Proline-Rich Synapse-Associated Protein-1/Cortactin Binding Protein 1 (ProSAP1/CortBP1) Is a PDZ-Domain Protein Highly Enriched in the Postsynaptic Density (original) (raw)
Abstract
The postsynaptic density (PSD) is crucially involved in the structural and functional organization of the postsynaptic neurotransmitter reception apparatus. Using antisera against rat brain synaptic junctional protein preparations, we isolated cDNAs coding for proline-rich synapse-associated protein-1 (ProSAP1), a PDZ-domain protein. This protein was found to be identical to the recently described cortactin-binding protein-1 (CortBP1). Homology screening identified a related protein, ProSAP2. Specific antisera raised against a C-terminal fusion construct and a central part of ProSAP1 detect a cluster of immunoreactive bands of 180 kDa in the particulate fraction of rat brain homogenates that copurify with the PSD fraction. Transcripts and immunoreactivity are widely distributed in the brain and are upregulated during the period of synapse formation in the brain. In addition, two short N-terminal insertions are detected; they are differentially regulated during brain development. Confocal microscopy of hippocampal neurons showed that ProSAP1 is predominantly localized in synapses, and immunoelectron microscopy in situ revealed a strong association with PSDs of hippocampal excitatory synapses. The accumulation of ProSAP1 at synaptic structures was analyzed in the developing cerebral cortex. During early postnatal development, strong immunoreactivity is detectable in neurites and somata, whereas from postnatal day 10 (P10) onward a punctate staining is observed. At the ultrastructural level, the immunoreactivity accumulates at developing PSDs starting from P8. Both interaction with the actin-binding protein cortactin and early appearance at postsynaptic sites suggest that ProSAP1/CortBP1 may be involved in the assembly of the PSD during neuronal differentiation.
Keywords: rat brain, synapse, postsynaptic density (PSD), PDZ domain, synaptogenesis, actin-based cytoskeleton, development, synamon
The postsynaptic density (PSD) is a specialized structure beneath the postsynaptic membrane that is crucially involved in the organization of the neurotransmitter receptive apparatus and the adhesion of the postsynapse to presynaptic terminals (Ziff, 1997). It constitutes a structural matrix that anchors and clusters neurotransmitter receptors, synaptic cell adhesion molecules, and components of intracellular signaling pathways exactly opposite to the neurotransmitter release site and thus must be considered as a general organizer of the postsynaptic signal transduction machinery (Ziff, 1997; Kennedy, 1998; O’Brien et al., 1998). It has been suggested that important mechanisms of synaptic plasticity, including activity-dependent changes in postsynaptic signal transduction, may have a structural basis in the PSD (Siekevitz, 1985;Lismann and Goldring, 1988). Consequently, major efforts have been undertaken to identify the protein components of this structure (Walsh and Kuruc, 1992; Langnaese et al., 1996; Kennedy, 1998).
Cytoskeletal elements including actin (Fifkova and Morales, 1992), fodrin (Carlin et al., 1983), or α-actinin-2 (Wyszynski et al., 1997) are abundant components of PSDs. This suggests that the actin-based cytoskeleton constitutes the basic filamentous meshwork of the postsynaptic cytomatrix and plays an important role in the organization of PSDs (Adam and Matus, 1996).
A number of PDZ-domain proteins have been identified as central elements for clustering synaptic membrane proteins at the postsynapse and anchoring them to the cytoskeleton. For example, membrane-associated guanylate kinases SAP90/PSD-95, Chapsyn/PSD-93, SAP97, SAP102, and Drosophila DlgA bind and cluster ion channels, including NMDA, kainate receptors, and K+channels, and cell adhesion molecules, including neuroligin and fasciclin II, as well as ephrins and their receptors at synaptic sites (Kim et al., 1995, 1996; Kornau et al., 1995; Lau et al., 1996;Müller et al., 1996; Irie et al., 1997; Tejedor et al., 1997;Thomas et al., 1997; Zito et al., 1997; Garcia et al., 1998; Torres et al., 1998). In addition, PDZ domains of these proteins recruit intracellular signaling proteins, like neuronal nitric oxide-synthase or SynGAP, to the postsynaptic membrane (Brenman et al., 1996a,b; Chen et al., 1998; Kim et al., 1998). Several other PDZ-domain proteins, e.g., GRIP (Dong et al., 1997), ABP (Srivastava et al., 1998), CCIP (Kurschner et al., 1998), or S-SCAM (Hirao et al., 1998), were also reported to be involved in the assembly of postsynaptic structures.
Using antisera against synaptic protein preparations, we cloned a collection of cDNAs by expression screening encoding components of central synapses (Kistner et al., 1993; Langnaese et al., 1996). Here we report on one of these components, the proline-rich synapse-associated protein-1 (ProSAP1). ProSAP1 is a 180 kDa protein that carries a PDZ domain at its N terminus. Biochemical analysis and ultrastructural localization studies showed that ProSAP1 is a component of the PSD. Multiple developmentally and spatially regulated processing variants code for ProSAP1 isoforms, one of which is identical to the cortactin-binding protein-1 (CortBP1) recently identified as a Cortactin SH3 domain interacting protein (Du et al., 1998). Electron microscopic localization studies on the developing cortex suggest a role for ProSAP1 in early steps of synaptic assembly.
MATERIALS AND METHODS
Cloning of rat ProSAP1 cDNA. The initial cDNA clone sap24e was isolated from a λlgt11 expression library with polyclonal antibodies generated against a rat brain synaptic junction preparation as described previously (Kistner et al., 1993; Langnaese et al., 1996). Overlapping cDNA clones were obtained by several rounds of screening of λZAPII (Stratagene, La Jolla, CA) rat hippocampal and total brain cDNA libraries with the 32P-labeled cDNA fragments.
Antibody production. Partial cDNAs of the ProSAP1 cDNA [encoding amino acids (aa) 355–509 and aa 826–1259] were subcloned into the bacterial expression vector pGEX-1T (Pharmacia Biotech, Uppsala, Sweden). A 45 and a 95 kDa glutathione_S_-transferase (GST)-ProSAP1 fusion protein was expressed in_Escherichia coli_ XL 1 Blue and purified on glutathione-Sepharose 4B as recommended by the manufacturer (Pharmacia Biotech). The fusion proteins were used to generate ProSAP1 antibodies in guinea pigs and rabbits.
Immunohistochemistry. Immunocytochemical staining was performed using 7 μm microtome sections from rat brains, which were fixed by immersion in Bouin’s fluid for 48 hr, dehydrated, and embedded in paraplast. ProSAP1 was detected with the C-terminal rabbit anti-ProSAP1 polyclonal antibody diluted 1:600 using the peroxidase–anti-peroxidase method (Sternberger et al., 1970). Antibody binding was visualized by incubating sections first with porcine anti-rabbit IgG (Dako, Hamburg, Germany) for 30 min and with rabbit peroxidase–anti-peroxidase complex (Dako) for 30 min. Subsequently, the detection reagent 3,3-diaminobenzidine/H2O2(DAB) (Sigma, Munich, Germany) was applied. Some sections were counterstained with hematoxylin for morphological orientation. Controls were performed as follows: (1) preabsorption of the antibody with the antigen and (2) omitting the primary or secondary antibody. No staining was observed under either of these conditions.
Electron microscopy was performed using vibratome sections (50 μm) from rat brains fixed by perfusion with Karnowsky’s solution, i.e., 2.2% glutaraldehyde, 2% paraformaldehyde in 0.1 mphosphate buffer, pH 7.35. Cortical tissue from rat brains at postnatal day 5 (P5), P8, and P10 was fixed by immersion with the same fixative. Floating sections were stained with the Vectastain ABC Staining Kit (Vector, Burlingame, CA) according to the manufacturer’s instructions. After color reaction with DAB, sections were extensively washed in 0.05m Tris/HCl buffer, pH 7.4, (twice) and 0.1 mcacodylate buffer, pH 7.4, (twice) before being fixed in 2.5% glutaraldehyde in 0.1 m cacodylate buffer, pH 7.4, for 2 hr (4°C). Subsequently sections were further washed in 0.1 mcacodylate buffer, pH 7.4, (twice) and doubly distilled water (ddH2O) (four times).
Silver enhancement of the DAB product was performed as follows: solution A, 3% hexamethylenetetramine in ddH2O; solution B, 5% silver nitrate (AgNO3) in ddH2O; solution C, 2.5% disodium tetraborate in ddH2O; solution D, 0.05% tetrachloroauric(III) acid in ddH2O; solution E, 2.5% sodium thiosulfate in ddH2O. First, sections were incubated for 10 min at 60°C in premade mixed solution, 5 ml (A) + 250 μl (B) + 500 μl (C), and then washed in distilled water (dH2O) (three times for 3 min). Thereafter sections were incubated in solution D at room temperature for 3 min, washed with dH2O (three times for 3 min), incubated in solution E for 3 min, and washed again for three times for 3 min in dH2O. Subsequently, the sections were post-fixed in 1% OsO4, dehydrated in ethanol, and embedded in epon. Parallel semithin sections were stained with toluidine blue for morphological orientation; ultrathin sections were contrasted with uranyl acetate/lead citrate before analysis with a Philips electron microscope.
For double- or triple-immunofluorescence staining the animals were perfusion-fixed transcardially with 4% paraformaldehyde in 0.1m PBS, pH 7.4 (Richter et al., 1996). After post-fixation overnight in the same fixative, brains were cut into 50-μm-thick frontal sections on a vibratome and pretreated with a mixture of methanol/PBS (1:1) and subsequently incubated with 5% goat serum in PBS. After further washing steps in PBS, free-floating sections were incubated in a mixture of two or three primary antisera containing 0.4% Triton X-100 for 36 hr. The selection of the primary antibodies and the detection systems from different species ensured cross talk-free stainings in the colocalization experiments:ProSAP1/Bassoon: guinea pig anti-ProSAP1 1:500, anti-guinea pig-CY3 1:100; rabbit anti-Bassoon 1:500 (tom Dieck et al., 1998), anti-rabbit-CY5 1:100; ProSAP1/Synaptophysin: rabbit anti-ProSAP1 1:1000, anti-rabbit-CY3 1:100, mouse monoclonal anti-synaptophysin (Dako) 1:500, anti-mouse CY5 1:100;ProSAP1/Bassoon/MAP2: guinea pig anti-ProSAP1 1:500, anti-guinea pig-CY3 1:100; rabbit anti-bassoon 1:500, anti-rabbit FITC 1:100 (Sigma Aldrich, Deisenhofen, Germany), mouse monoclonal anti-MAP2 (Sigma Aldrich) 1: 500, anti-mouse CY5 1:100;ProSAP1/Synapsin/MAP2: guinea pig anti-ProSAP1 1:500, anti-guinea pig-CY3 1:100, rabbit anti-synapsin (Sigma Aldrich), anti-rabbit FITC 1:100 (Sigma Aldrich), mouse monoclonal anti-MAP2 (Sigma Aldrich) 1:500, anti-mouse CY5 1:100. CY3- and CY5-labeled secondary antibodies were purchased from Jackson Laboratories (Bar Harbor, ME).
