Whole Genome Amplification of DNA from Laser Capture-Microdissected Tissue for High-Throughput Single Nucleotide Polymorphism and Short Tandem Repeat Genotyping (original) (raw)
Abstract
Genome-wide screening of genetic alterations between normal and cancer cells, as well as among subgroups of tumors, is important for establishing molecular mechanism and classification of cancer. Gene silencing through loss of heterozygosity is widely observed in cancer cells and detectable by analyzing allelic loss of single nucleotide polymorphism and/or short tandem repeat markers. To use minute quantities of DNA that are available through laser capture microdissection (LCM) of cancer cells, a whole genome amplification method that maintains locus and allele balance is essential. We have successfully used a ø29 polymerase-based isothermal whole genome amplification method to amplify LCM DNA using a proteinase K lysis procedure coupled with a pooling strategy. Through single nucleotide polymorphism and short tandem repeat genotype analysis we demonstrate that using pooled DNA from two or three separate amplification reactions significantly reduces any allele bias introduced during amplification. This strategy is especially effective when using small quantities of source DNA. Although a convenient alkaline lysis DNA extraction procedure provided satisfactory results from using 1500 to 3000 LCM cells, proteinase K digestion was superior for lower cell numbers. Accurate genotyping is achieved with as few as 100 cells when both proteinase K extraction and pooling are applied.
The revelation that cancer is a genomic disease along with the availability of draft human genome sequence have motivated the development of high-throughput technologies that can detect genetic alterations between normal and cancer cells, as well as differences among subgroups of tumors.1 Recently, gene expression profiling research on molecular classification of cancer highlighted the advantage of a genome-wide perspective on genetic variations.2–4 The altered mRNA levels in cancer genomes are often related to gene amplification of growth factor receptors, or the loss of functional tumor suppressor genes through homozygous deletion or loss of heterozygosity.5,6 Detection of such gene copy number changes has been achieved by comparative genomic hybridization analysis.7 However, because loss of heterozygosity may be accompanied by chromosome multiplicity or duplication of a dysfunctional allele, the most reliable approach to identify loss of heterozygosity is through detection of locus-specific genotype loss using a panel of single nucleotide polymorphism (SNP) markers and short tandem repeat (STR) or microsatellite markers. Additionally, STR genotyping may reveal the presence of microsatellite instability in cancer cells,8 while SNP genotyping may identify potential cancer risk and drug effects that are attributable to SNPs.9
To ensure accurate data interpretation, it is desirable to use laser capture microdissection (LCM) to separate cancer cells from surrounding normal cells in tumor lesions.10 To perform genotyping assays in a genome-wide manner, a whole genome amplification (WGA) method that amplifies DNA from a small number of LCM cells while maintaining locus and allele balance is necessary. To date, several WGA methods, including degenerated oligonucleotide primed-polymerase chain reaction (DOP-PCR),11–13 primer extension preamplification (PEP),14–16 and linker-adaptor ligation-based SCOMP,17,18 have been used to amplify DNA extracted from LCM cells. Each of these methods has limitations in terms of incomplete genomic coverage, low extent of amplification, or complex experimental manipulations. Moreover, these methods have been applied primarily for comparative genomic hybridization analysis that has somewhat lenient requirements for gene coverage and allele bias. To the best of our knowledge, there has been no previous report on using WGA products for SNP genotyping using a TaqMan platform, which is known for its accuracy, ease of operation, and amenability to automation.19
Recently, an isothermal WGA method using the strand-displacing ø29 polymerase was developed and successfully applied to amplify DNA from cell culture and blood.20 This method, termed “whole genome multiple strand displacement amplification (MDA),” demonstrated a high-amplification potential (up to 104-fold) and excellent loci representation (less than threefold bias).20 To evaluate this method for amplification of DNA from LCM cells, we used Amersham’s ø29 polymerase-based GenomiPhi kit (Amersham Biosciences, Piscataway, NJ) and assessed the quality of amplified products relative to unamplified DNA using SNP- and STR-genotyping assays. Using LCM cells collected from cancer and normal samples of colon and prostate, we found that DNA isolated from LCM cells generated more pronounced allelic bias than cell culture DNA, and that this bias seemed to inversely correlate with template quantity. Decreased allele imbalance was observed when proteinase K digestion was used to isolate DNA instead of the standard alkaline lysis method, probably because of a more complete digestion of cellular proteins and better release of DNA.15 In addition, we found that accurate genotype calling rates were much higher when amplified DNA products were pooled from two or three separate amplification reactions before analysis, and that this effect was especially dramatic when less cells were used. With pooling, greater than 95% of SNP and STR accurate genotype calling rates were achieved from alkaline lysis of ∼1500 cells, whereas proteinase K digestion rendered effective performance with as few as 100 cells.
Materials and Methods
WGA of Isolated DNA
Isolated human DNA samples from six cell lines representing two sets of trios (mother, father, and offspring) were obtained from the Coriell Cell Repositories (Camden, NJ). Amplification was performed using the GenomiPhi WGA kit (Amersham Biosciences, Piscataway, NJ) according to kit instructions. Briefly, 5 ng of DNA (1 μl) was added to 9 μl of sample buffer and the mixture was heat-denatured at 95°C for 3 minutes. After cooling on ice for 5 minutes, 10 μl of reaction mix (9 μl of reaction buffer plus 1 μl of ø29 polymerase) was added and the resulting mixture was incubated at 30°C for 16 hours. The polymerase was heat-inactivated at 65°C for 10 minutes. Each sample was diluted twofold using 1× TE (10 mmol/L Tris-HCl pH 8.0, 1 mmol/LEDTA) (pH 8.0) and its DNA concentration measured using the Hoechst dye assay (Bio-Rad, Hercules, CA). Typically 3 to 6 μg of amplified DNA was obtained from 5 ng of input DNA in a 20-μl WGA reaction.
