Reducing dynamin 2 (DNM2) rescues DNM2-related dominant centronuclear myopathy (original) (raw)
Significance
Centronuclear myopathies are rare and severe congenital muscle diseases. Here we hypothesized that reducing dynamin 2 (DNM2) may rescue the pathophysiology observed in _DNM2_-related dominant centronuclear myopathy. The total DNM2 expression was reduced in a faithful murine model (_Dnm2_RW/+ mice) using two different methods targeting both mutated and wild-type Dnm2, adeno-associated virus-shRNA, or antisense oligonucleotides, leading to a restoration of muscle mass, histopathology, and muscle ultrastructural features to wild-type levels. This provides a therapeutic strategy for treating this disease. We also propose that targeting both alleles in dominant diseases due to a mutation in only one of the alleles can successfully rescue the phenotypes.
Keywords: congenital myopathy, myotubular myopathy, dynamin 2, antisense oligonucleotides, adeno-associated virus
Abstract
Centronuclear myopathies (CNM) are a group of severe muscle diseases for which no effective therapy is currently available. We have previously shown that reduction of the large GTPase DNM2 in a mouse model of the X-linked form, due to loss of myotubularin phosphatase MTM1, prevents the development of the skeletal muscle pathophysiology. As DNM2 is mutated in autosomal dominant forms, here we tested whether DNM2 reduction can rescue _DNM2_-related CNM in a knock-in mouse harboring the p.R465W mutation (_Dnm2_RW/+) and displaying a mild CNM phenotype similar to patients with the same mutation. A single intramuscular injection of adeno-associated virus-shRNA targeting Dnm2 resulted in reduction in protein levels 5 wk post injection, with a corresponding improvement in muscle mass and fiber size distribution, as well as an improvement in histopathological CNM features. To establish a systemic treatment, weekly i.p. injections of antisense oligonucleotides targeting Dnm2 were administered to _Dnm2_RW/+mice for 5 wk. While muscle mass, histopathology, and muscle ultrastructure were perturbed in _Dnm2_RW/+mice compared with wild-type mice, these features were indistinguishable from wild-type mice after reducing DNM2. Therefore, DNM2 knockdown via two different strategies can efficiently correct the myopathy due to DNM2 mutations, and it provides a common therapeutic strategy for several forms of centronuclear myopathy. Furthermore, we provide an example of treating a dominant disease by targeting both alleles, suggesting that this strategy may be applied to other dominant diseases.
Centronuclear myopathies (CNM) are rare congenital myopathies characterized by a severe and generalized muscle weakness, associated with fiber hypotrophy (1, 2). Patient muscle biopsies exhibit a characteristic mislocalization of myonuclei internalized within fibers in the absence of excessive regeneration. To date, no specific therapies are available.
Mutations in several genes have been identified to cause CNM, and both the genetic and clinical spectrums of this group of myopathies have rapidly expanded in recent years (3). Mutations in dynamin 2 (DNM2) were first associated with an adult-onset autosomal dominant form of CNM, characterized by a moderate yet progressive muscle weakness (4, 5) [autosomal dominant CNM (ADCNM), Online Mendelian Inheritance in Man (OMIM) 160150]. More recently, DNM2 mutations were associated with severe neonatal-onset cases (6). DNM2 encodes for dynamin 2, a large ubiquitously expressed GTPase mechanoenzyme implicated in membrane remodeling and endocytosis (7–9), as well as cytoskeleton organization (10, 11). Several in vitro studies suggest that DNM2 mutations associated with CNM increase dynamin GTPase activity and oligomerization (12, 13), suggesting that _DNM2_-CNM mutations induce a gain-of-function. This hypothesis is supported by in vivo studies, where overexpression of wild-type DNM2 in wild-type mice recapitulates a CNM-like phenotype (14, 15). A knock-in mouse model of the most common _DNM2_-CNM human mutation, p.R465W, was created. While homozygous mice die at birth, heterozygous R465W knock-in mice (_Dnm2_RW/+) are viable and progressively develop muscle weakness as observed in patients (16). This model recapitulates the myopathic phenotype observed in human disease and provides a valid tool for testing therapeutic strategies.
In addition to DNM2, the main mutated genes linked with CNM are MTM1, encoding the lipid phosphatase myotubularin, for the X-linked form (OMIM 310400) (17); and BIN1, encoding the membrane remodeling protein amphiphysin 2 for autosomal forms (OMIM 255200) (18, 19). Mutations in these genes are believed to be mainly loss-of-function (18, 20, 21), and we have previously shown that reduction of DNM2 expression to 50% by genetic crosses with a Dnm2+/− mouse rescue the lifespan, muscle force, and histopathology of _Mtm1_-/y and _Bin1_−/− mice (20, 22). These results combined suggest BIN1 and MTM1 as negative regulators of DNM2 and identify DNM2 as a potential therapeutic target for these CNM forms. A translated approach was developed using antisense oligonucleotides (ASO) that target the nuclear pre-mRNA of Dnm2. Repeated injections of ASO targeting Dnm2 into _Mtm1_-/y mice efficiently reduced DNM2 and improved lifespan while reducing disease pathology (23). Most recently, using adeno-associated virus to express shRNA targeting Dnm2 in _Mtm1_-/y mice, we demonstrated a robust DNM2 knockdown and disease rescue, providing an alternative strategy to reduce DNM2 (24). Therefore, targeting DNM2 was shown to be a valid potential therapy for _MTM1_-CNM using different approaches.
We hypothesized that reducing DNM2 may also rescue the pathophysiology observed in _DNM2_-related CNM, as observed in the models of MTM1- and _BIN1_-CNM. In this study, we reduced total DNM2 expression in _Dnm2_RW/+ mice through several methodologies and analyzed the impact on CNM pathology and DNM2 function. We highlight here that targeting the total pool of a gene in a nonallele-specific manner efficiently rescued an autosomal dominant myopathy due to gain-of-function mutations.
Results
Reducing DNM2 by Intramuscular Adeno-Associated Virus-shRNA Delivery in _Dnm2_RW/+ Mice Improves Muscle Mass.
