Inhibition of p53-dependent transcription by BOX-I phospho-peptide mimetics that bind to p300 (original) (raw)
Abstract
The N-terminal BOX-I domain of p53 containing a docking site for the negative regulator MDM2 and the positive effector p300, harbours two recently identified phosphorylation sites at Thr18 or Ser20 whose affect on p300 is undefined. Biochemical assays demonstrate that although MDM2 binding is inhibited by these phosphorylations, p300 binding is strikingly stabilized by Thr18 or Ser20 phosphorylation. Introducing EGFP-BOX-I domain peptides with an aspartate substitution at Thr18 or Ser20 induced a significant inhibition of endogenous p53-dependent transcription in cycling cells, in irradiated cells, as well as in cells transiently co-transfected with p300 and p53. In contrast an EGFP-wild-type BOX-I domain peptide stimulated p53 activity via inhibition of MDM2 protein binding. These results suggest that phosphorylation of p53 at Thr18 or Ser20 can activate p53 by stabilizing the p300–p53 complex and also identify a class of small molecular weight ligands capable of selective discrimination between MDM2- and p300-dependent activities.
INTRODUCTION
The link between the role of p53 as a tumour suppressor and its activity as a transcription factor has been well documented (Oren, 1999). Recent efforts have concentrated on the post-translational events upstream of p53 that activate its tumour suppressor function. Such studies have highlighted two key proteins that play a role in regulating p53 function. The MDM2 protein forms a component of a negative regulatory pathway that facilitates ubiquitin-dependent degradation of p53 through the proteasome (Lohrum and Vousden, 2000). The transcriptional adaptor protein, p300, forms a component of a positive regulatory pathway that facilitates the induction of p53-dependent gene expression (Goodman and Smolik, 2000). The balance between the ability of p300 and MDM2 proteins to compete for binding to the same N-terminal BOX-I domain of p53 has the potential to modulate the specific activity of p53 as a tumour suppressor.
Post-translational modification of the N-terminal BOX-I domain of p53 via kinase phosphorylation provides a potential mechanism for regulating the binding of MDM2 and p300 proteins and therefore influences the specific activity of p53 as a tumour suppressor. The role of phosphorylation in modulating p53 function becomes apparent during senescence, quiescence or after exposure to DNA damaging agents, where steady-state phosphorylation of p53 increases at Ser15 (Webley et al., 2000). Phosphorylation at Ser15 increases binding to CBP (Lambert et al., 1998) and p300 (Dumaz and Meek, 1999), and simultaneously decreases binding to MDM2 (Shieh et al., 1997), highlighting the physiological importance of this phosphorylation event in yielding a transcriptionally competent form of p53. One other newly identified phospho-acceptor site at Thr18 in the BOX-I domain is modified in human breast cancers (Craig et al., 1999b), induced during senescence (Webley et al., 2000) or transiently following ionizing radiation (Sakaguchi et al., 2000). The second newly-identified phospho-acceptor site at Ser20 is modified constitutively in normal human fibroblasts and oxidant stresses, including radiolabelling with [32P]orthophosphate, can result in de-phosphorylation at this site (Craig et al., 1999a). In addition, an intact Ser20 residue is required for effective p53 activity (Unger et al., 1999) and the ionizing irradiation-induced form of p53 protein is phosphorylated at Ser20 by a Chk2-dependent pathway (Shieh et al., 2000). The overlap of the MDM2-binding site and the p300-binding site within the BOX-I domain of p53 complicates our understanding of the role of the Thr18 and Ser20 phosphorylation sites in p53 function. It was therefore the primary purpose of this study to determine the role for the Thr18 and Ser20 phosphorylation sites in regulating p53 function.
