Regulation of p53 Function and Stability by Phosphorylation (original) (raw)
Abstract
The p53 tumor suppressor protein can be phosphorylated at several sites within the N- and C-terminal domains, and several protein kinases have been shown to phosphorylate p53 in vitro. In this study, we examined the activity of p53 proteins with combined mutations at all of the reported N-terminal phosphorylation sites (p53N-term), all of the C-terminal phosphorylation sites (p53C-term), or all of the phosphorylation sites together (p53N/C-term). Each of these mutant proteins retained transcriptional transactivation functions, indicating that phosphorylation is not essential for this activity of p53, although a subtle contribution of the C-terminal phosphorylation sites to the activation of expression of the endogenous p21Waf1/Cip1-encoding gene was detected. Mutation of the phosphorylation sites to alanine did not affect the sensitivity of p53 to binding to or degradation by Mdm2, although alteration of residues 15 and 37 to aspartic acid, which could mimic phosphorylation, resulted in a slight resistance to Mdm2-mediated degradation, consistent with recent reports that phosphorylation at these sites inhibits the p53-Mdm2 interaction. However, expression of the phosphorylation site mutant proteins in both wild-type p53-expressing and p53-null lines showed that all of the mutant proteins retained the ability to be stabilized following DNA damage. This indicates that phosphorylation is not essential for DNA damage-induced stabilization of p53, although phosphorylation could clearly contribute to p53 stabilization under some conditions.
The p53 tumor suppressor protein plays an important role in the cellular response to stress such as DNA damage and hypoxia, and loss of p53 is associated with genomic instability and tumor development (28). Activation of p53 can lead to cell cycle arrest, which can be reversed under some circumstances, or apoptotic cell death (3). Both of these responses prevent replication of cells with damaged DNA and are thereby thought to protect cells from an accumulation of potentially oncogenic genetic alterations. The ability of other signals, such as hypoxia or deregulated proliferation, to activate a p53 response may allow the elimination of cells which are already dividing under abnormal conditions.
One of the principal mechanisms by which p53 functions is as a transcription factor, and many cellular genes have been identified which are transcriptionally regulated by p53. Broadly, these genes can be divided into those likely to contribute to activation of a cell cycle arrest, such as the gene encoding the cyclin-dependent kinase inhibitor p21Waf1/Cip1, and those which are more likely to play a role in mediating the apoptotic response, such as the genes encoding Bax and KILLER/DR5 (3). Another important p53 target gene is Mdm2. Although the Mdm2 protein does not play a direct role in mediating the p53 response, it is crucially involved in negatively regulating p53 protein levels (26). Mdm2 binds directly to p53 and targets it for degradation through the ubiquitin-dependent proteolytic pathway (19, 24). In this way, Mdm2 is responsible, at least in part, for maintaining low levels of p53 under conditions of normal cell growth (5), although Mdm2-independent degradation targeted by kinase-inactive JNK has also recently been described (13).
Under most circumstances, signals which activate the p53 response lead to rapid elevation of the p53 protein, principally through stabilization of the protein, and activation of the DNA binding function of p53. Regulation of p53 stability could be achieved by regulating the ability of Mdm2 to target p53 for degradation, and the DNA binding activity of p53 can be regulated independently by alterations in protein conformation. In particular, the extreme C terminus of p53 has been implicated in playing a negative regulatory role, maintaining p53 in a latent form which can be activated by several mechanisms which modify this C-terminal region (15, 16, 20, 44).
Posttranslational modification of p53 by phosphorylation has been proposed to be an important mechanism by which p53 stabilization and function are regulated (34). p53 has been described as a substrate for many kinases in vitro, and phosphorylation of numerous serine and threonine residues within the N- and C-terminal regions of the protein has been shown (40). Despite substantial information on kinases which can phosphorylate p53 in vitro, comparatively little is known about which sites in p53 are phosphorylated and by which kinases in vivo. In recent studies using phosphospecific antibodies, serines 15 and 37 were identified as sites of phosphorylation in cells following DNA damage (45, 46), with evidence that the ATM kinase may participate in the phosphorylation of serine 15 in response to some, but not all, activating signals (1, 6, 46). Phosphorylation of serine 33 in response to UV or ionizing radiation has also been shown recently (43) with evidence that N-terminal phosphorylation of p53 can direct C-terminal acetylation of the protein. Phosphorylation at the CK2 site (serine 392) has also been confirmed in vivo with evidence that this site is phosphorylated following UV irradiation but not gamma irradiation (4, 23).
Although there is substantial evidence that p53 can be phosphorylated at several sites, the consequences of such phosphorylation for p53 function are still very poorly understood. Analyses of p53 proteins from a number of different species carrying mutations at either one or several of the potential phosphorylation sites have been carried out in a number of cellular systems. Many of these studies have shown contradictory and confusing results, probably due to variability in the cell types and p53 proteins used. For example, mutation of the CK2 site had no apparent effect on the growth-suppressive function of p53 (7, 10, 12), while mutation of this site reduced the ability of p53 to suppress transformation (7, 37). There is also evidence that phosphorylation of this site is important to maintain the transcriptional activity of mouse p53 in contact-inhibited cells (17). Similarly, although loss of phosphorylation of serine 15 in human p53 has been shown to extend the half-life of p53 (9), phosphorylation of this residue has also been suggested to play a role in preventing interaction with Mdm2, leading to the prediction that loss of this site might prevent stabilization of the protein (38, 45). Phosphorylation of p53 by MEKK1/JNK has also been reported to stabilize p53 by inhibiting interaction with Mdm2 (14). In general, functional studies such as these have shown only subtle defects in phosphorylation site mutant p53, although defects in the transcriptional activity of a p53 protein containing several N-terminal phosphorylation site mutations have been reported (32).
Since p53 has been shown to be phosphorylated at several sites within the N- and C-terminal regions, it is possible that mutation of a number of sites in combination is required for any global effects on the regulation of p53 or on p53 activity itself. To address this problem and to determine the requirement of phosphorylation for p53 function, we created three human p53 mutant proteins in which all of the potential N-terminal phosphorylation sites, all of the C-terminal phosphorylation sites, or all of the N- and C-terminal sites were altered in combination to nonphosphorylatable residues within the full-length protein. We then analyzed these mutant proteins for the ability to be phosphorylated in vivo, the ability to function in transcriptional assays, and stability in response to DNA-damaging signals.
MATERIALS AND METHODS
Construction of mutant p53 cDNAs.
The human p53 N-terminal (p53N-term) and C-terminal (p53C-term) phosphorylation mutant proteins were generated independently by PCR mutagenesis. To generate the p53N-term mutant protein, an initial PCR using forward primer 5′-GCCATGGAGGAGCCGCAGGCAGATCCTGCCGTCGAGCCCCCTCTGGCTCAGGAAGCATTTGCAGACC-3′ (the underlined sequences are the mutated codons) and reverse primer 5′-GATGGCCATGGCGCGGACGCG-3′ was performed. This was subcloned into human Sp65p53Pro at the _Nco_I site and oriented by using _Pvu_II. This construct was used as the template in a second PCR using forward primer 5′-ATAGAATCCGGGAGCAGGTAGCTGCTGGG-3′ and reverse primer 5′-ATCATCCATTGCTTGGGCCGGCAAGGGGGCCAGAACG-3′. The PCR product was subcloned into Sp65p53Δ210 at the _Eco_RI/_Pfl_MI sites. The _Nco_I fragment from the resulting construct was subcloned into either Sp65p53Pro or Sp65p53C-term (described below) to generate Sp65p53N-term and Sp65p53N/C-term, respectively. Sequencing analysis (ABI prism) was carried out to confirm that all mutations had been made correctly.