Heterozygous R465W knock-in mice (_Dnm2_RW/+) are viable with a normal lifespan and body weight and progressively develop a myopathic phenotype similar to patients (16), whereas the heterozygous knockout Dnm2 +/− mice are viable and do not present with any obvious clinical phenotypes (22). To decrease DNM2 expression, we first crossed Dnm2 +/− mice with _Dnm2_RW/+ mice to produce _Dnm2_RW/− mice expressing only the mutated allele. A strong reduction in the proportion of pups with this genotype was noted (SI Appendix, Table S1; 3% in lieu of expected 25%), with only a few _Dnm2_RW/− mice surviving more than a week and dying before weaning at 3 wk. This indicates that selective loss of the wild-type Dnm2 allele does not rescue the myopathy in _Dnm2_RW/+ mice and thus that a wild-type DNM2 protein is necessary for embryonic and perinatal survival.
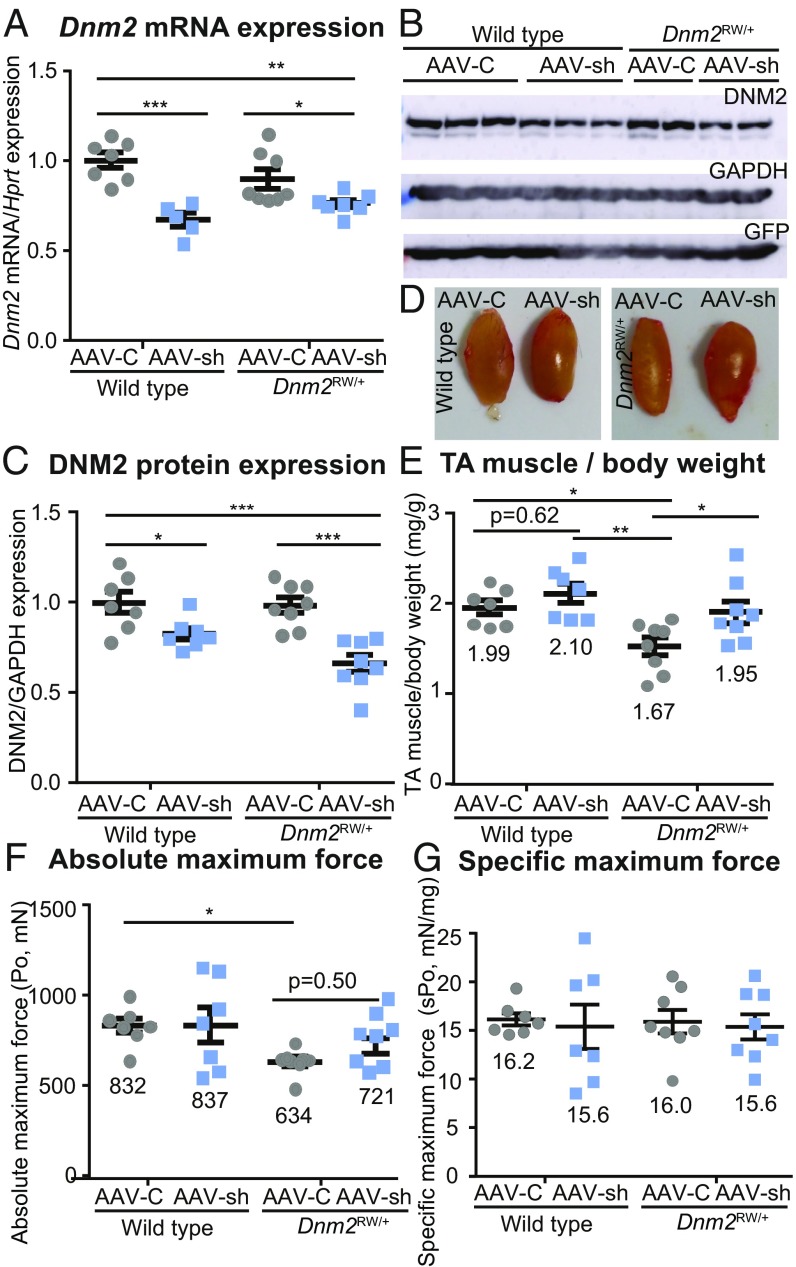
We therefore aimed to reduce the total pool of DNM2 after birth. Adeno-associated virus (AAV) vectors have previously been used effectively to repress gene expression by encoding for shRNA complementary to the target mRNA (25). We previously screened several Dnm2 shRNA sequences that provide robust targeting of Dnm2 mRNA for degradation in vitro and in vivo (24). The best shRNA-Dnm2 sequence (AAV-sh) as well as a scrambled control sequence (AAV-C) were selected and evaluated here for their potential to knock down Dnm2 RNA in the _Dnm2_RW/+ mice. Intramuscular injections of AAV-sh or AAV-C were administered into tibialis anterior (TA) muscles of 3-wk-old wild-type or _Dnm2_RW/+ mice (1.2 × 1010 viral genome per TA) (SI Appendix, Table S2). Mice were killed 5 wk post injection, and DNM2 expression was quantified from TA muscles. Of note, _Dnm2_RW/+ muscle expresses Dnm2 mRNA and protein at the same level as wild-type littermates (Fig. 1 A_–_C), suggesting that the mutation does not alter protein stability. In wild-type and _Dnm2_RW/+ mice treated with AAV-sh, RT-qPCR detected a significant reduction of Dnm2 mRNA expression relative to AAV-C–treated animals (Fig. 1_A_). A corresponding 30–40% reduction in DNM2 protein expression was observed by Western blot (Fig. 1_B_) and densitometry (Fig. 1_C_). Muscle transduction was confirmed by GFP expression from the same AAV constructs (Fig. 1_B_).
Fig. 1.
Reducing DNM2 by intramuscular AAV-shRNA delivery in _Dnm2_RW/+ mice improves muscle mass. (A) Dnm2 mRNA expression quantified by RT-qPCR, relative to Hprt expression, in wild-type and DNM2 R465W knock-in (_Dnm2_RW/+) mice, treated with AAV-shRNA targeting Dnm2 (AAV-sh) or AAV-scrambled control (AAV-C). (B) Immunoblot for protein expression of DNM2, GAPDH (protein loading control), and GFP (AAV-control). (C) DNM2 protein expression quantified relative to GAPDH loading control. (D) TA representative images. (E) TA muscle mass from wild-type and _Dnm2_RW/+ mice, treated with AAV-sh or AAV-C as a ratio to body weight. (F) Absolute maximum force (Po) and (G) specific maximum force (sPo) (relative to TA muscle weight). Graphs represent mean ± SEM. Each point represents one mouse; n > 5 per group. *P < 0.05, **P < 0.01, ***P < 0.001.
