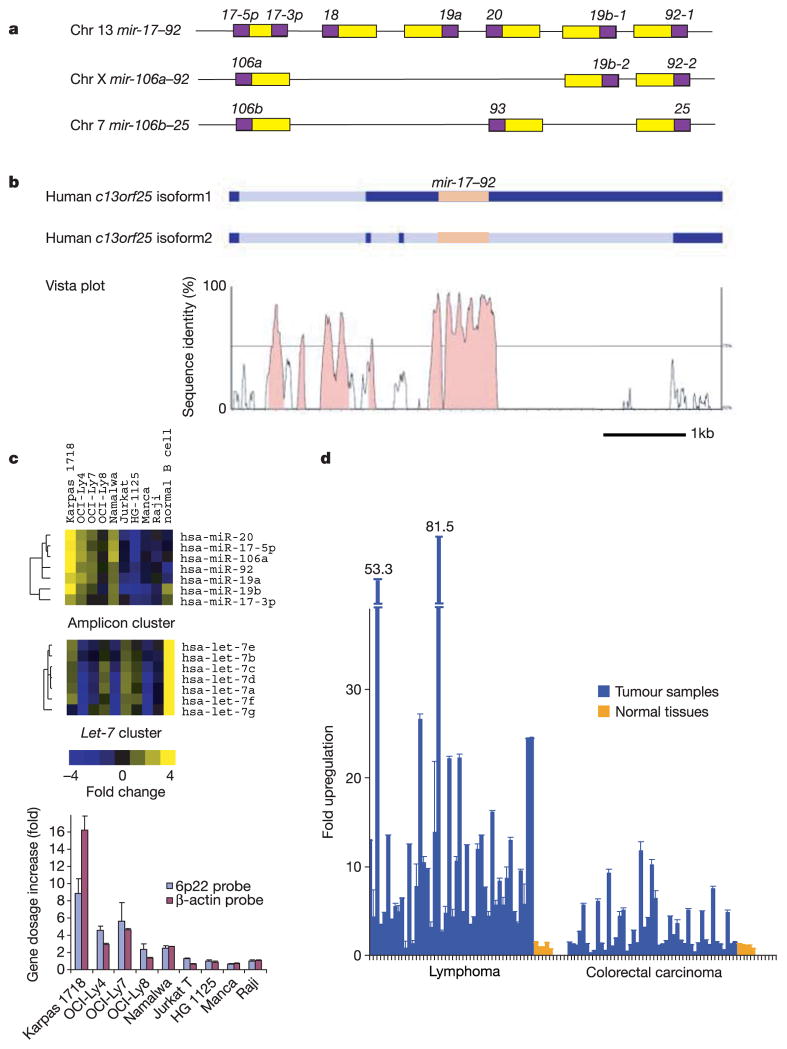
Figure 1. The _mir_-17_–_92 cluster shows increased expression in B-cell lymphoma samples and cell lines.
a, Genomic organization of three polycistronic miRNA clusters is shown. There are five paralogous groups located in three homologous clusters (mir-17–92, mir-106a–92 and mir-106b–25) with a conserved order: miR-17-5p/miR-106a/miR-106b; miR-18; miR-19a/miR-19b-1/miR-19b-2; miR-20/miR-93; and miR-92-1/miR-92-2/miR-25 (yellow boxes, pre-miRNAs; purple boxes, mature miRNAs). b, The level of conservation between human and mouse homologues is represented using an mVista plot28. Two alternative isoforms have been detected for the human gene, and these are shown schematically1 (dark blue, exons; light blue, introns; orange, the mir-17_–_92 cluster). c, MicroRNA expression levels in cell lines carrying the 13q31-q32 amplicon (including Karpas 1718, OCI-Ly4 and OCI-Ly7) were compared to those in leukaemia and lymphoma cell lines lacking this genetic lesion, and to normal B-cells isolated from cortical blood (top panel). We included in this analysis the OCI-Ly8 cell line, which has previously been identified as a cell line carrying the 13q31-32 amplicon, but showed no gene dosage increase at the c13orf25 locus in our study. Normalized one-channel measurements for 191 human miRNAs were hierarchically clustered for all miRNA genes represented on the array. An excerpt of the data is shown, and the full cluster analysis is presented in Supplementary Fig. 1. The expression map node that correlates with the amplification is shown. The let-7 cluster node is also shown for comparison (middle panel). In the cell lines examined, expression levels of the mature microRNAs from the mir-17_–_92 polycistron correlate with the copy number at the mir-17_–_92 locus (bottom panel). d, The level of mir-17_–_92 pri-miRNA was determined by real-time quantitative PCR in 46 lymphomas and 47 colorectal carcinomas, and compared to levels found in corresponding normal tissues from five individuals. In c and d, error bars indicate standard deviation (s.d.).
