Analysis of Primary Structural Determinants That Distinguish the Centromere-Specific Function of Histone Variant Cse4p from Histone H3 (original) (raw)
Abstract
Cse4p is a variant of histone H3 that has an essential role in chromosome segregation and centromere chromatin structure in budding yeast. Cse4p has a unique 135-amino-acid N terminus and a C-terminal histone-fold domain that is more than 60% identical to histone H3 and the mammalian centromere protein CENP-A. Cse4p and CENP-A have biochemical properties similar to H3 and probably replace H3 in centromere-specific nucleosomes in yeasts and mammals, respectively. In order to identify regions of Cse4p that distinguish it from H3 and confer centromere function, a systematic site-directed mutational analysis was performed. Nested deletions of the Cse4p N terminus showed that this region of the protein contains at least one essential domain. The C-terminal histone-fold domain of Cse4p was analyzed by changing Cse4p amino acids that differ between Cse4p and H3 to the analogous H3 residues. Extensive substitution of contiguous Cse4p residues with H3 counterparts resulted in cell lethality. However, all large lethal substitution alleles could be subdivided into smaller viable alleles, many of which caused elevated rates of mitotic chromosome loss. The results indicate that residues critical for wild-type Cse4p function and high-fidelity chromosome transmission are distributed across the entire histone-fold domain. Our findings are discussed in the context of the known structure of H3 within the nucleosome and compared with previous results reported for CENP-A.
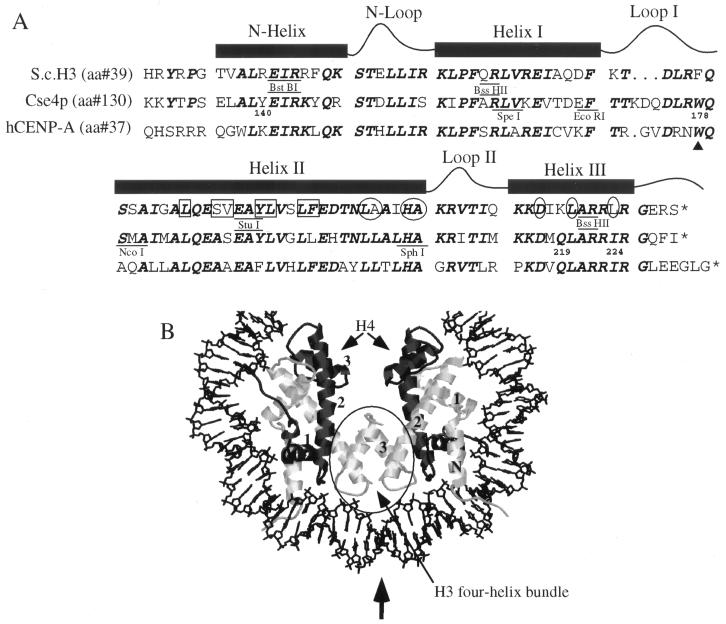
The nucleosome core is composed of an evolutionarily conserved octamer of four core histone proteins, H2A, H2B, H3, and H4, and 145 to 147 bp of DNA (26). Histone-histone and histone-DNA interactions that form the core nucleosome are mediated through a globular, highly evolutionarily conserved histone-fold domain found in all four core histones. The histone-fold domain is composed of a long central α-helix (helix II) flanked by two short α-helices (helix I and helix III) (Fig. 1). The three α-helices are separated by two β-strand loops (loop I and loop II). H3-H4 and H2A-H2B dimers form in an antiparallel fashion such that the helix II α-helices cross each other and juxtapose loop I with loop II from the two different molecules (1, 11). This arrangement creates three regions in each dimer that bind the minor groove of DNA, two loop I-loop II regions and a region formed from the two adjacent helix I α-helices. In the case of H3, an additional N-terminal helix also makes DNA contacts (11). During nucleosome assembly, two H3-H4 heterodimers form a tetramer through an H3-H3′ interaction called the four-helix bundle (Fig. 1). To complete the octamer, two H2A-H2B heterodimers bind to the tetramer through a four-helix bundle interaction involving H4 and H2B (11).
FIG. 1.
(A) Alignment of the histone-fold domains of S. cerevisiae H3, Cse4p, and human CENP-A. Residues that are identical in two or three of the proteins are indicated in boldface italic type. The α-helices and β-loops in the histone-fold domain of H3 are shown above the residues involved in those structures. The amino acids in H3 involved in dimer formation with H4 are boxed, and the residues involved in the H3-H3′ four-helix bundle interaction are circled. The highly conserved phenylalanine in loop I of H3 that is replaced by a tryptophan (W178) in Cse4p and CENP-A is noted (▴). Specific Cse4p amino acids mentioned in the text are numbered. The positions in the protein sequences of H3 and Cse4p corresponding to the restriction sites used for domain exchanges are indicated (Materials and Methods). (B) The (H3-H4)2 tetramer initiates formation of a standard nucleosome by binding DNA (50 bp are shown). The DNA helix is black, the two copies of H4 are in dark gray, and the two copies of H3 are light gray. The H3-H3′ four-helix bundle is circled, and the vertical arrow indicates the dyad axis. Helix I, II, and III of H3 and H4 are numbered 1, 2, and 3, respectively, and the H3 N-helix is marked with an “N.” The figure was generated by using RasMac version 2.6 with X-ray coordinates from the Protein Data Bank, Brookhaven National Laboratory (ID code 1aoi) (11).
The kinetochore is a complex of specialized centromeric chromatin and proteins that mediates interactions between the chromosomal DNA and spindle fiber during mitosis and meiosis. Proper assembly of the kinetochore on centromeric chromatin and attachment of the kinetochore to the spindle fiber are essential for accurate chromosome segregation. Studies in a variety of organisms support the idea that epigenetic mechanisms, as well as primary DNA sequences, impart identity and function to centromeric DNA (10), indicating a critical role for histone proteins and other chromatin components in determining the functional state of a centromere.
The organization of centromeric chromatin and the assembly of functional centromeres are specified at least in part by the H3-like histone variants Cse4p and CENP-A, discovered in Saccharomyces cerevisiae and mammals, respectively (16, 23, 27). Both proteins have uniquely different N termini and C-terminal histone-fold domains that are more than 60% identical to the histone-fold domain of H3 (Fig. 1) (23). In humans, tandem arrays of AT-rich α-satellite DNA are found in the inner kinetochore plate where CENP-A localizes. CENP-A copurifies with core histones in nucleosome-like structures (16) and binds predominantly to α-satellite DNA in phased nucleosomal arrays (25). Immunolocalization studies of CENP-A proteins expressed from transfected DNA in the presence of endogenous CENP-A show that the histone-fold domain mediates targeting of the protein to the mammalian centromere and that the unique N terminus is not required for centromere localization (24). Similar assays on mutant CENP-A proteins containing H3 substitutions in analogous sites in the histone-fold domain show that residues throughout the histone-fold domain of CENP-A cooperate to localize the protein to the mammalian centromere (19).
