Arabidopsis ETA2, an Apparent Ortholog of the Human Cullin-Interacting Protein CAND1, Is Required for Auxin Responses Mediated by the SCFTIR1 Ubiquitin Ligase (original) (raw)
Abstract
Auxin response in Arabidopsis thaliana requires the SCFTIR1 ubiquitin ligase. In response to the hormone, SCFTIR1 targets members of the auxin/indoleacetic acid (Aux/IAA) family of transcriptional regulators for ubiquitin-mediated proteolysis. To identify additional regulators of SCFTIR1 activity, we conducted a genetic screen to isolate enhancers of the tir1-1 auxin response defect. Here, we report our analysis of the eta2 mutant. Mutations in ETA2 confer several phenotypes consistent with reduced auxin response. ETA2 encodes the Arabidopsis ortholog of human Cullin Associated and Neddylation-Dissociated (CAND1)/TIP120A, a protein recently identified as a cullin-interacting factor. Previous biochemical studies of CAND1 have suggested that it specifically binds to unmodified CUL1 to negatively regulate SCF assembly. By contrast, we find that ETA2 positively regulates SCFTIR1 because Aux/IAA protein stability is significantly increased in eta2 mutants. Modification of CUL1 by the RUB1/NEDD8 ubiquitin-like protein has been proposed to free CUL1 from CAND1 and promote SCF assembly. We present double mutant analyses of eta2 axr1 plants indicating that liberating CUL1 from ETA2/CAND1 is not the primary role of the RUB modification pathway in the regulation of SCF activity. Our genetic and molecular analysis of SCFTIR1 function in eta2 mutants provides novel insight into the role of CAND1 in the regulation of SCF ubiquitin-ligase activity.
INTRODUCTION
The hormone auxin regulates many aspects of plant growth and development, including embryonic patterning, lateral root development, vascularization, and tropic growth responses (Gray and Estelle, 2000). Previous genetic and molecular studies in Arabidopsis thaliana have identified the SCFTIR1 ubiquitin ligase as a positive regulator of auxin signaling. Mutations in TIR1 confer several phenotypes consistent with a reduced ability to respond to auxin (Ruegger et al., 1998). The TIR1 gene encodes an F-box protein that interacts with a SKP1-like protein (ASK1 or ASK2), the cullin CUL1, and the RBX1 RING domain protein to form an SCF-type ubiquitin ligase (Gray et al., 1999, 2002).
F-box proteins act as recognition factors that recruit specific substrates to the SCF for ubiquitination. The SCFTIR1 complex regulates auxin response, at least in part, by targeting members of the auxin/indoleacetic acid (Aux/IAA) family of transcriptional regulators for ubiquitin-mediated proteolysis in response to an auxin stimulus (Gray et al., 2001; Zenser et al., 2001). Dominant gain-of-function mutations conferring reduced auxin response have been isolated in several Aux/IAA genes (Rouse et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000; Rogg et al., 2001). All of these mutations affect a highly conserved motif termed domain II that functions as a degradation signal that targets the Aux/IAA protein to the SCFTIR1 complex (Ramos et al., 2001). These mutant Aux/IAA derivatives are unable to interact with SCFTIR1 and consequently exhibit increased stability compared with their wild-type counterparts (Gray et al., 2001; Ouellet et al., 2001). Auxin promotes the interaction between wild-type Aux/IAA proteins and TIR1; however, the molecular mechanisms underlying this hormonal regulation are unclear (Gray et al., 2001; Dharmasiri et al., 2003a).
Modification of the CUL1 subunit by the covalent attachment of the ubiquitin-related protein RUB1/NEDD8 is required for normal SCF ubiquitin ligase activity (Lammer et al., 1998; del Pozo and Estelle, 1999; Podust et al., 2000; Kawakami et al., 2001). Mutations affecting components of the RUB-conjugation pathway, including AXR1, ECR1, and RCE1, result in decreased SCFTIR1 activity and a dramatic reduction in auxin response (Lincoln et al., 1990; Gray et al., 2001; del Pozo et al., 2002; Dharmasiri et al., 2003b). Although genetic studies clearly indicate that RUB/NEDD8 modification plays an important role in the regulation of SCF ubiquitin-ligase activity, the precise function of this modification is unclear. Some biochemical studies of mammalian SCF complexes suggest that RUB/NEDD8 modification promotes an increase in SCF activity in vitro, perhaps by increasing the affinity of CUL1 for the ubiquitin conjugating enzyme (Read et al., 2000; Kawakami et al., 2001).
In contrast with mutations in the RUB-conjugation pathway, overexpression of RBX1 and mutations affecting the COP9 signalosome (CSN) result in enhanced CUL1 modification (Lyapina et al., 2001; Schwechheimer et al., 2001; Gray et al., 2002). In addition to its role in the SCF complex, RBX1 has also been proposed to function as the RUB/NEDD8 ligase. The CSN copurifies with SCF complexes from both Arabidopsis and animal cells, and biochemical studies have demonstrated that purified CSN inhibits SCF ubiquitin ligase activity in vitro (Lyapina et al., 2001; Schwechheimer et al., 2001; Yang et al., 2002). Insight into the mechanism underlying this observation was recently provided by the demonstration that the CSN5/JAB1 subunit of the CSN possesses an isopeptidase activity capable of cleaving RUB/NEDD8 from CUL1 (Cope et al., 2002). Surprisingly, the RBX overexpression lines and the CSN mutants are also defective in auxin response, including the degradation of AUX/IAA proteins (Schwechheimer et al., 2001; Gray et al., 2002). These findings suggest that RUB/NEDD8 modification of CUL1 is a dynamic process, with both RUB conjugation and cleavage being required for normal SCFTIR1 ubiquitin ligase activity.
Support for the hypothesis that cycles of RUB/NEDD8 conjugation and cleavage are required for proper SCF function was recently provided by the characterization of the human Cullin Associated and Neddylation-Dissociated (CAND1)/TIP120A protein (Liu et al., 2002; Zheng et al., 2002a; Hwang et al., 2003; Min et al., 2003; Oshikawa et al., 2003). CAND1 was originally identified several years ago as a TATA binding interacting protein (Yogosawa et al., 1996). More recently, CAND1 was identified by several groups as a Cul1-interacting protein. Interestingly, CAND1 was found to specifically bind unmodified Cul1, and in vitro RUB/NEDD8 modification of Cul1 preassembled with CAND1 dissociated the interaction between the two proteins (Zheng et al., 2002a). CAND1 binding requires both the N-terminal domain of Cul1, which is the site of Skp1 binding, and the C-terminal domain, which is the site of RUB/NEDD8 modification (Zheng et al., 2002a). Unmodified Cul1 and Rbx1 coimmunoprecipitated with CAND1, but Skp1 and the F-box protein subunits did not, indicating that CAND1 and Skp1 binding to Cul1 are mutually exclusive (Liu et al., 2002; Zheng et al., 2002a; Min et al., 2003; Oshikawa et al., 2003). Consequently, CAND1 has been proposed to negatively regulate SCF activity by sequestering unmodified Cul1 away from Skp1-F-box protein complexes, thus preventing assembly of the SCF complex. These findings also suggest that the function of RUB/NEDD8 modification in the control of SCF ubiquitin ligase activity may be to relieve this negative regulation by CAND1. Genetic support for such a model is lacking, however, because no CAND1 mutants have been described in any species to date.
In an effort to identify additional genes required for SCFTIR1-mediated auxin response, we have isolated several novel mutations that enhance the relatively weak auxin response defect conferred by the tir1-1 mutation (Gray et al., 2003). Here, we report our identification and analysis of enhancer of tir1-1 auxin resistance (eta2-1), a mutation in the Arabidopsis gene encoding CAND1.