A principal consequence of 13q31-q32 amplification could be an increase in the abundance of mature miRNA species from the _mir_-17_–_92 cluster. We acquired four cell lines previously described as carrying amplifications in the 13q31-q32 region1 and confirmed the gene dosage increase at the c13orf25 locus in three of those cell lines using quantitative polymerase chain reaction (PCR) analysis. The abundance of 191 mature miRNAs was assessed in these four cell lines and compared to normal B-cells, and to five leukaemia and lymphoma cell lines lacking the amplicon (Fig. 1c and Supplementary Fig. 1). Using a significance analysis of microarrays (SAM) analysis15, we identified six miRNAs for which high-level expression correlated with increased gene dosage of c13orf25 (see Supplementary Table 1). Five were from the _mir_-17_–_92 cistron, and the sixth, miR-106a, was identified as a probable result of cross-hybridization to miR-17-5p, from which it differs at only two terminal nucleotides (Fig. 1c). This hypothesis is supported by the observation that the mir-106a_–_92 locus does not show copy number alterations in these cell lines (not shown). In each cell line, expression levels correlated with the copy number of the mir-17_–_92 locus (Fig. 1c, lower panel).
We also examined the expression of pri-_mir_-17_–_92 in a series of human tumour samples comprising both lymphomas and colorectal carcinomas. Of 46 lymphoma samples, including 13 diffuse large B-cell lymphomas and 6 follicular lymphomas, we saw significant (greater than fivefold) overexpression of pri-_mir_-17_–_92 in 65% of the samples. Considering all of the B-cell lymphoma samples analysed, the average increase in pri-miRNA expression was about tenfold (Fig. 1d). In contrast, colorectal carcinomas rarely showed over-expression of the pri-miRNA. Increases in expression from this locus were less common (15% of samples showed greater than fivefold upregulation), and the degree of overexpression was substantially lower (Fig. 1d).
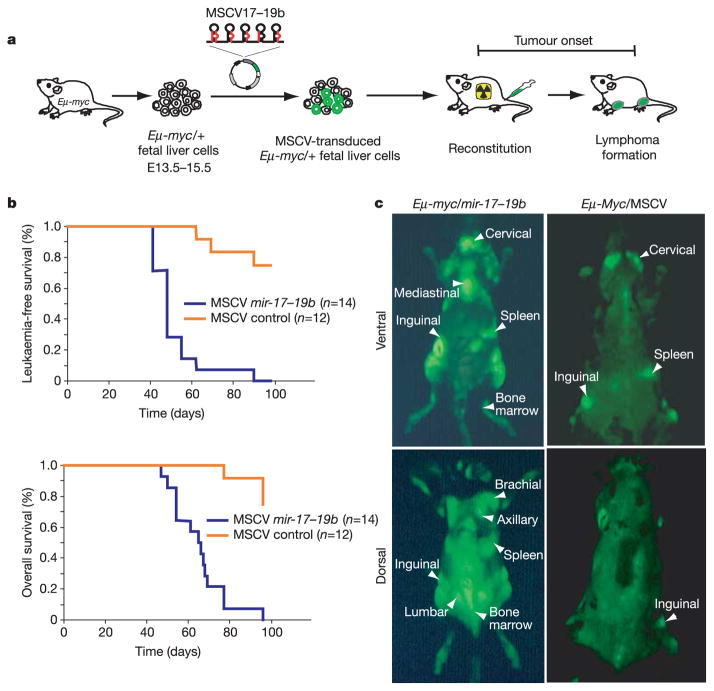
Considered together, our data prompted the hypothesis that _mir_-17_–_92 might contribute to tumour development. To test this idea directly, we used a mouse model of human B-cell lymphoma. Transgenic animals carrying a _c_-myc oncogene, driven by the immunoglobulin heavy-chain enhancer (Eμ), develop B-cell lymphomas by 4–6 months of age16. Similarly, haematopoietic stem cells (HSCs) derived from fetal livers of _Eμ_-myc transgenic mice generate B-cell lymphomas with comparable latency when transplanted into lethally irradiated recipients17–20 (Fig. 2a). We therefore infected _Eμ_-myc/+ HSCs with a murine stem cell virus (MSCV) retrovirus that directs expression of a truncated cluster comprising _mir_-_17_–_19b_-1 (hereafter _mir_-17_–_19b), the vertebrate-specific portion of the _mir_-17_–_92 miRNA cistron (Fig. 1a). This virus also contained a green fluorescent protein (GFP) transgene, allowing us to follow infected stem cells in vitro and in vivo (Fig. 2a). Mice reconstituted