ACE2: from vasopeptidase to SARS virus receptor (original) (raw)
Abstract
The zinc metallopeptidase angiotensin-converting enzyme 2 (ACE2) is the only known human homologue of the key regulator of blood pressure angiotensin-converting enzyme (ACE). Since its discovery in 2000, ACE2 has been implicated in heart function, hypertension and diabetes, with its effects being mediated, in part, through its ability to convert angiotensin II to angiotensin-(1–7). Unexpectedly, ACE2 also serves as the cellular entry point for the severe acute respiratory syndrome (SARS) virus and the enzyme is therefore a prime target for pharmacological intervention on several disease fronts.
It is approaching 50 years since the discovery of the dual role of the zinc metallopeptidase angiotensin-converting enzyme (ACE) in the production of the vasoconstrictor angiotensin II (Ang II) and the destruction of the vasodilator bradykinin 1, 2. It is perhaps surprising, therefore, that the discovery of its homologue, ACE2, did not take place until almost half a century later. It took the application of two independent genomics-based approaches to reveal the presence of ACE2 in the human genome 3, 4. Given the widespread use of ACE inhibitors (ACEIs) in the treatment of hypertension and other cardiovascular conditions, there was immediate interest in ACE2 as a potential therapeutic target. This possibility was emphasized by the relatively high levels of ACE2 expression in the human heart and kidney 3, 4. The first surprises emerged from the basic characterization of the nature of ACE2 activity. Despite its close similarity to ACE and conservation of many of the key active site features, ACE2 displayed a distinct preference for substrates, operating exclusively as a carboxypeptidase, removing single amino acids, unlike ACE, which removes dipeptides from the C-terminus of a peptide 3, 4. Hence, ACE2 could not convert angiotensin I (Ang I) to Ang II and was unable to inactivate bradykinin. Furthermore, ACE2 was not susceptible to inhibition by any ACEIs tested. What has been revealed about the pharmacology of ACE2 in the subsequent three years?
1. ACE2 and vasopeptide metabolism
A key role for ACE2 is emerging in the conversion of the octapeptide Ang II to its metabolite angiotensin-(1–7) (Ang1–7). For example, a compensatory increase in Ang1–7 levels in failing human heart ventricles has been correlated with upregulation of ACE2 [5], and cardiac myocyte Ang1–7 levels are increased in ischaemic cardiomyopathy [6]. The physiological role of Ang1–7 has been controversial but it generally appears to oppose the pressor, proliferative and pro-fibrotic actions of Ang II [7], and acts through its own G-protein-coupled receptor. This would suggest that ACE and ACE2 might act as counterbalances in the renin–angiotensin system (RAS) (Figure 1). The successful production of Ace2 null (Ace2−/−) mice supports this hypothesis [8]. Ace2 knockout mice show major cardiac contractility defects and increased levels of Ang II [8] and, overall, studies in these mice suggest that ACE2 is a key regulator of cardiac function, although the mechanism by which this is mediated is unclear. However, double knockouts, in which both Ace and Ace2 genes have been deleted, do not show the cardiac defects of the Ace2 knockout alone [8], emphasizing the Yin–Yang nature of ACE and ACE2 expression. When Ace2 is transgenically overexpressed in mouse heart, cardiac defects are again observed, most notably a lethal ventricular arrhythmia, which is associated with disruption of gap junction formation [9]. The high incidence of sudden death in these mice correlated with the levels of Ace2 transgene expression. Surviving older mice showed a spontaneous downregulation of the transgene and restoration of normal cardiac function. There are also indirect associations of Ace2 expression with hypertension. In particular, Crackower et al. [8] noted that, in the rat, the Ace2 gene maps to a defined quantitative trait locus associated with hypertension and, additionally, two single nucleotide polymorphisms in the Ace2 gene locus have been associated with human cardiovascular disease [10]. Another disease model in which ACE2 expression has recently been studied is diabetes. In streptozotocin-induced diabetes in the rat, expression of renal tubule ACE2 mRNA and protein was substantially reduced whereas an increase in ACE2 protein expression was seen in the diabetic glomeruli [11]. The significance of these changes, and the underlying mechanisms involved, remain to be elucidated. Clearly, searches for possible correlations of ACE2 mRNA and protein over- or under-expression with human cardiovascular and renal disease are warranted to determine whether these correlations mirror the effects observed in rodent models. Several potent and relatively selective inhibitors of ACE2 have been described 12, 13, and the modelling of the active site of ACE2 [14], based on the recently reported structure of human ACE [15], will facilitate the development of new classes of specific inhibitors. The active site model also provides an explanation for the differences in substrate specificity and inhibitor sensitivity between ACE and ACE2 [14].
Figure 1.