To generate the p53C-term mutant protein, a PCR was carried out by using forward primer 5′-ATAGAATCCGGGAGCAGGTAGCTGCTGGG-3′, reverse primer 5′-CCATGGATCCGTCTGCGTCAGGCCCTTCTG-3′, and the p53 cDNA template pCB6+371A/376A/378A. The resulting PCR product was subcloned into Sp65 at the _Eco_RI/_Bam_HI sites. This construct was used as a backbone to subclone the _Eco_RI/_Stu_I fragment from pGem4Zp53-315A to generate the final p53C-term mutant protein. Sequencing analysis (ABI prism) was carried out to confirm that all mutations had been made correctly.
The 15A/37A, 15A/37D, 15D/37A, and 15D/37D mutant combinations were generated in a similar manner by using forward primer 5′-GCCATGGAGGAGCCGCAGTCAGATCCTAGCGTCGAGCCCCCTCTGGCTCAG-3′ or 5′-GCCATGGAGGAGCCGCAGTCAGATCCTAGCGTCGAGCCCCCTCTGGATCAG-3′ and the appropriate cDNA template (pCB6+p53-37A or pCB6+p53-37D). Sequencing analysis (ABI prism) was carried out to confirm that all mutations had been made correctly. Similarly, PCR mutagenesis was performed to generate the 18A, 18D, 20A, and 20D point mutations, which were subsequently subcloned into pCB6+ at the _Eco_RI/_Bam_HI sites.
To generate the ΔII mutant combinations, the _Nco_I and _Pfl_MI fragments from pGem4Zp53ΔII were subcloned into Sp65p53C-term or Sp65p53N-term, respectively, to generate Sp65p53ΔIIC-term and Sp65p53ΔIIN-term. The Sp65p53ΔII-15A/37A and Sp65p53ΔI-ΔII combination mutant cDNAs were generated in a similar manner. All ΔII mutant combinations were confirmed by sequencing analysis (ABI prism).
Plasmids and antibodies.
The pCB6+ expression plasmid, containing the cytomegalovirus (CMV) early promoter (27), was used to generate all expression constructs. The human p53ΔI, p53ΔII, p53Δ370 (C-terminal truncation), p53-37A, p53-37D, and p53-315A mutant cDNAs were previously generated by N. Marston et al. (30). The mouse Mdm2 expression plasmid has already been described (pCOC Mdm2 X2) (18), as have the luciferase reporter plasmids containing the p21Waf1/Cip1 promoter sequence pWWP-luc (8), the Mdm2 promoter sequence pGL2NA(mdm2)-luc (21), and the Bax promoter sequence pGL3Bax-luc (11). The pEGFP-N1 plasmid was purchased from Clontech. p53-specific monoclonal antibodies PAb1801 and D0-1 have been described previously (2, 48). A p21Waf1/Cip1-specific antibody was purchased from Oncogene Science. Monoclonal antibodies to green fluorescent protein (GFP) and actin were purchased from Clontech and Chemicon International, respectively, and used in Western blot analysis to assess protein levels where indicated.
Cell culture and transfections.
The Saos-2 human osteosarcoma line (null for wild-type p53), the MCF-7 human breast tumor line (expressing wild-type functional p53), and p53-null mouse embryo fibroblasts (MEFs) were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (FCS) at 37°C in an atmosphere of 10% CO2 in air.
For transient transfection of Saos-2 cells and MEFs, 8 × 105 cells were plated in 10-cm2 dishes and transfected the following day by calcium chloride precipitation with 50 ng to 10 μg of either a vector control (pCB6+) or a human wild-type p53 or mutant p53 expression plasmid where indicated. For luciferase reporter assays, cells were cotransfected with 5 μg of each reporter construct. To assess Mdm2 effects, cells were cotransfected with 9 μg of mouse Mdm2. To assess transfection efficiency and normalize protein levels, transfections also included either 1 μg of pEGFP-N1 or 5 μg of pCB6+βgal where indicated. Cells were washed on the day following transfection, harvested 24 h later, and prepared for Western blotting, flow cytometry, or luciferase and β-galactosidase analyses as previously described (41, 42). For stable transfection of MCF-7 cells, 5 × 105 cells were plated in 10-cm2 dishes and transfected as for transient transfection with 10 μg of either a vector control (pCB6+) or a human p53ΔII or p53ΔII mutant expression plasmid where indicated. Cells were split at 1:20 into 15-cm2 dishes (in duplicate) 24 h after washing and selected with G418 (600 μg/ml) on the following day. Selected clones were isolated and expanded for further analysis.
In vivo 32P-phosphate labeling of cells.
To assess the relative phosphorylation of the mutant proteins in vivo, Saos-2 cells were transiently transfected with 5 μg of either a pCB6+ control plasmid or a wild-type p53, p53-15A, p53N-term, p53C-term, or p53N/C-term expression plasmid as described above. Approximately 4 h prior to harvesting, the cells were washed twice in 5 ml of phosphate-free medium (per 10-cm2 dish) and then incubated at 37°C in 3 ml of phosphate-free medium supplemented with 10% dialyzed FCS containing 1 mCi of 32P-labeled sodium orthophosphate for 4 h. Cells were lysed in radioimmunoprecipitation assay lysis buffer (20 mM Tris [pH 8.0], 137 mM NaCl, 0.5 mM EDTA, 10% glycerol, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate [SDS], 1% deoxycholate) (ice cold), and p53 protein was immunoprecipitated with PAb1801-protein A Sepharose beads. Samples were separated by SDS–10% polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose, and phosphorylated protein was assessed by PhosphorImager analysis (Storm 860 PhosphorImager; Molecular Dynamics Inc.). To assess immunoprecipitated p53 levels, blots were probed with PAb1801. Additionally, for each experiment, phosphorylated proteins were assessed directly from SDS–10% PAGE by PhosphorImager analysis.
In vitro association assays.