RESULTS AND DISCUSSION
Full-length p300 binds preferentially to small phospho-peptides derived from the BOX-I domain of p53
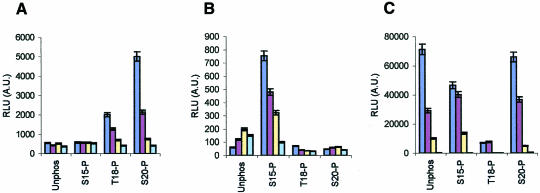
The BOX-I domain is subject to two key protein–protein interactions with cellular components: MDM2 and p300. There are unresolved data demonstrating that either Thr18 and/or Ser20 phosphorylation events have no effect on p53 activity (Ashcroft et al., 1999) or that Ser20 phosphorylation is required for p53 activity (Unger et al., 1999). We therefore wished to determine whether covalent modification at the phospho-acceptor sites (Thr18 or Ser20) had the most striking effect on MDM2 binding to p53 and/or p300 binding to p53. Full-length MDM2 and p300 proteins were produced and used in an ELISA-based peptide binding assay for quantitating the specific activity of MDM2 and p300 proteins. A titration of the peptide phosphorylated at Ser20 did not inhibit p300 interaction with the BOX-I domain, but rather promoted stabilization of the p300–peptide complex, which is in contrast with the reduced ability of p300 protein to bind to the unphosphorylated BOX-I domain peptides (Figure 1A). The Thr18 phospho-peptide similarly displayed a significant binding affinity for p300 protein, but not with the same potential observed with the Ser20 phospho-peptide (Figure 1A).
Fig. 1. Phosphorylation stabilizes p300-p53 BOX-I peptide complexes. The binding of: (A) full-length p300 protein, (B) truncated p300 (1135–2414) and (C) full-length MDM2 protein, to the indicated biotinylated-peptides was analysed by ELISA as described in the Methods. The amount of p300 or MDM2 protein bound are represented as relative light units (RLUs). The amount of biotinylated peptide titrated onto streptavidin-coated ELISA surfaces is indicated in this Figure, and the other Figures where ELISA is used, as follows: dark blue bars, 1 ng; brown bars, 0.1 ng; yellow bars, 0.01 ng and light blue bars, 0 ng.
Surprisingly, full-length p300 protein exhibited a relatively low affinity for the Ser15 phospho-peptide (Figure 1A) despite previous observations demonstrating that phosphorylation of p53 by DNA-PK increased acetylation by CBP in vitro (Lambert et al., 1998). These differences may simply reflect that _K_d alterations of full-length p300 towards small phospho-peptides and full-length phosphorylated p53 protein, since at higher concentrations of full-length p300 protein in the ELISA, Ser15 phospho-peptide binding was observed (data not shown). Additionally, an N-terminal deletion of p300 protein to produce the variant p300 (1135–2414) stabilized the binding of the protein to the Ser15 phospho-peptides (Figure 1B). More strikingly, the deleted variant of p300 protein abrogates its ability to bind to the Ser20 and Thr18 phospho-peptides (Figure 1B), suggesting that the N-terminal domain of p300 contains a regulatory motif that is essential for stabilizing the binding of p300 to the Ser20 and Thr18 phosphorylated BOX-I region. Under conditions where neither full-length p300 or p300 (1135–2414) could bind the unphosphorylated BOX-I domain peptide, full-length MDM2 protein bound with highest affinity to the unphosphorylated BOX-I domain peptide (Figure 1C). As reported previously (Craig et al., 1999b; Sakaguchi et al., 2000), the most potent inhibitor of MDM2 binding was the Thr18 phospho-substitution, while the Ser15 or Ser20 phospho-substituted peptides displayed the most ineffective inhibition of MDM2 protein binding (Figure 1C). MDM2 and p300 proteins display similar affinities for the Ser20 phospho-substituted peptide in a direct peptide-competition binding assay when equivalent amounts of protein are titrated (see supplementary Figure S1, available at EMBO reports Online). This suggests that Ser20 or Thr18 phosphorylation may serve as a regulatory switch for p53 discrimination between p300 and MDM2 binding.