with _Eμ_-myc/+ HSCs carrying a control MSCV vector developed lymphomas after the expected latency (3–6 months), with incomplete penetrance (Fig. 2b). Similarly, we examined >40 animals reconstituted with _Eμ_-myc/+ HSCs expressing subsets of 96 different, single microRNAs (see Supplementary Table 2). Although we did not confirm miRNA overexpression for every case, we did not observe any significantly accelerated onset of disease. In contrast, 100% of the animals co-expressing the _mir_-17_–_19b polycistron and _c_-myc developed leukaemias at an average of 51 days following transplantation (s.d. ±13 days, P < 0.0001 compared with MSCV controls using the log rank test), and died of B-cell lymphomas at an average age of 65 days (s.d. ±13 days, P < 0.0001 compared to MSCV controls; Fig. 2b). In all but one case, primary lymphomas could be visualized by virtue of the linked GFP marker (Fig. 2c and Table 1). The mature miRNAs from the _mir_-17_–_19b cluster show high-level expression in these tumours, compared with miRNAs from the paralogous _mir_-106a_–_92 locus, and also have similar _mir_-17_–_19b expression levels compared to the Karpas 1718 lymphoma cell line, which has increased c13orf25 gene dosage (see Supplementary Fig. 2).
Figure 2. Overexpression of the _mir_-17_–_19b cluster accelerates _c_-_myc_-induced lymphomagenesis in mice.
a, Schematic representation of the adoptive transfer protocol using _Eμ_-myc HSCs. b, Mice reconstituted with HSCs expressing _mir_-17_–_19b in an MSCV retroviral vector (MSCV _mir_-17_–_19b) or infected with a control MSCV virus were monitored by blood smear analysis starting 5 weeks after transplantation. The Kaplan-Meier curves represent the percentage of leukaemia-free survival or overall survival. c, External GFP imaging of tumour-bearing mice with _Eμ_-myc/_mir_-17_–_19b or _Eμ_-myc/MSCV shows the overall distribution of tumour cells. _Eμ_-myc/_mir_-17_–_19b tumours show a more disseminated phenotype compared with control tumours. These animals are representative of their genotype.
Table 1.
Phenotypic analysis of a subset of _Eμ_-myc/_mir_-17_–_19b tumours
| Animal | GFP | Immunophenotyping | Cell type | Ki67 staining | Apoptosis* | Pathological features |
|---|---|---|---|---|---|---|
| 1 | + | B220+, Thy1.2−, IgM−, CD19+, CD4−, CD8− | Pre-B cell | ++(70–80%) | Low | Tumour cells invaded liver and spleen; mild infiltrations observed in lung and kidney; spleen enlarged; hindlimb paralysis |
| 2 | + | B220+, Thy1.2low, IgM−, CD19+, CD4−, CD8− | Pre-B cell | ++(80–90%) | Low | Tumour cells invaded liver, lung and spleen; spleen enlarged |
| 3 | + | B220+, Thy1.2−, IgM−, CD19+, CD4−, CD8− | Pre-B cell | ++(80–90%) | Low | Tumour cells invaded liver, lung and spleen; spleen enlarged |
| 4 | + | B220+, Thy1.2low, IgM−, CD19+, CD4−, CD8− | Pre-B cell | ++(70–80%) | Low | Tumour cells invaded liver and spleen; spleen enlarged |
| 5 | + | B220+, Thy1,2−, IgM−, CD19+, CD4−, CD8− | Pre-B cell | ++(80–90%) | Slightly less than control | Highly disseminated lymphoma; tumour cells invaded liver, spleen, lung and kidney; spleen enlarged |