The S. cerevisiae centromere contains three conserved DNA elements, CDE I, CDE II, and CDE III, which are sufficient and required for high-fidelity mitotic and meiotic chromosome segregation (4, 5). In vivo all three CDE elements are packaged into a 150- to 220-bp nuclease-resistant chromatin structure that is flanked by phased nucleosomal arrays (3, 6). Depletion of either histone H4 or H2B renders the CDE elements accessible to nucleases, showing that core histone proteins play a critical role in centromere structure (18). Cse4p is a component of yeast centromeric chromatin. Mutant cse4 alleles cause cell cycle arrest at G2-M, increased chromosome missegregation and disrupt centromere chromatin structure (23, 14). Cse4p associates with yeast centromere DNA in vivo and is an integral component of yeast chromatin with biochemical properties similar to H3 (14, 23). Genetic evidence supports direct interactions between Cse4p and H4. Overexpression of CSE4 suppresses the temperature-sensitive phenotype of a mutant H4 allele (hhf1-20) that causes a G2-M cell cycle arrest and increased chromosome missegregation (22). In addition, overexpression of CSE4 rescues the defective centromeric chromatin structure observed in hhf1-20 cells (14), further supporting a direct interaction between Cse4p and H4 at the centromere. Interestingly, the hhf1-20 mutations are located in a region of H4 which, according to the nucleosome crystal structure, would be adjacent to loop I of Cse4p, a region of Cse4p that diverges significantly from H3.
By analogy to the structure of H3 in standard nucleosomes, the N terminus of Cse4p would be located outside the nucleosome core, while the histone-fold domain would be located in the core of the nucleosome. We reason that some regions of Cse4p have functions similar to those of histone H3 (e.g., nucleosome assembly), while other regions are involved in centromere specific functions (e.g., kinetochore assembly, sister chromatid cohesion). As a first step toward understanding how Cse4p functions in organizing centromeric chromatin, we conducted a systematic mutagenic analysis of the protein. Deletions were made in the Cse4p N terminus, and Cse4p histone-fold domain amino acids that differ between Cse4p and yeast H3 were changed to the H3 counterparts. We found that the N terminus of Cse4p, which differs significantly in length and primary amino acid sequence from H3, is required for cell viability. Analysis of the histone-fold domain revealed that extensive substitution by H3 amino acids is lethal but that any single amino acid in the Cse4p histone fold can be changed to the analogous H3 residue without disrupting essential Cse4p function. Many cse4 mutations, though viable, caused elevated rates of mitotic chromosome loss. The results are discussed in the context of the known three-dimensional structure of H3 in the nucleosome and compared with findings for CENP-A.
MATERIALS AND METHODS
Media and strains.
The medium for yeast growth was described previously (20). Rich medium for yeast cultures (YPD) contains 2% glucose, 1% peptone, and 0.5% yeast extract supplemented with 50 mg of filter-sterilized tryptophan per liter. Color medium contains 0.6% yeast nitrogen base without amino acids (Difco), 0.5% Casamino Acids, 2% glucose, and 30 μg of uracil and 4.5 μg of adenine per ml. 5-Fluoroorotic acid (FOA) media contains 0.5 μg of FOA per ml supplemented with 20 μg of uracil per ml. All yeast strains were grown at 30°C unless otherwise noted. Genetic analysis was performed by standard methods described previously (23).
Yeast strains used in this study are listed in Table 1. KC100 is a meiotic product of SB961 (CSE4/cse4::HIS3) carrying wild-type CSE4 on the low-copy-number plasmid pCL1 (23). KC115 was obtained by transforming KC100 with pHCC4, which contains the _Cla_I-_Dra_III CSE4 DNA fragment cloned into YEp351 (9), and plating it on medium containing FOA to select for cells that have lost pCL1 (Ura−). Cured cells were then transformed with an _Eco_RI-linearized CEN3 substitution vector (pJUP/3B14) containing wild-type CEN3 (7, 13). Genomic integration events were verified by Southern hybridization. Yeast strain KC115, which contains properly integrated URA3-SUP11-CEN3 in chromosome III, was crossed with KC150 to produce the diploid strain KC405. To study chromosome segregation, plasmids containing cse4 mutant alleles were transformed into KC405 cells, which were then cured of pHCC4.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| SB961 | MATa/MATα cen130-3-URA3-SUP11/CEN3 ade2-101/ade2-101 his3-11,15/his3-11,15 LEU2/leu2-3 LYS2/lys2-801 trp1Δ901/trp1Δ901 ura3-52/ura3-52 CSE4/cse4::HIS3 | 23 |
| KC100 | MATα ade2-101 his3-11,15 leu2-3 lys2-801 trp1Δ901 ura3-52 cse4::HIS3 plus pCL1 (CEN4 URA3 CSE4) | This study |
| KC115 | MATα CEN3-URA3-SUP11 ade2-101 his3-11,15 leu2-3 lys2-801 trp1Δ901 ura3-52 cse4::HIS3 plus pHCC4 (CSE4 in YEp351) | This study |
| KC150 | MATa ade2-101 his3-11,15 leu2-3 trp1Δ901 ura3-52 cse4::HIS3 plus pHCC4 (CSE4 in YEp351) | This study |
| KC405 | MATa/MATα CEN3-URA3-SUP11/CEN3 ade2-101/ade2-101 his3-11,15/his3-11,15 leu2-3/leu2-3 LYS2/lys2-801 trp1Δ901/trp1Δ901 ura3-52/ura3-52 plus pHCC4 | This study |
CSE4 mutants.
CSE4 N-terminal deletion alleles were constructed by using plasmid pRB190, which contains the _Eco_RI-_Pst_I fragment of CSE4 inserted into the polylinker of pRS314 (21). The CSE4 N-terminal coding region in pRB190 was modified by recombinant PCR to encode a hexahistidine tag between codons 1 and 2, introducing a unique _Nsi_I site at the initiator methionine codon (ATGCAT). By using a wild-type CSE4 clone as a template, PCR was carried out with an upstream primer that annealed at the desired deletion endpoint and a downstream primer located in the 3′-untranslated DNA. An in-frame _Nsi_I site was incorporated into the upstream primer. The PCR product was cleaved with _Nsi_I and _Sph_I, the latter site located at codon 208, and used to replace the corresponding segment in pRB190. Thus, the resulting _cse4_Δ alleles all encode Met-His at their extreme N termini. Hemagglutinin (HA) epitope-tagged versions of the deletion alleles were constructed identically except that the template for PCR was plasmid pCSE4HA (see below).
