Testing for Population Subdivision and Association in Four Case-Control Studies (original) (raw)
Abstract
Population structure has been presumed to cause many of the unreplicated disease-marker associations reported in the literature, yet few actual case-control studies have been evaluated for the presence of structure. Here, we examine four moderate case-control samples, comprising 3,472 individuals, to determine if detectable population subdivision is present. The four population samples include: 500 U.S. whites and 236 African Americans with hypertension; and 500 U.S. whites and 500 Polish whites with type 2 diabetes, all with matched control subjects. Both diabetes populations were typed for the PPARg Pro12Ala polymorphism, to replicate this well-supported association (Altshuler et al. 2000). In each of the four samples, we tested for structure, using the sum of the case-control allele frequency χ2 statistics for 9 STR and 35 SNP markers (Pritchard and Rosenberg 1999). We found weak evidence for population structure in the African American sample only, but further refinement of the sample, to include only individuals with U.S.-born parents and grandparents, eliminated the stratification. Our examples provide insight into the factors affecting the replication of association studies and suggest that carefully matched, moderate-sized case-control samples in cosmopolitan U.S. and European populations are unlikely to contain levels of structure that would result in significantly inflated numbers of false-positive associations. We explore the role that extreme differences in power among studies, due to sample size and risk-allele frequency differences, may play in the replication problem.
Introduction
Association studies for mapping disease-related genes have gained in popularity over traditional family-based linkage analyses. In principle, for an equivalent number of genotypes and for common disease alleles, population-based tests of association offer far greater power to detect the presence of genes whose effects (or relative risks) are minor than do pedigree-based designs in which a greater fraction of the genome is correlated among closely related pedigree members (Risch and Merikangas 1996). Nevertheless, population-based studies are regarded with considerable skepticism, which is frequently vindicated by apparently false or nonreplicable associations (Editorial 1999; Weiss et al. 2001). A common explanation for these false associations is that unrecognized population stratification produces spurious associations at loci of which the allele frequencies differ coincidentally among the subpopulations comprising the case and control subjects (Lander and Schork 1994; Risch 2000; Cardon and Bell 2001; Peltonen et al. 2001). Specifically, false-positive associations may arise if case and control subjects are drawn differentially from two or more subpopulations in which marker allele frequencies and disease prevalence differ across the subpopulations. For this reason, tremendous effort has been devoted over the past decade to the development of family-based tests of both association and linkage. The transmission/disequilibrium test (TDT) (Spielman et al. 1993), which uses untransmitted parental chromosomes as controls, pioneered this approach. Numerous extensions for various family-based designs are now available. The primary drawbacks of family-based association designs are that use of closely related controls reduces power considerably and that recruitment of family members is often difficult or impossible, particularly for late-onset diseases.
To date, the extent of population structure in actual population-based case-control studies has not been examined carefully. There is scant evidence that stratification is to blame for the perceived excess of false-positive associations in case-control studies, and theoretical considerations seem to put strict limits on the contribution that stratification can make (Wacholder et al. 2000). Furthermore, it is not clear that failures to replicate reported associations are lower for family-based versus population-based tests of association (Ioannidis et al. 2001). Although population stratification has been emphasized as an explanation for failures to replicate reported associations, other factors may turn out to be more important. Chief among these may be the overinterpretation of marginal findings, inadequate (or too restrictive) corrections for multiple hypothesis testing, publication biases toward positive results, differing or inadequate clinical diagnostic criteria, inadequate sample sizes, differences in risk-allele frequency among studies, and genuine locus or allelic heterogeneity among different population samples (Gambaro et al. 2000; Cardon and Bell 2001; Ioannidis et al. 2001; Vieland 2001).
In an effort to recoup some of the statistical power lost to family-based study designs, a recent trend has focused on recognizing when structure may affect a case-control study and correcting for it when it is present (Devlin and Roeder 1999; Pritchard and Rosenberg 1999; Pritchard et al. 2000_a,_ 2000_b;_ Reich and Goldstein 2001). When case and control samples are drawn from different subpopulations, allele frequencies will tend to differ among the subpopulations for most randomly chosen loci. Population subdivision can then be detected by genotyping a number of unlinked markers and looking for systematic differences in allele frequency. A single measure of collective difference in allele frequency can be used to detect an overall deviation between populations or subpopulations (Pritchard and Rosenberg 1999). In the event that structure is detected, two general approaches have been proposed to correct for it. One method, commonly known as “genomic control,” uses the observed patterns of variation at unlinked loci to calculate empirical test statistic distributions for use in evaluating the validity of an association (Pritchard and Rosenberg 1999; Devlin and Roeder 1999; Reich and Goldstein 2001). A second take on the problem uses data from random, unlinked markers to identify cryptic subpopulations (Pritchard et al. 2000_b;_ Satten et al. 2001; Thornsberry et al. 2001). Tests of association are then essentially performed in each of the identified subpopulations.
