Autophagy in Yeast: Mechanistic Insights and Physiological Function (original) (raw)
Abstract
Unicellular eukaryotic organisms must be capable of rapid adaptation to changing environments. While such changes do not normally occur in the tissues of multicellular organisms, developmental and pathological changes in the environment of cells often require adaptation mechanisms not dissimilar from those found in simpler cells. Autophagy is a catabolic membrane-trafficking phenomenon that occurs in response to dramatic changes in the nutrients available to yeast cells, for example during starvation or after challenge with rapamycin, a macrolide antibiotic whose effects mimic starvation. Autophagy also occurs in animal cells that are serum starved or challenged with specific hormonal stimuli. In macroautophagy, the form of autophagy commonly observed, cytoplasmic material is sequestered in double-membrane vesicles called autophagosomes and is then delivered to a lytic compartment such as the yeast vacuole or mammalian lysosome. In this fashion, autophagy allows the degradation and recycling of a wide spectrum of biological macromolecules. While autophagy is induced only under specific conditions, salient mechanistic aspects of autophagy are functional in a constitutive fashion. In Saccharomyces cerevisiae, induction of autophagy subverts a constitutive membrane-trafficking mechanism called the cytoplasm-to-vacuole targeting pathway from a specific mode, in which it carries the resident vacuolar hydrolase, aminopeptidase I, to a nonspecific bulk mode in which significant amounts of cytoplasmic material are also sequestered and recycled in the vacuole. The general aim of this review is to focus on insights gained into the mechanism of autophagy in yeast and also to review our understanding of the physiological significance of autophagy in both yeast and higher organisms.
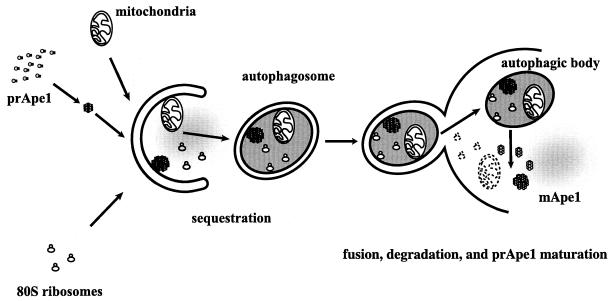
Unicellular eukaryotic organisms must be capable of rapid adaptation to changing environments. While such changes do not normally occur in the tissues of multicellular organisms, developmental and pathological changes in the environment of cells often require adaptation mechanisms not dissimilar from those found in simpler cells. Autophagy is a catabolic membrane-trafficking phenomenon that occurs in response to dramatic changes in the nutrients available to yeast cells, for example during starvation for nitrogen or carbon (111, 116) or on challenge with rapamycin, a macrolide antibiotic whose effects mimic starvation (80). Autophagy also occurs in animal cells that are serum starved or challenged with specific hormonal stimuli. In macroautophagy, the form of autophagy commonly observed, cytoplasmic material is sequestered in double-membrane vesicles called autophagosomes. Autophagosomes fuse with the lytic compartment, the vacuole (or lysosome in cells from higher eukaryotes), whereupon vesicles called autophagic bodies are released into the lumen of this organelle. These vesicles are degraded, allowing vacuolar hydrolases to access the cargo and recycle cellular building blocks (Fig. 1 shows a schematic description of yeast macroautophagy). While autophagy is induced only under specific conditions, salient mechanistic aspects of autophagy are functional in a constitutive fashion. In Saccharomyces cerevisiae, induction of autophagy subverts a constitutive membrane-trafficking mechanism called the cytoplasm-to-vacuole targeting (Cvt) pathway (Fig. 2) from a specific mode, in which it carries the resident vacuolar hydrolase, aminopeptidase I (Ape1), to a nonspecific bulk mode, in which significant amounts of cytoplasmic material are also sequestered and recycled in the vacuole.
FIG. 1.
Overview of membrane trafficking in yeast macroautophagy. During autophagy, various components of the cytoplasm, such as mitochondria, ribosomes, specific cargo such as prApe1 (see Fig. 2), and soluble cytoplasmic material (depicted as diffuse gray), are sequestered into autophagosomes, which are large (300- to 900-nm) double-bilayer vesicles. Once formed, autophagosomes fuse with the vacuole, releasing a single-bilayer-bound autophagic body into the lumen of this organelle. Vacuolar hydrolases act on the limiting membrane of the autophagic body, releasing its contents, which are degraded into biosynthetic building blocks.
FIG. 2.
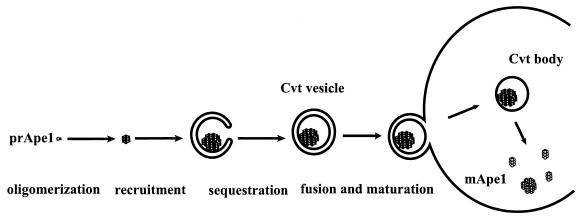
Discrete steps in cytoplasm-to-vacuole targeting of aminopeptidase I. Aminopeptidase I (Ape1), a resident vacuolar hydrolase, is synthesized as a soluble zymogen on cytoplasmic ribosomes. It then undergoes homo-oligomerization to form dodecamers, which further assemble into large, membrane-associated complexes. The membrane-associated form is sequestered into an autophagosome-like structure called the Cvt vesicle (140 to 160 nm diameter). Cvt vesicles fuse with the vacuole, giving rise to an intravacuolar Cvt body that is degraded, releasing prApe1 (61 kDa) into the lumen, where it is processed to the mature form (mApe1; 50 kDa).
A number of recent comprehensive reviews have covered many aspects of autophagy (25, 53, 59). The general aim of this review is to focus on insights gained into the mechanism of autophagy by using yeast as a model system and also to review our understanding of the physiological significance of autophagy in both yeast and higher organisms. Finally, we discuss future questions and hypotheses that remain to be examined within this rapidly expanding field.
MACROAUTOPHAGY: A UNIQUE, INDUCIBLE VESICULAR TRAFFICKING MECHANISM
Autophagy is a unique membrane transport event. It was first described by Clark (19) as an unusual presence of cytoplasmic organelles (mitochondria, in that instance) in the lumen of degradative vacuoles of developing kidney cells. Ashford and Porter (5) then discovered that glucagon can induce autophagy in perfused rat liver, and this led to a number of mechanistic studies by De Duve and colleagues (22, 23). These mostly morphological studies showed that during macroautophagy, the more commonly observed form of autophagy, cellular membranes enwrap and sequester cytoplasmic material in double-membrane-bound compartments. The feature that distinguishes macroautophagy from the less commonly studied form of autophagy, microautophagy (see below), is that the initial sequestration event takes place away from the lysosome/vacuole. The resulting cytoplasmic compartments, called autophagosomes, then take on lysosomal characteristics such as acidification, the presence of typical lytic enzymes, etc. These analyses of macroautophagy in mammalian cells led to immunoelectron microscopy studies by Dunn (26, 27), which traced the origin of autophagosomes to the endoplasmic reticulum and demonstrated that the nascent autophagosomes gradually take on endosomal and lysosomal characteristics. These findings were not universally accepted, however, and alternative theories have persisted regarding the origins of the autophagosomal membrane, including the idea that a specialized novel organelle was involved (110).
The relative significance of autophagy in comparison with more standard trafficking phenomena was not established, most probably due to the lack of a genetic approach to test its physiological significance. Thus, the discovery of macroautophagy in yeast by Ohsumi and colleagues (111) and the subsequent identification of autophagy mutants (116, 117) and their characterization and cloning are landmarks in our understanding of the system. In yeast starved for nitrogen and/or carbon, the vacuole, the yeast equivalent of the lysosome, accumulates membrane-enclosed material derived from the cytoplasm under conditions where vacuolar hydrolases are inhibited using the serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF) or due to mutations in the structural genes encoding these enzymes. As in mammalian systems, this autophagic pathway entails sequestration of cytoplasmic contents into double-membrane-enclosed vesicles called autophagosomes (300 to 900 nm diameter [Fig. 1]), and the autophagosomes then dock and fuse with the vacuole (8). After this fusion event, the internal vesicle of the autophagosome is released into the vacuolar lumen, and this intravacuolar vesicle is called the autophagic body. Vacuolar hydrolases then degrade the limiting membrane of the autophagic body, allowing access to its cytoplasm-derived content and leading to its recycling into molecular building blocks. Ohsumi and colleagues went on to identify an autophagy mutant, apg1, by morphological screening of PMSF-treated and nitrogen-starved yeast cells (117). Analysis of the apg1 phenotype demonstrated that the mutant quickly lost viability upon starvation, and this fact was used to rapidly screen a large number of mutants, which were then subjected to a secondary, morphological examination. In this fashion, a collection of 16 complementation groups of apg mutants was isolated. Working independently, Thumm et al. isolated aut mutants based on their inability to degrade fatty acid synthase in the vacuole following starvation (116).
While autophagy is clearly a form of membrane trafficking, several properties set it apart from more conventional trafficking phenomena. Membrane-trafficking studies of eukaryotic cells commonly tackle issues that derive from the work of George Palade and colleagues. In those studies, it was shown that the eukaryotic cell is composed of membrane-bound compartments and that interchange of material between these compartments occurs constantly, through vesicular trafficking (81). The standard view of the vesicular trafficking process has two basic features. First, cellular compartmentalization is conserved: material that derives from the lumen of the donor compartment does not exit into the cytoplasm, and cytoplasmic material does not enter the lumen of an organelle. Second, the process conserves membrane topology: material that derives from the cytoplasmic leaflet of the donor compartment is directed to the cytoplasmic leaflet of the target compartment, and material derived from the lumenal leaflet of the donor finds itself facing the lumen of the target compartment.
Because all intracellular compartments are interconnected by vesicular trafficking pathways or related membrane-trafficking events, a cellular steady state implies that vesicular trafficking must also be at steady state to ensure that compartments do not disappear or overexpand. This in turn implies that vesicular trafficking pathways are constitutive. We know of exceptions to this principle; however, these exceptions do not entail the induction of novel trafficking routes but, rather, are the result of kinetic control over preexisting routes. Thus, Golgi dispersion during mitosis involves inhibition of homotypic fusion between COP-I vesicles while budding continues unchecked (68). Likewise, regulated exocytosis during synaptic transmission or secretory granule release in mast cells involves the removal of an inhibition to the fusion of predocked vesicles.
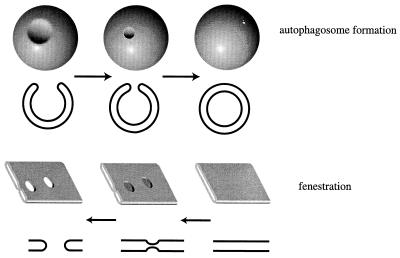
Autophagy, on the other hand, is a vesicular trafficking phenomenon that forms an exception to these rules. It is a membrane-trafficking process in which cellular topology is not conserved: cytoplasmic material is introduced into the lumen of the vacuole or lysosome. In addition, we do not have evidence for large-scale autophagy occurring in logarithmically growing, unstimulated yeast cells; hence, it is an inducible, not a constitutive, process. Should we therefore distinguish it from classical vesicular trafficking pathways? Several lines of evidence suggest that autophagy utilizes basic membrane transformations that have been described in more standard trafficking pathways. It has been shown, for example, that the fusion of autophagosomes with the vacuole requires Vam3, the yeast vacuolar syntaxin homologue, as well as other components of the vesicular trafficking machinery (21, 98). The topologically distinct step in autophagy is the sequestration of cytoplasm into autophagosomes. From the membrane fusion aspect, this is simply homotypic fusion between cellular membranes with the added complication that the fusion partners are already joined on a continuous sheet of membrane. However, the three-dimensional picture is actually the reverse of a fenestration event. For example, in higher eukaryotic cells, the Golgi cisternae form multilamellar stacks. Some of these cisternae are fenestrated, and this is thought to occur by fusion events between the luminal faces of the membrane bilayer that delimits the cisternae (87). As shown in Fig. 3, the formation of a fenestration is the topological equivalent of the reverse reaction of the closure of the autophagosome. The only difference is that in autophagy we are dealing with a spherical “cisterna” instead of a planar one (this could, in principle, be derived from a deformation of a planar cisterna). Thus, the membrane transformations that occur during autophagosome formation have known analogues in conventional vesicular trafficking and membrane homeostasis.