To determine if reducing DNM2 has an impact on the phenotypes, muscle mass and force were analyzed. While wild-type muscles appeared similar in size regardless of the AAV construct injected (Fig. 1_D_, Left, and Fig. 1_E_), muscle mass was clearly reduced in _Dnm2_RW/+ muscles (Fig. 1_D_, AAV-C, Right, and Fig. 1_E_), in accordance with previously published data (16). Analysis of the TA muscle mass/body-weight ratio showed that the reduction of TA muscle mass observed in _Dnm2_RW/+ mice was rescued to a wild-type level by reducing DNM2 expression (Fig. 1 D and E). Absolute TA muscle force was significantly reduced in _Dnm2_RW/+ mice (Fig. 1_F_). A trend toward improvement was observed when mice were treated with AAV-sh, although this was not statistically significant. No difference in specific muscle force (absolute force relative to muscle mass) or in time to fatigue was observed (Fig. 1_G_ and SI Appendix, Table S3). These results indicate that shRNA can efficiently reduce Dnm2 mRNA and protein expression in vivo in _Dnm2_RW/+ mice and show that reducing DNM2 after a single intramuscular injection of AAV-sh targeting Dnm2 efficiently improves muscle mass in this model of dominant CNM.
Reducing DNM2 by AAV-shRNA in _Dnm2_RW/+ Mice Corrects Histological Defects.
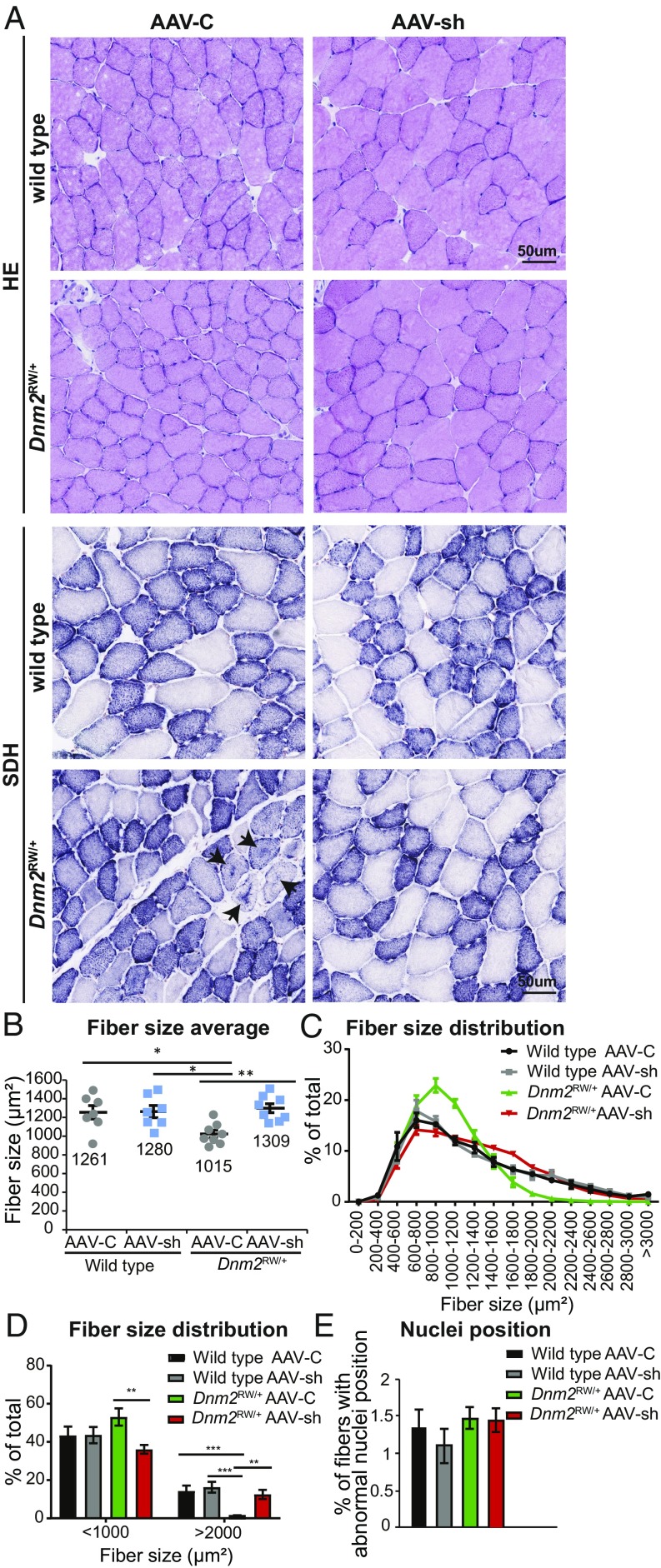
To determine if the functional improvement in _Dnm2_RW/+ muscle force was linked to an amelioration of muscle histology, transversal TA sections were stained with hematoxylin and eosin (H&E) and succinate dehydrogenase (SDH) (Fig. 2_A_, Upper and Lower, respectively). A significant reduction in fiber size was observed in _Dnm2_RW/+ muscles compared with wild type (Fig. 2 A and B), with a reduction in the size heterogeneity normally observed in TA fibers (Fig. 2 C and D). Fiber size and distribution were restored to wild-type levels by treatment with AAV-sh targeting Dnm2. Unlike patients, no mislocalization of nuclei was observed in _Dnm2_RW/+ mice (Fig. 2_E_), as described previously (16), and the absence of myofiber degeneration and centralized nuclei confirms that AAV treatment was not toxic to the muscles. The most striking histological abnormality previously observed in these mice was the abnormal central accumulation of SDH (labeling mainly mitochondria oxidative activity) within fibers. Here we confirm that observation, with ∼5% of fibers with abnormal SDH staining in _Dnm2_RW/+ muscles injected with AAV-C (Fig. 2_A_, arrows; SI Appendix, Table S4). This was dramatically improved in _Dnm2_RW/+ mice with reduced DNM2, with almost all fibers displaying SDH staining akin to wild-type muscle. Therefore, reducing DNM2 by AAV-shRNA intramuscular injection in _Dnm2_RW/+ mice ameliorates fiber size and distribution of oxidative activity to wild-type levels. Taken together with the improvement in muscle mass and function, reducing DNM2 by intramuscular injections of AAV-shRNA was able to fully rescue the CNM phenotype in TA muscles from the _Dnm2_RW/+ mouse model.
Fig. 2.