Here, we implement some of these recently proposed methods to test for the presence of structure in four independent case-control sample sets, all selected from the Global Repository at Genomics Collaborative, Inc. (GCI), as part of ongoing association studies. In two of these case-control samples, we also test whether we can detect a well-supported association with the PPARg Pro12Ala polymorphism and type 2 diabetes (DM2) (Altshuler et al. 2000), and we examine the possible impact of structure on the association observed in these two replicate samples.
Methods
Case-Control Samples
Four separate case-control samples were selected from samples residing in the Global Repository at GCI. The repository contains globally collected DNA samples from >100,000 individuals enrolled in studies of multifactorial disease, as well as samples from healthy individuals enrolled as control subjects. In addition to extensive clinical data, all samples in the repository have data on self-reported ethnicity for three generations, place and country of birth with current living place, location and country of birth for both parents, country of birth for all four grandparents, and languages spoken in the home. For all samples, controls were matched individually with cases, by use of multiple criteria including age (± 5 years), sex (exact match), BMI (± 2.5 units), and individual and family disease history. Demographic information used in matching control subjects varied but included self-reported ethnicity, for each index individual, and, in the U.S. samples, geographic region of current residence (on the basis of a four-region breakdown). No birth-country information was used in matching case subjects to control subjects. The four study populations were (1) 500 white case subjects with hypertension and 500 white control subjects, from the United States; (2) 236 African American case subjects with hypertension and 236 African American control subjects; (3) 500 white case subjects with DM2 and 500 white control subjects, from the United States; and (4) 500 white case subjects with DM2 and 500 white control subjects, from Poland. All samples from Poland came from a single location. All U.S. whites were non-Hispanic. Parents and grandparents of the individuals in the African American sample were also required to be African American. Clinical information used to define each case sample varied among the specific studies; however, the two diabetes patient samples were chosen specifically as replicates of one another and thus identical clinical criteria were used. Individuals with BMI >35 kg/m2 were excluded from both DM2 case samples, and inclusion preference was given to individuals with one first-degree relative who also had DM2. All case individuals were defined as having at least one fasting blood glucose measurement >140 mg/dL at diagnosis.
Genotyping
The PPARg Pro12Ala polymorphism was genotyped in the two diabetic case-control samples (Poland and U.S. whites), by use of Sequenom’s Mass Array technology (Ross et al. 1998). To assess structure, 40 biallelic SNPs were chosen from a public SNP reference database (SNP Consortium Web site). Each included SNP was chosen to have a minor allele frequency of at least 0.2 in the African American and white population samples reported in the database. Firm estimates of the level of structure that a given collection of markers can detect are difficult to obtain with incomplete information on allele frequencies in the various subpopulations; however, simulations (Pritchard and Rosenberg 1999) indicate that 30 biallelic markers should have reasonable power to detect stratification in the subpopulations that have diverged only recently (within the past 10,000 years), such as the main population groups we compare here. To minimize the likelihood that any two SNPs were in linkage disequilibrium (LD), we chose one SNP per chromosome arm (where precise mapping information was available), with the exception of the Y chromosome. Suitable SNPs were not available at the time of choosing on chromosomes 17, 19, 21, and 22; and only one suitable SNP was found on chromosome 9. To reach a total of 40 SNPs, 3 each were chosen on the three largest chromosomes—1, 2, and 3. Genotyping of SNPs was performed by mass spectroscopy performed with Qiagen’s Masscode system (Kokoris et al. 2000). Additionally, a panel of 10 tetranucleotide STR markers (Profiler Plus, Applied Biosystems) are typed on all DNA samples in the repository as part of the routine quality-control process. These markers show large differences in population allele frequency, since the panel is used for identifying humans. One of the markers is a sex-chromosome marker, and the remaining nine are autosomal. As this information already existed, genotypic information from the nine autosomal loci were also used in our analyses. The average number of alleles for these loci is 17.9, and the average heterozygosity for the nine markers is 81.1%.