FIG. 3.
Autophagosomal sequestration is analogous to a reverse fenestration. It is thought that during fenestration, as occurs in Golgi lamellae in higher eukaryotes (bottom panel), the lumenal faces of the limiting lamellar membrane fuse, creating a pore that then expands to form a fenestration. In the formation of the autophagosome, a similar process is occurring but does so in the opposite direction (top panel). Here, a circular pore is being closed to form a closed lamellar sheet, with the added complication that this sheet is spherical.
Microautophagy: Variation on a Theme
While the majority of our knowledge of autophagic uptake comes from observations of macroautophagy, studies with mammalian and plant systems (6, 25, 72) and the methylotrophic yeast Pichia pastoris (118) revealed an alternative scheme for autophagic degradation of cytosolic material. In this pathway, called microautophagy, the limiting membrane of the vacuole invaginates, finally pinching off to form an internal vacuolar vesicle that contains material derived from the cytoplasm, akin to the autophagic bodies formed in macroautophagy. Microautophagic uptake of peroxisomes into the vacuole, called “micropexophagy,” has been demonstrated to be important in peroxisome degradation in P. pastoris (see “Pexophagy: organelle-specific housecleaning through autophagy” below). More recently, microautophagy has also been described in S. cerevisiae. Horst et al. first described starvation-dependent in vitro uptake of proteins into isolated vacuoles (38). These authors purified vacuoles from growing and starved yeast cells and assayed the ability of these isolated vacuoles to degrade proteins from radiolabeled yeast extracts. They found a fivefold increase in this activity in vacuoles derived from starved cells compared to those growing in rich medium. Pretreatment of vacuoles with trypsin abrogated the degradation of cytosolic proteins, suggesting a requirement for protein components on the vacuole surface. Finally, they showed that uptake depended on cytosolic Hsp70. This study was followed by an investigation by Mayer and coworkers (73, 93), who described the formation and pinching of autophagic tubes in isolated vacuoles from S. cerevisiae as well as an in vitro reconstitution of protein uptake into the vacuole using this pathway. According to these studies, mutations in genes required for autophagy attenuated but did not completely abolish this microautophagic uptake, indicating that microautophagy and macroautophagy are not mechanistically identical in S. cerevisiae.
Cvt Pathway: a Constitutive Biosynthetic Analog of Macroautophagy in Yeast
To state that a given phenomenon is inducible, one needs to define a basal, uninduced state. In mammalian cells we are hampered by the fact that autophagy is defined primarily through morphological assays, which are not readily quantifiable. In S. cerevisiae, on the other hand, we are provided with a specific biochemical marker that allows us to determine the degree of inducibility of autophagy under nutrient depletion relative to rich-medium conditions.
It was shown that the vacuolar hydrolase, aminopeptidase I (Ape1), is transported to the vacuole in yeast in a manner analogous to autophagosomal delivery. Ape1 is synthesized as an inactive zymogen (prApe1, 61 kDa) in the cytosol, homo-oligomerizes into a large complex, and is transported to the vacuole without translocation into the lumen of the endoplasmic reticulum (54, 57). In the vacuole, a proteolytic processing event trims the protein to the enzymatically active, 50-kDa form. Biochemical analyses demonstrated the existence of prApe1-containing membrane-bound transport intermediates both in the cytoplasm and in the vacuolar lumen. This was confirmed by immunoelectron microscopy studies, which also found that the cytoplasmic intermediate was a double-membrane-bound vesicle, termed the Cvt vesicle, while the intravacuolar intermediate, the Cvt body, was bound by a single membrane (7, 98). Most significant, however, were genetic studies in which mutants defective in prApe1 processing were identified and the corresponding genes were cloned. It was found that the majority of cvt mutants (defective in cytoplasm-to-vacuole targeting) were allelic with apg and aut mutants (32, 100). Thus, not only do autophagy and the Cvt pathway share physical attributes, but also they require the function of the same group of proteins, implying that they share very basic mechanistic themes. Importantly, however, there exist proteins required only in the Cvt pathway but not in autophagy, and vice versa (7) (see below).
Underscoring the close relationship between the Cvt pathway and autophagy, prApe1 is specifically targeted not only into Cvt vesicles but also into autophagosomes. This specificity underlies the much higher rate of uptake of prApe1 by autophagosomes compared with other cytosolic marker proteins (100) (see “Sorting of specific cargo into Cvt vesicles and autophagosomes” below).
The similarity between the Cvt pathway and autophagy (discussed in more detail below) allows the use of specific tools that were derived in the investigation of Ape1 trafficking in the analysis of mutant phenotypes. There are six experimentally defined steps in Ape1 biogenesis (Fig. 2): synthesis, oligomerization, recruitment to membranes, engulfment by membranes (formation of Cvt vesicles), fusion of Cvt vesicles with the vacuole, and breakdown of Cvt bodies within the vacuole. One can assay the formation of Cvt vesicles in cvt mutants by probing the accessibility of prApe1 to exogenous proteinase K in permeabilized cells (54), and one can test for the attachment to membranes by simple fractionation procedures (33). Thus, by analyzing the point at which prApe1 trafficking is blocked as a result of mutation in a given protein, one may try and deduce the point of function of this protein in autophagy. There is another way in which the Cvt pathway allows the analysis of autophagy in more depth. The existence of proteins required specifically in the Cvt pathway but not in autophagy means that the maturation of prApe1 can be used as a marker for autophagy in genetic backgrounds that are specifically defective in Cvt trafficking but not in autophagy. Thus, in specific mutants that accumulate prApe1, induction of autophagy results in prApe1 maturation concomitant with appearance of autophagic bodies in the vacuole and in a fashion that depends on autophagy factors (1, 3, 101). The existence of such autophagy-dependent maturation of prApe1 is also a direct demonstration of the very low degree to which autophagy occurs in unstimulated yeast cells.
COMMON MECHANISTIC THEMES BETWEEN AUTOPHAGY AND THE CVT PATHWAY
A majority of the genes required for the Cvt pathway are also required for autophagy. These genes can mostly be viewed as required for distinct subprocesses, as enumerated below (Table 1).
TABLE 1.
Partial glossary of proteins required in autophagy and cytoplasm-to-vacuole targeting
| Protein | Aliases | Known biochemical relationship or function | Mammalian homolog |
|---|---|---|---|
| Apg1 | Cvt10, Aut3 | Protein kinase | ULK1 |
| Apg5 | Undergoes isopeptide linkage to Apg12 at lysine 149 | hAPG5 | |
| Apg7 | Cvt2 | El-like ATP-dependent enzyme which activates Apg12 and Aut7 for covalent conjugation reactions with Apg5 and phosphatidylethanolamine, respectively | hsGSA7 |
| Apg9 | Aut9, Cvt7 | Integral membrane protein | |
| Apg10 | E2-like enzyme which catalyzes formation of an isopeptide bond between Apg12 and Apg5 | ||
| Apg12 | Undergoes a ubiquitin-like isopeptide link between the C-terminal glycine and lysine 149 of Apg5 | hAPG12 | |
| Apg13 | Regulatory subunit of Apg1 protein kinase; undergoes dephosphorylation on induction of autophagy | ||
| Apg14 | Subunit of Vps34 PI3-kinase holoenzyme that is required for autophagy and the Cvt pathway | ||
| Apg16 | Binds the Apg5-Apg12 conjugate to form multimers through self-association that is mediated by coiled-coil domains | ||
| Apg17 | Autophagy-specific subunit of Apg1 protein kinase complex | ||
| Aut1 | Apg3 | E2-like enzyme that mediates amidation of Aut7 to phosphatidylethanolamine | |
| Aut2 | Apg4 | Cysteine peptidase; hydrolyzes C-terminal arginine of Aut7 to expose a glycine residue that then reacts with Apg7; also required for hydrolysis of Aut7-phosphatidylethanolamine conjugate | |
| Aut7 | Cvt5 | Undergoes lipidation through amidation of the penultimate glycine residue to phosphatidylethanolamine | GATE-16, MAP LC-3 |
| Cvt17 | Aut5 | Homologous to triglyceride lipases; integral membrane | |
| Cvt19 | A specificity factor for cargo sorting into Cvt vesicles | ||
| Cvt9 | A Cvt pathway specific subunit of Apg1 protein kinase complex | ||
| Fkb1 | Fpr1, Rbp1 | Rapamycin binding protein; immunophillin | FKBP |
| Tlg2 | Syntaxin homologue required for formation of Cvt vesicles but not autophagosomes; integral membrane protein | Syntaxin | |
| Vps45 | Sec1 homologue required for formation of Cvt vesicles but not autophagosomes | VPS45A, VPS45B | |
| Tor1 Tor2 | Protein kinases; inhibition by Fpr1-rapamycin complex leads to induction of autophagy | mTor | |
| Vac8 | Cvt pathway-specific subunit of Apg1 protein kinase; contains armadillo repeats | ||
| Vps30 | Apg6 | Subunit of Vps34 PI3-kinase holoenzyme | Beclin 1 |
| Vps15 | Protein kinase; forms a complex with Vps34 and is required for Vps34 membrane attachment and function | PIK3R4 (PI3-kinase adaptor protein) | |
| Vps34 | End12 | PI3-kinase; required in autophagy, Cvt pathway, and vacuolar protein sorting | Type III PI3-kinase |
Apg1 Protein Kinase Activity
The first autophagy-defective mutant isolated, apg1, was found to be allelic with cvt10 (66, 100, 117). Induction of autophagy has been demonstrated to entail a change in Apg1 catalytic activity, which can be monitored as an increase in the in vitro activity of the protein in kinase assays (46). Furthermore, this change is thought to result from changes in the association of Apg1 with specific regulatory proteins (46, 101). However, because Apg1 activity is also required for Cvt trafficking, this change in activity may not simply be a quantitative effect. Indeed, as we discuss in later sections, different regulatory subunits may associate with Apg1 under these different conditions.