RESULTS
Identification of the eta2-1 Mutant
We have previously described a genetic screen designed to identify mutations that enhance the relatively weak auxin resistance phenotype of tir1-1 seedlings. This screen led to the identification of a mutant allele of the SGT1b/ETA3 gene, which encodes an SCF accessory factor of unknown function (Gray et al., 2003). Several additional eta mutants were isolated from this screen, including a single allele of a gene we designated as ETA2. The eta2-1 tir1-1 M2 plant was backcrossed to tir1-1, and auxin response of the F2 progeny was assessed by examining root growth on media containing 0.25 μM 2,4-D, a concentration inhibitory to tir1-1 seedlings. Sixty-eight of 281 of the F2 seedlings exhibited auxin-resistant root growth, indicating that eta2-1 was a recessive mutation of a single locus (3:1; P < 0.01). When eta2-1 tir1-1 plants were crossed to the wild type, 17/319 F2 plants were resistant to 0.25 μM 2,4-D (15:1; P < 0.01). However, when assayed on media containing 0.085 μM 2,4-D, 181 of 444 F2 seedlings were resistant, indicating that the eta2-1 mutation confers a weak auxin resistance phenotype independent of tir1-1. This possibility was confirmed when PCR-based genotyping determined that several of the resistant segregants were TIR1+/TIR1+.
Like the original eta2-1 tir1-1 mutant, the eta2-1 segregants from backcrosses to tir1-1 and Columbia exhibited a dramatic dwarf phenotype, which continued to cosegregate with the eta2-1 auxin resistance phenotype through several additional backcrosses (Figures 1A and 1B). The severity of the eta2-1 mutation was largely unaffected by the tir1-1 mutation (Figure 1B), although eta2-1 tir1-1 adult plants were slightly shorter with reduced internode lengths compared with eta2-1 single mutants. In addition to the dwarf phenotype, eta2-1 plants develop an excess number of rosette leaves (Figure 1C), form aerial rosettes, and exhibit reduced apical dominance. The eta2-1 mutation also conferred delayed senescence (data not shown).
Figure 1.
The Arabidopsis eta2-1 Mutant.
(A) and (B) Adult phenotype of Col, tir1-1, eta2-1, and eta2-1 tir1-1 plants. Bars = 5 cm.
(C) Rosette leaves from 30-d-old Col and eta2-1 plants. Bar = 1 cm.
Characterization of the eta2-1 Auxin Response Defect
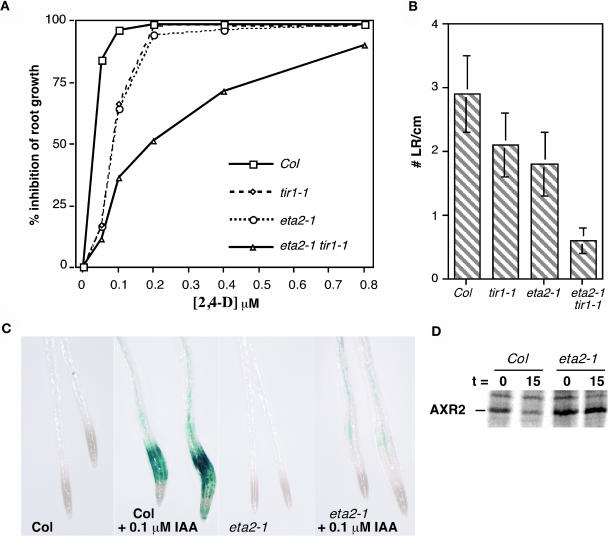
The auxin response defect conferred by the eta2-1 mutation was quantified in a dose–response assay measuring auxin inhibition of root elongation (Figure 2A). In the absence of exogenous auxin, eta2-1 roots grew slightly slower than wild-type controls (data not shown). In the presence of applied auxin, however, eta2-1 seedlings exhibited a modest auxin resistance phenotype similar to that of tir1-1 seedlings. eta2-1 tir1-1 seedlings were significantly more resistant than either single mutant line, suggesting that ETA2 and TIR1 interact synergistically.
Figure 2.
eta2-1 Exhibits Reduced Auxin Response.
(A) Inhibition of root elongation by increasing concentrations of the synthetic auxin 2,4-D. Seedlings were grown for 4 d on unsupplemented media and then transferred to media containing 2,4-D and grown an additional 4 d. Data points are from the averages of 12 seedlings, and standard deviations for all data points were <10% of the mean.
(B) Lateral root (LR) initiation was assessed in 10-d-old seedlings grown on unsupplemented nutrient medium (n = 12).
(C) Transgenic Col and eta2-1 seedlings carrying the _PS-IAA4/5_-GUS reporter BA3. Eight-day-old seedlings were induced with 0.1 μM IAA for 12 h before histochemical staining.
(D) AXR2 pulse-chase assay. AXR2 protein was immunoprecipitated from 7-d-old wild-type (Col) or eta2-1 seedlings labeled with 35S-Met. Precipitations were performed immediately after labeling (t = 0) or after a 15-min chase with medium containing unlabeled Met and cycloheximide (t = 15).
To further explore the eta2-1 auxin response defect, lateral root development and auxin-inducible gene expression were examined. eta2-1 seedlings developed fewer lateral roots than the wild type, and the eta2-1 mutation enhanced the tir1-1 lateral root defect (Figure 2B). Auxin-inducible gene expression was examined using the BA3-β-glucuronidase (GUS) reporter construct consisting of auxin-responsive regulatory elements from the PS-IAA4/5 genes fused to GUS (Oono et al., 1998). Consistent with previous reports, no GUS expression was detected in untreated seedlings by histochemical staining (Figure 2C). Treatment of wild-type seedlings with 0.1 μM IAA resulted in strong GUS expression in the root elongation zone. By contrast, similar treatment of eta2-1 seedlings promoted only a slight increase in GUS staining (Figure 2C).
We next examined whether the eta2-1 mutation affected SCFTIR1 ubiquitin ligase activity by monitoring the stability of the AXR2/IAA7 protein in a pulse-chase assay. Protein extracts were prepared from metabolically labeled wild-type and eta2-1 seedlings, and the AXR2 protein immunoprecipitated at the end of the labeling period or following a 15-min chase with an excess of unlabeled amino acids (Figure 2D). Quantitative analysis of the immunoprecipitates indicated AXR2 was significantly more stabile in eta2-1 seedlings than in the wild type. The average AXR2 half-life determined from three independent experiments was 25.7 ± 4.5 min in eta2-1 seedlings compared with 11.65 ± 1.6 min in wild-type seedlings.
Additional eta2-1 Phenotypes
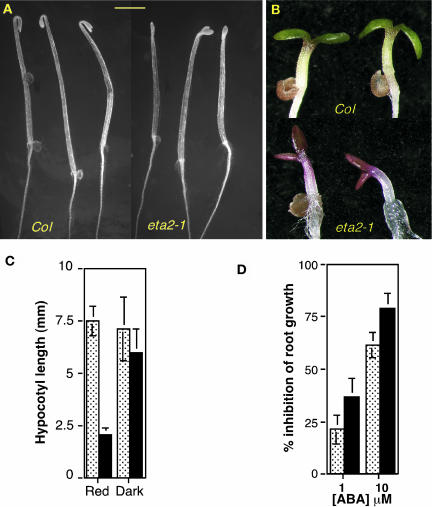
Further analysis of eta2-1 plants revealed several additional phenotypes associated with the mutation. Dark-grown eta2-1 seedlings displayed a weak constitutive photomorphogenesis (COP) phenotype. Hypocotyl length was not dramatically affected by the mutation, but eta2-1 seedlings lacked the strong apical hook characteristic of dark-grown wild-type seedlings (Figure 3A). Additionally, young eta2-1 seedlings were highly anthocyanic, which is also characteristic of cop/det/fus mutants (Figure 3B) (Schwechheimer and Deng, 2000).