To determine the ability of each of the mutant proteins to bind to Mdm2 in vitro, an in vitro association assay was performed by using in vitro-translated proteins. The wild-type p53, p53N-term, p53C-term, and p53N/C-term proteins were translated in the presence of 35S-Pro-Mix (Amersham) and human flag-Mdm2 was translated in the absence of 35S-Pro-Mix (Amersham) by using a reticulocyte lysate-based in vitro translation kit as directed by the manufacturer (Promega). A 5-μl volume of hot, in vitro-translated wild-type p53, p53N-term, p53C-term, or p53N/C-term protein was mixed with 5 μl of cold, in vitro-translated flag-Mdm2 protein and incubated for 30 min at 30°C. A 500-μl volume of LSAB buffer (100 mM NaCl, 100 mM Tris [pH 8.0], 1% Nonidet P-40) supplemented with 3% bovine serum albumin was then added to each sample. Samples were spun at full speed for approximately 5 min at room temperature and transferred to a fresh microcentrifuge tube. A 5-μl volume of anti-flag antibody was added, and samples were incubated at 4°C for 2 h to overnight with gentle rotation. Flag-Mdm2 complexes were precipitated by the addition of 50 μl of protein G Sepharose beads, and samples were incubated for 1 h at 4°C with gentle rotation. Immunoprecipitated complexes were washed five times with LSAB buffer, resuspended in 50 μl of 2 × SDS sample buffer, boiled for 10 min, and then assessed by SDS–10% PAGE. To assess 35S-labeled protein, gels were dried and exposed to Kodak film.
Induction of p53 proteins in MCF-7 cells and p53-null MEFs.
To assess DNA damage-induced stabilization of endogenous wild-type p53 and exogenous mutant p53 in MCF-7 clonal lines stably expressing the ΔII mutant combinations, cells were plated at 8 × 105 into 10-cm2 dishes. At least 24 h after plating, cells were treated with actinomycin D (5 nM) or irradiated with UV light at 50 J/m2. Cells were harvested at 10 to 16 h for actinomycin D treatment and at 6 h for UV treatment. Cells were harvested and lysed directly by the addition of 500 μl of 2 × SDS sample buffer. Samples were boiled, analyzed by SDS–10% PAGE, and assessed by Western blotting using PAb1801 and D01. Stabilization of mutant p53 in p53-null MEFs was carried out by transiently transfecting cells with 500 ng of DNA and carrying out the DNA damage protocols described above at 24 h posttransfection.
The half-life of the p53 protein in MCF-7 cells stably expressing the ΔII mutant combinations was determined after treatment with or without actinomycin D (5 nM, 16 h) by radioactive pulse-labeling of cells for 30 min and a chase in unlabeled medium for 0, 1, 2, or 4 h. Endogenous wild-type p53 and exogenous p53 were then immunoprecipitated by using monoclonal antibody PAb1801 or PAb421 (the two gave similar results). Quantification of labeled p53 protein was carried out by using the Storm 860 PhosphorImager (Molecular Dynamics Inc.).
RESULTS
Phosphorylation site mutant p53.
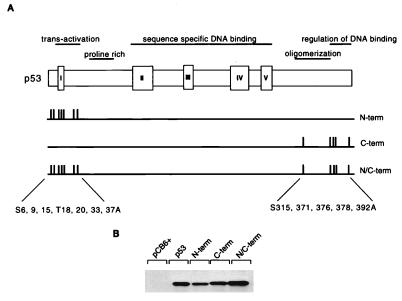
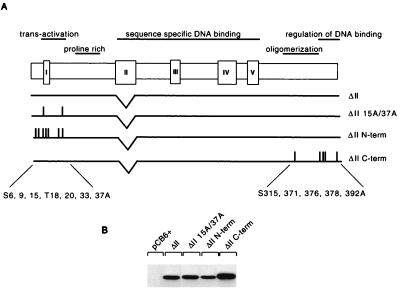
Site-directed mutagenesis was used to construct mutant p53 carrying alanine substitutions at the known or predicted phosphorylation sites within the p53 N and C termini (Fig. 1A). The N-terminal amino acids serine 6, serine 9, serine 15, threonine 18, serine 20, serine 33, and serine 37 were replaced in the mutant protein p53N-term. The mutant protein p53C-term carried substitutions of serine 315, serine 371, serine 376, serine 378, and serine 392. A further mutant protein, for which both the p53N- and p53C-term mutant proteins were combined, was also created and called p53N/C-term. All of the mutant proteins were efficiently expressed to levels comparable to that of wild-type p53 following transient transfection into p53-null Saos-2 cells (Fig. 1B).
FIG. 1.
(A) Schematic representation of the human p53 protein indicating the important functional domains (transactivation, proline rich, sequence-specific DNA binding, oligomerization, and regulation of DNA binding), conserved box regions I to V, and potential sites of phosphorylation within the N- and C-terminal regions. To generate N-terminal phosphorylation mutant p53 (N-term), Ser6, Ser9, Ser15, Thr18, Ser20, Ser33, and Ser37 were mutated to Ala in combination. To generate C-terminal phosphorylation mutant p53 (C-term), Ser315, Ser371, Ser376, Ser378, and Ser392 were mutated to Ala in combination. N- and C-terminal phosphorylation mutant p53 (N/C-term) was generated by a combination of the above. (B) Western blot analysis (using PAb1801) of Saos-2 cells transiently expressing wild-type p53 and the p53N-term, p53C-term, and p53N/C-term mutant proteins after transient transfection with 5 μg of the respective plasmid. Cells were harvested at 24 h.
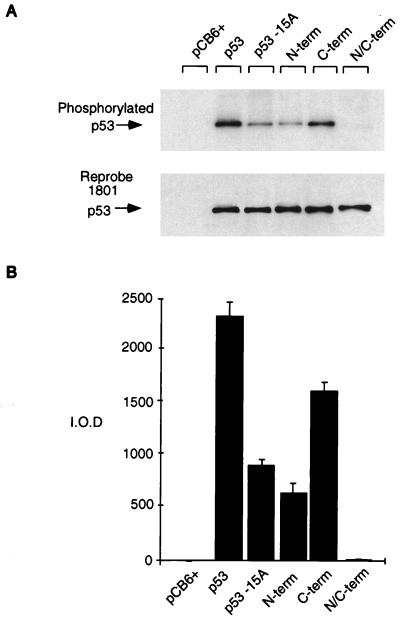
In order to determine whether all of the major phosphorylation sites had been altered in these mutant proteins, we carried out orthophosphate labeling following transient transfection in Saos-2 cells. Efficient phosphorylation of the wild-type protein was achieved under these conditions (Fig. 2), with each of the mutants showing a reproducible reduction in phosphate incorporation in vivo. The p53N/C-term mutant protein, carrying substitutions of all the candidate phosphorylation sites under investigation, showed no detectable phosphorylation, indicating that all of the major kinase sites had been lost. Mutation of the N- or C-terminal sites resulted in a decrease, but no total loss, of phosphorylation, confirming that sites within both of these regions are targets for kinases in vivo. These results indicated, however, that the N terminus of p53 is more extensively phosphorylated than the C terminus. Recent results have shown that serine 15 is a major phosphorylation site of p53 in vivo, and in agreement with this, we have found that mutation of only this residue in the full-length protein greatly reduces phosphorylation to almost the same extent as mutation of all of the other potential N-terminal phosphorylation sites together. However, there appears to be at least one other phosphorylation site within the N-terminal region, in agreement with a recent study showing three phosphorylation sites within the N-terminal 24 amino acids of p53 (46).
FIG. 2.