Statistical Methods
Within each of the four case-control samples, we computed χ2 tests of Hardy-Weinberg equilibrium (HWE) in the combined case and control sample for the 40 SNPs and 9 STR markers. We computed a contingency χ2 statistic comparing case and control allele frequencies for each marker (Pritchard and Rosenberg 1999), grouping STR alleles with <1% frequency in the four combined samples. Under the null hypothesis that there are no allele-frequency differences between the case and control subjects, the statistic for each marker has a χ2 distribution with degrees of freedom equal to 1 less than the number of alleles. Under the null hypothesis that the populations have the same allele frequencies, the sum of the statistics for all of the markers has a χ2 distribution with degrees of freedom equal to the sum of the degrees of freedom from all of the markers. In effect, this statistic tests for an overall difference in allele frequencies between the case and control subjects. We tested for stratification, using the SNPs and STRs combined, and separately, as the marker types may differ in mutation rate and history. Combining case and control subjects, we tested for an overall difference in allele frequencies between the two U.S. white samples, between the combined U.S. white samples (DM2 and HTN) and the Polish DM2 sample, and between each of these samples and the African American HTN sample, using the same contingency χ2 statistic used to test for stratification.
Association tests for PPARG
We tested for association between the PPARG SNP genotype or allele frequencies and diabetes case/control status separately in the Polish and U.S. samples, using a Pearson χ2 statistic. We computed the Mantel-Haenszel statistic and common estimate of the odds ratio (Mantel and Haenszel 1959) controlling for sample in the combined U.S. and Polish sample. We used the Breslow-Day test (1994) to test for homogeneity of the odds ratios in the two samples.
Population structure
We used the computer program structure (Pritchard et al. 2000_a_) to attempt to identify clusters of genetically similar diploid individuals from multilocus genotypes. We did not define population affinities when clustering individuals and used the correlated allele-frequency option to examine our Polish and U.S. white samples for undetected structure that may affect the PPARG association, using 34 SNPs and 9 STRs. We omitted one of our X-linked SNPs for these analyses, as the two X-linked SNPs were closely linked (∼30 kb) and in strong LD in our samples. To confirm that the 34 SNPs and 9 STRs provided enough power to identify distinct ethnic groups, we also ran structure using the combined U.S. white and African American HTN samples.
Results
HWE
We genotyped 40 SNPs in 3,472 individuals. For quality-control purposes, eight additional individuals were repeatedly genotyped for each SNP: four individuals were repeatedly genotyped 22 times, and four individuals were repeatedly genotyped 20 times, for a total of 168 repeated genotypes per SNP. A reproducibility score was constructed as the number of consistent genotypes divided by the total number of attempted genotypes. Low reproducibility can reflect either a high “no call” rate (i.e., the software could not confidently call the genotype) or a high genotyping-error rate. We eliminated one SNP because of low (86%) reproducibility: for this SNP, there were 20 “no calls” and three discordant genotypes. The sample genotypes had a “no call” rate of >10% as well. All other SNPs had reproducibility scores of ⩾92%, and 33/40 had reproducibility ⩾95%.
Four SNPs had extreme deviations from HWE (P<.0001), in which heterozygotes and only a single species of homozygote were observed in all of the samples, but the allele frequency was high enough that a considerable number of the other homozygote species had been expected. Neither the assay with low reproducibility score nor the four assays with extreme HWE deviations had significant allele or genotype frequency differences in any of the four case-control samples. Further, the HWE test P values for case and control subjects for each sample were very similar. We therefore excluded these five assays from further analyses, leaving a total of 35 SNPs for study of population stratification. We did not reject HWE for the nine STR markers or the 35 SNPs in any of the four samples.
Population Stratification
Table 1 shows the results of the population stratification tests for the four samples and two sets of markers. There is no evidence for stratification using the STR markers or the combined STR and SNP markers. However, with the SNP markers alone, there is some evidence (_P_=.01) of stratification in the African American sample. The large χ2 value is due primarily to large differences in allele frequency between case and control subjects at 3 SNPs, rather than smaller differences over all 35 SNPs. Our African American sample contains a heterogeneous mix of individuals. Some of these individuals have parents or grandparents born outside of North America (e.g., the Caribbean or Africa). For 79% of the cases and 73% of the controls, we know with certainty that the parents and all four grandparents were born in North America. Among the case and control subjects in this subgroup, there is no evidence for stratification (_P_=.26 for SNPs, _P_=.93 for STRs).