Laser Capture Microdissection (LCM)
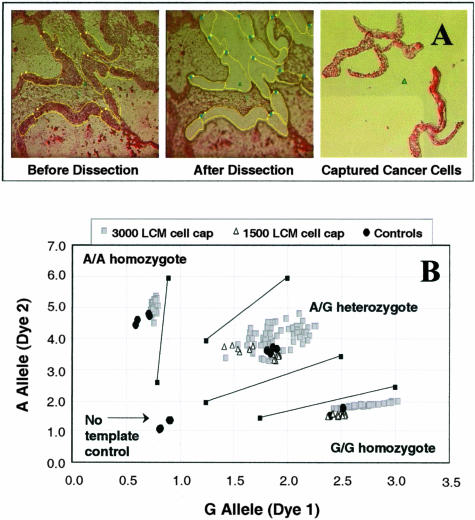
Normal and matching primary prostate cancer samples from eight patients were obtained from Clinomics (Pittsfield, MA). Normal and cancer colon samples of two individuals were obtained from the Cooperative Human Tissue Network. Fresh-frozen samples were embedded in OCT, sections 8-μm thick were cut in cryostat at − 23°C and mounted on slides covered by polyethylene membrane (PEN slides; C. Zeiss, Thornwood, NY). The resulting tissue slides were stored at −80°C until microdissection. Each slide was rinsed with sterile water for 1 minute, 70% ethanol for 1 minute, and stained in Mayer’s hematoxylin (Sigma Chemical Co., St. Louis, MO) for 30 seconds. After twice rinsing with deionized water, the slide was treated with staining bluing reagent (Harleco, EM Science, Gibbstown, NJ) for 1 minute, 70% ethanol for 1 minute, 95% ethanol for 1 minute, 1% Eosin Y in alcohol (Harleco) for 20 seconds, 95% ethanol for 1 minute, twice more with fresh 95% ethanol for 1 minute, and 100% ethanol for 3 minutes. After allowed to air-dry for 5 minutes with the airflow turned on, the stained slides were microdissected within 2 hours. A pathologist (GD) performed LCM using a P.A.L.M. Robot-Microbeam System (Oberkochen, Germany) following the manufacturer’s recommendations. First, the cell density on a standard hematoxylin and eosin-stained coverslipped slide was determined by counting the number of cells in an area of 1000 μm2 (×400 magnification) using software provided by the manufacturer of the P.A.L.M. This result was then used to calculate how many square microns are needed for a desired number of cells to be collected. Typical LCM images before and after microdissection are shown in Figure 1A (×100 to ×200 magnification). Immediately after the capture of cells, the cap containing the cells was placed on top of its matching Eppendorf tube and centrifuged for 1 minute at 14,000 rpm (16,000 × g). The cap/tube was snap-frozen in a dry ice/ethanol bath, and the collection of LCM cells contained within was hereafter referred to as a LCM cell cap or a cap. The cells were stored at −80°C for up to 2 months before DNA extraction.
Figure 1.
A: Representative LCM images of a colon cancer cell microdissection. B: TaqMan SNP-genotyping assay plot for G/A1182 of the EDNRB gene with WGA products from 70 different 3000 prostate LCM cell caps and WGA products from 12 different 1500 colon LCM cell caps. Assays were done in replicates. Arbitrary fluorescence units are shown on axes.
DNA Extraction from LCM Cells
DNA was extracted from LCM cells using either an alkaline lysis or a proteinase K lysis protocol. When using the alkaline lysis method, 5 μl of 1× phosphate-buffered saline was added to the cap containing LCM cells, followed by 5 μl of lysis buffer (400 mmol/L KOH, 100 mmol/L dithiothreitol, 10 mmol/L ethylenediaminetetraacetic acid). The mixture was pipetted multiple times to facilitate the collection of all cells from the cap. The 10-μl suspension was transferred to its matching Eppendorf tube and vortexed for 1 minute. After incubation on ice for 10 minutes, the solution was neutralized by addition of 5 μl of neutralization buffer containing 0.4 mol/L HCl and 0.6 mol/L Tris-HCl, pH 7.5. When using the proteinase K lysis protocol, 15 μl of a proteinase K buffer containing 0.4 mg/ml of proteinase K from Qiagen (Valencia, CA), 1× TE (pH 8.0), and 1% Tween-20 was added to a LCM cell cap, and mixed with the cells by pipetting multiple times. The 15-μl suspension was transferred to its matching Eppendorf tube, vortexed for 1 minute, and incubated at 55°C for 3 hours.
The amount of DNA from a typical 3000 cell LCM cap was measured by quantitative PCR analysis using primers targeting PGK1 and TFRC genes. Five μl of lysed DNA was added to assay reagents to obtain a 70-μl PCR mix containing 1× ABI TaqMan master mix, 4 μmol/L 5′ primer, 4 μmol/L 3′ primer, and 2 μmol/L probe. For each sample, triplicates of 20 μl of PCR reactions were run and a standard curve used to determine the amount of DNA containing the probed locus as described in the quantitative PCR and relative copy number section (see below). The calculated concentrations of the PGK1 and TFRC loci in μg/μl were averaged and used to calculate the total μg of isolated DNA per LCM cap.
WGA of DNA from LCM Cells
MDA was performed using the GenomiPhi WGA kit (Amersham) according to kit instructions with some modifications for LCM samples. Five μl of lysed DNA from LCM cells was added to 20 μl of sample buffer, heated at 95°C for 3 minutes, and then cooled on ice for 5 minutes. Twenty-five μl of reaction mix (22.5 μl of reaction buffer plus 2.5 μl of ø29 polymerase) was added and the resulting samples were incubated at 30°C for 16 hours. After heat denaturation at 65°C for 10 minutes, each sample was diluted twofold using 1× TE (pH 8.0) and its DNA concentration was measured using the Hoechst dye assay (Bio-Rad). Typically 12 to 16 μg of amplified DNA product was obtained from a 50-μl reaction mixture.
TaqMan SNP Genotyping
TaqMan genotyping was performed using an ABI PRISM 7900HT sequence detection system following the manufacturer’s instructions (Applied Biosystems, Foster City, CA). Assays targeting 12 different genetic loci were designed through ABI’s Assay-by-Design service and used for genotyping analysis. Duplicate reactions were run for each assay with a 384-well plate, with control DNA samples run on the same plate. PCR was performed with 20 ng of input DNA, 1× ABI TaqMan master mix, 1 μmol/L of each primer, and 0.25 μmol/L of each probe. Fluorescence intensities (arbitrary units) of the two probes were plotted and genotype calling was performed using predefined calling parameters. Calling parameters for each genotype were determined beforehand based on the clustering of a panel of 101 genomic DNA samples obtained from Coriell. Briefly, we first defined the plot regions that contained allele 1, allele 2, or heterozygous genotypes using lines with lower and upper intensities and slopes (eg, as shown in Figures 1B, 3, and 4A). Genotyping data points that fell within these defined regions were called, data points that fell outside these regions, and those below intensity values as defined by no template controls were considered no calls. Miscalls were assigned to data points that were in contradiction to a genotype call made by either an unamplified sample of the same tissue or a consensus call based on all of the replicate assays performed on the same amplified tissue sample (pooled, unpooled, different cell numbers, and so forth). The consensus calls based on amplified material were always in agreement with the accurate genotype calls from unamplified samples when both sets of data were available. Calling rates (Tables 1 and 3) were the percentage of calls (accurate calls plus miscalls where applicable) out of all assays performed. Accurate calling rates (Table 2) were the percentage of correct genotype calls out of all of the genotyping assays performed.