Figure 3.
Additional Phenotypes Associated with eta2-1.
(A) Four-day-old dark-grown Col and eta2-1 seedlings. Bar = 2 mm.
(B) Four-day-old light-grown Col and eta2-1 seedlings.
(C) Hypocotyl lengths of 4-d-old Col (speckled bars) and eta2-1 (black bars) seedlings grown under 10 μM/m2/s constant red light or total darkness.
(D) Root growth assay on medium containing abscisic acid. Five-day-old Col (speckled) and eta2-1 (black) seedlings were transferred to media containing 0, 1, or 10 μM abscisic acid and grown for an additional 5 d. Error bars in (C) and (D) indicate standard deviation from the mean.
We also detected a light hypersensitivity phenotype of eta2-1 mutants. This phenotype was particularly striking under low fluence red light, where eta2-1 hypocotyls were dramatically shorter than wild-type control seedlings (Figure 3C). Lastly, assays examining responses to other phytohormones revealed that eta2-1 seedlings were hypersensitive to abscisic acid in both root elongation (Figure 3D) and germination assays (data not shown).
Double Mutant Analysis
The ASK1 and AXR1 gene products are also required for auxin response (Lincoln et al., 1990; Gray et al., 1999). ASK1 encodes a SKP1 subunit of the SCFTIR1 complex, and AXR1 encodes a subunit of the RUB1 activating enzyme that is required for the modification of the CUL1 subunit. eta2-1 plants were crossed to each of these lines to generate double mutants.
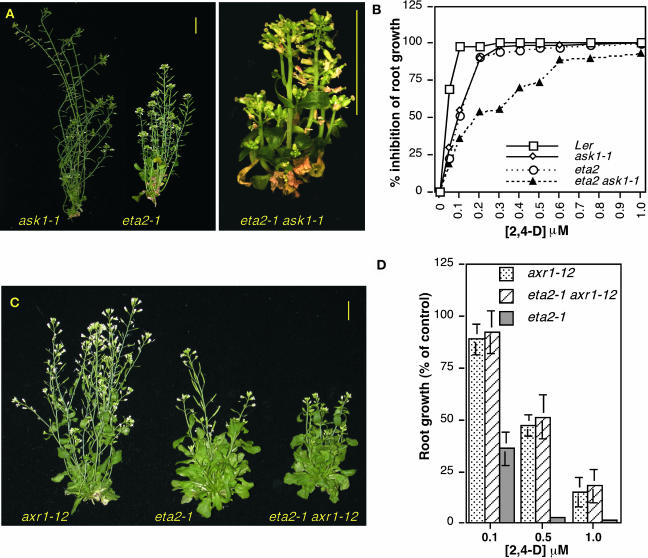
A strong genetic interaction between the eta2-1 and ask1-1 mutations was observed, with most double mutants dying when transplanted to soil. Surviving double mutants exhibited a severe dwarf phenotype (Figure 4A), with adult plants reaching a height of only ∼2 cm. Auxin response in the double mutant was reduced in comparison to the eta2-1 and ask1-1 single mutants (Figure 4B). This heightened reduction in auxin response cannot account for the severe double mutant morphological phenotype, however, because the auxin response defect of eta2-1 ask1-1 seedlings is comparable to that of eta2-1 tir1-1 mutants (Figure 2A).
Figure 4.
Genetic Interactions with ask1-1 and axr1-12.
(A) Adult phenotypes of ask1-1, eta2-1, and eta2-1 ask1-1 double mutants.
(B) Root growth assay on medium containing the synthetic auxin 2,4-D. Standard deviations for all data points were ≤10% of the mean.
(C) Adult phenotypes of axr1-12, eta2-1, and double mutant plants. Bars in (A) and (C) = 1 cm.
(D) Inhibition of root growth by 2,4-D. Root growth assays shown in (B) and (D) were performed by transferring 4-d-old seedlings to media containing 2,4-D and measuring root growth after an additional 4 d. Error bars indicate standard deviation from the mean (n = 12).
By contrast, only a modest genetic interaction was detected between eta2-1 and axr1-12 (Figure 4C). Double mutants were slightly smaller than eta2-1 plants, and the reduction in fertility conferred by the axr1-12 mutation was further enhanced by eta2-1. Analysis of auxin response in the double mutant revealed that axr1-12 was largely epistatic to the eta2-1 mutation because there was no significant difference in root growth between double mutant and axr1-12 seedlings on hormone-supplemented media (Figure 4D).
ETA2 Encodes the Arabidopsis CAND1 Ortholog
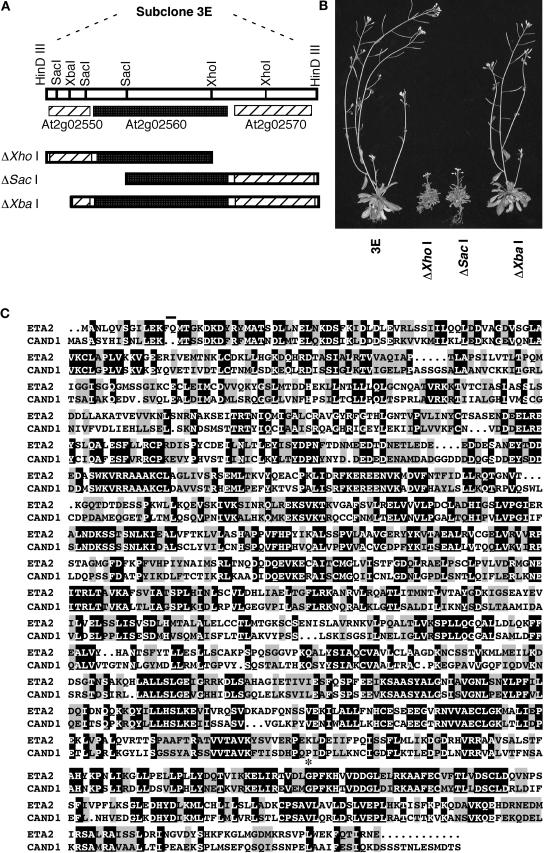
A map-based cloning strategy was used to isolate the ETA2 gene. The eta2-1 mutation was initially mapped between markers rga and ciw2 on the north end of chromosome 2. Additional mapping narrowed the location of ETA2 to an ∼78-kb interval spanning BACs T8K22 and T20F6. These two BACs were partially digested with Hin_DIII and subcloned into the plant transformation vector pCLD04541. The resulting subclones were then used to transform eta2-1 plants. A single T8K22 subclone, designated clone 3E, was identified that complemented both the auxin-resistant root and dwarf phenotypes of the eta2-1 mutation. This construct carried a 16.4-kb insert that harbored three genes: At2g02550, At2g02560, and At2g02570 (Figure 5A). Three deletion derivatives of clone 3E were generated and transformed into eta2-1 plants. Only the Δ_Xba I construct was able to complement the eta2-1 mutation, suggesting that At2g02560 encoded ETA2 (Figure 5B). To confirm this possibility, the coding regions of the three genes were PCR amplified from eta2-1 plants and sequenced. We detected no sequence differences between eta2-1 and Columbia for At2g02550 or At2g02570, but we did identify a single base pair change in an exon of At2g02560. Based on the complementation and sequence analyses, we conclude that At2g02560 encodes ETA2.
Figure 5.
ETA2 Encodes CAND1.
(A) Restriction map of the complementing 3E subclone from BAC T8K22. The positions of the three genes contained in this subclone are indicated as well as the deletion derivatives used in the complementation analysis.