Apg9: an Integral Membrane Protein Required for Both Autophagy and the Cvt Pathway
It is interesting that of the proteins required for the formation of autophagosomes and Cvt vesicles, only Apg9 is an integral membrane protein, containing five potential transmembrane domains (78). Cells lacking Apg9 are completely defective in both the Cvt pathway and autophagy, and they accumulate protease-sensitive prApe1, indicating that the protein is required prior to closure of the double-bilayer vesicle. A green fluorescent protein-Apg9 fusion protein appears to localize to punctate structures in the cell, and these were shown to be distinct from known intracellular compartments as marked by Kex2 (late Golgi), Pma1p (plasma membrane), Pho8 (vacuole), Pep12 (late endosome), and Sec12 (endoplasmic reticulum). Unlike Aut7 (see below), Apg9 is not localized to the autophagosome or Cvt vesicle itself and is excluded from the vacuolar lumen. Thus, its localization may mark the sites of assembly for Cvt vesicles and autophagosomes. It was also shown that in _vps4_Δ cells the punctate structures stained by Apg9 do not coalesce into the class E compartment, suggesting that the Apg9-stained structure is not identical to the late endosome. More recently, a novel soluble factor, Apg2, was shown to physically interact with Apg9 (121). Like Apg9, Apg2 function is required prior to the formation of Cvt vesicles and autophagosomes and also shows a punctate distribution that may at least partly coincide with that of Apg9.
Apg12 Protein Conjugation Machinery
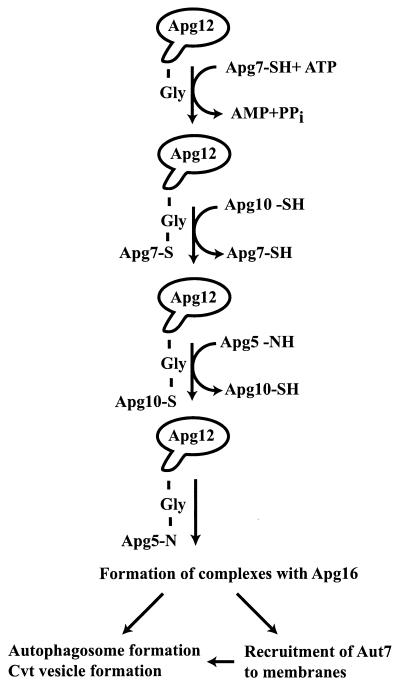
It was recently discovered (70, 71) that five of the APG genes have related functions. Apg12 is found as two species on sodium dodecyl sulfate-polyacrylamide gel electrophoresis blots prepared from cell extracts: a low-molecular-mass band at the size of the expected Apg12 monomer, but also a larger, 70-kDa band. It was shown that the 70-kDa band is the result of the formation of an isopeptide bond between the C-terminal glycine of Apg12 and an internal lysine residue of a second protein, Apg5 (70). Thus, while Apg12 has no clear homology to ubiquitin, it is being conjugated in an analogous fashion, although the only known conjugation partner for Apg12 is Apg5. Sequence similarity to Uba1p suggested that Apg7 is an Apg12-activating enzyme that requires ATP (50, 113). Apg7 forms a labile thioester linkage with the C-terminal glycine of Apg12 and transfers the activated Apg12 to an active-site cysteine on Apg10, which acts in a fashion analogous to an E2 enzyme, in transferring Apg12 to Apg5. Finally, Apg16 is a protein that specifically binds to the 70-kDa Apg12-Apg5 adduct and is required for its function (Fig. 4). A coiled-coil region of Apg16 mediates self-multimerization, which in turn results in the formation of a large multisubunit complex consisting of Apg16 and the Apg12-Apg5 adduct (69).
FIG. 4.
Apg12 undergoes a ubiquitin-like conjugation to Apg5. Apg12 is activated by the E1-like protein, Apg7, by formation of a labile thioester bond between the carboxyl group of the Apg12 C-terminal glycine residue and a thiol group from an internal cysteine (Cys 507) of Apg7. The activated Apg12 is transferred to a cysteine group on Apg10 in a fashion reminiscent of ubiquitin transfer from E1 proteins to E2. Apg10 then transfers Apg12 to lysine 149 of Apg5 by the formation of a more stable amide bond. The Apg12-Apg5 conjugate recruits Apg16 dimers. The divalent nature of Apg16 results in the formation of large multimeric complexes, and these are thought to play a role in the nucleation of both autophagosomes and Cvt vesicles. In addition, it has been shown that the Apg12-Apg5 complex is required for the recruitment of Aut7 to membranes (see Fig. 5).
Both the Cvt pathway and autophagy require all five of these conjugation proteins. Using the Cvt pathway model, it was possible to determine the stage at which these proteins function. In yeast defective in the machinery required for Apg12 conjugation, prApe1 is able to undergo homo-oligomerization and attachment to the membrane, but this membrane is not closed, as determined by protease sensitivity assays (30, 50). Thus, it was determined that the conjugation machinery is required for the formation of Cvt vesicles but not for earlier events. While the Apg12-conjugation system is an attractive candidate for a control mechanism over autophagic activity, current experimental evidence does not support this: the relative amounts of conjugated and unconjugated Apg12, as well as the cell fractionation properties of these proteins, are not affected by stimuli that induce autophagy (69).
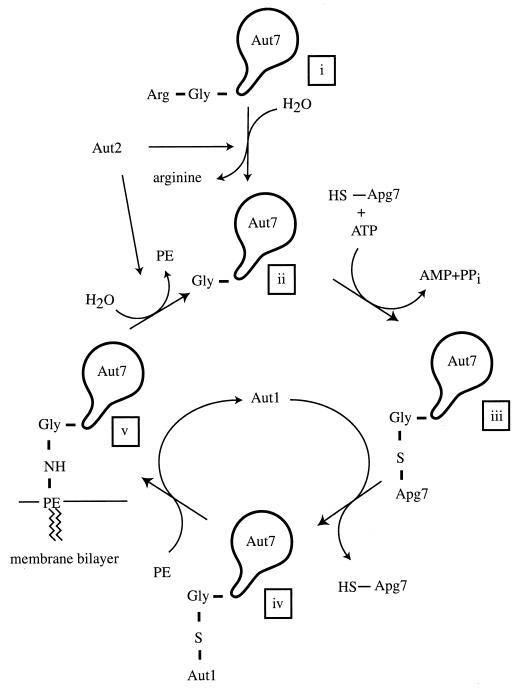
Conjugation of Aut7 to Phosphatidylethanolamine
The aut7 mutant is allelic to the apg8 and cvt5 mutants (39, 55). Although it was originally thought to function in the tethering of autophagosomes to microtubules (59), later studies contested this (see below). Green fluorescent protein-tagged Aut7 localizes, at least in part, to the lumen of autophagosomes and Cvt vesicles and is transported to the vacuole, where it is degraded (55). As a consequence, the level of Aut7 is very low in growing cells. Upon induction of autophagy, Aut7 undergoes a transcriptional induction that leads to a more than 20-fold increase in the amount of protein (39). Remarkably, Aut7 undergoes a conjugation reaction that is surprisingly similar to that of Apg12 and other ubiquitin-like proteins (42) (Fig. 5). Aut2, which is identical to Apg4, is a cysteine protease that clips off the C-terminal amino acid of Aut7 to uncover a C-terminal glycine (56). This C-terminal glycine is activated in an ATP-dependent fashion by formation of a thioester with Cys507 of Apg7 (the same E1-like activating enzyme required in the conjugation of Apg12 to Apg5) and is then transferred to the conjugating enzyme Aut1 (Apg3), which catalyzes the formation of an amide bond between the C-terminal glycine and phosphatidylethanolamine. While full-length Aut7 is a soluble protein, the conjugated form is tightly bound to membranes as a result of this amidation to the lipid moiety. It was also reported that other components of the Apg12-conjugation machinery, Apg5, Apg10, Apg16, and Apg12 itself, are also required for the attachment of Aut7 to membranes (51).
FIG. 5.
Aut7 undergoes ubiquitin-like conjugation to phosphatidylethanolamine. Aut7 is synthesized with a C-terminal arginine residue encoded in the reading frame of the gene (i). The cysteine protease Aut2/Apg4 hydrolyzes the peptide bond to the C-terminal arginine to create form (ii), which has a C-terminal glycine residue. Form (ii) is activated by forming a thioester link with cysteine 507 of Apg7 (form iii) and is then transferred to a cysteine residue on an E2 analog, Aut1 (form iv). Aut1 then transfers Aut7 to phosphatidylethanolamine (PE), via the formation of an amide bond, to create the tightly membrane bound form (v). Aut7 is released from the membrane by hydrolysis of the amide bond, a reaction again catalyzed by Aut2, to regenerate form (ii). In the absence of Aut2, ectopic expression of form (ii) does not completely rescue the Cvt and autophagy defects of the _aut2_Δ mutant, indicating that a complete cycle is required for Aut7 function.
Strikingly, ectopic expression of the truncated (glycine-terminated) version of Aut7 does not completely complement the loss of Aut2. Indeed, while Aut7 is tightly bound to membranes in these cells, Ape1 is not fully matured and autophagy is not normal. It was found using in vitro reconstitution that Aut2 is required not only for the initial clipping of Aut7 but also for a second hydrolytic event that frees Aut7 from its membrane anchor, presumably phosphatidylethanolamine, and this second event is also required in both prApe1 transport and autophagy (56). Thus, Aut7 cycles between soluble and membrane-anchored states, and this cycling, not just membrane binding per se, is required for function.
Phosphatidylinositol 3-Kinase Activity Requirement
Yeast are known to contain only one phosphatidylinositol (PI) 3-kinase, Vps34 (36, 95), homologous to mammalian type III PI 3-kinase. The vps34 mutant, which was isolated in a screen for carboxypeptidase Y (Prc1) missorting, is allelic with the end12 mutant, which was isolated in a screen for endocytosis mutants (74). Vps34 function is regulated by the protein kinase activity of Vps15, with which it forms a stable, membrane-associated complex under normal conditions (107, 108). The lipid kinase activity associated with Vps34 is thought to create lipid patches of PI 3-phosphate at specific _trans_-Golgi locations, and these patches then function in protein sorting into vesicles that traffic from the Golgi to the endosome (109). PI 3-phosphate is also thought to act as a second messenger that regulates the interaction of Vac1 (also known as Pep7) as well as Vps27 with endosomal membranes (12, 82, 106). More recently, PI 3-kinase activity has been shown to be required for autophagy and autophagy-related phenomena in a number of systems (48, 83).
Vps30/Apg6 is also required for vacuolar delivery through the secretory pathway as well as autophagy and the Cvt pathway. In addition, Vps30/Apg6 physically interacts with Apg14, a protein required for autophagy and the Cvt pathway but not for Prc1 targeting or other vacuolar trafficking pathways. Kihara et al. (49) demonstrated that the Vps15-Vps34 complex controls autophagy and the Cvt pathway in yeast via physical association with Vps30 and Apg14. Loss of this activity results in the accumulation of prApe1 in a protease-accessible form, indicating a function in the formation of Cvt vesicles and autophagosomes. On the other hand, Vps34 functions in Prc1 trafficking through an alternate, and more abundant, complex of Vps15, Vps34, Vps38, and Vps30. They also showed that Vps38 was mediating the interaction of Vps30 with the Vps34-Vps15 subcomplex. Finally, formation of these complexes requires Vps15 protein kinase activity, similar to the result shown by Stack et al. for the binary complex of Vps34 and Vps15 (107). How does PI 3-kinase activity affect the molecular machinery of autophagy and the Cvt pathway? This question is unresolved. Generally speaking, three types of hypotheses can be suggested. PI 3-kinase activity may be needed tonically; i.e., it is prerequisite as a constitutive background but is not modulated during induction of autophagy. Second, PI 3-kinase activity may be an upstream regulator of a central mechanism such as Apg1 kinase activity or a protein conjugation event mediated by Apg7. Finally, PI 3-kinase activity may be downstream of the other players and may be regulated by their function.