The Cse4p/H3 hybrid alleles were constructed by using several different strategies. The starting plasmids were pRB190, pCSE4, which contains a _Cla_I-_Dra_I fragment carrying wild-type CSE4 inserted into the polylinker of pRS314 (21), or pCSE4HA, which is identical to pCSE4 except that it encodes an HA-tagged version of Cse4p (23). No detectable differences between these plasmids were observed with respect to CSE4 expression or function. Silent mutations were introduced into CSE4 and HHT2 (H3) to generate unique restriction sites (indicated in Fig. 1). These restriction sites were then used, in one or two steps, to exchange regions of CSE4 with the corresponding regions of H3 (cse4-261, cse4-268, cse4-270, and cse4-271). Boundaries of the exchanged regions were narrowed or extended by using recombinant PCR (i.e., the two halves of the desired construct, overlapping 18 to 21 bp, were obtained in separate PCRs and then joined in a third PCR by using outside primers). The recombinant PCR products were cut with the appropriate enzyme to generate restriction fragments carrying the desired replacement allele (cse4-278 and cse4-279). Helix III changes were introduced by incorporating the desired mutation(s) into PCR primers that also contain an in-frame _Bss_HII site. Amplified DNA was cleaved with _Bss_HII and used to replace CSE4 DNA either upstream (cse4-276, cse4-277, cse4-284, and cse4-285) or downstream (cse4-327, cse4-337, cse4-338, cse4-350, and cse4-369) of the helix III _Bss_HII site. Substitutions in the C-terminal end of helix II (cse4-286 and cse4-287) were made in an analogous fashion except that the mutations were incorporated into PCR primers 3′ of an in-frame _Nsp_I site, and the amplified DNA used to replace CSE4 sequences upstream of the naturally occurring _Sph_I site. Allele cse4-290 was constructed from cse4-278 and cse4-279 by exchanging DNA 3′ of the common _Bss_HII site. Allele cse4-425 was constructed from cse4-278 and cse4-350 by exchanging DNA 3′ of the _Sph_I site. Mutagenesis of the helix I-loop I-helix II region (cse4-259, cse4-260, cse4-262, cse4-263, cse4-264, cse4-265, and cse4-266) was accomplished by inserting oligonucleotide linkers into _Spe_I-_Nco_I, _Eco_RI-_Nco_I, or _Nco_I-_Sph_I sites, as noted in Fig. 1.
Alleles cse4-70, cse4-71, cse4-72, cse4W-F, cse4W-H, and cse4W-Y were made by using the Clontech Transformer Site-Directed Mutagenesis kit according to vendor’s instructions and with pCSE4HA as template. The other W178 mutants were made by PCR by using the degenerate oligonucleotide KK316 (5′-GAT TTA GGT (A,C,G)(A,C,T)(A,C) CAG TCA ATG GCG-3′) and Pfu polymerase (Stratagene).
In all cases, mutated cse4 alleles were sequenced to confirm that the intended changes were present. For constructions involving PCR, regions derived from PCR-amplified DNA were sequenced to check for unintentional sense mutations.
Assays for Cse4p expression and function.
The cse4 mutant alleles were tested for their ability to complement a cse4 null mutation by using a plasmid shuffle assay. The test strain was KC100 (cse4::HIS3) carrying pCL1 (CEN-ARS-URA3-CSE4) (23). Since CSE4 is an essential gene, this strain is pCL1 dependent; the plasmid-borne CSE4 is required to complement the chromosomal cse4::HIS3 null allele. When introduced into the test strain, mutant cse4 alleles that complement cse4::HIS3 allow mitotic loss of pCL1 and reversion to a Ura− FOA-resistant phenotype. The cse4 alleles to be tested were cloned into pRS314 (CEN-ARS-TRP1) and transformed into KC100. Two or three independent KC100 transformants were picked, suspended in 0.5 ml of water, and 15 μl of 1:40 and 1:400 dilutions were plated on medium lacking tryptophan and on medium containing FOA. Plates were photographed after 4 days. In many cases, both native and epitope-tagged versions of the cse4 mutant proteins were tested. In no case did the epitope tag have a detectable effect on the complementation phenotype.
Protein expression from mutant cse4 alleles was assayed by immunoblotting. Epitope-tagged versions of the mutant alleles were obtained either by constructing the mutation in pCSE4HA or by replacing DNA encoding the Cse4p N terminus with a DNA fragment encoding the HA-tagged version (by using the native _Bst_BI or _Nde_I site). In all cases, the triple-HA epitope was located at Cse4p codon 81 (23). For immunoblot analysis, yeast strains carrying plasmids with cse4 mutant alleles were grown in Trp− dropout medium. Cell pellets were resuspended at a concentration equivalent to an optical density at 600 nm of 50 in sodium dodecyl sulfate (SDS) gel sample buffer containing β-mercaptoethanol and boiled for 1 min. A 20- to 40-μl sample was loaded directly onto SDS–12% polyacrylamide (Laemmli) gels. After electrophoresis, proteins were transferred to Immobilon (Millipore) membranes, and the HA epitope was detected by anti-HA monoclonal antibody (Boehringer Mannheim) and peroxidase-conjugated goat anti-mouse immunoglobulin G (Gibco BRL) by chemiluminescence (Renaissance; Dupont NEN). Primary and secondary antibodies were diluted 1:800 and 1:3,000, respectively; otherwise, reaction conditions and buffers were as recommended by the respective manufacturers.
Mitotic chromosome loss assays.
Mitotic loss events involving marked copies of chromosome III were quantified by using fluctuation assays as described previously (8) with the following modifications. A pink colony was picked and grown for 4 to 6 h at 30°C in medium that selects for the cse4 mutant plasmid (TRP1) and the marked chromosome (URA3). Approximately 50 cells were plated onto color medium and incubated at 30°C for 18 to 20 h. Eight equal-sized colonies were picked, pooled into one microcentrifuge tube, and vortexed; half the volume of each tube was then spread onto color indicator plates. A total of five pools were plated for each strain. The plates were incubated at 30°C for 5 days and at 4°C for 4 to 6 days to allow colony colors to develop fully. The total number of colonies and the total number of red colonies were counted, values were doubled to reflect the total number of cells originally picked, and the rate of mitotic chromosome loss was determined (8).
RESULTS
The N terminus of Cse4p is essential.
Cse4p differs from CENP-A and histone H3 by having an extended 135-amino-acid N terminus. An N terminus of this length is novel among H3 molecules and bears no significant homology to other proteins in the GenBank protein database. To determine whether the extended N terminus of Cse4p is essential, we tested a series of deletions which removed the first 15, 27, 39, or 50 amino acids (Fig. 2A). The ability of the various deletion alleles to complement a lethal cse4 null mutation was tested by using the plasmid shuffle assay described in Materials and Methods. Briefly, the cse4 mutant alleles were introduced into a haploid shuttle strain (KC100) carrying a lethal cse4 null allele (cse4::HIS3) on the chromosome but maintained by wild-type CSE4 contained on a URA3 plasmid. Complementation of cse4::HIS3 by the test allele allows mitotic loss of the CSE4-URA3 plasmid as revealed by growth on medium containing FOA, which is lethal to cells expressing URA3.
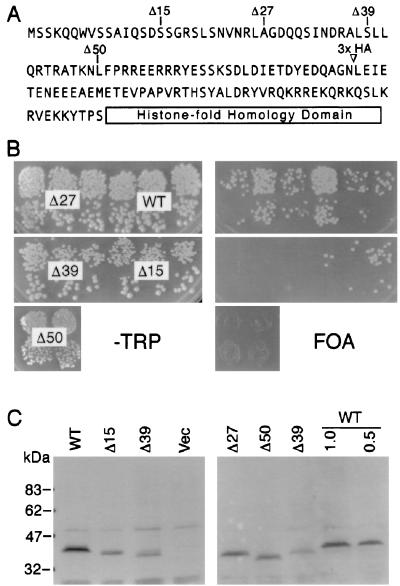
FIG. 2.