Image acquisition by confocal laser scanning microscopy. The hippocampal regions CA1 and CA3 of immunostained sections were scanned with a confocal laser scanning microscope (Leica TCS 4D, Leica Bensheim) equipped with an Argon-Krypton-Ion laser (488/568/647 nm) and an acousto-optical tunable filter (AOTF) for selection and intensity adaptation of laser lines. The configuration of the system [excitation beam splitter, TD (488/568/647); detector beam splitter, RSP 580; barrier filter, BP 535 (channel 1); detector beam splitter, RSP 660; barrier filter, BP 600 (channel 2); and barrier filter, 0G 665 (channel 3)] allowed a rapid simultaneous detection of the CY3 fluorescence in channel 2 (indicative for ProSAP1) and CY5 in channel 3 (indicative for bassoon or synaptophysin) without an appreciable cross talk. Along the_z_-axis, usually 9–16 thin optical sections with a z-resolution of 0.5–1 μm (focus depth) were scanned in a 512 × 512 or 1024 × 1024 pixel format. Images were taken at various magnifications, usually with a Fluotar 40× oil, NA 1.0–0.5, Plan Apo 63× oil, NA 1.4, or Plan Apo 100× oil, NA 1.4 objective lens at various zoom factors (1–4) as indicated in the legends. Subsequently, maximum intensity projections (extended focus images) were calculated from each fluorescence channel of the image stack and stored as RGB images together with original image stacks. For further image analysis and restoration files were transferred to an Apple Macintosh computer, where image processing (contrast enhancement) was performed with Adobe Photoshop (Version 5.0; Adobe Systems, Mountain View, CA). Color prints of the CLSM images from individual focal planes were printed on a Pictrography 3000 printer (Fuji, Tokyo, Japan).
Hybridization. In situ hybridization was performed essentially as described previously (Kreutz et al., 1997). Transcripts encoding ProSAP1 were detected with a cDNA antisense oligonucleotide purchased from MWG-Biotech (Ebersberg, Germany) directed against the 3′-end of the mRNA: 5′-TTC TTA CTG TCT GTA GAG TTG GCT GGT TGG CTG GAG TTC-3′ (bp 3155–3113).
The expression of the N-terminal insertions was detected with the following antisense oligonucleotides: presence of insert A (at bp 1050), 5′-CGG TTT ATC CTT CTT CTT CCG GAC TGA GGC TTT ATC C-3′ (bp 1079–1044); absence of insert A (at bp 1050), 5′-CGG TTT ATC TTT ATC CAC GAG TTC CTC CAA TTC CGC TGT-3′ (bp 1079–1020 without insert A); presence of insert B (at bp 1287), 5′-GCT GTC TAT CGA TTT CTG CCT TCG CAT CGT ACC TCG AGG-3′ (bp 1329–1281); absence of insert B (at bp 1287), 5′-TCT GCT GTC TCG AGG GAG GCC CAG AAA TGG GCC TTT CGG-3′ (bp 1331–1257 without insert B).
Controls were performed as follows: (1) omission of the antisense oligonucleotide, (2) posthybridizational washing steps above the calculated melting point of the hybrid, (3) hybridization with the corresponding sense oligonucleotide, and (4) hybridization in the presence of 100-fold excess of unlabeled oligonucleotide. In no case was any specific labeling observed.
Isolation of subcellular protein fractions and Western blot analysis. Tissue from adult rats (total brain, heart, liver, kidney, thymus, testis, spleen) was homogenized in 20 mmTris buffer, pH 7.4, containing either 2 mmCaCl2 or 1 mm EDTA and protease inhibitor mixture (Boehringer Mannheim, Mannheim, Germany). Soluble proteins were obtained as the supernatant after 100,000 × _g_centrifugation. After detergent extraction of the remaining pellet with 1% Triton X-100, the detergent-insoluble pellet was extracted with 1% SDS to obtain a fraction of cytoskeletal proteins.
Tissue fractionation was performed essentially as described by Carlin et al. (1980) with some modifications (tom Dieck et al., 1998). Brains of 30-d-old rats were homogenized in homogenization buffer (5 mm HEPES, pH 7.4; 320 mm sucrose) containing protease inhibitor mixture (Boehringer Mannheim); cell debris and nuclei were removed by 1000 × g centrifugation. The supernatant was spun for 20 min at 13,000 × g, resulting in supernatant S2 and pellet P2 (crude membrane fraction). S2 was centrifuged at 100,000 × g for 1 hr, and the resulting supernatant was taken as cytoplasmic fraction (S100). The P2 pellet was further fractionated by centrifugation in a sucrose step gradient as described by Carlin et al. (1980). For isolation of synaptic junctional proteins (PSD fraction), the synaptosomal fraction of the first gradient was diluted with 320 mm sucrose (60 ml/10 gm wet tissue) and an equal volume of 1% Triton X-100, 320 mm sucrose, and 12 mm Tris-HCl, pH 8.1. The suspension was kept on ice for 15 min and was centrifuged for 30 min at 32,800 × g. The pellet was resuspended in 320 mm sucrose, 1 mm NaHCO3 (6 ml/10 gm wet tissue), and an equal volume of 1% Triton X-100; 320 mm sucrose was added, and synaptic junctional proteins were pelleted by a 2 hr centrifugation at 201,800 × g. All steps were performed at 4°C.
To study the association of ProSAP1 with the cytoskeleton during postnatal development, P2 fractions from brains of 1-, 4-, 7-, 10-, 14-, 21-, 30-, and 90-d-old rats were rehomogenized in lysis buffer (0.5 mm NaHCO3, 2.5 mmTris/HCl, 0.5% Triton X-100, pH 8.1), incubated for 1 hr at 4°C, and subsequently spun for 1 hr at 100,000 × g. Pellets were washed once with lysis buffer (1 hr, 100,000 × g) and finally resuspended in 500 μl Laemmli buffer. Proteins (20 μg/lane) were separated by SDS-PAGE on 5–20% gels under fully reducing conditions and transferred onto nitrocellulose. For immunodetection, Western blots were incubated overnight either with the polyclonal ProSAP1 antiserum (dilution 1:3000) or monoclonal antisera generated against SAP90/PSD95 (clone P43520, dilution 1:250; Transduction Laboratories, Mamhead, UK) or the NR1 subunit of the NMDA receptor (clone 54.1, dilution 1:250, PharMingen, San Diego, CA). Immunoreactivity was visualized using the ECL detection system (Amersham Buchler, Braunschweig, Germany).
Extraction experiments of P2 pellets were performed with the following agents: (1) 5 mm HEPES, pH 7.4; 320 mm sucrose; (2) 25 mm Tris-HCl, pH 8.0; (3) 0.5 m NaCl; (4) 1 m NaCl; (5) 1 m Tris-HCl; (6) 1% Triton X-100 in 25 mm Tris-HCl; (7) 2.5% CHAPs in 25 mm Tris-HCl; (8) 1% SDS; (9) 0.1% SDS; (10) 8m urea; (11) 3 m potassium rhodanite (tom Dieck et al., 1998). P2 pellets were resuspended in homogenization buffer, aliquoted into six samples (200 μg protein each), and centrifuged at 15,000 × g for 20 min. Each pellet was then resuspended in 0.5 ml of one of the extraction agents, incubated for 15 min at 4°C with gentle shaking, and centrifuged again for 15 min at 100,000 × g. The resulting pellets were washed in homogenization buffer and dissolved in 80 μl gel-loading buffer (Laemmli, 1970). The supernatants were precipitated with trichloroacetic acid, and the resulting pellets were dissolved in 80 μl loading buffer. Proteins were separated by SDS-PAGE on 5–20% gels under fully reducing conditions and transferred onto nitrocellulose. For immunodetection, Western blots were incubated overnight with primary antiserum (dilution 1:2000), and immunoreactivity was visualized using the ECL detection system (Amersham Buchler). Subsequently Western blots were analyzed for the ability of the different extraction agents to solubilize ProSAP1.
RESULTS
From a collection of cDNA clones isolated by expression screening with antisera against a rat brain synaptic protein preparation (Langnaese et al., 1996), one cDNA clone, sap24e, encoded a protein fragment of 80 amino acids (Boeckers et al., 1998). Northern blot analysis revealed that corresponding transcripts are primarily expressed in the brain (data not shown). The sap24e clone was used to isolate a set of 13 overlapping cDNAs ranging in size from 1.3 to 6.9 kb. Eventually, cDNAs covering the N terminus of encoded protein were isolated using a 5′-terminal fragment of the 6.9 kb clone. Full-length cDNA (7.8 kb) was constructed taking advantage of a _Hin_dIII restriction site at nucleotide 1391 of the assembled sequence (accession no. AJ131899). The cDNA has an open reading frame coding for a proline-rich (12% prolines) protein of 1259 amino acid residues, which we named ProSAP1. Analysis of several cDNA clones revealed the existence of at least three processing variants of ProSAP1 (Fig.1A,B), one of which is identical with the recently published cortactin-binding protein CortBP1 (Du et al., 1998).
Fig. 1.