Relative phosphorylation of p53 mutant proteins in vivo. Phosphorylation site mutant p53 proteins (5 μg) were transiently transfected into Saos-2 cells. At approximately 4 h prior to harvesting (at 24 h), the cells were washed and incubated in 5 ml of phosphate-free medium (supplemented with 10% dialyzed FCS) containing 1 mCi of 32P-labeled sodium orthophosphate (per 10-cm2 dish). Cells were lysed with radioimmunoprecipitation assay lysis buffer, and p53 protein was immunoprecipitated with PAb1801-protein A Sepharose beads. (A) Phosphorylated protein was assessed by SDS–10% PAGE (top), and blots were reprobed with PAb1801 to assess immunoprecipitated p53 levels (bottom). (B) Phosphorylated protein was quantified by PhosphorImager analysis and represented graphically by using absolute integrated optical density units (I.O.D.). The graph represents the results of four independent experiments.
Activity of phosphorylation site mutant proteins.
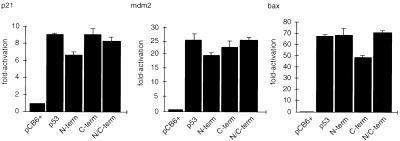
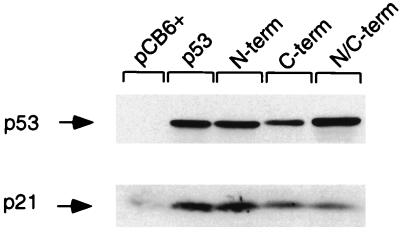
Many previous studies have examined the effect of mutating one of several phosphorylation sites within p53 on the biological activity of the p53 protein, with varied results. Some analyses showed no effect of loss of phosphorylation sites compared to the wild-type protein; others showed subtle alterations in the ability to modulate cell cycle progression or inhibition of cell growth. However, none of those previous studies examined p53 mutant proteins that were simultaneously mutated at all phosphorylation sites and were therefore unable to address the consequences of complete loss of phosphorylation for p53 function. We examined the activity of our mutant proteins in transactivation assays by using a series of p53-responsive promoters (Fig. 3). Each of the mutant proteins was as efficient as wild-type p53 in activating transcription from the human p21Waf1/Cip1, Mdm2, and bax promoters in luciferase reporter constructs in these transient assays. By using Western blot analysis and flow cytometry to estimate p53 expression levels and the percentage of cells transfected, we estimated that p53 proteins are overexpressed approximately fivefold per cell following transient transfection with 500 ng of a p53 expression plasmid compared with the levels seen following DNA damage of a wild-type p53-expressing cell (data not shown). Each of the mutant proteins was also able to elevate levels of endogenous p21Waf1/Cip1 protein following transient transfection into Saos-2 cells (Fig. 4), although slightly reduced activation of p21Waf1/Cip1 by p53 proteins with C-terminal phosphorylation site mutations was seen in several repeats of this experiment. However, under the conditions used in this assay, we were unable to detect any differences between the phosphorylation site mutant proteins and wild-type p53 in the activation of cell cycle arrest or apoptosis (data not shown).
FIG. 3.
Transcriptional activation by p53 phosphorylation mutant proteins assessed by using a luciferase reporter assay. p53 phosphorylation mutant proteins (50 ng) were cotransfected into Saos-2 cells with 5 μg of either a human p21Waf1/Cip1, Mdm2, or bax luciferase reporter construct. Five micrograms of pCB6+βgal was included to assess transfection efficiency. Cells were harvested at 24 h, lysed, and assessed for luciferase and β-galactosidase activities. Each graph shows fold activation relative to that of the pCB6+ control harvested at 24 h.
FIG. 4.
Wild-type p53 and p53 phosphorylation mutant proteins (5 μg) were transiently transfected into Saos-2 cells and the cells were harvested at 24 h for Western blot analysis to assess p21Waf1/Cip1 protein levels by using an anti-p21 antibody.
Regulation of p53 stability.
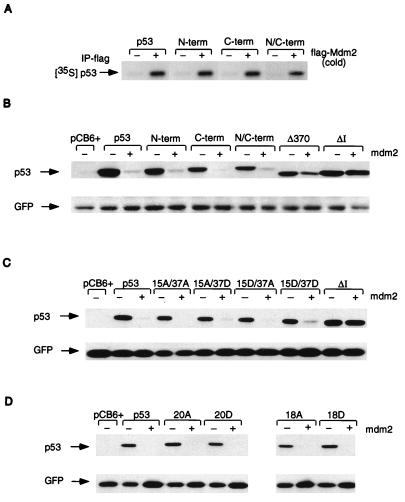
Although phosphorylation of p53 does not appear to be essential for p53 function, several studies have suggested that phosphorylation of p53 plays an important role in regulating p53 activities such as stability and DNA binding. p53 stability has recently been shown to be regulated through interaction with Mdm2 (19, 24), and in vitro analysis showed that each of the phosphorylation site mutant proteins was able to bind to Mdm2 (Fig. 5A). We then analyzed the sensitivity of each of the mutant proteins to Mdm2-mediated degradation in a transient assay (Fig. 5B) in which each mutant protein was expressed in a p53-null cell line with or without the addition of exogenous Mdm2. As previously reported, wild-type p53 was efficiently degraded following expression of exogenous Mdm2, and mutations within the N terminus of p53 which prevent interaction with Mdm2 (ΔI) (24) or deletions within the C terminus of p53 which do not interfere with Mdm2 binding (Δ370) (25) rendered the p53 protein resistant to degradation by Mdm2. All of the phosphorylation site mutant proteins retained sensitivity to Mdm2-mediated degradation in a manner essentially indistinguishable from that of wild-type p53. It has recently been shown in an in vitro association assay that phosphorylation of serines 15 and 37 by dsDNA-PK reduces the ability of p53 to bind to Mdm2 (38, 45). This study also indicated that alanine substitution at these sites enhances the binding of Mdm2 in vitro, and it is therefore not surprising that the phosphorylation site mutant proteins under investigation here (which also have alanine substitutions) remain susceptible to Mdm2-mediated degradation. We therefore assessed whether replacement of serines 15 and 37 with aspartic acid (which mimics the charge of a phosphorylation group) would render the protein resistant to Mdm2-mediated degradation when transiently cotransfected into Saos-2 cells. Although individual alteration of serine 15 or 37 to alanine or aspartic acid did not significantly affect the susceptibility of the p53 protein to Mdm2-mediated degradation, mutation of both serines 15 and 37 to aspartic acid rendered the protein very slightly resistant to degradation (Fig. 5C), consistent with the previous reports that phosphorylation at these sites reduces Mdm2 binding. This effect was very subtle, even when titration of smaller amounts of Mdm2 was used (data not shown). Individual mutations of other phosphorylation sites in or close to the Mdm2 binding site (threonine 18 and serine 20) also failed to affect the sensitivity of p53 to Mdm2-mediated degradation in this assay (Fig. 5D).
FIG. 5.