N-terminal Cse4p deletions reveal an essential domain. (A) Amino acid sequence of the Cse4p N terminus showing deletion endpoints. Mutant alleles initiate with methionine, followed by a histidine residue, and continue with the amino acid indicated. 3× HA indicates the location of the inserted triple HA epitope tag. (B) Plasmid shuffle complementation assays for the cse4 deletion mutants (see Materials and Methods). (C) Immunoblot analysis of yeast strains carrying cse4 deletion alleles. Cells carrying the HA-tagged wild-type allele (WT) were loaded at 1× and 0.5× amounts (all other lanes are 1×) to allow an estimation of signal strength. Lane Vec was loaded with cells carrying the expression vector lacking a CSE4 insert.
Deleting 15 or 27 residues from the Cse4p N terminus resulted in no detectable phenotype (Fig. 2B). FOA-resistant colonies arose at the same frequency and were of the same size as those produced by cells transformed with a control plasmid carrying wild-type CSE4. Larger N-terminal deletions, Δ39 and Δ50, were lethal since no FOA-resistant colonies were produced (Fig. 2B). Immunoblot analysis of epitope-tagged derivatives showed that each of the four mutant proteins were expressed, although at various steady-state levels (Fig. 2C). The lethality of the Δ39 and Δ50 cse4 alleles was unlikely to be due to the decreased expression levels since other cse4 alleles were expressed at equally low levels and nonetheless supported wild-type growth (see below). These results demonstrate that the first 27 amino acids of Cse4p are dispensable for cell viability and that the N terminus contains at least one domain beyond residue 27 that is essential for Cse4p function.
Residues in the histone-fold domain of Cse4p that differ from those in yeast H3 are important for Cse4p function.
The existence of an essential domain in the novel N terminus of Cse4p potentially explains why neither H3 nor CENP-A are able to fulfill the centromere-specific function of Cse4p (23). However, hybrid proteins consisting of the N terminus of Cse4p fused to the histone-fold domains of yeast H3 or CENP-A were also unable to rescue the cse4 null mutant (data not shown), implying that the Cse4p histone-fold domain, in addition to the unique N terminus, contributes to the protein’s specialized function. This finding prompted a directed mutational analysis of the Cse4p histone-fold domain in hopes of identifying specific amino acids or structural elements that functionally distinguish Cse4p from H3. Our directed mutational strategy took advantage of the fact that the three-dimensional structure of H3 within the nucleosome is known (Fig. 1) (11).
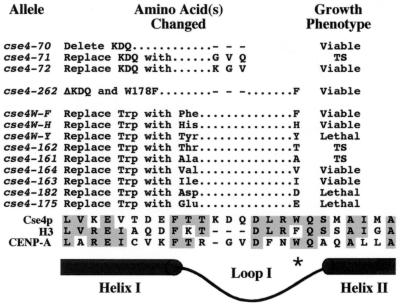
Loop I in the Cse4p histone-fold diverges from that of H3 in two ways, both of which may potentially confer Cse4p specificity to centromere DNA (Fig. 1). Both Cse4p and CENP-A have additional amino acids in loop I and have a tryptophan in place of a highly conserved phenylalanine in H3. This region of CENP-A has been shown to be critical for centromere targeting (19). Loop I of Cse4p was mutagenized to determine whether the additional residues (KDQ) and the tryptophan (W178) are important for Cse4p function. Four KDQ alleles were generated (Fig. 3): cse4-70 has all three KDQ amino acids deleted, cse4-71 and cse4-72 have two of the three residues replaced with GV to resemble loop I of CENP-A, and cse4-262 has the KDQ deletion combined with a W178-to-F substitution, resembling H3. Surprisingly, all four alleles were viable (Fig. 3), although cse4-71 caused temperature sensitivity and cse4-262 caused elevated mitotic chromosome loss rates (see below). We also tested cse4 alleles in which W178 was changed to nine different amino acids (Fig. 3). Alleles replacing W178 with an acidic amino acid (D or E) or tyrosine (Y) were lethal, but changes to aliphatic (V or I) or aromatic residue (F and H) were not. Two mutants, W178T (cse4-162) and W178A (cse4-161), were temperature sensitive (38°C). These results indicate that while loop I KDQ residues and W178 may optimize Cse4p function, they are not essential, suggesting that other regions of the histone-fold domain are also involved in specifying centromere function.
FIG. 3.
Functional analysis of cse4 alleles with mutations in loop I. The loop I region of Cse4p is aligned with H3 and CENP-A at the bottom of the figure to show the KDQ insert and the position of W178. Mutant cse4 alleles were tested for their ability to complement a cse4 null mutation by using the plasmid shuffle assay (see Materials and Methods). Alleles exhibiting a temperature-sensitive growth phenotype (38°C) are noted (TS).