(B) eta2-1 plants transformed with the various subclones.
(C) Sequence alignment of the ETA2 (At2g02560) and human CAND1 proteins. Amino acids 13 and 14 of the ETA2 sequence, which were missing from some of our ETA2 cDNA clones, have a line above them. The site of the eta2-1 Gly → Asp missense mutation is indicated by an asterisk.
A BLAST search of the National Center for Biotechnology Information database identified a full-length 4.1-kb cDNA for the ETA2 gene (NM_126312). ETA2 contains 28 exons and encodes a 1219–amino acid protein that is closely related (43% identity; 64% similarity) to the human CAND1/TIP120A protein (Figure 5C). CAND1 was recently identified as a CUL1 binding protein that has been proposed to regulate SCF ubiquitin ligase activity (Liu et al., 2002; Zheng et al., 2002a; Min et al., 2003). Unlike the human genome, which contains a second gene (TIP120B) that is closely related to CAND1, ETA2 is the only CAND1-like gene in the Arabidopsis genome.
We used primers flanking the ETA2 open reading frame to amplify the coding sequence from RNA prepared from Columbia plants and sequenced the cloned RT-PCR products. Of the nine ETA2 RT-PCR clones analyzed, five were identical to the database cDNA entry, but four were missing the codons for amino acids 13 and 14 (Figure 5C). Analysis of the ETA2 genomic sequence revealed the presence of a cryptic/alternative splice acceptor site upstream of exon 3. Sequence alignments of the ETA2 amino acid sequence with human CAND1 and predicted CAND1 orthologs from several other species indicated that these two amino acids are not present in most CAND1 proteins.
The eta2-1 mutation causes a Gly → Asp missense mutation at position 1069 of the ETA2 protein. This residue is absolutely conserved in all of the predicted CAND1 homologs we could identify in database searches, including mammals, Drosophila melanogaster, Caenorhabditis elegans, Dictyostelium, Schizosaccharomyces pombe, and several plant species (data not shown).
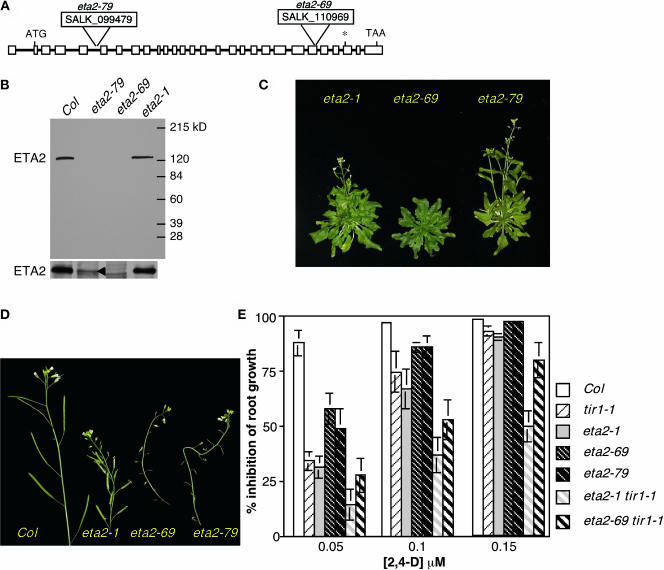
Analysis of eta2 T-DNA Lines
A search of the SALK Institute's SIGnAL T-DNA database (Alonso et al., 2003) identified two lines containing T-DNA insertions within the ETA2 locus. SALK_110969, designated eta2-69, carries a T-DNA within exon 23 of the ETA2 gene, and SALK_099479, designated eta2-79, has a T-DNA insertion in the intron between exons 5 and 6 (Figure 6A). We confirmed the T-DNA insertion sites by sequencing PCR products and identified plants that were homozygous for the insertions. Protein gel blot analysis with an antibody we raised against an internal fragment of the ETA2 protein detected an ∼125-kD protein in wild-type extracts. This band was absent in extracts prepared from eta2-69 plants, demonstrating that the antibody recognizes the ETA2 protein and that this insertion mutation is an apparent null allele (Figure 6B, top). A weak band was detected in eta2-79 extracts (Figure 6B, bottom). Because the T-DNA in this line is in an intron, we suspect that the T-DNA may be spliced out of the ETA2 mRNA at low efficiency, allowing a small amount of ETA2 protein to be translated. By contrast, the ETA2 protein was present at wild-type levels in eta2-1 extracts, consistent with the finding that eta2-1 is a missense mutation.
Figure 6.
Analysis of T-DNA Alleles of ETA2.
(A) Position of the SALK_099479 and SALK_110969 T-DNA insertions within the ETA2 gene. Exons are indicated as boxes, and introns are indicated by lines. The asterisk indicates the position of the eta2-1 mutation.
(B) α-ETA2 protein gel blot analysis of floral extracts. Bottom panel shows a longer exposure of the region containing the ETA2 protein. A small amount of ETA2 is detectable in eta2-79 extracts (arrowhead).
(C) Thirty-eight-day-old eta2-1, eta2-69, and eta2-79 plants.
(D) Influorescences of wild-type and eta2 mutant plants.
(E) Inhibition of root growth by the synthetic auxin 2,4-D. Four-day-old seedlings were transferred to media containing 2,4-D and grown an additional 4 d.
Several aspects of the eta2-69 and eta2-79 plants resembled the eta2-1 mutant phenotype (Figure 6C). Like eta2-1, both insertion mutants were dwarves with increased numbers of rosette leaves in comparison to the wild type. All three eta2 alleles also conferred a wrinkly leaf phenotype, hypersensitivity to red light and abscisic acid, and delays in flowering time and senescence. Unlike the eta2-1 point mutant, both T-DNA alleles also conferred a dramatic reduction in fertility. Whereas eta2-1 plants exhibited only a slight reduction in seed set, eta2-69 and eta2-79 plants were almost completely sterile (Figure 6D). Genetic analysis of eta2-69 and eta2-79 backcrosses to Columbia (Col) confirmed that all of these phenotypes were linked to the T-DNA insertions within the ETA2 gene. Additionally, both T-DNA alleles failed to complement the eta2-1 mutation, confirming that all three mutations are allelic (data not shown). eta2-1/eta2-69 and eta2-1/eta2-79 heterozygotes were indistinguishable from eta2-1 homozygotes, indicating that the eta2-1 mutant protein retains some functional activity.
Curiously, whereas eta2-1 and eta2-79 mutations conferred only a slight delay in flowering time (∼4 to 6 d), eta2-69 plants were severely delayed, with plants flowering ∼3 weeks later than wild-type controls under long-day conditions. Feng et al. (2004) report that an independently isolated null allele of ETA2 (cand1-1) also confers a severe delay in flowering time similar to that of eta2-69. We suspect that the eta2-79 mutation exhibits a less severe flowering time defect because of the residual ETA2 expression we observe in these plants. Feng and colleagues also report that eta2-79 (cand1-2) plants are late flowering. This discrepancy with our analysis of eta2-79 is likely because of differences in growth conditions between the two laboratories.
Root growth assays on auxin containing media, as well as AXR2 pulse-chase assays, indicated that the eta2 T-DNA alleles conferred a reduction in auxin response. Surprisingly, however, this reduction was not as severe as that obtained with the eta2-1 point mutant (Figure 6E). Similarly, double mutant analysis revealed that the eta2-69 mutation enhanced the auxin response defect of tir1-1 but not to the extent of the eta2-1 mutation. These findings suggest that, although eta2-1 is recessive, the eta2-1 protein possesses a novel function that perturbs SCFTIR1 function to a greater extent than the complete loss of ETA2.