Figure 3.
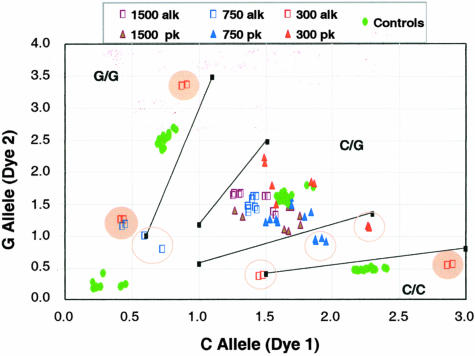
Effect of lysis procedure on a TaqMan SNP-genotyping assay for C/G16996 of the ERBB3 gene. LCM colon cell caps containing 1500 cells (8 caps), 750 cells (12 caps), and 300 cells (8 caps) were lysed by a proteinase K (pk) or an alkaline (alk) lysis protocol. All samples should give a heterozygote call. No calls are highlighted by open circles and miscalls are highlighted by filled circles. Shown on axes are arbitrary fluorescence units.
Figure 4.
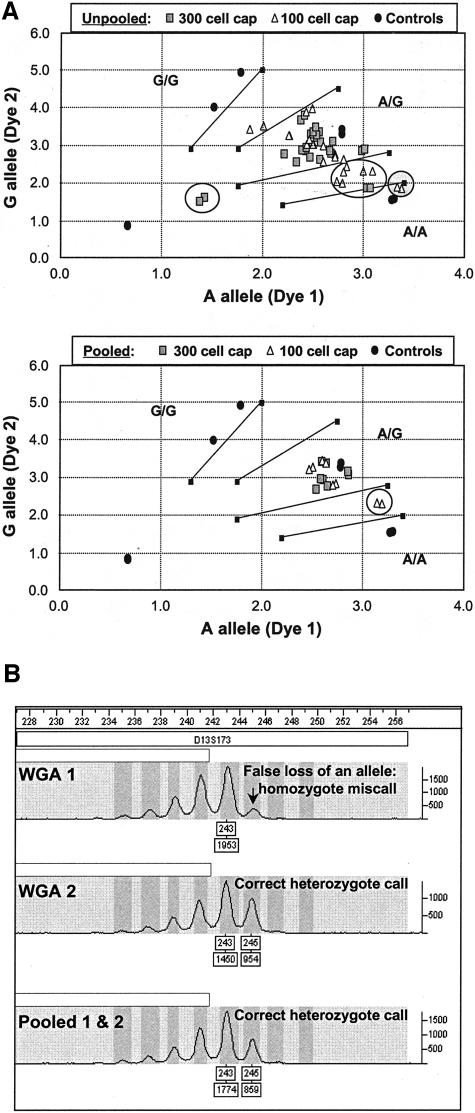
A: Effect of pooling WGA products from a 300 or a 100 LCM prostate cell cap on TaqMan SNP-genotyping assay for A/G36177 of the CYP3A5 gene: 24 WGA products were assayed in replicates for each cell number to afford 96 unpooled data points; 8 individual WGA products were assayed in replicates for each cell number to afford 32 pooled data points. No calls are highlighted by open circles and miscalls are highlighted by filled circles. Shown on axes are arbitrary fluorescence units. B: Effect of pooling WGA products from a 300 colon cell cap on a STR-genotyping assay using a 2-bp repeat marker D13S173.
Table 1.
Effect of LCM Cell Number and Lysis Method on Genotyping Performance of Whole Genome Amplified DNA using φ29 Polymerase-Based GenomiPhi Reagents
| No. of colon cells/cap | Lysis method | No. of amp. rxn./cap | SNP analysis | STR analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of loci assayed | Performance data | No. of loci assayed | Performance data | |||||||
| % Calling | % Cap-to-cap concordance | % Replicate concordance | % Calling | % Cap-to-cap concordance | % Replicate concordance | |||||
| 3000 | Alkaline | 2 | 4 | 96.9 | 100.0 | 95.8 | 10 | 99.0 | 100.0 | 98.3 |
| Proteinase K | 2 | 4 | 97.9 | 100.0 | 93.8 | 10 | 100.0 | 100.0 | 100.0 | |
| 1500 | Alkaline | 2 | 9 | 92.0 | 90.0 | 97.5 | 10 | 95.6 | 97.5 | 95.0 |
| Proteinase K | 2 | 6 | 90.5 | 92.9 | 100.0 | 9 | 98.8 | 100.0 | 96.7 | |
| 750 | Alkaline | 2 | 12 | 79.6 | 71.4 | 88.9 | 10 | 84.2 | 70.0 | 96.7 |
| Proteinase K | 2 | 8 | 95.9 | 94.1 | 97.2 | 10 | 95.0 | 98.3 | 97.5 | |
| 300 | Alkaline | 2 | 12 | 49.0 | 33.3 | 95.8 | 10 | 68.1 | 80.0 | 83.8 |
| Proteinase K | 2 | 8 | 83.0 | 88.2 | 97.0 | 10 | 96.7 | 95.0 | 91.3 | |
| 100 | Alkaline | 3 | 0 | nd* | nd* | nd* | 0 | nd* | nd* | nd* |
| Proteinase K | 3 | 7 | 75.0 | 100.0 | 97.6 | 9 | 97.9 | 91.7 | 91.7 |
Table 3.
