The Prioritization of Cancer Antigens: A National Cancer Institute Pilot Project for the Acceleration of Translational Research (original) (raw)
. Author manuscript; available in PMC: 2018 Jan 23.
Abstract
The purpose of the National Cancer Institute (NCI) pilot project to prioritize cancer antigens was to develop a well-vetted ranked prioritized list of cancer vaccine target antigens based on pre-defined and pre-weighted objective criteria. An additional aim was for the NCI to test a new approach for prioritizing translational research opportunities based on an Analytic Hierarchy Process for dealing with complex decisions. Antigen prioritization involved developing a list of “ideal” cancer antigen criteria/characteristics, assigning relative weights to those criteria using pair-wise comparisons, selecting 75 representative antigens for comparison and ranking, assembling information on the pre-defined criteria for the selected antigens, and ranking the antigens based on the pre-defined, pre-weighted criteria. Using the pair-wise approach, the result of criteria weighting, in descending order was: (1) Therapeutic function, (2) Immunogenicity, (3) Role of the antigen in oncogenicity, (4) Specificity, (5) Expression level and percent of antigen positive cells, (6) Stem cell expression, (7) Number of patients with antigen positive cancers, (8) Number of antigenic epitopes and (9) Cellular location of antigen expression. None of the 75 antigens had all of the characteristics of the “ideal” cancer antigen. However, 46 were immunogenic in clinical trials and 20 of them had suggestive clinical efficacy in the “Therapeutic function” category. These findings reflect the current status of the cancer vaccine field, highlight the possibility that additional organized efforts and funding would accelerate the development of therapeutically effective cancer vaccines, and accentuate the need for prioritization.
Introduction
Virtually any mutant, over- or abnormally-expressed protein in cancer cells can serve as a target for cancer vaccines and/or T cell therapy [1–75]. Scores of cancer vaccines have been shown to be immunogenic in clinical trials, and many of them have demonstrated efficacy in at least small numbers of patients. No cancer vaccine has yet been approved by the FDA, despite extensive developmental efforts by academia and industry. Nevertheless, there is consensus that optimally designed cancer vaccine trials combining the best antigens with the most effective immunotherapy agents might yield positive clinical results.
Cancer vaccine development is limited by several factors, including funding constraints. Limited resources mandate transparent methods to prioritize developmental opportunities with the least possible bias. An NCI immunotherapy agent workshop was held in July 2007 to rank agents with high potential to serve as immunotherapeutic drugs1. The ranking was based on the likelihood for efficacy in cancer therapy and was exceedingly well-vetted, with broad and substantial input from academia, industry and the government. Many of the ranked immunotherapeutic agents have been shown to be effective as components of cancer vaccine regimens in pre-clinical models, but this abundance of promising opportunities raises immediate questions as to which antigen or sets of antigens is/are most appropriate for co-development. Our current effort to prioritize cancer antigens represents the logical next step in attempting to focus translational efforts on cancer vaccine regimens with the highest potential for success.
The task of ranking cancer antigens is immense, and the number of potential cancer antigens almost limitless. At present, investigator-initiated funding of science dictates innovation, i.e., that each investigator discovers and develops his/her own antigens. This leads to an ever increasing number of potential vaccine targets as well as validation of those targets through pre-clinical and early clinical cancer vaccine development. Few investigators have both the financial and organizational resources to advance their vaccines past early developmental stages.
The NCI, recognizing the untapped potential of therapeutic cancer vaccines as well as many other novel therapies, embarked on a new approach to the identification, prioritization, and funding of translational cancer research based on recommendations of the Translational Research Working Group (TRWG) 2. The primary objective is to identify specific translational cancer research projects that warrant a dedicated effort to accelerate progress through focused collaborations. This process requires a mechanism for identifying high priority translational research projects based on scientific validity, clinical need, and technical feasibility. The NCI’s initial endeavor to implement the TRWG’s recommendation for prioritization of translational opportunities has focused on evaluation of a method to select cancer antigens for subsequent development through the Immune Response Modifier (IRM) Pathway, one of the six TRWG Pathways leading from fundamental laboratory discoveries to definitive testing in clinical trials [76, 77].
The IRM Pathway was selected as the pilot effort for several reasons. It is the most complex of the TRWG Pathways and successful application of a prioritization process in this context is expected to be generalizable to other TRWG pathways. In addition, the immunology community had already prioritized immunotherapy agents at the NCI Immunotherapy Agent Workshop3, an experience that greatly facilitated implementation of this pilot project.
The methodology for prioritization of cancer antigens was based on the Analytic Hierarchy Process (AHP), a structured technique and mathematical model for dealing with complex decisions. AHP has been refined since its initial description by Thomas L. Saaty in the 1970’s [78], and has been used throughout the world in a wide variety of decision settings spanning government, business, industry, healthcare, and education. AHP is considered most useful to teams contending with complex problems that involve human perception and judgment [79]. The process breaks down a complex problem into a hierarchy of sub-problems that can be compared to each other on a pair-wise basis. It has unique advantages where major decision elements are difficult to quantify or compare, or where communication among team members is impeded by their different specializations, terminologies, or perspectives. For the current project, criteria for cancer vaccines were determined. The criteria were then broken down into sub-criteria for greater granularity within each higher level criterion. A panel of cancer vaccine experts used pair-wise comparisons to weight first the criteria, then the sub-criteria within the criteria. The AHP converted the weighted criteria into numerical values that could be analyzed and compared for the ranking of antigens and to permit the comparison of rankings based on hypothetical alternative weightings.
The AHP generated primary and alternative priority rankings of 75 cancer antigens on the basis of criteria pre-identified and weighted by a broadly constituted panel of cancer vaccine experts. These rankings are dynamic, given that priorities change as knowledge accrues from new studies. The associated lists of weighted criteria inform investigators as to what experimental evidence is required to advance antigens to higher priority levels. Above all, the rankings provide a basis for deciding which antigens are most likely to pay off on investments to generate cancer vaccines for testing in later-stage clinical trials.
Materials and Methods
Decision Lens, Inc. provided the AHP methodology as a web-based tool with four modules4. The first phase of the process focused on identifying the participants, criteria, and alternatives to be prioritized. In the second phase, criteria essential to the decision were identified, grouped, compared, and weighted using the Build Model and Compare Criteria modules. The third phase focused on the Evaluate Alternatives module, wherein alternatives (antigens) were compared to each of the weighted criteria to determine their benefit or value using customized rating scales. The Reporting module provided a flexible tool for the analysis of information to facilitate informed decision-making.
Phase I: Decision Preparation
The key objective of the decision preparation phase was to gather the critical data needed to make the decision and to define expectations for key participants regarding the decision process. There were three distinct steps to the process.
The first step was to determine who would be participating in the prioritization process. The NCI selected investigators who participated in the Immunotherapy Agent Workshop. The Workshop participants had been selected on the basis of recommendations from the American Association for Cancer Research (AACR), American Association of Immunologists (AAI), American Society of Clinical Oncology (ASCO), American Society of Hematology (ASH), Cancer Vaccine Consortium (CVC), International Society for Biological Therapy of Cancer (iSBTc), and NCI intramural and extramural program staff. Experts from this group were used to contribute to the criteria determination, weighting, and evaluation steps of the process (List of participants available as Supplements A, B and C).
The 19 investigators listed in Supplement A provided the criteria used to evaluate cancer antigens. Top-down and bottom-up approaches were used. For the top-down approach, approximately half of the experts were asked to submit via e-mail what they regarded to be characteristics of an “ideal” cancer antigen. For the bottom-up approach, the remaining experts were asked which characteristics made the following antigens good or poor candidates for therapeutic development: (1) mutated segment of p53, (2) MUC1, (3) MAGE-A3, (4) HER-2/neu, (5) gp100, and (6) mutated proteins unique to each patient. The two lists were vetted, combined and structured into a list of criteria and sub-criteria. Using the same information source, definitions for each criterion and sub-criterion were developed. The final criteria and definitions are shown in Table 1, columns 1 and 2.
TABLE 1.
