Single-cell dynamics determines response to CDK4/6 inhibition in triple negative breast cancer (original) (raw)
. Author manuscript; available in PMC: 2018 Oct 8.
Abstract
Purpose
Triple negative breast cancer (TNBC) is a heterogeneous subgroup of breast cancer that is associated with poor prognosis. We evaluated the activity of CDK4/6 inhibitors across TNBC subtypes, and investigated the mechanisms of sensitivity.
Experimental design
A panel of cell lines representative of TNBC were tested for CDK4/6 sensitivity_, in vitro_ and in vivo. A fluorescent CDK2 activity reporter was used for single cell analysis in conjunction with time-lapse imaging.
Results
The luminal androgen receptor (LAR) subtype of TNBC was highly sensitive to CDK4/6 inhibition both in vitro [p<0.001 LAR vs. basal-like] and in vivo in MDA-MB-453 LAR cell line xenografts. Single cell analysis of CDK2 activity demonstrated difference in cell cycle dynamics between LAR and basal-like cells. Palbociclib-sensitive LAR cells exit mitosis with low levels of CDK2 activity, into a quiescent state that requires CDK4/6 activity for cell cycle re-entry. Palbociclib-resistant basal-like cells exit mitosis directly into a proliferative state, with high levels of CDK2 activity, bypassing the restriction point and the requirement for CDK4/6 activity. High CDK2 activity post-mitosis is driven by temporal deregulation of cyclin E1 expression. CDK4/6 inhibitors were synergistic with PI3 kinase inhibitors in PIK3CA mutant TNBC cell lines, extending CDK4/6 inhibitor sensitivity to additional TNBC subtypes.
Conclusion
Cell cycle dynamics determines response to CDK4/6 inhibition in TNBC. CDK4/6 inhibitor, alone and in combination, are a novel therapeutic strategy for specific subgroups of TNBC.
Introduction
The CDK4/6 – RB1 axis controls transition through the restriction point in the G1 phase of the cell cycle, and cancers frequently subvert the regulation of this axis to promote proliferation[1, 2]. CDK4/6 inhibition is a proven therapeutic strategy for oestrogen receptor positive (ER+ve) breast cancers [3, 4], with selective CDK4/6 inhibitors (palbociclib and ribociclib) demonstrating substantial improvements in progression free survival (PALOMA1[3], PALOMA2[5], PALOMA3[4] and MONALEESA-2[6]) in phase two and three clinical trials.
Triple negative breast cancer (TNBC) is an aggressive subtype of breast cancer associated with poor prognosis. Although TNBC may be sensitive to chemotherapy there is a substantial need to identify novel targeted therapeutic strategies. TNBC are a heterogeneous group of tumours with gene expression profiling identifying distinct subgroups [7, 8], including luminal androgen receptor (LAR), mesenchymal stem like (MSL), mesenchymal (MES), and basal-like [7]. The majority of TNBC fall within the dominant basal-like and MES subgroups. TNBC are highly proliferative tumours enriched for high expression of cell cycle genes [7], yet as a heterogeneous group are considered to be largely resistant to CDK4/6 inhibition [9], as are many other tumour types.
The determinants of sensitivity to CDK4/6 inhibition are poorly understood. Loss of retinoblastoma protein (RB1) causes resistance to CDK4/6 inhibition [10], however for the majority of cancers, the factors that determine sensitivity or resistance to CDK4/6 inhibitors are unclear. Recent studies of cell cycle dynamics have redefined our understanding of the mitosis-S phase transition in asynchronously dividing cells [11–13], with cells at mitotic exit entering either a quiescent or an active-proliferative state [12, 13]. Here we show that cell cycle exit into a quiescent or proliferative state is a major factor determining sensitivity to CDK4/6 inhibitors. We identify subgroups of TNBC that are highly sensitive to CDK4/6 inhibition, and using a CDK2 activity live-cell reporter [12] we show that CDK2 activity after mitotic exit dictates sensitivity to CDK4/6 inhibition.
Methods
Cell lines
Cell lines were obtained from ATCC or Asterand and maintained according to the manufacturer's instructions. Cell lines were banked in multiple aliquots on receipt, identity confirmed by STR profiling with the PowerPlex 1.2 System (Promega) and tested for mycoplasma every two weeks.