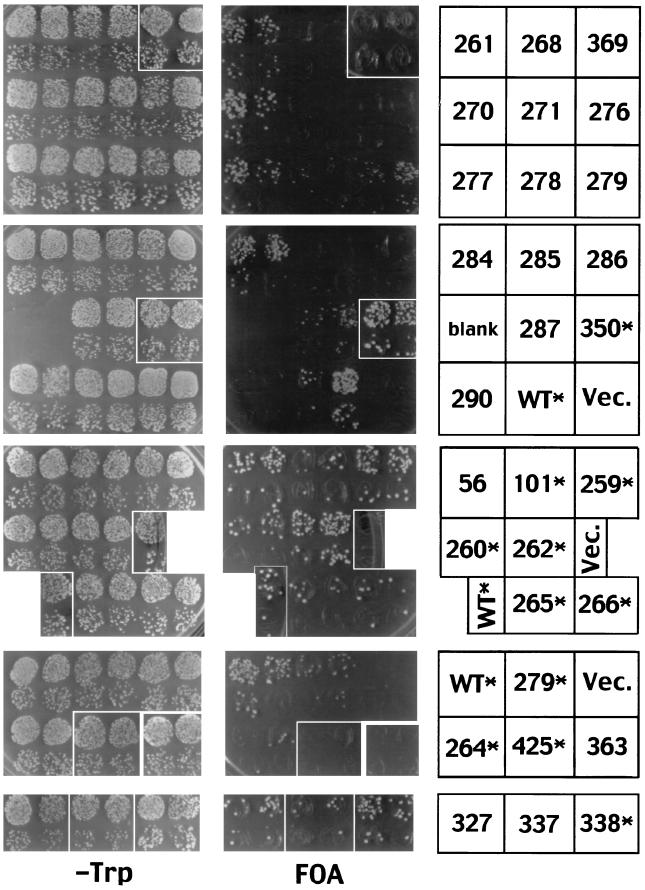
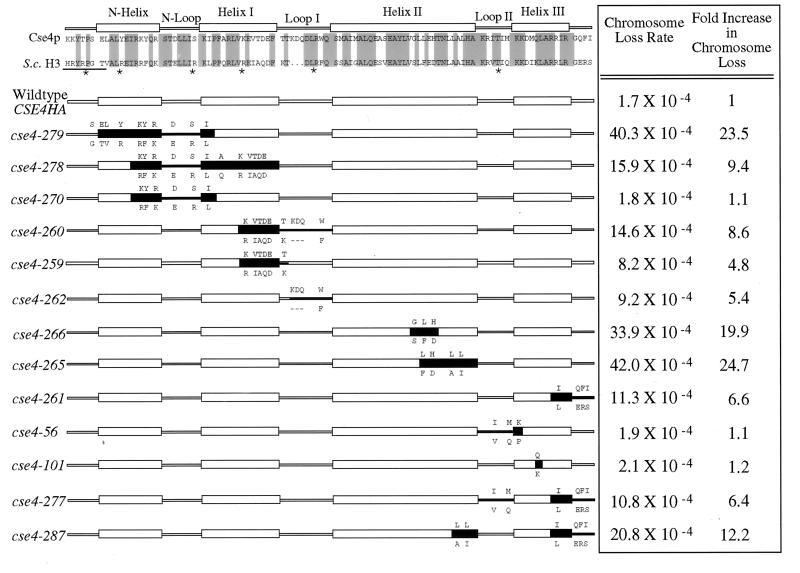
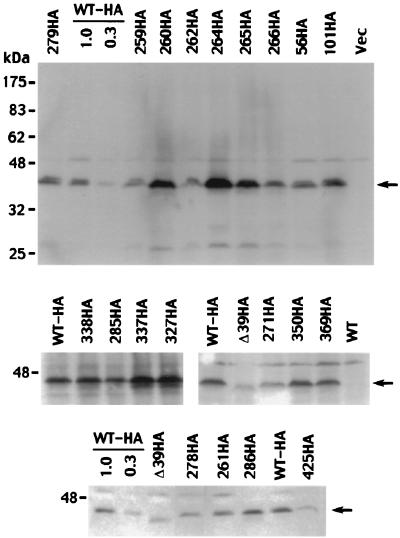
The resilience of Cse4p to wholesale alterations of loop I to resemble H3 led us to systematically replace other segments of Cse4p with the corresponding H3 residues (Fig. 4). This mutagenesis strategy potentially allowed us to identify Cse4p residues involved in centromere-specific functions without disrupting regions of Cse4p that interact with other core histones to form nucleosomes. The mutant cse4 alleles were tested in the shuffle assay for their ability to complement a cse4 null allele (Fig. 5), and several viable alleles were assayed for defects in mitotic chromosome loss (Fig. 6). Steady-state expression levels of most of the mutant alleles were determined by immunoblotting (Fig. 7).
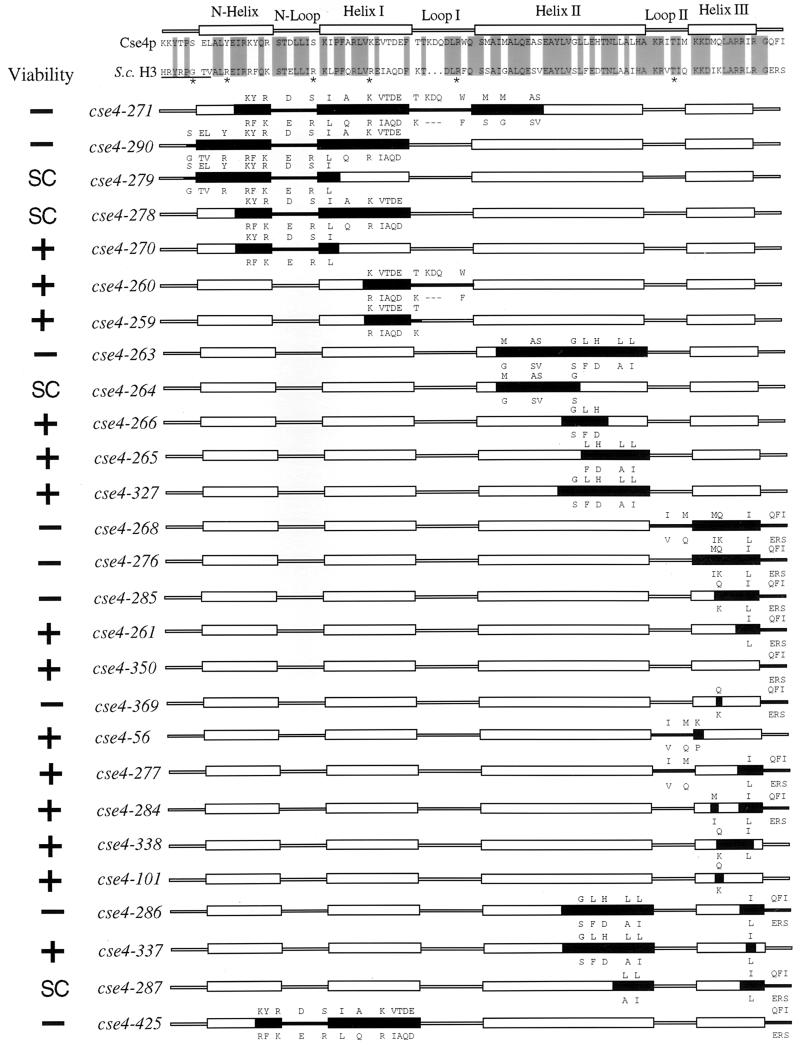
FIG. 4.
Cse4p histone-fold domain mutants. Regions of the histone-fold domain of Cse4p were substituted with the analogous residues from H3. A sequence alignment of the S. cerevisiae H3 and Cse4p proteins is shown at the top of the figure, with identical residues shaded. The positions of the α-helices and β-loops are noted above the residues. Underlined amino acids and asterisks denote regions in H3 that contact DNA in the nucleosome crystal structure. For each mutant allele, the black boxes represent the regions containing substituted H3 residues. The altered Cse4p amino acids are shown above the helical and loop structures, and the substituted H3 amino acids expressed in the mutant are shown below. The viability of the cse4 mutant alleles was determined by using the plasmid shuffle assay. SC, cse4 alleles that reproducibly produce small FOA-resistant colonies in the plasmid shuffle assay (see Materials and Methods).
FIG. 5.
Functional complementation by cse4 histone-fold domain swap mutations. The plasmid shuffle assays (TRP− and FOA plates are as indicated) were performed on cells transformed by the cse4 allele indicated on the right. Asterisks indicate epitope-tagged alleles, WT denotes the wild-type CSE4 allele, and Vec denotes a vector lacking a CSE4 gene. Each of the four rows shows single plates from three independent assays (except for cse4-363, which was taken from a separate plate). The top two sets of plates comprised a single assay, while the bottom sets of plates were separate, independent assays. Growth on Trp− but no growth on FOA indicates lethal cse4 mutations that fail to complement the cse4 null, while growth on FOA plates indicates viable cse4 alleles that can complement the cse4 null. Results from the shuffle assays are summarized in Fig. 4. Several alleles produce small colonies on the FOA plates, a finding indicative of a slower growth rate. These are denoted “SC” in Fig. 4.