Analysis of ETA2 Expression
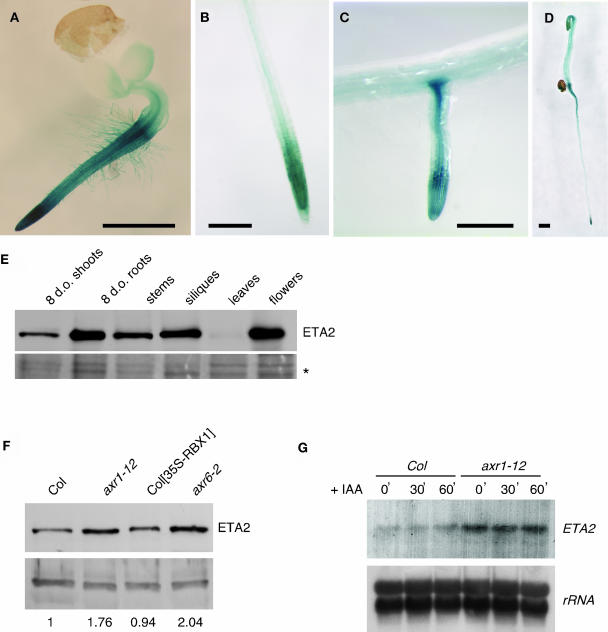
We constructed an ETA2 promoter-GUS fusion construct to examine the pattern of ETA2 expression. ETA2 was expressed throughout young seedlings, with expression being very high in the root and weaker in the shoot (Figure 7A). In older seedlings, ETA2-GUS expression was strongest in root tips, within the root vasculature, and at the tip and base of lateral roots (Figures 7B and 7C). In addition, dark-grown seedlings exhibited strong ETA2-GUS expression in the elongating hypocotyl (Figure 7D). Expression was not affected by treatment with auxin or other plant hormones (data not shown).
Figure 7.
Analysis of ETA2 Expression.
(A) to (D) Col seedlings carrying a PETA2-GUS reporter gene. Bars = 1 mm.
(A) Three-day-old seedling.
(B) Root tip of a 10-d-old seedling.
(C) Lateral root from a 10-d-old seedling.
(D) Four-day-old dark-grown seedling.
(E) α-ETA2 protein gel blot with 25 μg of crude extracts prepared from the indicated tissues. A portion of the Ponceau S-stained blot is shown as a loading control (asterisk).
(F) α-ETA2 protein gel blot of 6-d-old cotyledon extracts prepared from different genetic backgrounds. Band in the bottom panel is a cross-reacting protein used as a loading control. The relative abundance of ETA2 to the cross-reacting protein is indicated below the lanes.
(G) ETA2 RNA gel blot of Col and axr1-12 seedlings treated with 10 μM IAA for 0, 30, or 60 min.
We also analyzed ETA2 expression patterns by RNA gel blot and protein gel blot analyses using various tissues. ETA2 was expressed in all tissues examined, with levels being highest in roots and lowest in mature rosette leaves (Figure 7E). ETA2 protein levels were also examined in several mutant and transgenic lines with impaired SCFTIR1 function. Mutations in TIR1, ASK1, and ETA3/SGT1B did not affect ETA2 protein levels (data not shown). Because ETA2 interacts with CUL1 and CAND1 has been proposed to regulate CUL1 incorporation into the SCF complex, we were especially interested in what affect mutations affecting the RUB modification state of CUL1 might have on ETA2 levels. We detected a slight but consistent increase in ETA2 abundance in axr1-12 and axr6-2 extracts (Figure 7F). axr6-2 is a dominant mutation in the gene encoding CUL1 (Hellmann et al., 2003). Curiously, both the axr1-12 and axr6-2 mutations result in an increase in the ratio of unmodified to modified CUL1 (del Pozo et al., 2002; Hellmann et al., 2003). By contrast, overexpression of RBX1, which results in increased RUB modification of CUL1 (Gray et al., 2002), did not affect the level of ETA2 protein. At least in the case of axr1-12, the increase in ETA2 levels appears to occur at the transcriptional level. Like ETA2 protein levels, ETA2 transcripts were slightly more abundant in axr1-12 plants than in the wild type (Figure 7G). Figure 7G also demonstrated that ETA2 transcription is not regulated by auxin.
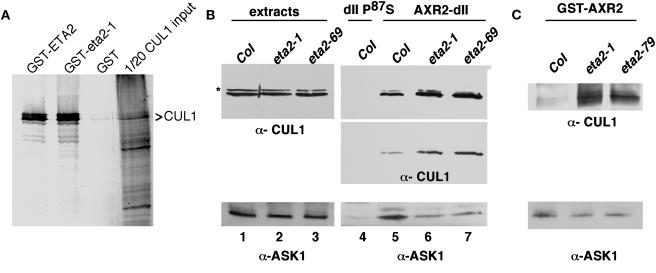
ETA2 Interacts with CUL1 in Vitro
Our genetic studies implicating ETA2 in SCFTIR1-mediated auxin response and the previously published reports that human CAND1 binds unmodified CUL1 prompted us to examine whether ETA2 interacts with the CUL1 subunit of the SCFTIR1 complex. We detected a strong interaction in vitro between a recombinant glutathione _S_-transferase (GST)-ETA2 fusion protein and CUL1 expressed in a reticulocyte expression system (Figure 8A). By contrast, GST alone failed to interact with CUL1 in this assay. The eta2-1 point mutation had no apparent affect on CUL1 binding in vitro. We attempted to coimmunoprecipitate CUL1 and ETA2 from plant extracts using antibodies raised against either protein. However, although both CUL1 and ETA2 were precipitated by their respective antibodies, we were unable to detect coimmunoprecipitation of the reciprocal protein (data not shown).
Figure 8.
ETA2 Interacts with CUL1 and Modulates SCF Assembly.
(A) One microgram of GST, GST-ETA2, or a GST-eta2-1 fusion protein was added to a reticulocyte lysate expressing CUL1. 35S-labeled CUL1 was visualized by autoradiography.
(B) AXR2-dII pull-down assay. One microgram of a 6xHis fusion protein containing the AXR2 domain II degron (amino acids 71 to 100) was incubated with, and subsequently purified from, 2 mg of 7-d-old seedling extracts supplemented with 50 μM IAA. A derivative containing the axr2-1 mutation (dII-P87S), which abolishes TIR1 binding, was used as a negative control. Pull downs were immunoblotted with antisera against CUL1 and ASK1. The top two panels are different exposures of the same α-CUL1 blot. CUL1-RUB is indicated with an asterisk.
(C) GST-AXR2 pull-down assay as in (B), with the exception that a full-length GST-AXR2 fusion protein was used as the bait.
Mutations in ETA2 Perturb the SCFTIR1 Complex
Our finding that eta2 mutants exhibit reduced SCFTIR1 ubiquitin ligase activity prompted us to investigate the possibility that SCF assembly might be affected. We examined the SCFTIR1 complex in eta2 mutants by performing pull-down assays with a 6xHis-tagged AXR2 domain II fusion protein. Protein gel blot analysis with α-CUL1 and α-ASK1 antisera revealed that the eta2-1 and eta2-69 mutations conferred no change in CUL1, CUL1-RUB, or ASK1 levels in crude extracts (Figure 8B, lanes 1 to 3). In the AXR2 pull downs, however, we detected a consistent modest reduction in the amount of ASK1 that copurified with the AXR2 fusion protein (Figure 8B, lanes 4 to 7 bottom panel). At the same time, we observed a slight increase in the amount of CUL1 pulled down from eta2 mutant extracts (Figure 8B, lanes 4 to 7, middle and top panels). We repeated this analysis using a GST full-length AXR2 fusion protein and obtained similar results (Figure 8C).