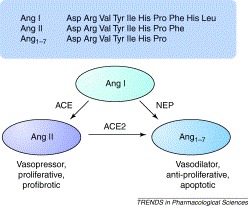
The counter-actions of angiotensin-converting enzyme (ACE) and angiotensin-converting enzyme 2 (ACE2) in angiotensin metabolism. In the classical renin–angiotensin pathway, the decapeptide Ang I (i.e. Ang1–10), derived from angiotensinogen via the action of renin, is converted by ACE to the vasoactive Ang II (Ang1–8) through the removal of the C-terminal dipeptide His-Leu 1, 2. There is increasing evidence that Ang1–7 can counterbalance some of the actions of Ang II [7], being formed from Ang II by the action of ACE2, or possibly from Ang I by the action of neprilysin (NEP). Although ACE2 can convert Ang I to Ang1–9, which can in turn be converted to Ang1–7 by ACE or NEP, this pathway is unlikely to occur at significant levels in vivo because the metabolism of Ang I by ACE2 is kinetically unfavourable compared with its conversion of Ang II (G.I. Rice. et al., unpublished) and hence is not shown.
2. ACE2 as a gateway to SARS
During several months of 2003, the virus that caused the newly identified illness severe acute respiratory syndrome (SARS) spread rapidly from China throughout Asia to Canada and beyond, causing almost 800 deaths and disrupting travel, economics and even scientific conventions. The death rate following infection approached almost 10%. Almost as mysteriously as it appeared, perhaps as a result of public health measures, the disease faded away only to re-emerge in China this year. The speed with which the infection spread and the severity of the symptoms led to a massive effort to identify the agent responsible. Within months it had been identified as a positive strand RNA virus, classified as a member of the coronavirus family (SARS-CoV); its genome was sequenced 16, 17 and the search was underway to identify the cellular receptor for the virus. As with other coronaviruses, it is the N-terminal portion (S1 domain) of the viral spike (S) glycoprotein that mediates the initial high-affinity binding to a receptor on the surface of susceptible cells. The spike sits in the viral envelope and projects outwards to give a ‘corona’-like appearance to the virus, hence its name. The replication strategy of the virus is summarized in Figure 2, and Figure 3depicts the coronavirus-infected cell.
Figure 2.
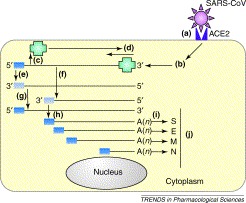
The replication strategy of the severe acute respiratory syndrome coronavirus (SARS-CoV). The replication and genome expression strategy of coronaviruses is complex. (a) The SARS-CoV spike glycoprotein recognizes angiotensin-converting enzyme 2 (ACE2) as a receptor on the cell surface. (b) Upon entering the cytoplasm, the virus core particle, which contains the genomic RNA bound to the virus nucleoprotein, is released. (c) The 5′ two-thirds of the genomic RNA is translated by host ribosomes to generate the virus replicase polyprotein (green; RNA-dependent RNA polymerase and other proteins). (d) The replicase attaches to the 3′ end of the input genome and begins (e) replication of a full-length anti-genome and (f) synthesis of negative strand subgenomic RNAs that serve as templates for the synthesis of (g) new genomic RNA and (h) viral subgenomic mRNAs (sgRNAs), respectively. All coronavirus mRNAs have conserved 5′ ends (dark blue box) and are 3′ co-terminal and polyadenylated. There are thought to be eight sgRNAs produced in SARS-CoV infected cells; (i) some of these encode the viral structural proteins (shown here). (j) Release of virus occurs after processing and assembly of virus particles in the Golgi apparatus and rough endoplasmic reticulum. Although primary replication occurs in the cytoplasm, recent evidence suggests that coronaviruses and related viruses use nuclear factors to facilitate the replication process [27].
Figure 3.
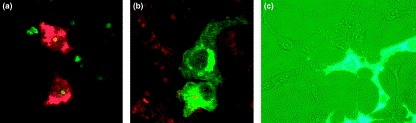
The corona virus-infected cell. (a) A confocal microscope image shows the infection of primary cells by coronaviruses. Viral proteins are shown in red and nucleolar fibrillarin is shown in green. Most viral proteins remain in the cytoplasm, although nucleoprotein can localize to the nucleolus and redistribute fibrillarin. Studies of coronavirus infection have been helped greatly by the expression of individual proteins (and mutants) and, following their cellular localization; examples of the localization of (b) membrane protein and (c) nucleoprotein are shown. (b) Membrane protein is shown in green and nucleic acid is stained with propidium iodide (red). (c) Phase-contrast image of cells expressing a green fluorescence protein–nucleoprotein fusion protein (green). Images (a) and (b) are courtesy of Torsten Wurm, and image (c) is courtesy of Jae-Hwan You.