Cancer antigen pilot prioritization: criteria & sub-criteria definitions & weightings
| THERAPEUTIC FUNCTION (Weight of Criteria - 0.32) | ||
|---|---|---|
| Sub-criteria | Definition | Weight of Sub-criteria |
| Controlled vaccine trial suggestive_Data ranked as being superb, very_ strong, adequate, or fair. | Clinical trial data showing that avaccine induced clinical responses inat least a small number of patients, orprovided suggestive evidence ofbenefit vs. controls | |
| Superb Data Controlled vaccine trialsuggestive | 100.0% (1.0) | |
| Very Strong Data Controlled vaccinetrial suggestive | 93.0% (0.93) | |
| Adequate Data Controlled vaccine trialsuggestive | 85.0% (0.85) | |
| Fair Data Controlled vaccine trialsuggestive | 75.0% (0.75) | |
| Responses in T cell therapy | 65.0% (0.65) | |
| Pre existent immunity/survivalcorrelation | 15.0% (0.15) | |
| Positive appropriate animal models | 10.0% (0.1) | |
| Not applicable (N/A) | 0.0% (0.0) | |
| IMMUNOGENICITY (Weight of Criteria - 0.17) | ||
| Immunogenic in clinical trials | T cell and/or Ab responses elicited inclinical trials | 100.0% (1.0) |
| T cell immunity observed | Spontaneous T cell responsesobserved in some patients | 39.0% (0.39) |
| Immunogenic in appropriate animalmodels | Immunogenic in animal models withnatural levels of antigen expressionsimilar to humans | 11.0% (0.11) |
| Ab immunity observed | Spontaneous Ab observed in somepatients | 10.0% (0.1) |
| Not applicable (N/A) | 0.0% (0.0) | |
| ONCOGENICITY (Weight of Criteria - 0.15) | ||
| Oncogenic “self” protein | Associated with oncogenic process,i.e., oncogenic “self” protein | 100.0% (1.0) |
| Persistent viral Ag | Persistently expressed viral antigen | 34.0% (0.34) |
| Function uncertain, correlated todecreased survival | Uncertain function, but increasedexpression correlated with decreasedsurvival and/or more aggressive oradvanced disease | 25.0% (0.25) |
| Tissue differentiation, not oncogenic | Associated with tissue differentiation,but not “oncogenic” | 12.0% (0.12) |
| Tumor-related stroma | Expression on tumor-related stroma,but not on malignant cells | 12.0% (0.12) |
| Not applicable (N/A) | 0.0% (0.0) | |
| SPECIFICITY (Weight of Criteria - 0.15) | ||
| Absolute specificity | Absolutely specific, e.g., mutatedoncogene, idiotype protein or viralprotein | 100.0% (1.0) |
| Onco-fetal antigen | Antigens expressed in fetus with no orlittle expression in adult tissue.Includes cancer testis antigens | 54.0% (0.54) |
| Over expressed in cancer | Over expressed in cancer, butexpressed in some normal adulttissues | 35.0% (0.35) |
| Abnormal post-translationalmodification | Core protein expressed in normaltissue, but expressed in cancer withunique post-translational changes,e.g. glycosylation or phosphorylation | 23.0% (0.23) |
| Tissue specific (expendable tissue) | Tissue specific expression in normaladult tissue relatively expendable forsurvival, e.g., prostate andmelanocytes | 21.0% (0.21) |
| Unique random mutations | Unique random mutations specific toeach patient | 10.0% (0.1) |
| Tumor stroma antigen | Normal antigen expressed on tumorstroma | 10.0% (0.1) |
| Not applicable (N/A) | 0.0% (0.0) | |
| EXPRESSION LEVEL & % POSITIVE CELLS (Weight of Criteria - 0.07) | ||
| High level, All cancer cells | Highly expressed on all cancer cells inpatients designated for treatment | 100.0% (1.0) |
| High level, Most cancer cells | Highly expressed on most cancer cellsin patients designated for treatment | 37.0% (0.37) |
| Lower level, All cancer cells | Lower level of expression on all cancercells in patients designated fortreatment | 23.0% (0.23) |
| Lower level, Most cancer cells | Lower level of expression on mostcancer cells in patients designated fortreatment | 8.0% (0.08) |
| Not applicable (N/A) | 0.0% (0.0) | |
| STEM CELL EXPRESSION (Weight of Criteria - 0.05) | ||
| Stem cell expression, presumptive | Evidence for expression on putativecancer stem cells | 100.0% (1.0) |
| No info concerning stem cells - but onall stages from premalignant tometastatic | Present at all stages of tumordevelopment, from premalignant tometastatic cancer cells, but withoutinformation about putative stem cells | 66.0% (0.66) |
| No info concerning stem cells, but onmost cancer cells | Expression on the all or most cancercells, but without information aboutputative stem cells | 20.0% (0.2) |
| Not applicable (N/A) | 0.0% (0.0) | |
| NUMBER OF PATIENTS WITH ANTIGEN POSITIVE CANCERS (Weight of Criteria - 0.04) | ||
| Many patients, high level | High level of expression in manypatients with a particular tumor type | 100.0% (1.0) |
| Many patients, lower level | Low level of expression in manypatients with a particular tumor type | 16.0% (0.16) |
| All patients/ unique antigens | Unique antigens from randommutations presumed to be present inall patients | 14.0% (0.14) |
| Few patients, high level | High level of expression in a smallsubset of patients with a particulartumor type | 11.0% (0.11) |
| Not applicable (N/A) | 0.0% (0.0) | |
| NUMBER OF EPITOPES (Weight of Criteria - 0.04) | ||
| Longer antigen | Longer antigen with multiple epitopesand the potential to bind to mostMHC molecules | 100.0% (1.0) |
| Short antigenic segment | Short antigenic segment with one orfew epitopes and the potential to bindto only selected MHC molecules | 13.0% (0.13) |
| CELLULAR LOCATION OF EXPRESSION (Weight of Criteria - 0.02) | ||
| Cell surface expression; no or littlecirculating Ag | Normally expressed on the cell surfacewith no or little circulating antigen | 100.0% (1.0) |
| Internal with MHC presentation | Internal only with MHC presentation | 95.0% (0.95) |
| Cell surface expression; andcirculating Ag | Normally expressed on the cell surfacewith substantial circulating antigen | 25.0% (0.25) |
| Not applicable (N/A) | 0.0% (0.0) |
The cancer antigens to be prioritized were determined through a search of the PubMed database over the last 5 years using the terms “cancer vaccine target”. One hundred of the most frequently mentioned antigens were selected and submitted to the participating experts for categorization according to the pre-defined criteria and sub-criteria. Eighty investigators (listed in Supplement B) with expertise in one or several of the cancer antigens were asked to categorize the one or several antigens according to the criteria and sub-criteria. These experts were typically corresponding authors on published articles concerning the specific antigens. In certain cases, when necessary and where appropriate, experts not directly involved with the particular antigen were asked to categorize select antigens based on the pre-defined criteria. For some antigens, several experts were asked to categorize the antigen. A few experts did not respond and certain antigens were no longer under development. In the final analysis, 75 antigens were scored according to the pre-defined criteria.
Differences in scoring were debated and voted on at the face-to-face “assessment of alternatives” meeting described below. An example of the Antigen Information Form sent to the antigen experts is provided in Supplement D.
Phase II: Criteria refinement and weighting
The criteria and sub-criteria were used as the basis for discussion during a web-facilitated remote meeting using the Decision Lens model. They were discussed and definitions refined based on the combined expertise of the 19 expert participants (Supplement A). The criteria were then compared in a pair-wise fashion to determine the experts’ cumulative judgment’ of their relative importance to each other. The relative importance of each criterion to each of the other criterion was voted on by each expert and the relative importance of each was given a numerical rating on a scale from minus 9 to plus 9. The sub-criteria within each criterion were then compared in a similar pair-wise fashion by the same process.
Each expert participant had a single vote of equal weight. Participants who were unable to complete their pair-wise comparisons during the facilitated meeting were able to complete the process on line at a later date. Thirty-six pair-wise comparisons were employed to assess the relative priority of the 9 criteria. Similar pair-wise comparisons of sub-criterion within each criterion were determined to generate the relative weight of each sub-criterion to other sub-criterion. Sub-criteria were compared only to sub-criteria within their parent criteria. The cumulative results of the ratings of all of the experts were converted to a set of priority ratios for the criteria and sub-criteria. The results were nonlinear in their value differences.
Phase III: Assessment of alternatives
The weighted criteria and sub-criteria, which were used as rating scales, were used to assess the relative priority of each of the 75 cancer antigens at a face-to-face meeting of 16 participants that was hosted by the NCI (Supplement C). The information provided by up to 3 experts (Supplement B) per antigen on the Antigen Information Sheet (Supplement D) was entered in the Decision Lens software tool. The sub-criteria/rating scales were ordered from highest to lowest weight, but information on the relative weights of each criterion and sub-criterion was shared with participants only after the evaluation was completed. Each antigen was assigned to a meeting participant who acted as a reviewer and led the discussion of that antigen.
Each antigen was categorized according to the criteria and sub-criteria. If an antigen fulfilled more than one sub-criterion within a criteria, the sub-criteria with the highest value was selected. If a difference of opinion among participants was noted, it was discussed and then voted upon. Often consensus was not reached. When consensus was not reached the votes ended up with a value between the two sub-criteria. The value scores were calculated by taking the average of the ratings, then multiplying it by the weight of the criterion to cumulate to an aggregate score. The participants voted using a radio-frequency keypad and each vote had equal weight. Participants who were unable to complete antigen prioritization during the facilitated meeting were able to complete the process on line at a later date.
Results
Weighting of Criteria
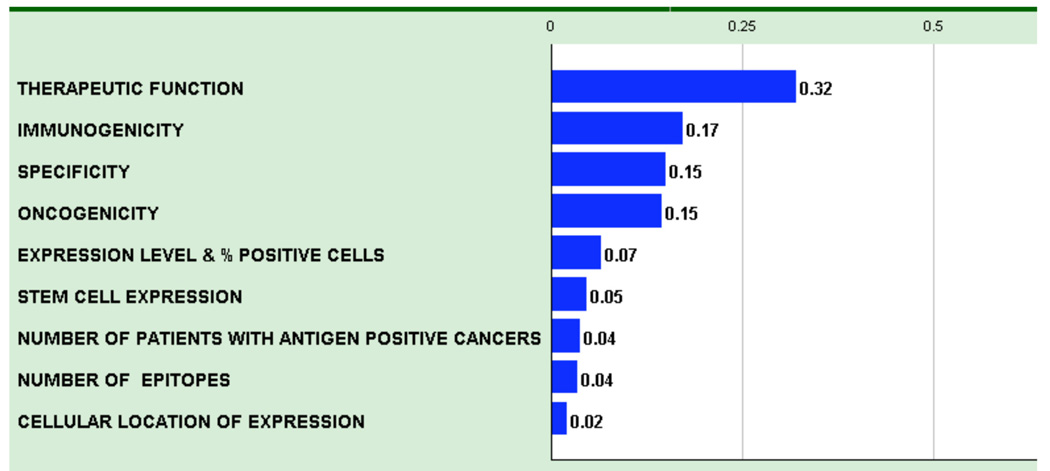
The AHP pair-wise comparison process resulted in a weighted model where the criteria relative weights reflect the derived priorities of the group of participants (Figure 1 and Table 1). The numerical values reflect the relative priorities of each criterion. As an example, pair-wise comparisons of criteria determined that “Therapeutic Function” represented 32% of the weight and “Immunogenicity” represented 17% of the weight, whereas “Cellular Location of Expression” represented only 2% of the weight. Thus, “Therapeutic Function” was deemed to be approximately twice as important as “Immunogenicity” and approximately 16-times more important than the “Cellular Location of Expression”.
Figure 1.
Criteria for an idea cancer antigen were weighted by pair-wise comparison and the resulting relative weights are indicated. Therapeutic function was considered the most important criteria, and was more than twice (0.32/0.15) as important as specificity or oncogenicity.
In some cases, there was considerable variation in response during the pair-wise comparison process. The participants were asked to explain their positions so that their implicit knowledge could become explicit and possibly result in readjustment of votes. However, the final weighting did not require and often did not achieve consensus.