Table 1.
χ2 Tests for Differences in Allele Frequency between Case and Control Subjects and for Allele Frequency between Selected Sample Populations
| Values for STR Markers | Values for SNP Markers | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Population | T | df | P | No. ofSTRs with P<.05 | T | df | P | No. ofSNPs with P<.05 | P for Combined SNP and STR |
| Within samples: | |||||||||
| Hypertension: | |||||||||
| African American | 53.1 | 72 | .95 | 0 | 58.1 | 35 | .01 | 3 | .37 |
| U.S. white | 71.1 | 72 | .51 | 0 | 40.9 | 35 | .23 | 1 | .35 |
| Diabetes: | |||||||||
| U.S. white | 75.9 | 72 | .35 | 0 | 24.4 | 35 | .91 | 0 | .66 |
| Poland | 78.3 | 72 | .28 | 0 | 29.0 | 35 | .75 | 1 | .47 |
| Between samples: | |||||||||
| White/hypertension vs. white/diabetes | 67.3 | 72 | .63 | 0 | 35.1 | 35 | .46 | 1 | .61 |
| U.S. white/diabetes vs. Polish/diabetes | 193.4 | 72 | 5×10-13 | 5 | 150.8 | 35 | 2×10-16 | 13 | <10−20 |
| White/hypertension vs. African American/hypertension | 1261.1 | 72 | <10−20 | 9 | 4002.6 | 35 | <10−20 | 30 | <10−20 |
Between-Population Comparisons
The two U.S. white samples did not have significantly different allele frequencies overall (table 1). The U.S. white and U.S. African American HTN samples had significantly different allele frequencies, as did the U.S. and Polish white DM2 samples (table 1). For a small subset (_N_=46) of the U.S. DM2 sample, all known parents and grandparents were born in central Europe. Neither the SNP nor the STR frequencies were significantly different between this subset and the Polish DM2 sample (SNPs: χ2=32.8, df 35, _P_=.58; STRs: χ2=45.5, df 51, _P_=.69; combined: χ2=78.3, df 86, _P_=.71).
PPARG Test of Association
Genotyping rates of 96.2% and 92.6% were obtained for the PPARg Pro12Ala polymorphism in the Polish case and control subjects, and genotyping rates of 99.0% and 99.2% were obtained for the U.S. case and control subjects. HWE was not rejected in any of the subsamples (HWE test statistic _P_=.40 and _P_=.25 for the Polish case and control subjects, respectively; _P_=.17 and _P_=.54 for the U.S. case and control subjects, respectively). In the Polish sample, there is strong evidence for an association between case status and genotype (Pearson χ2=16.04, df 2, _P_=.0003) (table 2), and case status and allele frequency (χ2=16.37, df 1, _P_=5×10-5) (table 2). The odds ratio for the C allele in cases versus controls is 0.60 (95% CI 0.47–0.77). In the U.S. sample, no such association is evident for genotype (χ2=0.70, df 2, _P_=0.71) (table 2), or allele frequency (χ2=0.19, df 1, _P_=.66) (table 2). The odds ratio for the C allele in this sample is 0.94 (95% CI 0.70–1.26). The odds ratios in the two samples are significantly different (χ2=5.14, df 1, _P_=.02). The Mantel-Haenszel estimate of the common odds ratio and CI for the two samples combined is 0.72 (95% CI 0.60–0.87), which is very similar to the common odds ratio and CI calculated in a recent meta-analysis (Altshuler et al. 2000).
Table 2.
