Differentiation-induced replication-timing changes are restricted to AT-rich/long interspersed nuclear element (LINE)-rich isochores (original) (raw)
Abstract
The replication timing of some genes is developmentally regulated, but the significance of replication timing to cellular differentiation has been difficult to substantiate. Studies have largely been restricted to the comparison of a few genes in established cell lines derived from different tissues, and most of these genes do not change replication timing. Hence, it has not been possible to predict how many or what types of genes might be subject to such control. Here, we have evaluated the replication timing of 54 tissue-specific genes in mouse embryonic stem cells before and after differentiation to neural precursors. Strikingly, genes residing within isochores rich in GC and poor in long interspersed nuclear elements (LINEs) did not change their replication timing, whereas half of genes within isochores rich in AT and long interspersed nuclear elements displayed programmed changes in replication timing that accompanied changes in gene expression. Our results provide direct evidence that differentiation-induced autosomal replication-timing changes are a significant part of mammalian development, provide a means to predict genes subject to such regulation, and suggest that replication timing may be more related to the evolution of metazoan genomes than to gene function or expression pattern.
It is generally presumed that early replication is a necessary (albeit not sufficient) condition for transcription, whereas late replicating sequences are assembled into transcriptionally inactive chromatin; however, the evidence for this assumption has been far from conclusive (1, 2). Recent whole-genome studies have confirmed a positive correlation between early replication and the probability of gene expression (3, 4), but have also identified many transcriptionally active and silent genes replicating at all times during S phase. The extent to which replication timing is regulated during development has been even more difficult to substantiate. Very few genes have been demonstrated to replicate at different times in different cell lines, and most genes analyzed do not change replication timing in different cell types (5). Most of these genes have been analyzed in established, often karyotypically unstable cell lines, so the extent to which these differences in replication timing could have resulted from chromosome rearrangements is unclear. Moreover, no examples of programmed changes in replication timing have been observed in a cultured-cell-differentiation system other than those that accompany X-chromosome inactivation (6, 7). These limitations have precluded the ability to directly demonstrate a relationship between replication timing and developmentally regulated patterns of gene expression; thus, it has not been possible to predict how many or what types of genes might be subject to replication timing control. Nonetheless, because chromatin proteins are reassembled at each round of replication, it is reasonable to presume that DNA replication plays some role in epigenetic regulation of gene expression (8).
Addressing the significance of replication timing to developmentally regulated programs of gene expression will require cultured-cell-differentiation systems in which changes in replication control can be elicited in homogeneous cell populations. The recent development of systems for directed changes in cell fate (9, 10) prompted us to search for autosomal genes that are subject to replication timing changes during the differentiation of mouse embryonic stem cells (ESCs) to neural precursors (neural stem cells, NSCs). Here, we evaluated the replication timing of 54 tissue-specific genes in mouse ESCs before and after differentiation to neural precursors. This analysis identified four genes that switch from early to late replication upon transcriptional down-regulation and five genes that switch from late to early replication upon activation, providing direct evidence for differentiation-induced changes in autosomal replication timing. Intriguingly, differentiation-induced replication timing changes were restricted to isochores rich in AT and long interspersed nuclear elements (LINEs), suggesting a relationship between replication timing and the evolution of metazoan genomes.
Materials and Methods
Cell Culture, Neural Differentiation, and BrdUrd Labeling. ESCs (46C) were routinely grown without feeders on a 0.1% gelatin-coated flask in a leukemia inhibitory factor-supplemented Glasgow minimum essential medium. Differentiation in N2B27 is described in ref. 9. For BrdUrd labeling, both undifferentiated and differentiated cells were incubated with 50 μM BrdUrd for 1 h before trypsinization and ethanol fixation.
Cell Cycle Fractionation and Isolation of BrdUrd-Labeled DNA. BrdUrd-labeled, fixed cells were resuspended in PBS (2.0 × 106 cells per ml), stained with propidium iodide (50 μg/ml) for 30 min in the presence of RNaseA (0.5 mg/ml) and then sorted into two cell cycle fractions (early and late S phase) (Fig. 1 A and B) by flow cytometry, as described in refs. 11 and 12. Isolation of BrdUrd-labeled DNA has been described (13, 14). Briefly, 40,000 cells were lysed in SDS-PK buffer (1 M NaCl/10 mM EDTA/50 mM Tris·HCl, pH 8.0/0.5% SDS/0.2 mg/ml PK/50 μg/ml glycogen), and DNA was extracted by phenol/chloroform extraction followed by ethanol precipitation. Extracted DNA was sonicated, incubated for 20 min at room temperature with anti-BrdUrd antibody (BD Biosciences) in 1× immunoprecipitation buffer (10 mM sodium phosphate, pH 7.0/0.14 M NaCl/0.05% Triton X-100), then added to 35 μg of rabbit anti-mouse IgG (Sigma) for another 20-min incubation. DNA–protein complex was precipitated by centrifugation, washed once with 1× immunoprecipitation buffer, and resuspended in digestion buffer (50 mM Tris·HCl, pH 8.0/10 mM EDTA/0.5% SDS/0.25 mg/ml PK) for overnight protein digestion at 37°C. Finally, immunoprecipitated DNA was collected by ethanol precipitation and resuspended in Tris-EDTA at a concentration of 250 cell equivalents per μl.