Palbociclib-resistant MFM223pR cells were generated by chronic exposure to increasing concentrations of palbociclib (100, 250, 500, 1000nmol) over 4 months. Drug treatments were replaced every 3-4 days with fresh media.
Antibodies, reagents and constructs
Phospho-RB1 S807/811 (8516), RB1 (9313), Cyclin E1 (HE12; 4129), Cyclin E2 (4132), CDK2 (2546), phospho-CDK2 T160 (2561); CDK4 (12790), Androgen Receptor (3202) were all Cell Signalling Technology, Danvers, MA; p16 F-12 (SC-1661, Santa Cruz), β-actin (A5441, Sigma); Cyclin E1 (ab33911) and c-myc (ab32072) were Abcam. Western blot analysis was performed using pre-cast 4-12% SDS gels, as described previously [14]. Densitometry analysis was performed on western blot films using ImageJ software (National Institute of Health, USA), and expressed relative to their corresponding loading control.
Palbociclib (PD-0332991; SelleckChem) was used at 500nmol, pictilisib (GDC-0941; SelleckChem) at 200nmol, and taselisib (GDC-0032; Genentech) at 100nmol, unless otherwise stated. Palbociclib 500nmol was used for the majority of experiments as previously [10]. No increase in effect on clonogenic growth was observed with doses above 500nmol (Fig. 1A).
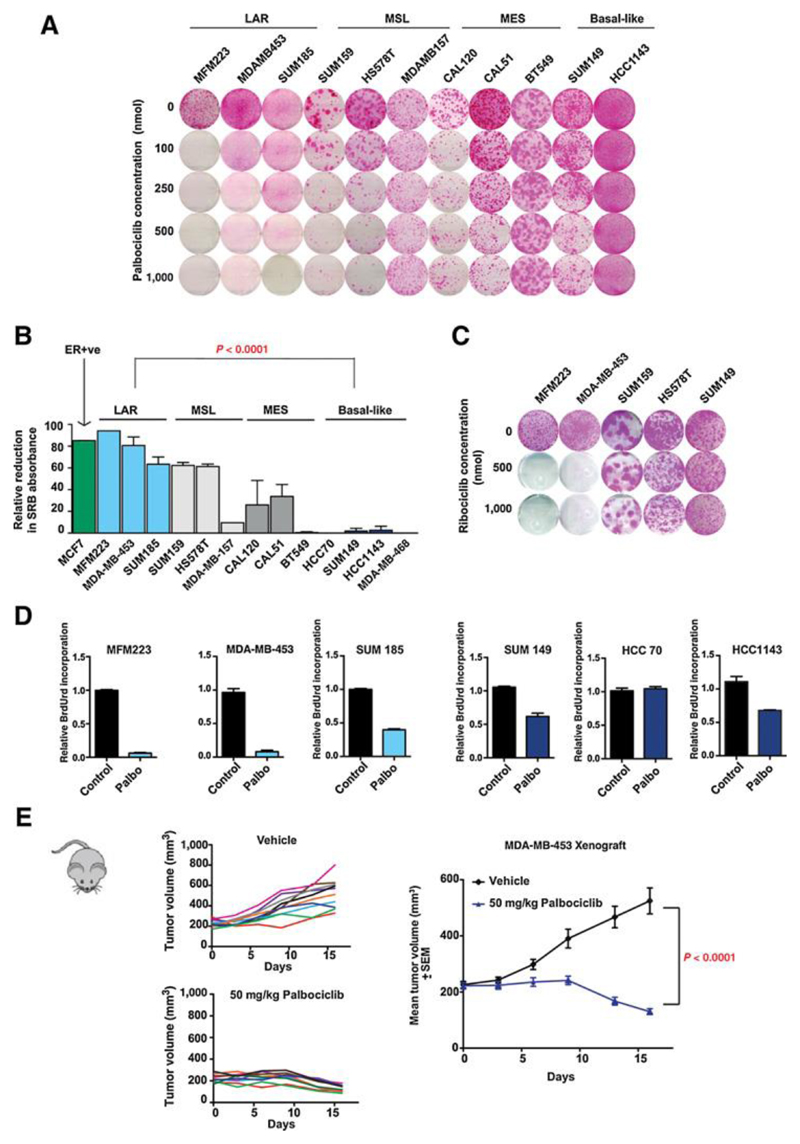
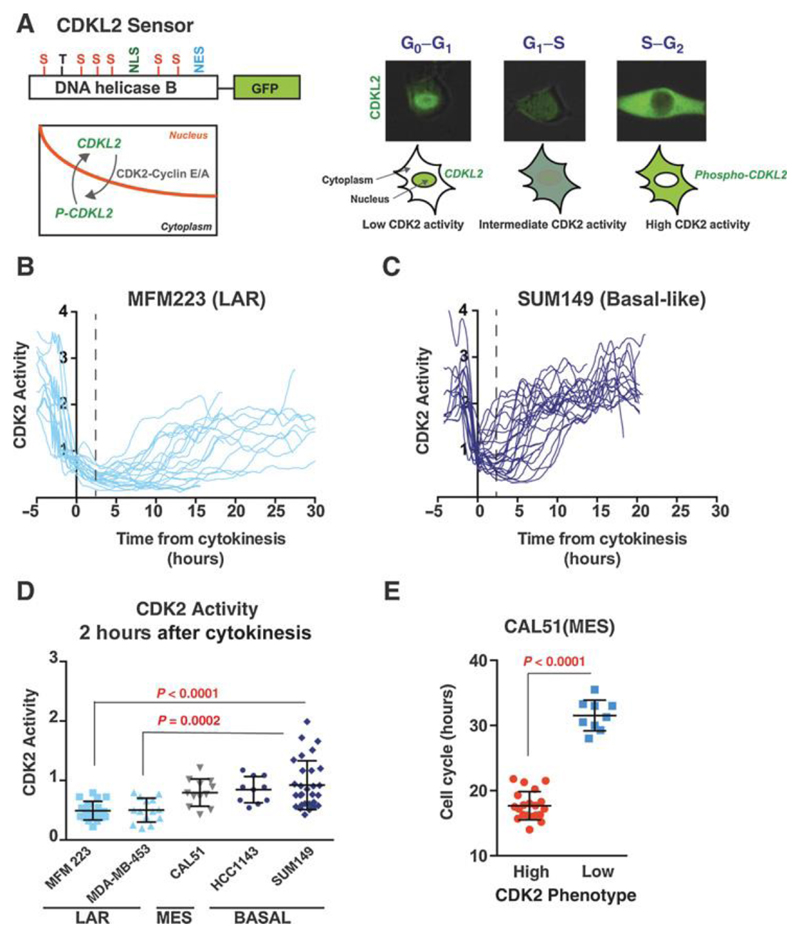
Figure 1. Luminal androgen receptor subgroup (LAR) of TNBC is sensitive to CDK4/6 inhibition.
A. Clonogenic assays of triple negative breast cancer (TNBC) cell lines grown continuously in increasing concentrations of palbociclib, divided by gene expression subtypes: LAR=luminal androgen receptor, MSL=mesenchymal stem like, MES=mesenchymal and basal-like. All cell lines are RB1 wild-type except RB1 mutant BT549.
B. Sensitivity to 500nmol palbociclib across 13 TNBC cell lines in clonogenic assays from part A. LAR subtype is highly sensitive to CDK4/6 inhibition [p<0.0001 Student’s T test LAR vs. basal-like]. ER+ve MCF7 cells are shown as positive control of sensitivity to palbociclib.
C. Clonogenic assays of 5 TNBC cell lines treated with the CDK4/6 inhibitor ribociclib.
D. Relative BrdU incorporation after 72 hours with or without 500nmol palbociclib for LAR cell lines (MFM223, MDAMB453 and SUM185), and basal-like cell lines (SUM149, HCC70 and HCC1143).
E. Xenografts of LAR MDA-MB-453 cells, treated daily with vehicle (n=10) or palbociclib (n=10) for 2 weeks. p<0.0001 Student’s T test vehicle vs. palbociclib at end of treatment..
siRNAs were from Thermofisher Scientific: siCCNE1 (#4390824, #4390824, #1299001), siCON1 (#4390843), siCON2 (#4390846) and siUBB (# 4390824).
The CDK2 activity live-cell sensor (CDK2L-GFP) was generated by cloning the C-terminal PSLD region of Human DNA Helicase B (DHB; 957-1087 amino acids) into pIRES-GFP puromicin as described previously[11]. PCNA was tagged at the N terminus with the modified LSS2-mKate fluoro-phore to generate LSS2-mKate-PCNA[15].
Cell Transfections
For CDK2 activity sensor experiments, cells were transfected with CDK2L-GFP using Lipofectamine® 2000 according to the manufacturer’s instructions, 48-72 hours prior imaging. For siRNA knockdown experiments, SUM149 cells were transfected with CDK2L-GFP on day one, then GFP-positive cells (3000 cells/well) were FACS-sorted into 384-well plate on day 3, and next siRNA transfection using siCON1 and siCCNE1 was performed with Lipofectamine® RNAiMAX on day 6, followed by time-lapse imaging on days 9-11. As previously demonstrated, the CDK2L-GFP sensor is not phosphorylated by CDK4/6 [11], nor by CDK1 [12].
Time-lapse microscopy
CDK2L-GFP positive cells were FACS sorted into 384 well plates, with 1000 to 3000 cells seeded per well, 24 hours prior time-lapse imaging. Unless specified, drugs or vehicle were added immediately prior time-lapse experiments. Images were taken on a High-content Opera Spinning Disk confocal microscope (PerkinElmer) with 40X water objective, every 10 minutes, in a NA 0.9 humidified environmental chamber at 37°C and 5% CO2.
To assess the impact of palbociclib on S phase entry in SUM149 cells (Fig. 3A), palbociclib or vehicle (DMSO) was added 8 hours after initiation of CDK2L-GFP time-lapse imaging, with continued imaging for a further 48 hours. Analysis was restricted to cells that underwent mitosis 1-3 hours prior to adding palbociclib or vehicle (DMSO).
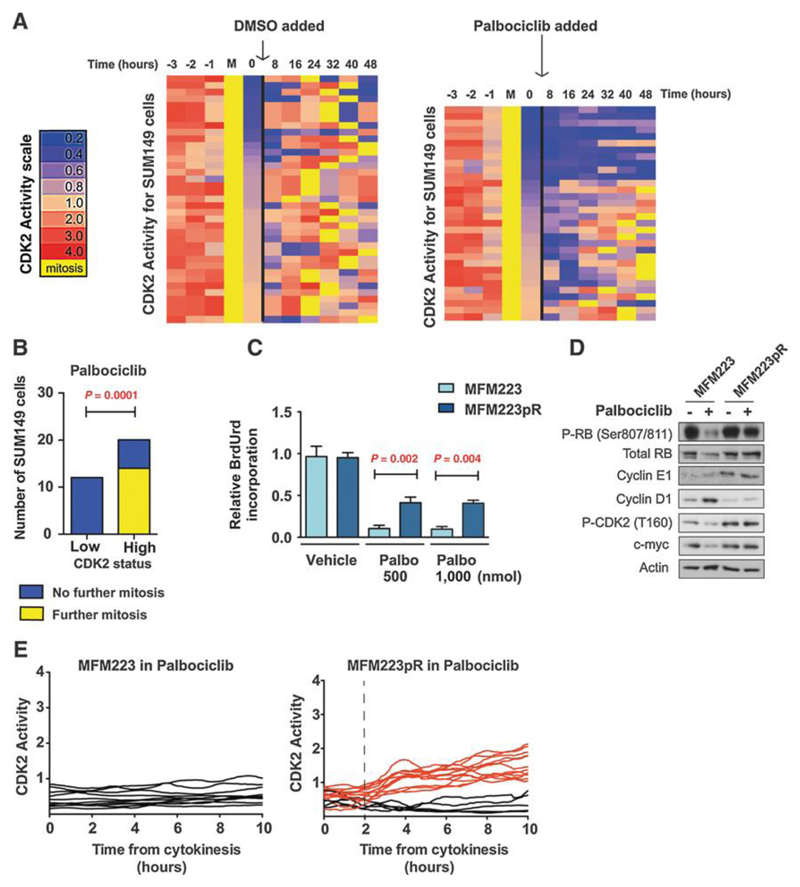
Figure 3. The proliferative CDK2high subpopulation drives resistance to CDK4/6 Inhibition at the single cell level.