(A) In vitro association assay using in vitro-translated, unlabeled, human flag-Mdm2 (cold) and 35S-labeled, in vitro-translated wild-type p53 and p53N-term, p53C-term, and p53N/C-term mutant proteins. Complexes were immunoprecipitated (IP) with anti-flag antibody in the presence of protein G Sepharose beads, and radioactively labeled p53 proteins were assessed by SDS–10% PAGE. (B) Western blot analysis (using PAb1801) of Saos-2 cells after transient cotransfection of the phosphorylation mutant p53 (3 μg), mouse Mdm2 (9 μg), and pEGFP-N1 (1 μg). Cells were harvested at 24 h. The ΔI mutant protein (deletion of conserved box I, the region containing the Mdm2 binding site), which is resistant to Mdm2-mediated degradation, and the Δ370 mutant protein (C-terminal truncation mutant protein), which is partially resistant to Mdm2-mediated degradation, were used as controls. Relative transfection efficiency was monitored by expression of cotransfected GFP. (C) Western blot analysis (using PAb1801) of Saos-2 cells after transient cotransfection of the p53-15A/37A, -15A/37D, -15D/37A, and -15D/37D combination mutant proteins (3 μg), mouse Mdm2 (9 μg), and pEGFP-N1 (1 μg). Cells were harvested at 24 h. Relative transfection efficiency was monitored by expression of cotransfected GFP. (D) Western blot analysis (using PAb1801) of Saos-2 cells after transient cotransfection of the p53-20A, -20DD, -18, and -18D combination mutant proteins (3 μg), mouse Mdm2 (9 μg), and pEGFP-N1 (1 μg). Cells were harvested at 24 h. Relative transfection efficiency was monitored by expression of cotransfected GFP.
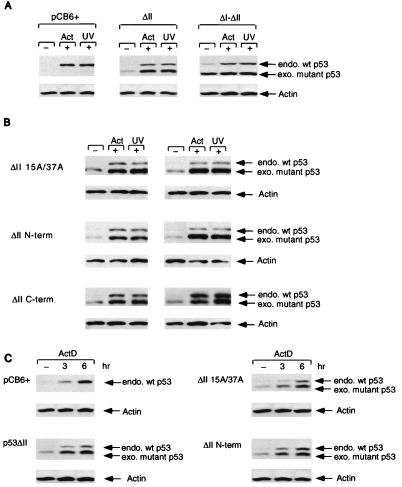
One attractive hypothesis is that phosphorylation of p53 plays an important role in allowing the protein to become resistant to degradation, with the prediction that phosphorylation site mutant proteins would show a defect in the ability to become stabilized following exposure to signals which normally activate a p53 response, such as DNA damage. In our transient assays, there was no clear indication that the phosphorylation site mutant proteins were less stable or were expressed at lower levels than the wild-type protein. We were concerned, however, that high levels of transient expression were masking a more subtle regulation of p53 stability and therefore turned to the analysis of cells stably expressing the phosphorylation site mutant proteins. Since wild-type p53 and all of the mutant proteins under consideration in this study induce cell cycle arrest and apoptosis, we took advantage of the observation that deletion of conserved regions in the DNA binding domain of p53 renders the protein unable to arrest cell growth but still responsive to normal regulation of stability through Mdm2 (35). The N-term, C-term, and 15A/37A phosphorylation site mutant proteins were therefore made in the context of a deletion of conserved region II (ΔII) (Fig. 6A), which abolishes the growth-inhibitory functions of p53 (7). All of the ΔII combination mutant proteins were expressed to similar levels when transiently transfected into Saos-2 cells (Fig. 6B), and we therefore stably transfected each of these mutant proteins into MCF-7 cells, which express endogenous wild-type p53. Initial analysis of polyclonal cell lines showed that each of the exogenous mutant proteins was expressed to low levels in MCF-7 cells and that, like that of endogenous wild-type p53, the level of each mutant protein increased following DNA damage by actinomycin D or UV irradiation (data not shown). Clones of cells expressing the mutant p53 proteins were then isolated and subjected to closer analysis. Expression of the mutant protein was slightly higher than that of the endogenous protein, possibly because the mutant protein is expressed from the efficient CMV promoter. It was clear that either actinomycin D treatment or UV irradiation resulted in rapid elevation of both endogenous wild-type p53 and exogenous p53ΔII which retained all of the wild-type phosphorylation sites (Fig. 7A). The p53ΔI-ΔII mutant protein used as a control, which is unable to bind Mdm2, was stable in untreated cells, and protein levels were not further increased by either actinomycin D or UV treatment (Fig. 7A). Importantly, this observation indicates that the increase in p53ΔII after actinomycin D treatment and UV irradiation is not due to enhanced activation of the CMV promoter at these time points. We did note that stable expression of the p53ΔI-ΔII mutant protein in untreated cells occasionally resulted in a very slight elevation of endogenous p53 protein levels compared to those seen in other uninduced lines (Fig. 7A).
FIG. 6.
(A) Schematic representation of the conserved box II deletion mutant proteins. (B) Western blot analysis (using PAb1801) of Saos-2 cells transiently expressing p53ΔII and the p53ΔII 15A/37A, p53ΔII N-term, and p53ΔII C-term mutant proteins after transient transfection with 5 μg of the respective plasmid. Cells were harvested at 24 h.
FIG. 7.
Stable expression of p53ΔII combination mutant proteins in MCF-7 cells which express wild-type (wt) p53. Stable clones were isolated, and p53 protein levels were assessed by Western blot analysis using PAb1801 or D01 after treatment with either 5 nM actinomycin D (Act; harvested at 10 h) or 50-J/m2 UV (harvested at 6 h). (A) Western blot analysis of MCF-7 stable cell lines expressing either the pCB6+ control (leftmost blot), p53ΔII (middle blot), or p53ΔI-ΔII (rightmost blot). Equal protein loading was confirmed by actin expression. endo., endogenous; exo, exogenous. (B) Western blot analysis of two independent MCF-7 stable cell lines (clone 1, left; clone 2, right) expressing either p53ΔII15A/37A (top), p53ΔIIN-term (middle), or p53ΔIIC-term (bottom). Equal protein loading was confirmed by actin expression. (C) Western blot time course analysis of actinomycin D (ActD) treatment (0, 3, and 6 h) of MCF-7 cells stably expressing either pCB6+ (control), p53ΔII (clone 1), p53ΔII15A/37A (clone 1), and p53ΔIIN-term (clone 1). All blots were reprobed with an antiactin monoclonal antibody to assess protein levels. Equal protein loading was confirmed by actin expression.