Reducing DNM2 by AAV-shRNA in _Dnm2_RW/+ mice corrects histological features of disease. (A) Transverse muscle sections from wild-type and DNM2 R465W knock-in (_Dnm2_RW/+) mice, treated with AAV-shRNA targeting Dnm2 (AAV-sh) or AAV-scrambled control (AAV-C), stained with H&E or SDH. Arrows indicate abnormal central accumulation of oxidative staining. (Scale bar: 50 µm.) (B) Fiber size average and (C and D) fiber size distribution were quantified from H&E images in A. (E) Fibers with internal or centralized nuclei (“abnormal nuclei position”) were quantified from H&E images in A. n = 7–8 mice per group. (B_–_E) More than 500 fibers per mouse were analyzed. Graphs represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Systemic Reduction of DNM2 by ASO Improves Muscle Mass in _Dnm2_RW/+ Mice.
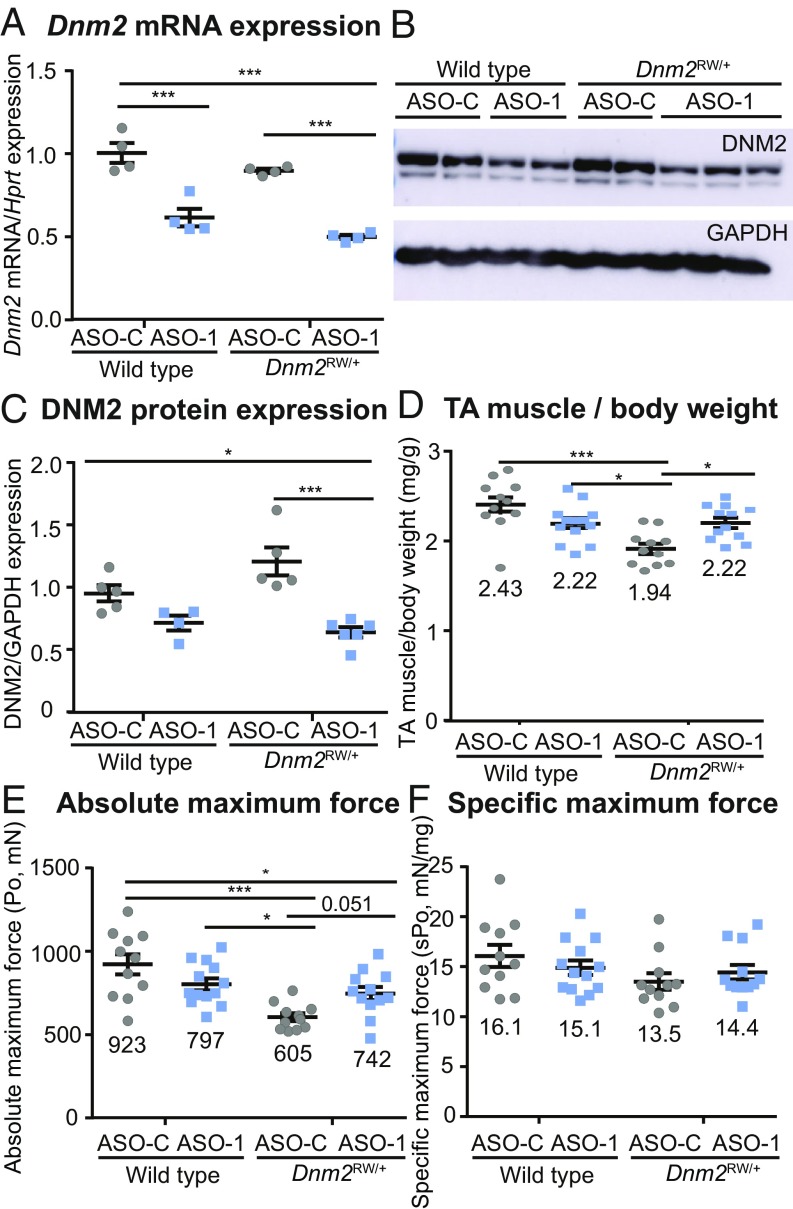
As shRNA targeting Dnm2 was able to locally rescue the CNM phenotype, we next tested a second approach to reduce DNM2, to confirm its therapeutic potential as a therapeutic target for _DNM2_-related CNM and validate a systemic treatment. ASO-mediated knockdown represents another promising therapeutic approach for neuromuscular diseases. We recently found that i.p. injections of ASO targeting Dnm2 in _Mtm1_-/y mice result in improved lifespan, muscle force, and histology (23). Here we applied a similar approach to _Dnm2_RW/+ mice. To assess if systemic ASO delivery could rescue the CNM phenotype, weekly injections of 25 mg/kg of ASO targeting Dnm2 (ASO-1) were administered to wild-type or _Dnm2_RW/+ mice from 3 to 7 wk of age by i.p. injections, and these were compared with littermate mice treated with 25 mg/kg of ASO control (not targeting any known mouse genes, ASO-C) (SI Appendix, Table S2). Of note, we targeted a different sequence and exon compared with the shRNA approach as a way to control for potential off-target effects. We observed a similar concentration of ASO uptake in wild-type and _Dnm2_RW/+ mouse muscle tissue, indicating efficient delivery to the target tissue (SI Appendix, Table S5). The genotypes did not alter the ASO uptake. Notably, and similarly to the AAV-shRNA intramuscular approach, ASO systemic administration resulted in an ∼50% reduction in Dnm2 RNA and protein expression in muscle (Fig. 3 A_–_C). A significant improvement in TA muscle mass relative to body weight, and a nonsignificant improvement in absolute muscle force (P = 0.051), was observed in ASO-1–treated _Dnm2_RW/+ compared with mice treated with ASO-C (Fig. 3 D and E). Importantly, these values were indistinguishable from wild-type mice treated with ASO-1. Again, no significant difference in specific maximal force or relaxation time was observed between groups (Fig. 3_F_ and SI Appendix, Table S3). Therefore, ASO-1 administration can efficiently reduce Dnm2 mRNA and protein expression in vivo in _Dnm2_RW/+ mice, and this correlated with normalization of muscle mass.
Fig. 3.
Reducing DNM2 by systemic ASO injection in _Dnm2_RW/+ mice improves muscle mass. (A) Dnm2 mRNA expression quantified by RT-qPCR analysis, relative to Hprt expression, in wild-type and DNM2 R465W knock-in (_Dnm2_RW/+) mice, treated with ASO-1–targeting Dnm2 (ASO-1) or ASO-control (ASO-C). (B) Immunoblot for protein expression of DNM2 and GAPDH (protein loading control). (C) DNM2 protein expression quantified relative to GAPDH loading control. (D) TA muscle mass from wild-type and _Dnm2_RW/+ mice, treated with ASO-1 or ASO-C, represented as a ratio to body weight. (E) Absolute maximum force (Po) and (F) specific maximum force (sPo) (relative to TA muscle weight). Each point represents one mouse; n ≥ 5 per group. Graphs represent mean ± SEM. *P < 0.05, ***P < 0.001.