WGA Methods for Amplification of LCM DNA: A Comparison Based on References and Our Results
| WGA method | No. of LCM cells/cap | Amp. yield as no. of assays | SNP analysis | STR analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Platform | Performance data | Platform | Performance data | |||||||
| % Calling | % Wrong | Total no. of assays run | % Calling | % Wrong | Total no. of assays run | |||||
| DOP-PCR* | 100¶ | ns‡‡ | nd§§ | nd§§ | nd§§ | nd§§ | Slap gel/EtBr | 85.0 | 0 | 20 |
| PEP* | 250 | 10 SNP or 30 STR | RFLP: Slap gel/EtBr | 94.7 | 0 | 19 | Slap gel/EtBr | 98.8 | 0 | 82 |
| PEP† | 5000 | 150 STR | nd§§ | nd§§ | nd§§ | nd§§ | ABI/florescence | nd§§ | nd§§ | ns‡‡ |
| SCOMP‡ | 48∥ | 32 STR | nd§§ | nd§§ | nd§§ | nd§§ | Slap gel/SYBR green | 94.4 | 5.6 | 18 |
| Pooled MDA§ | 100–200** | 2000 SNP or 4000 STR | TaqMan/florescence | 87.1 | 0 | 124 | ABI/florescence | 99.1 | 0 | 232 |
| Pooled MDA§ | 300–600†† | 2000 SNP or 4000 STR | TaqMan/florescence | 93.9 | 0 | 164 | ABI/florescence | 97.5 | 0.6 | 362 |
| Not amplified§ | 3000–6000 | 50 SNP or 50 STR | TaqMan/florescence | 58.9 | 0 | 56 | ABI/florescence | 97.5 | 0 | 40 |
Table 2.
SNP and STR Genotyping Results from Pooling Two or Three Amplification Reactions Starting with Proteinase K-Lysed Colon or Prostate Cells
| Tissue no. of patients | No. of LCM cells/cap | Avg. amp. fold | No. of WGA rxn. pooled | SNP analysis | STR analysis | ||||
|---|---|---|---|---|---|---|---|---|---|
| No. of loci assayed | Total no. of assays | % Accurate calling | No. of loci assayed | Total no. of assays | % Accurate calling | ||||
| Prostate | |||||||||
| 2 | 1500 | 4.5 × 103 | 3 | 6 | 48 | 95.8 | 10 | 80 | 100.0 |
| 4 | 750 | 1.0 × 104 | 3 | 6 | 96 | 94.8 | 10 | 160 | 95.0 |
| 4 | 300 | 2.3 × 104 | 3 | 6 | 96 | 99.0 | 10 | 160 | 96.9 |
| 4 | 100 | 6.4 × 104 | 3 | 6 | 96 | 89.6 | 10 | 160 | 100.0 |
| Colon | |||||||||
| 2 | 1500 | 4.5 × 103 | 2 | 6 | 48 | 92.9 | 9 | 72 | 100.0 |
| 2 | 750 | 1.0 × 104 | 2 | 12 | 96 | 100.0 | 10 | 80 | 90.6 |
| 2 | 300 | 2.3 × 104 | 2 | 12 | 96 | 94.1 | 10 | 80 | 97.5 |
| 2 | 100 | 6.4 × 104 | 3 | 7 | 56 | 81.5 | 9 | 72 | 97.2 |
STR Genotyping
STR genotyping was performed using Applied Biosystems Linkage Mapping Set of fluorescence-labeled primer pairs. A total of 45 different assays targeting various loci were used for this study. For each assay, replicate reactions were run for every DNA sample. PCR reactions were run with a 7.5-μl total volume containing 10 ng DNA, 1× ABI True Allele master mix, and 0.2 μl ABI primers. Reactions were incubated at 95°C for 12 minutes, cycled 15 times at 95°C for 15 seconds, 55°C for 15 seconds, 72°C for 30 seconds, and cycled an additional 25 times at 89°C for 15 seconds, 55°C for 15 seconds, 72°C for 30 seconds, with a final extension at 72°C for 10 minutes. PCR products were pooled (for multiplexing) and run on an ABI PRISM 3700 DNA analyzer. Data analysis was performed using ABI PRISM Genotyper Software Version 3.7. Peak height ratios between 0.4 and 6 were called as heterozygotes, peak height ratios between 0.1 and 0.4 and that between 6 and 10 were defined as no calls. Accurate calling rates were the percentage of correct genotype calls of all of the genotyping experiments.
Quantitative PCR and Relative Copy Number Calculations
The relative copy numbers of four genes, GAPD, TBP, TFRC, and PPIA, were determined for WGA products from three different origins. The first set of samples (×4) were prepared by pooling three WGA products from 1500 LCM colon cell caps. The second set of samples (×4) were pooled from three WGA products originating from 100 LCM colon cell caps. The final set of samples (×6) were from WGA reactions using 5 ng of genomic DNA isolated from six different cell lines as templates. The concentration of each amplified DNA product was determined by a Picogreen fluorescence assay (Molecular Probes, Eugene, OR), and a stock solution at 3 ng/μl was prepared for each sample. Fifteen ng (5 μl) of each amplified DNA sample was added to assay reagents to obtain a 70-μl PCR mix containing 1× ABI TaqMan master mix, 4 μmol/L 5′ primer, 4 μmol/L 3′ primer, and 2 μmol/L probe. For each sample, triplicates of 20 μl of PCR were run. All PCRs were run in a 384-well plate using an ABI PRISM 7900HT detection system. The cycling program was one cycle of 50°C for 2 minutes, one cycle of 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute. A standard curve was generated for each genetic locus using a human genomic DNA standard from Promega (catalog no. G3041; Promega, Madison, WI). This curve was then used to determine the amount of DNA containing the probed locus in each sample. The relative copy number of each gene was calculated by dividing the quantity of genomic DNA measured using TaqMan by the quantity of total DNA determined using Picogreen. The average relative copy number for the four samples of pooled WGA LCM DNA or the six samples of amplified genomic DNA was used for comparison.
Results
WGA of Purified Genomic DNA
The GenomiPhi kit was first evaluated for WGA of high-quality DNA templates. Two sets of trio DNA isolated from cultured cells were amplified and a consistent amplification from 5 ng to 5 μg (103-fold) was observed. Allele representation was assessed by both SNP- and STR-genotyping assays using PCR-based high-throughput fluorescence detection systems. The genotyping results from six different WGA products assayed in replicates (data not shown) demonstrated an excellent concordance with that from unamplified DNA. Both sets of trio samples presented allelic calls that were consistent with hereditary predictions.
Isolation of Genomic DNA from LCM Cells
Cancer and normal cells were isolated by LCM from colon and prostate to ensure homogeneous cell populations. Representative microscopic images taken during the LCM process are shown in Figure 1A. To extract DNA from cultured cells or blood, an alkaline lysis protocol was recommended by both Dean and colleagues20 and the GenomiPhi kit to yield 5 μl of DNA sample. For LCM cells, it was necessary to slightly modify the lysis procedure and minimize the lysis volume to a practical limit of 15 μl. To accommodate this larger extraction volume, the amount of GenomiPhi’s sample buffer was optimized and only one third of the lysis mixture (5 μl) was directly used for the subsequent WGA reaction. Attempts to purify or concentrate DNA from lysis mixtures, including ethanol precipitation and affinity column chromatography, led to inconsistent WGA results (data not shown). Using the modified alkaline lysis procedure, both 3000 cell caps and 1500 cell caps provided satisfactory results for SNP and STR genotyping. A representative SNP assay is shown in Figure 1B.