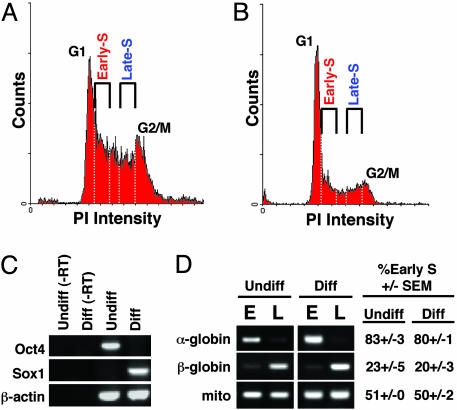
Fig. 1.
Experimental scheme of replication-timing analysis before and after differentiation. (A and B) Undifferentiated (A) and neural differentiated (B) ESCs were pulse-labeled with BrdUrd and then separated into populations from early or late S phase by flow cytometry. The white dotted lines represent the sorting windows for fractions. (C) Differentiation state as determined by RT-PCR. Oct4 (ESC-specific) is down-regulated after differentiation, whereas neural precursor-specific Sox1 is up-regulated. Samples marked -RT did not contain reverse transcriptase in the reaction. (D) Replication-timing analysis of control genes, α-globin (early-replicating in ESC), β-globin (late-replicating in ESC), and mitochondrial DNA (mito; replicates throughout the entire cell cycle) (28). The average and SEM of early S phase abundance (percentage of the total) were measured as described in Materials and Methods. Undiff., undifferentiated ESC; diff., differentiated ESC; E, early S-phase fraction; L, late S-phase fraction; PI, propidium iodide.
Replication-Timing Analysis by PCR. A control PCR (35 cycles) experiment was performed to confirm the enrichment of α-globin, β-globin, and mitochondrial DNA sequences in the expected fractions of immunoprecipitated BrdUrd-labeled DNA samples, early S phase, late S phase, and both fractions, respectively. We then determined whether a gene replicated in the first half or the second half of S phase based on conventional PCRs (35–42 cycles, determined empirically for each primer set) and subsequent ethidium bromide gel staining. For α-globin, β-globin, and mitochondrial DNA, a 23-cycle PCR followed by Southern hybridization (5) gave results comparable to ethidium bromide gel staining (data not shown). To quantify the relative enrichment in each fraction, we measured the intensity of the PCR products by using the image processing software imagej (http://rsb.info.nih.gov/ij). The relative abundance was determined by calculating the early S-phase percentage of total: (intensity of early S phase)/(intensities of early and late S phase combined). A percentage >60% and <40% in early S fraction was defined as early replicating (E) and late replicating (L), respectively. We categorized genes that showed 40–60% enrichment in early S phase as early replicating (E) because cells are synchronized after the completion of a 1-h BrdUrd pulse-labeling and genes that replicated in early middle S phase move into late S phase by the end of the pulse-labeling period.
Gene Expression Analysis by RT-PCR. RNA isolation was done by using a RNeasy RNA extraction kit (Qiagen, Valencia, CA). Extracted RNA was treated with DNaseI to eliminate genomic DNA, extracted with phenol/chloroform, and ethanol precipitated. Reverse transcription was performed by using random hexamer (Invitrogen) and SuperScript III (Invitrogen). For every primer set, we initially used serial dilutions of cDNA for PCR and stopped the reaction at every two or three cycles between 21 and 30 cycles to determine the cycle numbers that fall within the exponential range of the PCR, where the amount of PCR products of 2-fold serial dilutions should stay in a linear range. After cycle number determination, cDNA derived from undifferentiated and differentiated ESCs was analyzed by PCR and quantified by using imagej as in the replication timing analysis. For the GC-low (GCL) genes and the GC-high (GCH) genes analyzed in this study, a difference of >2.5- and 1.5-fold was defined as tissue-specific, respectively (GCL genes showed a higher average fold-change than GCH genes).