Having established that the MCF-7 cells could be used to measure DNA damage-induced protein stabilization of both endogenous and exogenous p53, we examined several independently derived clones of cells expressing the phosphorylation site mutant proteins (Fig. 7B). In each case, levels of the mutant p53 proteins were clearly elevated in response to either actinomycin D treatment or UV irradiation, to levels similar to those seen with the ΔII mutant protein alone. Similar results were also obtained for treatment with gamma irradiation (data not shown). Treatment of the cells with a proteasome-calpain inhibitor (_N_-acetyl-Leu-norleuceinal) stabilized both the wild-type and mutant p53 proteins in the absence of DNA damage to levels which were not further elevated by actinomycin D treatment or UV irradiation (data not shown). In an attempt to distinguish more modest differences in the rate of stabilization of the phosphorylation site mutant proteins, the time course of induction was analyzed. All of the exogenous p53 mutant proteins were induced at similar rates over the same time course as endogenous wild-type p53 (Fig. 7C). To confirm that the increase in the wild-type and mutant proteins following DNA damage was due to an increase in stability, the half-lives of the p53ΔII, p53ΔI-ΔII, and p53ΔIIN-term mutant proteins and the respective endogenous wild-type proteins for each line were assessed by radioactive pulse-labeling of the cells in the absence of actinomycin D and after treatment with actinomycin D (Table 1). The results show that both the p53ΔII and p53ΔIIN-term proteins are stabilized to comparable degrees by an increase in half-life following treatment, from approximately 1 to 3 h, indicating that mutation of all of the phosphorylation sites within the N-terminal region of p53 does not impair stabilization of the protein after DNA damage. Furthermore, these results also confirm that the elevated levels of p53ΔI-ΔII protein observed in undamaged cells (Fig. 7A, rightmost blot) is due to increased stability compared with the control, as shown previously for p53ΔI (24), and that the half-life of the p53ΔI-ΔII protein does not increase after DNA damage.
TABLE 1.
Half-lives of endogenous wild-type p53 and exogenously expressed p53 mutant proteins in MCF-7 cells in the absence of or after treatment with actinomycin Da
| Protein | Half-life (min) | |||
|---|---|---|---|---|
| Expt 1 | Expt 2 | |||
| − ActD | + ActD | − ActD | + ActD | |
| p53ΔII | ||||
| Endo wt p53 | 34 | 105 | 64 | 129 |
| Exo mutant p53 | 76 | 163 | 85 | 148 |
| p53ΔIIN-term | ||||
| Endo wt p53 | 49 | 184 | 52 | 188 |
| Exo mutant p53 | 84 | 202 | 75 | 202 |
| p53ΔI-ΔII | ||||
| Endo wt p53 | 34 | 104 | 53 | 148 |
| Exo mutant p53 | >240 | >240 | >240 | >240 |
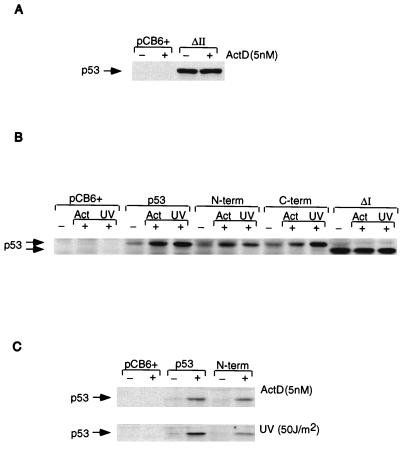
Since p53 forms tetramers, it is possible that stabilization of the mutant proteins expressed in MCF-7 cells may be enhanced by hetero-oligomerization with the stabilized wild-type protein after DNA damage. The observation that overexpression of the exogenous p53ΔI-ΔII protein had only a very slight effect on the stability of endogenous p53 in these cells (Fig. 7A) indicated that this was unlikely to be a significant factor in these studies. However, to rule out this possibility, we analyzed the stability of mutant p53 in cells lacking endogenous wild-type p53. Preliminary evidence suggested that Saos-2 cells do not retain a normal DNA damage response, and we therefore analyzed the stability of mutant p53 in p53-null MEFs (Fig. 8). As expected, the transcriptionally inactive p53ΔII mutant protein was stably expressed in these cells even in the absence of DNA damage (Fig. 8A), consistent with its inability to activate expression of Mdm2. We therefore analyzed the expression of the p53 phosphorylation site mutant proteins containing a wild-type DNA binding domain (Fig. 1), which retained transcriptional activity, in transient assays. These studies confirmed that both the p53N-term and p53C-term mutant proteins could be stabilized in response to actinomycin D treatment and UV irradiation, like wild-type p53, while the p53ΔI mutant protein was stable under all conditions (Fig. 8B and C). Interestingly, although induction of mutant p53 levels in response to actinomycin D treatment was essentially identical to that of the wild-type protein, stabilization of the p53N-term mutant protein in response to UV irradiation was slightly less than that observed for wild-type p53 (Fig. 8C).
FIG. 8.
Stable and transient expression of p53 proteins in p53-null MEFs. (A) Western blot analysis of MEFs stably expressing either a vector control (pCB6+) or p53ΔII with or without treatment with 5 nM actinomycin (ActD). (B) Western blot analysis of MEFs transiently expressing a vector control (pCB6+) or the indicated p53 proteins without DNA damage or following treatment with actinomycin D (Act; 5 nM) or UV irradiation (50 J/m2). (C) Western blot analysis of MEFs transiently expressing a vector control (pCB6+), wild-type p53, or p53N-term without DNA damage or following treatment with actinomycin D (ActD; 5 nM) or UV irradiation (50 J/m2).
DISCUSSION
In this study, we have asked whether phosphorylation of p53 is essential for the transcriptional activation, cell cycle arrest, or apoptotic function of the protein. To address this question, we generated a series of human p53 phosphorylation site mutant proteins which were unable to become phosphorylated within the N-terminal and/or C-terminal regions of the protein. By using transient overexpression assay systems, we showed that the transcriptional activation function was retained by all phosphorylation site mutant proteins, including one mutant protein which completely lacked any detectable phosphorylation, although a reduced activity of p53 proteins mutated at the C-terminal phosphorylation sites could be detected. These results are consistent with suggestions that C-terminal phosphorylation enhances p53 DNA binding activity (33), although another study has recently shown an association of decreased C-terminal phosphorylation with increased sequence-specific DNA binding by p53 (49). Our results show that phosphorylation of p53 is not essential for transcriptional activation by p53, and indeed, numerous reports over the last 5 years have contained similar findings for mutations at single or double sites within the N- or C-terminal region of the protein (reviewed in reference 36). However, it seems likely that phosphorylation of p53 modulates p53 activities in a more subtle manner. Previous studies have suggested that loss of phosphorylation sites results in reduction, but not loss, of activity (7, 9) or that loss of phosphorylation sites differentially affects functions such as transcriptional activation in a cell type-specific manner (29). Furthermore, evidence from one study suggests that mutation of certain combinations of sites may be important for a clear phenotype (32). While we have shown that phosphorylation is not essential for the transcriptional function of p53, it is possible that subtle levels of regulation of p53 function through phosphorylation would not be seen in these transient overexpression assays. We believe that our mutant proteins will be useful in future studies using stable expression systems to address this question.