BOX-I domain phospho-mimetic peptides selectively inhibit endogenous p53-dependent transcription in vivo
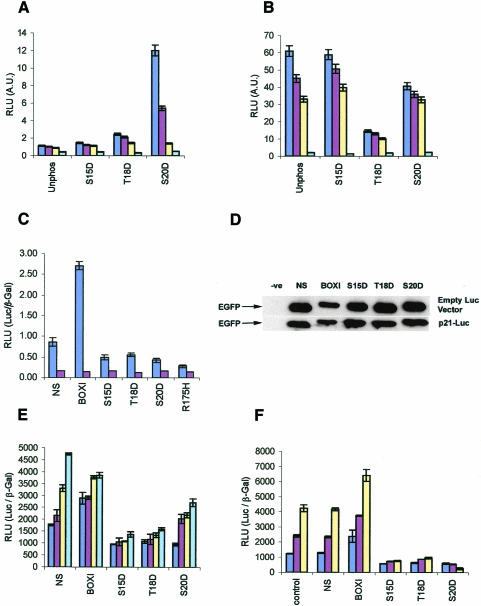
The biochemical data now suggest that the primary role of phosphorylation of p53 at Ser20 is to stabilize the p300–p53 complex (Figure 1), rather than inhibit MDM2 protein binding. As site-directed mutagenesis of p53 has often failed to support a clear role for Ser20, Thr18 or even Ser15 phosphorylation in promoting p53 activity (Ashcroft et al., 1999), we took an alternative approach to determine whether this cluster of phosphorylation events affects p300 or MDM2 protein function in vivo by developing BOX-I phospho-peptide mimetics for intracellular expression. In order to produce a phospho-peptide mimetic for in vivo use, phosphorylation was mimicked by the use of aspartate-substituted peptides at the Ser15, Thr18 or Ser20 residues. Binding of p300 to the BOX-I domain was stabilized by the Asp20 or Asp18 substitutions (Figure 2A), while MDM2 protein was most inhibited by the Asp18 substitution mutant (Figure 2B). The specific activity of full-length p300 protein (in RLUs) in binding to phospho-peptides or aspartate-substituted peptides can be compared directly (Figure 1A versus Figure 2A) and it is clear that the aspartate substitution does not fully compensate for the phosphate moiety. Nevertheless, these data indicate that a single point mutation in the BOX-I motif can convert the domain into a p300-binding ligand, highlighting the suitability of the use of aspartate mutants in vivo as relatively effective phosphate mimetics.
Fig. 2. In vivo inhibition of endogenous p53-dependent transcription by phospho-peptide mimetics. (A) p300 and (B) MDM2 binding in vitro to aspartate-substituted BOX-I domain peptides. The binding of p300 protein and MDM2 protein, to biotinylated-peptides substituted with aspartate at the indicated positions was as described in Figure 1. (C). EGFP-Asp18 and Asp20 peptide fusion proteins inhibit p53-dependent transactivation in vivo. EGFP-constructs or the mutant p53HIS175 allele (100 ng) were transiently transfected with 2 µg p21-Luc or 2 µg control-Luc and 1 µg control-β-Gal-reporter into cycling A375 cells, and the cells harvested 24 h post-transfection. p53-dependent activity (RLUs) is expressed as a ratio of p21-luciferase activity (dark blue bars) or control-luciferase activity (brown bars) to the internal transfection control (β-Gal). (D). Expression levels of EGFP-peptide fusion proteins in A375 cells. Lysates from cells transfected with the indicated EGFP-peptide fusion constructs, as described in Figure 2C, were immunoblotted with antibodies to EGFP to determine the relative level of each fusion protein expressed in cells transfected with the 2 µg p21-Luc or control-Luc vectors. (E) p300 can recover p53 activity in cells co-transfected with the inhibitory EGFP-Asp18 and Asp20 peptide fusion proteins. A375 cells were co-transfected with increasing amounts of the p300 gene (dark blue bars, 0 µg; brown bars, 1 µg; yellow bars, 2 µg and light blue bars, 5 µg), and fixed levels of p21-Luc (2 µg), β-Gal-reporter (1 µg) and the EGFP-peptide fusion vectors (100 ng), the cells were then processed for analysis of p53 activity as described in the legend for Figure 2C. (F) EGFP-Asp18 and Asp20 peptide fusion proteins inhibit p53-dependent transactivation in irradiated cells. EGFP-constructs (100 ng) were transiently transfected with 2 µg p21-Luc or 2 µg control-Luc and 1 µg control-β-Gal-reporter into A375 cells that were either untreated (dark blue bars), damaged with 20 J/m2 UV-C (brown bars), or with 5 Gy ionizing radiation (yellow bars). The cells were processed for analysis of p53 activity as described in the legend for Figure 2C.
A series of phospho-peptide mimetics were then constructed by fusion with enhanced green fluorescent protein (EGFP) for intracellular synthesis to determine whether the peptides can affect transcription from a p53-dependent reporter gene. EGFP-BOX-I domain phospho-peptide mimetics were produced by incorporating a gene encoding the amino acids 11–30 of human p53, incorporating the BOX-I domain, either with no modifications (EGFP-BOX-I) or an aspartate substitution at Ser15 (EGFP-S15D), Thr18 (EGFP-T18D) or Ser20 (EGFP-S20D) fused to the C-terminus of EGFP-NS. Proliferating A375 cells were transiently co-transfected with EGFP-NS, EGFP-BOX-I, EGFP-S15D, EGFP-T18D or EGFP-S20D and control or p21-luciferase reporter constructs. Alterations in the basal p53-dependent transcription activity were quantitated 24 h post-transfection. All three of the EGFP-aspartate-substituted fusion proteins inhibited basal p53-dependent transactivation of the p21 promoter relative to controls, with the EGFP-S20D inhibiting p53 activity to the highest degree (Figure 2C). EGFP-S15D and EGFP-T18D fusion proteins both reduced significantly the basal p53-dependent transactivation of the p21 promoter (Figure 2C). As a control, the dominant-negative mutant p53 encoded by the HIS175 allele gave rise to the highest level of inhibition of p53-dependent activity (Figure 2C). The EGFP-BOX-I fusion protein yielded the expected increase in p53-dependent transactivation of the p21 promoter relative to the control (Figure 2C), consistent with previously published data showing that fusion proteins expressing the BOX-I domain sequesters MDM2 protein, thus increasing p53-dependent transcription (Bottger et al., 1997). A titration of each EGFP-peptide construct into both A375 cells and the HCT116 colorectal cancer cell line shows that distinct cell models with a wild-type p53 pathway can give rise to similar changes in the specific activity of p53 (see supplementary Figure S2A and S2B, respectively, available at EMBO reports Online).
One control performed to address the specificity of the inhibition of the EGFP-phospho-peptide mimetics was to examine cells for changes in the steady-state level of the EGFP fusion protein by immunoblotting after transfection (Figure 2D). It is possible for example, that the higher degree of inhibition by the EGFP-phospho-peptide mimetics is due to higher expression that may non-specifically affect transcription. However, the only major difference observed in the levels of the EGFP fusion proteins were the reduced levels of EGFP-BOX-I protein, presumably due to the ability of the BOX-I peptide-fusion protein to be degraded by the binding of MDM2 (Haupt et al., 1997). These data indicate that the specific activity of each EGFP-peptide fusion protein as an inhibitor or an activator of p53-dependent transcription is correctly reflected in the RLUs used to quantitate p53 activity shown in Figure 2C. A second control was set up to examine whether p53-dependent activity could be restored in A375 cells containing inhibitory amounts of the EGFP-phospho-peptide mimetics by co-transfection with increasing amounts of the p300 gene. A dose-dependent increase in p53 activity from the p21 promoter is observed in cells co-transfected with the p300 gene, with the most striking recovery observed using the most potent p53-inhibitor encoded by the EGFP-S20D construct (Figure 2E).
The potency of the EGFP-T18D and EGFP-S20D fusion proteins in blocking basal p53-dependent transactivation from the p21 promoter in cycling cells was a promising observation and it was therefore important to determine if the peptide-fusion proteins would be as effective in blocking p53-dependent transactivation in an irradiated cell. Cells were transiently co-transfected with EGFP-NS, EGFP-BOX-I, EGFP-S15D, EGFP-T18D or EGFP-S20D and p21-luciferase reporter constructs, followed by treatment with ionizing radiation (5 Gy) or UV-C (20 J/m2). Two hours or 5 h after treatment with ionizing radiation or UV-C, respectively, there was an increase in p21-luciferase reporter activity, indicating that this transient transfection assay can detect radiation-dependent activation of p53 (Figure 2F). p53-dependent gene expression in the irradiated cells was most potently inhibited by the EGFP-S20D construct titration (Figure 2F), while the EGFP-BOX-I peptide stimulated p53 activity in the irradiated cell.
BOX-I domain phospho-mimetic peptides selectively inhibit ectopically-expressed p53-dependent transcription in vivo
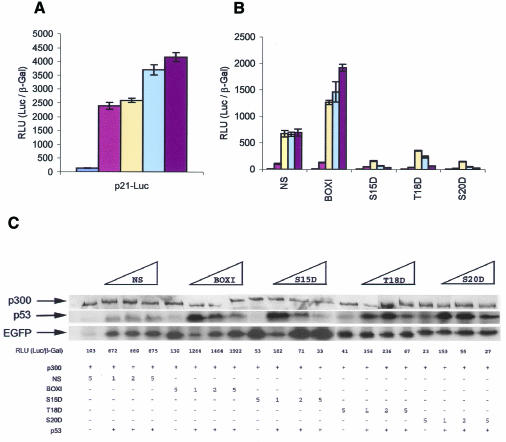
Another model used to examine whether the phospho-peptide mimetic peptides inhibit p53 activity involved the use of the p53-null cell line, Saos-2, co-transfected with p300, p53, and the indicated reporter or expression vectors (Figure 3). Transient co-transfection of increasing amounts of p300 with p53 into Saos-2 cells lead to maximal stimulation of p53-dependent gene expression from the p21 promoter (Figure 3A) or the bax promoter (see supplementary Figure S3A, available at EMBO reports Online). Using the levels of the p300 and p53 genes that give rise to the highest level of transactivation, co-transfection with increasing amounts of the EGFP-S20D peptide inhibits transactivation from the p21 promoter (Figure 3B) or the bax promoter (see supplementary Figure S3B, available at EMBO reports Online). As a control, the EGFP-BOX-I peptide stimulated p53 activity under the same conditions, distinguishing the effects of the BOX-I fusion proteins from the phospho-peptide mimetics.
Fig. 3. In vivo inhibition of ectopically expressed p53 from the p21 promoter by phospho-peptide mimetic fusion proteins. (A) Stimulation of p53 activity by co-transfection with p300. Saos-2 cells were transiently co-transfected with 1 µg pCMV-p53, 2 µg p21-Luc, 1 µg pCMVβ-Gal and increasing amounts of pCMV-βp300: brown bars, 0 ng; yellow bars, 1 µg; light blue bars, 2 µg and purple bars, 5 µg. As a negative control, (dark blue bars) 5 µg pCMV-βp300, 2 µg p21-Luc and 1 µg pCMVβ-Gal were co-transfected. The cells were harvested 30 h post-transfection and the relative activity is expressed as a ratio of luciferase activity to β-Gal activity. (B) EGFP-S20D peptide inhibits p300 induction of p53-dependent gene expression. Saos-2 cells were transiently co-transfected with 1 µg pCMV-p53, 2 µg p21-Luc, 5 µg pCMVβp300, 1 µg pCMVβ-Gal and increasing amounts of EGFP-constructs as indicated (yellow bars, 1 µg; light blue bars, 2 µg and purple bars, 5 µg). To function as a critical control, 5 µg of each EGFP construct and 5 µg of pCMVβp300 were co-transfected with 2 µg p21-Luc and pCMVβGal (brown bars), and as a negative control (dark blue bars), 5 µg pCMV-βp300, 2 µg p21-Luc and 1 µg pCMVβ-Gal co-transfected. The cells were processed as described in the legend of Figure 3A. (C) Immunoblots of p53 and EGFP-fusion proteins in transfected Saos-2 cells. Lysates from transfected Saos-2 cells (as described in Figure 3B) were normalized for protein content by Bradford assay and loading for immunoblots was confirmed by Red Ponceau staining. The constructs transfected (in µg) are highlighted by the legend above the Figure (increasing amounts of EGFP fused to NS, BOX-I, S15D, T18D, and S20D BOX-I domain peptides) and are described below the Figure as ‘+’. The levels of p300, p53 and EGFP proteins are in the top, middle or bottom panels, respectively. p53-dependent activity (in RLUs) from Figure 3B is listed below the immunoblots for direct comparison of p53 activity to p53 protein and EGFP protein levels.
In Saos-2 cells co-transfected with varying combinations of DNA encoding the EGFP peptide-fusions with p53 and p300, we examined the levels of p53 protein and EGFP-peptide fusion protein by immunoblotting (Figure 3C) in comparison to p21 reporter activity. The highlights of the data are two-fold. First, the inhibitory EGFP-T18D and EGFP-S20D peptide fusion proteins were expressed at similar levels relative to the EGFP-NS control in the co-transfected Saos-2 cells (Figure 3C, bottom panel), indicating that the ability of the phospho-mimetic peptides to inhibit p53 reflect changes in the specific activity of the aspartate-substituted peptide fusion protein (Figure 3B). The EGFP-BOX-I peptide fusion protein was expressed at slightly lower levels at the lowest point in the titration (1 µg DNA; Figure 3C), consistent with its lowered expression in A375 cells (Figure 2D). However, at the higher levels of the EGFP-BOX-I construct titration (2 and 5 µg; Figure 3C) where p53 activity is stimulated (Figure 3B), the levels of EGFP fusion were similar to the EGFP-NS controls. The exception in this analysis was the expression level of the EGFP-S15D construct, where the mutation at position 15 increased expression of the fusion protein relative to all other vectors (Figure 3C). These latter data indicate that the specific activity of the EGFP-S15D peptide fusion protein as a p53-inhibitor (Figure 3B) is significantly lower than the other phospho-peptide mimetics. Additionally, these data are consistent with the reduced ability of p300 to bind to the Asp15 phospho-peptide (Figure 2A).
The second set of experiments designed to compare p53 protein levels under the varying conditions demonstrated that p53 protein levels were increased after titrations of 1 µg of the EGFP-BOX-I, EGFP-S15D, EGFP-T18D and EGFP-S20D constructs, relative to the EGFP-NS control (Figure 3C, middle panel). These data suggest that there may be some degree of inhibition of MDM2 binding and subsequent stabilization of p53 protein by all BOX-I variants. However, the induced form of p53 protein is stimulated only using the EGFP-BOX-I construct, as the aspartate-substituted fusion proteins presumably bind avidly to p300 and prevent transactivation being triggered by the induced form of p53. Thus, if p53 protein levels in EGFP-transfected cells are plotted as a function of p21-luciferase reporter activity, then the phospho-peptide mimetics prove to be even higher affinity inhibitors when the data are normalized (data not shown).
CONCLUSION
Regulation of p53-dependent transactivation occurs by competition between MDM2 or p300 binding to the BOX-I domain of p53 which may be further regulated, in turn, by phosphorylation. It was the purpose of this study to define the role of the recently identified BOX-I phosphorylation sites at Thr18 and Ser20 as predominantly positive or negative effectors of p300- or MDM2-regulation of p53. Scaffold proteins fused to small peptide regulators have been used in vivo as reagents to dissect regulatory steps in many pathways including cyclin-dependent cdk2 isoforms (Mendelsohn and Brent, 1999). We report here that scaffold proteins fused to phospho-peptide mimetics of the BOX-I domain of p53 can be used to inhibit p300-coactivation of p53-dependent transcription. These data suggest that the predominant role of the Thr18 or Ser20 phosphorylation events are to switch p53 from an MDM2 binding protein to a p300 binding protein, consistent with the cellular models showing that Thr18 or Ser20 phosphorylation events are associated with the activation of p53.
METHODS
Plasmids, antibodies and cells
EGFP-peptides were constructed by ligating double-stranded oligonucleotides encoding amino acids 11–30 of human p53 (EGFP-BOX I) and with a codon encoding an aspartate mutant at Ser15 (EGFP-S15D), Thr18 (EGFP-T18D or Ser20 (EGFP-S20D), into _Xho_I–_Xba_I digested EGFP-C3 plasmid (Clontech). An EGFP-NS control plasmid with non-specific amino acids was created by ligating _Xho_I–_Xba_I ends of EGFP-C3. All EGFP constructs were confirmed by DNA sequencing. p21-Luc, Bax-Luc and pGL3-Basic constructs were a gift from M. Oren (Weizmann Institute of Science, Israel). pCMV-βGal was a gift from M.G. Luciani (University of Dundee, UK). Full-length p300 and p300 (1135–2414) baculovirus were a gift from N.B. La Thangue [(Shikama et al., 1999) University of Glasgow, UK]. pC53–175 expression plasmid was a gift form B. Vogelstein (The Johns Hopkins Oncology Center, USA). pCMVβ-p300 was a gift from M. Giacca (ICGEB, Italy). Full-length MDM2 protein was purified as described (Burch et al., 2000). Full-length p300 and FLAG-p300 (1135–2414) were purified and quantitation determined empirically as described previously (Shikama et al., 1999).
A375 and HCT116 cells both containing a wild-type p53 pathway were maintained in DMEM and McCoy’s 5A (Gibco BRL), respectively, supplemented with 10% FBS and incubated at 37°C with an atmosphere of 10% CO2. For transient transfections, 3 × 105 cells were plated out in 6-well dishes and transfected with LipofectAMINE 2000 (Gibco BRL). The exact quantity of DNA transfected is indicated in each experiment and where necessary, carrier DNA was incorporated to keep the same quantity of DNA consistent in each transfection. To monitor transfection efficiency, pCMV-βGal was included in each transfection. Unless otherwise stated, cells were harvested 24 h post-transfection and lysed in Reporter Lysis Buffer and the corresponding luciferase and β-Gal assay carried out according to the manufacturer’s protocol (Promega).
Immunochemical assays
Biotinylated peptides were immobilized to streptavidin-coated 96-well plates (Dynex Microlite 2) with a titration of 0–1 ng/well as indicated previously (Craig et al., 1999b). Essentially, non-reactive sites were blocked in 5% Milk/50 mM NaF/5 mM βPG in PBST20 (0.1% v/v) and emperically-determined levels of p300, FLAG-p300 (1135–2414) or full-length MDM2 proteins were incubated for 1 h at 4°C. Plates were rigorously washed three times with PBST20 (0.1% v/v) to reduce the non-specific binding. Primary antibodies anti-p300 (N-15) (Santa Cruz Biotechnology Inc.), anti-FLAG (Sigma) and anti-MDM2 (2A10) were used and the appropriate secondary antibody cross-linked to HRP. The signal detected by enhanced chemiluminescence was developed using Fluoroskan Ascent FL. Immunoblots for p53 were performed using DO-1 as described previously (Craig et al., 1999a) and antibodies to EGFP (Anti-EGFP; Clontech) were used according to the manufacturer’s suggestion.
Supplementary data
Supplementary data for this paper are available at EMBO reports Online.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Marcus Achison (University of Dundee, UK) for critical reading of the manuscript. D.D. is the recipient of a BBSRC PhD studentship. T.R.H. is supported by a Programme Grant from the Cancer Research Campaign, a Career Establishment Grant from the UK Medical Research Council, and the Association for International Cancer Research.
REFERENCES
- Ashcroft M., Kubbutat, M.H. and Vousden, K.H. (1999) Regulation of p53 function and stability by phosphorylation. Mol. Cell. Biol., 19, 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottger A., Bottger, V., Sparks, A., Liu, W.L., Howard, S.F. and Lane, D.P. (1997) Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr. Biol., 7, 860–869. [DOI] [PubMed] [Google Scholar]
- Burch L.R., Midgley, C.A., Currie, R.A., Lane, D.P. and Hupp, T.R. (2000) Mdm2 binding to a conformationally sensitive domain on p53 can be modulated by RNA. FEBS Lett., 472, 93–98. [DOI] [PubMed] [Google Scholar]
- Craig A.L., Blaydes, J.P., Burch, L.R., Thompson, A.M. and Hupp, T.R. (1999a) Dephosphorylation of p53 at Ser20 after cellular exposure to low levels of non-ionizing radiation. Oncogene, 18, 6305–6312. [DOI] [PubMed] [Google Scholar]
- Craig A.L., Burch, L., Vojtesek, B., Mikutowska, J., Thompson, A. and Hupp, T.R. (1999b) Novel phosphorylation sites of human tumour suppressor protein p53 at Ser20 and Thr18 that disrupt the binding of mdm2 (mouse double minute 2) protein are modified in human cancers. Biochem. J., 342, 133–141. [PMC free article] [PubMed] [Google Scholar]
- Dumaz N. and Meek, D.W. (1999) Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J., 18, 7002–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman R.H. and Smolik, S. (2000) CBP/p300 in cell growth, transformation, and development. Genes Dev., 14, 1553–1577. [PubMed] [Google Scholar]
- Grossman S.R., Perez, M., Kung, A.L., Joseph, M., Mansur, C., Xiao, Z.X., Kumar, S., Howley, P.M. and Livingston, D.M. (1998) p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol. Cell., 2, 405–415. [DOI] [PubMed] [Google Scholar]
- Haupt Y., Maya, R., Kazaz, A. and Oren, M. (1997) Mdm2 promotes the rapid degradation of p53. Nature, 387, 296–299. [DOI] [PubMed] [Google Scholar]
- Lambert P.F., Kashanchi, F., Radonovich, M.F., Shiekhattar, R. and Brady, J.N. (1998) Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem., 273, 33048–33053. [DOI] [PubMed] [Google Scholar]
- Lohrum M.A. and Vousden, K.H. (2000) Regulation and function of the p53-related proteins: same family, different rules. Trends Cell Biol., 10, 197–202. [DOI] [PubMed] [Google Scholar]
- Mendelsohn A.R. and Brent, R. (1999) Protein interaction methods–toward an endgame. Science, 284, 1948–1950. [DOI] [PubMed] [Google Scholar]
- Oren M. (1999) Regulation of the p53 tumor suppressor protein. J. Biol. Chem., 274, 36031–36034. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K., Saito, S., Higashimoto, Y., Roy, S., Anderson, C.W. and Appella, E. (2000) Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J. Biol. Chem., 275, 9278–9283. [DOI] [PubMed] [Google Scholar]
- Shieh S.Y., Ikeda, M., Taya, Y. and Prives, C. (1997) DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell, 91, 325–334. [DOI] [PubMed] [Google Scholar]
- Shieh S.Y., Ahn, J., Tamai, K., Taya, Y. and Prives, C. (2000) The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev., 14, 289–300. [PMC free article] [PubMed] [Google Scholar]
- Shikama N., Lee, C.W., France, S., Delavaine, L., Lyon, J., Krstic-Demonacos, M. and La Thangue, N.B. (1999) A novel cofactor for p300 that regulates the p53 response. Mol. Cell., 4, 365–376. [DOI] [PubMed] [Google Scholar]
- Unger T., Juven-Gershon, T., Moallem, E., Berger, M., Vogt Sionov, R., Lozano, G., Oren, M. and Haupt, Y. (1999) Critical role for ser20 of human p53 in the negative regulation of p53 by mdm2. EMBO J., 18, 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webley K., Bond, J.A., Jones, C.J., Blaydes, J.P., Craig, A., Hupp, T. and Wynford-Thomas, D. (2000) Posttranslational modifications of p53 in replicative senescence overlapping but distinct from those induced by DNA damage. Mol. Cell. Biol., 20, 2803–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3