Systemic ASO Treatment in _Dnm2_RW/+ Mice Corrects Fiber Size and Histological Abnormalities.
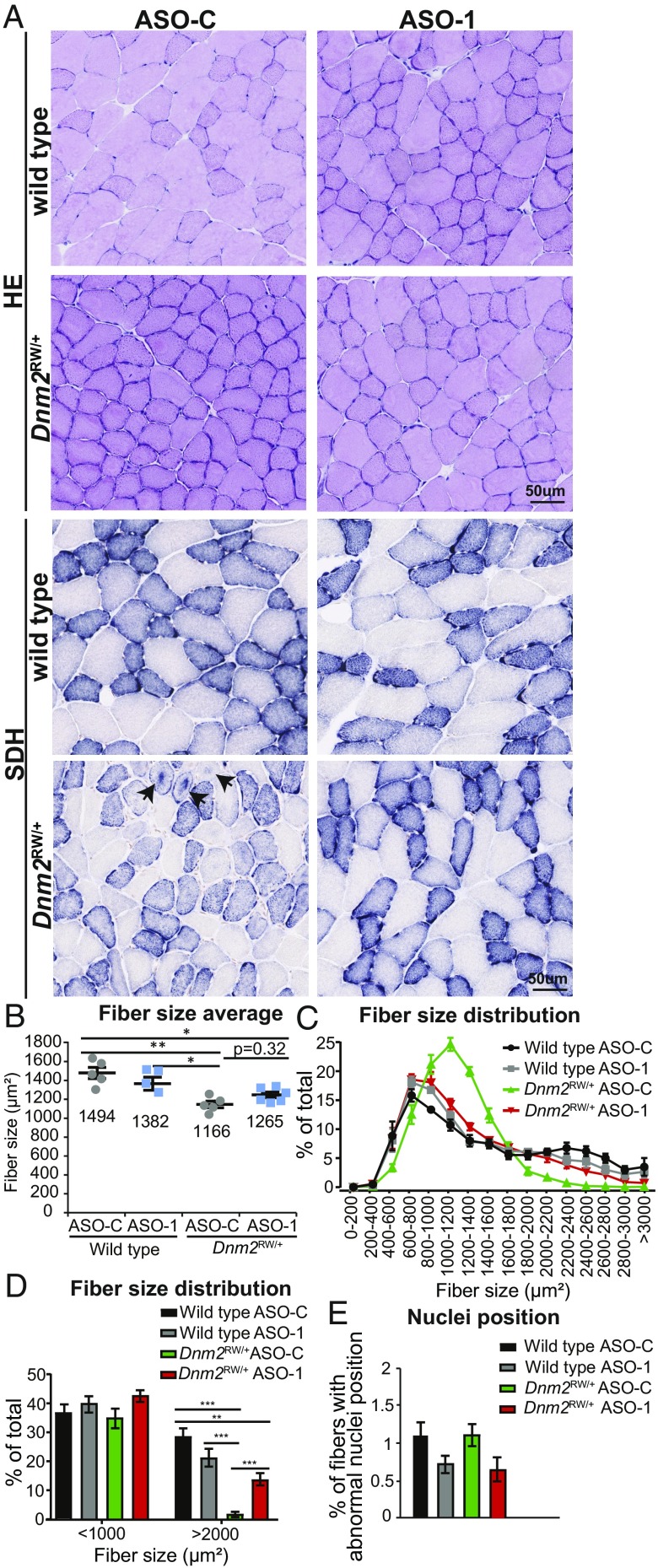
To determine if an amelioration in muscle histopathology was also observed by systemic ASO-1 administration, transversal TA sections were stained with H&E and SDH (Fig. 4_A_, Upper and Lower, respectively). A reduced mean fiber size was observed in _Dnm2_RW/+ muscles compared with wild type and was not corrected by administration of ASO-1 targeting Dnm2 (Fig. 4 A and B). However, the fiber size distribution was clearly restored to the wild-type distribution by administration of ASO-1 targeting Dnm2 (Fig. 4 C and D). This is explained by the fact that the _Dnm2_RW/+ muscles have an abnormal enrichment of middle-sized fibers while the wild-type and ASO-1–treated _Dnm2_RW/+ muscles display the typical heterogeneous fiber distribution with small, medium, and large fibers; indeed, the absence of large fibers in the _Dnm2_RW/+ muscle was strongly rescued by lowering DNM2 (Fig. 4 C and D). Nuclei position was unaffected in _Dnm2_RW/+ mice (Fig. 4_E_). The abnormal accumulation of SDH centrally within _Dnm2_RW/+ fibers was confirmed, and this was completely rescued in _Dnm2_RW/+ mice with reduced DNM2 (Fig. 4_A_, Lower, and SI Appendix, Table S4). Therefore, reducing DNM2 by systemic ASO injection rescues the physiological and histological defects due to the Dnm2 mutation to wild-type levels.
Fig. 4.
Reducing DNM2 by systemic ASO injection in _Dnm2_RW/+ mice corrects histological features of disease. (A) Transverse muscle sections from wild-type and DNM2 R465W knock-in (_Dnm2_RW/+) mice, treated with ASO-1 targeting Dnm2 (ASO-1) or ASO-control (ASO-C), stained with H&E or SDH. Arrows indicate abnormal central accumulation of oxidative staining. (Scale bar: 50 µm.) (B) Fiber size average and (C and D) fiber size distribution were quantified from H&E images in A. (E) Fibers with internal or centralized nuclei (“abnormal nuclei position”) were quantified from H&E images in A. n = 5–6 mice per group. (B_–_E) More than 500 fibers per mouse were analyzed. Graphs represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Ultrastructure Morphology of _Dnm2_RW/+ Muscles After DNM2 Reduction by ASO.