Weighting of Sub-Criteria/rating scales
The sub-criteria were similarly weighted by pair-wise comparisons. Weighting is presented in Table 1. The sub-criteria, which served as the rating scales for each criterion, are also nonlinear. The top sub-criterion for each antigen received full value for the criterion. Other sub-criteria received less value for the criteria with the level dependent upon the pre-determined weighting. For example, for the criterion “Specificity”, an antigen deemed to have “Absolutely Specificity” received 100% of the value for that criterion, whereas an antigen that was “Over-expressed in Cancer” as the highest ranking within this category only received 35% of that value. The experts agreed that top sub-criterion for each criterion approximately portrayed an “Ideal Cancer Antigen” (Table 2).
TABLE 2.
Characteristics of an “ideal” cancer antigen
| CRITERIA | TOP SUB-CRITERIA |
|---|---|
| THERAPEUTIC FUNCTION | Superb Data Controlled vaccine trial suggestive |
| IMMUNOGENICITY | T cell and/or Ab responses elicited in clinicaltrials |
| ONCOGENICITY | Associated with oncogenic process, i.e.,oncogenic “self” protein |
| SPECIFICITY | Absolutely specific, e.g., mutated oncogene,idiotype protein or viral protein |
| EXPRESSION LEVEL & % POSITIVE CELLS | Highly expressed on all cancer cells in patientsdesignated for treatment |
| STEM CELL EXPRESSION | Evidence for expression on putative cancer stemcells |
| NUMBER OF PATIENTS WITH ANTIGEN POSITIVE CANCERS | High level of expression in many patients with aparticular tumor type |
| NUMBER OF EPITOPES | Longer antigen with multiple epitopes and thepotential to bind to most MHC molecules |
| CELLULAR LOCATION OF EXPRESSION | Normally expressed on the cell surface with noor little circulating antigen |
The criterion “Therapeutic Function” carried the most weight in the prioritization process. This category also generated substantial debate regarding the assessment of available information. The basis of the criterion was defined as “clinical trial data showing that a vaccine induced clinical responses in at least a small number of patients, or provided suggestive evidence of benefit vs controls”. The quality of published or publicly reported data was often disputed by the panel members. In anticipation of this discussion, the sub-category “Controlled vaccine trials suggestive” was subdivided before the meeting into the following sub-criteria:
- Superb data suggesting therapeutic benefit in a controlled vaccine trial,
- Very strong data suggesting therapeutic benefit in a controlled vaccine trial,
- Adequate data suggesting therapeutic benefit in a controlled vaccine trial,
- Fair data suggesting therapeutic benefit in a controlled vaccine trial.
Although subjective, these four sub-criteria parallel the evaluation process commonly used to assess NIH grant applications and emphasized the need for expert evaluation at all stages of the process. The other sub-criteria within the criterion of “Therapeutic Function” were - Responses in T cell therapy trial,
- Pre existent immunity/survival correlation, and
- Positive data in appropriate animal models.
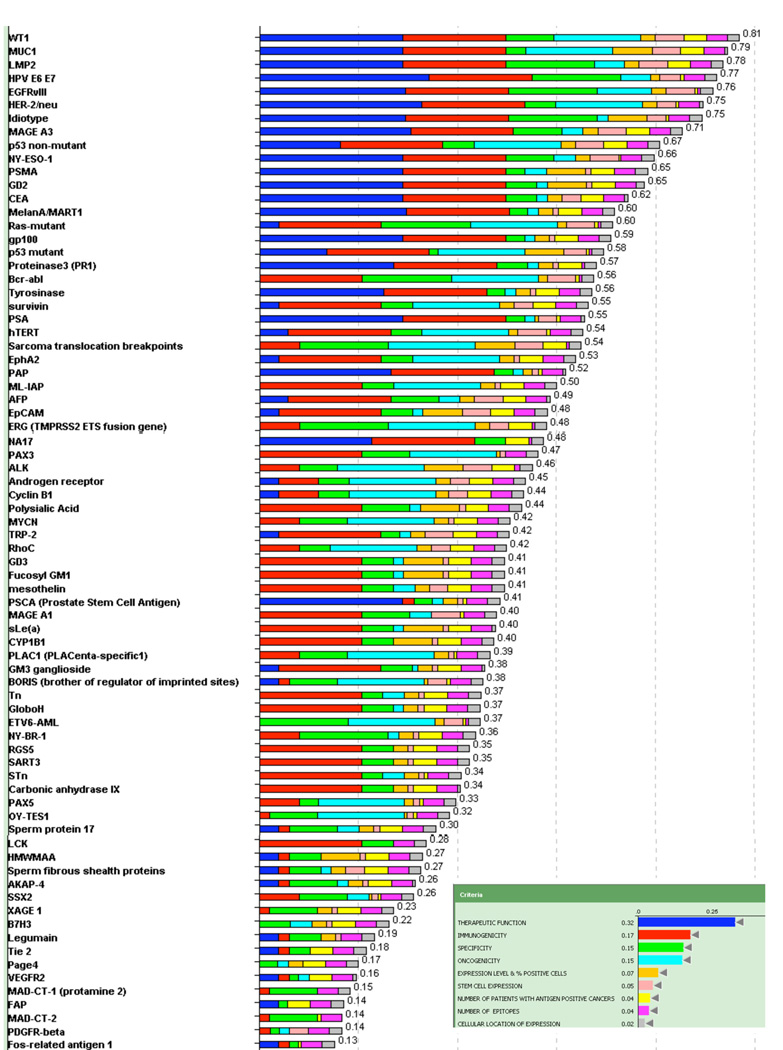
The results of the evaluation and weighting of the 75 cancer antigens are presented in Table 3. The results presented in Figure 2 show the cumulative score for each antigen. The color-coded bars indicate the relative contribution of each criterion.
TABLE 3.
Cancer antigen pilot prioritization: ranking based on predefined & preweighted criteria
| CRITERIA► | CUMMU-LATIVESCORE | THERAPEUTICFUNCTION(0.32) | IMMUNO-GENICITY(0.17) | ONCO-GENICITY(0.15) | SPECIFICITY(0.15) | EXPRES-SION LEVEL& %POSITIVECELLS(0.07) | STEM CELLEXPRES-SION(0.05) | NUMBEROFPATIENTSWITHANTIGENPOSITIVECANCERS(0.04) | NUMBEROFEPITOPES(0.04) | CELLULARLOCATIONOF EXPRES-SION(0.02) |
|---|---|---|---|---|---|---|---|---|---|---|
| ANTIGENS▼(Rank/Referencenumber &Name) | ||||||||||
| 1. WT1 | 0.81 | 0.75Fair data | 1.0Imm in trials | 1.0Oncogenic | 0.54Oncofetal | 0.37High most | 1.0Stem cells | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 2. MUC1 | 0.79 | 0.75Fair data | 1.0Imm in trials | 1.0Oncogenic | 0.23Post-translationalchanges | 1.0High all | 1.0Stem cells | 1.0Many patientshigh level | 1.0Multiple | 0.25SurfaceCirculating |
| 3. LMP2 | 0.78 | 0.75Fair data | 1.0Imm in trials | 0.34Persistent viral | 1.0Absolute | 0.37High most | 1.0Stem cells | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 4. HPV E6 E7 | 0.77 | 0.89Very Strong-8Adequate-6Fair-1 | 1.0Imm in trials | 0.34Persistent viral | 1.0Absolute | 0.23Low all | 0.73All stages-11Stem cells-3 | 0.16Many patientslower level | 1.0Multiple | 0.95Internal MHC |
| 5. EGFRvIII | 0.76 | 0.76Fair-12Adequate-2 | 1.0Imm in trials | 0.62Oncogenic-8Uncertain butdecreasedsurvival-7 | 1.0Absolute | 0.37High most | 1.0Stem cells | 0.11High level -Small subset | 0.13Single | 1Surface LittleCirculationMHC |
| 6. HER-2/neu | 0.75 | 0.85Adequate | 1.0Imm in trials | 1.0Oncogenic | 0.35Overexpressed | 0.37High most | 0.66All stages | 0.11High level -Small subset | 1.0Multiple | 0.25SurfaceCirculating |
| 7. Idiotype | 0.75 | 0.76Fair-13Adequate-2 | 1.0Imm in trials | 0.12Differentiation | 1.0Absolute | 1.0High all | 0.66All stages | 0.14 Unique | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 8. MAGE A3 | 0.71 | 0.79Fair-9Adequate-6 | 1.0Imm in trials | 0.25Uncertain butdecreasedsurvival | 0.54Oncofetal | 0.37High most | 1.0Stem cells | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 9. p53 non-mutant | 0.67 | 0.42Fair-9N/A-7 | 1.0Imm in trials | 1.0Oncogenic | 0.35Overexpressed | 0.37High most | 1.0Stem cells | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 10. NY-ESO-1 | 0.66 | 0.75Fair data | 1.0Imm in trials | 0.25Uncertain butdecreasedsurvival | 0.54Oncofetal | 0.37High most | 1.0Stem cells | 0.11.0High level -Small subset | 1.0Multiple | 0.95Internal MHC |
| 11. PSMA | 0.65 | 0.75Fair data | 1.0Imm in trials | 0.25Uncertain butdecreasedsurvival | 0.21Tissuespecific | 1.0High all | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 12. GD2 | 0.65 | 0.75Fair data | 1.0Imm in trials | 0.12Differentiation | 0.35Overexpressed | 1.0High all | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 0.62Surface withindeterminatecirculating |
| 13. CEA | 0.62 | 0.75Fair data | 1.0Imm in trials | 0.12Differentiation | 0.35Overexpressed | 0.37High most | 0.66All stages | 1.0Many patientshigh level | 1.0Multiple | 0.25SurfaceCirculating |
| 14.MelanA/MART1 | 0.60 | 0.77Fair-11Superb-1.0Adequate-1T cell Tx-1 | 1.0Imm in trials | 0.12Differentiation | 0.21Tissuespecific | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 15. Ras-mutant | 0.60 | 0.1Animal | 1.0Imm in trials | 1.0Oncogenic | 1.0Absolute | 0.23Low all | 1.0Stem cells | 0.16Many patientslower level | 0.13Single | 0.95Internal MHC |
| 16. gp100 | 0.59 | 0.75Fair data | 1.0Imm in trials | 0.12Differentiation | 0.21Tissuespecific | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 17. p53 mutant | 0.58 | 0.35N/A-8Fair-7 | 1.0Imm in trials | 1.0Oncogenic | 0.1Uniquerandommutations | 1.0High all | 0.77 Allstages-10Stem cells-5 | 0.14 Unique | 0.13Single | 0.95Internal MHC |
| 18. Proteinase3(PR1) | 0.57 | 0.7Fair-15N/A-1 | 1.0Imm in trials | 0.12Differentiation | 0.35Overexpressed | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 0.13Single | 0.95Internal MHC |
| 19. Bcr-abl | 0.56 | 0.00 | 1.0Imm in trials | 1.0Oncogenic | 1.0Absolute | 0.23Low all | 1.0Stem cells | 0.16Many patientslower level | 0.13Single | 0.95Internal MHC |
| 20. Tyrosinase | 0.56 | 0.65T cell Tx | 1.0Imm in trials | 0.12Differentiation | 0.21Tissuespecific | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 21. Survivin | 0.55 | 0.1Animal | 1.0Imm in trials | 1.0Oncogenic | 0.35Overexpressed | 0.37High most | 0.66All stages | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 22. PSA | 0.55 | 0.75Fair data | 1.0Imm in trials | 0.12Differentiation | 0.21Tissuespecific | 0.08Low most | 0.66All stages | 0.16Many patientslower level | 1.0Multiple | 0.25SurfaceCirculating |
| 23. hTERT | 0.54 | 0.15Preexistent | 1.0Imm in trials | 1.0Oncogenic | 0.35Overexpressed | 0.23Low all | 1.0Stem cells | 0.16Many patientslower level | 1.0Multiple | 0.95Internal MHC |
| 24. Sarcomatranslocationbreakpoints | 0.54 | 0.00 | 0.39T cellobserved | 1.0Oncogenic | 1.0Absolute | 1.0High all | 1.0Stem cells | 1.0Many patientshigh level | 0.13Single | 0.95Internal MHC |