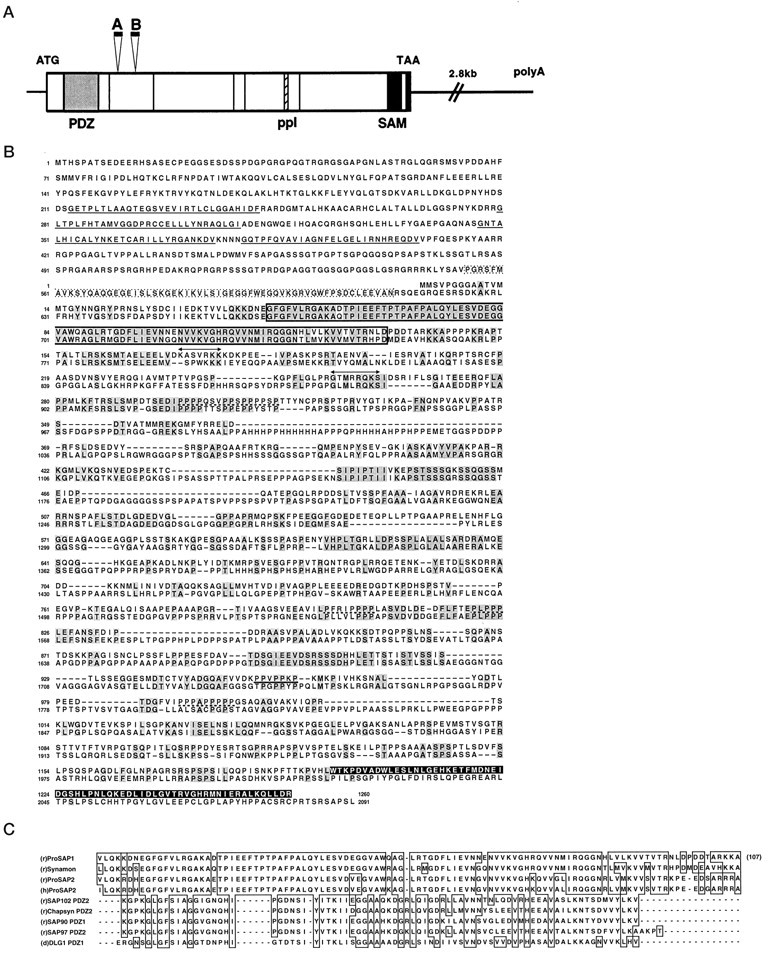
Primary structure of ProSAP1. A, Physical map of the rat ProSAP1 cDNA. The protein coding region is_boxed_. The PDZ and SAM domain are indicated in_light_ and dark gray, respectively. The SH3 interaction module (ppI) is_hatched_; proline-rich elements (5) are indicated by_thin lines_. The positions of two alternatively processed inserts (A and B) are marked. In the 3′-untranslated region a polyA-tail (beginning at nucleotide 7688) is found (compare accession no. AJ131899). B, Alignment of the amino acid sequences of rat ProSAP1 (as deduced from the cDNA, top sequence) and synamon (EMBL/GenBank accession no. AF102855, bottom sequence). Regions of high homology are shaded in_gray_; PDZ domain is lined in black; insertions A and B in ProSAP1 are indicated by double-headed arrows; the SH3-binding motif and proline-rich elements are underlined with_bold_ and broken lines, respectively; the N-terminal SAM domain (displayed as inverted characters) is not conserved between the two proteins. The ankyrin repeats of synamon are underlined, and the SH3 domain of synamon is_boxed_ in broken lines. C, ProSAP1, ProSAP2, and synamon PDZ domains define a new subfamily. Shown is alignment of the PDZ domains of rat ProSAP1, rat synamon, rat and human ProSAP2 (accession nos. AJ131899, AF102855, AJ133120, AC000050, and AC000036) with examples of a more distantly related PDZ domain of synaptic membrane-associated guanylate kinases of SAP90/PSD-95 (Kistner et al., 1993), SAP97 (Müller et al., 1995), SAP102 (Müller et al., 1996), Chapsyn-110/PSD-93 (Kim et al., 1996; Brenman et al., 1996a,b), and DLG1 (Woods and Bryant, 1989).
Sequence analysis predicts several structural domains of ProSAP1/CortBP1 (Fig. 1A,B). These include an N-terminal PDZ domain (aa 38–144), a proline-rich SH3 binding motif (ppI motif, aa 954–960) that has been shown previously to interact with the SH3 domain of the actin-binding protein cortactin (Du et al., 1998), and a C-terminal SAM domain (aa 1193–1257) consisting of four short helices linked by loops (Ponting, 1995) [for a more detailed analysis of the SAM domain of ProSAP1/CortBP1, see Du et al. (1998)]. Moreover, prolines frequently occur as clusters of three or more residues (Fig. 1A,B).
The N-terminal PDZ domain found in ProSAP1 shows only moderate similarity with previously described PDZ domains (Fig. 1C). The highest degree of identity was found to synapse-associated PDZ-domain proteins, e.g., PDZ2 of SAP102 (31%), PDZ2 of Chapsyn-110/PSD-93 (29%), PDZ1 of SAP90/PSD-95 (27%), and PDZ2 of SAP97 (27%). The first PDZ domain of the Drosophila disks large tumor suppressor protein (DlgA) is 28% identical. The GLGF motif, a hallmark of most PDZ domains that plays an important functional role in binding the C termini of interaction partners (Kornau et al., 1997) is substituted by GFGF in ProSAP1. A search for ProSAP1-related proteins by homology screening with PDZ-domain probes identified cDNAs for a new protein, ProSAP2 (accession no. AJ133120), with a PDZ domain that is 80% identical with that of ProSAP1 (Fig.1C). Genomic sequences of the human homolog of ProSAP2 were found in public databases (Cosmid Clones; accession nos. AC000050 andAC000036; mapped at chromosome 22). There is a very high degree of sequence identity (96%) between rat and human ProSAP2 PDZ domains. Like ProSAP1, ProSAP2 is a proline-rich protein. However, regions of high similarity between the two proteins are restricted to the ppI motif and the SAM domain (Winter C, Kreutz MR, Smalla KH, Bockmann J, Garner CC, Gundelfinger ED, and Boeckers TM, unpublished data). Another PDZ domain highly related to that of ProSAP1 and ProSAP2 is found in synamon, a synaptic protein recently included in public databases (Fig.1B,C).
Sequencing of several different ProSAP1 cDNA clones suggested the expression of alternatively spliced transcripts affecting the N-terminal part of the protein. Sequence insertions/deletions were found at nucleotide 1050 (insert A; 21 bp) and nucleotide 1287 (insert B; 27 bp). Several independent clones showed the presence of either one of these insertions or of both inserts. In none of the sequenced clones the absence of both exons could be observed. The originally described CortBP1 is missing insert A but contains insert B (Du et al., 1998). ProSAP1 has several putative phosphorylation sites for cAMP and cGMP-dependent protein kinases (1), casein kinase II (20), tyrosine kinase (1), and protein kinase C (20). Interestingly, both insertions add several positive charges to the protein (Fig. 1B) and carry putative protein kinase C phosphorylation sites (insertion A: aa 175–178 SVR; insertion B: aa 254–257 TMR).
Characterization and expression pattern of ProSAP1
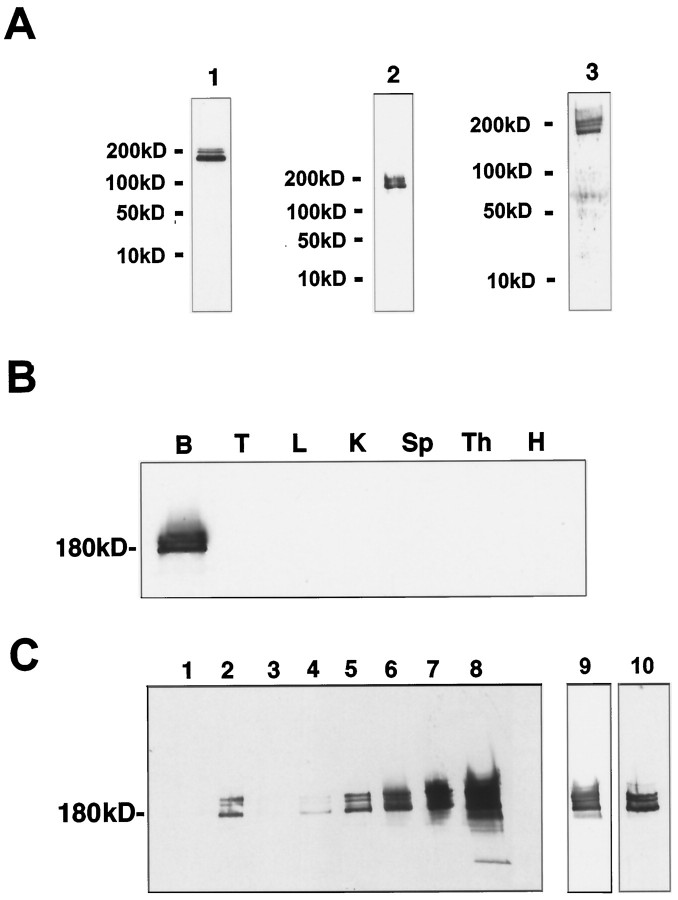
Recombinant GST-fusion proteins including either part of the central region or the C-terminal region of ProSAP1 were used to raise polyclonal antisera against ProSAP1 in rabbits and guinea pigs. In accordance with the staining results obtained by Du et al. (1998), all antisera specifically detect a cluster of protein bands at a molecular weight in the range of 180–220 kDa on immunoblots of brain protein preparations (Fig. 2A), but not of other tissue homogenates, including testes, liver, kidney, spleen, thymus, and heart (Fig. 2B). Thus ProSAP1 migrates in SDS-PAGE slower than expected from the calculated molecular weight, a behavior that is frequently observed for cytoskeletal proteins. None of the bands were recognized in total brain homogenates by either of the preimmune sera or after preabsorption with the antigen (data not shown). Because ProSAP1 has been cloned using antisera directed against the PSD fraction, the subcellular distribution of the protein was investigated. ProSAP1 immunoreactivity is present in the crude membrane fraction (P2) of rat brain (Fig. 2C,lane 2) but not in the soluble protein fraction (Fig.2C, lane 1). During the further steps of subcellular fractionation by differential centrifugation, ProSAP1 immunoreactivity essentially cofractionates with PSDs (Fig.2C). To investigate the association of ProSAP1 with the cytoskeleton, attempts were made to solubilize the protein from the crude membrane fractions (tom Dieck et al., 1998). A partial extraction of immunopositive bands was achieved with high salt (1 mNaCl, 1 m Tris-HCl) or 0.1% SDS. Virtually complete solubilization was observed when using strongly denaturing conditions (1% SDS or 8 m urea). Treatment with 0.5 mNaCl, the detergents CHAPS (2.5%), or Triton X-100 (1%) or with the chaotropic agent potassium rhodanite (3 m) did not result in a substantial release of ProSAP1 from the pellet. All ProSAP1 isoforms represented by the different immunoreactive bands displayed a similar extraction behavior. These solubilization characteristics of the protein indicate a close cytoskeletal association of ProSAP1.