Genotypes and Allele Frequencies for the PPARg Pro12Ala Polymorphism in the Polish and U.S. DM2 Samples, Odds Ratios and 95% CIs for C Allele in Cases, and P Values of Association Tests
| No. of Subjects with Genotype | Allele Frequencies | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| PopulationandSubjects | CC | GC | GG | Total | C | G | Odds Ratio | 95% CI | Pa |
| Poland: | .60 | .47–.77 | .00005 | ||||||
| Case | 8 | 108 | 365 | 481 | .129 | .871 | |||
| Control | 22 | 139 | 302 | 463 | .198 | .802 | |||
| U.S. | .94 | .70–1.26 | .66 | ||||||
| Case | 2 | 92 | 401 | 495 | .097 | .903 | |||
| Control | 4 | 94 | 398 | 496 | .103 | .897 | |||
| Combined | .72 | .60–.87 | .0007 |
Analysis of Structure
Genetic differentiation within the two DM2 populations was low, and Fst values for individual loci were not greater for the U.S. population than for Poland, ranging from 0.0009 to 0.0752 for both populations. To detect the presence of cryptic population structure, we used the program structure (Pritchard et al. 2000_a_). This program attempts to identify subpopulations by grouping individuals in a way that minimizes Hardy-Weinberg and linkage disequilibrium among unlinked markers. A single population fit both the U.S. and Polish sample data best, as well as the combined U.S./Polish sample. Although our U.S. and Polish DM2 samples have statistically significantly different allele frequencies, the actual differences in allele frequencies are relatively small (range 0.03–0.06 for the 13 SNPs with differences significant at the 0.05 level). Thus, the combined U.S. and Polish sample exhibits neither more Hardy-Weinberg disequilibrium within markers than expected nor more LD between unlinked markers than expected, and structure is unable to identify subpopulations. Much larger sample sizes would be required to detect Hardy-Weinberg disequilibrium and/or linkage disequilibrium under these conditions. In contrast, structure easily identified the white and African American subgroups of our combined hypertension samples (analyses not shown).
Discussion
In the four case-control collections we examined, we found no evidence for significant cryptic population structure for any of the marker sets within any of the three white samples. With the combination of 35 SNPs and 9 STR markers, we should have high power to detect strong stratification in these samples if it exists (Pritchard and Rosenberg 1999). Thus, it is unlikely that there is strong hidden structure in these particular samples. Some low-level structure may nevertheless be present in the populations from which these samples were drawn, and, moreover, the prevalence of DM2 and HTN is known to vary substantially across populations, even within Europe (see, e.g., Van Den Hoogen et al. 2000; World Health Organization Web site), but the fact that we do not detect any stratification in three of the four independent case-control samples selected here is very encouraging.
One exception to this was the African American sample for which the SNP markers, although not the STR markers, did show evidence for stratification in the case subjects versus the control subjects (table 1). The African American population currently residing in the United States is known to be admixed, primarily with whites, and several studies have examined the extent of admixture in populations from different U.S. regions. These studies reveal a complex situation with levels of admixture differing throughout the United States and the Caribbean (Chakraborty et al. 1992; Parra et al. 1998; Destro-Bisol et al. 1999). In this sample, we therefore used more-extensive demographic information (ethnicity information for three generations). We did not initially use any of the information available on birthplace for individuals or their parents and grandparents. On examining birthplace information, however, we found that the African American sample contained recent Caribbean and African immigrants, in addition to third-generation North Americans. The simplest approach to this sample was to remove all the known recent immigrants from both the case and control subjects, leaving ∼79% of the case subjects and ∼73% of the control subjects for whom all three generations were born in North America. Although varying and unknown degrees of finer-level structure must remain in this sample, there was no evidence for population structure in either the SNPs or STRs within this subgroup after taking this measure, suggesting that careful matching using demographic parameters, such as region of U.S. residence and recency of immigration, may help to reduce stratification bias in African Americans to nominal levels, at least for moderate sample sizes. Nevertheless, because the extent of bias caused by population structure will increase with sample size (Pritchard and Donnelly 2001), which provides greater power to detect both real associations and any weak spurious associations, it may be imperative to type additional markers in larger studies so that such finer-level structure can be detected.