A. Cells with low CDK2 activity post-mitosis do not re-enter the cell cycle when exposed to palbociclib. Heat-map of CDK2 activity in individual SUM149 cells treated with vehicle (left; n=37) or palbociclib (right; n=32) added 1-3 hours post-cytokinesis. CDK2 activity was synchronized and aligned in silico to point of cytokinesis (mitosis, M, yellow). T=0 hours corresponds to CDK2 activity post-cytokinesis and prior to palbociclib/vehicle addition. Each row represents one single cell. Heat map is ordered by CDK2 activity post-mitosis.
B. Quantification of cells undergoing a second mitosis in SUM149 cells exposed to palbociclib 1-3 hours post-mitosis as in part A, post-mitosis CDK2 activity CDK2high versus CDK2low. Cells that underwent a second mitosis (yellow) versus those that did not underwent a second mitosis (blue) during the tracking period (p=0.0001 Fisher’s exact test).
C. Relative BrdU incorporation in MFM223 and MFM223pR treated with vehicle, palbociclib 500nmol and 1000nmol for 72 hours (Student’s T-test p<0.002 and p<0.004).
D. Western blot of MFM223 and MFM223pR cells treated with vehicle or palbociclib 500nmol for 72 hours and probed with the indicated antibodies.
E. Single cell CDK2 activity traces of cells grown in 500nmol Palbociclib Left: parental MFM223, Right: Palbocicilb-resistant MFM223pR. Red: Cells with CDK2 activity>1 at 10 hours (proliferating). Black: Cells with CDK2 activity <1 at 10 hours (quiescent).
To study stability of CDK2 activity in individual cell clones (Fig. 5D and Supplementary Fig. 4A), a single cell was FACS-sorted into each well of the 96 well plate, with single cell sorting confirmed by bright field microscopy. Plates were incubated for 4 weeks to generate clonal populations. Individual wells were transfected with CDK2L-GFP, and 48 hours later imaged using time-lapse microscopy. All examined clonal populations were STR typed.
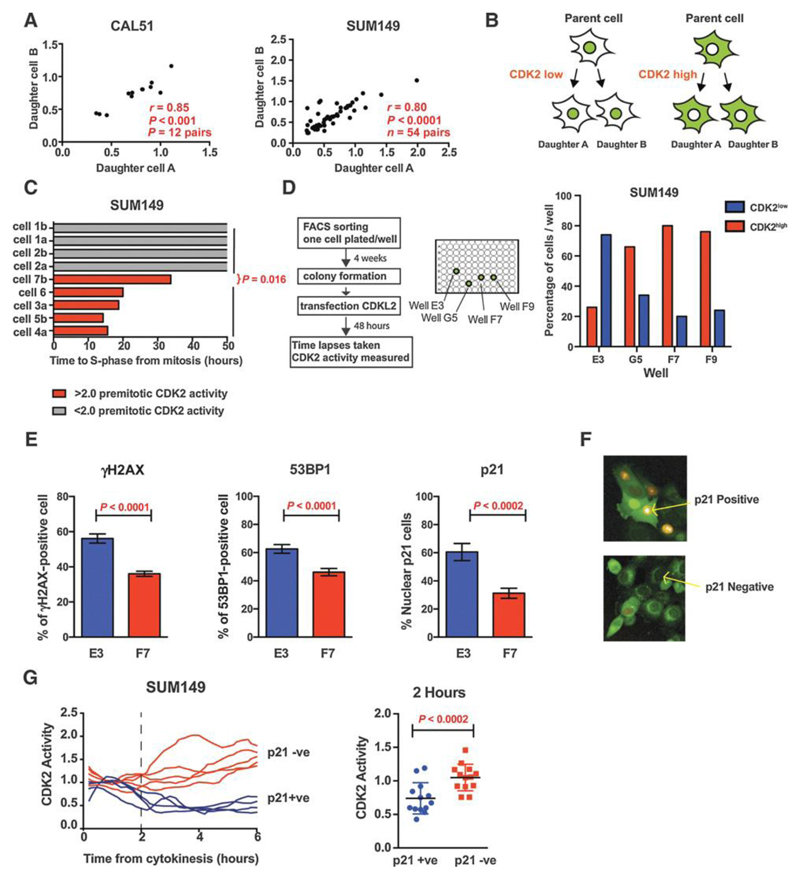
Figure 5. Post-mitotic CDK2 activity is determined pre-mitosis.
A. Correlation of CDK2 activity 2 hours post-cytokinesis between the daughter cell pairs generated in the same mitosis, in CAL51 (left) and SUM149 (right).
B. Schematic illustrating shared levels of CDK2 activity between daughter cells.
C. Time to S phase entry for SUM149 cells which divided between 0-2 hours after palbociclib was added. Pre-mitotic CDK activity (-2hrs) was measured in cells before palbociclib addition. Red: cells with pre-mitotic CDK2 activity >2; Grey: cells with pre-mitotic activity <2.0, none of which entered S phase by time-point 50 hours, post cytokinesis (p=0.016 Mann-Whitney).
D. SUM149 cells were single cell sorted by FACS and clones expanded from single cells for 4 weeks before transfection with CDK2L-GFP sensor (left). Percentage of CDK2low and CDK2high SUM149 cells per well, 50 cells tracked per well (right).
E. Percentage of cell positive for gamma H2AX (E3=total 860 cells; F7 =total 1508), 53BP1 (E3=total 719 cells; F7 =total 1298) and p21 (E3=total 275 cells; F7 =total 539) by immunofluorescence in the predominately CDK2low E3 clones and CDK2high F7 clone.
F. Immunofluorescence staining of p21 (yellow) in SUM149 cells transfected with CDK2 biosensor (green)
G. CDK2 activity in SUM149 cell in relation to the p21 staining status at 6 hours post-mitosis in SUM149 cells. Left: CDK2 activity traces and Right: CDK2 activity at 2 hours post cytokinesis, in p21 positive and p21 negative cells.
Image analysis
Time frames of cells transfected with CDK2L-GFP or LSS2-mKate-PCNA were captured at 10 minutes intervals over a 48-72 hours period. Dynamic changes of CDK2L-GFP or LSS2-mKate-PCNA were manually tracked in individual cells using Volocity (PerkinElmer). To quantify CDK2 activity, both nuclear and peri-nuclear cytoplasmic CDK2L-GFP intensity were measured simultaneously. CDK2 activity for each time point was calculated as a ratio of mean GFP fluorescent intensity in the cytoplasm divided by the mean GFP fluorescent intensity in the nucleus. For representation, asynchronous single cell CDK2 activity traces were aligned in silico to time of cytokinesis as T0 (time-point = 0). The CDK2 activity traces were smoothed with a window of 4 data points minimize background noise.
Cell cycle length (hours) was calculated as the time from first cytokinesis to the second cytokinesis. Unless stated otherwise, post-mitotic CDK2 activity was assessed as the CDK2 activity at 2 hours post-cytokinesis, as the first time-point across imaged cell lines that allowed reliable quantification of CDK2 activity after reformation of the nuclear membrane and cell flattening [11]. Cells with CDK2 activity <0.6 at 2 hours post cytokinesis were defined as CDK2low, and cells with CDK2 activity >0.6 at 2 hours points post cytokinesis were defined as CDK2high, as previously described [12]. In PCNA tracking experiments S-phase entry was defined a sharp increase in PCNA intensity with the appearance of nucleoli, as previously described [11]. Accumulation of CDK2 activity to a ratio of 1 was used as surrogate for cell cycle entry, as previously demonstrated [12].
Immunofluorescence
Immediately after time lapse, cells were fixed in 4% paraformaldehyde, washed three times in PBS, permeabilized with 0.2% Triton X-100, and stained at room temperature using mouse and rabbit primary antibodies, detected with corresponding fluorescent secondary antibodies: anti-mouse Alexa Fluor-555 and anti-rabbit Alexa Fluor-647. Nuclear pixels were measured as an output of intensity for each cell, using Columbus™ image data storage and analysis system.
Nuclear cyclin E1 protein levels in individual LAR MDAMB453 cells and basal-like SUM149 cells 1-2 hours after mitosis (Fig. 4D), was determined by time-lapse imaging of CDK2L-GFP positive cells (24 hours), followed by immunofluorescence for cyclin E1. Nuclear cyclin E1 protein levels were quantified by measuring the intensity of the immunofluorescence signal with Columbus™ imaging software, specifically in cells that had undergone mitoses 1-2 hours prior to fixation.