DISCUSSION
The SCFTIR1 ubiquitin ligase plays a central role in auxin signaling by targeting members of the Aux/IAA family of transcriptional regulators for ubiquitin-mediated proteolysis in response to auxin. In an effort to identify factors that regulate SCFTIR1 activity, we screened for mutations that enhance the weak auxin response defect of tir1-1 mutants. This analysis identified a novel mutation in the ETA2 gene, encoding the Arabidopsis ortholog of the recently identified human CUL1 binding protein CAND1.
ETA2 Is Required for Normal SCFTIR1 Function
We find that mutations in eta2 confer several phenotypes consistent with reduced auxin response, including auxin-resistant root elongation, diminished apical dominance, and reductions in lateral root development and auxin-induced gene expression. Pulse-chase analysis of an SCFTIR1 substrate, the AXR2/IAA7 protein, suggests that these phenotypes are the result of reduced SCFTIR1 ubiquitin ligase activity because AXR2 stability was significantly increased in eta2 seedlings. These findings indicate that ETA2 positively regulates SCFTIR1 activity in vivo.
By contrast, biochemical studies with mammalian CAND1 have suggested that CAND1 negatively regulates SCF activity by sequestering a fraction of the CUL1 pool, preventing it from assembling into an SCF complex (Liu et al., 2002; Zheng et al., 2002a). This discrepancy between the in vivo and in vitro data is reminiscent of studies of the CSN. Purified CSN inhibits ubiquitin ligase activity in an in vitro p27kip1 ubiquitination assay (Yang et al., 2002). Several genetic studies, however, indicate that reductions in CSN activity compromise SCF ubiquitin ligase activity in vivo (Schwechheimer et al., 2001; Cope et al., 2002; Doronkin et al., 2003; Feng et al., 2003). This apparent paradox has been suggested to be the result of a dynamic cycle of RUB/NEDD8 conjugation and deconjugation to Cul1 (Cope and Deshaies, 2003). The finding that CAND1 specifically binds to unmodified Cul1 suggests that RUB/NEDD8 conjugation and cleavage dynamics might drive a cycle of assembly and disassembly of the SCF complex. Modification frees Cul1 from CAND1, promoting SCF assembly. Removal of the RUB/NEDD8 modifier by the CSN promotes disassembly by allowing CAND1 to bind Cul1 and strip it from the SCF complex. Such a cyclical regulatory mechanism may be required in vivo to allow a cell's entire complement of F-box proteins to assemble into SCF complexes. For example, the Arabidopsis genome encodes >700 predicted F-box proteins (Gagne et al., 2002), all of which presumably compete for access to the common SCF core subunits. Cycles of SCF assembly and disassembly would prevent the cell's SCF complexes from becoming locked-up with a fixed set of F-box proteins at the exclusion of other F-box proteins. A cyclical model could also facilitate the reequilibration of the cell's CUL1 pool into new SCF complexes in response to a stimulus.
Our finding that eta2 mutants display reduced SCFTIR1 activity suggests that, like the CSN, ETA2/CAND1 is required to sustain SCF activity over time in vivo, perhaps by facilitating cycles of assembly and disassembly of the SCF complex. The observation that eta2-1 seedlings exhibit a weak cop− phenotype is consistent with the notion that ETA2 and the CSN play related roles in the regulation of ubiquitin ligase activity. Given such a model, it is tempting to speculate that the function of RUB/NEDD8 modification is to free CUL1 from the clutches of ETA2/CAND1. Consistent with this possibility, modification was shown to dissociate Cul1 from preformed CAND1-Cul1 complexes in vitro (Liu et al., 2002). This model predicts that mutations in ETA2/CAND1 should suppress, or at least be epistatic to, mutations in the RUB/NEDD8 conjugation pathway because modification should not be required in the absence of negative regulation by ETA2/CAND1. Our double mutant analysis of eta2-1 axr1-12 plants does not support this hypothesis, however, because axr1-12 was largely epistatic to eta2-1, indicating that the RUB modification pathway is still required by eta2-1 plants. We have obtained similar results with eta2-69 axr1-12 double mutant plants (data not shown). Because RUB/NEDD8 modification may regulate SCF activity at multiple levels, however, we cannot eliminate the possibility that one of the functions of the modification is to promote the release of CUL1 from ETA2/CAND1. Nonetheless, our finding that SCF function in eta2 mutants is still highly dependent on AXR1 indicates that dissociation of CUL1 from ETA2/CAND1 is not the essential function of the RUB/NEDD8 pathway as has been previously suggested (Liu et al., 2002).
Curiously, the auxin response defect of the eta2-1 point mutant is significantly more severe than that of the two T-DNA insertion mutants we characterized, including the eta2-69 null allele. Similarly, whereas eta2-69 seedlings are also hypersensitive to red light, this phenotype is also not as severe as that observed with eta2-1 seedlings (data not shown). These findings are especially puzzling because eta2-1 is a recessive mutation. The fact that essentially all aspects of the eta2-1 mutant phenotype are similar to those conferred by the T-DNA alleles strongly suggests that eta2-1 is a loss-of-function mutation. Nonetheless, it is equally apparent that the point mutation confers some novel effect on the ETA2 protein. Within the context of the cycling model of SCF assembly discussed above, one possible explanation for the increased severity of the eta2-1 mutation is if the mutant protein binds to CUL1 and causes a block in the cycle. For example, if the mutation disrupts the regulated dissociation from CUL1, this could result in the formation of a poisoned complex that prevents the assembly of an active SCF complex. The mutation could still be recessive if wild-type ETA2 binds to CUL1 with a higher affinity than the mutant protein.
Equally interesting is the fact that certain aspects of the eta2 null phenotype are more severe than the eta2-1 phenotype. For example, the eta2 T-DNA mutants are nearly completely sterile, whereas eta2-1 plants only exhibit a modest reduction in seed set. Together, these findings suggest that the eta2-1 point mutation may only affect a subset of the functions of the ETA2 protein.
ETA2 Interacts with CUL1 to Regulate SCF Assembly
We detected a strong in vitro interaction between ETA2 and CUL1 but were unable to detect this interaction in plant extracts. This suggests that the in vivo interaction may be unstable or that only a relatively small fraction of the CUL1 pool is in a complex with ETA2. Consistent with this notion, Feng et al. (2004) detected a weak interaction with CUL1 in plant extracts, as well as in a yeast two-hybrid system. Like human CAND1, Arabidopsis ETA2/CAND1 appears to specifically interact with unmodified CUL1.
If ETA2/CAND1 prevents CUL1 from incorporating into an SCF complex as described above, one would expect that mutations in ETA2 would alter SCF assembly dynamics. Using AXR2 pull-down assays, we found that mutations in ETA2 resulted in a mild reduction in the amount of ASK1 that copurifies with the AXR2 bait protein. Surprisingly, this reduction was coupled with a slight increase in the amount of CUL1 present in the pull downs. Based on several studies of SCF ubiquitin ligases, including crystal structure analysis of the human SCFSkp2 complex (Zheng et al., 2002b), ASK1 association with the AXR2 bait protein should be dependent upon the TIR1 and TIR1-like F-box proteins that recognize the SCF substrate. Similarly, the presence of CUL1 in the pull downs should be dependent upon its interaction with ASK1. Therefore, a likely explanation for the reduction in ASK1 levels is that TIR1 levels are diminished in eta2 mutant seedlings. Indeed, several F-box proteins are known to be destabilized by their incorporation into the SCF complex (Zhou and Howley, 1998; Galan and Peter, 1999; Wirbelauer et al., 2000). Furthermore, Zheng et al. (2002a) reported that silencing of CAND1 in HeLa cells resulted in increased Skp1 binding to Cul1 and a reduction in the level of the Skp2 F-box protein. We were unable to directly examine TIR1 stability in eta2 plants because of the lack of a TIR1 antibody. We have used a TIR1-myc transgene in past studies, but we were unable to isolate eta2 lines, by either transformation or crosses with TIR1-myc plants, that expressed this construct at detectable levels.