Because LCM is a laborious process, it is desirable to reduce the number of cells needed for WGA. Furthermore, most biopsy samples are limited in quantity and in many cases it is impractical to obtain 1500 to 3000 cells of a homogeneous cell population. To identify the lower limit of LCM cells necessary for whole genome MDA, we performed cell number titration experiments using LCM colon cell caps ranging from 100 to 3000 cells. To standardize the procedure for generating LCM caps containing desired cell numbers, the cells within a typical 1000-μm2 area of an 8-μm-thick tissue sample were counted and used as a reference to determine the area required to generate a cap containing the desired number of cells. To assess the accuracy of our cell collection procedure, DNA samples isolated from five different 3000 cell caps were quantified using TaqMan quantitative PCR. The results showed that the amount of extracted DNA ranged from 21.0 ng to 54.4 ng, with an average of 38.4 ng. This value is slightly higher than, but reasonably close to, the expected 21.3 ng calculated based on the estimation that there is 7.1 pg of DNA per diploid human cell.21
WGA of Genomic DNA from LCM Cells
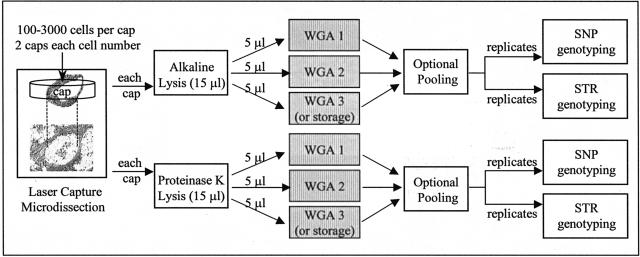
To detect potential allele bias or allele dropout that might have occurred during WGA, the amplified materials were directly compared to the corresponding unamplified DNA isolated in parallel. To ensure a meaningful assessment, we focused on highly heterozygous SNP and STR loci where the heterozygosity rates in the general population were estimated to be greater than 30% for SNP assays and greater than 70% for STR assays. Because proteinase K cell lysis conditions are milder for nucleic acids and have been found advantageous for amplifying DNA from LCM cells,15 we performed a series of experiments to compare the quality of SNP and STR genotyping data using WGA products originating from 3000, 1500, 750, 300, and 100 colon cell caps that were lysed by either KOH or proteinase K. The sample preparation and data analysis process is illustrated in Figure 2.
Figure 2.
Schematic sample preparation and data analysis.
Colon samples from two different patients were used for LCM, except for the 3000 cell caps, in which only one patient’s tissue was tested. As illustrated in Figure 2, for each tissue sample, two caps were microdissected for each cell number: one cap was lysed with the alkaline lysis method, the other with the proteinase K lysis method. Each lysis mixture (15 μl) was divided into three equal aliquots (3 × 5 μl), two or three of which were separately amplified, as specified. Thus, only a third of the lysed cells from each cap were present in each WGA reaction mixture, eg, from a 3000 cell cap only 1000 cells were added to each WGA reaction. An optional pooling step after WGA will be addressed in detail later. WGA products were analyzed in replicates for each SNP and STR assay targeted to specific genomic loci. This entire process, starting from LCM and ending with genotyping analysis, was performed in duplicate to evaluate the cap-to-cap reproducibility.
Table 1 summarizes experiments performed and the corresponding SNP- and STR-genotyping performance data. The number of genomic loci tested by SNP and STR assays for each sample is listed. SNP genotype calls were made based on data clustering patterns obtained using a panel of control genomic DNA. STR genotype calls were made based on observed peak ratios between the two alleles. The concordance between the two caps and two replicate assays, and the accurate calling rates were calculated according to the genotypes determined by consensus data and/or an unamplified sample. At low cell numbers, the proteinase K lysis method consistently showed better performance statistics. For example, when 300 cell caps were used, proteinase K gave an 83.0% SNP calling rate and a 96.7% STR calling rate; whereas the corresponding rates for alkaline lysis were only 49.0% and 68.1%, respectively. Importantly, the concordance between replicate assays was consistent throughout irrespective of the lysis method used (>80% in every case), indicating that these genotyping assays are highly reproducible. However, the cap-to-cap concordance seemed to be affected by the specific lysis method applied, eg, the cap-to-cap concordance dropped to as low as 33.3% for 300 cell caps when the alkaline lysis method was used. This decrease in concordance rate correlates with the lower percentage of calling rate for these samples (49.0%) and suggests that the observed assay failures may be mainly because of the lysis inconsistencies of the alkaline method. These failures appeared to be caused almost exclusively by a loss of allelic balance, as illustrated in Figure 3 for a SNP-genotyping assay in which the colon sample is a C/G heterozygote. Alkaline lysis (open squares in Figure 3) of 750 and 300 cell caps resulted in G/G or C/C homozygote miscalls for the C/G heterozygote samples. In contrast, proteinase K lysis did not produce any miscalls, although a few no calls were generated.
Pooling of WGA Products Improves Genotyping Accuracy
Because the allele bias introduced during WGA may arise from arbitrary events such as biased priming at an exponential amplification phase, we investigated whether pooling individual WGA reactions would minimize some of these differences and afford more reproducible and accurate allele representation. Thus, following the sample preparation process illustrated in Figure 2 and using DNA extracted from proteinase K-lysed microdissected prostate and colon cells, two or three separate WGA reactions were run and pooled, and the effect on genotyping was examined. Table 2 summarizes the information on the experiments performed, the corresponding amplification yields, and the SNP- and STR-genotyping performance statistics. Two or four different patients’ tissue samples were used for each cell number shown. Each cell number entry also represents duplicate caps and all assays were done in replicates (see Figure 2). A total of 48 to 160 individual assays were performed for each starting cell number, taking into account the number of samples, replicates, and loci analyzed. The accurate SNP calling rate ranged from 82 to 100% and the accurate STR calling rates are between 91% and 100%. Comparing the same cell number data highlighted in Tables 1 and 2 (percent calling in bold), it is clear that the pooling resulted in better calling rates for TaqMan SNP analysis. For example, when 750 cell caps were used, pooled WGA gave a 100.0% accurate SNP calling rate (Table 2) whereas the total calling rate (including a few miscalls) for an unpooled sample was 95.9% (Table 1).