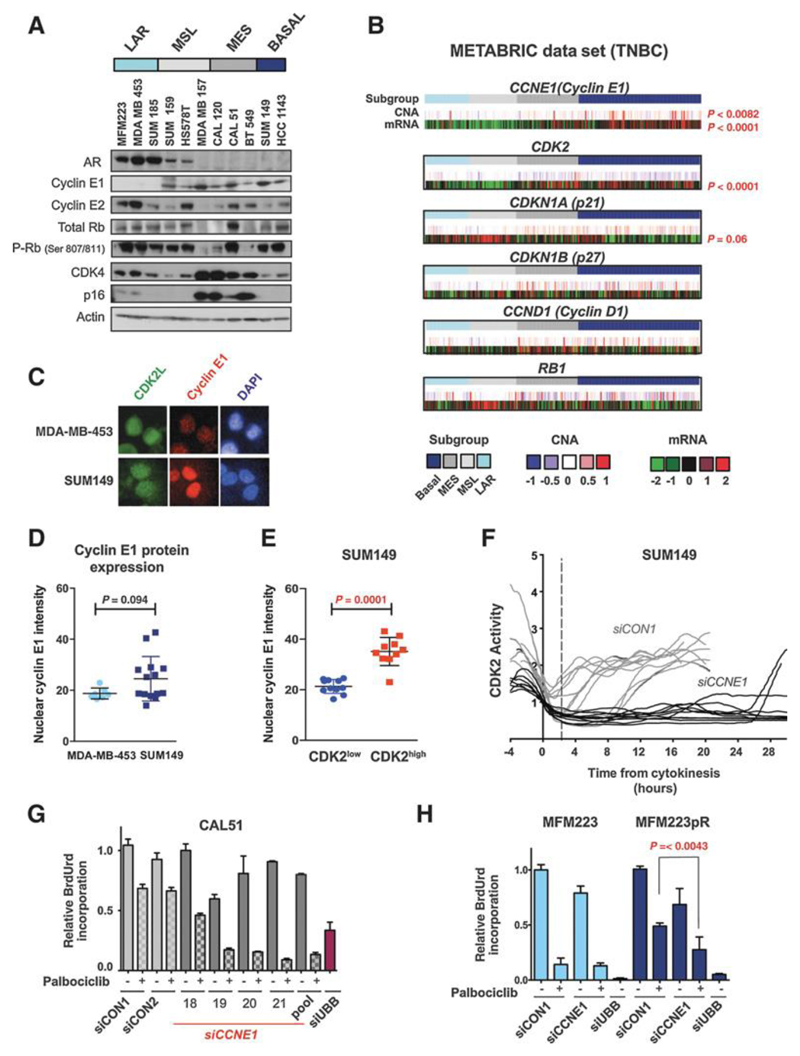
Figure 4. High cyclin E1 expression post-mitosis drives high CDK2 activity and CDK4/6 inhibitor resistance.
A. Western blot of lysates from a panel of TNBC cell lines (n=11 cell lines), probed with the indicated antibodies (Not shown: Densitometry for LAR vs. non-LAR for androgen receptor (AR) p=0.01; Cyclin E1 p= 0.02; Mann-Whitney test).
B. Copy number aberrations (CNA upper track) and gene expression (mRNA lower track) from 219 primary triple negative breast cancers, clustered according to gene expression subtypes: LAR (n=35); MSL (n=38); MES (n=49); Basal-like (n=97). P values generated from multiple t-tests (1% false discovery rate) for gene expression comparisons LAR vs. basal-like TNBC subtypes: Cyclin E1 (CCNE1) p<0.0001; CDK2 p<0.0001; CDKN1A (p21) p=0.06; CDKN1B (p27) p=0.23. Copy number gain LAR vs. basal-like: CCNE1 p=0.0082. Red: statistical significance.
C. Immunofluorescent staining of nuclear cyclin E1 (red) in CDK2L positive (GFP) MDA-MB-453 cells and SUM149 cells 1 hour after mitosis. DAPI (blue).
D. Cyclin E1 nuclear intensity assessed by immunofluorescence and quantified with Columbus software in individual cells 1-2 hours post-mitosis, in LAR MDAMB453 and Basal-like SUM149 cell lines (p= 0.094 Student’s T-test).
E. Cyclin E1 nuclear intensity assessed by immunofluorescence in SUM149 cells 1-2 hours post-cytokinesis in CDK2low versus CDK2high cells (p value <0.0001 Student’s T-test).
F. Single cell CDK2 activity traces of proliferating SUM149 cells transfected 72 hours earlier with siCON1 or siCCNE1.
G. Relative BrdU incorporation in CAL51 cells transfected 72 hours earlier with siCON1, multiple individual CCNE1 siRNAs and siCCNE1 pool, treated with vehicle or palbociclib 500nmol. H. Relative BrdU incorporation in MFM223 and MFM223pR cells transfected with the indicated siRNAs and treated with vehicle or palbociclib.
Immunohistochemistry
For immunohistochemical analysis of MDAMB453 xenografts, tumours were extracted 4 hours post-dose and formalin fixed for immunohistochemistry (IHC). Each sampling time point includes 4 animals /treatment group. IHC was performed on 4um thick formalin-fixed, paraffin-embedded tissue sections mounted on glass slides. For cleaved caspase 3 (Cell Signaling Technologies, Danvers, MA), staining was performed on a DAKO autostainer. Sections were treated with DAKO Target Retrieval (Dako; Carpinteria, CA), incubated with primary antibody at 0.12ug/ml overnight at 4°C followed by biotinylated goat anti-rabbit IgG (Vectorlabs, Burlingame, CA) and detected with Vectastain ABC-HRP (Vectorlabs, Burlingame, CA). For phospho-S6, IHC was performed on the Ventana Discovery XT Autostainer platform (Ventana Medical Systems Inc, Tucson, AZ). The slides were pre-treated with CC1, standard time, followed by anti-phospho-S6 (Cell Signaling Technologies, Danvers, MA), incubated at 0.26ug/ml for 32 minutes at 37°C. The antibody was detected with anti-rabbit-UltraMap (Ventana Medical Systems Inc, Tucson, AZ). Staining was visualized with DAB. Sections were counter stained with hematoxylin, dehydrated, cleared and cover-slipped for viewing.
Assessment of viability and proliferation
All clonogenic assays were conducted in triplicates of 6-well plates, with 1000 to 5000 cells seeded per well 24 hour prior to exposure to the indicated drug concentrations, or vehicle. Wells were treated continuously for at least 2 weeks replacing media/drug every 3-4 days. Plates were fixed with tricyclic acid (10%), stained with sulforhodamine B (SRB) and absorbance measured. Absorbance for drug treated wells was expressed relative to the control wells, with subtraction of the background SRB absorbance from an empty well. The mean of at least three replicate wells was calculated for each dose/combination.
Synergy was assessed using SRB absorbance from long-term clonogenic assays. Wells were treated every 3-4 days with palbociclib: 0, 100, 250, 500 or 750nmol, and/or pictilisib: 0, 100, 200, 400, 500 or 1000nmol. For AZD2014, wells were treated every 3-4 days with palbociclib: 0, 100, 250, 500 or 750nmol, and/or AZD2014: 0, 50, 100, 200, 400 and 750nmol. Assessment of compound synergy was conducted using Bliss independence score. A Bliss additivity score of <-1.0 was consider synergistic for that combination of drug concentrations. A cell line was considered to show drug synergy if at least 3 different combinations of drug concentrations were synergistic.
S phase fraction was assayed after 24 or 72 hours exposure to compounds, with the addition of 10 M BrdU for 2 hours prior to fixing. BrdU incorporation was assessed with Cell Proliferation chemiluminescent ELISA-BrdU assay (Roche 11 669 915 001) according to manufacturer’s instructions and adjusted for viable cells in parallel wells assessed with CellTiter-Glo®.