Gene Selection, GC Content Calculation, and Primer Design. For gene selection, we used three existing microarray databases of genes enriched in mouse ESCs and animal-derived NSCs (15–17) and selected genes that were reproducibly defined as either ESC-specific or NSC-specific in at least two of the three studies. For the ESC-GCL, ESC-GCH, NSC-GCL, and NSC-GCH categories, 36, 32, 24, and 25 primer sets, respectively, were initially designed, and a total of 54 genes presented in this study gave PCR results. For GC content calculations, 400 kb of surrounding DNA [200 kb upstream and downstream of the transcription start position (txStart)] was analyzed, based on the October 2003 mouse genome assembly (build 32) of the University of California, Santa Cruz, genome browser (http://genome.ucsc.edu). For high-throughput GC content calculations of large numbers of genes, 20-kb windows containing the txStart and transcription end positions were analyzed. The txStart and transcription end positions can be found on the University of California, Santa Cruz, genome browser. For those genes analyzed out to 400-kb windows, 20-kb windows were found to be representative of the entire isochore (Tables 2 and 3, which are published as supporting information on the PNAS web site). Primers were designed to amplify 200- to 650-bp fragments by primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi).
Analysis of Point Mutation Rate, LINE Density, and Gene Frequency Versus GC Content. A data set containing normalized 4-fold substitution rates of 14,790 mouse/human orthologous genes was kindly provided by H. Li and J. Chuang (University of California, San Francisco) (18). We were able to identify the GC content of a 20-kb window surrounding the txStart position for 8,097 genes, and these genes were used for the analysis. For LINE density, chrN_rmsk.txt files were downloaded from the University of California, Santa Cruz, genome browser (build 32, October 2003), and the percentage of repetitive sequences per 400 kb surrounding the txStart position was calculated for each gene and plotted against average GC content of the surrounding 400-kb region. For gene frequency versus GC content, 16,652 National Center for Biotechnology Information Reference Sequence (RefSeq) genes and GC content value of the 20-kb windows surrounding the txStart position were used.
Results and Discussion
Predicting Genes That Switch Replication Timing. Because there are so few examples of genes that change their replication timing, one challenge was how to predict which genes, if any, might be subject to developmental control over DNA replication in any given differentiation system. We reasoned that clues might be derived from a comparison of the α-globin and β-globin genes. These genes encode subunits of the same protein complex and are coordinately regulated, yet their replication timing regulation is very different: α-Globin is early-replicating and euchromatic in all tissues, whereas β-globin is late-replicating and heterochromatic in most tissues and early-replicating in erythroid cells (19–22). This example renders unlikely any hypotheses that relate replication-timing switches to gene function or expression pattern. One notable difference between these genes is that α-globin is in a GC-rich isochore, whereas β-globin is in an AT-rich isochore, which prompted us to examine the GC content of DNA surrounding genes whose replication timing has been determined in cell lines derived from different tissues (Table 2) (5, 23, 24). Strikingly, from this sampling, every gene (eight of eight) found to change replication timing was AT-rich (<43% GC; see Table 2).
Although it has long been suspected that early-replicating DNA is more GC-rich than late-replicating DNA (1, 2, 25), there have also been significant contradictory studies (26, 27). Recently, a genome-wide study of replication timing in a human lymphoblastoid cell line revealed a strong positive correlation between early replication and GC content (4). Hence, we considered the possibility that genes residing within AT-rich isochores are typically rendered late-replicating and must switch to early replication to be expressed. This hypothesis predicts that, during the differentiation of ESCs to NSCs, cell-type-specific genes localized within GC-rich isochores would replicate early in both cell types, whereas many genes within AT-rich isochores will change their replication timing in response to differentiation.
To monitor the differentiation of ESCs to NSCs, we took advantage of a cell line (46C) in which the expression of the GFP is under the control of the native, neural precursor-specific Sox1 promoter (9). When differentiated in N2B27 medium (9), 86–95% of 46C cells were GFP-positive by day 6 by visual inspection under the microscope, providing a convenient read-out for successful differentiation. Sox1 marks the proliferating neural progenitors before they exit the cell cycle, and the differentiation protocol used maintains these cells in the proliferating state, as confirmed by the DNA histograms of ESCs before (Fig. 1 A) and after differentiation (Fig. 1_B_). The up-regulation of Sox1 and down-regulation of Oct4 (an ESC-specific gene), respectively, also confirmed successful differentiation (Fig. 1_C_). To analyze replication timing, nascent DNA was pulse-labeled with BrdUrd, and cells were retroactively sorted by flow cytometry into populations from either early or late S phase (5, 11, 12, 14). After isolation of total genomic DNA from each fraction, nascent (BrdUrd-substituted) DNA was immunoprecipitated with anti-BrdUrd antibodies. The relative amounts of the genes of interest were then determined by PCR. To provide controls, we monitored the replication of α-globin and β-globin genes, previously shown to be early- and late-replicating in ESCs, respectively (28), and mitochondrial DNA, which replicates throughout the cell cycle (28) and is equally represented in early and late S-phase nascent DNA preparations (Fig. 1_D_).