Fig. 2.
Tissue distribution and subcellular co-partitioning of ProSAP1 in rat brain A antisera generated against ProSAP1 (1, polyclonal rabbit antiserum, C-Term aa 826–1259; 2, polyclonal guinea pig antibody, C-Term aa 826–1259; 3, polyclonal rabbit antiserum, central part, aa 355–509) detect a major protein band at 180 kDa and two weaker bands at ∼200 and 220 kDa in rat brain protein preparations.B, ProSAP1 immunoreactivity is only detectable on immunoblots of rat brain protein preparations and not found in testis (T), liver (L), kidney (K), spleen (Sp), thymus (Th), or heart muscle (H). Protein extracts were obtained from 8-week-old rats. Western blots were loaded with 50 μg per slot of detergent-insoluble cytoskeletal protein. C, ProSAP1 is highly enriched in synaptic junctional protein preparation. Synaptic proteins were prepared according to Carlin et al. (1980). Western blots (15 μg protein per lane) of the soluble protein fraction (lane 1), the crude membrane fraction P2 (lane 2), the myelin fraction (lane 3), the light membranes fraction (lane 4), the synaptosomal fraction (lane 5), detergent extracted synaptosomes (lane 6) [i.e., One Triton, Kennedy (1997)], postsynaptic density fraction obtained from the detergent extracted fraction in a second gradient (lane 7), and the twice Triton X-100-extracted PSD fraction (lane 8) [Two Triton, Kennedy (1997)] were probed with the rabbit anti-ProSAP1 antibody using a chemiluminescent detection system. Lane 9 shows the twice Triton X-100-extracted PSD fraction (lane 8) with a shorter exposition time; lane 10 displays a Western blot of the twice Triton X-100-extracted PSD fraction with less protein loaded (3 μg). Note that the distribution of other synaptic proteins, including the presynaptic cytomatrix proteins Bassoon and Piccolo, the PSD proteins SAP102, and the synaptic vesicle protein synaptophysin in the presented subcellular fractionation experiment, has been assessed previously (tom Dieck et al., 1998).
To examine the spatial distribution of the ProSAP1 transcripts we performed in situ hybridization studies to horizontal brain sections (Fig. 3A). An antisense oligonucleotide directed against the C-terminal part of the mRNA and thus recognizing all known variants of ProSAP1 transcripts can be localized to many parts of the brain with high levels of expression in the olfactory bulb, cerebral cortex, cerebellum, central gray, and hippocampus. The caudate putamen and thalamic nuclei as well as the brain stem are moderately labeled. In control experiments, the use of a sense oligonucleotide, competition with 100-fold excess of unlabeled oligonucleotide, as well as washing steps above the calculated melting temperature of the hybrid yielded no labeling above background (data not shown). This broad expression pattern is reflected by the light-microscopic localization of ProSAP1 immunoreactivity in rat brain (Fig. 3B,C) showing an intense labeling of cerebral cortex, molecular layer of the cerebellum, hippocampal formation, thalamic nuclei, and basal ganglia. At higher magnification of the hippocampus, a representative light micrograph (Fig. 3D) shows a punctate staining pattern of hippocampal neuropil in the stratum oriens and stratum radiatum (CA2/CA3 region). Ultrastructural investigation of the hippocampal CA3 region identifies the close association of the antigen with the PSD (Fig. 3E,F). Although axon terminals do not show any labeling above background, the antigen can be found in dendrites with a highly significant increase of staining toward spines and PSDs. Double- and triple-immunofluorescent stainings of hippocampal sections for ProSAP1 and various marker proteins for different neuronal compartments further document the primarily synaptic localization of ProSAP1. Figure 4_A,B_illustrates the punctate distribution and close apposition of ProSAP1-immunoreactive structures and the presynaptic cytomatrix protein Bassoon (tom Dieck et al., 1998) at many synapses of the CA1 region. The virtually complementary localization of ProSAP1 and the synaptic vesicle protein synaptophysin or Bassoon at mossy fiber boutons of the CA3 region can be seen in Figure 4C,D. Triple staining with the dendritic marker protein MAP2, the presynaptic markers Bassoon or synapsin, and ProSAP1 illustrates the dense clustering of ProSAP1 on dendrites of hippocampal neurons. Large clusters are seen at the mossy fiber terminals in the stratum lucidum, whereas smaller PSDs decorate dendrites in the stratum radiatum (Fig.4E,F).
Fig. 3.
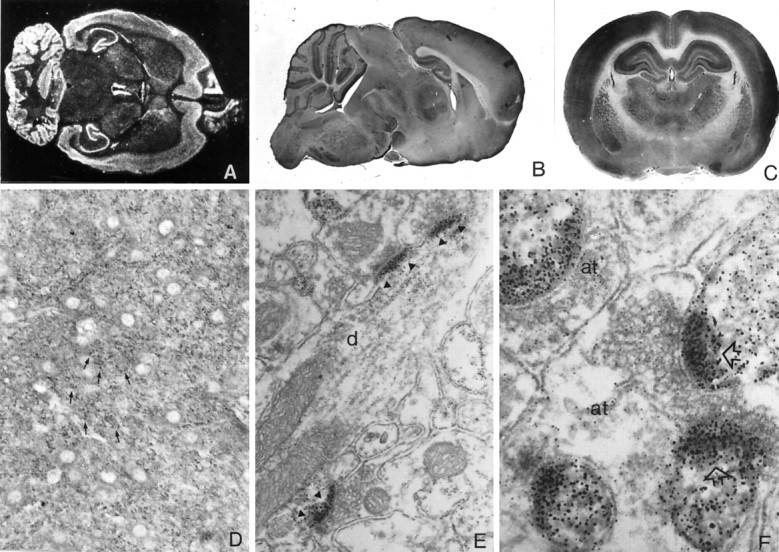
Distribution of ProSAP1 in the adult rat brain.A, Distribution of mRNA transcripts. In situ hybridization to a horizontal section from brain with the35S-labeled ProSAP1 antisense oligonucleotide shows the overall expression of the transcript in the adult brain. Intense labeling is observed in cerebral cortex, cerebellum, hippocampus, and olfactory bulb, whereas putamen, thalamic nuclei, and brainstem show a moderate staining. Magnification: 2.5×. B,C, Overview of spatial distribution of ProSAP1 protein in rat brain by immunohistochemistry with ProSAP1 antisera. Sagittal (B) and frontal (C) sections are shown. Strong ProSAP1 immunoreactivity is detected in cerebral cortex, hippocampus, and the molecular layer of the cerebellum. Furthermore, the thalamic nuclei, the putamen, and to a much lesser extent the hypothalamus are labeled. Further enlargement of the hippocampal CA2/CA3 region (D) illustrates that cell nuclei and cell bodies are free of staining, whereas in the neuropil a punctate staining (arrows) can be observed. Magnifications: B, C, 2.5×; D, 450×. Electron micrographs (E, F) were taken from hippocampal CA3 sections. Silver enhancement of the DAB reaction product results in the punctate appearance of the immunoreactivity. Note that labeling is relatively weak in dendrites (d), enhanced toward dendritic spines (sp), and very strong at PSDs (arrowheads, arrows). Axon terminals (at) are essentially unlabeled. Magnification: F, 45,000×; G, 85,000×.
Fig. 4.
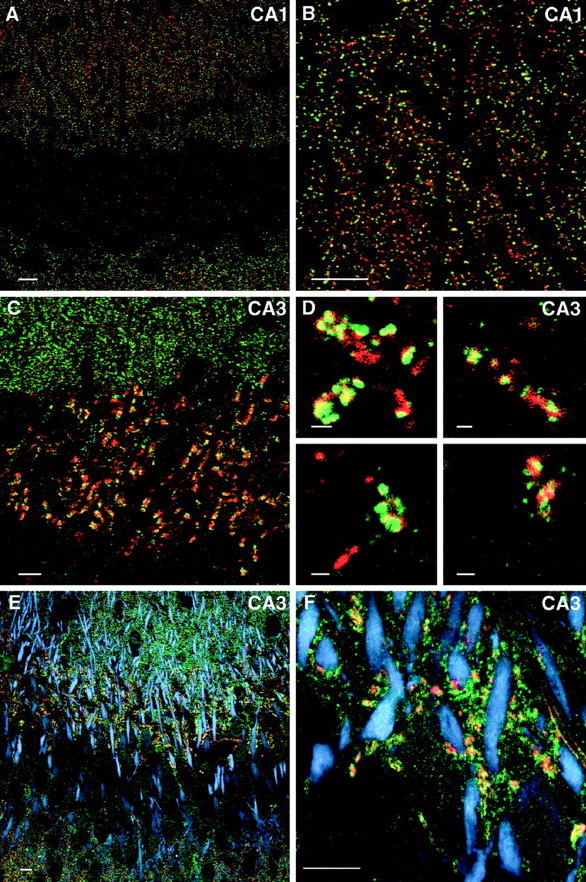
Distribution of ProSAP1 in hippocampal neurons as revealed by double- and triple-immunofluorescence labeling.A_–_D, Double immunofluorescence of hippocampal neurons in the CA1 and CA3 region with the rabbit antibody directed against ProSAP1 (CY3, green) and a monoclonal antibody directed against the presynaptic protein Bassoon (A, B, D, CY4, red) or synaptophysin (C, CY4, red). Note that the antigens are largely co-distributed at hippocampal synapses. At higher magnifications of mossy fiber terminals in the stratum lucidum of the CA3 region (C, D), the close apposition of ProSAP1 with both presynaptic marker proteins can be seen. The staining with the Bassoon antibody especially illustrates the close spatial relationship of the two proteins because Bassoon is mainly restricted to the active zone of the presynapse (tom Dieck et al., 1998; Richter et al., 1999). Confocal images of triple immunofluorescence ProSAP1 (green), Bassoon (red), and MAP2 (blue) (E) as well as ProSAP1 (green), synapsin (red), and MAP2 (blue) (F) show the localization of synaptic structures on dendrites of hippocampal CA3 neurons. Please note that labeled shaft and spine synapses are discernible that decorate the MAP2-positive dendritic trees. Scale bars: A–C, E, F, 10 μm; D (all_insets_), 1 μm.