We do know that cells lacking Vps30/Apg6 do not show a change in the intracellular distribution of Apg9 (78) and are able to normally dephosphorylate Apg13 after challenge with rapamycin (H. Abeliovich, unpublished data). It remains to be determined whether PI 3-kinase activity is required in any way for the covalent modifications of Aut7 or Apg12 or for other molecular events in the Cvt/autophagy system. An interesting recent observation made by Kovacs et al. with murine epithelial cells is that inhibition of PI 3-kinase with wortmannin or 3-methyladenine in cells that were preinduced for autophagy resulted in regression of autophagic sequestration, suggesting that PI 3-kinase is required for increasing the size of the sequestering membrane, presumably through fusion events (60). Two facts complicate this analysis, however. First, 3-methyladenine and wortmannin inhibit both class I and class III types of mammalian PI 3-kinase, and Petiot et al. demonstrated that these subtypes had opposite effects on autophagy (83): class I enzymes inhibit autophagic sequestration, while class III enzymes (homologous to yeast Vps34) stimulate autophagic sequestration. Second, the studies described by Kovacs et al. were unusual in that autophagy was induced by exposing the cells to low concentrations of Triton X-100, a nonspecific reagent.
If the involvement of PI 3-phosphate is achieved through its effect on membrane fusion events, as suggested for endosomal trafficking (12, 82, 106), then the requirement for Vps34 may reflect specific, PI 3-phosphate-dependent recruitment of membrane fusion factors to preautophagosomal membranes.
Membrane Docking and Fusion with the Vacuole
Our present model of both autophagy and the Cvt pathway envisions two membrane fusion steps that occur in both trafficking modes: one occurs at the formation of the Cvt vesicle or autophagosome, and the other occurs when the Cvt vesicle or autophagosome fuses with the limiting membrane of the vacuole. As a result, all molecules that are required for general docking and fusion at the vacuolar membrane are also required for both Cvt and autophagic trafficking. This includes Vam3, the vacuolar t-SNARE (21); the SNARE protein Vti1, which interacts with Vam3 but also with Tlg2 (28, 37) (see below); Ypt7, the yeast vacuolar Rab homologue (50); and members of the class C/HOPS complex (91, 92, 98, 102). Not surprisingly, then, temperature-sensitive mutants with mutations in these genes that are incubated at nonpermissive temperature, and also null mutants, accumulate prApe1 in a form that is resistant to degradation in protease protection assays, reflecting the fact that it is sequestered either in Cvt vesicles (under vegetative conditions) or in autophagic vesicles (under starvation conditions or after treatment with rapamycin). Similarly, electron microscopy was used to show that _vam3_tsf cells accumulate cytoplasmic autophagosomes at nonpermissive temperature when starved for nitrogen (21) or challenged with rapamycin (2) and can serve as a morphological assay for the formation of autophagosomes in various mutant backgrounds that contain the _vam3_tsf allele.
Breakdown of Cvt Vesicles and Autophagosomes in the Vacuole
In both the Cvt pathway and autophagy, a double-membrane-bound vesicle is formed, either the Cvt vesicle or the autophagosome. This vesicle then fuses with the vacuole, releasing a single-bilayer-bound vesicle, the Cvt body and autophagic body, respectively (7). This vesicle is then degraded by vacuolar enzymes. Under conditions that inhibit proteinase B activity, cells accumulate intact Cvt vesicles or autophagosomes in the vacuolar lumen (33, 111). These conditions include deletion or mutation of the PRB1 gene, mutations in genes required for proteinase B trafficking to the vacuole (as happens in many vps mutants), and administration of PMSF. A second gene product required for intravacuolar vesicle breakdown is Cvt17 (115). Cells deficient in the CVT17 gene are defective in both autophagy and the Cvt pathway, accumulate prApe1 in an intravacuolar membrane-bound form (98), and accumulate intravacuolar autophagic bodies on starvation (32). Recently, the CVT17 gene was cloned, and it was found that the Cvt17 protein is homologous to a family of triglyceride lipases and that the putative active-site serine of Cvt17, as predicted from the homology, is essential for function (115). While Cvt17 is a glycoprotein that localizes to some part of the secretory pathway, it does not appear to predominantly localize to the vacuole, leaving the way in which it affects breakdown of intravacuolar vesicles a mystery at present.
MECHANISTIC DIFFERENCES BETWEEN THE CVT PATHWAY AND MACROAUTOPHAGY
While a large contingent of Cvt proteins are required for autophagy, there exist proteins that are required for one process but not the other. Understanding the way these proteins function is crucial to understanding the relationship between the Cvt pathway and autophagy. The following are such proteins, required specifically for one pathway or the other and grouped according to their functional assignments.
Factors Involved in Apg1 Function: Cvt9, Apg17, Apg13, and Vac8
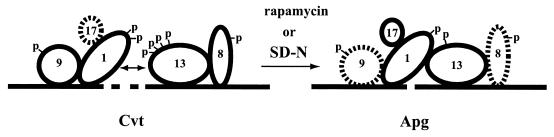
The cvt9 mutant and the as yet uncharacterized cvt3 mutant are the two original mutations that were identified as being required for Cvt trafficking but not for autophagy (32). More recently it was shown that vac8, originally isolated in a screen for mutants defective in vacuole inheritance (122, 123), is defective in the Cvt pathway but not for autophagy. Scott and coworkers showed that Vac8 exists in a complex with Apg13. They also showed that Cvt9 and Apg13 associate with Apg1, a protein kinase required for both the Cvt and autophagy pathways, although not necessarily in the same complex (52, 101). In addition, a novel gene, APG17, was identified in a two-hybrid screen for Apg1-interacting proteins. Strikingly, Apg17 is required for autophagy but not for the Cvt pathway, mirroring the fact that Cvt9 is required for the Cvt pathway but not autophagy (46). A crucial piece of evidence was the demonstration by Kamada et al. (46) that the association of Apg13 with Apg1 is modulated in response to stimuli that induce autophagy and that this modulation appears to be the result of Apg13 dephosphorylation: the coimmunoprecipitation efficiency of Apg13 and Apg1 is increased on treatment of cells with rapamycin, with a concomitant increase in the protein kinase activity measured in these immunoprecipitates. Overexpression of Apg1 suppresses the autophagy defect of _apg13_Δ, apg13-1, and _apg17_Δ cells, supporting the view that Apg1 function is downstream from these proteins (29, 46). Because Vac8 interacts with Apg13 (although not necessarily in the same complex with Apg1) and because Cvt9 and Apg17 also interact with Apg1, it is possible that Apg1 exists in multiple complexes as a catalytic kinase subunit and that its association with different combinations of regulatory subunits such as Apg17, Apg13, and Cvt9 determines whether the cell engages in Cvt trafficking or autophagy (Fig. 6). The switch between the Cvt trafficking mode and autophagy occurs independently of de novo protein synthesis. This is also true of Apg13 dephosphorylation, which is independent of Apg1 itself (2). Thus, it appears that the mechanism that controls Apg1 function in response to rapamycin and nitrogen starvation is a direct signal transduction event that modulates membrane trafficking in a way that induces a dormant membrane-trafficking pathway, autophagy.
FIG. 6.
Apg1 activity is modulated by different regulatory proteins under different physiological conditions. In logarithmically growing cells, Apg13 is highly phosphorylated and can be weakly coprecipitated with Apg1. Under these conditions, Vac8 can be coprecipitated with Apg13. In addition, two-hybrid data show an interaction of Apg1 with Apg17 and Cvt9. On induction of autophagy, Apg13 undergoes partial dephosphorylation, concomitant with an increase in its coprecipitation efficiency with Apg1 and an increase in in vitro Apg1 protein kinase activity. However, Vac8 and Cvt9 are dispensible for autophagy, suggesting that they may be lost from the complex upon induction of autophagy. Likewise, Apg17 is not required for Cvt trafficking and thus may require activation of autophagy to bind tightly to the complex. Thus, in the complex on the left, Apg17 is designated by a dashed circle, as are Vac8 and Cvt9 in the complex on the right. Proteins are indicated by number as follows: Apg1, 1; Apg13, 13; Apg17, 17; Cvt9, 9; Vac8, 8. Additional proteins may also be present in this complex. Phosphorylation is indicated by “p”. The phosphorylation state of Apg17 is not known. SD-N, minimal medium lacking nitrogen. Modified from reference 101 with permission of the publisher.
Factors Required for Membrane Fusion Events: Tlg2 and Vps45
As mentioned above, the formation of the Cvt vesicle and the formation of the autophagosome require membrane fusion events. In eukaryotic organisms, protein-protein interactions control intracellular membrane fusion at several levels. Thus, small GTP binding proteins of the Rab family mediate reversible tethering events in what might be a proofreading step. This tethering is suggested to be prerequisite for an irreversible interaction that requires the formation of a protein complex between integral proteins of the vesicle membrane and integral proteins of the target membrane, in what is called the SNARE complex (67, 84, 88). Target membrane SNARE proteins (t-SNAREs) interact with vesicle membrane SNARE proteins (v-SNAREs) to form a highly stable complex that spans both lipid bilayers. There are two families of t-SNARE proteins: SNAP-25 homologues and syntaxin homologues (124). In yeast, the vacuolar/endocytic system contains at least three syntaxin homologues: Vam3, Pep12, and Tlg2. These three proteins constitute a subfamily of yeast syntaxins related by sequence similarity. Vam3 is located at the vacuolar membrane and is required for all membrane trafficking into the vacuole (21). In contrast, Pep12 is located on endosomal membranes and is specifically required for biosynthetic trafficking that traverses the endosome, through the Prc1 pathway (10). Tlg2 is localized to endosomal and late Golgi membranes and is required for efficient endocytosis (3, 103). Sec1-like proteins interact with syntaxin homologues and are thought to control their function. Vps45 is a Sec1 homologue that interacts with both Pep12 (13) and Tlg2 (3, 76) in separate complexes. It was discovered that both _tlg2_Δ cells and _vps45_tsf cells are defective for prApe1 maturation and that these cells accumulated prApe1 in a protease-sensitive form. This is significant because _tlg2_Δ cells are not blocked in any other biosynthetic trafficking pathways, implying specificity, which is supported by the cvt phenotype of the _vps45_tsf allele. When either _vps45_tsf or _tlg2_Δ cells are challenged with rapamycin, however, they rapidly mature the prApe1, and this maturation requires an intact autophagic machinery. In addition, _tlg2_Δ _pep4_Δ cells accumulate autophagic bodies in the vacuolar lumen in response to rapamycin. Thus, these cells are not defective in autophagy. These results support a model where the membrane transport requirements of the Cvt pathway and autophagy are fundamentally distinct.