Shown in Figure 4A are representative TaqMan assay results obtained using unpooled and pooled WGA products from 300 cell caps and 100 cell caps. The dramatic improvement in data clustering because of the pooling is evident. Although two miscalls and many no calls were made for the unpooled WGA samples, there were no miscalls and only two no calls for the pooled samples. The improvement on STR calling is not as obvious because the calling rates were already very high without pooling. However, the benefit of pooling was observed occasionally by virtue of more clearly defined allele peaks. Illustrated in Figure 4B is an example in which an unpooled sample generated a homozygote miscall, whereas the pooled sample produced the correct heterozygote call. It is worth noting that wrong SNP or STR genotype calls were very rare for pooled WGA products from proteinase K-lysed cells, and that the vast majority of failed assays were because of no calls. In addition to the results listed in Table 2, we have genotyped 16 prostate tissue samples using pooled WGA products from 3000 alkaline-lysed cell caps under a high-throughput setting. Accurate calling rates of 97% and 98% were achieved for 23 SNP assays and 21 STR assays, respectively.
Locus Representation of WGA Products
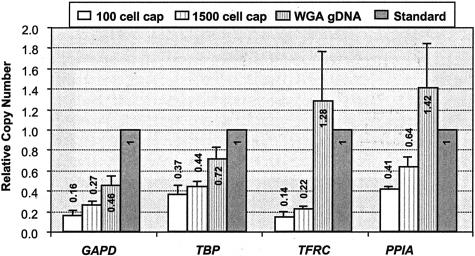
TaqMan quantitative PCR experiments targeting four different genetic loci were used to assess locus representation of pooled WGA products. WGA products from four 100 cell caps (pooled 3 × 33 cell WGA) and four 1500 cell caps (pooled 3 × 500 cell WGA), as well as WGA products from six different 5-ng genomic DNA samples isolated from cultured cells, were analyzed and compared to an unamplified genomic DNA standard. The resulting relative copy numbers for GAPD, TBP, TFRC, and PPID genes for each sample are illustrated in Figure 5, with the copy number of the standard set to 1. The WGA products of DNA isolated from cultured cells showed gene copy numbers that are close to the standard unamplified DNA. However, the genes appeared to be consistently underrepresented in amplified LCM samples (0.14 to 0.64), and the 100 cell samples seem to have lower genetic content than the 1500 cell samples (comparing white bars to neighboring striped bars). Nonetheless, the data shows that for each WGA sample, less than or equal to threefold variation was observed among the genes tested (comparing the bars of the same fill pattern), similar to what was reported for whole genome MDA of DNA from cultured cells.20
Figure 5.
Relative gene copy number analysis using an unamplified genomic DNA as a standard. Pooled WGA products from microdissected colon cells were analyzed using TaqMan quantitative PCR assays and compared with a WGA product from 5 ng of genomic DNA isolated from cultured cells. The copy numbers are averages of four or six amplified samples, and the error bars show corresponding SD.
Discussion
Studies on genomic aberrations in cancer require large quantities of DNA from pure cancer cell populations. Thus, a WGA method that can greatly amplify the minute quantities of DNA available from laser capture microdissected cells is an important prerequisite for genome-wide genetic evaluations of clinical tissue samples. The successful application of ø29 polymerase in WGA of linear genomic DNA was the first WGA method that did not rely on thermal cycling.20 By using DNA from cultured cells and blood as templates, 104-fold amplification products from as low as 0.3 ng of source DNA showed excellent locus representation and supported successful SNP analysis by RFLP. A commercial WGA kit based on the same strand displacing amplification principle has been developed by Amersham and was used in the present study.
After the completion of the present study, a report on applying this WGA method to analyze various clinical samples, excluding LCM tissue samples, was published.22 Because DNA isolated from LCM cells often sustains various lesions during processing and dissection, and ø29 polymerase-based WGA was believed compromised by damaged DNA,20 there could be significant hurdles for its successful application. To this end, we have developed a strategy that utilizes a commercially available ø29-based WGA kit to amplify LCM DNA, and demonstrated that high-throughput SNP- and STR-genotyping analysis are supported by up to 6 × 104-fold WGA starting from as few as 100 LCM cells.
WGA of LCM Samples for SNP and STR Genotyping
To whole genome amplify LCM DNA, we devised and implemented several modifications to published protocols. First, because of low quantities of LCM DNA, cell lysis mixtures were used directly for WGA without any purification. Consequently, it was necessary to minimize the lysis volume and to apply only a fraction of the lysed cells to each WGA reaction. Secondly, it was necessary to use proteinase K lysis in place of alkaline lysis when less than 1500 LCM cells were available. With alkaline lysis, allelic imbalance was observed in both SNP- and STR-genotyping assays, as evidenced by genotype miscalls. We speculate that under alkaline lysis conditions random segments of genomic DNA might remain bound to cellular proteins after digestion, which could impede primer binding to those segments and result in locus- or allele-biased amplification when only a small number of DNA templates are present (eg, 100 cells).
We found that the pooling of replicate or triplicate WGA reactions was very beneficial for subsequent genotyping analysis, and through pooling any allele bias relating to limited template quantity was consistently remedied. For example, pooled WGA products originating from a 300 cell cap (Table 2; 94.1% SNP and 97.5% STR accurate calling rates) consistently outperformed that from unpooled products from a 300 cell cap (Table 1; 83.0% SNP and 96.7% STR total calling rates). In practice, a 100 cell cap (ie, pooled 3 × 33 cells) may be a lower limit for this WGA method because a moderate 80% SNP accurate calling rate was observed. In agreement with our observation, a previous report indicated that at least 0.3 ng of DNA (ie, ∼42 cells) was needed to achieve a minimal (threefold) genomic locus bias.20 The effectiveness of pooling likely reflects the randomness of primer hybridization and WGA initiation, suggesting that additional division of the lysed cells may produce even better results. For example, a 150-cell sample can be divided into 5 × 30 cell amplification reactions. Another pragmatic outcome of the pooling was that a larger quantity of DNA product was generated (usually more than 30 μg) from the available LCM cells. This should be particularly useful for genome-wide genetic analysis involving needle biopsy samples in which only hundreds of cells may be available.
Our experiments also showed that the slide preparation and LCM procedure used here seemed to underestimate the number of captured cells by approximately twofold. This could be because of the presence of more than one single layer of cells in the 8-μm-thick tissue sections. Attempts at dissecting fresh-frozen tissue samples to thinner sections were not always successful. Because we consistently performed the cell extraction process in a similar manner, this approximately twofold underestimation should not change the general conclusions of the present study.