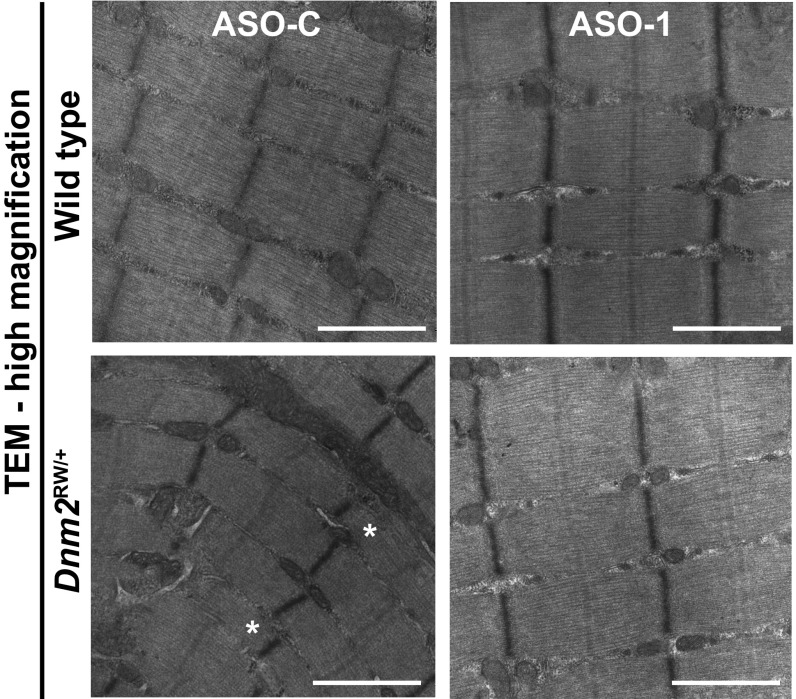
We next analyzed TA muscles at the ultrastructural level by transmission electron microscopy. Longitudinal images of _Dnm2_RW/+ muscles presented overall well-organized fibers, with a notable reduction in myofibril width (Fig. 5, asterisks; SI Appendix, Fig. S1_A_), as observed previously (16). Measurement of sarcomere length and myofibril width confirmed a tendency for the _Dnm2_RW/+ muscles to have thinner myofibrils (SI Appendix, Fig. S1 B and C). This feature was ameliorated by ASO-1 administration targeting Dnm2.
Fig. 5.
Ultrastructure morphology of _Dnm2_RW/+ muscles after reducing DNM2 by systemic ASO injection. TA ultrastructure in wild-type and _Dnm2_RW/+ mice, treated with ASO control (ASO-C) or ASO targeting Dnm2 (ASO-1) at high magnification. (Scale bar: 1 µm.) Low-magnification images can be found in SI Appendix. Images are representative of two mice per genotype. Asterisks (*) indicate abnormally small myofibrils that were not observed in _Dnm2_RW/+ after ASO-1 administration.
Structural and Functional Assessment of Isolated Fibers.
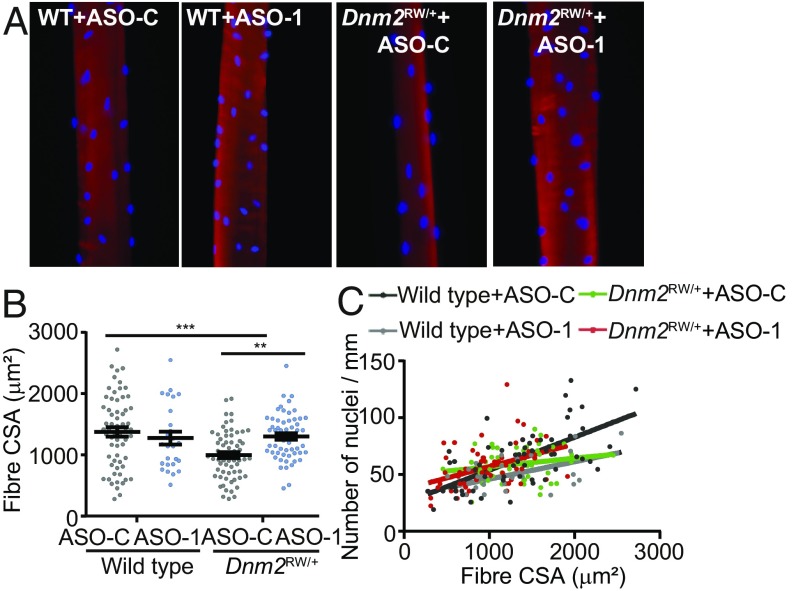
To explore further the impact of the DNM2 mutation on myofiber function and the rescue efficacy of DNM2 down-regulation, fibers were isolated from the TA muscles, skinned, and stained for actin (phalloidin, red) and nuclei (DAPI, blue) (Fig. 6_A_). Individual fiber size and force were analyzed. No difference in specific force produced from isolated fibers relative to the cross-sectional area was observed (SI Appendix, Table S6) in wild-type vs. _Dnm2_RW/+ fibers, confirming our whole muscle analysis (Fig. 3_F_). However, the cross-sectional area of isolated fibers was significantly reduced in _Dnm2_RW/+ mice, and this was fully rescued by administration of ASO-1 (Fig. 6_B_), confirming analysis of fiber size distribution from whole TA muscles (Fig. 4_C_). In addition, when comparing the nuclear number to fiber size, no obvious abnormality was noted in _Dnm2_RW/+ mice (Fig. 6_C_). Overall, the combination of in vitro fiber analysis and in vivo muscle analysis supports that _Dnm2_RW/+ muscles lack large myofibers and that their fibers are usually smaller with reduced myofibril size, leading to a decrease in overall muscle mass and force. Isolated fiber analysis confirms that reducing DNM2 by ASO in _Dnm2_RW/+ muscles corrects fiber hypotrophy to wild-type levels.
Fig. 6.
ASO treatment in _Dnm2_RW/+ mice improves fiber size in isolated fibers. (A) TA fibers from wild type (WT) and _Dnm2_RW/+ mice, treated with ASO targeting Dnm2 (ASO-1) or ASO-control (ASO-C), were isolated, skinned, and stained for actin (phalloidin, red) and nuclei position (DAPI, blue). (B) Cross-sectional area of isolated fibers (µm2), estimated from fiber width and depth. (C) Comparison of the nuclei position (number of nuclei/mm), relative to fiber size (µm2). A total of 18–22 fibers per genotype were analyzed. A one-way ANOVA statistical test was performed. Graphs represent mean ± SEM, with individual values shown. **P < 0.01, ***P < 0.001.
Discussion
In this study, we demonstrate that DNM2 knockdown, through either AAV intramuscular or ASO systemic delivery, can correct the CNM phenotype observed in _Dnm2_RW/+ mice by restoring muscle mass and structure. These results provide insights on the impact of DNM2 mutations in CNM and support a therapeutic application.