Expression of ProSAP1 during early postnatal period
To assess whether ProSAP1 may be involved in synaptic assembly during development or whether its function is restricted to preformed synapses, we investigated the expression of ProSAP1 during the period of synaptogenesis. To this end we performed _in situ_hybridization, immunocytochemistry studies, and Western blot analyses. Hybridization to brain sections of days P5, P9, and P18 indicate an increase of ProSAP1 transcripts during early postnatal brain development, especially in the caudate putamen and thalamic nuclei (Fig. 5A). ProSAP1 expression in the cerebral cortex, the hippocampus, and the cerebellum is moderate to high already at P5 and shows a stable expression throughout development. Immunohistochemical staining of cortical neurons during the early postnatal period (P5, P8, P10) demonstrates a striking change in the localization of the antigen from being localized mainly in the cytoplasm of cell bodies and neurites to a close association of the protein with postsynaptic structures (Figs. 5, 6). On P5, ProSAP1 immunoreactivity is seen in the cytoplasms of densely packed cortical neurons (Figs. 5B,a). At the ultrastructural level the antigen appears localized in small processes and lamellopodia (Fig.6A,B). Interestingly, already at that stage of development the antibody detects ProSAP1 only in a subset of neurites that are in close contact with unlabeled neuritic structures. On P8 the antigen can be localized in the cytoplasm of cell bodies and in larger outgrowing neurites (Fig.5B,b). Ultrastructural investigations revealed that ProSAP1 is already localized in the now apparent PSD of early synapses (Fig.6C–E). On P10 a punctate staining can be recognized at the light microscopic level (Fig. 5B,c) that reflects the specific labeling of PSDs of cortical synapses (Fig.6F,G). At this stage of development the staining pattern does not differ from that in adult animals.
Fig. 5.
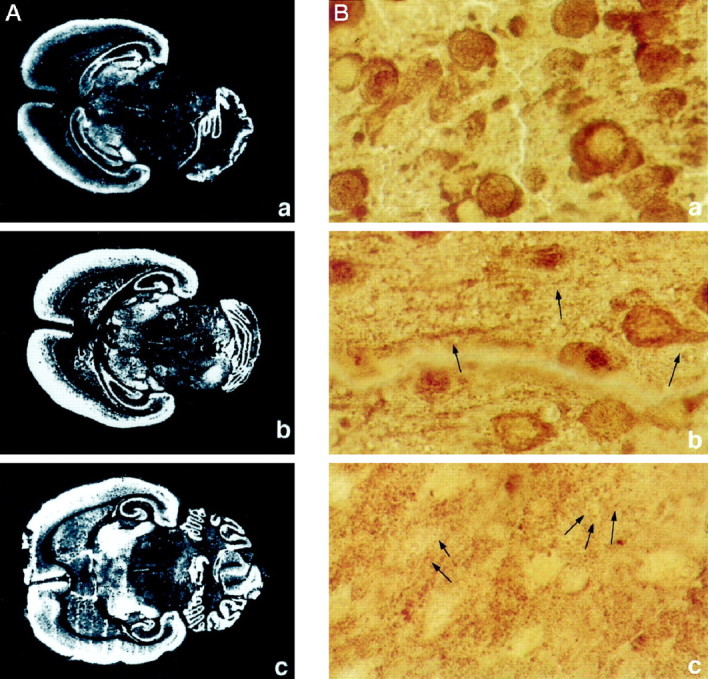
A, Distribution of the ProSAP1 transcripts and protein in the developing rat brain. In situ hybridizations of horizontal brain sections from 5 (a)-, 9 (b)-, and 18-d-old (c) rats. X-ray film images of in situ hybridizations with the ProSAP1 antisense oligonucleotide show the dense expression of the transcript in cortex, cerebellum, and hippocampus at these developmental stages. The transcript is especially upregulated during development in the thalamic nuclei and the caudate putamen. Magnification: 3.5×. B, Immunohistochemical staining of cortical neurons at P5 (a), P8 (b), and P10 (c). Note the strong labeling of cytoplasm and small outgrowing neurites at P5 (a), whereas the neuropil appears largely unstained. On P8 the cytoplasmic staining is reduced, and larger neurites (arrows) are clearly labeled (b). Two days later (P10) the staining pattern changes to a punctate labeling (arrows) in the neuropil of the developing cortex (c). Magnification: 500×.
Fig. 6.
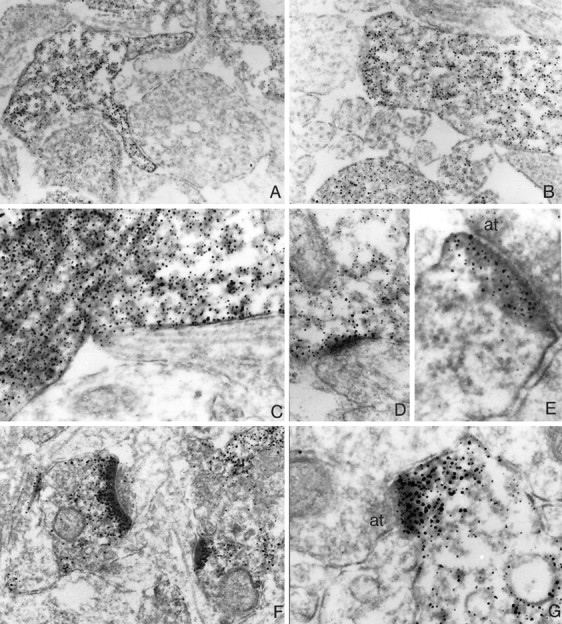
Ultrastructural localization of ProSAP1 in the developing rat cortex. Electronmicroscopy of immunostained cortical sections at P5 (A, B) shows the primarily cytoplasmic localization of ProSAP1 in a subset of outgrowing neurites. Note the clearcut differentiation between ProSAP1-positive and -negative neurites. In A, a ProSAP1-positive neurite with putatively pathfinding lamellopodia is displayed. _B_shows the close contact between a ProSAP1-positive and -negative neurite. At P8, strong labeling can be found in the cytoplasm of growing neurites (C); synaptic contacts show strong ProSAP1 immunoreactivity in the now appearing PSDs (D, E). At P10 (F, G), differentiation of brain tissue has advanced, and ProSAP1 immunoreactivity is primarily found in spines and in particular at PSDs. Magnification: A, B, 46,000×; C, D, 70,000×; E, 90,000×.at, Axon terminal.
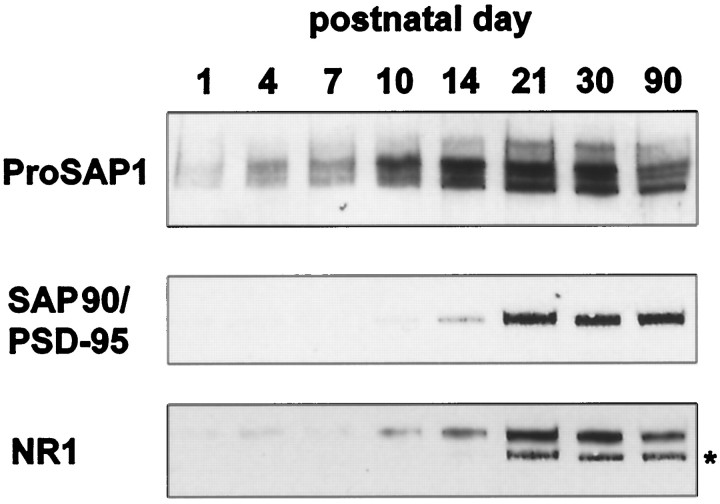
These localization data are consistent with the assumption that ProSAP1 is one of the first protein components assembling into developing PSDs. If so, one prediction would be that ProSAP1 becomes anchored to the subsynaptic cytoskeleton earlier than other known components of the PSD protein fraction, e.g., SAP90/PSD-95 or NMDA receptors. To test this hypothesis, we analyzed on immunoblots the appearance during development of ProSAP1, SAP90/PSD-95, and the NR1 subunit of NMDA receptors in cytoskeletal protein fractions that should be enriched for PSD components. As shown in Figure 7, the association of ProSAP1 with the PSD-enriched fraction is strongly enhanced between P7 and P10, consistent with the period of its appearance at postsynaptic sites of cortical synapses (Fig. 6). In contrast, enhanced appearance of NR1 and SAP90/PSD-95 immunoreactivity in PSD-enriched protein fractions is observed only later during development, i.e., from P10 and P14 onward, respectively.
Fig. 7.
Developmental association of ProSAP1, SAP90/PSD-95, and the NR1 subunit of the NMDA receptor with cytoskeletal protein fractions. Western blot analysis of a fraction enriched for PSD elements (20 μg/lane) during postnatal development shows that moderate amounts of ProSAP1 are already detectable at P1 with a significant increase of the signal intensity between P7 and P10. Association of SAP90/PSD-95 with the protein fraction strongly increases from P14 onward. The detection of the NR1 subunit of the NMDA receptor is detectable from P1, but a strong increase in immunoreactivity is seen only between P10 and P21. The_star_ indicates the band resulting from the SAP90/PSD-95 staining. The blot has been reprobed for the detection of the NR1 subunit of the NMDA receptor.