To select ESC- and NSC-specific genes, we initially relied on existing microarray comparisons of gene expression in mouse ESC and animal-derived NSC (15–17). Because many discrepancies between the three existing studies have been noted (17), we selected genes that were reproducibly defined as either ESC-specific or NSC-specific in at least two of the three studies. Genes were then defined as residing within AT-rich (GCL) or GC-rich (GCH) isochores by analyzing 400 kb of surrounding DNA (Tables 2 and 3), which is sufficient to constitute an isochore, as defined in mammals (27). Finally, we carried out RT-PCR analysis to confirm the tissue-specific expression pattern of these genes during ESC differentiation, resulting in the adjustment of the actual expression pattern for some of the genes, which could be due to either errors in microarray or differences in the cell types and sources. This process narrowed down the number of gene candidates to 37 ESC- and NSC-specific genes, with 17 genes expressed in both cell types.
AT-Rich But Not GC-Rich Genes Switch Replication Timing. Fig. 2 shows the replication timing analysis for all 22 GCH genes. Regardless of expression pattern, every one of these genes replicated in the first half of S phase. Although the resolution of our study cannot rule out subtle replication-timing changes during early S phase (in fact, some of the genes fluctuate in their degree of early S-phase enrichment), our results strongly suggest that GC-rich genes do not undergo large changes in replication timing during differentiation. Moreover, because most cell types replicate euchromatin throughout the first half of S phase, as measured by the spatial distribution of replication sites (29, 30), changes in replication timing within this period are probably less consequential than those that cross the boundaries between euchromatin and heterochromatin replication.
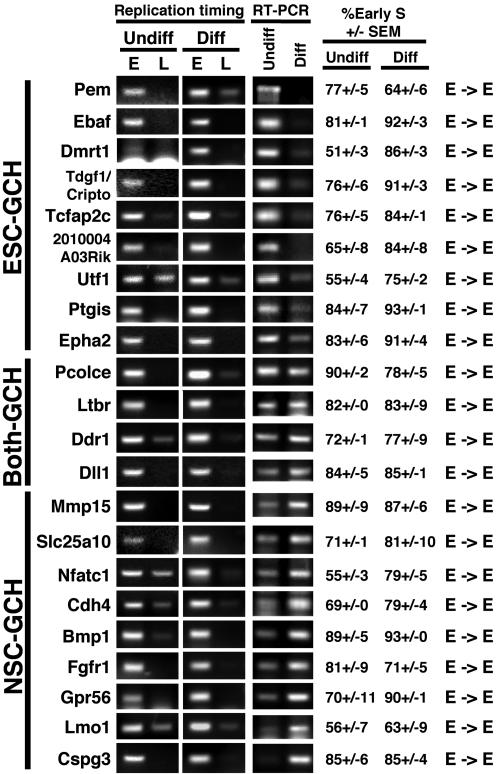
Fig. 2.
Replication-timing and RT-PCR analyses of GC-rich (GCH) genes. Replication-timing and RT-PCR analyses of genes residing in GC-rich isochores were carried out by using samples prepared from undifferentiated (Undiff.) and differentiated (Diff.) ESCs and categorized by gene expression patterns ESC-specific (ESC-GCH), NSC-specific (NSC-GCH), or expressed in both (Both-GCH). None of the primer sets used gave any PCR product without reverse transcription (data not shown). Similar results were obtained from multiple analyses of two independent experiments in which all genes were analyzed and two additional experiments in which a subset of genes was further analyzed. The mean and the SEM of early S phase abundance (percentage of the total) were measured as described in Materials and Methods. Shown are the gel photographs that were closest to the mean value. E→E indicates early replication before and after differentiation.