Tumour xenografts
In vivo efficacy and pharmacodynamic studies were approved by Genentech’s Institutional Animal Care and Use Committee (IACUC) and adhered to the ILAR Guide for the Care and Use of Laboratory Animals. Naïve female C.B-17 SCID mice (Charles River Laboratories, San Diego, CA) were inoculated into the right 2/3 mammary fat pad with 20 million MDAMB453 cells suspended in a 1:1 ratio of HBSS and phenol red-free matrigel (BD Biosciences, San Jose, CA). Once tumours reached a mean volume of about 300 mm3, mice with similarly sized tumours were distributed into treatment cohorts (n=10/group). Mice were dosed daily and orally, with vehicle [0.5% methylcellulose/0.2% tween-80 (MCT)], 5 mg/kg taselisib (GDC-0032), 50 mg/kg palbociclib or the combination of taselisib and palbociclib for 21 consecutive days. Length (l) and width (w) of each tumour were measured using digital calipers (Fred V. Fowler Company, Inc., Newton, MA) and tumour volumes were calculated based on the following formula: tumour volume = l x w2 x 0.5.
Analysis of publically - available data sets
The METABRIC dataset (n=1991) was obtained by application to the European Genome-phenome archive [16]. 320 putative TNBC samples were normalised using the beadarray package [17] and classified using TNBC type [18]. 5 samples were then removed as putative ER positive samples leaving 315 for analysis. Segmented (CBS) copy number logR ratios were downloaded and used for copy number analysis, with gain/loss thresholds as previously defined [16]. Heat maps representing key cell cycle genes were generated in R.
For 102 TCGA samples representing TNBC, level 3 RNA-seq data (raw gene counts) was downloaded from the TCGA web site for these samples. The gene counts were normalised using edgeR packages. Copy number, mutation and RPPA data was extracted from cBioPortal using the CGDS-R (http://www.cbioportal.org/cgds_r.jsp). Additional data was downloaded from the Cancer Proteome Atlas project [17].
Statistical analysis
For in vitro studies, all statistical tests were performed with GraphPad Prism version 6.0. _P_-values were two-tailed and considered significant if P < 0.05. Error bars represent Standard deviation or SEM of three experiments. Assessment of compound synergy was conducted using Bliss independence score.
Results
Luminal Androgen Receptor (LAR) subtype of triple negative breast cancer is sensitive to CDK4/6 inhibition
We investigated whether the different molecular subgroups of TNBC were sensitive to CDK4/6 inhibition. Clonogenic assays were performed on a panel of 12 RB1 wild-type TNBC cell lines and one RB1 mutant cell line (BT549), with the CDK4/6 inhibitor palbociclib (Fig. 1A and Fig. 1B). Cell lines from the dominant basal-like and mesenchymal (MES) subgroups of TNBC were resistant to palbociclib, whereas the LAR TNBC cell lines were highly sensitive to palbociclib (p<0.0001 basal-like vs. LAR), and ribociclib (Fig. 1C), with sensitivity similar to the oestrogen positive cell line MCF7 (Fig. 1B). In BrdU proliferation assays, palbociclib had a substantially greater effect on S phase entry in LAR compared to basal-like cell lines (Fig. 1D). The sensitivity of LAR tumours to palbociclib was investigated in vivo in MDAMB453 xenografts. Tumour reductions were observed after the initial nine days of consecutive dosing (palbociclib oral 50mg/kg), with reduction in tumour size observed in 7 out of 10 mice in the palbociclib treatment arm (Fig 1E).
Palbociclib sensitive cell lines have low post-mitotic CDK2 activity
We hypothesized that sensitivity to CDK4/6 inhibition would be determined by the level of CDK2 activity post-mitosis [12, 13]. To investigate this, we used a live cell fluorescent sensor to measure CDK2 activity (CDK2L) in five RB1 wild type TNBC models (Fig. 2A). The CDK2L sensor specifically reports CDK2 activity, and is not phosphorylated by CDK4/6 and nor CDK1 (Methods).
Figure 2. Palbociclib-sensitive cell lines have low post-mitotic CDK2 activity.
A. Left: Schematic of CDK2 activity live-cell sensor (CDK2L-GFP). CDK2 consensus phosphorylation sites shown: S=serine and T=threonine. NLS=nuclear localization signal. NES= nuclear export signal. Left. Phosphorylation of CDK2L-GFP sensor by active CDK2 leads to export from the nucleus to the cytoplasm. Right. Schematic of changes in sensor localisation through the cell cycle. CDK2 activity is at its highest during S/G2 phase of the cell cycle. Fluorescence images correspond to CDKL2-GFP in a single MFM223 cell.
B. and C. Single cell CDK2 activity traces of B. palbociclib-sensitive LAR MFM223, n=20 cells, 20% CDK2high and C. palbocicilb-resistant basal-like SUM149, n=30 cells, 74% CDK2high.
D. Post-mitotic (2 hours post-cytokinesis) CDK2 activity in individual cells from five TNBC cell lines. LAR MFM223 vs. basal-like SUM149 cell lines (Student’s T-test p<0.0001). LAR MDAMB453 vs. basal-like SUM149 (p<0.0002). LAR=luminal androgen receptor subgroup (light blue), MES=mesenchymal (grey) and BASAL=basal-like (dark blue).
E. Cell cycle lengths (hours) in post-mitotic CDK2high vs. CDK2_low_ subpopulations in CAL51 (p<0.0001 Student’s T test).
We measured CDK2 activity in individual cells as they transitioned through mitosis and into the next cell cycle. Individual cells in the LAR palbociclib-sensitive cell lines MFM223 and MDAMB435, exited mitosis with low levels of CDK2 activity, which continued to fall during the first two hours post-mitosis (Fig. 2B and Supplementary Fig. 1A). The dynamics of CDK2 activity after mitosis in the LAR cells were similar to CDK2 activity levels seen in the ER +ve MCF7 cell line (Supplementary Fig. 1B). In contrast, the palbociclib-resistant basal-like cell lines SUM149 and HCC1143 (Fig. 2C and Supplementary Fig. 1C), and the MES cell line CAL51 (Supplementary Fig. 1D), were predominately composed of cells that exited mitosis and started the next cell cycle with high levels of CDK2 activity, that rapidly accumulated higher levels of CDK2 activity.
To compare CDK2 activity between cell lines, CDK2 activity levels were quantified 2 hours post-cytokinesis, the earliest time-point that allowed accurate quantification after nuclear envelope reformation. Basal-like TNBC cell lines had significantly higher CDK2 activity compared to the LAR cell lines _(p<0.0001;_ Fig. 2D). Cells which exited mitosis with CDK2 activity <0.6 at 2 hours post cytokinesis were defined as CDK2low, and cells with CDK2 activity >0.6 at 2 hours post cytokinesis were defined as CDK2high [12]. Whereas LAR cells exited mitosis with a relatively homogeneous CDK2low phenotype, basal-like cells existed mitosis with heterogeneous CDK2 activity, with a large proportion of CDK2high cells (Fig. 2D). In basal-like and MES cell lines, CDK2high cells had substantially shorter cell cycles compared to CDK2low cells (p<0.0001; Fig. 2E and Supplementary Table 1). These results suggested that TNBC cell lines resistant to CDK4/6 inhibition were predominantly composed of actively proliferating CDK2high cells, whereas TNBC cell lines sensitive to CDK4/6 inhibition were predominantly composed of more quiescent CDK2low cells.
The proliferative CDK2high subpopulation drives resistance to CDK4/6 inhibition at the single cell level
We hypothesised that the CDK2high subpopulation would be resistant to CDK4/6 inhibitors. The TNBC basal-like SUM149 model had a heterogeneous mix of CDK2 mitotic exit phenotypes with predominately CDK2high cells and a smaller fraction of CDK2low cells, and thus represented a good model to test this hypothesis. To assess the effect of palbociclib on the two populations, SUM149 cells expressing CDKL2 were imaged for 8 hours prior to the addition of palbociclib or vehicle, and only cells that completed cytokinesis 1-3 hours prior to palbociclib addition were tracked. CDK2 activity was then assessed in individual cells for a further 48 hours post-treatment (methods). We observed that SUM149 cells that exited mitosis with a CDK2low phenotype were blocked from entering the cell cycle by palbociclib over the 48 hours duration of time lapse imaging (Fig. 3A), whereas cells that exited mitosis in a CDK2high state successfully entered the cell cycle despite palbociclib treatment and subsequently underwent a second mitosis (Fig. 3A and Fig. 3B). Therefore, although the SUM149 cell line was intrinsically resistance to palbociclib in long-term clonogenic assays, the sub-fraction of CDK2low cells were sensitive to palbociclib. Palbociclib did prolong cell cycle length in CDK2high cells (mean cell cycle length 20 hours vehicle versus 38 hours palbociclib, Fig. 3A) suggesting that CDK4/6 did facilitate, but was not essential for S-phase entry in CDK2high cells. After 14 days chronic treatment of SUM149 cells with palbociclib the length of the cell cycle returned towards pre-treatment levels, suggesting adaption to CDK4/6 inhibition (Supplementary Fig. 2A and 2B). Collectively this data demonstrates that post-mitotic CDK2 activity dictates sensitivity to CDK4/6 inhibition. CDK2high cells have sufficient CDK2 activity to bypass the restriction point, the point where CDK4/6 activity is necessary for cell cycle re-entry, resulting in CDK4/6 inhibitor resistance.