Michael Farzan and colleagues [18] demonstrated that the S1 domain of the SARS-CoV S protein bound to the African Green monkey kidney cell line Vero E6, which is permissive for viral replication. They were then able to co-immunoprecipitate the protein responsible for viral binding and entry. This SARS-CoV receptor, a glycoprotein of Mr 110 000, was identified by mass spectrometry as ACE2. Its identity was confirmed by showing that, when ACE2 was overexpressed in human cells non-permissive for viral infection, SARS-CoV entry and replication were facilitated; this process was blocked by an ACE2 antibody [18]. The tissue distribution of ACE2 also appears to show some correlation with the sites of SARS-CoV infection and disease pathology. Independently, Xiao et al. [19] confirmed that ACE2 was a SARS-CoV receptor and showed that the receptor-binding domain was probably located between residues 272 and 537 of the spike glycoprotein. Farzan's group [20] has now established the recognition region within a 193 amino acid sequence (residues 318–510) and smaller fragments were unable to bind ACE2. A crucial aspartic acid in this region was essential for binding with ACE2. Modelling of the ACE2 structure based on the known structure of ACE has provided another approach to predicting potential binding contacts between ACE2 and the SARS-CoV S protein [21] but ultimately experimental studies are essential to identify the nature of the interactions between these two proteins. The recent solution of the structure of ACE2 [22], which supports much of the earlier modelling of the active site of the enzyme [14], should facilitate studies of ACE2 and S protein interactions. Two unexpected features of ACE2 inhibitor binding arise from the structural studies [22]. First, inhibitor binding induced a large conformational change in the enzyme that aligns crucial residues for catalysis. Second, the inhibitor binds to the enzyme in an inverse orientation from that predicted from the binding of the ACEI lisinopril to the active site of ACE [22].
Another zinc peptidase, aminopeptidase N (APN), acts as the receptor for several other coronaviruses [23]. Again, with this receptor, its distribution can be correlated with sites of infection. However, apart from the zinc-binding region, APN and ACE2 bear no discernible sequence homology and show a different membrane topology. Several features probably facilitate the usurping of plasma membrane peptidases as viral receptors. They exist as ectoenzymes in which the bulk of the protein, including the catalytic domain, faces the extracellular space, and they are heavily glycosylated. They are also abundantly expressed on certain cell types: for example, APN constitutes >5% of the membrane protein of the kidney brush border. The catalytic domain itself is not responsible for coronaviral interaction in either ACE2 or APN because mutations of key active site residues do not affect viral infection. This probably reflects the fact that the catalytic site of ACE2 is buried deep in a narrow cleft within the protein and is accessible only to the oligopeptide substrates of the enzyme.
3. Future perspectives
In a relatively short space of time, ACE2 has gained prominence in several disease states and not just those of cardiovascular origin as originally predicted from its close similarity to ACE. Other physiological correlations for ACE2 are also appearing. Recently, for example, ACE2 immunoreactivity in the kidney has been shown to double during pregnancy in rats, leading to the suggestion that ACE2 might contribute to the localized overproduction of Ang1–7 in the kidney observed in pregnancy, which might protect against rises in blood pressure [24]. It is therefore conceivable that deficient expression of ACE2 might underlie pathological conditions of pregnancy such as pre-eclampsia.
Perhaps too much attention to date has focused on the ability of ACE2 to produce Ang1–7 from Ang II. Future analysis of the physiological roles of ACE2 needs to bear in mind that it can metabolize a range of biologically active peptides other than angiotensin-related peptides 14, 25. For example, it can hydrolyse (des-Arg9)-bradykinin, which is the endogenous ligand of the bradykinin B1 receptor. In this context, the availability of potent and selective inhibitors of ACE2 12, 13 will provide valuable pharmacological tools for exploring the physiology and pathology of the enzyme. The therapeutic potential of ACE2 inhibitors in cardiovascular-related diseases, however, is questionable given the apparent cardioprotective nature of ACE2 activity; much further study is needed in this area. Indeed, mechanisms for selectively increasing cardiac or renal ACE2 levels might be desirable in some conditions; this will require an understanding of the factors that regulate its tissue-specific expression.
The biggest surprise, however, was the identification of ACE2 as a viral receptor [18] and, although current data are consistent with ACE2 as a SARS-CoV receptor, there might be other receptors or co-receptors for this virus that are yet to be discovered. Effective therapies against SARS are urgently required should further major outbreaks occur. From studies on other coronaviruses, a vaccination approach might be viable, although not without limitations [26]. Hence, soluble ACE2 fragments or ACE2 antibodies might provide alternative anti-viral therapeutic approaches, although the possible side-effects of blocking ACE2 are unknown. Indeed, infection by SARS-CoV might affect ACE2 function adversely, which could in turn contribute to some of the pathology of the disease. Given the ability of viruses to usurp cell-surface peptidases and the prevalence of ACE in respiratory and other tissues, ACE itself might have a role as a receptor for an as yet unidentified virus.