SORTING OF SPECIFIC CARGO INTO CVT VESICLES AND AUTOPHAGOSOMES
The Cvt pathway-specific proteins discussed above have one thing in common: under growing conditions in nitrogen-rich media, they accumulate prApe1, while in nitrogen-poor media or on administration of rapamycin, they rapidly revert and process prApe1 to the mature form. One can compare the rate of uptake of prApe1 with that of the recombinant alkaline phosphatase (Pho8) derivative Pho8Δ60 (79). Pho8Δ60 lacks the N-terminal membrane-spanning domain of Pho8 and as a result is found as a soluble cytosolic zymogen, whereas the native protein is transported through the early secretory pathway and into the vacuole, where it is activated. On induction of autophagy, Pho8Δ60 is sequestered within autophagosomes, delivered to the vacuole, and matured. Thus, under these conditions, maturation of Pho8Δ60 is an assay for autophagy. Scott et al. compared the rate of uptake of prApe1 by autophagy with the rate of uptake of a resident cytosolic protein Pho8Δ60 and suggested that the much faster uptake of prApe1 could be explained by the presence of a prApe1 receptor protein (100). One expects, however, that if this receptor mechanism is common to both Cvt and autophagy, yeast lacking this receptor protein will not mature prApe1 under either condition although they will still be competent for nonspecific autophagic uptake and consequently will be able to survive nitrogen starvation. Recently, α-mannosidase (Ams1) was identified as a second biosynthetic cargo molecule for the Cvt pathway (40). Like prApe1, Ams1 is synthesized as a soluble, oligomeric protein that is imported into the vacuole by a mechanism dependent on Apg and Cvt genes.
The perusal of a comprehensive database of yeast protein-protein interactions (119) yielded one protein, subsequently named Cvt19 (99), which showed a two-hybrid interaction with prApe1. A null mutant with a mutation in CVT19 was unable to form mature prApe1 in synthetic rich medium and was also defective in maturation of prApe1 under nitrogen starvation. In contrast, survival of the _cvt19_Δ strain under nitrogen starvation and the rates of pexophagy were similar to those in the wild type. The accumulated prApe1 was shown to be more easily washed from membranes than with the wild type. Cvt19 was shown to coprecipitate with prApe1 and is taken up by the Cvt pathway in the process. Interestingly, cells lacking Cvt19 are also defective for vacuolar delivery of Ams1, suggesting that it is part of a more general cargo recruitment mechanism. The existence of a single cargo-specific receptor mechanism for both autophagy and the Cvt pathway underscores the basic similarity between the two modes of transport. In addition, it is a first demonstration of the long-suspected specific recruitment of substrates for autophagic sequestration, which has been difficult to prove due to the high degree of nonspecific background.
RELATIONSHIP BETWEEN THE CVT PATHWAY AND AUTOPHAGY: SPECULATIONS, HYPOTHESES, AND FACTS
In principle, two different types of relationships may be hypothesized between the Cvt pathway and autophagy. The simplest explanation would be that there is only one pathway: under rich nutrient conditions, small vesicles (Cvt) are formed, while under nitrogen limitation or on challenge with rapamycin, large vesicles (autophagosomes) are formed by enlargement of the small vesicles without significant changes in other parameters. A second hypothesis is that these are two different pathways, which share mechanistic attributes but also have fundamental differences.
If there were only a single pathway, why would we have a group of proteins that are required for one pathway but not the other? Strikingly, all proteins that are required for Cvt trafficking but not autophagy are blocked before Cvt vesicle formation and completion. Thus, the autophagosomes that arise in these cells cannot be derived from Cvt vesicles, because these mutants are unable to form them. In addition, the single-pathway model implies that expansion of Cvt vesicles is synonymous with autophagy. However, it has been clearly shown that induction of autophagy can be uncoupled from vesicle expansion without erasing the distinction between autophagic and Cvt trafficking: that is, in specific mutants or under specific conditions, one can induce autophagosomes that are approximately the same size as Cvt vesicles, but these are still distinguished from Cvt vesicles by their capacity to bypass a block in Cvt trafficking on induction of autophagy (1). From a strictly genetic viewpoint, a simple, one-pathway model seems less capable of explaining these data convincingly. In addition, the nature of the proteins required only for Cvt trafficking is instructive: they are either molecules that are components of putative signaling complexes, such as Cvt9 and Vac8, or molecules that are involved in membrane trafficking, such as Tlg2 and Vps45. The latter also strongly suggests, but does not prove, a fundamental difference in the membrane origins of Cvt vesicles and autophagosomes.
SIGNALING EVENTS IN THE SWITCH BETWEEN AUTOPHAGY AND THE CVT PATHWAY
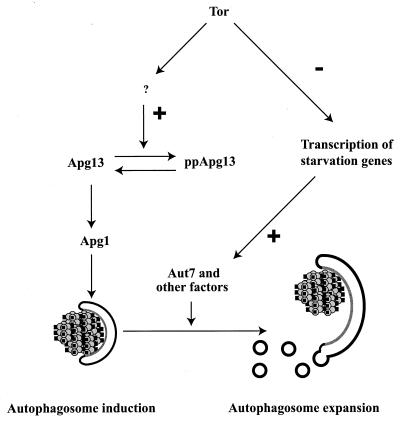
If the induction of autophagy involves a switch between two trafficking modes, one can make experimental predictions about such a switch. The stimuli that cause autophagy, nitrogen starvation and rapamycin, are known to cause very dramatic changes in transcription and translation patterns in the cell (9, 11, 17, 34, 85, 129). It might be hypothesized that induction of autophagy is a direct result of changes in the amounts of specific gene products. Indeed, the levels of both Aut7 and Ape1 are drastically increased on both nitrogen starvation and administration of rapamycin (2, 39, 56, 100). However, a direct experimental test of this hypothesis showed that induction of autophagy is not simply a matter of turning genes on and off. In the absence of protein synthesis, one can still observe autophagy-dependent rescue of a Cvt trafficking block (2). This event, which is indicative of a change in trafficking patterns between the two modes, is therefore the result of a signaling cascade. Nonetheless, the autophagosomes that are formed in the absence of protein synthesis are abnormally small. Thus, one may distinguish between two aspects of autophagosome formation: an initial step dependent on a dedicated signaling pathway, which switches the system from a Cvt to an autophagy mode, and a secondary event, dependent on changes in gene expression, which is required for the formation of normal-sized autophagosomes.
How would such an initial signal be propagated, and how does it impinge on membrane behavior? We know very little of this process, but several points along this signaling route have been marked out. Clearly, inhibition of the Tor protein kinase by the rapamycin-binding protein (RBP)-rapamycin complex (35) is required for the rapamycin-dependent induction of autophagy (80), and this has been accepted as evidence for a requirement for Tor inhibition in the induction of autophagy by nitrogen starvation as well. What occurs downstream of Tor is unclear. Many processes that depend on Tor inactivation are inhibited by the phosphorylated form of Tap42 (11, 44, 94), which is a substrate for Tor and a regulatory subunit of a number of protein phosphatase 2A-like catalytic subunits (24, 44). In the absence of Tor activity. Tap42 is dephosphorylated and uncouples from the catalytic subunits, which then associate with different regulatory subunits, leading to changes in activity that mediate downstream events. However, Kamada et al. demonstrated that inactivation of a temperature-sensitive allele of Tap42 does not result in the induction of autophagy, in apparent contradiction of any dominant role for Tap42 phosphorylation in the inhibition of autophagy (46). Because the Tap42-mediated signal is the only pathway characterized immediately downstream of Tor in yeast, it remains to be seen how the signal is propagated from Tor to the autophagic machinery. What we do know, as alluded to above, is that inhibition of Tor, as well as nitrogen starvation, results in the dephosphorylation of Apg13, a regulatory subunit of Apg1, and that this dephosphorylation event increases the affinity of Apg13 for Apg1 as well as the kinase activity of Apg1 as assayed in vitro. Indeed, this dephosphorylation is independent of de novo protein synthesis and also occurs in the absence of Apg1 itself (2), consistent with it being an upstream signaling event that controls Apg1 function. How, then, does a change in Apg1 activity affect membrane homeostasis? To date, no Apg1-dependent molecular event has been correlated with the induction of autophagy, and the identification of such an event remains an important goal.
While the change in Apg1 activity is required for the diversion of the Cvt pathway machinery from the formation of Cvt vesicles to the formation of autophagosomes, by itself it is not sufficient for the formation of normal autophagosomes. In the absence of de novo protein synthesis or in mutants lacking the AUT7 gene, whose product is up-regulated by rapamycin or starvation, one observes abnormally small autophagosomes (2) (Fig. 7). One expects that such abnormally small autophagosomes might not be able to shoulder the physiological burden imposed by starvation conditions or rapamycin, and this is indeed borne out by the (relatively weak) autophagy defect of _aut7_Δ cells. These facts also suggest that the function of the Aut7 protein, and other associated proteins such as Aut2 and Aut1, is specifically in controlling the size of the autophagosome. It was recently demonstrated that Aut7 associates with at least two yeast v-SNAREs, Bet1 and Nyv1 (62). Because SNARE proteins are required for most intracellular membrane fusion events and because one would predict that an increase in the size of the membrane entails fusion, an attractive hypothesis is that Aut7 functions by specifically promoting these fusion events. Finally, a subset of these fusion events, required in the Cvt pathway but not in autophagy, may be identical to events mediated by Tlg2 and Vps45 in the formation of the Cvt vesicle.
FIG. 7.
Induction of autophagy can be divided into nucleation and expansion steps. Tor activity prevents dephosphorylation of Apg13, by an unknown mechanism. Inhibition of Tor by the rapamycin-RBP complex then results in rapid dephosphorylation of Apg13. Dephosphorylated Apg13 acts on Apg1 to mediate autophagosome nucleation. In the absence of new protein synthesis, this results in abnormally small autophagosomes. Under normal conditions, however, inhibition of Tor also up-regulates the transcription of starvation- and autophagy-specific genes, and the ensuing increase in levels of specific proteins, such as Aut7, allows the expansion of autophagosomes. Reprinted from reference 2 with permission of the publisher.
Relationship of Autophagy to Other Starvation-Induced Responses in Yeast
Haploid yeast defective in autophagy are unable to survive starvation for nitrogen in the presence of either a fermentable (111) or a nonfermentable (116) carbon source. Interestingly, diploid yeast normally undergo sporulation in response to starvation for nitrogen in the presence of nonfermentable sugars but are unable to do so if they are defective in autophagy. Hence, both haploid and diploid cells defective in autophagy are unable to survive starvation (111). A third type of starvation-dependent response, induced by starvation for nitrogen in the presence of fermentable sugars, is filamentous growth. While the conditions for diploid filamentous growth are identical for those inducing autophagy in haploids, two points should be noted. First, in contrast to filamentous growth, autophagy is not inhibited in the presence of a nonfermentable carbon source, and second, at least a subset of the genes required for filamentous growth signaling appear dispensable for autophagy (H. Abeliovich, unpublished). At least two signaling pathways are required for filamentous growth in diploids: a mitogen-activated protein (MAP) kinase cascade that shares components with the pheromone response of haploid cells, and the nutrient-sensing cyclic AMP (cAMP) pathway. In the currently proposed scheme of events (reviewed comprehensively in reference 63), activation of Gpa2, a receptor-dependent GTP binding protein, is activated by GDP-GTP exchange catalyzed by a seven-transmembrane domain protein, Gpr1. This results in the activation of adenylyl cyclase as well as the MAP kinase cascade. Noda and Ohsumi (80) tested the effects of artificially clamping cAMP levels in yeast on the activation of autophagy in response to nitrogen starvation (in the presence of glucose) and rapamycin. They found that while changes in cAMP levels do not result in activation of autophagy, a high level of cAMP is inhibitory to autophagy induced by both rapamycin and nitrogen starvation.