| 25. EphA2 | 0.53 | 0.1Animal | 1.0Imm in trials | 1.0Oncogenic | 0.35Overexpressed | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 1.0Surface LittleCirculation MHC |
| 26. PAP | 0.52 | 0.69Fair-9Adequate-4Animal-2 | 1.0Imm in trials | 0.12Differentiation | 0.21Tissuespecific | 0.23Low all | 0.2Most cancercells | 0.16Many patientslower level | 1.0Multiple | 0.25SurfaceCirculating |
| 27. ML-IAP | 0.50 | 0.00 | 1.0Imm in trials | 1.0Oncogenic | 0.35Overexpressed | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 28. AFP | 0.49 | 0.15Preexistent | 1.0Imm in trials | 0.24Uncertain butdecreasedsurvival-11Differenetiation-1 | 0.54Oncofetal | 0.37High most | 1.0Stem cells | 1.0Many patientshigh level | 1.0Multiple | 0.25SurfaceCirculating |
| 29. EpCAM | 0.48 | 0.1Animal | 1.0Imm in trials | 0.12Differentiation | 0.35Overexpressed | 1.0High all | 1.0Stem cells | 1.0Many patientshigh level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 30. ERG(TMPRSS2ETS fusiongene) | 0.48 | 0.00 | 0.39T cellobserved | 1.0Oncogenic | 1.0Absolute | 0.37High most | 0.66All stages | 1.0Many patientshigh level | 0.13Single | 0.95Internal MHC |
| 31. NA17 | 0.48 | 0.59Fair-11N/A-3 | 1.0Imm in trials | 0.00 | 0.35Overexpressed | 0.00 | 0.00 | 1.0Many patientshigh level | 0.13Single | 0.95Internal MHC |
| 32. PAX3 | 0.47 | 0.00 | 1.0Imm in trials | 1.0Oncogenic | 0.54Oncofetal | 0.08Low most | 0.2Most cancercells | 0.00 | 1.0Multiple | 0.95Internal MHC |
| 33. ALK | 0.46 | 0.00 | 0.39T cellobserved | 1.0Oncogenic | 0.42Overexressed-6 Absolute-3N/A-3 | 1.0High all | 1.0Stem cells | 1.0Many patientshigh level | 0.27Single-8Multiple-2 | 0.95Internal MHC |
| 34. Androgenreceptor | 0.45 | 0.1Animal | 0.39T cellobserved | 1.0Oncogenic | 0.35Overexpressed | 0.37High most | 0.66All stages | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 35. Cyclin B1 | 0.44 | 0.1Animal | 0.39T cellobserved | 1.0Oncogenic | 0.35Overexpressed | 0.32High most-7Low all-4 | 0.66All stages | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 36. PolysialicAcid | 0.44 | 0.00 | 1.0Imm in trials | 0.12Differentiation | 0.54Oncofetal | 1.0High all | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 37. MYCN | 0.42 | 0.00 | 0.39T cellobserved | 1.0Oncogenic | 0.54Oncofetal | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 38. RhoC | 0.42 | 0.00 | 0.39T cellobserved | 1.0Oncogenic | 0.35Overexpressed | 0.37High most | 0.66All stages | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 39. TRP-2 | 0.42 | 0.1Animal | 1.0Imm in trials | 0.12Differentiation | 0.21Tissuespecific | 0.37High most | 1.0Stem cells | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 40. GD3 | 0.41 | 0.00 | 1.0Imm in trials | 0.12Differentiation | 0.35Overexpressed | 1.0High all | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 41. FucosylGM1 | 0.41 | 0.00 | 1.0Imm in trials | 0.12Differentiation | 0.35Overexpressed | 1.0High all | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 42. Mesothelin | 0.41 | 0.00 | 1.0Imm in trials | 0.25Uncertain butdecreasedsurvival | 0.35Overexpressed | 0.37High most | 0.66All stages | 1.0Many patientshigh level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 43. PSCA | 0.41 | 0.75Fair data | 0.11Animal | 0.12Differentiation | 0.21Tissuespecific | 0.37High most | 0.2Most cancercells | 0.16Many patientslower level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 44. MAGE A1 | 0.40 | 0.00 | 1.0Imm in trials | 0.25Uncertain butdecreasedsurvival | 0.54Oncofetal | 0.00 | 1.0Stem cells | 0.1.06Many patientslower level | 1.0Multiple | 0.95Internal MHC |
| 45. sLe(a) | 0.40 | 0.00 | 1.0Imm in trials | 0.12Differentiation | 0.35Overexpressed | 1.0High all | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 0.25SurfaceCirculating |
| 46. CYP1B1 | 0.40 | 0.00 | 1.0Imm in trials | 0.00 | 0.35Overexpressed | 1.0High all | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 47. PLAC1 | 0.39 | 0.00 | 0.39T cellobserved | 1.0Oncogenic | 0.54Oncofetal | 0.37High most | 0.2Most cancercells | 0.11High level - Small subset | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 48. GM3 | 0.38 | 0.1Animal | 1.0Imm in trials | 0.12Stroma | 0.35Overexpressed | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 0.25SurfaceCirculating |
| 49. BORIS | 0.38 | 0.1Animal | 0.11Animal | 0.1Oncogenic | 0.54Oncofetal | 0.08Low most | 0.66All stages | 0.16Many patientslower level | 1.0Multiple | 0.95Internal MHC |
| 50. Tn | 0.37 | 0.00 | 1.0Imm in trials | 0.25Uncertain butdecreasedsurvival | 0.23Post-translationalchanges | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 51. GloboH | 0.37 | 0.00 | 1.0Imm in trials | 0.12Differentiation | 0.35Overexpressed | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 52. ETV6-AML | 0.37 | 0.00 | 0.00 | 1.0Oncogenic | 1.0Absolute | 0.23Low all | 0.66All stages | 0.11High level -Small subset | 0.13Single | 0.95Internal MHC |
| 53. NY-BR-1 | 0.36 | 0.00 | 0.39T cellobserved | 0.12Differentiation | 1.0Absolute | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 54. RGS5 | 0.35 | 0.00 | 1.0Imm in trials | 0.00 | 0.35Overexpressed | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 55. SART3 | 0.35 | 0.00 | 1.0Imm in trials | 0.00 | 0.35Overexpressed | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 56. STn | 0.34 | 0.00 | 1.0Imm in trials | 0.25Uncertain butdecreasedsurvival | 0.23Post-translationalchanges | 0.37High most | 0.2Most cancercells | 0.16Many patientslower level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 57. Carbonicanhydrase IX | 0.34 | 0.00 | 1.0Imm in trials | 0.00 | 0.35Over expressed | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 0.25SurfaceCirculating |
| 58. PAX5 | 0.33 | 0.00 | 0.39T cellobserved | 1.0Oncogenic | 0.21Tissuespecific | 0.23Low all | 0.2Most cancercells | 0.16Many patientslower level | 1.0Multiple | 0.95Internal MHC |
| 59. OY-TES1 | 0.32 | 0.00 | 0.1Ab observed | 1.0Oncogenic | 0.54Oncofetal | 0.08Low most | 0.2Most cancercells | 0.16Many patientslower level | 1.0Multiple | 0.95Internal MHC |
| 60. Spermprotein 17 | 0.30 | 0.1Animal | 0.11Animal | 0.25Uncertain butdecreasedsurvival | 0.54Oncofetal | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 61. LCK | 0.28 | 0.00 | 1.0Imm in trials | 0.00 | 0.35Overexpressed | 0.00 | 0.00 | 0.00 | 1.0Multiple | 0.95Internal MHC |
| 62. HMWMAA | 0.27 | 0.1Animal | 0.11Animal | 0.00 | 0.35Overexpressed | 1.0High all | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 63. AKAP-4 | 0.26 | 0.1Animal | 0.11Animal | 0.12Differentiation | 0.54Oncofetal | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 0.25SurfaceCirculating |
| 64. SSX2 | 0.26 | 0.00 | 0.39T cellobserved | 0.25Uncertain butdecreasedsurvival | 0.54Oncofetal | 0.08Low most | 0.2Most cancercells | 0.11High level -Small subset | 1.0Multiple | 0.95Internal MHC |
| 65. XAGE 1 | 0.23 | 0.00 | 0.1Ab observed | 0.00 | 0.54Oncofetal | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 66. B7H3 | 0.22 | 0.00 | 0.00 | 0.25Uncertain butdecreasedsurvival | 0.35Overexpressed | 0.37High most | 0.2Most cancercells | 1.0Many patientshigh level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 67. Legumain | 0.19 | 0.1Animal | 0.11Animal | 0.00 | 0.35Overexpressed | 0.37High most | 0.2Most cancercells | 0.00 | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 68. Tie 2 | 0.18 | 0.1Animal | 0.11Animal | 0.00 | 0.23Post-translationalchanges | 0.00 | 0.00 | 1.0Many patientshigh level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 69. Page4 | 0.17 | 0.00 | 0.00 | 0.12Differentiation | 0.21Tissuespecific | 0.37High most | 0.00 | 1.0Many patientshigh level | 1.0Multiple | 0.95Internal MHC |
| 70. VEGFR2 | 0.16 | 0.1Animal | 0.11Animal | 0.12Stroma | 0.1Stromal | 0.00 | 0.00 | 1.0Many patientshigh level | 1.0Multiple | 0.25SurfaceCirculating |
| 71. MAD-CT-1 | 0.15 | 0.00 | 0.1Ab observed | 0.00 | 0.54Oncofetal | 0.00 | 0.00 | 0.00 | 1.0Multiple | 0.95Internal MHC |
| 72. FAP | 0.14 | 0.1Animal | 0.00 | 0.00 | 0.1Stromal | 0.00 | 0.00 | 1.0Many patientshigh level | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 73. PDGFR-beta | 0.14 | 0.00 | 0.11Animal | 0.12Stroma | 0.1Stromal | 0.00 | 0.66All stages | 0.00 | 1.0Multiple | 1.0Surface LittleCirculationMHC |
| 74. MAD-CT-2 | 0.14 | 0.00 | 0.1Ab observed | 0.00 | 0.54Oncofetal | 0.00 | 0.00 | 0.16Many patientslower level | 1.0Multiple | 0.00 |
| 75. Fos-relatedantigen 1 | 0.13 | 0.1Animal | 0.11Animal | 0.00 | 0.1Stromal | 0.00 | 0.00 | 0.11High level -Small subset | 1.0Multiple | 1.0Surface LittleCirculationMHC |
Figure 2.