| 6 | + | B220+, Thy1,2−, IgM−, CD19+, CD4−, CD8− | Pre-B cell | ++(80–90%) | Low | Highly disseminated lymphoma; tumour cells invaded liver, spleen, lung and kidney; spleen enlarged |
| 7 | + | B220+, Thy1.2−, IgM−, CD19+, CD4−, CD8− | Pre-B cell | ++(70–80%) | Low | Spleen enlarged; mild infiltration of tumour cells into liver only |
| 8 | + | B220+, Thy1.2low, IgM−, CD19+, CD4−, CD8− and B220+, Thy1.2low, IgM−, CD19+, CD4−, CD8−† | Pre-B cell and mature B cell | ++(70–80%) | Low | Highly disseminated lymphoma; tumour cells invaded liver, spleen, lung and kidney; enlarged spleen |
| 9 | − | ND | ND | ++(80–90%) | Low | Tumour cells invaded liver, lung and spleen; enlarged spleen |
The full _mir_-17_–_92 cistron was also tested in a small cohort of animals. Although similar results were obtained compared to those animals reconstituted with HSCs expressing _mir_-17_–_19b, studies in cell lines indicated that the construct used to express the entire cluster gave lower levels of mature miRNAs. We therefore focused most of our study on the truncated _mir_-17_–_19b cluster. In these ongoing studies, we have yet to find any individual miRNA from the _mir_-17_–_19b cluster that can accelerate tumour formation to the extent seen with the intact polycistron (not shown).
The _Eμ_-myc/_mir_-17_–_19b lymphomas are true malignancies rather than hyperplasias, because primary tumour cells, when transplanted into C57B6/J recipients, induce B-cell lymphomas in 2–3 weeks that result in lethality after 4–5 weeks (data not shown). The secondary tumours show pathological features indistinguishable from the original tumours, and retain tumorigenic potential after two additional rounds of serial transplantation (data not shown). Therefore, a microRNA cluster can accelerate _Eμ_-myc induced tumorigenesis in mice.
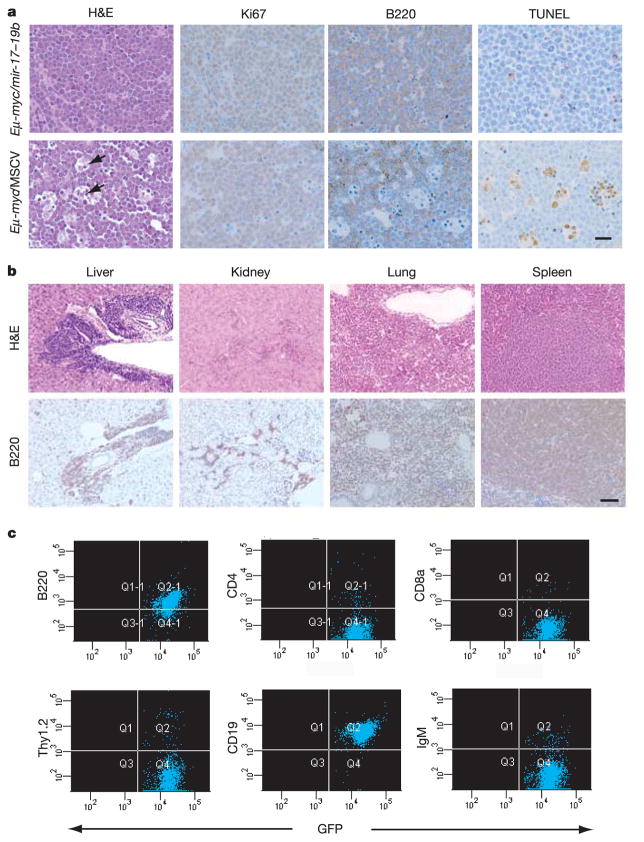
The pathological hallmarks of _Eμ_-myc/_mir_-17_–_19b mosaic animals included massive enlargement of lymph nodes, splenic hyperplasia, infiltration of the thymus by lymphoma cells, and leukaemia (Fig. 2c). Animals with advanced lymphomas showed extramedullary haematopoiesis due to functional failure of the bone marrow. Furthermore, 6 out of 14 animals showed hind limb paralysis, associated with substantial tumour growth at the lumbar node. Tumours resulting from combined _c_-myc and _mir_-17_–_19b expression consistently invaded visceral organs outside the lymphoid compartment, including liver, lung and occasionally kidney (Figs 2c, 3b and Table 1). Additionally, _Eμ_-myc/_mir_-17_–_19b lymphomas show a high mitotic index without extensive apoptosis (Fig. 3a). This contrasts with the _Eμ_-myc/MSCV tumours lacking the microRNA cluster, which show a high degree of apoptosis (Fig. 3a). These findings indicate that cooperation between _Eμ_-myc and _mir_-17_–_19b gives rise to highly malignant, disseminated lymphomas capable of evading normal apoptotic responses to inappropriate proliferation.