FIG. 6.
Cse4p histone-fold domain mutants are defective in chromosome segregation. Viable cse4 histone-fold domain mutants were assayed for mitotic loss of chromosome III. Loss rates were determined by fluctuation assays as described in Materials and Methods and are reported as fold increases relative to the wild-type CSE4 control strain.
FIG. 7.
Detection of HA-tagged Cse4 mutant proteins by immunoblotting. Representative blots for viable and lethal cse4 alleles are shown, including alleles used in the mitotic chromosome loss assays. The upper panel shows an entire blot, while the lower panels show only the Cse4p region. The various mutants are identified by the allele numbers above the gel lanes, with HA designating HA-tagged derivatives. Samples of HA-tagged wild-type (WT) cells were loaded at 1× and 0.3× amounts (all other lanes, 1×) to allow an estimation of signal strength. The lane labeled Vec was loaded with cells carrying the expression vector lacking a CSE4 insert. The mobility of protein size markers is indicated at the left, and arrows mark the position of the Cse4p band.
The first set of Cse4p-H3 swap mutants exchanged amino acids in the N helix, N loop, helix I, and loop I regions of Cse4p. Alleles involving changes spanning all of these domains, cse4-271 and cse4-290, were lethal (Fig. 4 and 5). This region was divided into smaller overlapping mutations, cse4-279, cse4-278, and cse4-270, all of which were viable. Comparison of mutant and wild-type colony size indicates that cse4-279 and cse4-278 alleles lead to slow growth (Fig. 5). Likewise, altering amino acids in the C-terminal half of helix I alone (cse4-259) or in combination with the loop I substitution (cse4-260) were also viable mutations. These results demonstrate that the N-terminal half of the Cse4p histone-fold domain clearly contributes to an essential function of the protein. However, no individual residue or short contiguous stretch of amino acids, if substituted with the corresponding H3 residues, was found to be essential for Cse4p function.
Mutations replacing Cse4p helix II or helix III with the analogous regions of H3 were lethal (cse4-263 and cse4-276, respectively) (Fig. 4 and 5). In the case of helix II, overlapping substitutions changing three to five residues were viable (cse4-264, cse4-265, cse4-266, and cse4-327), although cse4-264 gave rise to a slow-growth phenotype (Fig. 5). Analysis of the helix III swap mutants revealed, unexpectedly, that the extreme C terminus of Cse4p was an important structural determinant. Any mutation that combined the Q-to-K change at position 219 of helix III with the substitution of the three C-terminal Cse4p residues (QFI) with those of H3 (ERS) resulted in lethality (cse4-268, cse4-276, cse4-285, and cse4-369). Neither the Q219-to-K substitution by itself (cse4-101) nor the QFI-to-ERS change (cse4-350) by itself caused a detectable phenotype. Also, other single or pairwise substitutions of helix III residues (cse4-261, cse4-277, cse4-284, and cse4-338) and the entire swap of loop II (cse4-56) were without discernible effects. An effect of the ERS C terminus substitution was also observed in combination with mutations altering other regions of the molecule. Combining the QFI-to-ERS mutation with viable mutations in helix II or in the region encompassing the N helix, N loop, and helix I resulted in lethality (cse4-286 and cse4-425, respectively), and a smaller helix II mutation in combination with the ERS substitution resulted in a slow-growth phenotype (cse4-287).
Mutations in the histone-fold domain of Cse4p cause increased mitotic chromosome loss.
To determine whether mutations altering the Cse4p histone-fold domain affected chromosome segregation, we measured mitotic chromosome loss of a marked chromosome in cells expressing various cse4 mutant alleles from plasmids as their sole source of Cse4p (Fig. 6). Cells carrying a plasmid expressing wild-type CSE4 tagged with the HA epitope (CSE4HA) exhibited a chromosome III loss rate of 1.7 × 10−4 per mitotic division, similar to that observed for isogenic cells carrying wildtype CSE4 without the HA epitope (data not shown). The alleles altered in the N helix, N loop, and helix I cse4-279 and cse4-278 caused 23.5- and 9.4-fold increases in chromosome III loss rates, respectively, while cse4-270, containing a smaller subset of these mutations, did not increase chromosome loss (1.1-fold). A helix I allele, cse4-259, and a loop I allele, cse4-262, increased loss rate by 4.8- and 5.4-fold, respectively, while cse4-260, which combines both mutations, exhibited an additive increase in chromosome loss (8.6-fold). The integrity of helix II is also important for chromosome segregation, since H3 swap mutations affecting this region, cse4-265 and cse4-266, elevated chromosome loss rates significantly (24.7- and 19.9-fold, respectively). The helix III-C terminus substitution in cse4-261 caused only a moderate increase in chromosome loss (6.6-fold), but when combined with two amino acid changes in helix II (cse4-287) the loss rate increased to 12.2-fold. Substitutions in loop II (cse4-56) or the single Q219 to K change in helix III (cse4-101) did not significantly increase chromosome loss rates (1.1- and 1.2-fold, respectively). There is a general correlation between the cse4 slow-growth and chromosome loss phenotypes. Mutants exhibiting the small-colony phenotype (cse4-278, cse4-279, and cse4-287) displayed large increases in chromosome loss rate (9.4-fold or higher), although the converse was not always true (e.g., the helix II mutants, cse4-265 and cse4-266, caused high chromosome loss rates but did not exhibit slow growth). Interestingly, while cse4-265 and cse4-266 appeared to support a wild-type growth rate on FOA plates (Fig. 5), they reproducibly gave FOA-resistant colonies at significantly reduced frequencies (data not shown), possibly suggesting that the latent growth defect was readily suppressed in the subpopulation of FOA-resistant cells which eventually formed colonies.
Protein expression of Cse4p mutants.
Expression of the mutant Cse4p proteins was determined by immunoblot analysis of cells expressing HA epitope-tagged derivatives. Figure 7 shows typical results. Wide variability was observed in the steady-state protein levels produced by the different mutant alleles. There did not appear to be a correlation between the severity of the mutant phenotypes and the observed levels of mutant proteins. Lethal alleles were found to be expressed at wild-type levels (cse4-285, cse4-286, and cse4-369) and reduced levels (cse4_Δ_39, cse4-271, and cse4-425). Likewise, alleles causing strong chromosome loss phenotypes had elevated expression levels (cse4-260, cse4-264, and cse4-265), lower expression levels (cse4-278), and wild-type expression levels (cse4-279 and cse4-266). Importantly, mutant phenotypes could not be accounted for by low expression per se. Mutants cse4Δ15 (Fig. 2) and cse4-425 (Fig. 7) are expressed at about the same reduced level (30 to 50% of the wild type), yet the former has no detectable growth phenotype whereas the latter is lethal. Furthermore, we found that the expression of some mutant constructs was lower in the presence of wild-type CSE4. Thus, protein extracts from cells expressing lethal alleles may exhibit lower steady-state protein levels due to the presence of wild-type untagged Cse4p. Another potential source of variability is plasmid copy number. The mutant alleles were carried on CEN-based plasmids and, due to the effects of the cse4 mutations, CEN plasmids would be prone to missegregation, increasing or decreasing the average plasmid copy number per cell. Indeed, requiring the mutant allele to provide Cse4p function (wild-type CSE4 absent) may actually select a population of cells carrying multiple copies of the mutant cse4 allele.