Expression of ProSAP1 isoforms during postnatal development
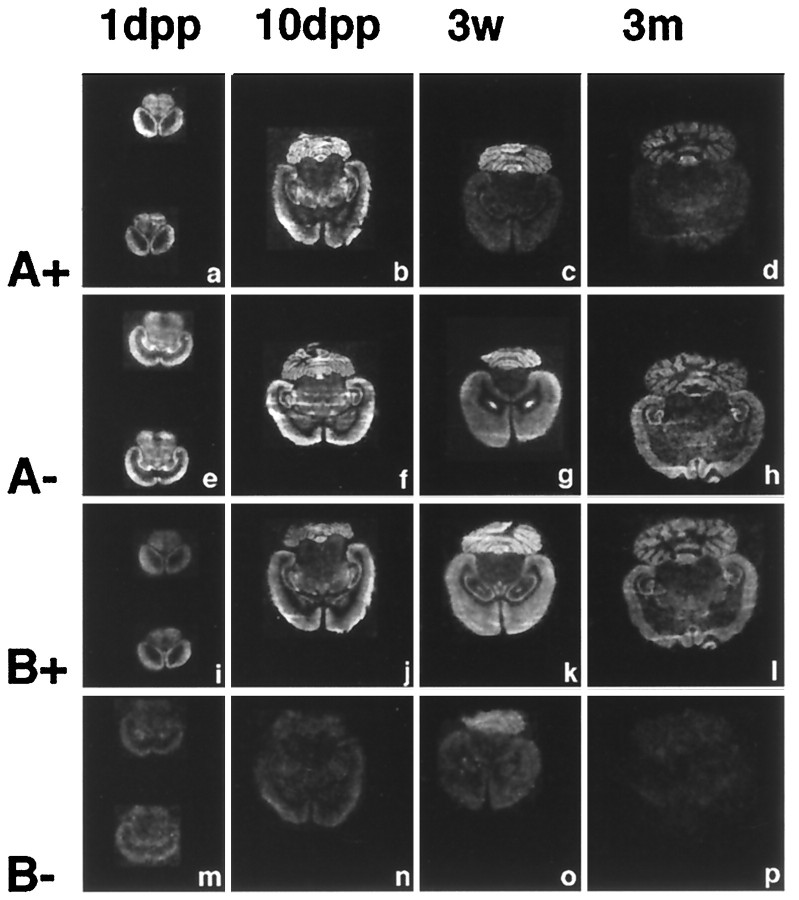
To examine whether the expression of ProSAP1 isoforms encoded by the identified processing variants of ProSAP1 transcripts are subject to temporal or spatial regulation, we performed _in situ_hybridizations at different developmental stages with four different oligonucleotides. The probes were designed to detect the presence or absence of inserts A and B (Fig. 1, compare A,B). Insert A-containing transcripts are predominantly expressed during early stages of brain development (Fig.8a–d, A+). On days P1 and P10 it is found in cortical brain areas as well as in the cerebellum, hippocampus, and thalamic nuclei. After 3 weeks and in the adult brain the hybridization signal intensity is reduced, and the mRNA is found almost exclusively in the cerebellum. Transcripts without insert A (Fig. 8e–h, A−) display stable expression at all postnatal stages in all brain regions expressing ProSAP1 (compare Figs.3A, 5A).
Fig. 8.
Expression of processing variants of ProSAP1 transcripts in the brain during postnatal development. During early postnatal development, insert A (a, b, A+) shows a wide distribution throughout the brain. At later stages the hybridization signal intensity decreases and becomes mainly restricted to the cerebellum (c, d, A+). ProSAP1 transcripts without insert A show a wide expression in rat brain at all developmental stages investigated (e–h, A−). Nearly identical results were obtained with an antisense oligonucleotide designed to recognize insert B (i–l, B+). In contrast, ProSAP1 mRNA without insert B shows a very weak expression on P1 and P10; it is largely restricted to the cerebellum after 3 weeks and cannot be detected in the adult rat brain. Therefore in most brain regions the_A−/B+_ transcript seems to be regularly expressed, whereas during development, especially in the cerebellum, transcripts with insert A but without insert B can also be detected.
On the contrary, insert B is expressed throughout the rat brain from day 1 onward with a pattern similar to the A− hybridization signals (Fig. 8i–l, B+). ProSAP1 transcripts without insert (Fig. 8m–p, B−) show only a very weak expression, mainly in cortex and cerebellum on days P1 through P10. At 3 weeks there is a somewhat stronger hybridization signal in the cerebellum. In the adult brain no B−transcript levels above background could be detected.
DISCUSSION
ProSAP1 originally has been isolated as a protein contained in synaptic junctional protein preparations from rat brain (Kistner et al., 1993; Langnaese et al., 1996, Boeckers et al., 1998). Biochemical analysis and ultrastructural localization studies revealed that ProSAP1 is indeed a component of the PSD of excitatory brain synapses. Analysis of the primary structure of ProSAP1 identified several sequence motifs typical for proteins of the membrane-associated cytoskeleton. While this study was in progress, CortBP1, which is identical with ProSAP1, has been identified as an interaction partner of the actin-binding protein cortactin (Du et al., 1998). This indicates that ProSAP1/CortBP1 may be one of the elements that links the postsynaptic signaling apparatus to the actin-based cytoskeleton within the PSD.
PDZ domains of ProSAP1 and ProSAP2 define a new subfamily of PDZ domains
ProSAP1 has various structural motifs that are known to be involved in protein–protein interactions. As shown by Du et al. (1998), ProSAP1/CortBP1 is able to interact with the cortactin SH3 domain via a ppI motif in the central part of the protein. Additional proline-rich domains identified in ProSAP1 may be involved in similar interactions with other proteins. A hallmark of the N-terminal part of ProSAP1 is a new type of PDZ domain. PDZ domains are protein interaction modules that mediate the binding of distinct cell surface and intracellular proteins to the cortical cytoskeleton (Kornau et al., 1997). PDZ domains related to that of ProSAP1 include those of ProSAP2, a protein that is also primarily expressed in the brain (Winter, Kreutz, Smalla, Bockmann, Garner, Gundelfinger, and Boeckers, unpublished data), and of the synaptic SAPAP-interacting protein synamon (accession no. AF102855). PDZ domains of ProSAP1, ProSAP2, and synamon share >80% sequence identity, whereas PDZ domains of previously known proteins, such as SAP90/PSD-95, SAP97, Chapsyn-110/PSD-93, and SAP102, are only ∼30% identical with this new subfamily of PDZ domains (Fig. 1C).
The sequence similarity between ProSAP1 and ProSAP2 is striking in the PDZ and SAM domains; the degree of sequence similarity in other regions of the two proteins is relatively low. The ppI motif is conserved between the two proteins, suggesting that ProSAP2 is also a SH3 domain-binding protein. A database search for ProSAP1-related proteins revealed several brain cDNAs and genomic DNA fragments encoding human ProSAP1 (accession nos. M86079, H41098, and HSU73633/chromosome 11) and human ProSAP2 (accession nos. AC000050, AC000036/chromosome 22). Synamon is a 2091 aa synaptic protein with four amino-terminal ankyrin repeats, a central SH3 domain next to a PDZ domain (GenBank accession no. AF102855). The similarity between ProSAP1 and synamon is not restricted to the PDZ domain, but clusters of high sequence identity are distributed throughout the C-terminal part of synamon and the entire length of ProSAP1. However, no SAM domain is found in synamon (Fig. 1B).
ProSAP1 is highly enriched in PSDs in the adult rat brain
The actin-based cytoskeleton is thought to play a crucial role in the regulation of dendritic spine morphology and the assembly of postsynaptic structures, including the PSD. Thus, dendritic spines and in particular the PSD are extremely rich in distinct isoforms of actin (Matus et al., 1982; Cohen et al., 1985; Fifkova and Morales, 1992;Kaech et al., 1997). Spine mobility and expression of synaptic plasticity appear to be intimately associated with the modulation of the actin cytoskeleton (Fifkova and Morales, 1992; Fischer et al., 1998). Various elements of the actin cytoskeleton, including brain spectrin/fodrin (Carlin et al., 1983), dystrophin (Kim et al., 1992), α-adducin (Seidel et al., 1995), drebrin (Hayashi et al., 1996), and α-actinin-2 (Wyszynski et al., 1997) have been shown to be components of the PSD. The functional significance of the cortical cytoskeleton is underscored by the fact that postsynaptic NMDA receptor activity depends critically on the integrity of actin filaments (Rosenmund and Westbrook, 1993). Moreover, NMDA receptor linkage to the postsynaptic actin cytoskeleton appears to be mediated by α-actinin-2 and is regulated by a Ca2+/calmodulin-dependent mechanism (Wyszinski et al., 1997).
ProSAP1 is likely to be a component of the actin-based cytomatrix of the PSD. First, ProSAP1/CortBP1 is linked to the actin cytoskeleton via cortactin (Du et al., 1998). Second, it is specifically expressed in brain tissue, and solubilization experiments as well as Western blot analysis after subcellular fractionation of brain tissue revealed that ProSAP1 is a cytoskeletal protein that is highly enriched in the PSD fraction. Moreover, in situ hybridization and immunohistochemical studies at the light and electron microscopic level revealed that ProSAP1 is widely expressed in neurons and mainly located in the submembraneous matrix of the PSD. It occurs primarily at asymmetric type 1 synapses, which are thought to be excitatory (Peters et al., 1991; Ziff, 1997). These data indicate that ProSAP1 is part of the highly specialized cytoskeleton at the PSD, which anchors neurotransmitter receptors, cell adhesion molecules, and intracellular signal transduction pathways to the postsynaptic site (Ziff, 1997;Craven and Bredt, 1998; O’Brien et al., 1998). Cortactin that links ProSAP1/CortBP1 to the actin cytoskeleton is substrate for the nonreceptor protein tyrosine kinase Src (Wu and Parsons, 1993). It is an F-actin binding protein that is thought to mediate aspects of cell signaling associated with the cortical cytoskeleton (Du et al., 1998). Thus both proteins may contribute to the enormous dynamic potential of the postsynaptic cytoskeleton supposed to provide the basic mechanisms for synaptic plasticity (Buchs and Muller, 1996).
ProSAP1 isoforms may be functionally involved in the assembly of the postsynaptic cytomatrix during development
It is still unknown which mechanisms govern the formation of the PSD beneath the postsynaptic membrane. On theoretical grounds the initial formation requires the docking of several proteins, including neurotransmitter receptors, protein kinases and phosphatases, adaptor proteins, and filamentous cytoskeletal proteins, to fulfill the morphological and functional criteria of building the PSD. At least two processes have to occur in parallel: (1) clustering of receptor molecules in apposition to the active zone of the presynaptic membrane providing the structural basis for an excitable membrane and (2) anchoring of proteins involved in intracellular signal transduction to this membrane. Currently we do not know anything about the initiation of these two processes.