Cancer antigen pilot prioritization: representation of ranking based on predefined and preweighted criteria and sub-criteria. Inset indicates the color used to designate each criterion and its relative weight. The number at the end of each bar indicates the relative rank of that antigen.
No antigen exhibited all of the top sub-criteria (Table 2). By this assessment, no antigen, among those selected, satisfied the criteria for an “Ideal” cancer antigen. The dominant criterion was “Therapeutic Function” and the top 14 antigens all have significant contributions from that criterion, namely “Fair” to “Very Strong Data Controlled vaccine trial”. Altogether, 20 antigens were deemed to have at least “Fair Data Controlled vaccine trial suggestive”. None were deemed to have “Superb Data” by any of the experts.
The second dominant criterion was “Immunogenicity”. In all, 46 of the 75 antigens – including the top 14 – had documented immunogenicity in human clinical trials. The total weight of “Therapeutic Function” plus “Immunogenicity” was 0.49. The dominance of “Therapeutic Function” and “Immunogenicity” biased the ratings towards antigens already in analyzable clinical trials, i.e., antigens further along in the developmental process.
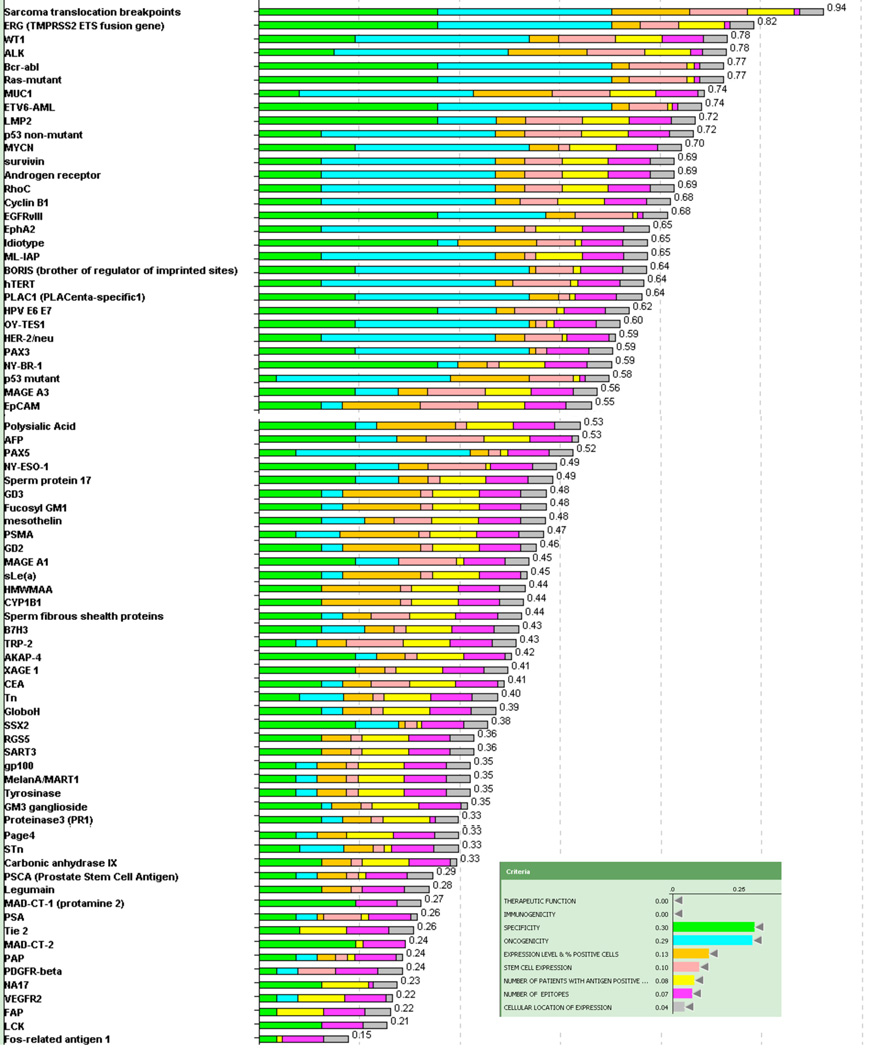
To assess priorities without bias toward already having been in clinical trials, the antigens were re-ranked, excluding “Therapeutic Function” and “Immunogenicity” (Figure 3). After excluding these top 2 criteria, the antigen ranking was dominated by the criteria of “Oncogenicity”, “Specificity” and “Stem Cell Expression”. In this alternative model the break point region of translocated fusion genes (Ewing’s sarcoma and alveolar rhabdomyosarcoma; ALK, bcr-abl and ETV6-AML) and mutant oncogenes (ras) rose to the top. The method of reporting data in Table 3 allows re-prioritization of the antigens and development of alternative rankings based on alternative assessment or weighting of criteria and sub-criteria of interest.
Figure 3.
Representation of ranking following exclusion of “therapeutic efficacy” and “immunogenicity”. Inset indicates the color used to designate each criterion and its relative weight. The number at the end of each bar indicates the relative rank of that antigen.
Discussion
This study developed a well vetted, priority-ranked list of cancer vaccine target antigens based on pre-defined and pre-weighted objective criteria developed by a panel of content experts. The AHP method and Decision Lens platform provided the framework to catalogue and weight vaccine development decision criteria, and to rank 75 selected antigens. This process was done in three stages by three panels of cancer vaccine experts with overlapping members. The first panel defined the criteria to be ranked for priority. The second panel weighted the criteria. The third panel ranked the 75 antigens according to the pre-defined pre-weighted criteria based on information on the antigens provided by researchers familiar with the individual antigens. The broad nature of the input underpinning the list of criteria should facilitate subsequent NCI or other funding agency discussions as to which antigens to test in subsequent focused translational/clinical studies.
This study is termed a “Pilot Prioritization Project” with the emphasis on “Pilot”. One of the goals was to determine whether the methods employed could be used to rank priorities for subsequent efforts to accelerate translational research. The finding that 20 of the 75 evaluated antigens had some clinical efficacy and that 46 of them had validated immunogenicity in human clinical trials documents the extent and vigor of the cancer vaccine field, but also accentuates the need for prioritization. Notably, there are cancer antigens under development that were not included in this prioritization effort, again, accentuating the breadth of the opportunities in the field.
Of the 46 antigens with validated immunogenicity and 20 with suggestive clinical data, none are FDA-approved for general use. It is generally assumed that development of any of the top antigens will require concerted collaboration on the part of experts in cancer vaccine development. We anticipate that the prioritization of Immunotherapy Agents with High Potential for Cancer Therapy 5 and the current ranking of cancer antigens will jointly lay the foundation for such focused collaborations.
The final scoring and ranking was necessarily done with incomplete knowledge. Much more was known about some antigens than others. Antigens that had undergone the most prior research had a marked advantage in the ranking. The experts ranked “Therapeutic Efficacy” and “Immunogenicity” as the top criteria. Within these categories there were many types of trials with different endpoints and different patient selection criteria. Thus, it was necessary to further divide the criteria according to the level of the data. The sub-categorization was not precise, and was subjective along the scale of Fair to Superb Data. The experts had varying opinions about the quality of the data and the panel had no opportunity to examine raw data from any trials. An in-depth analysis of primary clinical data for the antigens would be required to substantiate the results before any definitive action could be taken. Furthermore, the ranking at best represents the current state of our knowledge and will change as new information becomes available.