Our ability to detect population structure when it is present is demonstrated in this African American case-control sample (table 1) and also by the comparisons between populations (table 1). The latter represent an artificially extreme example of stratification, in which one group comprises members of a single “self-defined ethnicity” and the comparison group comprises members of another. Our comparisons between these samples indicate the ability to detect deliberate structure, with both sets of markers showing very significant differences between groups when the U.S. whites are compared with the African American sample (table 1). The comparison between the two separate U.S. white samples shows no significant difference in allele frequencies. In contrast, the comparison between the U.S. white DM2 and the Polish DM2 populations clearly indicates that these populations have significantly different allele frequencies. A comparison of birth locations over three generations revealed that both samples have some heterogeneity in their origins. However, the Polish group, which was collected from a single site in southern Poland, is considerably more homogeneous. Of the Polish sample, 74.5% are Polish by birth, with Polish-born parents and/or grandparents. An additional 8.8% have at least one Polish-born parent. The remaining 16.7% are primarily Central and Eastern European in origin. In contrast, the sample of U.S. whites with DM2 is composed of individuals for whom the two previous generations were born in the U.S. (46%) and numerous first- and second-generation immigrants from throughout Europe. Distribution of birth country among index cases, parents, and grandparents not born in the United States is similar in case and control subjects. However, more than twice as many control subjects were born outside of the United States than case subjects (12.2% vs. 4.6%). The differences in allele frequency between these two populations may thus reflect the greater heterogeneity and more-diverse European origins of the U.S. sample.
In addition to the ability to assess population structure within a case-control sample, the key question of interest is whether such stratification can contribute significantly to association—specifically, to reported positive associations. The only association we had typed at the time of assessing stratification in these four samples was the common Pro12Ala allele of PPARg in the two DM2 case-control samples (Poland and the U.S. whites), in which the proline allele has been reproducibly, although variably, associated with cases and with an increased risk of DM2 (Altshuler et al. 2000). In our two populations, we were able to replicate this strong association in the Polish sample, whereas, in the U.S. sample, we found no evidence for an association. Despite this difference, comparison of the estimated risk in each of these two populations with the meta-analysis of Altshuler and colleagues (2000) demonstrates that both are individually consistent with the earlier studies and with the modest effect previously described. A sample size insufficient for reliable detection of the association has been suggested to be one key factor in the variability in prior studies (Altshuler et al. 2000), and here too we find that the combined population of 1,000 case subjects and 1,000 control subjects gives stronger support for the association and is closer to the modest across-population effect demonstrated in the meta-analysis in that study (table 2).
Although it is unlikely that the positive association observed here in the Polish population is the result of hidden substructure, a reason we may have failed to detect an association in the U.S. sample could be that subtle stratification or population heterogeneity masks the effect in this sample. Recent studies have shown that, by use of high-resolution statistical methods such as those in structure (Pritchard et al. 2000_a,_ 2000_b_), on multilocus genotypes, it is practical to distinguish related populations of recent common ancestry (Rosenberg et al. 2001). Rosenberg et al. (2001) were able to distinguish a distinct genetic signature in a Libyan Jewish population, although the population had likely been relatively secluded from the others tested. Here, despite a comparable number of markers, we were unable to detect genetically distinguishable subpopulations within either DM2 case-control sample. This may not be surprising, given the likely continued mixing of European white populations. Recent recommendations suggest that structure may require hundreds of markers to reliably identify very closely related subpopulations and populations with a high degree of admixture (Pritchard et al. 2000_a,_ 2000_b;_ Wilson et al. 2001; Pritchard and Donnelly 2001). It is important to distinguish between heterogeneity and structure within a case-control sample. As long as the heterogeneity is equivalent in case and control subjects (i.e., the two groups have the same mix of ethnic/genetic subgroups), stratification bias will not occur. Nevertheless, it could occur in repeated samplings from the same population if the underlying heterogeneity in each sampling is not well matched with the others. Moreover, stratification bias will decrease with an increasing number of subpopulations, since biases will tend to cancel each other out (Wacholder et al. 2000). As far as can be determined, given the demographic data, the country-of-ancestry distribution is similar in the U.S. DM2 case subjects and control subjects, although the control subjects tend to be more recent immigrants than the case subjects. Thus, if population structure or heterogeneity remains in one sample here, it will require many more markers to detect it and thus correct for it.
These two diabetes populations exemplify the difficulties in reproducibly detecting risk factors of modest effect in case-control studies, where significant between-study heterogeneity in findings is frequent and the results of an initial study may correlate only modestly with subsequent studies of the same association (Editorial 1999; Altshuler et al. 2000; Ioannidis 2001). Although replication has become an accepted standard, variation in the strength of an association is common, even between studies of seemingly similar populations, such as the two presented here. Both populations are European in origin, cases were ascertained identically in the two populations, case and control subjects were matched with the same parameters in both, the two samples are equal in size, and neither had detectable population stratification, yet the results of an allelic association differ between the two. Small sample size is one predictor of study discrepancy (Ioannidis 2001), and our combined sample clearly gives a more accurate estimate of the population effect of the allele, suggesting that sample size is a factor. Additionally, a gene effect may be genuinely stronger in one subpopulation than in another. The ability to access large sample numbers and to share data for meta-analyses may be key to proper assessment of the validity of an association.