At first glance, the corresponding increase in CUL1 abundance in the AXR2 pull downs seems paradoxical. However, if ETA2 facilitates cycles of SCF assembly and disassembly as discussed above, ASK1 would be predicted to be present in a subcomplex with TIR1 in addition to the intact SCF complex. Thus, AXR2 pull downs with wild-type extracts would contain SCFTIR1 as well as the ASK1-TIR1 subcomplex. In eta2 mutants, CUL1 can freely assemble into an SCF complex, thus reducing the pool of free ASK1-TIR1. Therefore, we predict that there is less TIR1 protein in eta2 plants; however, a higher percentage of TIR1 should be present in the assembled SCF complex in comparison to the wild type. This then begs the question of why eta2 mutants exhibit reduced SCFTIR1 activity. A simple answer is not immediately evident. Perhaps a cycle of disassembly and reassembly is needed to recruit naïve TIR1-ASK after ubiquitination of the substrate. Although we concede that these findings are open to additional interpretation, they strongly suggest that SCF assembly dynamics are altered by mutations in ETA2.
Whereas CUL1, the CSN, and the RUB/NEDD8 modification pathway are all required for the viability of higher eukaryotes, ETA2 is not an essential gene in Arabidopsis. Nonetheless, eta2 mutants exhibit several severe developmental defects, indicating that it is required for normal growth and development. The pleiotropic nature of eta2 mutations as well as the strong genetic interaction we observed with ask1-1 indicate that ETA2 is not specifically defective in auxin signaling but rather acts as a global regulator of SCF ubiquitin ligase activity. Additionally, CAND1 interacts with multiple cullin family members (Liu et al., 2002; Min et al., 2003), raising the distinct possibility that other types of cullin-containing ubiquitin ligases are impaired in eta2 mutants. Our identification of ETA2/CAND1 mutants of Arabidopsis provides a new set of powerful genetic tools for elucidating the function of this highly conserved protein in eukaryotic cells.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana lines employed in this study are in the Col ecotype, with the exception of ask1-1, which is in Landsberg erecta (L_er_). For double mutant analysis with ask1-1, the eta2-1 mutation was introgressed into L_er_ by three successive backcrosses. Seedlings were grown under sterile conditions on vertically oriented ATS nutrient medium (Lincoln et al., 1990). Adult plants were grown under long-day lighting. Conditions for the mutagenesis and screen for eta− mutants have been previously described (Gray et al., 2003).
Map-Based Isolation of the ETA2 Gene
A total of 609 auxin-resistant F2 seedlings from a cross between eta2-1 and L_er_ were used to prepare DNA for PCR-based mapping with codominant cleaved-amplified polymorphic sequence and simple sequence length polymorphic markers. We initially mapped the eta2-1 mutation to an interval between markers rga and ciw2 (http://www.arabidopsis.org). For fine mapping, we generated several additional markers in this interval using the Cereon Arabidopsis polymorphism collection (Jander et al., 2002). Markers defining our final mapping interval were CER461163 (5′- GCGCGAGTATATCAATGAAC-3′ and 5′-GCTCTAATTATGGATGAAG-3′), which amplifies an simple sequence length polymorphic of 174 bp (Col) and 164 bp (L_er_), and CER447661 (5′-GTGGTGTACTTGATGAACTTC-3′ and 5′-CAATTCCATTTGACAGCCATG-3′), which amplifies a 374-bp cleaved-amplified polymorphic sequence marker that contains an Hpa_I site in L_er but not in Col. These and additional markers in the mapping interval are available upon request.
BAC clones T8K22 and T20F6 were partially digested with _Hin_DIII and the resulting fragments subcloned into the _Hin_DIII site of the plant transformation vector pCLD04541 (Tao and Zhang, 1998). Subclones were introduced into Agrobacterium tumefaciens by triparental mating, which was then used to transform eta2-1 plants.
Antibodies
A 462-bp _Eco_RI-_Xho_I fragment encoding amino acids 926 to 1080 of the ETA2 protein was cloned into the pET30A Escherichia coli expression vector (Novagen, Madison, WI). Expression was induced and the fusion protein purified on nickel-nitrilotriacetic acid agarose agarose using standard protocols (Gray et al., 1999). The fusion protein was eluted with imidazole and used to immunize a New Zealand white rabbit (Cocalico Biological, Reamstown, PA). α-ETA2 antisera was immunopurified using nitrocellulose-bound antigen (Pringle et al., 1989). CUL1 and ASK antibodies have been previously described (Gray et al., 1999).
Pull-Down Assays, Pulse-Chase Assays, and Protein Gel Blot Analysis
AXR2 pulse-chase assays were conducted with 7-d-old seedling extracts as previously described (Gray et al., 2001). AXR2 half-life was determined using a Molecular Dynamics PhosphorImager and ImageQuant software (Sunnyvale, CA) to quantify labeled AXR2 protein present in the immunoprecipitates. GST-ETA2 pull-down assays were performed by adding ∼1 μg of purified fusion protein to 25 μL of a TNT (Promega, Madison, WI) reticulocyte lysate reaction expressing CUL1. Reactions were diluted with 300 μL of buffer C (Gray et al., 1999) containing 10 μM MG132, and incubated at 30°C for 2 h. Pull downs were washed three times with 1 mL of buffer C and analyzed by SDS-PAGE and autoradiography. GST-AXR2 and 6xHis-AXR2-dII pull downs were conducted with 1 μg of purified fusion protein and 2 mg of seedling extracts supplemented with 50 μM IAA. GST-AXR2 pull downs were washed three times with buffer C. 6xHis-AXR2-dII pull downs were washed three times with buffer C supplemented with 20 mM imidazole. Protein extraction and protein gel blot procedures have been described previously (Gray et al., 1999). ETA2 immunoblots were quantitated using NIH Image with ECL exposures on preflashed autoradiography film.
GUS Histochemical Staining
A 2.7-kb fragment containing genomic sequence from upstream of the ETA2 locus through the ATG start codon was cloned in frame with the GUS coding sequence of pBI101.2 (Clontech, Palo Alto, CA). Seedlings were stained for GUS activity as previously described (Stomp, 1991).