We next established whether CDK2 activity at mitotic exit changed after acquired resistance to CDK4/6 inhibitors in LAR cell lines. We generated palbociclib-resistant cells from the MFM223 cell line (MFM223pR) via four months of chronic palbociclib exposure. In clonogenic assays, the MFM223pR were resistant to palbociclib, with a greater proportion of cells in S-phase during palbociclib treatment compared to the parental palbociclib sensitive cell line MFM223 (Fig. 3C). The MFM223pR model acquired higher protein levels of cyclin E1 and of the activating CDK2 T160 phosphorylation (Fig. 3D). In MFM223pR cells a new CDK2high proliferative subpopulation emerged (Fig. 3E), suggesting that the mechanism of acquired resistance to palbociclib was due to a higher proportion of cells adopting the CDK2high phenotype.
Temporal dysregulation of Cyclin E1 expression in TNBC cells drives higher CDK2 activity post mitosis
To explore the molecular determinants of CDK2high cells, we profiled our panel of TNBC cell lines. The LAR cell lines had both high expression of the androgen receptor (AR) (p=0.01) and absent/low expression of cyclin E1 (p=0.02) (Fig. 4A). We profiled TNBC tumours using publically available datasets. In both the METABRIC [16] and TCGA [19] datasets TNBC LAR tumours had significantly lower transcriptomic expression levels of CCNE1 (p<0.0001) and CDK2 (p<0.0001), with higher CDKN1A (p21) levels, as compared to basal-like TNBC (p=0.06) (Fig. 4B, Supplementary Figures 3A and 3B). Basal-like TNBC tumours had more frequently increased CCNE1 gene copy number than LAR tumours (p=0.008 Fisher’s exact test, Fig. 4B). There was a high correlation between cyclin E1 mRNA with protein levels (r=0.89; Supplementary Fig. 3C and 3D).
Cyclin E1 is tightly regulated nuclear protein, periodically expressed during the cell cycle, with highest levels occurring during late G1 and early S-phase in non-cancer models [20]. We investigated the temporal regulation of cyclin E1 in palbociclib sensitive and resistant cell lines, in order to assess whether deregulation of cyclin E1 expression in early G1 promoted the CDK2high population. To test this we assessed the expression of nuclear cyclin E1 protein in individual cells 1-2 hours post-mitosis by dual immunofluorescence with CDK2L sensor (Methods). Basal-like SUM149 cells had aberrant high cyclin E1 expression post-mitosis (Fig. 4C-4D and Supplementary Fig. 3E) compared to LAR MDAMB453 cells that had uniform low cyclin E1 expression. This was substantially earlier than the time point where SUM149 cells typically entered S-phase, confirmed using a PCNA sensor (Supplementary Fig. 3F). CDK2high SUM149 cells had higher cyclin E1 protein expression than CDK2low SUM149 cells (Fig. 4E). Silencing of cyclin E1 in SUM149 cells resulted in the loss of the CDK2high population post-mitosis, inducing a CDK2low phenotype (Fig. 4F and Supplementary Fig. 3G). Furthermore, silencing of cyclin E1 sensitised CAL51 (Fig. 4G and Supplementary Fig. 3H), SUM149 (Supplementary Fig. 3I) and MFM223pR to palbociclib, with minimal effects upon parental MFM223 cells that had low cyclin E1 expression (Fig. 4H). Cyclin E1 silencing, without CDK4/6 inhibition, did not substantially reduce BrdU incorporation, likely due to redundancy between different CDKs and cyclins. Our data suggested that aberrant cyclin E1 expression immediately post-mitosis, promoted the CDK2high phenotype and resistance to CDK4/6 inhibition.
The CDK2high phenotype is determined pre-mitosis
We next addressed whether post-mitotic activity was determined pre-mitosis, as has been shown in non-cancer models [12]. SUM149 and CAL51 sister cell pairs, generated from the same mitosis, shared similar post-mitotic CDK2 activity (Fig. 5A and Fig. 5B) suggesting that the level of CDK2 activity post-mitosis was determined prior to cytokinesis. SUM149 cells with higher pre-mitotic CDK2 activity (2 hours prior to mitosis), generated daughter cells that entered S-phase despite CDK4/6 inhibition (Fig. 5C), whereas cells with lower pre-mitotic CDK2 activity were arrested by CDK4/6 inhibition (p=0.016).
Having demonstrated that post-mitotic CDK2 activity was determined pre-cytokinesis, we investigated whether this was due to the existence of fixed sub-population of CDK2high cells, or whether CDK2high and CDK2low populations inter-converted. To test the long-term stability of the CDK2low and CDK2high subpopulations, we FACS sorted a single SUM149 cell (Fig. 5D) and CAL51 cell (Supplementary Fig. 4A) into 96 well plates, confirmed single-cell seeding by microscopy, and assessed CDK2 activity in the resulting single cell clones after four weeks of multiplication. In general, clonal populations recapitulated variability in CDK2high and CDK2low populations despite arising from a single cell (Fig. 5D and Supplementary Fig. 4). The fraction of CDK2high cells was the most prevalent phenotype (50 cells measured/well) across three of the four wells imaged in SUM149. In contrast we identified a clonal SUM149 population that was robustly CDK2low (well E3) at a single time point, 2 hours post-cytokinesis, although it was not addressed whether greater heterogeneity in CDK2 activity could develop over time. Overall these results suggest that post-mitotic CDK2high activity is determined pre-mitosis, but these CDK2high and CDK2low subpopulations are not distinct fixed populations, with cells interconverting between phenotypes over many generations.
The SUM149 cells have a BRCA1 mutation [21], which may have elevated levels of DNA damage. We hypothesized that the E3 clone, which maintained a large population of CDK2low cells, may have increased DNA damage. Immunofluorescence staining revealed that the SUM149 E3 clonal cell population had a higher percentage of gamma H2AX-, 53BP1- and p21-positive cells, as compared to the F7 clonal population (Fig. 5E and Fig. 5F). Parental SUM149 cells that exited mitosis with CDK2low phenotype were p21 positive by immunofluorescence (Fig. 5G), as previously observed in non-cancer models [12]. These results suggest that DNA damage and p21 expression may in part determine CDK2 activity state at mitotic exit [22].
Inhibition of PI3 kinase synergizing with CDK4/6 inhibitors in PIK3CA mutant TNBC
The non-basal, LAR and MSL subtypes of TNBC are substantially enriched with activating mutations in the PI3 kinase catalytic subunit PIK3CA gene [23], and therefore we investigated the therapeutic potential of PI3 kinase pathway inhibitors in TNBC. A synergistic interaction (Bliss additivity score < -1.0) was observed between the pan class I PI3 kinase inhibitor pictilisib (GDC0941) and palbociclib in PIK3CA mutant TNBC cell lines (Fig. 6A), as we recently reported in ER+ve cell lines [10]. Combination synergy was not observed in PIK3CA wild-type cell lines nor the RB1 mutant cell line BT549. Synergy was also observed with mTOR inhibitor AZD2014 - palbociclib combinations in PIK3CA mutant and some PIK3CA wild-type TNBC cell lines (Supplementary Fig. 5A). We further validated the efficacy of the drug combination with the α-selective PI3 kinase inhibitor taselisib (GDC0032), which substantially sensitised the MSL cell line SUM159 to palbociclib in clonogenic assays (Fig. 6B) and BrdU proliferation assays (Supplementary Fig. 5B). These data suggested that combinations of PI3 kinase pathway inhibitors and CDK4/6 inhibitors could extend the utility of CDK4/6 inhibitors outside LAR subtype cancers.