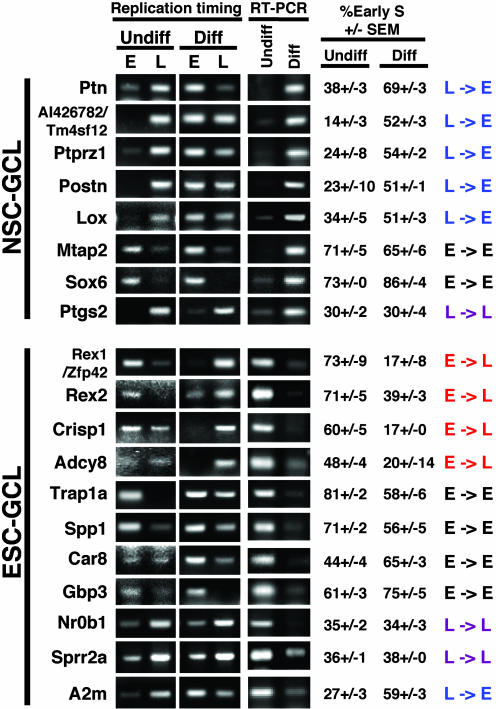
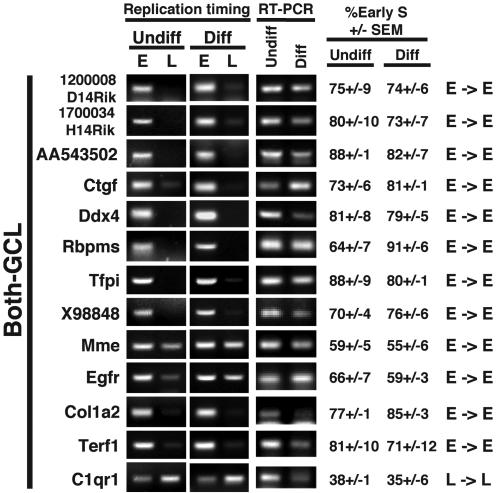
In striking contrast to GCH genes, five of the eight NSC-GCL genes analyzed changed their replication timing from late S phase to early S phase in parallel with gene activation (Fig. 3). Similarly, among the ESC-GCL genes, 4 of 11 genes changed their replication timing from early to late S phase, in parallel with gene repression. This number is almost certainly an underestimate of the percentage of ESC-GCL genes that may change replication timing during development. Genes that remain early-replicating after down-regulation may remain poised for expression or may be adjacent to genes that remain expressed. We have only examined one stage of differentiation, and these genes may switch to late replication in other tissues where they are repressed. Moreover, two of these genes (Trap1a and Car8) displayed changes in replication timing that were greater than those observed for most GCH genes, but, because they were confined to early S phase, these genes were categorized as early in both differentiation states for the reasons stated above. Among those GCL genes that were expressed in both undifferentiated and differentiated ESCs, all but one remained early-replicating (Fig. 4), and the mean difference in replication timing between differentiation states for these genes was significantly smaller than for GCL genes that did change expression (7 ± 2 versus 23 ± 3). These observations strongly suggest that genes residing within AT-rich isochores have a high propensity to switch replication timing coincident with changes in transcriptional state.
Fig. 3.
Replication-timing analysis of NSC- and ESC-specific AT-rich (GCL) genes analyzed as for Fig. 2. E, early-replicating; L, late-replicating.
Fig. 4.
Analysis of AT-rich genes that are expressed before and after differentiation (Both-GCL) as for Fig. 2. E, early-replicating; L, late-replicating.
Our analysis revealed five exceptional genes that were late-replicating and expressed (Fig. 3: Ptgs2, Nr0b1, Sprr2a, and A2m; Fig. 4: C1qr1). No such genes were found among the GCH group. Importantly, robust expression of late-replicating genes was primarily observed in ESCs (four of five genes) (Fig. 3: Nr0b1, Sprr2a, and A2m; Fig. 4: C1qr1). Because examples of late-replicating, expressed genes have been rare in the literature (Table 2), our observation raises the intriguing possibility that late-replicating regions are not as readily assembled into silent chromatin in pluripotent ESCs (31). Whole-genome analyses of replication timing versus gene expression in ESCs and NSCs should assess this possibility by determining whether the positive correlation identified in established Drosophila (3) and human (4) cell lines is less prominent in ESCs compared with NSCs.
Our results are summarized in Table 1. Strikingly, for genes that changed their transcriptional state, replication-timing changes were observed with 10 of 19 GCL genes and none of the 18 GCH genes (Fisher's exact test, P < 0.001). Because early replication is presumed to set up a permissive chromatin structure but is not sufficient for gene expression and some of these loci may be adjacent to expressed genes, a certain number of early-replicating nonexpressed GCL genes are expected. These results indicate that differentiation-induced changes in replication control are limited to genes that reside within AT-rich isochores.