Acknowledgments
We thank Cereon Genomics for access to its Arabidopsis polymorphism collection, Bill Crosby for the gift of α-ASK antisera, the Salk Institute Genomic Analysis Laboratory and the ABRC for providing ETA2 T-DNA insertion mutants, Min Ni for providing a red light source, and S. Feng and X.W. Deng for communicating results before publication. We would also like to acknowledge Neil Olszewski and Mark Estelle for helpful comments of the manuscript and Jane Gray for technical assistance. This work was supported by National Institutes of Health Grant GM067203 to W.M.G.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: William M. Gray (grayx051@tc.umn.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021923.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301**,** 653–657. [DOI] [PubMed] [Google Scholar]
- Cope, G.A., and Deshaies, R.J. (2003). COP9 signalosome: A multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114**,** 663–671. [DOI] [PubMed] [Google Scholar]
- Cope, G.A., Suh, G.S., Aravind, L., Schwarz, S.E., Zipursky, S.L., Koonin, E.V., and Deshaies, R.J. (2002). Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298**,** 608–611. [DOI] [PubMed] [Google Scholar]
- del Pozo, J.C., Dharmasiri, S., Hellmann, H., Walker, L., Gray, W.M., and Estelle, M. (2002). AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14**,** 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., and Estelle, M. (1999). The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. USA 96**,** 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., Jones, A.M., and Estelle, M. (2003. a). Auxin action in a cell-free system. Curr. Biol. 13**,** 1418–1422. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, S., Dharmasiri, N., Hellmann, H., and Estelle, M. (2003. b). The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J. 22**,** 1762–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronkin, S., Djagaeva, I., and Beckendorf, S.K. (2003). The COP9 signalosome promotes degradation of Cyclin E during early Drosophila oogenesis. Dev. Cell 4**,** 699–710. [DOI] [PubMed] [Google Scholar]
- Feng, S., Ma, L., Wang, X., Xie, D., Dinesh-Kumar, S.P., Wei, N., and Deng, X.W. (2003). The COP9 signalosome interacts physically with SCF COI1 and modulates jasmonate responses. Plant Cell 15**,** 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S., Shen, Y., Sullivan, J.A., Rubio, V., Xiong, Y., Sun, T.-p., and Deng, X.W. (2004). Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell 16**,** 1870–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J.M., Downes, B.P., Shiu, S.H., Durski, A.M., and Vierstra, R.D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99**,** 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan, J.M., and Peter, M. (1999). Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. USA 96**,** 9124–9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13**,** 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., and Estelle, I. (2000). Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem. Sci. 25**,** 133–138. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., Hellmann, H., Dharmasiri, S., and Estelle, M. (2002). Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell 14**,** 2137–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414**,** 271–276. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., Muskett, P.R., Chuang, H.W., and Parker, J.E. (2003). Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 15**,** 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann, H., Hobbie, L., Chapman, A., Dharmasiri, S., Dharmasiri, N., del Pozo, C., Reinhardt, D., and Estelle, M. (2003). Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis. EMBO J. 22**,** 3314–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, J.W., Min, K.W., Tamura, T.A., and Yoon, J.B. (2003). TIP120A associates with unneddylated cullin 1 and regulates its neddylation. FEBS Lett. 541**,** 102–108. [DOI] [PubMed] [Google Scholar]
- Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I.M., and Last, R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129**,** 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, T., Chiba, T., Suzuki, T., Iwai, K., Yamanaka, K., Minato, N., Suzuki, H., Shimbara, N., Hidaka, Y., Osaka, F., Omata, M., and Tanaka, K. (2001). NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20**,** 4003–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammer, D., Mathias, N., Laplaza, J.M., Jiang, W., Liu, Y., Callis, J., Goebl, M., and Estelle, M. (1998). Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 12**,** 914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2**,** 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Furukawa, M., Matsumoto, T., and Xiong, Y. (2002). NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol. Cell 10**,** 1511–1518. [DOI] [PubMed] [Google Scholar]
- Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Tsuge, T., Zhou, C., Wolf, D.A., Wei, N., and Deshaies, R.J. (2001). Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292**,** 1382–1385. [DOI] [PubMed] [Google Scholar]
- Min, K.W., Hwang, J.W., Lee, J.S., Park, Y., Tamura, T.A., and Yoon, J.B. (2003). TIP120A associates with cullins and modulates ubiquitin ligase activity. J. Biol. Chem. 278**,** 15905–15910. [DOI] [PubMed] [Google Scholar]
- Nagpal, P., Walker, L.M., Young, J.C., Sonawala, A., Timpte, C., Estelle, M., and Reed, J.W. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123**,** 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono, Y., Chen, Q.G., Overvoorde, P.J., Kohler, C., and Theologis, A. (1998). Age mutants of Arabidopsis exhibit altered auxin-regulated gene expression. Plant Cell 10**,** 1649–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshikawa, K., Matsumoto, M., Yada, M., Kamura, T., Hatakeyama, S., and Nakayama, K.I. (2003). Preferential interaction of TIP120A with Cul1 that is not modified by NEDD8 and not associated with Skp1. Biochem. Biophys. Res. Commun. 303**,** 1209–1216. [DOI] [PubMed] [Google Scholar]
- Ouellet, F., Overvoorde, P.J., and Theologis, A. (2001). IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell 13**,** 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust, V.N., Brownell, J.E., Gladysheva, T.B., Luo, R.S., Wang, C., Coggins, M.B., Pierce, J.W., Lightcap, E.S., and Chau, V. (2000). A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc. Natl. Acad. Sci. USA 97**,** 4579–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle, J.R., Preston, R.A., Adams, A.E., Stearns, T., Drubin, D.G., Haarer, B.K., and Jones, E.W. (1989). Fluorescence microscopy methods for yeast. Methods Cell Biol. 31**,** 357–435. [DOI] [PubMed] [Google Scholar]
- Ramos, J.A., Zenser, N., Leyser, O., and Callis, J. (2001). Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13**,** 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read, M.A., Brownell, J.E., Gladysheva, T.B., Hottelet, M., Parent, L.A., Coggins, M.B., Pierce, J.W., Podust, V.N., Luo, R.S., Chau, V., and Palombella, V.J. (2000). Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol. Cell. Biol. 20**,** 2326–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg, L.E., Lasswell, J., and Bartel, B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13**,** 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279**,** 1371–1373. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12**,** 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C., and Deng, X.W. (2000). The COP/DET/FUS proteins: Regulators of eukaryotic growth and development. Semin. Cell Dev. Biol. 11**,** 495–503. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X.W. (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292**,** 1379–1382. [DOI] [PubMed] [Google Scholar]
- Stomp, A.-M. (1991). Histochemical localization of β-glucuronidase. In GUS Protocols, S.R. Gallagher, ed (London: Academic Press), pp. 103–113.
- Tao, Q., and Zhang, H.B. (1998). Cloning and stable maintenance of DNA fragments over 300 kb in Escherichia coli with conventional plasmid-based vectors. Nucleic Acids Res. 26**,** 4901–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Q., and Reed, J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126**,** 711–721. [DOI] [PubMed] [Google Scholar]
- Wirbelauer, C., Sutterluty, H., Blondel, M., Gstaiger, M., Peter, M., Reymond, F., and Krek, W. (2000). The F-box protein Skp2 is a ubiquitylation target of a Cul1-based core ubiquitin ligase complex: Evidence for a role of Cul1 in the suppression of Skp2 expression in quiescent fibroblasts. EMBO J. 19**,** 5362–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., Menon, S., Lykke-Andersen, K., Tsuge, T., Di, X., Wang, X., Rodriguez-Suarez, R.J., Zhang, H., and Wei, N. (2002). The COP9 signalosome inhibits p27(kip1) degradation and impedes G1-S phase progression via deneddylation of SCF Cul1. Curr. Biol. 12**,** 667–672. [DOI] [PubMed] [Google Scholar]
- Yogosawa, S., Makino, Y., Yoshida, T., Kishimoto, T., Muramatsu, M., and Tamura, T. (1996). Molecular cloning of a novel 120-kDa TBP-interacting protein. Biochem. Biophys. Res. Commun. 229**,** 612–617. [DOI] [PubMed] [Google Scholar]
- Zenser, N., Ellsmore, A., Leasure, C., and Callis, J. (2001). Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 98**,** 11795–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J., Yang, X., Harrell, J.M., Ryzhikov, S., Shim, E.H., Lykke-Andersen, K., Wei, N., Sun, H., Kobayashi, R., and Zhang, H. (2002. a). CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol. Cell 10**,** 1519–1526. [DOI] [PubMed] [Google Scholar]
- Zheng, N., et al. (2002. b). Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416**,** 703–709. [DOI] [PubMed] [Google Scholar]
- Zhou, P., and Howley, P.M. (1998). Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell 2**,** 571–580. [DOI] [PubMed] [Google Scholar]