DISCUSSION
The centric H3 variants Cse4p and CENP-A have histone-fold domains that are more than 60% identical to the histone-fold domain of H3. The striking sequence homology and predicted structural similarities among these proteins, taken together with biochemical and genetic evidence (15, 22, 23), strongly supports the idea that Cse4p and CENP-A replace H3 in centromere-specific nucleosomes in S. cerevisiae and mammalian cells, respectively. The similarities among Cse4p, CENP-A, and H3 imply that these proteins carry out common functions, such as interacting with other core histones and nucleosome assembly. In the various Cse4p swap mutants, we incorporated all of the H3 residues that differ between the two proteins into mutant cse4 alleles that proved to be viable. Apparently, no single or small group of contiguous amino acids in the histone-fold domain of Cse4p acts alone to specify Cse4p function at the centromere. Rather, the results suggest that amino acids distributed throughout the histone-fold domain interact in combination to impart centromere function. A similar conclusion was reached for CENP-A (19). Furthermore, since all regions of the Cse4p histone-fold domain can be individually changed to H3 residues, it is probable that Cse4p assembles into the nucleosome core by a mechanism similar to that of H3.
Judged simply by the number of Cse4p residues that could be altered without abolishing function, certain regions of the Cse4p histone-fold domain are more tolerant of H3 substitutions, while other regions are more sensitive to substitutions. In general, Cse4p can accommodate fairly extensive substitutions of H3 residues in the N helix, N loop, helix I, and loop I. The N helix, N loop, helix I, and loop I in H3 make extensive interactions with the DNA. Although we did not design the Cse4p-H3 swap mutants to directly test Cse4p-DNA interactions and the mutations that alter DNA contacts also change noncontact sites, we did observe a correlation between the severity of the cse4 phenotypes and the number of putative DNA contact sites altered by the N helix, N loop, helix I, and loop I mutations. H3 substitutions in Cse4p that change four of the putative DNA contact sites in these regions are lethal (cse4-290). However, the cse4 allele that changes three possible contact sites is viable but exhibits a severe chromosome loss phenotype (23.5 fold; cse4-279); those changing only two sites show modest increases in chromosome loss rates (8.6- and 9.4-fold; cse4-260 and cse4-278), while those changing one putative contact site have little or no phenotype (1.1-, 4.8-, and 5.4-fold; cse4-270, cse4-259, and cse4-262, respectively). The N helix, N loop, helix I, and loop II may be critical for providing specificity between Cse4p and the centromere DNA.
Helix II and especially helix III of Cse4p appeared to be very sensitive to substitution with H3 amino acids. In H3-H4 heterodimers, the long central helix II in H3 crosses over helix II in H4, positioning loop I of each molecule with loop II of the other molecule. Two H3-H4 dimers interact through the H3-H3′ four-helix bundle to form a tetramer, which then initiates binding to the DNA helix (1, 11). Replacing most of the Cse4p helix II residues with H3 residues is lethal, but smaller helix II substitutions are viable, suggesting that the smaller mutations do not prevent Cse4p-H4 dimer formation. However, the binding affinities between the mutant Cse4p protein and H4 might be altered by these mutations, thereby affecting the efficiency of Cse4p-H4 dimer and (Cse4p-H4)2 tetramer assembly and possibly interactions with centromeric DNA. Such destabilizing changes could explain the lethal and high chromosome loss phenotypes observed for the helix II cse4 mutants.
The extreme C-terminal tail of Cse4p (ERS) is an important structural determinant of helix III. Substitution of H3 amino acids for the Cse4p C-terminal tail moderately increases the rate of mitotic chromosome loss (cse4-261) and can be lethal when combined with changes elsewhere in Cse4p. This is most dramatic in the case of the Q219-to-K mutation in helix III (cse4-101), which is viable on its own but lethal when combined with the H3 C-terminal tail (cse4-369). The C-terminal halves of the H3-H3′ helix III’s contact each other across the dyad axis of the nucleosome. By analogy, helix III of Cse4p would be expected to be critical for stabilizing the (Cse4p-H4)2 tetramer. Substitution of H3 residues in helix II and helix III in Cse4p might interfere with interactions between Cse4p-H4 dimers required to assemble (Cse4p-H4)2 tetramers. Substitution mutations that make helix II and helix III in Cse4p more like H3, such as lethal alleles cse4-286 or cse4-369, would be expected to disrupt Cse4p-Cse4p′ four-helix bundle interactions and reduce the number of (Cse4p-H4)2 tetramers available for centromere recognition.
Immunolocalization studies of CENP-A mutants containing H3 amino acid substitutions in the histone-fold domain have identified regions of CENP-A that are involved in centromere localization (19). These data, taken together with results from this study, indicate that Cse4p and CENP-A may impart centromere function by similar mechanisms involving multiple sites within their histone-fold domains. The N helix, loop I and both ends of helix II of CENP-A were found to be critical for localization to the centromere (19). The same regions of Cse4p were found to be important, since substituting them with H3 amino acids either was lethal (helix II substitution) or caused increased chromosome loss rates (N helix, N loop substitution, loop I substitution, and partial helix II substitutions). An obvious caveat to direct comparisons between the two studies is that the CENP-A experiments directly tested localization but did not analyze other possible functions of the protein. In addition, the CENP-A mutant proteins were studied in the presence of wild-type CENP-A. In contrast, the study reported here analyzed the viability and centromere function of mutant Cse4p but did not directly measure centromere localization. We infer that mutant Cse4p proteins are properly localized if they rescue the cse4 null mutation and confer wild-type or near-wild-type rates of mitotic chromosome loss. However, mutant phenotypes detected by our in vivo assays may reflect defects other than mislocalization.
Replacing the histone-fold domain of Cse4p with that of CENP-A cannot rescue the cse4 null phenotype, indicating that specific regions within the histone-fold domain probably provide specificity to the different centromeres. Interestingly, regions in the histone-fold domain of Cse4p that differ from H3 also differ between Cse4p and CENP-A (Fig. 1). These divergent regions may provide each protein with the specificity required for the distinctly different yeast and mammalian centromeres. Our results did reveal potential differences between the CENP-A and Cse4p histone-fold domains. We clearly show that helix III of Cse4p is essential in determining centromere-specific function. The sequence of helix III in CENP-A varies only minimally from human H3, so a helix III replacement mutant of CENP-A was not tested (19). The extreme C terminus of Cse4p is also an important determinant of Cse4p function. Exchanging this region caused increased rates of chromosome loss and, when combined with other viable alleles, caused synthetic lethality. Substituting the CENP-A C terminus with that of H3 did not affect centromere localization (19); however, combinatorial mutations, including the C-terminal substitution, were not tested. Altering the single helix III amino acid that differs between human H3 and CENP-A in combination with the C-terminus substitution might inactivate CENP-A, as we observed for Cse4p when the helix III Q219-to-K change was combined with the QFI-to-ERS C-terminus substitution. Finally, our results show that helix I is involved in Cse4p function, whereas this region of CENP-A is not required for centromere localization.