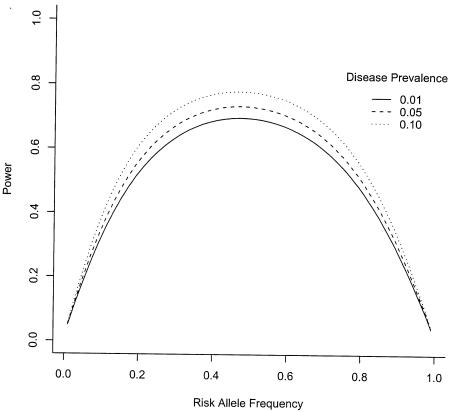
One issue that has not been rigorously addressed in the context of study replication is the variation in (putative) risk-allele frequencies across replication samples. Even when the genetic effect of a variant and the disease frequency are the same for two populations, samples of the same size from these two populations can have drastically different power to detect the association, depending on the frequency of the risk allele. For example, the allele frequency of the proline risk allele for PPARg is ∼0.90 in the U.S. population and ∼0.80 in the Polish population from which we sampled. For a genotype relative risk of 1.25 and a multiplicative model (Altshuler et al. 2000), the Polish 500 case, 500 control sample has 70% greater power than the U.S. sample (51% vs. 30% power) to detect an effect. Larger sample sizes make a difference: for 1,000 case subjects and 1,000 control subjects, the power of the Polish sample would be only 47% greater than the U.S. sample (81% vs. 55% power). Figure 1 illustrates that for the combination of (1) a weak genotypic risk ratio and (2) high (>0.80) or low (<0.20) risk-allele frequencies, small changes in risk-allele frequencies have large effects on power. The effect is much greater in magnitude than the effect of disease prevalence in a population. Thus, differing risk-allele frequencies across populations, resulting in drastic differences in power, are one more factor contributing to the difficulty in replicating association studies for complex phenotypes.
Figure 1.
Power to detect an allelic association for 500 case subjects and 500 control subjects under a multiplicative genetic model with genotypic risk ratio 1.25, by risk-allele frequency and disease prevalence. Allele frequencies for case and control subjects were determined under the given genetic models; power to detect the difference in allele frequencies was calculated for the exact test for a 2 × 2 contingency table, by the method of Walters (1979).
Our data show that, if matching is done carefully, using a reasonable amount of demographic data, then unanticipated genetic stratification can be kept to a minimum, at least for studies of similar size to our four examples here. Whether our results can be generalized to larger sample sizes depends on the unknown extent of remaining undetected structure. Clearly, investigators should confirm that new associations are not due to stratification, using appropriate methods. Our results should, however, be encouraging to investigators who have carefully matched their case and control subjects. Isolated populations are a popular target for association studies (Peltonen at al. 2001), because their homogeneity minimizes population structure, yet they may not always be appropriate for the disease or sample number of interest, and our results are encouraging for studies in more heterogeneous populations. Perhaps just as important in association-study design, however, are other causes of nonreplication in case-control studies. These may include different clinical definitions or different phenotypes being compared, different environmental and genetic backgrounds that should also be controlled for as carefully as possible, and differences in the power to detect an association.