The second point addressed by our study concerns the importance of phosphorylation for the regulation of p53 stability. Many recent studies have pointed to Mdm2 as a key regulator of p53 stability in normal cells, and inhibition of Mdm2-mediated degradation of p53 following DNA damage is likely to play an important role in activating a p53 response. Inhibition of the interaction of p53 with Mdm2 prevents degradation, and recent studies have shown that phosphorylation of serines 15 and 37 reduced the ability of p53 to bind to Mdm2, with the prediction that this might impair Mdm2-mediated degradation and thus lead to stabilization of p53 (38, 45). The observation that there are several phosphorylation sites either in or close to the Mdm2 binding domain of p53 makes this an extremely attractive model, since many kinases which might phosphorylate p53 at these sites could be activated by DNA damage or other forms of stress. Loss of the ATM kinase has been associated with an inability to stabilize p53 in response to certain signals, and recent studies have shown that ATM can phosphorylate p53 at serine 15 (1, 6) in response to DNA damage. Analysis of the sensitivity of our phosphorylation site mutant proteins to Mdm2 revealed no significant difference from the wild-type protein; each mutant protein bound Mdm2 in vitro and was sensitive to Mdm2-mediated degradation in transient assays in vivo. This might be an expected result when considering the model in which phosphorylation of p53 plays a role in allowing the protein to become resistant to Mdm2; each of our phosphorylation site mutant proteins with alanine substitutions would be constitutively sensitive to degradation. We therefore also examined mutant p53 with an aspartic acid substitution at serine residue 15, 18, 20, or 37 in order to mimic phosphorylation of these residues and potentially generate a stable protein. Although each of the single aspartic acid substitution mutant proteins retained full sensitivity to Mdm2-mediated degradation in our assay, the 15D/37D double mutant protein showed slight but reproducible resistance to Mdm2. Although the protein is clearly not completely resistant to Mdm2, like p53ΔI, this result suggests that multiple N-terminal phosphorylations could contribute to the stabilization of p53 in response to some activating signals.
A clear prediction for a role of phosphorylation in modulating sensitivity to Mdm2-mediated degradation in response to DNA damage is that the p53 proteins mutated at the critical phosphorylation sites should be unable to be stabilized in response to the normal activating signals. Our study shows that proteins mutated at the N- or C-terminal phosphorylation sites are stabilized following either actinomycin D treatment or UV irradiation to similar extents and with kinetics similar to those of the protein containing wild-type phosphorylation sites in stably expressing MCF-7 cells. Furthermore, this stabilization can be attributed to an increase in half-life after DNA damage, where the p53ΔIIN-term mutant protein is stabilized to the same degree as the p53ΔII control. While these results do not preclude a role for phosphorylation in the regulation of p53 stability, they show that stabilization of p53 in response to these DNA-damaging treatments can be carried out in the absence of any detectable N- or C-terminal phosphorylation of p53. One important point to consider when examining cells expressing both wild-type and mutant p53 proteins is that p53 is known to oligomerize, and hetero-oligomers of p53 proteins that can bind Mdm2 with proteins that have lost the ability to bind Mdm2 (such as p53ΔI) show reduced Mdm2 binding (31). This presents the possibility that stabilization of wild-type p53 in cells expressing the phosphorylation site mutant proteins could result in indirect stabilization of mutant p53 through oligomerization with the wild-type protein. This does not appear to be a significant concern in this system, however, since expression of the p53ΔI-ΔII protein, which fails to bind Mdm2 and is stable in the absence of any activating signal, does not markedly stabilize endogenous wild-type p53 protein levels (Table 1). Furthermore, the phosphorylation site mutant proteins were also shown to be stabilized in response to DNA damage in p53-null MEFs. It is of interest that although actinomycin D treatment consistently stabilized the p53 mutant proteins as efficiently as wild-type p53, analysis of p53-null MEFs provided some indication that loss of the N-terminal phosphorylation sites may reduce, although not abolish, stabilization in response to UV irradiation.
Taken together, our results show that phosphorylation of p53 is not absolutely required for regulation of p53 stability, although it may play a contributory role under some conditions, such as UV irradiation. The observation that p53 can be stabilized in response to DNA damage without any detectable N- or C-terminal phosphorylation demonstrates that other mechanisms which allow p53 to become resistant to degradation are likely to exist. This is likely to reflect resistance to Mdm2-mediated degradation, although resistance to other regulators of p53 stability, such as JNK (13), may also play a role. Studies of both p53 and Mdm2 have shown that the binding between the two proteins is necessary but not sufficient for degradation (24, 25) with evidence that regions of both proteins distant and distinct from the binding sites are important for degradation. This has been confirmed in recent studies showing that the p14ARF protein can inhibit Mdm2-mediated degradation of p53 without disrupting the p53-Mdm2 complex (22, 39, 47). Although p14ARF does not play a clear role in mediating the stabilization of p53 in response to DNA damage, it is possible that several proteins function like p14ARF to stabilize p53 in response to various types of stress. Our data suggest that such a mechanism may also contribute to the stabilization of p53 in response to DNA damage, either independently of or in cooperation with phosphorylation of the p53 protein.
ACKNOWLEDGMENTS
We are very grateful to all members of the Vousden lab for advice and encouragement. We also thank Thanos Halazonetis and Carol Prives for helpful discussions.
This work was sponsored by National Cancer Institute, DHHS, under contract with ABL.
REFERENCES
- 1.Banin S, Moyal L, Shieh S-Y, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 2.Banks L, Matlashewski G, Crawford L. Isolation of human p53 specific monoclonal antibodies and their use in the studies of human p53 expression. Eur J Biochem. 1986;159:529–534. doi: 10.1111/j.1432-1033.1986.tb09919.x. [DOI] [PubMed] [Google Scholar]
- 3.Bates S, Vousden K H. p53 in signalling checkpoint arrest or apoptosis. Curr Opin Genet Dev. 1996;6:1–7. doi: 10.1016/s0959-437x(96)90004-0. [DOI] [PubMed] [Google Scholar]
- 4.Blaydes J P, Hupp T R. DNA damage triggers DRB-resistant phosphorylation of human p53 at the CK2 site. Oncogene. 1998;17:1045–1052. doi: 10.1038/sj.onc.1202014. [DOI] [PubMed] [Google Scholar]
- 5.Böttger A, Böttger V, Sparks A, W.-L. L, Howard S F, Lane D P. Design of a synthetic Mdm2 binding mini protein that activates the p53 response in vivo. Curr Biol. 1997;7:860–869. doi: 10.1016/s0960-9822(06)00374-5. [DOI] [PubMed] [Google Scholar]