The order changed appreciably when re-analyzed without the top criteria, “Therapeutic Efficacy” and “Immunogenicity”. The leading criteria then became “Oncogenicity”, “Specificity” and “Stem Cell Expression”, and the priorities of the break point region of translocated fusion genes (Ewing’s sarcoma and alveolar rhabdomyosarcoma; ALK, bcr-abl and ETV6-AML) and mutant oncogenes (ras) rose to the top. Arguably, it will be harder for them to achieve therapeutic efficacy as these antigens require selective MHC presentation of a small and single epitope. Thus, there may be some underlying biologic justification for their lower ranking.
Knowledge within other categories was often also incomplete or inadequate. For example:
- “Stem cell expression” was deemed to be important, but the group recognized that the field of stem cell identification is rapidly evolving. Future thoughts and assessments about cancer stem cells could be markedly different.
- The criterion of “Oncogenicity” was important. However, many antigens not considered to be “oncogenic” are associated with a poor prognosis and are clearly involved in helping to sustain the malignant phenotype. Thus, the definition of “oncogenic” may be too restrictive. The outcome of immunologic pressure is often the evolution of antigen-negative variants. It would seem beneficial to target antigens, which if lost, resulted in diminished ability of the cancer cells to survive or thrive. Necessity for maintaining a malignant phenotype is a broader definition than “oncogenic” per se and might be more relevant.
- It was felt by the experts that antigens with “no or little circulating antigen” were substantially preferable to antigens with “circulating antigen”. However, the group did not have access to actual side-by-side data quantifying circulating antigen and did not define a threshold value discriminating between the two. Moreover, in certain cases the amount of circulating antigen was not well characterized in the literature.
No antigen exhibited all of the top sub-criteria. By this assessment, no antigen, among those selected, satisfied the criteria for an “ideal” cancer antigen. Some of the deficiencies such as stem cell expression are biologic and can’t be changed. Others such as immunogenicity and level of therapeutic efficacy can potentially be changed with additional experiments and more data and, most compellingly, by the use of more effective vaccine formulations and schedules of administration. For antigens too early in development to have garnered evidence of clinical efficacy or immunogenicity, the dominance of those criteria in the experts’ ratings provides a road-map for investigators by emphasizing that high quality data concerning these criteria are critical for prioritization of antigens for focused subsequent development.
Another question is whether there is “Ideal” cancer antigens left to be discovered. It can be assumed that the first antigens discovered would be amongst the most abundant and the most immunogenic. Abundance and immunogenicity are both major criteria. By extrapolation, it can be argued that many of the antigens left to be discovered would be less abundant and less immunogenic molecules.
Of the 75 antigens evaluated 46 were immunogenic in clinical trials and 20 of them had suggestive clinical efficacy in the “Therapeutic function” category with documented vaccine induced clinical responses in at least a small number of patients, or suggestive evidence of benefit vs. controls. However, none were deemed to have “superb data” in the category of “Therapeutic function”. The lack of “superb data” could be multifactorial, including inadequate trial design or patient selection, and inadequate vaccine formulation or regimens. These deficiencies can be overcome by more intelligent trial design based on assessment of past “productive failures”.
Two profound biologic issues limiting the efficacy of cancer vaccines are the strength of immunological tolerance and the intrinsic limitations on the ability of T cells to expand in number in response to antigenic stimulation. There are normally exceedingly strict biologic limits imporsed on the immune system to prevent excessive T-cell activation and expansion. The same biological restrictions limit cancar vaccines. Immunotherapeutic agents that can circumvent many of the biological restrictions have been invented and formulated and proven to be biologically active, including dendritic cell activators and growth factors, vaccine adjuvants, T-cell stimulators and growth factors, genetically-modified T cells, immune checkpoint inhibitors, and agents to neutralize or inhibit suppressive cells, cytokines, and enzymes. Unfortunately, few of these agents are broadly available for the development of effective multiple component cancer vaccine regimens. The tools needed to raise T-cell levels to extraordinary levels in vivo and to maintain T cell number for prolonged periods of time are at hand. A major problem facing immunotherapy today is a lack of broad availability of agents already in existence that could be effective in multiple component regimens as well as administrative difficulties funding and carrying out such multiple component regimens. It is highly likely that therapeutic regimens composed of optimal vaccine formulations with combinations of already invented immunotherapy agents in the above categories would lift the level of data into the “superb data” subcategory for many of the 20 antigens as well as others less studied. The current prioritization process, by validating that at least 20 antigens have “suggestive clinical efficacy”, highlights the need for an administrative and funding structure capable of translating these scientific discoveries into effective cancer therapies.
The AHP approach has several advantages over more standard evaluation and prioritization approaches. The AHP framework requires detailed discussion of the specific criteria in advance of the prioritization, permitting a comparison of individual perceptions and forcing the group to reach consensus on interpretations and definitions. This is presumed to improve the consistency of responses and has the effect of generating confidence in the results and “buy-in” among stakeholders. AHP allows the information to be evaluated quantitatively and qualitatively, using both subjective and objective ranking scales. The ability to apply non-linear weights to criteria and ranking scales was viewed as a distinct advantage over a system that simply averages the results. The Decision Lens platform provided an organized and consistent way to organize and view data, thereby facilitating evaluation. The transparency of the process was a benefit in that disagreements were quickly recognized and could be discussed. Finally, the web-based asynchronous approach was viewed as an efficient use of experts’ time.
The flexibility of the AHP/Decision Lens approach in permitting “what if” scenarios was exceptionally valuable in understanding how changing the weight of the criteria and sub-criteria would affect the outcome and helped to provide a comfort level with the generated priority list. The approach accommodates viewing the data with selected criteria given any proportion of the weighing, including zero. The flexibility of the system has the advantage of simplifying re-evaluation of alternatives when additional information becomes available, and allows for modification of criteria as more experience with generating cancer vaccines is gained. As one example, the flexibility will allow for alternative assessments of prioritization for the same antigen in different tumor types in circumstances where the antigen has markedly different expression patterns.
It must be noted that the AHP does not make decisions; rather it provides a way to analyze and prioritize alternatives. One of the limitations of AHP is that it only ranks degrees of positivity. In some cases there can be “deal breaking” negative information that needs to be assessed outside of the AHP. A list of ranked alternatives provides a rational basis for decisions at the executive level. This pilot prioritization study produced a ranked list of cancer antigens that can be used by the broad immunotherapy community when considering further investment in experimental research for individual antigens as they move toward the goal of translating the most promising cancer antigens into vaccines for cancer treatment or prevention.
Statement of Translational Relevance
We report on the development of a prioritized list of cancer vaccine target antigens using well-vetted criteria generated by expert panels. The elucidation and weighting of criteria to assess cancer antigens will assist investigators in the immunotherapy field in determining the characteristics and the experimental data required to select the most promising antigens for further development and testing in clinical trials.
Supplementary Material
1
Acknowledgments
We would like to thank Jennifer W. Kwok and Abdul Tawab-Amiri (Coordinating Center for Clinical Trials, Office of the Director, NCI, NIH) for project management and helpful suggestions, and Lada Krilov (NOVA Research Company) for transcription assistance.
The corresponding author was supported in part by grants from the NIH, NCI to M.A.C. UL1 RR 025014-01(P.I.: Disis, M.) and P30 CA 15704-35 (P.I.: Hartwell, L.).