ProSAP1/CortBP1 and cortactin are highly enriched in growth cones of hippocampal primary neurons before synaptogenesis (Du et al., 1998). This is consistent with our finding in situ that ProSAP1 immunoreactivity is found in lamellopodia of cortical neurons before synaptogenesis. A striking change in ultrastructural localization of ProSAP1 immunoreactivity occurs when synaptic contacts are formed. Then ProSAP1 is concentrated at sites where PSDs are thought to form. This early appearance at the differentiating postsynaptic membrane suggests that ProSAP1 could be involved in initial steps of PSD assembly. Interestingly, the association of ProSAP1 with the PSD appears to precede the anchoring of SAP90/PSD-95 and the NR1 subunit of the NMDA receptor. An important question to be answered in this context is whether ProSAP1 is actively involved in the formation of a postsynaptic cytoskeletal specialization at the synaptic junction or whether the protein is incorporated in the PSD after the structure has been formed. Of interest is the observation that during development the expression of splice variants containing insertion A as well as those lacking insertion B is restricted to the early postnatal period. These transcripts are predominantly expressed in the cerebellum of animals up to the age of ∼3 weeks. Interestingly, these inserts change the charge to the molecule and introduce additional potential phosphorylation sites for protein kinase C, an enzyme that is known to be involved in the regulation of synaptic assembly and plasticity (Shearman et al., 1991; Ben-Ari et al., 1992; Wang and Feng, 1992;Abeliovich et al., 1993; Klann et al., 1993; Moriya and Tanaka, 1994;Reymann and Staak, 1994; Pasinelli et al., 1995). Further studies must clarify whether these developmentally regulated processing variants are involved in early synaptogenesis and/or growth cone regulation.
Footnotes
This work was supported by grants from the Innovative Medizinische Forschung (IMF, WWW Muenster), the Land Sachsen-Anhalt, Deutsche Forschungsgemeinschaft, and Fonds der Chemischen Industrie to T.M.B., M.R.K., and E.D.G., and from National Institutes of Health to C.C.G (AG 12978-02). We thank A. Ahle, C. Borutzki, G. Gaede, U. Kaempf, A. Lewedag, S. Loheide, and B. Kracht for expert technical assistance.
The nucleotide sequences reported in this paper have been submitted to the GenBank/EMBL Data Bank with accession numbers AJ131899 (ProSAP1) and AJ133120 (ProSAP2).
Correspondence should be addressed to Dr. T. M. Boeckers, Department of Neurochemistry and Molecular Biology, Leibniz Institute for Neurobiology, P.O. Box D, 39118 Magdeburg, Germany.
Dr. Sanmarti-Vila’s present address: Memorial Sloan Kettering Cancer Center, Department of Neurology, New York, NY 10021.
Dr. Wex’s present address: Mount Sinai School of Medicine, Department of Human Genetics, New York, NY 10029-6514.
Dr. Langnaese’s present address: Institute of Human Genetics, University Magdeburg, 39120 Magdeburg, Germany.
REFERENCES
- 1.Abeliovich A, Chen C, Goda Y, Silva AJ, Stevens CF, Tonegawa S. Modified hippocampal long-term potentiation in PKC gamma-mutant mice. Cell 31 1993. 75:1253–1262. [DOI] [PubMed] [Google Scholar]
- 2.Adam G, Matus A. Role of actin in the organisation of brain postsynaptic densities. Brain Res Mol Brain Res. 1996;31:246–250. doi: 10.1016/s0169-328x(96)00177-5. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Ari Y, Aniksztejn L, Bregestovski P. Protein kinase C modulation on NMDA currents: an important link for LTP induction. Trends Neurosci. 1992;15:333–339. doi: 10.1016/0166-2236(92)90049-e. [DOI] [PubMed] [Google Scholar]
- 4.Boeckers TM, Kreutz MR, Bockmann J, Langnaese K, Sanmarti-Vila L, Garner CC, Gundelfinger ED. SAP24e, a novel postsynaptic density (PSD) protein. Soc Neurosci Abstr. 1998;24:2.1999. doi: 10.1523/JNEUROSCI.19-15-06506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenman JE, Christopherson KS, Craven SE, McGee AW, Bredt DS. Cloning and characterization of postsynaptic density 93, a nitric oxide synthase interacting protein. J Neurosci. 1996a;16:7407–7415. doi: 10.1523/JNEUROSCI.16-23-07407.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domain. Cell. 1996b;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 7.Buchs PA, Muller D. Induction of long-term potentiation is associated with major ultrastructural changes of activated synapses. Proc Natl Acad Sci USA. 1996;23:8040–8045. doi: 10.1073/pnas.93.15.8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlin RK, Grab DJ, Cohen RS, Siekievitz P. Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J Cell Biol. 1980;86:831–843. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlin RK, Bartelt DC, Siekevitz P. Identification of fodrin as a major calmodulin-binding protein in postsynaptic density preparation. J Cell Biol. 1983;96:443–448. doi: 10.1083/jcb.96.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H-J, Rojas-Soto M, Oguni A, Kennedy MB. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron. 1998;20:895–904. doi: 10.1016/s0896-6273(00)80471-7. [DOI] [PubMed] [Google Scholar]
- 11.Cohen RS, Chung SK, Pfaff DW. Immunochemical localization of actin in dendritic spines of the cerebral cortex using colloidal gold as a probe. Cell Mol Neurobiol. 1985;5:271–284. doi: 10.1007/BF00711012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craven SE, Bredt DS. PDZ proteins organize synaptic signaling pathways. Cell. 1998;39:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- 13.Dong H, O’Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL. GRIP. A synaptic PDZ domain containing protein that interacts with AMPA receptors. Nature. 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- 14.Du Y, Weed SA, Wen-Cheng X, Marshall TD, Parsons TJ. Identification of a novel cortactin SH3 domain-binding protein and its localization to growth cones of cultured neurons. Mol Cell Biol. 1998;18:5838–5851. doi: 10.1128/mcb.18.10.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fifkova E, Morales M. Actin matrix of dendritic spines, synaptic plasticity, and long term potentiation. Int Rev Cytol. 1992;139:267–307. doi: 10.1016/s0074-7696(08)61414-x. [DOI] [PubMed] [Google Scholar]
- 16.Fischer M, Kaech S, Knutti D, Matus A. Rapid actin-based plasticity in dendritic spines. Neuron. 1998;20:847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- 17.Garcia EP, Mehta S, Blair LA, Wells DG, Shang J, Fukushima T, Fallon JR, Garner CC, Marshall J. SAP90 binds and clusters kainate receptors causing incomplete desensitization. Neuron. 1998;21:727–739. doi: 10.1016/s0896-6273(00)80590-5. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi K, Ishikawa R, Ye LH, He XL, Takata K, Kohama K, Shirao T. Modulatory role of drebrin on the cytoskeleton within dendritic spines in the rat cerebral cortex. J Neurosci. 1996;16:7161–7170. doi: 10.1523/JNEUROSCI.16-22-07161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirao K, Hata Y, Ide N, Takeuchi M, Irie M, Yao I, Deguchi M, Toyoda A, Sudhof TC, Takai Y. A novel multiple PDZ domain-containing molecule interacting with N-methyl-d-aspartate receptors and neuronal cell adhesion proteins. J Biol Chem. 1998;14:21105–21110. doi: 10.1074/jbc.273.33.21105. [DOI] [PubMed] [Google Scholar]
- 20.Irie M, Hata Y, Takeuchi M, Ichtchenko K, Toyoda A, Hirao K, Takai Y, Rosahl TW, Sudhof TC. Binding of neuroligins to PSD-95. Science. 1997;277:1511–1515. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- 21.Kaech S, Fischer M, Doll T, Matus A. Isoform specificity in the relationship of actin to dendritic spines. J Neurosci. 1997;17:9565–9572. doi: 10.1523/JNEUROSCI.17-24-09565.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kennedy MB. The postsynaptic density at glutamatergic synapses. Trends Neurosci. 1997;20:264–268. doi: 10.1016/s0166-2236(96)01033-8. [DOI] [PubMed] [Google Scholar]
- 23.Kennedy MB. Signal transduction molecules at the glutamatergic postsynaptic membrane. Brain Res Brain Res Rev. 1998;26:243–257. doi: 10.1016/s0165-0173(97)00043-x. [DOI] [PubMed] [Google Scholar]
- 24.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 25.Kim E, Cho K-O, Rothschild A, Jan YN, Sheng M. Heteromultimerization and NMDA-receptor-clustering activity of chapsyn-110, a member of the PSD-95 family of proteins. Neuron. 1996;17:103–113. doi: 10.1016/s0896-6273(00)80284-6. [DOI] [PubMed] [Google Scholar]
- 26.Kim J-H, Liao D, Lau L-F, Huganir RL. SynGAP: a synaptic RasGAP that associates with the PSD95/SAP90 protein family. Neuron. 1998;20:683–691. doi: 10.1016/s0896-6273(00)81008-9. [DOI] [PubMed] [Google Scholar]
- 27.Kim TW, Wu K, Xu JL, Black IB. Detection of dystrophin in the postsynaptic density of rat brain and deficiency in a mouse model for Duchenne muscular dystrophy. Proc Natl Acad Sci USA. 1992;89:11642–11644. doi: 10.1073/pnas.89.23.11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kistner U, Wenzel BM, Veh RW, Cases LC, Garner AM, Appeltauer U, Voss B, Gundelfinger ED, Garner CC. SAP90, a rat presynaptic protein related to the product of the Drosophila tumor suppressor gene dlg-a. J Biol Chem. 1993;268:4580–4583. [PubMed] [Google Scholar]