Dominant mutations in the DNM2 gene lead to centronuclear myopathy in patients and in the mouse model (4, 16). Approximately 30% of ADCNM patients with DNM2 mutations present with the single-point p.R465W mutation in exon 11 (c.1393 A > T, p.R465W). While DNM2 expression is not increased in _Dnm2_RW/+ mice (Fig. 1), in vitro experiments showed that several _DNM2_-CNM mutations, including p.R465W, increase oligomerization and GTPase activity (12, 13). However, based on these in vitro data, it was unclear if these alterations lead to a gain-of-function or to a final impaired activity of the protein. In vivo work demonstrated that increased expression of wild-type DNM2 in mice promotes a CNM-like phenotype with muscle defects and centralization of nuclei, supporting that _DNM2_-CNM mutations are, at least in part, gain-of-function (14, 15). Based on this rationale, we targeted the overall DNM2 expression by two different approaches: shRNA and ASO-mediated knockdown. Importantly, we achieved knockdown of DNM2 to ∼50% at the RNA and protein levels by targeting both the mutated and wild-type allele, leading to a rescue in the CNM phenotype. These results support the hypothesis of a gain-of-function mechanism in the p.R465W _DNM2_-related CNM, as overall reduction of DNM2 is sufficient to rescue CNM features in mice. However, as knockout of wild-type Dnm2 in _Dnm2_RW mice is not viable (SI Appendix, Table S1), this suggests that the pathomechanisms may be more complex and may vary for different DNM2 mutations or that some wild-type DNM2 protein is needed for embryogenesis. We cannot also exclude an additive mechanism between a gain-of-function and a partial dominant-negative effect of CNM mutations. The role of DNM2 in muscle and the pathological mechanism of CNM mutations are not fully understood. Here, we provide data suggesting that DNM2 is important for fiber hypertrophy, most probably during postnatal development, as we noted a strong reduction in large fibers in the _Dnm2_RW/+ muscles, associated with a decrease in absolute force, when analyzing the mice at the end of the muscle postnatal development (8 wk). This is confirmed by the fact that decreasing DNM2 in a time window from 3 to 8 wk, corresponding to the period of postnatal muscle hypertrophy, rescued these phenotypes. However, the finding that specific force of isolated fibers and entire muscles is normal in _Dnm2_RW/+ mice suggests that the DNM2 mutation impairs myofiber hypertrophy but not muscle contraction, correlating with the rather mild myopathy associated with this mutation in patients, which could then be due mainly to fiber atrophy rather than contractile defect.
While complete loss of Dnm2 is embryonically lethal, Dnm2 heterozygous knockout mice do not present any obvious clinical phenotype (22, 26), supporting reducing total levels of DNM2 as a therapeutic strategy without deleterious impact. Recently, allele-specific knockdown targeting the p.R465W point mutation has shown therapeutic potential in the _Dnm2_RW/+ model (27). As only one allele is targeted in this approach, it may reduce potential effects of a massive inhibition of the DNM2 pool; however, it would require sufficient muscle targeting, and whether the allele specificity can be maintained in a clinical setting is unknown. Here we propose two approaches, adeno-associated virus or antisense oligonucleotides, to achieve reduction of the total DNM2 pool. DNM2 reduction by ASO allows injection and dosing to be adapted during the treatment. As ASOs from the same platform have now been accepted for use clinically for familial hypercholesterolemia [e.g., Mipomersen (Kynamro)] and for spinal muscular atrophy [Nursinersen (Spinraza)], this approach has clear advantages for clinical development. Alternatively, AAV provides good muscle tropism and long-term expression following a single injection. Several challenges remain, as enough vector or ASO must be produced and delivered to skeletal muscles without on-target toxicity or off-target effects, with the long-term benefit outweighing potential risks.
Interestingly, DNM2 protein expression is increased in X-linked CNM biopsies from patient muscles and in muscles from the Mtm1 -/y mouse model, and reducing Dnm2 also rescued disease features in this model (22, 23). Albeit a clear link to altered DNM2 expression in patients with BIN1 mutations has not yet been established, down-regulation of DNM2 level to 50% through a genetic cross in the _Bin1_−/− mouse also rescued the lifespan and muscle symptoms (20). Based on these different results and the present study, we hypothesize that the increase in DNM2 expression or function may be a key factor leading to the CNM phenotype in muscle in all these CNM forms, independent of the mutated genes. Our data validate the importance of DNM2 expression in CNM, and we propose that the form of centronuclear myopathy due to mutations in DNM2 may be treated by targeting the total pool of DNM2. DNM2 reduction is a strategy that may have therapeutic potential for several forms of centronuclear myopathies. Targeting both the wild-type and mutated allele without distinction, as was successfully done here, represents a different paradigm from the allele-specific therapies that were preferred to date in dominant diseases and may be tested in other diseases.
Materials and Methods
Materials.
Primary antibodies used were against glyceraldehyde-3-phosphate dehydrogenase (GAPDH, MAB374; Chemicon), rabbit anti-DNM2 (R2865) (14), and mouse anti-GFP antibodies made onsite at the antibody facilities of Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC). Secondary antibodies against mouse and rabbit IgG, conjugated with horseradish peroxidase, were purchased from Jackson ImmunoResearch Laboratories. Rhodamine phalloidin, TRITC-conjugated R415, and DAPI nuclear stain (D3571) were purchased from Molecular Probes. An ECL chemiluminescent reaction kit was purchased from Pierce. TRIzol reagent was purchased from MRCGENE. Superscript II reverse transcriptase was purchased from Invitrogen.
Generation of _Dnm2_RW/+ Mice.
This mouse line was created at the Mouse Clinical Institute (www.ics-mci.fr), as described previously (16). Further details can be found in SI Appendix, Supplementary Methods.
Animal Experiments.
Animals were housed in a temperature-controlled room (19–22 °C) with a 12:12-h light/dark cycle with free access to food. Animal experimentation was approved by the institutional ethical committee Com’Eth IGBMC-Institut Clinique de la Souris for 2017–5453 (ASO work), 2012–132/2017–5640 (AAV work), and 2013–034/01594.02 (Aurora in situ muscle contractile properties). Mice were humanely killed when required according to national and European legislations on animal experimentation. Male mice were analyzed in this study.
Statistics.
Statistical analysis was performed using a two-way ANOVA followed by post hoc Bonferroni unless otherwise stated. P values of <0.05 were considered significant.
Additional methods can be found in SI Appendix.