Footnotes
References
- 1.Oka Y, Tsuboi A, Oji Y, Kawase I, Sugiyama H. WT1 peptide vaccine for the treatment of cancer. Curr Opin Immunol. 2008;20:211–220. doi: 10.1016/j.coi.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Lepisto AJ, Moser AJ, Zeh H, et al. A phase I/II study of a MUC1 peptide pulsed autologous dendritic cell vaccine as adjuvant therapy in patients with resected pancreatic and biliary tumors. Cancer Ther. 2008;6:955–964. [PMC free article] [PubMed] [Google Scholar]
- 3.Khanna R, Moss D, Gandhi M. Technology insight: Applications of emerging immunotherapeutic strategies for Epstein-Barr virus-associated malignancies. Nat Clin Pract Oncol. 2005;2:138–149. doi: 10.1038/ncponc0107. [DOI] [PubMed] [Google Scholar]
- 4.Trimble CL, Peng S, Kos F, et al. A phase I trial of a human papillomavirus DNA vaccine for HPV16+ cervical intraepithelial neoplasia 2/3. Clin Cancer Res. 2009;15:361–367. doi: 10.1158/1078-0432.CCR-08-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sampson JH, Archer GE, Mitchell DA, Heimberger AB, Bigner DD. Tumor-specific immunotherapy targeting the EGFRvIII mutation in patients with malignant glioma. Semin Immunol. 2008;20:267–275. doi: 10.1016/j.smim.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weng WK, Czerwinski D, Timmerman J, Hsu FJ, Levy R. Clinical outcome of lymphoma patients after idiotype vaccination is correlated with humoral immune response and immunoglobulin G Fc receptor genotype. J Clin Oncol. 2004;22:4717–4724. doi: 10.1200/JCO.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Mittendorf EA, Holmes JP, Ponniah S, Peoples GE. The E75 HER2/neu peptide vaccine. Cancer Immunol Immunother. 2008;57:1511–1521. doi: 10.1007/s00262-008-0540-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brichard VG, Lejeune D. GSK's antigen-specific cancer immunotherapy programme: pilot results leading to Phase III clinical development. Vaccine. 2007;25(Suppl 2):B61–B71. doi: 10.1016/j.vaccine.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 9.Antonia SJ, Mirza N, Fricke I, et al. Combination of p53 cancer vaccine with chemotherapy in patients with extensive stage small cell lung cancer. Clin Cancer Res. 2006;12:878–887. doi: 10.1158/1078-0432.CCR-05-2013. [DOI] [PubMed] [Google Scholar]
- 10.Old LJ. Cancer vaccines: an overview. Cancer Immun. 2008;8(Suppl 1):1. [PubMed] [Google Scholar]
- 11.Olson WC, Heston WD, Rajasekaran AK. Clinical trials of cancer therapies targeting prostate-specific membrane antigen. Rev Recent Clin Trials. 2007;2:182–190. doi: 10.2174/157488707781662724. [DOI] [PubMed] [Google Scholar]
- 12.Wondimu A, Zhang T, Kieber-Emmons T, et al. Peptides mimicking GD2 ganglioside elicit cellular, humoral and tumor-protective immune responses in mice. Cancer Immunol Immunother. 2008;57:1079–1089. doi: 10.1007/s00262-007-0439-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gulley JL, Arlen PM, Tsang KY, et al. Pilot study of vaccination with recombinant CEA-MUC-1-TRICOM poxviral-based vaccines in patients with metastatic carcinoma. Clin Cancer Res. 2008;14:3060–3069. doi: 10.1158/1078-0432.CCR-08-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yee C, Thompson JA, Byrd D, et al. Adoptive T cell therapy using antigen-specific CD8+ T cell clones for the treatment of patients with metastatic melanoma: in vivo persistence, migration, and antitumor effect of transferred T cells. Proc Natl Acad Sci U S A. 2002;99:16168–16173. doi: 10.1073/pnas.242600099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toubaji A, Achtar M, Provenzano M, et al. Pilot study of mutant ras peptide-based vaccine as an adjuvant treatment in pancreatic and colorectal cancers. Cancer Immunol Immunother. 2008;57:1413–1420. doi: 10.1007/s00262-008-0477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith FO, Downey SG, Klapper JA, et al. Treatment of metastatic melanoma using interleukin-2 alone or in conjunction with vaccines. Clin Cancer Res. 2008;14:5610–5618. doi: 10.1158/1078-0432.CCR-08-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carbone DP, Ciernik IF, Kelley MJ, et al. Immunization with mutant p53- and K-ras-derived peptides in cancer patients: immune response and clinical outcome. J Clin Oncol. 2005;23:5099–5107. doi: 10.1200/JCO.2005.03.158. [DOI] [PubMed] [Google Scholar]
- 18.Rezvani K. PR1 vaccination in myeloid malignancies. Expert Rev Vaccines. 2008;7:867–875. doi: 10.1586/14760584.7.7.867. [DOI] [PubMed] [Google Scholar]
- 19.Bergman PJ, McKnight J, Novosad A, et al. Long-term survival of dogs with advanced malignant melanoma after DNA vaccination with xenogeneic human tyrosinase: a phase I trial. Clin Cancer Res. 2003;9:1284–1290. [PubMed] [Google Scholar]
- 20.Maslak PG, Dao T, Gomez M, et al. A pilot vaccination trial of synthetic analog peptides derived from the BCR-ABL breakpoints in CML patients with minimal disease. Leukemia. 2008;22:1613–1616. doi: 10.1038/leu.2008.7. [DOI] [PubMed] [Google Scholar]
- 21.Xiang R, Mizutani N, Luo Y, et al. A DNA vaccine targeting survivin combines apoptosis with suppression of angiogenesis in lung tumor eradication. Cancer Res. 2005;65:553–561. [PubMed] [Google Scholar]
- 22.Madan RA, Gulley JL, Schlom J, et al. Analysis of overall survival in patients with nonmetastatic castration-resistant prostate cancer treated with vaccine, nilutamide, and combination therapy. Clin Cancer Res. 2008;14:4526–4531. doi: 10.1158/1078-0432.CCR-07-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domchek SM, Recio A, Mick R, et al. Telomerase-specific T-cell immunity in breast cancer: effect of vaccination on tumor immunosurveillance. Cancer Res. 2007;67:10546–10555. doi: 10.1158/0008-5472.CAN-07-2765. [DOI] [PubMed] [Google Scholar]
- 24.Mackall CL, Rhee EH, Read EJ, et al. A pilot study of consolidative immunotherapy in patients with high-risk pediatric sarcomas. Clin Cancer Res. 2008;14:4850–4858. doi: 10.1158/1078-0432.CCR-07-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaguchi S, Tatsumi T, Takehara T, et al. Immunotherapy of murine colon cancer using receptor tyrosine kinase EphA2-derived peptide-pulsed dendritic cell vaccines. Cancer. 2007;110:1469–1477. doi: 10.1002/cncr.22958. [DOI] [PubMed] [Google Scholar]
- 26.Small EJ, Schellhammer PF, Higano CS, et al. Placebo-controlled phase III trial of immunologic therapy with sipuleucel-T (APC8015) in patients with metastatic, asymptomatic hormone refractory prostate cancer. J Clin Oncol. 2006;24:3089–3094. doi: 10.1200/JCO.2005.04.5252. [DOI] [PubMed] [Google Scholar]
- 27.Schmollinger JC, Vonderheide RH, Hoar KM, et al. Melanoma inhibitor of apoptosis protein (ML-IAP) is a target for immune-mediated tumor destruction. Proc Natl Acad Sci U S A. 2003;100:3398–3403. doi: 10.1073/pnas.0530311100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butterfield LH, Ribas A, Dissette VB, et al. A phase I/II trial testing immunization of hepatocellular carcinoma patients with dendritic cells pulsed with four alpha-fetoprotein peptides. Clin Cancer Res. 2006;12:2817–2825. doi: 10.1158/1078-0432.CCR-05-2856. [DOI] [PubMed] [Google Scholar]
- 29.Birebent B, Mitchell E, Akis N, et al. Monoclonal anti-idiotypic antibody mimicking the gastrointestinal carcinoma-associated epitope CO17-1A elicits antigen-specific humoral and cellular immune responses in colorectal cancer patients. Vaccine. 2003;21:1601–1612. doi: 10.1016/s0264-410x(02)00752-1. [DOI] [PubMed] [Google Scholar]
- 30.Tomlins SA, Laxman B, Varambally S, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trakatelli M, Toungouz M, Blocklet D, et al. A new dendritic cell vaccine generated with interleukin-3 and interferon-beta induces CD8+ T cell responses against NA17-A2 tumor peptide in melanoma patients. Cancer Immunol Immunother. 2006;55:469–474. doi: 10.1007/s00262-005-0056-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Himoudi N, Nabarro S, Yan M, Gilmour K, Thrasher AJ, Anderson J. Development of anti-PAX3 immune responses; a target for cancer immunotherapy. Cancer Immunol Immunother. 2007;56:1381–1395. doi: 10.1007/s00262-007-0294-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Passoni L, Gallo B, Biganzoli E, et al. In vivo T-cell immune response against anaplastic lymphoma kinase in patients with anaplastic large cell lymphomas. Haematologica. 2006;91:48–55. [PubMed] [Google Scholar]
- 34.Olson BM, McNeel DG. Antibody and T-cell responses specific for the androgen receptor in patients with prostate cancer. Prostate. 2007;67:1729–1739. doi: 10.1002/pros.20652. [DOI] [PubMed] [Google Scholar]
- 35.Krug LM, Ragupathi G, Ng KK, et al. Vaccination of small cell lung cancer patients with polysialic acid or N-propionylated polysialic acid conjugated to keyhole limpet hemocyanin. Clin Cancer Res. 2004;10:916–923. doi: 10.1158/1078-0432.ccr-03-0101. [DOI] [PubMed] [Google Scholar]
- 36.Kao H, Marto JA, Hoffmann TK, et al. Identification of cyclin B1 as a shared human epithelial tumor-associated antigen recognized by T cells. J Exp Med. 2001;194:1313–1323. doi: 10.1084/jem.194.9.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolchok JD, Yuan J, Houghton AN, et al. Safety and immunogenicity of tyrosinase DNA vaccines in patients with melanoma. Mol Ther. 2007;15:2044–2050. doi: 10.1038/sj.mt.6300290. [DOI] [PubMed] [Google Scholar]
- 38.Wenandy L, Sorensen RB, Svane IM, Thor Straten P, Andersen MH. RhoC a new target for therapeutic vaccination against metastatic cancer. Cancer Immunol Immunother. 2008;57:1871–1878. doi: 10.1007/s00262-008-0517-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Himoudi N, Yan M, Papanastasiou A, Anderson J. MYCN as a target for cancer immunotherapy. Cancer Immunol Immunother. 2008;57:693–700. doi: 10.1007/s00262-007-0409-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ragupathi G, Meyers M, Adluri S, Howard L, Musselli C, Livingston PO. Induction of antibodies against GD3 ganglioside in melanoma patients by vaccination with GD3-lactone-KLH conjugate plus immunological adjuvant QS-21. Int J Cancer. 2000;85:659–666. doi: 10.1002/(sici)1097-0215(20000301)85:5<659::aid-ijc11>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 41.Dickler MN, Ragupathi G, Liu NX, et al. Immunogenicity of a fucosyl-GM1-keyhole limpet hemocyanin conjugate vaccine in patients with small cell lung cancer. Clin Cancer Res. 1999;5:2773–2779. [PubMed] [Google Scholar]