Locus Representation of WGA-Amplified LCM Samples
Locus representation of pooled amplification products was examined using TaqMan quantitative PCR assays. The relative genetic locus bias of WGA products from 5 ng of isolated genomic DNA (unpooled) was within threefold, and the genomic content on average was similar to the unamplified DNA standard (0.97 versus 1). However, in amplified LCM samples the genes were underrepresented compared to the standard, with an average copy number of 0.27 for the 100 cell caps and 0.39 for the 1500 cell caps, respectively. This genetic underrepresentation is probably because of the formation of nonspecific, nongenomic artifacts promoted by the heat denaturation and annealing process, a phenomenon that was also observed previously by Dean and colleagues,20 and probably similar to that was reported for DOP-PCR amplification.11 It is worth noting that the observed underrepresentation was somewhat more exaggerated for the 100 cell caps than the 1500 cell caps. Considering that the underrepresentation was not as pronounced for WGA products from 5 ng of high-quality DNA, it is plausible that the nongenomic DNA products are from polymerization of cross-hybridized random primers that become relatively more dominant when less starting DNA template is available for hybridization. Despite this underrepresentation in overall genetic content, a well-balanced genetic distribution was observed for the amplified LCM samples. The relative gene copy numbers among the four genes were confined within a threefold range, ie, GAPD/TBP/TFRC/PPIA = 1.2/2.0/1/2.9 for 100 cell caps and 1.1/2.6/1/2.9 for 1500 cell caps, respectively, and did not seem to hamper the subsequent SNP or STR genotype analysis. We did not attempt to eliminate the heat denaturation step because we speculated that any heat-induced DNA damage was probably superseded by damage caused by various tissue manipulation processes including LCM.
Comparison to Other WGA Methods
One broadly used WGA method is DOP-PCR that uses partially degenerate primers for genomic amplification. When less than 100 source DNA cells are used, the fixed portion of the primers can promote preferential amplification of certain sequences over others resulting in loss of locus representation.11 In fact, efforts have been made to intentionally reduce genome complexity using custom-designed degenerate primer sets for genome-wide SNP genotyping through array hybridization.23 Theoretical calculations have also predicted that tagged PCR cannot provide complete genomic coverage at low cell numbers.24 Because MDA uses completely randomized primers, broader genomic coverage and less locus bias were achieved.20 A performance comparison between DOP-PCR and pooled MDA for LCM cells is shown in Table 3, based on our results and that from a widely cited reference.15 The limited STR data available for DOP-PCR products from cultured cells shows a mediocre performance level. In contrast, high-fidelity amplification of STR markers were observed in this study (Tables 1, 2, and 3), demonstrating that the whole genome MDA method can reliably amplify LCM DNA and provide suitable templates for robust microsatellite marker analysis. Whereas a recent report has demonstrated that the quality of DOP-PCR products are susceptible to long-term storage at −20°C,13 we have not observed any deterioration in performance after storing MDA products at −20°C for several months, as indicated by consistent SNP genotype calling before and after the storage.
PEP is another widely applied WGA method that utilizes completely degenerate primers, usually 15 nucleotides in length, and is theoretically regarded as a better choice for amplifying low amounts of source DNA.24 In practice, PEP typically generates only a moderate ∼30-fold amplification with a ∼100-fold locus bias.20 A recent report demonstrated that STR analysis on an ABI sequencer was achieved using PEP products from 5000 LCM cells (Table 3).16 However, a comparison with our data for unamplified LCM DNA (last row of Table 3) revealed that the effective amplification of this particular PEP protocol was not significant. It is worth noting that the same PEP procedure was also used to successfully analyze formalin-fixed paraffin-embedded tissue samples.16 Formalin-fixed tissue samples were not tested in the present study. A low-throughput slap-gel analysis of STR markers was reported to require PEP-amplified DNA from approximately eight cells per assay (250 cell cap for 30 STR assays) with very few data points.15 In comparison, pooled MDA provided an amplification from 100 to 600 cell caps that supports ∼4000 STR assays (ie, 7 to 40 assays per cell) with a reproducibly high success rate of 98% as calculated from hundreds of individual assay results. Considering that all PCR-based WGA methods are likely to generate stutter products caused by primer slippage over repetitive sequences, whole genome MDA may be the best-suited WGA method for downstream STR genotype analysis.
SCOMP is another method that has been used for amplifying DNA from LCM cells.18 This method focuses on amplification of DNA isolated from less than 50 cells and utilizes restriction digestion followed by linker-adaptor ligation and PCR amplification using primers containing linker sequences. This methodology has not been as widely used as DOP-PCR or PEP, probably because of its complicated cell isolation procedure and extra manipulations required by the adaptor ligation step. Its STR performance statistics are summarized in Table 3. Because the only data available for LCM cells was from formalin-fixed samples, it is not possible to make a direct comparison with our pooled MDA method. However, because of the random distribution of restriction sites in genomic DNA, a wide range of lengths of digested fragments is expected. Because PCR efficiency is inversely correlated with the length of an amplicon, locus bias is likely for SCOMP.
Biased amplification at heterozygous SNP loci introduced during WGA has not been systematically dealt with before the current study. Allele bias in PEP products has been reported,25 so has successful RFLP-based SNP analysis consuming ∼25 cells equivalent per assay (Table 3). RFLP seems to be a popular method for SNP analysis of WGA products even though it is not amenable to a high-throughput platform.15,20,26 In fact, RFLP may not be a sensitive method for allele bias assessment because a longer fragment may be overestimated as a result of increased staining or probe binding than the companion shorter fragment. We have found that TaqMan is a very convenient and flexible high-throughput SNP-genotyping platform to use even with its strict requirements on DNA quality. With our pooled MDA method, a high success rate of TaqMan SNP genotyping was achieved with ample amplification (Tables 1, 2, and 3). To our knowledge, this is the first successful application of TaqMan SNP genotyping using WGA-amplified DNA from LCM cells.
In conclusion, we have developed a practical solution for WGA of DNA from laser capture microdissected clinical samples of normal and cancer tissue. We validated the method by analyzing hundreds of SNP- and STR-genotyping assays in high-throughput platforms. Utilization of proteinase K for cell lysis and the subsequent split-pool MDA strategy seems to be by far the most productive and high-performing WGA method presently available and supports robust SNP and STR genotyping for LCM caps containing as few as 100 cells. If a faster alkaline lysis of LCM cells is desirable, our results indicate that a lower limit of ∼1500 LCM cell caps may be used for reliable genotype analysis. We anticipate that this pooling strategy will be applicable to a wide range of genome-wide or site-specific amplification reactions, including both PCR- and MDA-based nucleic acid amplifications.