Supplementary Material
Supplementary File
Acknowledgments
We thank Arnaud Ferry and Nadia Messaddeq for excellent technical assistance; and the animal house, histology platform, and imaging center of the IGBMC for support. This study was supported by the Grant ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the frame program Investissements d’Avenir ANR-10-IDEX-0002-02. This study was also supported by INSERM, CNRS, the University of Strasbourg, the Agence Nationale de la Recherche (14-CE12-0009), and Société d’Accélération du Transfert de Technologies (SATT) Conectus Alsace (2014) (J.L. and B.S.C.), and grants from the Medical Research Council (MR/N002768/1) (to J.O.).
Footnotes
Conflict of interest statement: H.T., J.L., and B.S.C. are inventors of a patent on targeting DNM2 for the treatment of centronuclear myopathies. J.L. and B.S.C. are scientific advisors for Dynacure.
This article is a PNAS Direct Submission.
References
- 1.Romero NB. Centronuclear myopathies: A widening concept. Neuromuscul Disord. 2010;20:223–228. doi: 10.1016/j.nmd.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Jungbluth H, Wallgren-Pettersson C, Laporte J. Centronuclear (myotubular) myopathy. Orphanet J Rare Dis. 2008;3:26. doi: 10.1186/1750-1172-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jungbluth H, et al. Congenital myopathies: Disorders of excitation-contraction coupling and muscle contraction. Nat Rev Neurol. 2018;14:151–167. doi: 10.1038/nrneurol.2017.191. [DOI] [PubMed] [Google Scholar]
- 4.Bitoun M, et al. Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat Genet. 2005;37:1207–1209. doi: 10.1038/ng1657. [DOI] [PubMed] [Google Scholar]
- 5.Böhm J, et al. Mutation spectrum in the large GTPase dynamin 2, and genotype-phenotype correlation in autosomal dominant centronuclear myopathy. Hum Mutat. 2012;33:949–959. doi: 10.1002/humu.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bitoun M, et al. Dynamin 2 mutations cause sporadic centronuclear myopathy with neonatal onset. Ann Neurol. 2007;62:666–670. doi: 10.1002/ana.21235. [DOI] [PubMed] [Google Scholar]
- 7.Warnock DE, Baba T, Schmid SL. Ubiquitously expressed dynamin-II has a higher intrinsic GTPase activity and a greater propensity for self-assembly than neuronal dynamin-I. Mol Biol Cell. 1997;8:2553–2562. doi: 10.1091/mbc.8.12.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Bliek AM, Meyerowitz EM. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- 9.Antonny B, et al. Membrane fission by dynamin: What we know and what we need to know. EMBO J. 2016;35:2270–2284. doi: 10.15252/embj.201694613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ochoa GC, et al. A functional link between dynamin and the actin cytoskeleton at podosomes. J Cell Biol. 2000;150:377–389. doi: 10.1083/jcb.150.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu C, et al. Direct dynamin-actin interactions regulate the actin cytoskeleton. EMBO J. 2010;29:3593–3606. doi: 10.1038/emboj.2010.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenniston JA, Lemmon MA. Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients. EMBO J. 2010;29:3054–3067. doi: 10.1038/emboj.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, et al. Dynamin 2 mutants linked to centronuclear myopathies form abnormally stable polymers. J Biol Chem. 2010;285:22753–22757. doi: 10.1074/jbc.C110.130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowling BS, et al. Increased expression of wild-type or a centronuclear myopathy mutant of dynamin 2 in skeletal muscle of adult mice leads to structural defects and muscle weakness. Am J Pathol. 2011;178:2224–2235. doi: 10.1016/j.ajpath.2011.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu N, et al. Mice lacking microRNA 133a develop dynamin 2–dependent centronuclear myopathy. J Clin Invest. 2011;121:3258–3268. doi: 10.1172/JCI46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durieux AC, et al. A centronuclear myopathy-dynamin 2 mutation impairs skeletal muscle structure and function in mice. Hum Mol Genet. 2010;19:4820–4836. doi: 10.1093/hmg/ddq413. [DOI] [PubMed] [Google Scholar]
- 17.Laporte J, et al. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat Genet. 1996;13:175–182. doi: 10.1038/ng0696-175. [DOI] [PubMed] [Google Scholar]
- 18.Nicot AS, et al. Mutations in amphiphysin 2 (BIN1) disrupt interaction with dynamin 2 and cause autosomal recessive centronuclear myopathy. Nat Genet. 2007;39:1134–1139. doi: 10.1038/ng2086. [DOI] [PubMed] [Google Scholar]
- 19.Böhm J, et al. Adult-onset autosomal dominant centronuclear myopathy due to BIN1 mutations. Brain. 2014;137:3160–3170. doi: 10.1093/brain/awu272. [DOI] [PubMed] [Google Scholar]
- 20.Cowling BS, et al. Amphiphysin (BIN1) negatively regulates dynamin 2 for normal muscle maturation. J Clin Invest. 2017;127:4477–4487. doi: 10.1172/JCI90542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laporte J, Kress W, Mandel JL. Diagnosis of X-linked myotubular myopathy by detection of myotubularin. Ann Neurol. 2001;50:42–46. doi: 10.1002/ana.1033. [DOI] [PubMed] [Google Scholar]
- 22.Cowling BS, et al. Reducing dynamin 2 expression rescues X-linked centronuclear myopathy. J Clin Invest. 2014;124:1350–1363. doi: 10.1172/JCI71206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tasfaout H, et al. Antisense oligonucleotide-mediated Dnm2 knockdown prevents and reverts myotubular myopathy in mice. Nat Commun. 2017;8:15661. doi: 10.1038/ncomms15661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tasfaout H, et al. Single intramuscular injection of AAV-shRNA reduces DNM2 and prevents myotubular myopathy in mice. Mol Ther. 2018;26:1082–1092. doi: 10.1016/j.ymthe.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimm D, Pandey K, Kay MA. Adeno-associated virus vectors for short hairpin RNA expression. Methods Enzymol. 2005;392:381–405. doi: 10.1016/S0076-6879(04)92023-X. [DOI] [PubMed] [Google Scholar]
- 26.Ferguson SM, et al. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell. 2009;17:811–822, and erratum (2010) 18:332. doi: 10.1016/j.devcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trochet D, et al. Allele-specific silencing therapy for Dynamin 2-related dominant centronuclear myopathy. EMBO Mol Med. 2017;10:239–253. doi: 10.15252/emmm.201707988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File