- 29.Klann E, Chen SJ, Sweatt JD. Mechanism of protein kinase C activation during the induction and maintenance of long-term potentiation probed using a selective peptide substrate. Proc Natl Acad Sci USA. 1993;15:8337–8341. doi: 10.1073/pnas.90.18.8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 31.Kornau HC, Seeburg PH, Kennedy MB. Interaction of ion channels and receptors with PDZ domain proteins. Curr Opin Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- 32.Kreutz MR, Böckers TM, Sabel BA, Hülser E, Stricker R, Reiser G. Expression and subcellular localization of a p42IP4/Centaurin-α, a-brain specific, high-affinity receptor for inositol 1,3,4,5-tetrakisphosphate and phosphatidylinositol 3,4,5-trisphosphate in rat brain. Eur J Neurosci. 1997;9:2110–2124. doi: 10.1111/j.1460-9568.1997.tb01378.x. [DOI] [PubMed] [Google Scholar]
- 33.Kurschner C, Mermelstein PG, Holden WT, Surmeier DJ. CIPP, a novel multivalent PDZ domain protein, selectively interacts with Kir4.0 family members, NMDA receptor subunits, neurexins, and neuroligins. Mol Cell Neurosci. 1998;11:161–172. doi: 10.1006/mcne.1998.0679. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Langnaese K, Seidenbecher C, Wex H, Seidel B, Hartung K, Appeltauer U, Garner A, Voss B, Mueller B, Garner CC, Gundelfinger ED. Protein components of a rat brain synaptic junctional protein preparation. Mol Brain Res. 1996;42:118–122. doi: 10.1016/s0169-328x(96)00147-7. [DOI] [PubMed] [Google Scholar]
- 36.Lau LF, Mammen A, Ehlers MD, Kindler S, Chung WJ, Garner CC, Huganir RL. Interaction of the N-methyl-d-aspartate receptor complex with a novel synapse associated protein, SAP102. J Biol Chem. 1996;271:21622–21628. doi: 10.1074/jbc.271.35.21622. [DOI] [PubMed] [Google Scholar]
- 37.Lisman JE, Goldring MA. Feasibility of long term storage of graded information by the Ca2+/calmodulin-dependent protein kinase molecules of the postsynaptic density. Proc Natl Acad Sci USA. 1988;85:5320–5324. doi: 10.1073/pnas.85.14.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matus A, Ackermann M, Pehling G, Byers HR, Fujiwara K. High actin concentration in brain dendritic spines and postsynaptic densities. Proc Natl Acad Sci USA. 1982;79:775–782. doi: 10.1073/pnas.79.23.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moriya M, Tanaka S. Prominent expression of protein kinase C (γ) mRNA in the dendrite-rich neuropil of mice cerebellum at the critical period for synaptogenesis. NeuroReport. 1994;5:929–932. doi: 10.1097/00001756-199404000-00019. [DOI] [PubMed] [Google Scholar]
- 40.Müller BM, Kistner U, Veh RW, Cases LC, Becker B, Gundelfinger ED, Garner CC. Molecular characterization and spatial distribution of SAP97, a novel presynaptic protein homologous to SAP90 and the Drosophila discs large tumor suppressor protein. J Neurosci. 1995;15:2355–2366. doi: 10.1523/JNEUROSCI.15-03-02354.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Müller BM, Kistner U, Kindler S, Chung WJ, Kuhlendahl S, Fenster SD, Lau LF, Veh RW, Huganir R, Gundelfinger E, Garner CC. SAP102, a novel postsynaptic protein that interacts with NMDA receptor complexes in vivo. Neuron. 1996;17:255–265. doi: 10.1016/s0896-6273(00)80157-9. [DOI] [PubMed] [Google Scholar]
- 42.O‘Brien RJ, Lau LF, Huganir RL. Molecular mechanisms of glutamate receptor clustering at excitatory synapses. Curr Opin Neurobiol. 1998;8:364–369. doi: 10.1016/s0959-4388(98)80062-7. [DOI] [PubMed] [Google Scholar]
- 43.Pasinelli P, Ramakers GM, Urban IJ, Hens JJ, Oestreicher AB, de Graan PN, Gispen WH. Long-term potentiation and synaptic protein phosphorylation. Behav Brain Res. 1995;23:53–59. doi: 10.1016/0166-4328(94)00124-x. [DOI] [PubMed] [Google Scholar]
- 44.Peters A, Palay SL, Webster H. The fine structure of the nervous system: neurons and their supporting cells. Oxford UP; New York: 1991. Synapses. pp. 138–211. [Google Scholar]
- 45.Ponting CP. SAM: a novel motif in yeast sterile and Drosophila polyhomeotic proteins. Protein Sci. 1995;4:1928–1930. doi: 10.1002/pro.5560040927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reymann KG, Staak S. Molecular mechanisms underlying long-term potentiation: postsynaptic glutamate receptors and protein kinase C. In: Canonico P, Scapagnini U, Pamparana F, Routtenberg A, editors. Protein kinase C in the CNS focus on neuronal plasticity, proceedings. Masson; Milano: 1994. pp. 31–56. [Google Scholar]
- 47.Richter K, Hamprecht B, Scheich H. Ultrastructural localization of glycogen phosphorylase predominantly in astrocytes of the gerbil brain. Glia. 1996;17:263–273. doi: 10.1002/(SICI)1098-1136(199608)17:4<263::AID-GLIA1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 48.Richter K, Langnaese K, Kreutz MR, Olias G, Zhai W, Scheich H, Garner CC, Gundelfinger ED. The presynaptic cytomatrix protein Bassoon is located at both excitatory and inhibitory synapses of rat brain. J Comp Neurol. 1999;408:437–448. doi: 10.1002/(sici)1096-9861(19990607)408:3<437::aid-cne9>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 49.Rosenmund C, Westbrook GL. Calcium-induced actin depolymerization reduces NMDA channel activity. Neuron. 1993;10:805–814. doi: 10.1016/0896-6273(93)90197-y. [DOI] [PubMed] [Google Scholar]
- 50.Seidel B, Zuschratter W, Wex H, Garner CC, Gundelfinger ED. Spatial and sub-cellular localization of the membrane cytoskeleton-associated protein alpha adducin in the rat brain. Brain Res. 1995;700:13–24. doi: 10.1016/0006-8993(95)00962-p. [DOI] [PubMed] [Google Scholar]
- 51.Shearman MS, Shinomura T, Oda T, Nishizuka Y. Synaptosomal protein kinase C subspecies: A. Dynamic changes in the hippocampus and cerebellar cortex concomitant with synaptogenesis. J Neurochem. 1991;56:1255–1262. doi: 10.1111/j.1471-4159.1991.tb11419.x. [DOI] [PubMed] [Google Scholar]
- 52.Siekevitz P. The postsynaptic density: a possible role in long-lasting effects in the central nervous system. Proc Natl Acad Sci USA. 1985;82:3494–3498. doi: 10.1073/pnas.82.10.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srivastava S, Osten P, Vilim FS, Khatri L, Inman G, States B, Daly C, DeSouza S, Abagyan R, Valtschanoff JG, Weinberg RJ, Ziff EB. Novel anchorage of GluR2/3 to the postsynaptic density by the AMPA receptor-binding protein ABP. Neuron. 1998;21:581–591. doi: 10.1016/s0896-6273(00)80568-1. [DOI] [PubMed] [Google Scholar]
- 54.Sternberger LA, Hardy PH, Cuculis JJ, Meyer HG. The unlabelled antibody-enzyme method of immunohistochemistry: preparation and properties of a soluble antigen-antibody complex (horseradish-peroxidase-antiperoxidase) and its use in identification of spirochetes. J Histochem Cytochem. 1970;18:315–333. doi: 10.1177/18.5.315. [DOI] [PubMed] [Google Scholar]
- 55.Tejedor FJ, Bokhari A, Rogero O, Gorczyca M, Zhang J, Kim E, Sheng M, Budnik V. Essential role for dlg in synaptic clustering of Shaker K+ channels in vivo. J Neurosci. 1997;17:152–159. doi: 10.1523/JNEUROSCI.17-01-00152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas U, Kim E, Kuhlendahl S, Koh YH, Gundelfinger ED, Sheng M, Garner CC, Budnik V. Synaptic clustering of the cell adhesion molecule fasciclin II by discs-large and its role in the regulation of the presynaptic structure. Neuron. 1997;19:787–799. doi: 10.1016/s0896-6273(00)80961-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.tom Dieck S, Sanmarti-Vila L, Langnaese K, Richter K, Kindler S, Soyke A, Wex H, Smalla K-H, Kaempf U, Fraenzer J-T, Stumm M, Garner CC, Gundelfinger ED. Bassoon, a novel zinc-finger CAG/glutamine-repeat protein selectively localized at the active zone of presynaptic nerve terminals. J Cell Biol. 1998;142:499–509. doi: 10.1083/jcb.142.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Torres R, Firestein BL, Dong H, Staudinger J, Olson EN, Huganir RL, Bredt DS, Gale NW, Yancopoulos GD. PDZ proteins bind, cluster, and synaptically colocalize with Eph receptors and their ephrin ligands. Neuron. 1998;21:1453–1463. doi: 10.1016/s0896-6273(00)80663-7. [DOI] [PubMed] [Google Scholar]
- 59.Walsh MJ, Kuruc N. The postsynaptic density: constituent and associated proteins characterized by electrophoresis, immunoblotting, and peptide sequencing. J Neurochem. 1992;59:667–678. doi: 10.1111/j.1471-4159.1992.tb09421.x. [DOI] [PubMed] [Google Scholar]
- 60.Wang JH, Feng DP. Postsynaptic protein kinase C essential to induction and maintenance of long-term potentiation in the hippocampal CA1 region. Proc Natl Acad Sci USA. 1992;1:2576–2580. doi: 10.1073/pnas.89.7.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woods DF, Bryant PJ. Molecular cloning of the lethal(1)Discs large-1 oncogene of Drosophila. Dev Biol. 1989;134:222–235. doi: 10.1016/0012-1606(89)90092-4. [DOI] [PubMed] [Google Scholar]
- 62.Wu H, Parsons JT. Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J Cell Biol. 1993;120:1417–1426. doi: 10.1083/jcb.120.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wyszynski M, Lin J, Rao A, Nigh E, Beggs AH, Craig AM, Sheng M. Competitive binding of alpha-actinin and calmodulin to the NMDA receptor. Nature. 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- 64.Ziff EB. Enlightening the postsynaptic density. Neuron. 1997;19:1163–1174. doi: 10.1016/s0896-6273(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 65.Zito K, Fetter RD, Goodman CS, Isacoff EY. Synaptic clustering of fascilin II and shaker: essential targeting sequences and role of Dlg. Neuron. 1997;19:1007–1016. doi: 10.1016/s0896-6273(00)80393-1. [DOI] [PubMed] [Google Scholar]