Table 1. Summary.
| Replication timing | |||||
|---|---|---|---|---|---|
| Gene category | L→E | L→L | E→L | E→E | Switches observed |
| ESC-GCL | 1 | 2 | 4 | 4 | |
| NSC-GCL | 5 | 1 | 0 | 2 | 10/19 (GCL) |
| ESC-GCH | 0 | 0 | 0 | 9 | |
| NSC-GCH | 0 | 0 | 0 | 9 | 0/18 (GCH) |
A Threshold LINE Density for Replication-Timing Switches. Are there predictive characteristics other than GC content that define the class of genes subject to developmentally regulated replication-timing switches? In the course of these studies, we noted that our GCL genes were much larger with more intronic sequences (Table 3). We therefore used genome sequencing data to examine the complete set of mouse GCH and GCL genes for these properties (Table 3). We found that the average length for 5,533 GCL (<43% GC) genes is 53,380 bp, whereas the average length for 6,003 GCH (>47% GC) genes is only 24,490 bp (genes were taken from RefSeq, a nonredundant, comprehensive data set of ≈17,000 mouse genes provided by National Center for Biotechnology Information). A significant component of this size difference is accounted for by the size of introns: average lengths of intronic sequence are 51,200 and 22,190 bp, respectively, for the GCL and GCH genes analyzed. We also noted that GCL genes often represent members of duplicated gene families. What are rarely found within GCL isochores are genes regarded as highly essential, such as housekeeping genes. Of the 575 human housekeeping genes defined by Eisenberg et al. (32), we identified 371 mouse counterparts and determined their GC content. Of these counterparts, we found that only 9% (33/371 genes) were AT-rich (<43% GC) and nearly two-thirds (217/371 genes) were GC-rich (>47% GC).
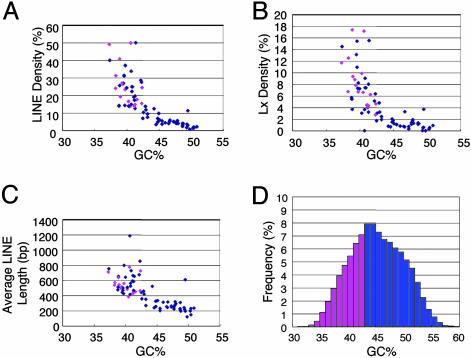
Because housekeeping genes are highly conserved and multigene families are indicative of functional diversification, these observations suggested a link between replication timing and the evolution of metazoan genomes. Because human genes are clustered into mutationally hot and cold domains (18), we plotted mutation frequency against the GC content of a 20-kb window surrounding the txStart of 8,097 genes (Fig. 6, which is published as supporting information on the PNAS web site). This analysis did not reveal any correlation between mutation frequency and GC content, implying that there is no correlation between replication timing and the accumulation of point mutations during evolution. However, recent studies suggest that the most significant changes in metazoan evolution have been mediated by changes in the regulation of genes, not in their protein-coding sequences (33, 34). The types of mutations that lead to rapid changes in gene regulation are those mediated by transposons, such as the LINEs. Unlike short interspersed repetitive elements (which are enriched in GC-rich regions), LINEs can shuffle exons, alter splicing patterns, or change developmental gene expression patterns; and recently they have been shown to affect transcriptional elongation efficiency and to introduce polyadenylation sites (35, 36). Because LINEs are enriched in AT-rich isochores (37) and are generally late replicating (4), we examined LINE density within our set of genes and within other genes whose replication timing values have been determined in different cell lines. This analysis revealed a striking prevalence of LINEs within the GCL class of genes, including the recently active subfamily of LINEs, Lx (38) (Fig. 5 A and B and Table 3). Intriguingly, as isochore GC content increased >45%, LINE and Lx density reached plateaus of <6% and <2%, respectively (Fig. 5 A and B). Importantly, no genes within this plateau were found to change replication timing, suggesting that a threshold LINE density is required for a gene to switch to late replication when it is not expressed. LINE length [only full-length LINEs can transpose (35)] was also inversely correlated with GC content (Fig. 5_C_) and showed a similar plateau.
Fig. 5.
Replication-timing changes are restricted to AT/LINE-rich isochores. (A–C) The density of LINE (A) and Lx (a young L1 subfamily) (B) and the average LINE length (C) were plotted against the average GC content of a 400-kb region surrounding the txStart position of 54 genes analyzed in this study as well as genes from the literature analyzed in Table 2. A negative correlation was observed for LINE parameters versus GC content (_R_2 values were 0.61, 0.53, and 0.52 for LINE density, average LINE length, and Lx density, respectively). (A–C) Pink dots represent genes that change replication timing either during differentiation (this study) or in different cell lines (Table 2). (D) A histogram plotting the frequency of genes in different GC content regions. The GC content of a 20-kb window surrounding the txStart of 16,652 RefSeq genes was analyzed. Because no genes at >43.4% GC (20-kb region surrounding the txStart) have been found to change replication timing (Tables 2 and 3), those genes with <43% GC, which represent 34% of all RefSeq genes, are potential candidates for genes subject to replication-timing changes at some point during development (pink bars, <43% GC). Most genes at >43% are probably not subject to replication-timing control and are presumably constitutively early-replicating (blue bars, ≥43% GC).