Acknowledgments
We thank R. S. Wells, L. Kruglyak, and J. R. Wakeley for advice and comments on the manuscript.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Global Repository at GCI, http://www.genomicsinc.com (for case-control samples)
- SNP Consortium, http://snp.cshl.org/allele_frequency_project/ (for biallelic SNPs)
- World Health Organization, http://www.who.int/ncd/dia/databases2.htm#t3
References
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES (2000) The common PPARg Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 26:76–80 [DOI] [PubMed] [Google Scholar]
- Breslow NE, Day NE (1994) Statistical methods in cancer research, volume II: the design and analysis of cohort studies. IARC Scientific Publications, No. 82, Oxford University Press, New York [PubMed] [Google Scholar]
- Chakraborty R, Kamboh MI, Nwankwo M, Ferrell RE (1992) Caucasian genes in American blacks: new data. Am J Hum Genet 50:145–155 [PMC free article] [PubMed] [Google Scholar]
- Destro-Bisol G, Maviglia R, Caglia A, Boschi I, Spedini G, Pascali V, Clark A, Tishkoff S (1999) Estimating European admixture in African Americans by using microsatellites and a microsatellite haplotype (CD4/Alu). Hum Genet 104:149–157 [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K (1999) Genomic control for association studies. Biometrics 55:997–1004 [DOI] [PubMed] [Google Scholar]
- Cardon LR, Bell JI (2001) Association study designs for complex diseases. Nat Rev Genet 2:91–99 [DOI] [PubMed] [Google Scholar]
- Editorial (1999) Freely associating. Nat Genet 22:1–2 [DOI] [PubMed] [Google Scholar]
- Gambaro G, Anglani F, D’Angelo A (2000) Association studies of genetic polymorphisms and complex disease. Lancet 355:308–311 [DOI] [PubMed] [Google Scholar]
- Ioannidis JPA, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG (2001) Replication validity of genetic association studies. Nat Genet 29:306–309 [DOI] [PubMed] [Google Scholar]
- Kokoris M, Dix K, Moynihan K, Mathis J, Erwin B, Grass P, Hines B, Duesterhoeft A (2000) High-throughput SNP genotyping with the Masscode system. Mol Diagn 5:329–340 [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork NJ (1994) Genetic dissection of complex traits. Science 265:2037–2048 [DOI] [PubMed] [Google Scholar]
- Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748 [PubMed] [Google Scholar]
- Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD (1998) Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet 63:1839–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K (2001) Use of population isolates for mapping complex traits. Nat Rev Genet 1:182–190 [DOI] [PubMed] [Google Scholar]
- Pritchard KJ, Donnelly P (2001) Case-control studies of association in structured or admixed populations. Theor Popul Biol 60:227–237 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA (1999) Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet 65:220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P (2000_a_) Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P (2000_b_) Association mapping in structured populations. Am J Hum Genet 67:170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Goldstein DB (2001) Detecting association in a case-control study while correcting for population stratification. Genet Epidemiol 20:4–16 [DOI] [PubMed] [Google Scholar]
- Risch NJ (2000) Searching for genetic determinants in the new millennium. Nature 405:847–856 [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Woolf E, Pritchard JK, Schaap T, Gefel D, Shpirer I, Lavi U, Bonne-Tamir B, Hillel J, Feldman MW (2001) Distinctive genetic signatures in the Libyan Jews. Proc Natl Acad Sci USA 98:858–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Hall L, Smirnov L, Haff L (1998) High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat Biotechnol 16:1347–1351 [DOI] [PubMed] [Google Scholar]
- Satten GA, Flanders WD, Yang Q (2001) Accounting for unmeasured population structure in case-control studies of genetic association using a novel latent-class model. Am J Hum Genet 68:466–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium; the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52:506–513 [PMC free article] [PubMed] [Google Scholar]
- Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES 4th (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28:286–289 [DOI] [PubMed] [Google Scholar]
- Van Den Hoogen CW, Feskens JM, Nagelkerke NJD, Menotti A, Nissinen A, Kromhout D (2000) The relation between blood pressure and mortality due to coronary heart disease among men in different parts of the world. N Engl J Med 342:1–8 [DOI] [PubMed] [Google Scholar]
- Vieland VJ (2001) The replication requirement. Nat Genet 29:244–245 [DOI] [PubMed] [Google Scholar]
- Wacholder S, Rothman N, Caporaso N (2000) Population stratification in epidemiologic studies of common genetic variants and cancer: quantification of bias. J Natl Cancer Inst 92:1151–1158 [DOI] [PubMed] [Google Scholar]
- Walters DE (1979) In defense of the arc sine approximation. The Statistician 28:219–222 [Google Scholar]
- Weiss ST, Silverman EK, Palmer LJ (2001) Case-control association studies in pharmacogenetics. Pharmacogenomics J 1:157–158 [DOI] [PubMed] [Google Scholar]
- Wilson JF, Weale ME, Smith AC, Gratrix F, Fletcher B, Thomas MG, Bradman N, Goldstein DB (2001) Population genetic structure of variable drug response. Nat Genet 29:265–269 [DOI] [PubMed] [Google Scholar]