- 42.Laheru D, Lutz E, Burke J, et al. Allogeneic granulocyte macrophage colony-stimulating factor-secreting tumor immunotherapy alone or in sequence with cyclophosphamide for metastatic pancreatic cancer: a pilot study of safety, feasibility, and immune activation. Clin Cancer Res. 2008;14:1455–1463. doi: 10.1158/1078-0432.CCR-07-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Hernandez Mde L, Gray A, Hubby B, Klinger OJ, Kast WM. Prostate stem cell antigen vaccination induces a long-term protective immune response against prostate cancer in the absence of autoimmunity. Cancer Res. 2008;68:861–869. doi: 10.1158/0008-5472.CAN-07-0445. [DOI] [PubMed] [Google Scholar]
- 44.Gribben JG, Ryan DP, Boyajian R, et al. Unexpected association between induction of immunity to the universal tumor antigen CYP1B1 and response to next therapy. Clin Cancer Res. 2005;11:4430–4436. doi: 10.1158/1078-0432.CCR-04-2111. [DOI] [PubMed] [Google Scholar]
- 45.van Baren N, Bonnet MC, Dreno B, et al. Tumoral and immunologic response after vaccination of melanoma patients with an ALVAC virus encoding MAGE antigens recognized by T cells. J Clin Oncol. 2005;23:9008–9021. doi: 10.1200/JCO.2005.08.375. [DOI] [PubMed] [Google Scholar]
- 46.Livingston PO, Hood C, Krug LM, et al. Selection of GM2, fucosyl GM1, globo H and polysialic acid as targets on small cell lung cancers for antibody mediated immunotherapy. Cancer Immunol Immunother. 2005;54:1018–1025. doi: 10.1007/s00262-005-0663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silva WA, Jr., Gnjatic S, Ritter E, et al. PLAC1, a trophoblast-specific cell surface protein, is expressed in a range of human tumors and elicits spontaneous antibody responses. Cancer Immun. 2007;7:18. [PMC free article] [PubMed] [Google Scholar]
- 48.Mkrtichyan M, Ghochikyan A, Loukinov D, et al. DNA, but not protein vaccine based on mutated BORIS antigen significantly inhibits tumor growth and prolongs the survival of mice. Gene Ther. 2008;15:61–64. doi: 10.1038/sj.gt.3303044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazorra Z, Mesa C, Fernandez A, Fernandez LE. Immunization with a GM3 ganglioside nanoparticulated vaccine confers an effector CD8(+) T cells-mediated protection against melanoma B16 challenge. Cancer Immunol Immunother. 2008;57:1771–1780. doi: 10.1007/s00262-008-0503-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilewski T, Ragupathi G, Bhuta S, et al. Immunization of metastatic breast cancer patients with a fully synthetic globo H conjugate: a phase I trial. Proc Natl Acad Sci U S A. 2001;98:3270–3275. doi: 10.1073/pnas.051626298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sabbatini PJ, Ragupathi G, Hood C, et al. Pilot study of a heptavalent vaccine-keyhole limpet hemocyanin conjugate plus QS21 in patients with epithelial ovarian, fallopian tube, or peritoneal cancer. Clin Cancer Res. 2007;13:4170–4177. doi: 10.1158/1078-0432.CCR-06-2949. [DOI] [PubMed] [Google Scholar]
- 52.Yotnda P, Garcia F, Peuchmaur M, et al. Cytotoxic T cell response against the chimeric ETV6-AML1 protein in childhood acute lymphoblastic leukemia. J Clin Invest. 1998;102:455–462. doi: 10.1172/JCI3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Theurillat JP, Zurrer-Hardi U, Varga Z, et al. NY-BR-1 protein expression in breast carcinoma: a mammary gland differentiation antigen as target for cancer immunotherapy. Cancer Immunol Immunother. 2007;56:1723–1731. doi: 10.1007/s00262-007-0316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boss CN, Grunebach F, Brauer K, et al. Identification and characterization of T-cell epitopes deduced from RGS5, a novel broadly expressed tumor antigen. Clin Cancer Res. 2007;13:3347–3355. doi: 10.1158/1078-0432.CCR-06-2156. [DOI] [PubMed] [Google Scholar]
- 55.Yajima N, Yamanaka R, Mine T, et al. Immunologic evaluation of personalized peptide vaccination for patients with advanced malignant glioma. Clin Cancer Res. 2005;11:5900–5911. doi: 10.1158/1078-0432.CCR-05-0559. [DOI] [PubMed] [Google Scholar]
- 56.Tarp MA, Clausen H. Mucin-type O-glycosylation and its potential use in drug and vaccine development. Biochim Biophys Acta. 2008;1780:546–563. doi: 10.1016/j.bbagen.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 57.Lucas S, Coulie PG. About human tumor antigens to be used in immunotherapy. Semin Immunol. 2008;20:301–307. doi: 10.1016/j.smim.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 58.Yan M, Himoudi N, Pule M, et al. Development of cellular immune responses against PAX5, a novel target for cancer immunotherapy. Cancer Res. 2008;68:8058–8065. doi: 10.1158/0008-5472.CAN-08-0153. [DOI] [PubMed] [Google Scholar]
- 59.Tammela J, Uenaka A, Ono T, et al. OY-TES-1 expression and serum immunoreactivity in epithelial ovarian cancer. Int J Oncol. 2006;29:903–910. [PubMed] [Google Scholar]
- 60.Chiriva-Internati M, Cobos E, Da Silva DM, Kast WM. Sperm fibrous sheath proteins: a potential new class of target antigens for use in human therapeutic cancer vaccines. Cancer Immun. 2008;8:8. [PMC free article] [PubMed] [Google Scholar]
- 61.Harashima N, Tanaka K, Sasatomi T, et al. Recognition of the Lck tyrosine kinase as a tumor antigen by cytotoxic T lymphocytes of cancer patients with distant metastases. Eur J Immunol. 2001;31:323–332. doi: 10.1002/1521-4141(200102)31:2<323::aid-immu323>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 62.Maciag PC, Seavey MM, Pan ZK, Ferrone S, Paterson Y. Cancer immunotherapy targeting the high molecular weight melanoma-associated antigen protein results in a broad antitumor response and reduction of pericytes in the tumor vasculature. Cancer Res. 2008;68:8066–8075. doi: 10.1158/0008-5472.CAN-08-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dubovsky JA, Albertini MR, McNeel DG. MAD-CT-2 identified as a novel melanoma cancer-testis antigen using phage immunoblot analysis. J Immunother. 2007;30:675–683. doi: 10.1097/CJI.0b013e3180de4d19. [DOI] [PubMed] [Google Scholar]
- 64.Chiriva-Internati M, Ferrari R, Yu Y, et al. AKAP-4: a novel cancer testis antigen for multiple myeloma. Br J Haematol. 2008;140:465–468. doi: 10.1111/j.1365-2141.2007.06940.x. [DOI] [PubMed] [Google Scholar]
- 65.Zhou Q, Guo AL, Xu CR, et al. A dendritic cell-based tumour vaccine for lung cancer: full-length XAGE-1b protein-pulsed dendritic cells induce specific cytotoxic T lymphocytes in vitro. Clin Exp Immunol. 2008;153:392–400. doi: 10.1111/j.1365-2249.2008.03724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen YW, Tekle C, Fodstad O. The immunoregulatory protein human B7H3 is a tumor-associated antigen that regulates tumor cell migration and invasion. Curr Cancer Drug Targets. 2008;8:404–413. doi: 10.2174/156800908785133141. [DOI] [PubMed] [Google Scholar]
- 67.Lewen S, Zhou H, Hu HD, et al. A Legumain-based minigene vaccine targets the tumor stroma and suppresses breast cancer growth and angiogenesis. Cancer Immunol Immunother. 2008;57:507–515. doi: 10.1007/s00262-007-0389-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luo Y, Wen YJ, Ding ZY, et al. Immunotherapy of tumors with protein vaccine based on chicken homologous Tie-2. Clin Cancer Res. 2006;12:1813–1819. doi: 10.1158/1078-0432.CCR-05-1990. [DOI] [PubMed] [Google Scholar]
- 69.Yokokawa J, Bera TK, Palena C, et al. Identification of cytotoxic T-lymphocyte epitope(s) and its agonist epitope(s) of a novel target for vaccine therapy (PAGE4) Int J Cancer. 2007;121:595–605. doi: 10.1002/ijc.22698. [DOI] [PubMed] [Google Scholar]
- 70.Niethammer AG, Xiang R, Becker JC, et al. A DNA vaccine against VEGF receptor 2 prevents effective angiogenesis and inhibits tumor growth. Nat Med. 2002;8:1369–1375. doi: 10.1038/nm1202-794. [DOI] [PubMed] [Google Scholar]
- 71.Hoeppner LH, Dubovsky JA, Dunphy EJ, McNeel DG. Humoral immune responses to testis antigens in sera from patients with prostate cancer. Cancer Immun. 2006;6:1. [PubMed] [Google Scholar]
- 72.Lee J, Fassnacht M, Nair S, Boczkowski D, Gilboa E. Tumor immunotherapy targeting fibroblast activation protein, a product expressed in tumor-associated fibroblasts. Cancer Res. 2005;65:11156–11163. doi: 10.1158/0008-5472.CAN-05-2805. [DOI] [PubMed] [Google Scholar]
- 73.Kaplan CD, Kruger JA, Zhou H, Luo Y, Xiang R, Reisfeld RA. A novel DNA vaccine encoding PDGFRbeta suppresses growth and dissemination of murine colon, lung and breast carcinoma. Vaccine. 2006;24:6994–7002. doi: 10.1016/j.vaccine.2006.04.071. [DOI] [PubMed] [Google Scholar]
- 74.Dubovsky JA, McNeel DG. Inducible expression of a prostate cancer-testis antigen, SSX-2, following treatment with a DNA methylation inhibitor. Prostate. 2007;67:1781–1790. doi: 10.1002/pros.20665. [DOI] [PubMed] [Google Scholar]
- 75.Luo Y, Zhou H, Mizutani M, et al. A DNA vaccine targeting Fos-related antigen 1 enhanced by IL-18 induces long-lived T-cell memory against tumor recurrence. Cancer Res. 2005;65:3419–3427. doi: 10.1158/0008-5472.CAN-04-3120. [DOI] [PubMed] [Google Scholar]
- 76.Hawk ET, Matrisian LM, Nelson WG, Dorfman GS, Stevens L, Kwok J, Viner J, Hautala J, Grad O Translational Research Working Group. The Translational Research Working Group developmental pathways: introduction and overview. Clin Cancer Res. 2008;14:5664–5671. doi: 10.1158/1078-0432.CCR-08-1268. [DOI] [PubMed] [Google Scholar]
- 77.Cheever MA, Schlom J, Weiner LM, Lyerly HK, Disis ML, Greenwood A, Grad O, Nelson WG Translational Research Working Group. Translational Research Working Group developmental pathway for immune response modifiers. Clin Cancer Res. 2008;14:5692–5699. doi: 10.1158/1078-0432.CCR-08-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.The Analytic Hierarchy Process: Planning, Priority Setting, Resource Allocation. McGraw-Hill: 1980. ISBN 0-07-054371-2. [Google Scholar]
- 79.Bhushan, Navneet, KanwalRai . Strategic Decision Making: Applying the Analytic Hierarchy Process. London: Springer-Verlag; 2004. Jan, ISBN 1-8523375-6-7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1