- 6.Canman C E, Lim D-S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 7.Crook T, Marston N J, Sara E A, Vousden K H. Transcriptional activation by p53 correlates with suppression of growth but not transformation. Cell. 1994;79:817–827. doi: 10.1016/0092-8674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 8.El-Deiry W, Tokino T, Velculescu V E, Levy D B, Parson V E, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumour suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 9.Fiscella M, Ullrich S J, Zambrano N, Shields M T, Lin D, Lees-Miller S P, Anderson C W, Mercer W E, Appella E. Mutation of the serine 15 phosphorylation site of human p53 reduces the ability of p53 to inhibit cell cycle progression. Oncogene. 1993;8:1519–1528. [PubMed] [Google Scholar]
- 10.Fiscella M, Zambrano N, Ullrich S J, Unger T, Lin D, Cho B, Mercer W E, Anderson C W, Appella E. The carboxy-terminal serine 392 phosphorylation site of human p53 is not required for wild-type activities. Oncogene. 1994;9:3249–3257. [PubMed] [Google Scholar]
- 11.Friedlander P, Haupt Y, Prives C, Oren M. A mutant p53 that discriminated between p53-responsive genes cannot induce apoptosis. Mol Cell Biol. 1996;16:4961–4971. doi: 10.1128/mcb.16.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs B, O’Connor D, Fallis L, Scheidtmann K H, Lu X. p53 phosphorylation mutants retain transcription activity. Oncogene. 1995;10:789–793. [PubMed] [Google Scholar]
- 13.Fuchs S Y, Adler V, Buschmann T, Yin Z, Wu X, Jones S N, Ronai Z. JNK targets p53 ubiquitination and degradation in nonstressed cells. Genes Dev. 1998;12:2658–2663. doi: 10.1101/gad.12.17.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs S Y, Adler V, Pincus M R, Ronai Z. MEKK1/JNK signaling stabilizes and activates p53. Proc Natl Acad Sci USA. 1998;95:10541–10546. doi: 10.1073/pnas.95.18.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 16.Halazonetis T D, Kandil A N. Conformational shifts propagate from the oligomerization domain of p53 to its tetrameric DNA binding domain and restore DNA binding to select p53 mutants. EMBO J. 1993;12:5057–5064. doi: 10.1002/j.1460-2075.1993.tb06199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao M, Lowy A M, Kapoor M, Deffie A, Liu G, Lozano G. Mutation of phosphoserine 389 affects p53 function in vivo. J Biol Chem. 1996;271:29380–29385. doi: 10.1074/jbc.271.46.29380. [DOI] [PubMed] [Google Scholar]
- 18.Haupt Y, Barak Y, Oren M. Cell type-specific inhibition of p53-mediated apoptosis by mdm2. EMBO J. 1996;15:1596–1606. [PMC free article] [PubMed] [Google Scholar]
- 19.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 20.Hupp T R, Meek D W, Midgley C A, Lane D P. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 21.Juven T, Barak Y, Zauberman A, George D L, Oren M. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene. 1993;8:3411–3416. [PubMed] [Google Scholar]
- 22.Kamijo T, Weber J D, Zambetti G, Zindy F, Roussel M F, Sherr C J. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapoor M, Lozano G. Functional activation of p53 via phosphorylation following DNA damage by UV but not γ-irradiation. Proc Natl Acad Sci USA. 1998;95:2834–2837. doi: 10.1073/pnas.95.6.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubbutat M H G, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 25.Kubbutat M H G, Ludwig R L, Ashcroft M, Vousden K H. Regulation of Mdm2 directed degradation by the C-terminus of p53. Mol Cell Biol. 1998;18:5690–5698. doi: 10.1128/mcb.18.10.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubbutat M H G, Vousden K H. Keeping an old friend under control: regulation of p53 stability. Mol Med Today. 1998;4:250–256. doi: 10.1016/s1357-4310(98)01260-x. [DOI] [PubMed] [Google Scholar]
- 27.Lechner M S, Mack D H, Finicle A B, Crook T, Vousden K H, Laimins L A. Human papillomavirus E6 proteins bind p53 in vivo and abrogate p53-mediated repression of transcription. EMBO J. 1992;11:3045–3052. doi: 10.1002/j.1460-2075.1992.tb05375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 29.Lohrum M, Scheidtmann K H. Differential effects of phosphorylation of rat p53 on transactivation of promoters derived from different p53 responsive genes. Oncogene. 1996;13:2527–2539. [PubMed] [Google Scholar]
- 30.Marston N J, Crook T, Vousden K H. Interaction of p53 with MDM2 is independent of E6 and does not mediate wild type transformation suppressor function. Oncogene. 1994;9:2707–2716. [PubMed] [Google Scholar]
- 31.Marston N J, Jenkins J R, Vousden K H. Oligomerisation of full length p53 contributes to the interaction with mdm2 but not HPV E6. Oncogene. 1995;10:1709–1715. [PubMed] [Google Scholar]
- 32.Mayr G A, Reed M, Wang P, Wang Y, Schwedes J F, Tegtmeyer P. Serine phosphorylation in the NH2 terminus of p53 facilitates transactivation. Cancer Res. 1995;55:2410–2417. [PubMed] [Google Scholar]
- 33.Meek D W. Multisite phosphorylation and the integration of stress signals at p53. Cell Signall. 1998;10:159–166. doi: 10.1016/s0898-6568(97)00119-8. [DOI] [PubMed] [Google Scholar]
- 34.Meek D W. Post-translational modification of p53. Semin Cancer Biol. 1994;5:203–210. [PubMed] [Google Scholar]
- 35.Midgley C A, Lane D P. p53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene. 1997;15:1179–1189. doi: 10.1038/sj.onc.1201459. [DOI] [PubMed] [Google Scholar]
- 36.Milczarek G J, Martinez J, Bowden G T. p53 phosphorylation: biochemical and functional consequences. Life Sci. 1997;60:1–11. doi: 10.1016/s0024-3205(96)00479-1. [DOI] [PubMed] [Google Scholar]
- 37.Milne D M, Palmer R H, Meek D W. Mutation of the casein kinase II phosphorylation site abolishes the anti-proliferative activity of p53. Nucleic Acids Res. 1992;20:5565–5570. doi: 10.1093/nar/20.21.5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pise-Masison C, Radonovich M, Sakaguchi K, Appella E, Brady J N. Phosphorylation of p53: a novel pathway for p53 inactivation in human T-cell lymphotropic virus type 1-transformed cells. J Virol. 1998;72:6348–6355. doi: 10.1128/jvi.72.8.6348-6355.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pomerantz J, Schreiber-Agus N, Liégeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, Cordon-Cardo C, DePinho R A. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 40.Prives C. Signaling to p53: breaking the MDM2-p53 circuit. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 41.Rowan S, Ludwig R L, Haupt Y, Bates S, Lu X, Oren M, Vousden K H. Specific loss of apoptotic but not cell cycle arrest function in a human tumour derived p53 mutant. EMBO J. 1996;15:827–838. [PMC free article] [PubMed] [Google Scholar]
- 42.Ryan K M, Vousden K H. Characterization of structural p53 mutants which show selective defects in apoptosis but not cell cycle arrest. Mol Cell Biol. 1998;18:3692–3698. doi: 10.1128/mcb.18.7.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaw P, Freeman J, Bovey R, Iggo R. Regulation of specific DNA binding by p53: evidence for a role of O-glycosylation and charged residues at the carboxy-terminus. Oncogene. 1996;12:921–930. [PubMed] [Google Scholar]
- 45.Shieh S-Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 46.Siliciano J D, Canman C E, Taya Y, Sakaguchi K, Appella E, Kastan M B. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stott F, Bates S A, James M, McConnell B B, Starborg M, Brookes S, Palmero I, Hara E, Ryan K M, Vousden K H, Peters G. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vojtesek B, Bartek J, Midgley C A, Lane D P. An immunochemical analysis of the human nuclear phosphoprotein p53. J Immunol Methods. 1992;151:237–244. doi: 10.1016/0022-1759(92)90122-a. [DOI] [PubMed] [Google Scholar]
- 49.Waterman M J, Stavridi E S, Waterman J L, Halazonetis T D. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat Genet. 1998;19:175–178. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]