Figure 3. Pathological and immunological analysis of lymphomas produced by cooperation between _mir_-17_–_19b and _c_-myc.
a, Haemotoxylin and eosin (H&E), Ki67, B220 and TUNEL staining of _Eμ_-myc/_mir_-17_–_19b lymph node tumours. The ‘starry sky’ morphology is a hallmark of cell clusters undergoing apoptosis (black arrows). Scale bar, 10_μ_m. b, Invasion of visceral organs (liver, spleen, lung and kidney) by _Eμ_-myc/_mir_-17_–_19b tumour cells, shown by H&E and B220 staining. Invasion was observed both perivascularly and parenchymally in liver. Scale bar, 50_μ_m. c, Immunophenotyping of _Eμ_-myc/_mir_-17_–_19b lymphomas. Tumour cells stained positively for the B-cell-specific marker B220, but not for the T-cell-specific markers CD4, CD8a and Thy1.2. Tumour cells bore cellular characteristics of pre-B cells, staining positively for CD19 but not for IgM, a marker of mature B-cells.
_Eμ_-_myc_-induced lymphomas originate from the B-lymphoid lineage. However, the developmental characteristics of these tumour cells are not stage-specific, as they can resemble either mature B cells or pre-B cells. To examine the cell lineage of the _Eμ_-myc/_mir_-17_–_19b lymphomas, we assessed the expression of cell-surface markers, including the B-cell-specific marker B220 and the T-cell-specific markers CD4 and CD8a. All tumours were found to be of B-cell origin, staining positive for B220 and negative for both CD4 and CD8a (Fig. 3c and Table 1). We next analysed these tumours for CD19 and IgM expression to distinguish pre-B from mature B cells. With one exception, _Eμ_-myc/_mir_-17_–_19b lymphomas were derived purely from the pre-B cell lineage (low to absent Thy1.2 staining, CD19+B220+IgM−) (Table 1), suggesting that overexpression of _mir_-17_–_19b strongly favours transformation of B-cell progenitors under our experimental conditions.
Exactly how overproduction of _mir_-17_–_19b might promote oncogenesis is unclear. At a minimum, studies of tumour pathology suggest that increased expression of this cluster mitigates the pro-apoptotic response to elevated myc expression in vivo. Furthermore, we have previously shown that this miRNA cistron is highly expressed in embryonic stem cells, with expression levels decreasing during embryonic development in mice21. It is therefore tempting to speculate that these miRNAs promote ‘stem cell’ properties or specify characteristics of early developmental lineages. A detailed, mechanistic understanding of how this non-coding RNA cluster acts as an oncogene is at present hampered by the lack of a validated biochemical strategy for identifying miRNA targets.
Previous circumstantial evidence has indicated the potential involvement of a number of miRNAs in tumorigenesis. Although miRNAs only represent 1% of the mammalian genome, more than 50% of miRNA genes are located within regions associated with amplification, deletion and translocation in cancer22. Expression studies of various tumour types have also revealed specific alterations in miRNA profiles22–25. For example, _mir_-15 and _mir_-16 are frequently deleted and/or downregulated in B-cell chronic lymphocytic leukaemia26, miR-143 and miR-145 show decreased abundance in colorectal neoplasia25, and miR-155 and its non-coding RNA host gene, BIC, are upregulated 100-fold in Burkitt’s lymphoma patients24. Here, we have shown that one miRNA polycistron is not only the subject of tumour-specific amplification, but that it is also overexpressed in tumours and tumour cell lines, and can act as an oncogene in vivo. Our results indicate that non-coding RNAs might act as integral parts of the molecular architecture of oncogene and tumour suppressor networks. Such oncogenic microRNAs might be designated ‘oncomiRs, with _mir_-17_–_92 being oncomiR-1.