Acknowledgements
A.J.T. and N.M.H. thank the MRC and the British Heart Foundation for support of their work on ACE2. J.A.H. thanks the BBSRC for support of his coronavirus research.
References
- 1.Turner A.J, Hooper N.M. The angiotensin-converting enzyme gene family: genomics and pharmacology. Trends Pharmacol. Sci. 2002;23:177–183. doi: 10.1016/s0165-6147(00)01994-5. [DOI] [PubMed] [Google Scholar]
- 2.Acharya K.R. ACE revisited: a new target for structure-based drug design. Nat. Rev. Drug Discov. 2003;2:891–902. doi: 10.1038/nrd1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tipnis S.R. A human homolog of angiotensin-converting enzyme: cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 4.Donoghue M. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 5.Zisman L.S. Increased angiotensin-(1-7)-forming activity in failing human heart ventricles. Evidence for up-regulation of the angiotensin-converting enzyme homologue ACE2. Circulation. 2003;108:1707–1712. doi: 10.1161/01.CIR.0000094734.67990.99. [DOI] [PubMed] [Google Scholar]
- 6.Averill D.B. Cardiac angiotensin-(1-7) in ischemic cardiomyopathy. Circulation. 2003;108:2141–2146. doi: 10.1161/01.CIR.0000092888.63239.54. [DOI] [PubMed] [Google Scholar]
- 7.Ferrario C.M. Counterregulatory actions of angiotensin-(1-7) Hypertension. 1997;30:535–541. doi: 10.1161/01.hyp.30.3.535. [DOI] [PubMed] [Google Scholar]
- 8.Crackower M.A. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 9.Donoghue M. Heart block, ventricular tachycardia, and sudden death in ACE2 transgenic mice with downregulated connexins. J. Mol. Cell. Cardiol. 2003;35:1043–1053. doi: 10.1016/s0022-2828(03)00177-9. [DOI] [PubMed] [Google Scholar]
- 10.Chen A.A. Two single nucleotide polymorphisms in the ACE2 locus are associated with cardiovascular disease. Gen. Epidemiol. 2002;23:272. [Google Scholar]
- 11.Tikellis C. Characterization of renal angiotensin-converting enzyme 2 in diabetic nephropathy. Hypertension. 2003;41:392–397. doi: 10.1161/01.HYP.0000060689.38912.CB. [DOI] [PubMed] [Google Scholar]
- 12.Dales N.A. Substrate-based design of the first class of angiotensin-converting enzyme-related carboxypeptidase (ACE2) inhibitors. J. Am. Chem. Soc. 2002;124:11852–11853. doi: 10.1021/ja0277226. [DOI] [PubMed] [Google Scholar]
- 13.Huang L. Novel peptide inhibitors of angiotensin-converting enzyme 2. J. Biol. Chem. 2003;278:15532–15540. doi: 10.1074/jbc.M212934200. [DOI] [PubMed] [Google Scholar]
- 14.Guy J.L. Angiotensin-converting enzyme-2 (ACE2): Comparative modeling of the active site, specificity requirements, and chloride dependence. Biochemistry. 2003;42:13185–13192. doi: 10.1021/bi035268s. [DOI] [PubMed] [Google Scholar]
- 15.Natesh R. Crystal structure of the human angiotensin-converting enzyme-lisinopril complex. Nature. 2003;421:551–554. doi: 10.1038/nature01370. [DOI] [PubMed] [Google Scholar]
- 16.Rota P.A. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 17.Marra M.A. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 18.Li W. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao X. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong S.K. A 193-amino-acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prabakaran P. A model of the ACE2 structure and function as a SARS-CoV receptor. Biochem. Biophys. Res. Commun. 2004;314:235–241. doi: 10.1016/j.bbrc.2003.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Towler, P. et al. ACE2 structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. (in press). [DOI] [PMC free article] [PubMed]
- 23.Yeager C.L. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brosnihan K.B. Enhanced renal immunocytochemical expression of ANG-(1-7) and ACE2 during pregnancy. Hypertension. 2003;42:749–753. doi: 10.1161/01.HYP.0000085220.53285.11. [DOI] [PubMed] [Google Scholar]
- 25.Vickers C. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J. Biol. Chem. 2002;277:14838–14843. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- 26.Cavanagh D. Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus. Avian Pathol. 2003;32:567–582. doi: 10.1080/03079450310001621198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiscox J.A. The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res. 2003;95:13–22. doi: 10.1016/S0168-1702(03)00160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]