PHYSIOLOGICAL IMPORTANCE OF AUTOPHAGY IN YEAST
The starvation response of haploid yeast, which involves arrest at the G0 step of the cell cycle (129), and the sporulation of diploid cells have been put forward as simple examples of differentiation processes in which the cell alters its transcription and translation programs in response to external cues (18, 61, 105). The requirement for autophagy during starvation may be rationalized through the need to recycle biological polymers (proteins, nucleic acids, carbohydrates, lipid bilayers, etc.) into building blocks for reuse under conditions where they may not be available outside the cell. One may also consider a second viewpoint that is not mutually exclusive with this: as the cell changes gears in response to these external signals, it is synthesizing new molecular components. The function of these components in supermolecular ensembles (organellar or otherwise) is likely to be incompatible with the function of preexisting macromolecular ensembles, optimized for function under the previous conditions. Thus, optimal cell survival depends not only on the synthesis of new components but also on the removal of preexisting ones.
To achieve such wholesale, quasi-selective turnover, one needs a mechanism that simultaneously has a vast capacity for hydrolysis of biological polymers yet at the same time will be specific enough to prevent the degradation of all intracellular components (e.g., the nucleus). In other words, one requires nonspecificity in that a wide spectrum of polymers is to be degraded, yet one also requires specificity in that not all supermolecular assemblages are targeted. While it is hard to imagine a cytoplasmic, molecular enzymatic mechanism that would meet these criteria, the quasi-selective targeting of cytosolic components into a degradative organelle is an elegant solution: the enzymes sequestered inside the vacuole or lysosome need not be selective in themselves, allowing for greater catalytic efficiency, while selective protection of complexes and organelles that need to be spared can also be achieved.
The involvement of autophagy genes in sporulation may reflect such a requirement for turnover on a grand scale; for example, RNA degradation may serve to replenish deoxynucleotide levels used for DNA synthesis. However, it should also be borne in mind that spore formation involves the engulfment of nuclear chromatin lobes by the prospore membrane, which fuses with itself to form immature spores bound by a double bilayer, a structure reminiscent of the autophagosome (75). The possible involvement of Apg proteins in this event awaits study.
Proteasome Function versus Autophagy: More of the Same or Very Different?
In eukaryotic cells, degradation of cytoplasmic proteins occurs by both vacuolar-lysosomal and cytoplasmic mechanisms. Chief among cytosolic degradation pathway enzymes is the 26S proteasome, a multisubunit enzymatic complex that degrades ubiquitinated and unfolded proteins. In various cell types and under different conditions, the relative weight of lysosomal-vacuolar protein degradation versus proteasomal degradation varies. In normally growing eukaryotic cells, as much as 80% of protein degradation depends on proteasomal function (20, 86), while under starvation conditions or in response to specific stimuli, autophagy becomes the primary agent for protein turnover.
In contrast to vacuolar enzymes, the proteasome is a highly specific protease, which recognizes polyubiquitinated or unfolded proteins (120). Thus, the proteasome serves two functions. First, it degrades unfolded proteins, both those that are recognized by the ubiquitination machinery and a subset that is not. In addition, it serves to degrade proteins that are targeted as a result of regulatory events that culminate in their ubiquitination and degradation, such as cell cycle regulators, various proto-oncogenes, and endocytosed antigen. In both housekeeping and regulatory functions of the proteasome, only specific proteins are targeted, even if the cellular implications may be far-reaching. Autophagy, in contrast, is not a simple housekeeping function: it is not constitutively active, and once it is triggered, it appears to be largely nonspecific on the molecular level. Most important, the vacuole contains a wide variety of hydrolytic activities in addition to proteases (for a review, see reference 58). Therefore, autophagy does not simply lead to proteolysis. Rather, it leads to the degradation and recycling of a wide spectrum of biological polymers, and it is this last property, coupled with the lack of steric constraint on degradation, that allows it to serve as a general mechanism for the degradation of ribonucleoprotein particles such as ribosomes and mRNA, complete organelles such as peroxisomes and mitochondria (see below), and other large polyprotein complexes, including the proteasome itself, that may not be good substrates for a specific and highly regulated enzyme such as the proteasome.
Pexophagy: Organelle-Specific Housecleaning through Autophagy
Certain organelles are needed in the cell to various degrees depending on the nutrient conditions. Because upkeep of organellar compartmentation is energetically costly, metabolic changes are accompanied by the turnover of organelles whose functions are not needed and may be counterproductive under inappropriate circumstances. The best known example of specific organelle number control is peroxisome biogenesis and turnover. The peroxisome number in the cell is modulated by two mechanisms. In the first mechanism, nutrient or catabolite repression, the synthesis of peroxisomal proteins is repressed in the presence of superior carbon sources such as glucose. The second mechanism provides a means for the disposal of preexisting organelles that become redundant for cell viability due to a change in conditions. Thus, yeast grown on fatty acids accumulate peroxisomes. When these cells are transferred to a preferred carbon source such as glucose or ethanol, there ensues a massive, vacuole-dependent degradation of peroxisomes, termed pexophagy.
It was found that in both mammalian and yeast cells, pexophagy occurs by a process that is topologically similar to autophagy: the organelle is sequestered by a cellular membrane and is then degraded in the vacuole or lysosome. In yeast, this process was found to depend on Apg proteins (41). However, pexophagy exhibits much faster kinetics than does general autophagy, suggesting the presence of specificity factors that target peroxisomes into autophagosomes under these conditions, analogous to the Cvt19-dependent uptake of prApel. While in most organisms studied pexophagy occurs by a process analogous to macroautophagy, P. pastoris cells utilize two distinct mechanisms depending on nutrient availability. One is directly related to macroautophagy, as defined above. The other, micropexophagy, is a type of microautophagy (118). In micropexophagy, peroxisomes are directly engulfed by the vacuolar-lysosomal membrane, which then involutes and pinches off into the lumen of the degradative organelle to form lumenal vesicles analogous to autophagic bodies (90). The relative prominence of the two routes varies between species and depends on the physiological conditions: While micropexophagy has not been found in S. cerevisiae, it is the dominant form in P. pastoris cells that are transferred from methanol medium to glucose, while macropexophagy is the dominant mechanism in P. pastoris cells that are transferred from methanol to ethanol (118). Genes required in P. pastoris micropexophagy have been identified and appear to be homologous to known APG and CVT genes from S. cerevisiae (52, 127).
The reports which suggest mechanistic differences between microautophagy and macroautophagy in S. cerevisiae (73, 93) are consistent with studies with P. pastoris in which only mutants that were blocked after the formation of autophagic bodies were common to both micropexophagy and macropexophagy (90, 118). Mutants that were blocked before the formation of autophagic bodies were specific for micropexophagic but not macropexophagic uptake. A caveat to this line of argument is the fact that APG7 and APG9, S. cerevisiae genes required for macroautophagy, have P. pastoris homologues, GSA7 and GSA14, respectively, which are required for micropexophagy (127; W. A. Dunn, Jr., unpublished data). One possible explanation is that in P. pastoris micro- and macropexophagy require different sets of related pexophagy genes. However, it should also be noted that few of the genes required for micropexophagy and none of those needed specifically for macropexophagy have been cloned in P. pastoris. Accordingly, it is difficult to draw conclusions about the distinctions between these two pathways.
Mitochondrial Recycling
Engulfment of mitochondria by autophagosomes is an observation as old as our knowledge of autophagosomes. In the purely morphological early studies on mammalian cells, mitochondria were the only distinctively cytoplasmic components that could be unambiguously ascribed to the strange new multilamellar bodies that came to be called autophagosomes (5, 19). While no direct experimental evidence exists, it has long been held that autophagy is the major pathway for mitochondrial recycling, and various theories suggest that a specific targeting of damaged mitochondria to vacuoles or lysosomes occurs by autophagy. Autophagy of mitochondria has been suggested as a step in the induction of apoptosis, potentially via the release of cytochrome c into the cytoplasm. Uptake of mitochondria into autophagosomes has also been observed in yeast (111). Thorsness and colleagues used a unique assay for the escape of mitochondrial genes into the nucleus to demonstrate a requirement for vacuolar proteases. Since they were also able to show that the process is cell-autonomous, i.e., does not proceed via cell lysis and uptake of mitochondrial DNA into neighboring cells (104), these data are consistent with the contention that autophagy is a major regulated pathway for mitochondrial breakdown (16). The fact that mutants that compromise mitochondrial function also increase mitochondrial DNA escape into the nucleus also suggests that defective mitochondria are selectively targeted.
PHYSIOLOGICAL SIGNIFICANCE OF AUTOPHAGY IN HIGHER EUKARYOTES
Autophagy in yeast and autophagy mammalian cells appear to share many characteristics, among them induction on starvation cues and morphology of intermediates. These do not, strictly speaking, imply complete conservation of mechanism at the molecular level. While molecular studies of the type undertaken in yeast are only in their first steps in the mammalian field, we already know of a number of yeast autophagy genes that have mammalian homologues. In several cases, these homologues are known to participate in autophagy in mammalian cells, as is the case for beclin-1, a homologue of the yeast Apg6 protein (64). Mammalian homologues have been found for Apg5, Apg7, Apg12, Aut7, and Apg1 (45, 89, 114). Interestingly, the human homologue of Apg5 is an apoptosis-specific protein (31). While cells in multicellular organisms enjoy a more constant environment, they also undergo changes in their surroundings that require autophagic responses, often as a result of hormone stimulation. For example, glucagon stimulates gluconeogenesis in liver cells. As a result, citric acid cycle intermediates are depleted, and their replenishment occurs by deamination and transamination of amino acids such as aspartate, glutamine, and glutamate, which are depleted in turn as well. Finally, autophagy is induced, allowing the replenishment of citric acid cycle intermediates from the breakdown of proteins (92). The following is a selective overview of our present knowledge about the involvement of autophagy in various physiological, pathological, and developmental phenomena in higher organisms.
Autophagy and Regulated Cell Death
In multicellular organisms, programmed cell death (PCD) is an active process by which a cell or class of cells is eliminated from tissues during both normal development and pathological events (reviewed in reference 14). The most commonly studied type of PCD is type I PCD, also known as apoptosis. Apoptosis is characterized by a number of features, namely, membrane blebbing, chromatin margination, and the breakdown of chromosomal DNA into nucleosome-sized fragments. A second type of programmed cell death, type II PCD, does not manifest these characteristics. Rather, type II PCD is inhibited by 3-methyladenine, a widely used inhibitor of autophagy in higher eukaryotes, and is correlated with the induction of autophagy. While it was originally suggested that type I and type II PCD are distinct, more recent data seems to imply that there is a mechanistic overlap such that the two archetypes represent extremes of a continuous spectrum of different shades of physiological cell death. Active autophagy appears to increase the tendency to undergo apoptosis, and both correlate with the mitochondrial permeability transition in certain cells. Xue et al. reported that in cultured sympathetic neurons, autophagy is activated by the apoptotic signaling machinery and in fact contributes to the apoptotic process (125). This is in agreement with other cases in which it was found that autophagy is required for efficient apoptosis (43). In a more recent report, the same group also reported that inhibition of caspase activity concomitant with an apoptotic stimulus resulted in disappearance of the mitochondria, possibly as a result of autophagy (126).