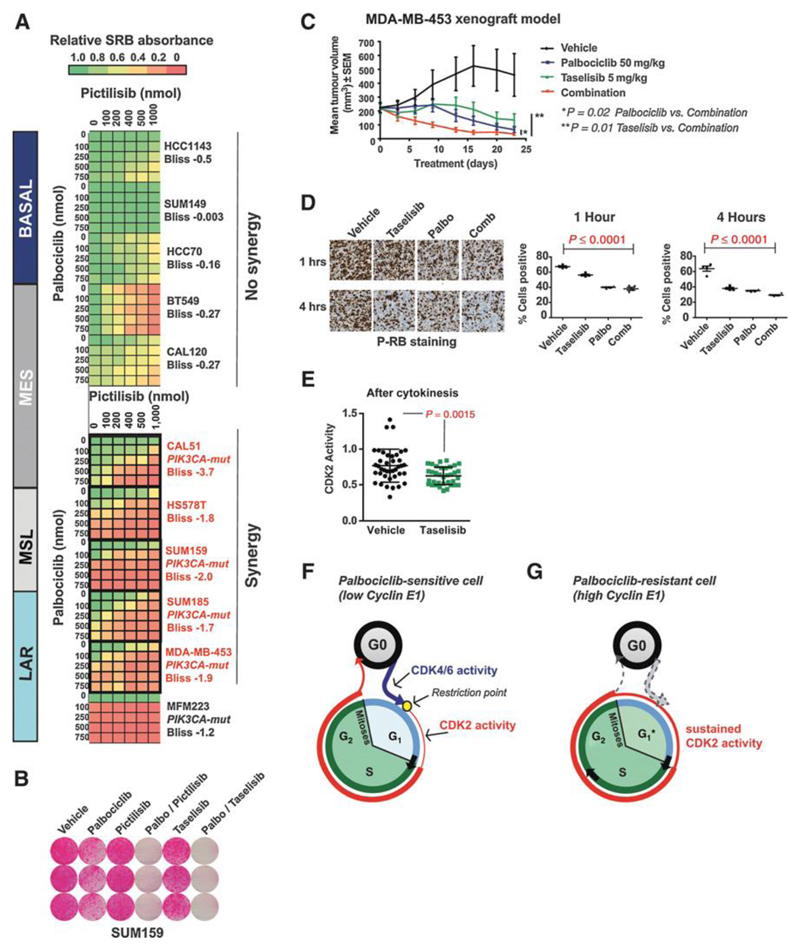
Figure 6. Inhibition of PI3 kinase signalling synergises with CDK4/6 inhibition in PIK3CA mutant TNBC.
A. Synergy heat maps matrices of clonogenic assays in TNBC cell lines treated with palbociclib and/or pictilisib (PI3 Kinase inhibitor) at increasing concentrations. Red: cell lines with combination synergy assessed using Bliss additivity score. PIK3CA mut = PIK3CA mutant. The lowest Bliss additivity score for each cell line is indicated.
B. Clonogenic assays in PIK3CA mutant SUM159 MSL TNBC cell line grown in palbociclib 500nmol, pictilisib (pan-PI3 kinase inhibitor) 200nmol, taselisib (alpha specific PI3 kinase inhibitor) 100nmol, and indicated combinations.
C. Mouse xenografts of LAR MDAMB453 cells, treated daily with vehicle (n=10), palbociclib (n=10), taselisib (n=10) or combination (n=10) (p=0.02 palbociclib vs. combination p=0.02; taselisib vs. combination p=0.01). Error bars are mean tumour volume and SD.
D. Left: Immunohistochemistry of MDAMB453 mouse xenografts stained for phosphorylated RB1 (Ser807/811) after 1 and 4 hours of indicated drug treatment(s). Right: Scatter plots with percentage of positively stained cells for phosphorylated RB1 in IHC sections (left) after 1 hour and 4 hours treatment.
E. CDK2 activity post-mitosis in PIK3CA mutant CAL51 treated with vehicle or taselisib 100nmol.
F. Model of cell cycle dynamics in CDK2low cells. Cells exits mitosis into a quiescent state with CDK4/6 activity (blue) necessary to pass through the restriction point (yellow circle) after which CDK2 activity (red) promotes S phase entry.
G. Model of cell cycle dynamics in CDK2high cells. Cells exit mitosis into an active proliferating state with high CDK2 activity, bypassing the restriction point, and CDK4/6 activity is not necessary to enter S phase. Consequently CDK2high cells, and cancers with a high proportion of CDK2high cells, are resistant to CDK4/6 inhibition.
The combination of palbociclib and taselisib demonstrated greater efficacy than either compound alone in xenografts from the PIK3CA mutant LAR MDAMB453 cell line (palbociclib vs. combination p=0.02; taselisib vs. combination p=0.01, Fig. 6C). Pharmacodynamic studies at 1 hour and 4 hours time-points in the mouse tumours from MDAMB453 xenografts treated with individual drugs or the combination, demonstrated significant reduction in phosphorylated RB1 compared to vehicle (Fig 6D; vehicle versus. taselisib 1 hour p= 0.0004, 4 hours p<0.0001; vehicle vs. palbociclib 1 hour p< 0.0001, 4 hours p< 0.0001; vehicle vs. combination 1 hour p< 0.0001, 4 hours p< 0.0001). The combination induced greater levels of apoptosis than either drug alone (Supplementary Fig. 5C).
Finally, we examined whether PI3 kinase inhibition with taselisib affected post-mitotic CDK2 levels, using the PIK3CA mutant MES CAL51 cell line. Taselisib decreased post-mitotic CDK2 activity with a greater proportion of cell that exit mitosis CDK2low (Fig. 6E). Pre-mitotic CDK2 levels influenced entry into the cell cycle post-mitosis (Supplementary Fig. 5D). These data suggested that the PI3 kinase inhibition sensitised to CDK4/6 inhibition, in part as PI3 kinase inhibition suppressed post mitotic CDK2 activity, inducing a CDK2low quiescent state where CDK4/6 activity was required to initiate the cell cycle.
Discussion
We have shown that the luminal androgen receptor (LAR) subgroup of triple negative breast cancers (TNBC) is highly sensitive to CDK4/6 inhibition in vitro and in vivo (Fig. 1). Sensitivity to CDK4/6 inhibition is dictated at the single cell level, with resistance to CDK4/6 inhibitors arising from cancer cells that exit mitosis directly into a CDK2high proliferative state, from which CDK4/6 is not necessary for cell cycle re-entry (Fig. 2C and Fig.3A).
Our results further extend prior work in non-cancer models that demonstrate a biphasic exit of cells into proliferative and quiescent states after mitosis [12, 13]. Cells that exit mitosis with a CDK2low phenotype enter into a quiescent state, requiring CDK4/6 activity to initiate re-entry into the cell cycle (Fig. 6F), and are hence sensitivity to CDK4/6 inhibition. In contrast, cells that exit mitosis with a CDK2high phenotype enter into a proliferative state, bypassing the restriction point (Fig. 6G), with short doubling times. Tumours with a high proportion of CDK2high cells are resistant to CDK4/6 inhibition. Palbociclib-sensitive LAR cancer cells typically exit into a quiescent CDK2low state post-mitosis, from which CDK4/6 is required to phosphorylate RB1 and pass the restriction point [2, 24–27]. In contrast, basal-like TNBC cells frequently enter a proliferative CDK2high state and are thus resistant to palbociclib treatment. This provides a mechanistic explanation for why basal-like, and potentially many other tumour types are resistant to CDK4/6 inhibition despite being RB1 wild type. CDK4/6 and PI3 kinase inhibitor combinations have substantial activity in PIK3CA mutant non-basal TNBC, of both LAR and mesenchymal-stem (MSL) subgroups (Fig. 6A), with such combinations having the potential to further expand the TNBC subgroups that could benefit from CDK4/6 inhibition
Cyclin E1 binds to and activates CDK2 [28, 29] and prior work has demonstrated that cyclin E1 expression mediates resistance to CDK4/6 inhibition [10, 30]. The classical view is that CDK2 and cyclin E are downstream of CDK4/6 activity [24]. Here we show that in palbociclib-resistant TNBC, cyclin E1 expression is dysregulated and expressed immediately post-mitosis (Fig. 4D). Dysregulated cyclin E1 promotes the CDK2high phenotype with CDK2-CyclinE complex active immediately post-mitosis resulting in short G1 phase (Fig. 6G). SUM149 harbour an inactivating FBXW7 mutation that disrupts the SCF complex and likely contributes to dysregulated cyclin E1 expression (Supplementary table 2). In other contexts, increased CCNE1 gene copy number probably contributes to dysregulation of cyclin E1 expression post-mitosis [10]. These observations suggest that measurement of cyclin E1 expression either at the RNA or protein level has the potential to be utilised as a predictive biomarker of resistance to CDK4/6 inhibition in breast cancer.