Another feature of the Cse4p primary structure that is decidedly different from CENP-A is the unique N terminus. The Cse4p N terminus has no amino acid homology with the N termini of either H3 or CENP-A and, while the N termini of H3 and CENP-A are both about 40 amino acids in length, the N terminus of Cse4p is much longer (135 amino acids). These facts suggest that the N terminus of Cse4p functions differently from the N termini of H3 and CENP-A. Indeed, we identified at least one region of the Cse4p N terminus that is essential for Cse4p function. In contrast, the CENP-A N terminus is not required for localizing CENP-A to the centromere, although it might be involved in kinetochore functions not revealed by the localization assay. The entire N terminus of yeast H3 can be deleted without affecting viability, although cells exhibit slow growth and defects in transcription. The H3 N terminus extends outside the nucleosome core, where it interacts with proteins involved in silencing and transcriptional regulation (11, 12). The N terminus of Cse4p may also extend outside the nucleosome core, making it accessible for interactions with proteins involved in a variety of centromere functions, including sister chromatid cohesion, chromosome separation, kinetochore assembly, and chromatin formation or modification. Although we have not yet fully characterized the essential region in the Cse4p N terminus, the spacing between the essential region and the histone-fold domain must be somewhat flexible, since insertion of the triple HA epitope tag (37 amino acids) in frame between amino acids 79 and 80 (Fig. 2) does not detectably affect Cse4p function.
Replacement of H3 with Cse4p or CENP-A in the nucleosome core confers two distinct advantages for packaging centromere DNA. First, since homotypic interactions within the octamer particle are restricted to the two H3 molecules, substitution of H3 with Cse4p would permit the formation of homotypic (Cse4p-H4)2 tetramers but not heterotypic tetramers containing one molecule each of Cse4p and H3. We propose that (Cse4p-H4)2 tetramers function to recognize the centromeric DNA and initiate assembly of a unique Cse4p nucleosome. Heterotypic (H4-Cse4p–H3-H4) tetramers might be defective in assembly or function at the centromere because of mislocalization of the tetramer to noncentromere sites. The second advantage is that homotypic (Cse4p-H4)2 tetramers would maximize the Cse4p-CEN DNA contact sites. Histone H3 is unique among the core histones in that it contacts the nucleosomal DNA at several distinct points along the 146 bp of DNA associated with the core octamer. The position of the (Cse4p-H4)2 tetramer determines the central dyad axis of the nucleosomal DNA, as well as the entry and exit points of the DNA helix on the histone octamer (11). By making multiple DNA-protein contacts along the centromere DNA, the two Cse4p proteins in the proposed homotypic Cse4p nucleosome are positioned to recognize the centromere, identify the central dyad axis, and initiate assembly.
ACKNOWLEDGMENTS
We thank Timothy J. Richmond for critical reading of the manuscript.
This work was supported by a grant to M.F.-H from the National Institutes of Health (GM54766) and by grants to R.E.B. from the National Science Foundation (MCB-9406050) and the Howard Hughes Genetics Initiative to the University of Massachusetts Medical School.
REFERENCES
- 1.Arents G, Burlingame R W, Wang B-C, Love W E, Moudrianakis E N. The nucleosomal core histone octamer at 3.1 Å resolution: a tripartite protein assembly and left-handed superhelix. Proc Natl Acad Sci USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baxevanis A D, Arents G, Moudrianakis E N, Landsman D. A variety of DNA-binding and multimeric proteins contain the histone-fold motif. Nucleic Acids Res. 1995;23:2685–2691. doi: 10.1093/nar/23.14.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloom K S, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 5.Fleig U, Beinhauer J D, Hegemann J H. Functional selection for the centromere DNA from yeast chromosome VII. Nucleic Acids Res. 1995;23:922–924. doi: 10.1093/nar/23.6.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Funk M, Hegemann J H, Philippsen P. Chromatin digestion with restriction endonucleases reveals 150-160 bp of protected DNA in the centromere of chromosome XIV in Saccharomyces cerevisiae. Mol Gen Genet. 1989;219:153–160. doi: 10.1007/BF00261171. [DOI] [PubMed] [Google Scholar]
- 7.Gaudet A, Fitzgerald-Hayes M. Alterations in the adenine-plus-thymine-rich region of CEN3 affect centromere function in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:68–75. doi: 10.1128/mcb.7.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hegemann J H, Shero J H, Cottarel G, Philippsen P, Hieter P. Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol Cell B. 1988;8:2523–2535. doi: 10.1128/mcb.8.6.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 10.Karpen G H, Allshire R C. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 11.Lugar K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 12.Mann R K, Grunstein M. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 1992;11:3297–3306. doi: 10.1002/j.1460-2075.1992.tb05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGrew J T, Xiao Z, Fitzgerald-Hayes M. Saccharomyces cerevisiae mutants defective in chromosome segregation. Yeast. 1989;5:271–284. doi: 10.1002/yea.320050407. [DOI] [PubMed] [Google Scholar]
- 14.Meluh P B, Yang P, Glowczewski L, Koshland D, Smith M M. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 15.Palmer D K, O’Day K, Trong H L, Charbonneau H, Margolis R L. Purification of the centromere-specific protein CENP-A and demonstration that it is a specific histone. Proc Natl Acad Sci USA. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer D K, O’Day K, Wener M H, Andrews B S, Margolis R L. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pluta A F, Mackay A M, Ainsztein A M, Goldberg I G, Earnshaw W C. The centromere: hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- 18.Saunders M J, Yeh E, Grunstein M, Bloom K. Nucleosome depletion alters the chromatin structure of Saccharomyces cerevisiae centromeres. Mol Cell Biol. 1990;10:5721–5727. doi: 10.1128/mcb.10.11.5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shelby R D, Vafa O, Sullivan K F. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherman F, Fink G, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. [Google Scholar]
- 21.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith M M, Yang P, Santisteban M S, Boone P W, Goldstein A T, Megee P C. A novel histone H4 mutant defective for nuclear division and mitotic chromosome transmission. Mol Cell Biol. 1996;16:1017–1026. doi: 10.1128/mcb.16.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoler S, Keith K C, Curnick K E, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 24.Sullivan K F, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone-fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vafa O, Sullivan K F. Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- 26.van Holde K E. Chromatin. New York, N.Y: Springer-Verlag; 1988. [Google Scholar]
- 27.Wolffe A P, Pruss D. Deviant nucleosomes: the functional specialization of chromatin. Trends Genet. 1995;12:58–62. doi: 10.1016/0168-9525(96)81401-6. [DOI] [PubMed] [Google Scholar]