Autophagy in Cancer and Human Disease
A number of workers have suggested a correlation between levels of autophagy and carcinogenesis in various cell types (reviewed in reference 96). Thus, hormone-dependent types of cancer cell lines undergo autophagic cell death on withdrawal of hormone, and tamoxifen, a drug used to treat breast cancer, causes autophagic death of cultured mammary carcinoma cells (15). It was also demonstrated that mutations in beclin, a mammalian homologue of the Apg6 protein, result in reduced autophagy in the mammary carcinoma cell line MCF-7. Overexpression of beclin in these cells restores autophagic activity in addition to reversing the transformed morphology of the cells and inhibiting their ability to grow on soft agar (64). Strikingly, beclin was shown to substitute for Apg6 in yeast that lack the endogenous protein, indicating that the mechanism of action is conserved between yeast and mammalian cells.
Tanaka et al. (112) reported that cells derived from mice deficient in the lysosomal membrane protein LAMP-2 accumulate immature autophagosomes, correlating with reduced protein turnover. This appears to be the result of an inability of autophagic vacuoles to fuse with lysosomal or endosomal compartments. In parallel, Nishino et al. reported that LAMP-2 deficiency is the cause of X-linked vacuolar cardiomyopathy and myopathy in Danon's disease (77). The primary cellular manifestation of Danon's disease is the accumulation of intracytoplasmic vacuoles that contain autophagic material and glycogen in skeletal and cardiac muscle cells. These findings suggest a role for autophagy in human disease and are also indicative of some function in development and muscle cell metabolism (see below). Autophagy is thought to play a role in a number of neurological pathologies, such as in Huntington's disease, where an overproduction of autophagosomes has been reported (47), and possibly in Parkinson's disease, where features of autophagic cell death have been seen in substantia nigra neurons from patients (4).
Autophagy in Development
PCD occurs widely during the development of multicellular organisms. While the majority of documented occurrences of PCD in development are type I, an increasing number of cases are being ascribed to autophagy. Autophagic cell death is prevalent in secretory cells or cells with an elaborate endoplasmic reticulum, such as the labial gland of the tobacco hornworm, Manduca sexta (97), as well as mammary gland cells (65, 128). In addition, it has long been known that specific events in insect metamorphosis, such as the death of intrasegmental muscle cells in moths, are the result of autophagic cell death.
CONCLUDING REMARKS
Autophagy is a unique membrane-trafficking phenomenon that is involved in normal development and physiology, as well as diseased states, of multicellular organisms. This mechanism is also highly conserved among eukaryotes, including the yeast S. cerevisiae. The facility of molecular genetic manipulation in yeast, together with the existence of a biosynthetic marker pathway for autophagy in this organism (the Cvt pathway), makes it an ideal system for addressing the molecular mechanisms that underlie autophagy.
While studies in yeast have yielded a rich harvest of molecular information on autophagy, important points remain to be established. We now know that a dedicated signal transduction pathway induces autophagy as a response to stimuli, although only two aspects of this signal are known: Inhibition of Tor can induce the process, and inhibition of Tor results in dephosphorylation of Apg13 and therefore in changes in Apg1 protein kinase activity. We do not know, however, how this signal is propagated from Tor to Apg13 and Apg1. We also do not know how Tor inhibition is achieved under normal nitrogen starvation, in the absence of rapamycin, or how the alteration of Apg1 activity induces autophagy. We know that autophagy and the Cvt pathway both require two unusual ubiquitin-like isopeptide and amide linkages, between Apg12 and Apg5 and between Aut7 and phosphatidylethanolamine, and that a single E1-like enzyme, Apg7, is required for both events. However, we do not know the role of these conjugations in the mechanism of formation of autophagosomes and Cvt vesicles or the membrane(s) of origin of these vesicles. Fusion of these double-membrane vesicles with the vacuole releases single-membrane subvacuolar vesicles inside the lumen, where they are subsequently degraded. We do not know how these vesicles are broken down without harming the limiting membrane of the vacuole and compromising its integrity. All these topics are the subject of ongoing research, and one hopes that answers will be forthcoming.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant GM53396 from the National Institutes of Health to D. J. Klionsky.
We thank William A. Dunn, Jr., for communicating unpublished results, Galia Lerech for help with illustrations, Per E. Stromhaug for critical reading of the manuscript, and members of the Klionsky laboratory for their encouragement and support.
REFERENCES
- 1.Abeliovich H, Darsow T, Emr S D. A t-SNARE/Sec1p complex composed of Tlg2p and Vps45p is required for cytoplasm to vacuole targeting of aminopeptidase I. EMBO J. 1999;18:6005–6016. doi: 10.1093/emboj/18.21.6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abeliovich H, Dunn W A, Jr, Kim J, Klionsky D J. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151:1025–1033. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abeliovich H, Grote E, Novick P, Ferro-Novick S. Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J Biol Chem. 1998;273:11719–11727. doi: 10.1074/jbc.273.19.11719. [DOI] [PubMed] [Google Scholar]
- 4.Anglade P, Vyas S, Javoy-Agid F, Herrero M T, Michel P P, Marquez J, Mouatt-Progent A, Ruberg M, Hirsch E C, Agid Y. Apoptosis and autophagy in nigral neurons of patients with Parkinson's disease. Histol Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- 5.Ashford T P, Porter K R. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aubert S, Gout E, Bligny R, Marty-Mazars D, Barrieu F, Alabouvette J, Marty F, Douce R. Ultrastructure and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: control of the supply of mitochondria with respiratory substrates. J Cell Biol. 1996;133:1251–1263. doi: 10.1083/jcb.133.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baba M, Osumi M, Scott S V, Klionsky D J, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbet N C, Schneider U, Helliwell S B, Stansfield I, Tuite M F, Hall M N. Tor controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becherer K A, Rieder S E, Emr S D, Jones E W. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beck T, Hall M N. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 12.Burd C G, Emr S D. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 13.Burd C G, Peterson M, Cowles C R, Emr S D. A novel Sec18p/NSF-dependent complex required for Golgi to endosome transport in yeast. Mol Biol Cell. 1997;8:1089–1104. doi: 10.1091/mbc.8.6.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bursch W, Ellinger A, Gerner C, Frohwein U, Schulte-Hermann R. Programmed cell death (PCD). Apoptosis, autophagic PCD, or others? Ann N Y Acad Sci. 2000;926:1–12. doi: 10.1111/j.1749-6632.2000.tb05594.x. [DOI] [PubMed] [Google Scholar]
- 15.Bursch W, Ellinger A, Kienzl H, Torok L, Pandey S, Sikorska M, Walker K, Schulte-Hermann R. Active cell death induced by the anti-estrogens tamoxifen and ICI 164384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis. 1996;17:1595–1607. doi: 10.1093/carcin/17.8.1595. [DOI] [PubMed] [Google Scholar]
- 16.Campbell C L, Thorsness P E. Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J Cell Sci. 1998;111:2455–2464. doi: 10.1242/jcs.111.16.2455. [DOI] [PubMed] [Google Scholar]
- 17.Cardenas M E, Cutler N S, Lorenz M C, Di Como C J, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. The transcriptional program of sporulation in yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 19.Clark S L. Cellular differentiation in the kidneys of newborn mice studied with the electron microscope. J Biophys Biochem Cytol. 1957;3:349–360. doi: 10.1083/jcb.3.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Craiu A, Gaczynska M, Akopian T, Gramm C F, Fenteany G, Goldberg A L, Rock K L. Lactacystin and clasto-lactacystin β-lactone modify multiple proteasome β-subunits and inhibit intracellular protein degradation and major histocompatibility complex class I antigen presentation. J Biol Chem. 1997;272:13437–13445. doi: 10.1074/jbc.272.20.13437. [DOI] [PubMed] [Google Scholar]
- 21.Darsow T, Rieder S E, Emr S D. A multispecificity syntaxin homolog, Vam3p, is essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deter R L, Baudhuin P, De Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967;35:C11–C16. doi: 10.1083/jcb.35.2.c11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deter R L, De Duve C. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J Cell Biol. 1967;33:437–449. doi: 10.1083/jcb.33.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Como C J, Arndt K T. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 25.Dunn W A., Jr Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 26.Dunn W A., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunn W A., Jr Studies on the mechanism of autophagy: maturation of the autophagic vacuole. J Cell Biol. 1990;110:1935–1945. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer von Mollard G, Stevens T H. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol Biol Cell. 1999;10:1719–1732. doi: 10.1091/mbc.10.6.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 30.George M D, Baba M, Scott S V, Mizushima N, Garrison B S, Ohsumi Y, Klionsky D J. Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol Biol Cell. 2000;11:969–982. doi: 10.1091/mbc.11.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammond E M, Brunet C L, Johnson G D, Parkhill J, Milner A E, Brady G, Gregory C D, Grand R J. Homology between a human apoptosis specific protein and the product of APG5, a gene involved in autophagy in yeast. FEBS Lett. 1998;425:391–395. doi: 10.1016/s0014-5793(98)00266-x. [DOI] [PubMed] [Google Scholar]
- 32.Harding T M, Hefner-Gravink A, Thumm M, Klionsky D J. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 33.Harding T M, Morano K A, Scott S V, Klionsky D J. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardwick J S, Kuruvilla F G, Tong J K, Shamji A F, Schreiber S L. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heitman J, Movva N R, Hall M N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 36.Herman P K, Emr S D. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6742–6754. doi: 10.1128/mcb.10.12.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holthuis J C M, Nichols B J, Dhruvakumar S, Pelham H R B. Two syntaxin homologs in the TGN/endosomal system of yeast. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horst M, Knecht E C, Schu P V. Import into and degradation of cytosolic proteins by isolated yeast vacuoles. Mol Biol Cell. 1999;10:2879–2889. doi: 10.1091/mbc.10.9.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang W P, Scott S V, Kim J, Klionsky D J. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- 40.Hutchins M U, Klionsky D J. Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J Biol Chem. 2001;276:20491–204918. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hutchins M U, Veenhuis M, Klionsky D J. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci. 1999;22:4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- 42.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimshoni Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 43.Jia L, Dourmashkin R, Allen P D, Gray A B, Newland A C, Kelsey S M. Inhibition of autophagy abrogates tumour necrosis factor α induced apoptosis in human T-lymphoblastic leukaemic cells. Br J Haematol. 1997;98:673–685. doi: 10.1046/j.1365-2141.1997.2623081.x. [DOI] [PubMed] [Google Scholar]