One enigma of evolutionary biology is how metazoa, which have substantially lower population numbers than single-celled eukaryotes, have evolved so rapidly (39). An attractive hypothesis is that metazoa have compartmentalized their genomes into segments harboring genes protected from transposons, and those that tolerate and, indeed, profit from their activity. The fact that mammalian LINEs predominate in AT-rich regions could either be explained by the preferred cleavage site of LINE endonuclease (TTTT/A) (40) or by a lower tolerance of GC-rich regions for LINE insertion (35). In either case, by restricting heritable transposition to an AT-rich compartment of the genome, specialized genes could be subject to high rates of transposon-mediated mutations, whereas housekeeping and other critical genes would be insulated within GC-rich regions. Most relevant to our findings is that, because transposition throughout somatic development would be detrimental both to host and transposon, metazoa may employ heterochromatin to silence AT-rich domains and their resident LINEs (41, 42), possibly by using late replication to maintain the heterochromatic state through somatic development (1). Those genes in AT-rich regions would then either switch to early replication in the tissue in which they are to be expressed or, in rare cases, evolve other means to be expressed in the context of late-replicating heterochromatin (43). This model describes an elegant evolutionary equilibrium that may have contributed significantly to the speciation of metazoa. An interesting prediction from this model is that the specific LINEs residing within any particular isochore may be most active in tissues where they have switched to early replication.
LINE density may also provide an explanation for why AT-rich isochores are generally late-replicating (4). The fact that LINEs bring with them their own promoter, have a weak polyadenylation site that can be bypassed, and are biased toward antisense-oriented insertion (44) suggests that they create the potential for double-stranded RNA-mediated silencing (41, 42). In fact, some evidence suggests that LINEs may serve as DNA signals, or “way stations” for propagating transcriptional inactivation of the X chromosome (45), which is exceptionally enriched in LINEs (37). Hence, a critical density of LINEs may seed the formation of large heterochromatic domains, which, because silent chromatin can delay replication timing (1, 2), provides a potential mechanistic link between LINE density and late replication. Whatever undefined mechanism reverses the formation of heterochromatin would necessarily have to overcome this LINE-mediated silencing. It will be interesting to determine whether this relationship bears out in other vertebrates that have different isochore structure and other types of transposons that may have different sequence preferences (46).
In summary, our results provide the first demonstration of replication timing changes induced by differentiation in culture, setting the stage for mechanistic studies of the relationship between replication timing and developmentally regulated patterns of gene expression. In doing so, we provide direct evidence for the longstanding notion that replication timing changes are a significant part of mammalian development, as more than one-third of all genes are potential candidates for such changes (Fig. 5_D_). Moreover, we also offer a means to predict which genes are subject to this type of regulation in any given mammalian differentiation system. Finally, this class of genes is rich in LINEs, and their structure implies that they have been subjected to rapidly evolving changes in developmentally regulated gene-expression patterns, a driving force in metazoan evolution.
Supplementary Material
Supporting Information
Acknowledgments
We thank A. Smith and M. Stavridis (University of Edinburgh, Edinburgh) for the 46C cell line and technical advice; H. Li and J. Chuang for providing mutational analysis data; Z. Lai and N. Gonchoroff for generously assisting with flow cytometry; S. Fiering and J. Goodier for helpful discussion; B. Knox, T. Sasaki, A. Fisher, and V. Azuara for comments on the manuscript; and J. Moran, H. Li, and J. Chuang for discussion and comments on the manuscript. This work was supported by National Institutes of Health Grant GM-57233-01 (to D.M.G.) and by a predoctoral fellowship from the Japan Society for the Promotion of Science (to I.H.).
Author contributions: I.H. and D.M.G. designed research; I.H. and A.L. performed research; I.H. and D.M.G. analyzed data; and I.H. and D.M.G. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LINE, long interspersed nuclear element; GCL, GC-low; GCH, GC-high; NSC, neural stem cell; ESC, embryonic stem cell; txStart, transcription start position.