METHODS
miRNA expression profiling
Five micrograms of total RNA was labelled with RNA ligase and a Cy3-conjugated dinucleotide, and hybridized to custom oligonucleotide microarrays as described in ref. 21. Cy3 median intensity values were filtered to remove data points that did not exceed background levels by twofold. Values were log2-transformed and median-centred by array. Clustering was performed with Cluster (Stanford University), using values that were median-centred by gene. Dendrograms and expression maps were generated using Treeview (Stanford).
Cell lines
The measurement of miRNA abundance was carried out using the following human cell lines: Karpas 1718 (derived from splenic lymphoma with villous lymphocytes, provided by A. Karpas), OCI-Ly4, OCI-Ly7 and OCI-Ly8 (derived from diffuse large B-cell lymphoma, provided by R. Dalla-Favera). The cell lines lacking the 13q31-q32 amplicon were Raji (B-cell, derived from Burkitt’s lymphoma, ATCC); Namalwa (B-cell, derived from Burkitt’s lymphoma, ATCC); HG 1125 (EBV-transformed human lymphoblastoid, provided by B. Stillman); Manca (lymphoblast-like, derived from chronic myelogenous leukaemia); Jurkat; proliferating B-cells (spleenic B-cells isolated from a C57B6/Ly5.2 mouse and stimulated to proliferate in culture with lipopolysaccharide, provided by I. Weissman); and normal B cells (derived from cord blood, Cambrex).
PCR and copy number analysis
Tumour samples were obtained from the Cooperative Human Tissue Network (http://www-chtn.ims.nci.nih.gov). Corresponding normal tissue RNA from five individuals was purchased from Biochain Institute Inc. For fluorogenic real-time PCR, primers that amplify _mir_-17_–_92 pri-miRNA and _β_-actin mRNA (control), and probes were designed using Primer Express software (V.2): _mir_-17_–_92 forward primer, 3′-CAGTAAAGGTAAGGAGAGCTCAATCTG-5′; reverse primer, 3′-CATACAACCACTAAGCTAAAGAATAATCTGA-5′; _mir_-17_–_92 probe, (6-FAM)-TGGAAATAAGATCATCATGCCCACTTGAGAC-(TAMRA); _β_-actin forward primer, 3′-GCAAAGACCTGTACGCCAACA-5′; reverse primer, 3′-TGCATCCTGTCGGCAATG-5′; _β_-actin probe, (6-FAM)-TGGCGGCACCACCATGTACC-(TAMRA). The ratios of RNA species detected by _mir_-17_–_92 primers and _β_-actin primers in each RNA sample were determined in triplicate by reverse-transcription, quantitative PCR using an ABI 7900HT Taqman sequence detector, following the ‘standard curve’ method. All values were subsequently normalized to the averaged ratio of the five corresponding normal samples. For DNA copy number determination using the ABI 7900HT sequence detector, we performed quantitative PCR analysis using the same _mir_-17_–_92 primer set described above, and normalized the data against a reference probe corresponding to chromosomal region 6p22 (forward primer, 3′-GGTCTCTATTTGCACTTGGCTGAT-5′; reverse primer, 3 ′-TTTTCATTGTTGACCAAGCTAGACA-5′; probe, (6-FAM)-TAGGGCATACTGCCTGCATATTTCCTGCT-(TAMRA)) or a _β_-actin probe. The reported values represent the ratios of DNA copy number at the _mir_-17_–_92 locus to the normal reference probe.
Adotptive transfer of _Eμ_-myc HSCs
Fetal liver-derived HSCs were isolated from _Eμ_-myc/+ mouse embryos at embryonic day (E)13.5–E15.5, and were transduced with MSCV alone or MSCV expressing the _mir_-17_–_19b cluster. To exclude the possibility that the observed acceleration of lymphomagenesis was due to insertional mutagenesis, independent experiments were carried out using fetal liver cells isolated from eight _Eμ_-myc/+ embryos. Cells from each isolation were separately infected with MSCV _mir_-17_–_19b or control MSCV. The MSCV retroviral vector used in our studies contains a PGK-puromycin-IRES-GFP (PIG) cassette18. Infection rates, assessed by fluorescence-activated cell sorting (FACS), were typically 40% of bulk fetal liver cells. HSCs infected with MSCV-_mir17_–_19b_-PIG and MSCV-PIG (control) were subsequently transplanted into 6–8-week-old, lethally irradiated C57BL/6 recipient mice17. Tumour onset was monitored by blood smear analysis, and tumour samples were either collected into 4% paraformaldehyde for histopathological studies, or prepared as single-cell suspensions for FACS.