- 44.Jiang Y, Broach J R. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kegel K B, Kim M, Sapp E, McIntyre C, Castano J G, Aronin N, DiFiglia M. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiel J A, Rechinger K B, van der Klei I J, Salomons F A, Titorenko V I, Veenhuis M. The Hansenula polymorpha PDD1 gene product, essential for the selective degradation of peroxisomes, is a homologue of Saccharomyces cerevisiae Vps34p. Yeast. 1999;15:741–754. doi: 10.1002/(SICI)1097-0061(19990630)15:9<741::AID-YEA416>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 49.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y-sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J, Dalton V M, Eggerton K P, Scott S V, Klionsky D J. Apg7p/Cvt2p is required for the Cvt, macroautophagy, and peroxisomal degradation pathways. Mol Biol Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J, Huang W-P, Klionsky D J. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J, Kamada Y, Stromhaug P E, Guan J, Hefner-Gravink A, Bevan A, Baba M, Scott S V, Ohsumi Y, Dunn W A, Jr, Klionsky D J. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J, Klionsky D. Autophagy, cytoplasm-to vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu Rev Biochem. 2000;69:303–342. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- 54.Kim J, Scott S V, Oda M, Klionsky D J. Transport of a large oligomeric protein by the cytoplasm to vacuole protein degradation pathway. J Cell Biol. 1997;137:609–618. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klionsky D J, Cueva R, Yaver D S. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klionsky D J, Herman P K, Emr S D. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev. 1990;54:266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klionsky D J, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 60.Kovacs A L, Rez G, Palfia Z, Kovacs J. Autophagy in the epithelial cells of murine seminal vesicles in vitro. Cell Tissue Res. 2000;302:253–261. doi: 10.1007/s004410000275. [DOI] [PubMed] [Google Scholar]
- 61.Kurtz S, Lindquist S. Subcellular differentiation in sporulating yeast cells. Cell. 1986;45:771–779. doi: 10.1016/0092-8674(86)90791-9. [DOI] [PubMed] [Google Scholar]
- 62.Legesse-Miller A, Sagiv Y, Gluzman R, Elazar Z. Aut7p, a soluble autophagic factor, participates in multiple membrane trafficking processes. J Biol Chem. 2000;275:32966–32973. doi: 10.1074/jbc.M000917200. [DOI] [PubMed] [Google Scholar]
- 63.Lengeler K B, Davidson R C, D'souza C, Harashima T, Shen W-C, Wang P, Pan X, Waugh M, Heitman J. Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev. 2000;64:746–785. doi: 10.1128/mmbr.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liang X H, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 65.Lockshin R A, Zakeri Z. Programmed cell death: early changes in metamorphosing cells. Biochem Cell Biol. 1994;72:589–596. doi: 10.1139/o94-078. [DOI] [PubMed] [Google Scholar]
- 66.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 67.McNew J A, Parlati F, Fukuda R, Johnston R J, Paz K, Paumet F, Sollner T H, Rothman J E. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407:153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- 68.Misteli T, Warren G. COP-coated vesicles are involved in the fragmentation of Golgi stacks in a cell-free system. J Cell Biol. 1994;125:269–282. doi: 10.1083/jcb.125.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George M D, Klionsky D J, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 71.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- 72.Moriyasu Y, Ohsumi Y. Autophagy in tobacco suspension culture cells in response to sucrose starvation. Plant Physiol. 1996;111:1233–1241. doi: 10.1104/pp.111.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Muller O, Sattler T, Flotenmeyer M, Schwarz H, Plattner H, Mayer A. Autophagic tubes: vacuolar invaginations involved in lateral membrane sorting and inverse vesicle budding. J Cell Biol. 2000;151:519–528. doi: 10.1083/jcb.151.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Munn A, Riezman H. Endocytosis is required for the growth of vacuolar H(+)-ATPase-defective yeast: identification of six new END genes. J Cell Biol. 1994;127:373–386. doi: 10.1083/jcb.127.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neiman A M. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J Cell Biol. 1998;140:29–37. doi: 10.1083/jcb.140.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nichols B J, Holthuis J C, Pelham H R. The Sec1p homologue Vps45p binds to the syntaxin Tlg2p. Eur J Cell Biol. 1998;77:263–268. doi: 10.1016/s0171-9335(98)80084-8. [DOI] [PubMed] [Google Scholar]
- 77.Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, Mora M, Riggs J E, Oh S J, Koga Y, Sue C M, Yamamoto A, Murakami N, Shanske S, Byrne E, Bonilla E, Nonaka I, DiMauro S, Hirano M. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 78.Noda T, Kim J, Huang W-P, Baba M, Tokunaga C, Ohsumi Y, Klionsky D J. Apg9/Cvt7 is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148:465–479. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- 80.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 81.Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 82.Peterson M R, Burd C G, Emr S D. Vac1p coordinates Rab and phosphatidylinositol 3-kinase signaling in Vps45p-dependent vesicle docking/fusion at the vacuole. Curr Biol. 1999;11:159–162. doi: 10.1016/s0960-9822(99)80071-2. [DOI] [PubMed] [Google Scholar]
- 83.Petiot A, Ogier-Denis E, Blommaart E F, Meijer A J, Codogno P. Distinct classes of phosphatidylinositol 3-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 84.Pfeffer S R. Transport vesicle docking: SNAREs and associates. Annu Rev Cell Dev Biol. 1996;12:441–461. doi: 10.1146/annurev.cellbio.12.1.441. [DOI] [PubMed] [Google Scholar]
- 85.Powers T, Walters P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 87.Rothman J E, Warren G. Implications of the SNARE hypothesis for intracellular membrane topology and dynamics. Curr Biol. 1994;4:220–233. doi: 10.1016/s0960-9822(00)00051-8. [DOI] [PubMed] [Google Scholar]
- 88.Rothman J E, Wieland F T. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 89.Sagiv Y, Legesse-Miller A, Porat A, Elazar Z. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 2000;19:1494–1504. doi: 10.1093/emboj/19.7.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sakai Y, Koller A, Rangell L K, Keller G A, Subramani S. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J Cell Biol. 1998;141:625–636. doi: 10.1083/jcb.141.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sato T K, Darsow T, Emr S D. Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol Cell Biol. 1998;18:5308–5319. doi: 10.1128/mcb.18.9.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sato T K, Rehling P, Peterson M R, Emr S D. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol Cell. 2000;6:661–671. doi: 10.1016/s1097-2765(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 93.Sattler T, Mayer A. Cell-free reconstitution of microautophagic vacuole invagination and vesicle formation. J Cell Biol. 2000;151:529–538. doi: 10.1083/jcb.151.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schmidt A, Beck T, Koller A, Kunz J, Hall M N. The Tor nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schu P V, Takegawa K, Fry M J, Stack J H, Waterfield M D, Emr S D. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 96.Schulte-Hermann R, Bursch W, Grasl-Kraupp B, Marian B, Torok L, Kahl-Rainer P, Ellinger A. Concepts of cell death and application to carcinogenesis. Toxicol Pathol. 1997;25:89–93. doi: 10.1177/019262339702500117. [DOI] [PubMed] [Google Scholar]
- 97.Schwartz L M, Smith S W, Jones M E, Osborne B A. Do all programmed cell deaths occur via apoptosis? Proc Natl Acad Sci USA. 1993;90:980–984. doi: 10.1073/pnas.90.3.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scott S V, Baba M, Ohsumi Y, Klionsky D J. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scott S V, Guan J, Hutchins M U, Kim J, Klionsky D J. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Scott S V, Hefner-Gravink A, Morano K A, Noda T, Ohsumi Y, Klionsky D J. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Scott S V, Nice D C, Nau J J, Weisman L S, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky D J. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 102.Seals D F, Eitzen G, Margolis N, Wickner W T, Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Seron K, Tieaho V, Prescianotto-Baschong C, Aust T, Blondel M O, Guillaud P, Devilliers G, Rossanese O W, Glick B S, Riesman H, Keranen S, Haguenauer-Tsapis R. A yeast t-SNARE involved in endocytosis. Mol Biol Cell. 1998;9:2873–2879. doi: 10.1091/mbc.9.10.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shafer K S, Hanekamp T, White K H, Thorsness P E. Mechanisms of mitochondrial DNA escape to the nucleus in the yeast Saccharomyces cerevisiae. Curr Genet. 1999;36:183–194. doi: 10.1007/s002940050489. [DOI] [PubMed] [Google Scholar]
- 105.Simchen G, Kassir Y. Genetic regulation of differentiation towards meiosis in the yeast Saccharomyces cerevisiae. Genome. 1989;31:95–99. doi: 10.1139/g89-018. [DOI] [PubMed] [Google Scholar]
- 106.Simonsen A, Lippe R, Christoforidis S, Gaullier J M, Brech A, Callaghan J, Toh B H, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- 107.Stack J H, DeWald D B, Takegawa K, Emr S D. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole. J Cell Biol. 1995;129:321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stack J H, Herman P K, Schu P V, Emr S D. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 1993;12:2195–2204. doi: 10.1002/j.1460-2075.1993.tb05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stack J H, Horazdovsky B, Emr S D. Receptor mediated protein sorting to the vacuole in yeast: roles for a protein kinase, a lipid kinase, and GTP binding proteins. Annu Rev Cell Dev Biol. 1995;11:1–33. doi: 10.1146/annurev.cb.11.110195.000245. [DOI] [PubMed] [Google Scholar]
- 110.Stromhaug P E, Berg T O, Fengsrud M, Seglen P O. Purification and characterization of autophagosomes from rat hepatocytes. Biochem J. 1998;335:217–224. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tanaka Y, Ghude G, Suter A, Eskelinen E L, Hartmann D, Lullmann-Rauch R, Janssen P M, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2 deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 113.Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homologue of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates, including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2000;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- 115.Teter S A, Eggerton K P, Scott S V, Kim J, Fischer A M, Klionsky D J. Degradation of lipid vesicles in the yeast vacuoles requires function of Cvt17, a putative lipase. J Biol Chem. 2000;276:2083–2087. doi: 10.1074/jbc.C000739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf D H. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 117.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 118.Tuttle D L, Dunn W A., Jr Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J Cell Sci. 1995;108:25–35. doi: 10.1242/jcs.108.1.25. [DOI] [PubMed] [Google Scholar]
- 119.Uetz P, Giot L, Cagney G, Mansfield T A, Judson R S, Knight J R, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg J M. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 120.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 121.Wang, C.-W., J. Kim, W.-P. Huang, H. Abeliovich, P. E. Stromhaug, W. A. Dunn, Jr., and D. J. Klionsky. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J. Biol. Chem., in press. [DOI] [PMC free article] [PubMed]
- 122.Wang Y X, Catlett N L, Weisman L S. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J Cell Biol. 1998;140:1063–1074. doi: 10.1083/jcb.140.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang Y X, Zhao H, Harding T M, Gomes de Mesquita D S, Woldringh C L, Klionsky D J, Munn A L, Weisman L S. Multiple classes of yeast mutants are defective in vacuole partitioning yet target vacuole proteins correctly. Mol Biol Cell. 1996;7:1375–1389. doi: 10.1091/mbc.7.9.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Weimbs T, Hui Low S, Chapin S J, Mostov K E, Bucher P, Hoffman K. A conserved domain is present in different family members of vesicular fusion proteins: a new superfamily. Proc Natl Acad Sci USA. 1997;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xue L, Fletcher G C, Tolkovsky A M. Autophagy is activated by apoptotic signalling in sympathetic neurons: an alternative mechanism of death execution. Mol Cell Neurosci. 1999;14:180–198. doi: 10.1006/mcne.1999.0780. [DOI] [PubMed] [Google Scholar]
- 126.Xue L, Fletcher G C, Tolkovsky A M. Mitochondria are selectively eliminated from eukaryotic cells after blockade of caspases during apoptosis. Curr Biol. 2001;11:361–365. doi: 10.1016/s0960-9822(01)00100-2. [DOI] [PubMed] [Google Scholar]
- 127.Yuan W, Stromhaug P E, Dunn W A., Jr Glucose-induced autophagy of peroxisomes in Pichia pastoris requires a unique E1-like protein. Mol Biol Cell. 1999;10:1353–1366. doi: 10.1091/mbc.10.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zakeri Z, Bursch W, Tenniswood M, Lockshin R A. Cell death. Programmed, apoptosis, necrosis, or other. Cell Death Differ. 1995;2:87–96. [PubMed] [Google Scholar]
- 129.Zaragoza D, Ghavidel A, Heitman J, Schultz M C. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol Cell Biol. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]