Acknowledgments
We thank Olga Charlat, William Wojcicki, Dr. Chris Leo, and Tina Tong for technical assistance with STR-genotyping assays, gene copy number assays, and laser capture microdissection; the scientists at Amersham who provided the GenomiPhi kits for this study before their official launch; and Dr. Tracy Zimmermann for her critical reading of this manuscript.
Footnotes
Address reprint requests Jia Liu Wolfe, Ph.D., Department of Molecular Biology, Massachusetts General Hospital, 50 Blossom St., Boston, MA 02114. E-mail: wolfe@molbio.mgh.harvard.edu.
Supported by Variagenics, Inc. and Nuvelo, Inc.
References
- Weber BL. Cancer genomics. Cancer Cell. 2002;1:37–47. doi: 10.1016/s1535-6108(02)00026-0. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, Lopez-Guillermo A, Grogan TM, Miller TP, LeBlanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM. Lymphoma/Leukemia Molecular Profiling Project: The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- Thiagalingam S, Foy RL, Cheng KH, Lee HJ, Thiagalingam A, Ponte JF. Loss of heterozygosity as a predictor to map tumor suppressor genes in cancer: molecular basis of its occurrence. Curr Opin Oncol. 2002;14:65–72. doi: 10.1097/00001622-200201000-00012. [DOI] [PubMed] [Google Scholar]
- Quon KC, Berns A. Haplo-insufficiency? Let me count the ways. Genes Dev. 2001;15:2917–2921. doi: 10.1101/gad.949001. [DOI] [PubMed] [Google Scholar]
- Forozan F, Karhu R, Kononen J, Kallioniemi A, Kallioniemi OP. Genome screening by comparative genomic hybridization. Trends Genet. 1997;13:405–409. doi: 10.1016/s0168-9525(97)01244-4. [DOI] [PubMed] [Google Scholar]
- Andrew SE, Peters AC. DNA instability and human disease. Am J Pharmacogenomics. 2001;1:21–28. doi: 10.2165/00129785-200101010-00003. [DOI] [PubMed] [Google Scholar]
- Roses AD. Genome-based pharmacogenetics and the pharmaceutical industry. Nat Rev Drug Discov. 2002;1:541–549. doi: 10.1038/nrd840. [DOI] [PubMed] [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- Cheung VG, Nelson SF. Whole genome amplification using a degenerate oligonucleotide primer allows hundreds of genotypes to be performed on less than one nanogram of genomic DNA. Proc Natl Acad Sci USA. 1996;93:14676–14679. doi: 10.1073/pnas.93.25.14676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Okita K, Shiraishi K, Kusano N, Furuya T, Oga A, Kawauchi S, Kondoh S, Sasaki K. Detection of genetic alterations in pancreatic cancers by comparative genomic hybridization coupled with tissue microdissection and degenerate oligonucleotide primed polymerase chain reaction. Oncology. 2002;62:251–258. doi: 10.1159/000059573. [DOI] [PubMed] [Google Scholar]
- Grant SF, Steinlicht S, Nentwich U, Kern R, Burwinkel B, Tolle R. SNP genotyping on a genome-wide amplified DOP-PCR template. Nucleic Acids Res. 2002;30:e125. doi: 10.1093/nar/gnf125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci USA. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietmaier W, Hartmann A, Wallinger S, Heinmoller E, Kerner T, Endl E, Jauch KW, Hofstadter F, Ruschoff J. Multiple mutation analyses in single tumor cells with improved whole genome amplification. Am J Pathol. 1999;154:83–95. doi: 10.1016/S0002-9440(10)65254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang VW, Bell DA, Berkowitz RS, Mok SC. Whole genome amplification and high-throughput allelotyping identified five distinct deletion regions on chromosomes 5 and 6 in microdissected early-stage ovarian tumors. Cancer Res. 2001;61:4169–4174. [PubMed] [Google Scholar]
- Klein CA, Schmidt-Kittler O, Schardt JA, Pantel K, Speicher MR, Riethmuller G. Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc Natl Acad Sci USA. 1999;96:4494–4499. doi: 10.1073/pnas.96.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklein NH, Erbersdobler A, Schmidt-Kittler O, Diebold J, Schardt JA, Izbicki JR, Klein CA. SCOMP is superior to degenerated oligonucleotide primed-polymerase chain reaction for global amplification of minute amounts of DNA from microdissected archival tissue samples. Am J Pathol. 2002;161:43–51. doi: 10.1016/S0002-9440(10)64155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Vega FM, Dailey D, Ziegle J, Williams J, Madden D, Gilbert DA. New generation pharmacogenomic tools: a SNP linkage disequilibrium map, validated SNP assay resource, and high-throughput instrumentation system for large-scale genetic studies. Biotechniques. 2002;Suppl:S48–S54. [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, Driscoll M, Song W, Kingsmore SF, Egholm M, Lasken RS. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Struhl K. New York: John Wiley & Sons,; Current Protocols in Molecular Biology Online. 2003 Table A. 1B. 1. [Google Scholar]
- Hosoro S, Faruqi AF, Dean FB, Du Y, Sun Z, Wu X, Du J, Kingsmore SF, Egholm M, Lasken RS. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003;13:954–964. doi: 10.1101/gr.816903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan B, Charest A, Dowd JF, Blumenstiel JP, Yeh RF, Osman A, Housman DE, Landers JE. Genome complexity reduction for SNP genotyping analysis. Proc Natl Acad Sci USA. 2002;99:2942–2947. doi: 10.1073/pnas.261710699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Arnheim N, Waterman MS. Whole genome amplification of single cells: mathematical analysis of PEP and tagged PCR. Nucleic Acids Res. 1995;23:3034–3040. doi: 10.1093/nar/23.15.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunio T, Reima I, Syvanen AC. Preimplantation diagnosis by whole-genome amplification, PCR amplification, and solid-phase minisequencing of blastomere DNA. Clin Chem. 1996;42:1382–1390. [PubMed] [Google Scholar]
- Snabes MC, Chong SS, Subramanian SB, Kristjansson K, DiSepio D, Hughes MR. Preimplantation single-cell analysis of multiple genetic loci by whole-genome amplification. Proc Natl Acad Sci USA. 1994;91:6181–6185. doi: 10.1073/pnas.91.13.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]