We also carried out a screen of 96 miRNAs, to look for miRNA(s) that accelerate Myc-induced lymphomagenesis. In this experiment, each pre-miRNA (and ~50 bp of flanking sequence 5′ and 3′ of the pre-miRNA) was cloned downstream of the cytomegalovirus (CMV) promoter in a MSCV vector containing SV40–GFP. Eight individual MSCV constructs, each overexpressing a specific miRNA, were pooled at equal concentrations. Twelve pools of DNA were used individually to produce virus to infect _Eμ_-myc/+ fetal liver cells for adoptive transfer into at least three recipient animals, as described above. Recipient animals were monitored for tumour growth for at least six months. For those that developed lymphoma, tumour cells were prepared from the enlarged lymph nodes and then subjected to FACS analysis for GFP expression.
Histopathology
Tissue samples were fixed in 4% paraformaldehyde, embedded in paraffin, sectioned into 5-_μ_m slices, and stained with haemotoxylin and eosin. For Ki67 detection (rabbit anti-Ki67 antibody, NovoCastra), representative sections were deparaffinized, rehydrated in graded alcohols, and processed using the avidin–biotin immunoperoxidase method. Sections were then subjected to antigen retrieval by microwave oven treatment using standard procedures. Diaminobenzidine was used as the chromogen and haematoxylin as the nuclear counterstain. For B220 immunohistochemistry, a rat anti-mouse CD45R/B220 antibody (clone RA3-6B2, BD Biosciences Pharmingen) was used; pretreatment for antigen retrieval was not required. Analysis of the rate of apoptotis by TUNEL assay was performed according to a published protocol27.
Supplementary Material
Supplemental
Acknowledgments
We thank members of the Hannon, Lowe and Hammond laboratories for discussions and input. We also thank Z. Xuan, N. Chen, N. Sheth and R. Sachidanandam for bioniformatic analysis. C. Perou and J. Leib provided advice and support for microarray methodologies, and A. Barnes and B. Boone gave assistance with mouse tissue procurement. We are grateful to H. Wendel, C. Scott, C. Marsden and C. Rosenthal, R. Karni, P. Moody and R. Whitaker, who provided advice and technical support. F. Slack coined the oncomiR nomenclature. L.H. and M.T.H. are Fellows of the Helen Hay Whitney Foundation. S.W.L. and C.C.-C. are supported by a program project grant from the NCI. G.J.H is supported by an Innovator Award from the US Army Breast Cancer Research Program and by grants from the DOD and NIH. S.M.H. is a General Motors Cancer Research Foundation Scholar, and J.M.T is a Frederick Gardner Cottrell Postdoctoral Fellow.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Microarray data have been deposited in NCBI-GEO under accession numbers GSM45026–GSM45065 and GSE-2399.
The authors declare no competing financial interests.
References
- 1.Ota A, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 7.Ruvkun G, Giusto J. The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature. 1989;338:313–319. doi: 10.1038/338313a0. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57:49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- 9.Chang S, Johnston RJ, Jr, Frokjaer-Jensen C, Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- 10.Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 11.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen S. M bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 12.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 13.Knuutila S, et al. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol. 1998;152:1107–1123. [PMC free article] [PubMed] [Google Scholar]
- 14.Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 15.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams JM, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 17.Schmitt CA, et al. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 18.Hemann MT, et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nature Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- 19.Hemann MT, et al. Suppression of tumorigenesis by the p53 target PUMA. Proc Natl Acad Sci USA. 2004;101:9333–9338. doi: 10.1073/pnas.0403286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wendel HG, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 21.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray plateform for analysis of microRNA gene expression. Nature Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 22.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calin GA, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosom Cancer. 2004;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 25.Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 26.Calin GA, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Cristofano A, De Acetis M, Koff A, Cordon-Cardo C, Pandolfi PP. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nature Genet. 2001;27:222–224. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
- 28.Mayor C, et al. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental