Dynamin Isoform-specific Interaction with the Shank/ProSAP Scaffolding Proteins of the Postsynaptic Density and Actin Cytoskeleton (original) (raw)
. Author manuscript; available in PMC: 2009 Jul 24.
Published in final edited form as: J Biol Chem. 2001 Oct 2;276(51):48458–48465. doi: 10.1074/jbc.M104927200
Abstract
Dynamin is a GTPase involved in endocytosis and other aspects of membrane trafficking. A critical function in the presynaptic compartment attributed to the brain-specific dynamin isoform, dynamin-1, is in synaptic vesicle recycling. We report that dynamin-2 specifically interacts with members of the Shank/ProSAP family of postsynaptic density scaffolding proteins and present evidence that dynamin-2 is specifically associated with the postsynaptic density. These data are consistent with a role for this otherwise broadly distributed form of dynamin in glutamate receptor down-regulation and other aspects of postsynaptic membrane turnover.
Dynamin is a 100-kDa GTPase (1, 2) that controls a variety of vesicular budding events including synaptic vesicle recycling, receptor-mediated endocytosis, caveolae internalization, phagocytosis, and secretory vesicle budding from the _trans_-Golgi network (3–9). It forms long spiral polymers around the necks of coated pits (10) and on lipid tubules (11), suggesting that the protein may directly function in membrane scission. Alternatively, dynamin has also been postulated to act as a GTPase switch by recruiting other endocytic factors to the neck and then activating them to sever the coated vesicle (12).
Dynamin contains an amino-terminal GTPase domain, followed by a central coiled-coil assembly domain (13), a pleckstrin homology domain, which binds to phosphoinositides and the βγ subunits of heterotrimeric GTPases (14, 15), and a carboxyl-terminal coiled-coil region (also called the assembly or GTPase effector domain) that is involved in self-association (13, 16, 17). At the extreme carboxyl terminus is a basic, proline-rich domain to which a number of Src homology 3 (SH3)1 domain-containing proteins, acidic phospholipids, and microtubules have been shown to bind (18–20).
Considerable insight into dynamin function at the synapse has come from genetic and morphological studies on the temperature-sensitive mutants of shibire, the dynamin ortholog in Drosophila (21, 22). Single point mutations in the GTPase domain of shibire cause paralysis at elevated temperatures, and ultrastructural analysis of nerve terminals under these conditions has revealed a depletion of synaptic vesicles, along with an accumulation of collared pits (23, 24).
In mammals, three closely related dynamin genes are expressed in a tissue-specific manner. Dynamin-1 is almost exclusively expressed in neurons (25). Dynamin-2 is found in the brain but is also widely expressed among other tissues (26–28). Dynamin-3 was initially identified in testis (29) but is also found in brain, lung, and heart (30). Differences in the subcellular distribution of the dynamin gene products and their alternative splice forms have been reported (30). Because of its restriction to neurons, dynamin-1 has been assumed to be the synaptic isoform. The function of dynamin-2 is less well understood, and a role in neurons has not been identified. Overexpression of a dominant inhibitory mutant form of dynamin-2 in cultured hippocampal neurons was recently reported to inhibit glutamate-induced down-regulation of _α_-amino-3-5-methyl-isoxazole-4-propionic acid receptors (31). This result is of considerable current interest in view of evidence that up- and down-regulation of glutamate receptors plays a role in long term potentiation and depression (32, 33). Whether dynamin-2, in particular, functions postsynaptically is uncertain from these experiments, because of the interchangeability of dynamin isoforms when overexpressed (Ref. 13; also see “Results”).
To learn more about the specific functions of dynamin-2, we have used the yeast two-hybrid assay to identify dynamin-2-specific interacting proteins. We report here that dynamin-2 interacts with at least two members of the Shank/ProSAP family, which have recently been described as components of the postsynaptic density (PSD) (34, 35) Dynamin-2 exhibits a synaptic distribution in cultured hippocampal neurons and is specifically enriched relative to dynamin-1 in the postsynaptic density. Our findings indicate a close association between the endocytic machinery and the PSD and provide new insight into the dynamics of this structure.
Experimental Procedures
Yeast Two-hybrid Screen and Shank Cloning
The entire dynamin-2bb cDNA was subcloned into the JK202 bait vector and used to screen an adult human brain cDNA library (Invitrogen, Inc.) for interactors in a LexA-based yeast two-hybrid assay utilizing the yeast strain, EGY191 (36). Potential positives, as deemed by their ability to both grow in the absence of leucine and activate _β_-galactosidase expression, were further analyzed using BLAST and SMART searches. To qualitatively evaluate the _β_-galactosidase activities, highly sensitive filter assays were carried out as described in the Matchmaker LexA two-hybrid system user manual (CLONTECH Laboratories, Inc.). For the domain mapping studies and interaction specificity analyses, carboxyl-terminal deletion mutants of dynamin-2 in JK202 were generated using PCR. The dynamin-1 two-hybrid bait construct in JK202 has been described previously (13), and the light intermediate chain-2 (LIC2) bait construct was generously provided by Dr. Jorge Garces.
The Shank1 cDNA was cloned from a rat brain cDNA library using reverse transcriptase-PCR with gene specific primers and LA Taq polymerase (Takara Shuzo Ltd.). The library was generated using the Marathon cDNA amplification kit (CLONTECH Laboratories, Inc.) and kindly provided by Dr. Atsushi Mikami. The various PCR fragments were initially cloned into pCR2.1 using the TOPO-TA cloning system (Invitrogen, Inc.) and subsequently assembled into a full-length cDNA in the mammalian expression vector, pcDNA3.1+ vector (Invitrogen, Inc.). Three consecutive Myc tags, which were required for the detection of the epitope in overexpressed cells, were added to the amino terminus of Shank1 by PCR. All PCR-generated constructs were sequenced for accuracy.
Antibodies
A Shank1 antibody was raised in rabbits against a His-tagged fusion protein consisting of the Shank1 region of a.a. 1792–2001. Carboxyl-terminal anti-dynamin-1 and -2 isoform-specific antibodies have been described previously (37) as have a dynamin-2-specific antibody prepared against an internal sequence (Oncogene; Ref. 38) and antibodies against cortactin-binding protein, a generous gift from Dr. Tom Parsons (39) and the Myc epitope (40, 41). The other antibodies used in this study and their sources are anti-cortactin (p80/85 Src60 substrate, Santa Cruz Biotechnology, Inc.), anti-synaptophysin (Roche Molecular Biochemicals), anti-PSD95 (Calbiochem), anti-_N_-methyl-d-aspartic acid receptor 1 (PharMingen), anti-calmodulin kinase II (Calbiochem), and anti-HA (Covance). All of the secondary antibodies used were obtained from Jackson ImmunoResearch, Inc.
Immunoprecipitation, GST Pull-downs, and Blot Overlays
For the immunoprecipitation assays in COS-7 cells, either dynamin-1 or -2 was co-transfected with Myc-tagged Shank2 or Shank1 using LipofectAMINE as suggested by the manufacturer (Invitrogen, Inc.), and the lysates were prepared in cold RIPA buffer as described previously (13). Immunoprecipitation assays were carried out with either the dynamin-1 or -2 isoform-specific antibody on protein A beads for 4 h at 4 °C with end-over-end rotation, after which the beads were extensively washed with cold RIPA buffer. The samples were then resolved by SDS-polyacrylamide gel electrophoresis (7.5%), transferred to polyvinylidene difluoride, and blotted with Myc or the dynamin isoform-specific antibodies. For the immunoprecipitation assays from rat brain tissue, the cytosolic extract was prepared as described previously (37), and the assays were carried out in the same manner as described for COS-7 cells (see above).
The GST pull-down assays were performed essentially as described (37). The bacterially expressed and purified recombinant GST fusion protein of a Shank2 fragment (a.a. 888–1180) that corresponds to the dynamin-2 binding region in Shank1 or the GST protein alone was immobilized on Sepharose 4B-glutathione beads (Amersham Pharmacia Biotech) in binding buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet 40, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 1 _μ_g/ml aprotinin, 1 _μ_g/ml leupeptin) for 30 min at room temperature. The beads were then washed three times with binding buffer. Baculo-virus expressed and purified recombinant full-length HA-tagged dynamin-2 was added, and binding was allowed to occur for an additional 1 h at room temperature with end-over-end rotation, after which the beads were washed four times with cold binding buffer. Bound HA-tagged dynamin-2 was eluted twice with elution buffer (50 mm reduced glutathione, 50 mm Tris-HCl, pH 8). The eluants were combined, resolved in 9% polyacrylamide-SDS gels, and processed for Western analysis. HA-dynamin-2 was assayed by immunoblotting with the anti-HA monoclonal antibody. Purification of the HA-tagged dynamin-2 protein was as reported previously (42).
For the blot overlay assays, cytosolic extracts of COS-7 cells transiently transfected with either Myc-tagged Shank1 or Myc-tagged dynein IC2C were prepared 48 h post-transfection in ice-cold RIPA buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm EDTA, 0.1% SDS, 1% sodium deoxycholate, 1% Triton X-100, 10 _μ_g/ml each of aprotinin, leupeptin, pepstatin A, E-64, 16 _μ_g/ml benzamidine, 1 mm 4-(2-aminoethyl)-benzenesulfonyl fluoride). The extracts were spun 20 min at 13,000 rpm at 4 °C, after which the overexpressed Myc-tagged proteins were immunoprecipitated with an anti-Myc polyclonal antibody for 2 h at 4 °C followed by an incubation with protein A-Sepharose beads (Amersham Pharmacia Biotech) for 1 h at 4 °C. Immunocomplexes were washed once with 1 m NaCl and 1% Nonidet P-40, twice with RIPA and 1 m urea, and once with RIPA. The proteins were resolved on a 7.5% acrylamide/SDS gel, transferred to polyvinylidene difluoride, cut into strips, and allowed to renature in renaturation buffer (TBST with 5% nonfat dry milk) at 4 °C for 16 h. The strips were then incubated for 1 h at room temperature in overlay binding buffer (TBST, 0.1% bovine serum albumin, 2 mm MgSO4) in the presence or absence of 0.5 _μ_m purified, recombinant HA-tagged dynamin-2 followed by two washes in wash buffer (TBST, 2 mm MgSO4) and then cross-linked to the membrane with 0.2% glutaraldehyde for 15 min at room temperature. After extensively washing the strips in wash buffer, they were then incubated for an hour with anti-HA monoclonal antibody in the overlay binding buffer. Enhanced chemiluminescence was used to process all of the above immunoblotting experiments.
Hippocampal Neuron Culture and Immunocytochemistry
Rat hippocampal neuronal cultures were prepared from 18–19-day-old rat embryos as previously described (43) and processed for immunofluorescence as essentially reported earlier (44). Briefly, after 19–21 days in culture, the neurons were fixed in 4% paraformaldehyde for 20 min and then extracted with 0.05% saponin in phosphate-buffered saline for 5 min at room temperature. Incubation in anti-dynamin-2, anti-dynamin-1, or anti-synaptophysin primary antibody was overnight at 4 °C followed by incubation in either Cy3- or fluorescein isothiocyanate-conjugated species-specific secondary antibody for 1 h at room temperature. Immunofluorescence was visualized with a Zeiss Axiophot microscope.
For the co-clustering experiments, COS7 cells were plated on coverslips that had been pretreated with poly-l-lysine and transiently transfected with HA-tagged dynamin-2, PSD95, Myc-tagged Shank1a, and/or GFP-tagged GKAP1a using LipofectAMINE Plus reagent (Invitrogen, Inc.). The cDNAs for PSD95 and GKAP1a were generously provided by Dr. Craig Garner (University of Alabama, Birmingham, AL). All of the constructs were made in pcDNA 3.1+ (Invitrogen, Inc.) with the exception of GFP-GKAP1a, which was subcloned into the pEGFP-N1 vector (CLONTECH, Inc.). After 24 h post-transfection, the cells were fixed in 4% paraformaldehyde for 20 min at room temperature and then extracted with ice-cold methanol for 5 min at −20 °C. HA, Myc, and PSD primary antibodies were applied to the cells for 1 h at 37 °C, after which they were briefly washed and then incubated with Cy3-, Cy5-, or Alexa Green-conjugated, species-specific secondary antibodies for an additional 1 h at 37 °C. All of the antibodies were diluted in 1% bovine serum albumin. The images were gathered and processed with a Leica DMIRBE immunofluorescent microscope interfaced to a CCD camera using the Metamorph software.
Subcellular Fractionation of Rat Brain Tissue
Rat brain homogenate and synaptosomal and postsynaptic density fractions were obtained by differential centrifugation following previously published protocol (45, 46) with the exception that 25 unstripped frozen whole rat brains (Pel-Freeze) were used. Twenty micrograms of each fraction was resolved by SDS-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride, and blotted for various synaptic proteins at the following dilutions: anti-synaptophysin, 1:200; anti-dynamin-2, 1:1000; anti-dynamin-1b, 1:6000; anti-dynamin-1a, 1:5000; anti-Shank1, 1:2000; anti-PSD95, 1:200; anti-calmodulin kinase II, 1:200; and anti-NMDAR1, 1:1000. All blots were processed for imaging using enhanced chemiluminescence.
Results
Dynamin-2 Binds to a Novel Family of PSD Proteins
To identify proteins that interact with dynamin-2, we used a full-length dynamin-2 construct as bait in a yeast two-hybrid screen of a human brain cDNA library. Of ∼3 × 107 transformants that were screened, 24 grew on Leu– medium and activated _β_-galactosidase. Two of the prey sequences coded for a 188-a.a. fragment of Shank1 (also known as ProSAP2 and synamon), a member of the recently discovered Shank/ProSAP PDZ domain-containing protein family that has been reported to act as scaffolding components of the PSD (35). Shank1 was initially identified on the basis of its interaction with GKAP/SAPAP/DAP1 (34, 35, 47), which in turn interacts with the well characterized PSD component PSD95/SAP90 (35,48,49; Fig. 1a). Besides its PDZ domain, Shank1 also contains an ankyrin repeat region, an SH3 domain, a polyhistidine segment, and a sterile α motif domain (Fig. 1a). Our 188-a.a. fragment mapped to a proline-rich region (a.a. 1603–1790) located between the latter two domains (Fig. 1, a and b).
Fig. 1. Dynamin isoform-specific association with Shank/ProSAP proteins and mapping of the binding sites via the yeast two-hybrid assay.
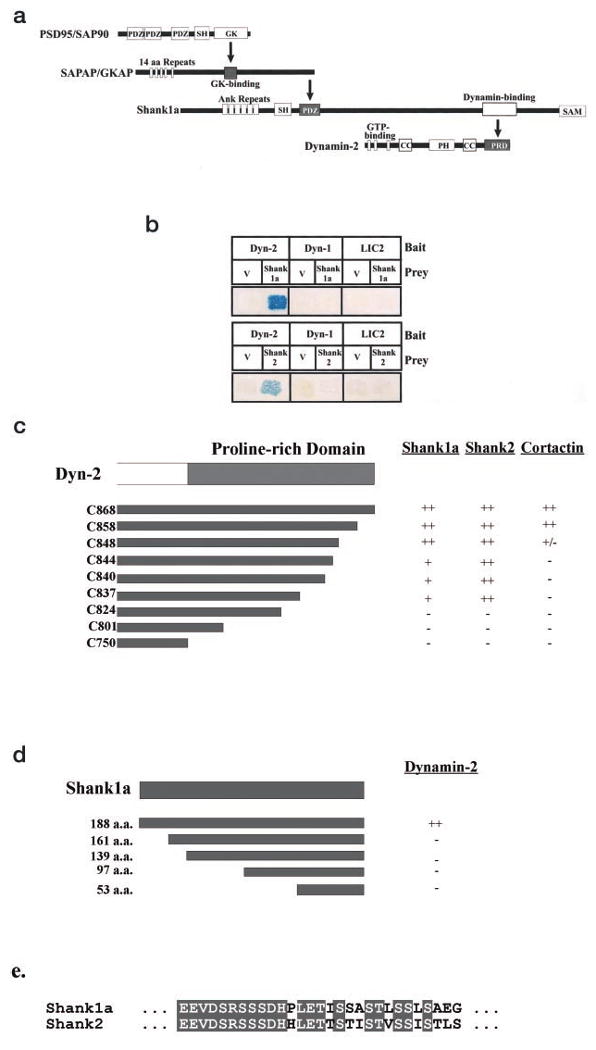
a, chain of interactions involving Shank1 and dynamin. Interaction domains identified in previous studies or in this paper are indicated by bold arrows. SH, SH3 domain; GK, guanylate kinase domain; Ank, ankyrin repeats; CC, coiled-coil domain; PRD, proline-rich domain. b, yeast two-hybrid assay for dynamin interactions. Bait constructs were full-length wild-type dynamin-2 (Dyn-2), dynamin-1 (Dyn-1), and cytoplasmic dynein light intermediate chain 2 (LIC2). Prey constructs were the initially isolated 188-a.a. Shank1 fragment, full-length Shank2, and the JG4–5 vector (V). c, mapping of the Shank1, Shank2, and cortactin binding sites within dynamin-2. Carboxyl-terminal deletions of the dynamin-2 proline-rich domain were used as bait in yeast two-hybrid assays with Shank1, Shank2, and cortactin as prey. ++, very strong interaction; +, strong interaction; +/–, weak but detectable interaction; −, undetectable interaction. d, mapping of the dynamin-2 binding sites in Shank1. Full-length and amino-terminal deletions of the original 188-a.a. Shank1 prey fragment were assayed for interaction with the dynamin-2 bait in a yeast two-hybrid assay. e, sequence comparison between Shank1 and Shank2 using the Clustal W sequence alignment method of the putative dynamin-2 binding site. Within this 29-a.a. region, 72% of the amino acids are identical as highlighted.
We also tested for an interaction between dynamin-2 and Shank2. Shank2 (50), also known as ProSAP1 (51), and cortactin-binding protein 1, or CortBP1 (39), has been shown to be present in PSDs, neuronal growth cones, and other actin-containing cytoskeletal structures (39) and exhibits 32% identity (53% similarity) to Shank1 within the dynamin-2 binding region we identified. As shown in Fig. 1b, clear evidence for an interaction with full-length Shank2 was observed. We also tested for dynamin isoform specificity in a directed two-hybrid assay. No interaction could be detected with dynamin-1 as bait (Fig. 1b). Taken together, these results reveal the identification of a novel class of isoform-specific dynamin-binding partners.
Because the dynamin-1 and -2 sequences differ most prominently within the carboxyl-terminal proline-rich domain, we reasoned that the binding site for the Shank/ProSAP proteins might lie within this region. To test this possibility, we used carboxyl-terminal dynamin-2 deletion mutants in a directed two-hybrid assay. Binding of both Shank1 and Shank2 persisted in constructs lacking up to 31 a.a. from the carboxyl terminus (Dyn-2 C837_–_C868; Fig. 1c). Removal of an additional 10 amino acids (Dyn-2 C824), however, completely abolished the interaction (Fig. 1c). At least two Shank family members (Shank2/CortBP1 and Shank3a) have been found to interact with the actin-binding protein, cortactin, which has recently also been reported to interact with dynamin-2 (35, 39, 52). We found that cortactin interacts with the carboxyl-terminal portion of dynamin-2 in our two-hybrid assay, consistent with previous results (52). However, binding was completely abolished in three dynamin-2 constructs that still exhibited a strong interaction with Shank1 and Shank 2 (Dyn-2 C837_–_C844; Fig. 1c).
Using the yeast two-hybrid assay, we also defined the region within the original Shank1 prey fragment responsible for dynamin-2 binding. Binding was abolished upon deletion of the amino-terminal 29 a.a. from the Shank1 188-a.a. prey fragment (Fig. 1c). This region is among the more highly conserved parts of the prey fragment (72% identity; 86% similar; Fig. 1c), consistent with a conserved functional role.
Biochemical Evidence for a Dynamin-2 Interaction with the Shank Proteins
To test further for an association between dynamin-2 and members of the Shank/ProSAP family, we overexpressed either Myc-tagged dynamin-1 or dynamin-2 with Shank2 in COS-7 cells. Full-length untagged Shank2 co-immunoprecipitated with either Myc-dynamin-1 or -2 (Fig. 2a). This result supports the two-hybrid data, but the co-immunoprecipitation of both dynamin isoforms with Shank2 appears to contradict the dynamin isoform specificity revealed by the two-hybrid analysis. However, we believe that co-immunoprecipitation of overexpressed dynamin-1 with Shank2 occurs through an interaction between dynamin-1 and dynamin-2, which is the predominant isoform expressed in COS7 and other non-neuronal cultured cell lines (data not shown). Direct evidence for the free interaction between dynamin isoforms can be observed in Fig. 2c (see below) and in our previous results (13), and we note that dynamin-2 co-immunoprecipitates Shank2 more efficiently than does dynamin-1. Both dynamin isoforms co-immunoprecipitated Myc-tagged Shank1 (Fig. 2b), and, again, overexpressed dynamin-2 performed better than dynamin-1.
Fig. 2. Co-immunoprecipitation of dynamin and Shank in COS7 cells.
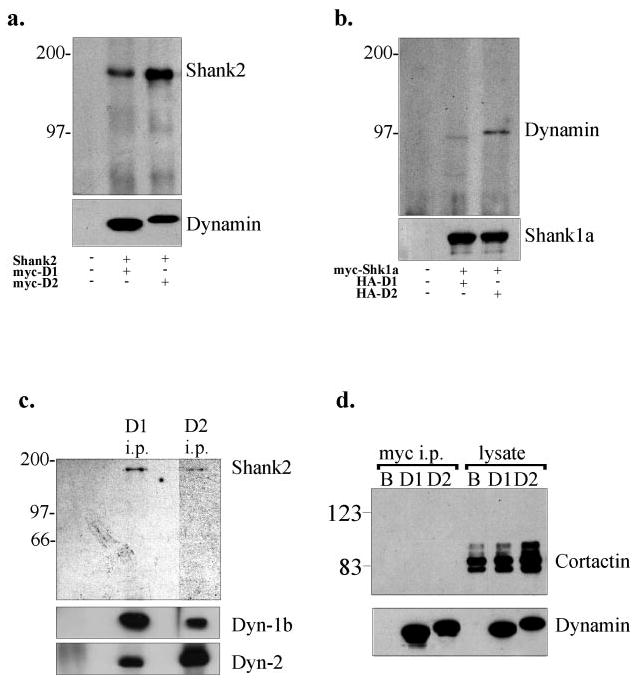
a, co-expression of full-length Shank2 with Myc-dynamin-1 or -2 in COS-7 cells. The dynamin isoforms were immunoprecipitated with anti-Myc, and the immunoprecipitates were immunoblotted with anti-Shank2 and anti-Myc. b, co-expression of Myc-tagged Shank1 with HA-tagged dynamin-1 or -2 in COS 7 cells. Shank1 was immunoprecipitated with anti-Myc, and the immunoprecipitates were blotted with anti-Myc and anti-HA antibodies. c, immunoprecipitation (i.p.) of dynamin-1 and -2 from rat brain cytosol using isoform-specific antibodies. The immunoprecipitates were blotted using anti-Shank2, anti-dynamin-1, and anti-dynamin-2 antibodies as indicated at right. d, anti-Myc immunoprecipitation of Myc-dynamin-1 and -2 singly expressed in COS-7 cells. The immunoprecipitates were detected using anti-Myc or anti-cortactin and showed no evidence for co-precipitation of cortactin with dynamin. B, beads alone control; D1, dynamin-1; D2, dynamin-2.
We also carried out co-immunoprecipitation assays using rat brain cytosol. Only Shank2 could be examined by this assay because of the insolubility of Shank1. Clear co-immunoprecipitation of Shank2 with dynamin was observed (Fig. 2c). Shank2 again co-immunoprecipitated with both dynamin-1 and -2, in this case using the carboxyl-terminal anti-dynamin antibodies, but we also note the clear ability of the two endogenous dynamin isoforms to co-immunoprecipitate with either antibody.
To test whether cortactin may play an intermediary role in the dynamin-2 and Shank/ProSAP interaction, dynamin-2 was overexpressed in wild-type COS-7 cells, and the immunoprecipitate was tested for endogenous cortactin. As shown in Fig. 2d, no cortactin was detected in these immunoprecipitates, although a significant amount of cortactin was present in these cells (lysate lanes in Fig. 2d). These results argued further that the interaction between dynamin-2 and Shank2 or Shank1 did not involve cortactin, although other intervening proteins could not be ruled out.
To ascertain whether the interaction between dynamin-2 and the Shank family is, indeed, direct, a GST fusion protein containing the region of Shank2 corresponding to the initially identified prey fragment was tested for its ability to bind purified dynamin-2. Clear binding by the GST-Shank2 fusion protein, but not by GST alone, was observed (Fig. 3a). Blot overlays using purified dynamin-2 also revealed specific binding to electrophoretically fractionated Shank1 (Fig. 3b).
Fig. 3. Dynamin-2 interacts directly with the Shank/ProSAP family members.
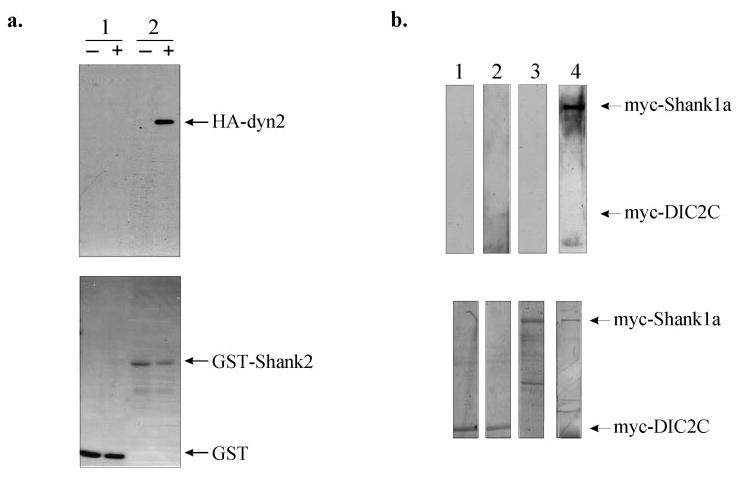
a, GST pull-down assays. Sepharose 4B beads, charged with either the GST-Shank2 fusion protein or GST alone, were incubated with purified recombinant HA-dynamin-2 and immunoblotted with an HA antibody. The upper panel shows that dynamin-2 directly interacts with GST-Shank2 (lane 2) but not with GST (lane 1), whereas the lower panel is the Coomassie-stained blot of the various purified GST proteins. −, absence of HA-dynamin-2; +, presence of HA-dynamin-2. b, blot overlay of Myc-tagged dynein intermediate chain 2C (DIC2C) or Myc-tagged Shank1 with purified recombinant HA-tagged dynamin-2. Shank1 and DIC2C were immunoprecipitated with an anti-Myc antibody and, after renaturation on the blot, overlaid with purified HA-dynamin-2 protein, which was detected with an anti-HA antibody. Upper panel, lanes 1 and 3, Myc-DIC2C and Myc-Shank1, respectively, in the absence of HA-dynamin-2; lanes 2 and 4, Myc-DIC2C and Myc-Shank1, respectively, overlaid with HA-dynamin-2. Lower panel, Coomassie-stained blots of the immunoprecipitated Myc-tagged proteins showing the location of each protein on the blot.
Association of Dynamin-2 with the Postsynaptic Density
Together our data indicate that dynamin-2 specifically associates with a family of known synaptic proteins. However, whether dynamin-2 is present in neurons and what its subcellular distribution may be are unknown. Immunofluorescence microscopy of cultured hippocampal neurons revealed punctate staining along dendrites with both the dynamin-2 and dynamin-1b carboxyl-terminal isoform-specific antibodies (Fig. 4). The dynamin-2 staining exhibited clear co-localization with the synaptic marker synaptophysin (Fig. 4).
Fig. 4. Immunofluorescence microscopy of dynamin isoforms in hippocampal neurons.
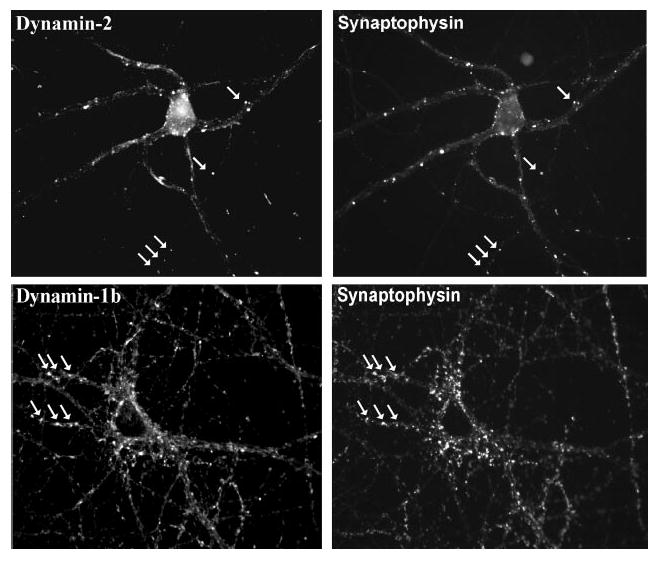
Rat hippocampal neurons (19 days in vitro) were double-labeled with anti-dynamin-1 or -2 antibodies versus anti-synaptophysin. Dynamin-2 exhibited punctate staining, which clearly overlapped with a subset of synaptophysin-positive spots (arrows). As a control, dynamin-1 also showed apparent co-localization with synaptophysin.
To examine whether dynamin-2 is present in PSDs, we tested a series of markers for biochemical co-fractionation with these structures. Both dynamin-1 and -2 isoforms were found in the synaptosomal fraction. However, dynamin-2 was clearly found in the purified PSD fraction, whereas dynamin-1 was present at low to undetectable amounts (Fig. 5). The results for dynamin-2 were confirmed using an independent isoform-specific antibody made against an internal unique dynamin-2 sequence (Ref. 38 and data not shown).
Fig. 5. Specific association of dynamin-2 with postsynaptic densities.
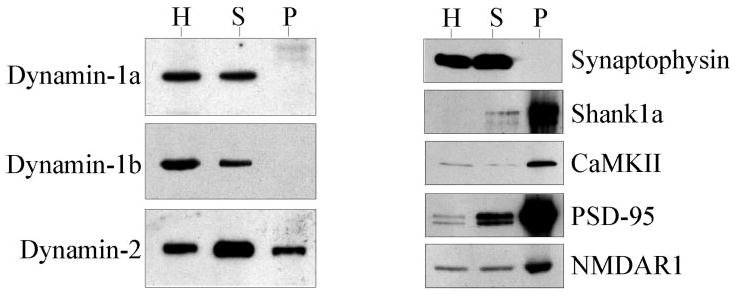
Adult rat brain homogenate (H), synaptosomal (S), and postsynaptic density (P) fractions were immunoblotted and probed with anti-dynamin-1a, -1b, and -2, anti-synaptophysin (a presynaptic marker), and anti-Shank1, anti-calmodulin kinase II, anti-PSD-95, and anti-NMDAR1 (PSD markers).
Shank1a/ProSAP2 has been shown to co-cluster with PSD95, Shaker K+ channels, and _N_-methyl-d-aspartic acid (NMDA) receptors in COS cells via its interaction with GKAP1a/SAPAP (Fig. 1a and Refs. 35, 53, and 54). To determine whether dynamin-2 might also be recruited to these clusters, we co-expressed it either with GKAP1a/SAPAP or with both GKAP1a/SAPAP and Shank1a/ProSAP in COS7 cells. When individually expressed, dynamin-2, GKAP1a/SAPAP, and Shank1a/ProSAP2 exhibited a diffuse or dispersed punctate staining pattern (data not shown). GKAP1a/SAPAP (Fig. 6) or Shank1a/ProSAP2 (data not shown) alone had no effect on dynamin-2. However, triple expression of dynamin-2 with both GKAP1a/SAPAP and Shank1a/ProSAP2 resulted in significant co-localization of the three proteins into clusters that were reminiscent of those formed by overexpressing PSD95 and Shaker K+ channels in COS cells (Fig. 6 and Ref. 53).
Fig. 6. Co-expression of PSD proteins.
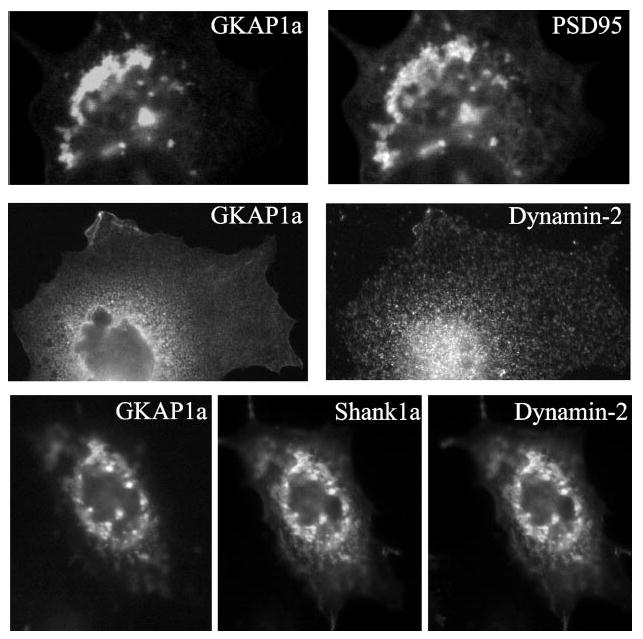
When dynamin-2 was co-expressed with two components of the postsynaptic density, GKAP1a and Shank1a/ProSAP, it co-localized into clusters that were mediated by its interaction with Shank1a/ProSAP.
Discussion
In this paper, we identify a novel interaction between dynamin-2 and the Shank/ProSAP family of PSD proteins. This is a new class of protein-protein interactions specific to dynamin-2 and provides strong support for a function for dynamin-2, in particular, in postsynaptic regulation.
The interaction with members of the Shank/ProSAP family contrasts with almost all reported dynamin interactions in not involving an SH3 domain. The Shank/ProSAP binding region identified in the current study does include a polyproline helix I sequence (55), which binds to various SH3 domains (37, 56, 57), and is one of three closely spaced sites implicated in cortactin binding (52). However, our current evidence demonstrates that the interaction between dynamin-2 and the Shank/ProSAP proteins is direct, as revealed using both blot overlay and GST pull-down assays. We also note that cortactin is absent from yeast and therefore cannot be responsible for mediating the two-hybrid interaction of Shank1 or Shank2 with dynamin-2 (Fig. 1). Furthermore, cortactin failed to co-immunoprecipitate with dynamin-2 in our hands (Fig. 2d). This result may reflect differences in the affinity of cortactin for the various alternatively spliced forms of dynamin-2 as alluded to in an earlier study (52). Thus, together our data strongly support a Shank/ProSAP interaction with dynamin-2. Whether cortactin participates in the formation of a common complex or, as suggested here, it represents an alternative binding partner for dynamin-2 remains to be further explored.
That the interaction between dynamin-2 and Shank/ProSAP is physiologically important is supported by co-fractionation of dynamin-2 with the PSD, a site to which Shank1 localizes almost exclusively, where it is anchored to GKAP/SAPAP and its interacting partner PSD-95 (49, 54, 58). This is a novel site of localization for dynamin and for dynamin-2 in particular. Dynamin-2 is widespread among vertebrate tissue and cell types. Within non-neuronal cells, it co-localizes to punctate, clathrin-containing structures at the plasma membrane and Golgi apparatus (7, 59), as well as to the cortical cytoskeleton underlying membrane ruffles (52). GTPase mutant forms of dynamin-2, like the equivalent mutations in dynamin-1, dominantly inhibit receptor-mediated endocytosis (31, 37), as well as exit from the Golgi apparatus (7, 8). The interpretation of these experiments is complicated by the ability of different dynamin isoforms to interact with each other (Ref. 13 and Fig. 2). However, endocytosis is inhibited in non-neuronal cells in which dynamin-2 is the predominant isoform (3, 4, 60, 61), suggesting that it must be responsible for this function in these cells.
A function for dynamin-2 in neurons has not been extensively explored. Dynamin-2 was reported to be present in primary dorsal root ganglion cultures but absent from purified dorsal root ganglion neurons, as judged by PCR (30). Albeit nonquantitative, these results suggested that, despite its otherwise widespread distribution, dynamin-2 might be absent from neurons. Our biochemical and immunocytochemical analysis indicates that dynamin-2, like dynamin-1, is abundant in whole synaptosomes and in PSDs. These results support a function for dynamin-2 in CNS neurons. Dynamin-2 was not enriched in the PSDs to the extent of Shank1, PSD95, or other PSD markers. This observation suggests that only a fraction of synaptosomal dynamin-2 is associated with PSDs, although the protein is clearly enriched relative to the presynaptic marker synaptophysin and to dynamin-1. In primary hippocampal neurons dynamin-2 appeared largely restricted to the synaptic region. Together with our biochemical data, these results suggest that dynamin-2 is, indeed, active in neurons, and that it performs an isoform-specific function in the postsynaptic compartment.
Our results are consistent with a recently published report that the endocytosis of glutamate receptors in cultured hippocampal and cerebellar Purkinje neurons is a clathrin-mediated, dynamin-dependent process (31, 62, 63). Extensive inhibition of _α_-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor internalization was observed in those hippocampal neurons overexpressing dominant inhibitory GTPase mutant forms of dynamin-1 or -2 (31, 62). However, as noted above, there is ample evidence that dynamin isoforms interact with each other physically and can dominantly inhibit each other's function (Fig. 2 and Ref. 37). Thus, our data add strong new support for a specific role for the dynamin-2 isoform in particular in postsynaptic membrane internalization (62, 63), and, by extension, in long term potentiation and depression.
It has also been reported that postsynaptic application of a proline-rich dynamin-1 peptide to cerebellar Purkinje neurons resulted in the rapid attenuation of long term depression as determined by electrophysiological studies (63). However, this treatment did not totally abolish postsynaptic clathrin-mediated endocytosis, and as the authors go on to suggest, this incomplete inhibitory effect may be attributed to the submaximal effect of the dynamin-1 peptide itself in their experiments (63). The dynamin-1 peptide sequence used to block postsynaptic endocytosis overlapped and is similar to the Shank/Pro-SAP binding sequence we identify in dynamin-2, suggesting that the peptide could also affect dynamin-2 function.
The extent to which postsynaptic membranes turn over is uncertain. Limited morphological evidence does exist for membrane internalization within dentritic spines via clathrin-coated pits (64). Invaginations can be observed adjacent to the PSD (64), but it is uncertain whether internalization can occur within the boundaries of this structure. What then, may be the function of a PSD-associated dynamin pool? Shank/ProSAP proteins, as well as their interacting partners, have been proposed to serve as scaffolds for the postsynaptic signaling machinery. Perhaps dynamin-2 is sequestered by the Shank/ProSAP proteins for use under conditions of intense endocytic activity. One function for a postsynaptic dynamin-2 pool could well be in glutamate receptor down-regulation as discussed above. Rapid turnover of the spines themselves, for which there is recent evidence (65), could also require a mechanism for rapid bulk membrane remodeling, a task for which the dynamins are well suited. Perhaps this is the explanation for so-called perforated PSDs observed by electron microscopy (64). Evidence exists for other endocytic hot spots where clathrin (66, 67) and dynamin (68, 69) have been found to be concentrated, in the latter case in association with DAP160, a member of the intersectin family (70). Because some members of the Shank/ProSAP family, such as Shank2/ProSAP1, are not completely restricted to the PSD, we suggest, finally, that the interactions identified in the current study may serve a more general dynamin-sequestering role.
Acknowledgments
We thank Drs. Stephen Lambert, Michele Jacob, Kristin Harris, and Denis Dujardin for helpful advice during the course of this study, Dr. Thomas Parsons for cDNA clones encoding CortBP1/Shank2 and cortactin and for the anti-CortBP1/Shank2 antibody, Dr. Atsushi Mikami for the rat brain cDNA library, Dr. Jorge Garces for the LIC2 bait construct, and Dr. Craig Garner for the PSD95 and GKAP1a/ SAPAP cDNAs.
Footnotes
*
This work was supported by National Institutes of Health Grant GM26701 (to R. B. V.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “_advertisement_” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
1
The abbreviations used are: SH3, Src homology 3; PSD, postsynaptic density; a.a., amino acid(s); PCR, polymerase chain reaction; GST, glutathione _S_-transferase; HA, hemagglutinin.
References
- 1.Obar RA, Collins CA, Hammarback JA, Shpetner HS, Vallee RB. Nature. 1990;347:256–261. doi: 10.1038/347256a0. [DOI] [PubMed] [Google Scholar]
- 2.Shpetner HS, Vallee RB. Nature. 1992;355:733–735. doi: 10.1038/355733a0. [DOI] [PubMed] [Google Scholar]
- 3.van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herskovits JS, Burgess CC, Obar RA, Vallee RB. J Cell Biol. 1993;122:565–578. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gold ES, Underhill DM, Morrissette NS, Guo J, McNiven MA, Aderem A. J Exp Med. 1999;190:1849–1856. doi: 10.1084/jem.190.12.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henley JR, Krueger EWA, Oswalk BJ, McNiven MA. J Cell Biol. 1998;141:85–99. doi: 10.1083/jcb.141.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones SM, Howell KE, Henley JR, Cao H, McNiven MA. Science. 1998;279:573–577. doi: 10.1126/science.279.5350.573. [DOI] [PubMed] [Google Scholar]
- 8.Kreitzer G, Marmorstein A, Okamoto P, Vallee R, Rodriguez-Boulan E. Nat Cell Biol. 2000;2:125–127. doi: 10.1038/35000081. [DOI] [PubMed] [Google Scholar]
- 9.van der Bliek AM. Trends Cell Biol. 1999;9:96–102. doi: 10.1016/s0962-8924(98)01490-1. [DOI] [PubMed] [Google Scholar]
- 10.Takei K, McPherson PS, Schmid SL, De Camilli P. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- 11.Sweitzer SM, Hinshaw JE. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 12.Sever S, Muhlberg AB, Schmid SL. Nature. 1999;398:481–486. doi: 10.1038/19024. [DOI] [PubMed] [Google Scholar]
- 13.Okamoto PM, Tripet B, Litowski J, Hodges RS, Vallee RB. J Biol Chem. 1999;274:10277–10286. doi: 10.1074/jbc.274.15.10277. [DOI] [PubMed] [Google Scholar]
- 14.Lin HC, Gilman AG. J Biol Chem. 1996;271:27979–27982. doi: 10.1074/jbc.271.45.27979. [DOI] [PubMed] [Google Scholar]
- 15.Lin HC, Barylko B, Achiriloaie M, Albanesi JP. J Biol Chem. 1997;272:25999–26004. doi: 10.1074/jbc.272.41.25999. [DOI] [PubMed] [Google Scholar]
- 16.Muhlberg AB, Warnock DE, Schmid SL. EMBO J. 1997;16:6676–6683. doi: 10.1093/emboj/16.22.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smirnova E, Shurland DL, Newman-Smith ED, Pishvaee B, van der Bliek AM. J Biol Chem. 1999;274:14942–14947. doi: 10.1074/jbc.274.21.14942. [DOI] [PubMed] [Google Scholar]
- 18.Herskovits JS, Shpetner HS, Vallee RB. Proc Natl Acad Sci U S A. 1993;90:11468–11472. doi: 10.1073/pnas.90.24.11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuma PL, Stachniak MC, Collins CA. J Biol Chem. 1993;268:17240–17246. [PubMed] [Google Scholar]
- 20.Schmid SL, McNiven M, De Camilli P. Curr Opin Cell Biol. 1998;10:504–512. doi: 10.1016/s0955-0674(98)80066-5. [DOI] [PubMed] [Google Scholar]
- 21.Chen MS, Obar RA, Schroeder CC, Austin TW, Poodry CA, Wadsworth SC, Vallee RB. Nature. 1991;351:583–586. doi: 10.1038/351583a0. [DOI] [PubMed] [Google Scholar]
- 22.van der Bliek AM, Meyerowitz EM. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- 23.Koenig JH, Saito K, Ikeda K. J Cell Biol. 1983;96:1517–1522. doi: 10.1083/jcb.96.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koenig JH, Ikeda K. J Neurosci. 1989;9:3844–3860. doi: 10.1523/JNEUROSCI.09-11-03844.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakata T, Iwamoto A, Noda Y, Takemura R, Yoshikura H, Hirokawa N. Neuron. 1991;7:461–469. doi: 10.1016/0896-6273(91)90298-e. [DOI] [PubMed] [Google Scholar]
- 26.Cook TA, Urrutia R, McNiven MA. Proc Natl Acad Sci U S A. 1994;91:644–648. doi: 10.1073/pnas.91.2.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sablin EP, Kull FJ, Cooke R, Vale RD, Fletterick RJ. Nature. 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- 28.Sontag J, Fykse EM, Yshkaryov Y, Liu J, Robinson PJ, Sudhof TC. J Biol Chem. 1994;269:4547–4554. [PubMed] [Google Scholar]
- 29.Nakata T, Takemura R, Hirokawa N. J Cell Sci. 1993;105:1–5. doi: 10.1242/jcs.105.1.1. [DOI] [PubMed] [Google Scholar]
- 30.Cao H, Garcia F, McNiven MA. Mol Biol Cell. 1998;9:2595–2609. doi: 10.1091/mbc.9.9.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carroll RC, Beattie EC, Xia H, C LS, Altschuler Y, Nicoll RA, Malenka RC, von Zastrow M. Proc Natl Acad Sci U S A. 1999;96:14112–14117. doi: 10.1073/pnas.96.24.14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll RC, Lissin DV, von Zastrow M, Nicoll RA, Malenka RC. Nat Neurosci. 1999;2:454–460. doi: 10.1038/8123. [DOI] [PubMed] [Google Scholar]
- 33.Luscher C, Xia H, Beattie EC, Carroll RC, von Zastrow M, Malenka RC, Nicoll RA. Neuron. 1999;24:649–658. doi: 10.1016/s0896-6273(00)81119-8. [DOI] [PubMed] [Google Scholar]
- 34.Boeckers TM, Winter C, Smalla KH, Kreutz MR, Bockmann J, Seidenbecher C, Garner CC, Gundelfinger ED. Biochem Biophys Res Commun. 1999;264:247–252. doi: 10.1006/bbrc.1999.1489. [DOI] [PubMed] [Google Scholar]
- 35.Naisbitt S, Kim EK, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 36.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 37.Okamoto PM, Herskovits JS, Vallee RB. J Biol Chem. 1997;272:11629–11635. doi: 10.1074/jbc.272.17.11629. [DOI] [PubMed] [Google Scholar]
- 38.Henley JR, McNiven MA. J Cell Biol. 1996;133:761–775. doi: 10.1083/jcb.133.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du Y, Weed SA, Xiong W, Marshall TD, Parsons JT. Mol Cell Biol. 1998;18:5838–5851. doi: 10.1128/mcb.18.10.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evan GI, Lewis GK, Ramsay G, Bishop JM. Molec Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gee MA, Heuser JE, Vallee RB. Nature. 1997;390:636–639. doi: 10.1038/37663. [DOI] [PubMed] [Google Scholar]
- 42.Warnock DE, Hinshaw JE, Schmid SL. J Biol Chem. 1996;271:22310–22314. doi: 10.1074/jbc.271.37.22310. [DOI] [PubMed] [Google Scholar]
- 43.Goslin GBAK, editor. Rat Hippocampal Neurons in Low-density Culture: Culturing Nerve Cells. MIT Press; Cambridge, MA: 1991. pp. 339–370. [Google Scholar]
- 44.Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- 45.Abbott MA, Wells DG, Fallon JR. J Neurosci. 1999;19:7300–7308. doi: 10.1523/JNEUROSCI.19-17-07300.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carlin RK, Grab DJ, Cohen RS, Siekevitz P. J Cell Biol. 1980;86:831–845. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao I, Hata Y, Hirao K, Deguchi M, Ide N, Takeuchi M, Takai Y. J Biol Chem. 1999;274:27463–27466. doi: 10.1074/jbc.274.39.27463. [DOI] [PubMed] [Google Scholar]
- 48.Satoh K, Yanai H, Senda T, Kohu K, Nakamura T, Okumura N, Matsumine A, Kobayashi S, Toyoshima K, Akiyama T. Genes Cells. 1997;2:415–424. doi: 10.1046/j.1365-2443.1997.1310329.x. [DOI] [PubMed] [Google Scholar]
- 49.Takeuchi M, Hata Y, Hirao K, Toyoda A, Irie M, Takai Y. J Biol Chem. 1997;272:11943–11951. doi: 10.1074/jbc.272.18.11943. [DOI] [PubMed] [Google Scholar]
- 50.Lim S, Naisbitt S, Yoon J, Hwang JI, Suh PG, Sheng M, Kim E. J Biol Chem. 1999;274:29510–29518. doi: 10.1074/jbc.274.41.29510. [DOI] [PubMed] [Google Scholar]
- 51.Boeckers TM, Kreutz MR, Winter C, Zuscharatter W, Smalla KH, Sanmarti-Vila L, Wex H, Langnaese K, Bockmann J, Garner CC, Gundelfinger ED. J Neurosci. 1999;19:6506–6518. doi: 10.1523/JNEUROSCI.19-15-06506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McNiven MA, Kim L, Krueger EW, Orth JD, Cao H, Wong TW. J Cell Biol. 2000;151:187–198. doi: 10.1083/jcb.151.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 54.Kim E, Naisbitt S, Hsueh YP, Rao A, Rothschild A, Craig AM, Sheng M. J Cell Biol. 1997;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng S, Chen JK, Yu H, Simon JA, Schreiber SL. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 56.Gout I, Dhand R, Hiles ID, Fry MJ, Panayotou G, Das P, Truong O, Totty NF, Hsuan J, Booker GW, Campbell ID, Waterfield MD. Cell. 1993;75:25–36. [PubMed] [Google Scholar]
- 57.Scaife R, Gout I, Waterfield MD, Margolis RL. EMBO J. 1994;13:2574–2582. doi: 10.1002/j.1460-2075.1994.tb06547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hunt CA, Schenker LJ, Kennedy MB. J Neurosci. 1996;15:1380–1388. doi: 10.1523/JNEUROSCI.16-04-01380.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao H, Thompson HM, Krueger EW, McNiven MA. J Cell Sci. 2000;113:1993–2002. doi: 10.1242/jcs.113.11.1993. [DOI] [PubMed] [Google Scholar]
- 60.Damke H, Baba T, Warnock DE, Schmid SL. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Damke H, Baba T, van der Bliek A, Schmid SL. J Cell Biol. 1995;131:69–80. doi: 10.1083/jcb.131.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Man YH, Lin JW, Ju WH, Ahmadian G, Liu L, Becker LE, Sheng M, Wang YT. Neuron. 2000;25:649–662. doi: 10.1016/s0896-6273(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 63.Wang YT, Linden DJ. Neuron. 2000;25:635–647. doi: 10.1016/s0896-6273(00)81066-1. [DOI] [PubMed] [Google Scholar]
- 64.Spacek J, Harris K. J Neurosci. 1997;17:190–203. doi: 10.1523/JNEUROSCI.17-01-00190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fischer M, Kaech S, Knutti D, Matus A. Neuron. 1998;20:847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- 66.Gaidarov I, Santini F, Warren RA, Keen JH. Nat Cell Biol. 1999;1:1–7. doi: 10.1038/8971. [DOI] [PubMed] [Google Scholar]
- 67.Gonzalez-Gaitan M, Jackle H. Cell. 1997;88:767–776. doi: 10.1016/s0092-8674(00)81923-6. [DOI] [PubMed] [Google Scholar]
- 68.Estes PS, Roos J, van der Bliek A, Kelly RB, Krishnan KS, Ramaswami M. J Neurosci. 1996;16:5443–5456. doi: 10.1523/JNEUROSCI.16-17-05443.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roos J, Kelly RB. Curr Biol. 1999;9:1411–1414. doi: 10.1016/s0960-9822(00)80087-1. [DOI] [PubMed] [Google Scholar]
- 70.Roos J, Kelly RB. J Biol Chem. 1998;273:19108–19119. doi: 10.1074/jbc.273.30.19108. [DOI] [PubMed] [Google Scholar]