Daughter cells share the same CDK2 activity state, suggesting that post-mitotic CDK2 activity is determined pre-mitosis (Fig. 5A). However, this is not the result of CDK2high and CDK2low cells, as colonies derived from individual cells largely recapitulate the same variability in CDK2 activity as the parental cell line (Fig. 5D). We provide some data to suggest that DNA damage, and the resulting induction of p21 expression, may also regulate CDK2 activity state post-mitosis (Fig. 5G). However, more research is required to further assess the potential role of DNA damaging signalling in this context.
Our results illustrate how single cell analysis can identify mechanisms of resistance to targeted therapies. Phenotypic heterogeneity between single cells may drive drug resistance, and this may be more clearly elucidated at the single cell level than through assessment of bulk cell populations. Through single cell approaches, we identify CDK2 activity post-mitosis as a key determinant of sensitivity to CDK4/6 inhibition, and highlight potential therapeutic strategies for triple negative breast cancer. An on going therapeutic trial is assessing the effectiveness of CDK4/6 inhibition in combination with PI3 kinase inhibition, in patients with _PIK3CA-_mutant TNBC (NCT02389842).
Supplementary Material
Supplementary Figures and Tables
Translational relevance.
Currently there are no effective targeted therapies for triple negative breast cancer (TNBC). We show that the Luminal Androgen Receptor subtype of TNBC is highly sensitive to CDK4/6 inhibition whilst basal-like TNBC tumours are resistant. Differential sensitivity of TNBC subtypes to CDK4/6 inhibition is shown to be a result of cells exiting mitosis into an active proliferating or a quiescent state. Basal-like TNBC have a high proportion of active proliferating cells, that are resistant to CDK4/6 inhibition, promoted by dysregulation of cyclin E1 expression. Our results identify novel therapeutic approaches for TNBC, and identify mechanisms of sensitivity to CDK4/6 inhibitors.
Acknowledgments
The authors acknowledge Frederick Wallberg and Rhadhika Patel for flow cytometry support services and Vicky Bousgouni for live imaging support.
Grant Support:
This research was funded by the Avon foundation (U.S. Asghar), Breast Cancer Now with generous support from the Mary-Jean Mitchell Green Foundation and Cancer Research UK C30746/A16642 (N.C. Turner). The authors also acknowledge NHS funding to the NIHR Biomedical Research Centre at The Royal Marsden and the ICR.
Footnotes
Author contributions:
U.S.A: Study conception, methodology and experimental design. Conducted _in vit_ro experiments including single cell analysis with sensors; recorded, organized and analysed data; prepared figures and manuscript.
A.R.B: Expanded CDK2 and PCNA sensors. Conducted experiments involving time-lapse imaging. Contributed to single cell analysis; provided input to figure preparation and manuscript. Organised data and generated Volocity libraries.
R.C: Performed the bioinformatic analysis of METABRIC and TCGA datasets to generate heat-maps and figures. Constructed our database.
M.B: Tracking of CDKL2 sensor for single cell data analysis.
I.B: Assisted with clonogenic assays and contributed to manuscript preparation.
DS, JG, JAL, LC, AY: In vivo mouse work, including both toxicity-efficacy and pharmacodynamic experiments with drugs in MDAMB453 xenografts. Staining of tumours for immunohistochemistry (IHC). Analysis of IHC. Generation of mouse in vivo figures and associated IHC data.
A.P: Contributed to study methodology and conception.
M.T-H: Supervision of study including conception, methodology and experimental design. Discussed data-analysis, contributed to preparation of figures and manuscript. Conducted the CDK2 activity analysis in MCF-7. Performer experiments required for the revised manuscript.
C.B: Contributed to study conception and preparation of manuscript.
N.C.T: Overall supervision of study including conception, methodology, and experimental design. Discussed data-analysis, contributed to preparation of figures and manuscript.
Conflict of Interest:
Nicholas C. Turner has received research funding from Pfizer and Roche and is a consultant/advisory board member for Novartis, Pfizer, and Roche. Deepak Sampath, Jennifer Arca Lacap, Lisa Crocker and Amy Young are employees of Genentech, Roche group. The other authors disclosed no potential conflicts of interest.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Asghar U, et al. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14(2):130–46. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finn RS, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16(1):25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 4.Turner NC, et al. Palbociclib in Hormone-Receptor-Positive Advanced Breast Cancer. N Engl J Med. 2015;373(3):209–19. doi: 10.1056/NEJMoa1505270. [DOI] [PubMed] [Google Scholar]
- 5.Finn RS, et al. Palbociclib and Letrozole in Advanced Breast Cancer. N Engl J Med. 2016;375(20):1925–1936. doi: 10.1056/NEJMoa1607303. [DOI] [PubMed] [Google Scholar]
- 6.Hortobagyi GN, et al. Ribociclib as First-Line Therapy for HR-Positive, Advanced Breast Cancer. N Engl J Med. 2016;375(18):1738–1748. doi: 10.1056/NEJMoa1609709. [DOI] [PubMed] [Google Scholar]
- 7.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burstein MD, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21(7):1688–98. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finn RS, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11(5):R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrera-Abreu MT, et al. Early Adaptation and Acquired Resistance to CDK4/6 Inhibition in Estrogen Receptor-Positive Breast Cancer. Cancer Res. 2016;76(8):2301–13. doi: 10.1158/0008-5472.CAN-15-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barr AR, H F, Zhang T, Bakal C, Novak B. A Dynamical Framework for the All-or-None G1/S Transition. Cell Systems. 2016 Jan 27;2:27–37. doi: 10.1016/j.cels.2016.01.001. 2, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spencer SL, C S, Tsai FC, Wesley Overton K, Wang CL, Meyer T. The Proliferation-Quiescence Decision Is Controlled by a Bifurcation in CDK2 Activity at Mitotic Exit. Cell. 2013;155(2):369–383. doi: 10.1016/j.cell.2013.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cappell SD, et al. Irreversible APC(Cdh1) Inactivation Underlies the Point of No Return for Cell-Cycle Entry. Cell. 2016;166(1):167–80. doi: 10.1016/j.cell.2016.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson A, et al. High-Level Clonal FGFR Amplification and Response to FGFR Inhibition in a Translational Clinical Trial. Cancer Discov. 2016 doi: 10.1158/2159-8290.CD-15-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piatkevich KD, et al. Monomeric red fluorescent proteins with a large Stokes shift. Proc Natl Acad Sci U S A. 2010;107(12):5369–74. doi: 10.1073/pnas.0914365107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunning MJ, et al. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23(16):2183–4. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 18.Chen X, et al. TNBCtype: A Subtyping Tool for Triple-Negative Breast Cancer. Cancer Inform. 2012;11:147–56. doi: 10.4137/CIN.S9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Network*, T.C.G.A. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Cam L, et al. Timing of cyclin E gene expression depends on the regulated association of a bipartite repressor element with a novel E2F complex. EMBO J. 1999;18(7):1878–90. doi: 10.1093/emboj/18.7.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elstrodt F, et al. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 2006;66(1):41–5. doi: 10.1158/0008-5472.CAN-05-2853. [DOI] [PubMed] [Google Scholar]
- 22.Barr AR, et al. DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression. Nat Commun. 2017;8:14728. doi: 10.1038/ncomms14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehmann BD, B J, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harbour JW, et al. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98(6):859–69. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 25.Ezhevsky SA, et al. Hypo-phosphorylation of the retinoblastoma protein (pRb) by cyclin D:Cdk4/6 complexes results in active pRb. Proc Natl Acad Sci U S A. 1997;94(20):10699–704. doi: 10.1073/pnas.94.20.10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18(2):753–61. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13(7):417–30. doi: 10.1038/nrclinonc.2016.26. [DOI] [PubMed] [Google Scholar]
- 28.Dulic V, Lees E, Reed SI. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257(5078):1958–61. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 29.Koff A, et al. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257(5077):1689–94. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 30.Taylor-Harding B, et al. Cyclin E1 and RTK/RAS signaling drive CDK inhibitor resistance via activation of E2F and ETS. Oncotarget. 2015;6(2):696–714. doi: 10.18632/oncotarget.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Tables