References
- 1.Gilbert, D. M. (2002) Curr. Opin. Cell Biol. 14**,** 377-383. [DOI] [PubMed] [Google Scholar]
- 2.Goren, A. & Cedar, H. (2003) Nat. Rev. Mol. Cell Biol. 4**,** 25-32. [DOI] [PubMed] [Google Scholar]
- 3.Schubeler, D., Scalzo, D., Kooperberg, C., van Steensel, B., Delrow, J. & Groudine, M. (2002) Nat. Genet. 32**,** 438-442. [DOI] [PubMed] [Google Scholar]
- 4.Woodfine, K., Fiegler, H., Beare, D. M., Collins, J. E., McCann, O. T., Young, B. D., Debernardi, S., Mott, R., Dunham, I. & Carter, N. P. (2004) Hum. Mol. Genet. 13**,** 191-202. [DOI] [PubMed] [Google Scholar]
- 5.Azuara, V., Brown, K. E., Williams, R. R., Webb, N., Dillon, N., Festenstein, R., Buckle, V., Merkenschlager, M. & Fisher, A. G. (2003) Nat. Cell Biol. 5**,** 668-674. [DOI] [PubMed] [Google Scholar]
- 6.Keohane, A. M., O'Neill, L. P., Belyaev, N. D., Lavender, J. S. & Turner, B. M. (1996) Dev. Biol. 180**,** 618-630. [DOI] [PubMed] [Google Scholar]
- 7.Brockdorff, N. (2002) Trends Genet. 18**,** 352-358. [DOI] [PubMed] [Google Scholar]
- 8.McNairn, A. J. & Gilbert, D. M. (2003) BioEssays 25**,** 647-656. [DOI] [PubMed] [Google Scholar]
- 9.Ying, Q. L., Stavridis, M., Griffiths, D., Li, M. & Smith, A. (2003) Nat. Biotechnol. 21**,** 183-186. [DOI] [PubMed] [Google Scholar]
- 10.Rathjen, J., Haines, B. P., Hudson, K. M., Nesci, A., Dunn, S. & Rathjen, P. D. (2002) Development (Cambridge, U.K.) 129**,** 2649-2661. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert, D. M. (1986) Proc. Natl. Acad. Sci. USA 83**,** 2924-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert, D. & Cohen, S. (1987) Cell 50**,** 59-68. [DOI] [PubMed] [Google Scholar]
- 13.Cimbora, D. M., Schubeler, D., Reik, A., Hamilton, J., Francastel, C., Epner, E. M. & Groudine, M. (2000) Mol. Cell. Biol. 20**,** 5581-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen, R., Canfield, T., Lamb, M., Gartler, S. & Laird, C. (1993) Cell 73**,** 1403-1409. [DOI] [PubMed] [Google Scholar]
- 15.Ramalho-Santos, M., Yoon, S., Matsuzaki, Y., Mulligan, R. C. & Melton, D. A. (2002) Science 298**,** 597-600. [DOI] [PubMed] [Google Scholar]
- 16.Ivanova, N. B., Dimos, J. T., Schaniel, C., Hackney, J. A., Moore, K. A. & Lemischka, I. R. (2002) Science 298**,** 601-604. [DOI] [PubMed] [Google Scholar]
- 17.Fortunel, N. O., Otu, H. H., Ng, H. H., Chen, J., Mu, X., Chevassut, T., Li, X., Joseph, M., Bailey, C., Hatzfeld, J. A., et al. (2003) Science 302**,** 393. [DOI] [PubMed] [Google Scholar]
- 18.Chuang, J. H. & Li, H. (2004) PLoS Biol. 2**,** E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furst, A., Brown, E. H., Braunstein, J. D. & Schildkraut, C. L. (1981) Proc. Natl. Acad. Sci. USA 78**,** 1023-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhar, V., Mager, D., Iqbal, A. & Schildkraut, C. L. (1988) Mol. Cell. Biol. 8**,** 4958-4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forrester, W. C., Epner, E., Driscoll, M. C., Enver, T., Brice, M., Papayan-nopoulou, T. & Groudine, M. (1990) Genes Dev. 4**,** 1637-1649. [DOI] [PubMed] [Google Scholar]
- 22.Brown, K. E., Amoils, S., Horn, J. M., Buckle, V. J., Higgs, D. R., Merken-schlager, M. & Fisher, A. G. (2001) Nat. Cell Biol. 3**,** 602-606. [DOI] [PubMed] [Google Scholar]
- 23.Hatton, K. S., Dhar, V., Brown, E. H., Iqbal, M. A., Stuart, S., Didamo, V. T. & Schildkraut, C. L. (1988) Mol. Cell. Biol. 8**,** 2149-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selig, S., Okumura, K., Ward, D. C. & Cedar, H. (1992) EMBO J. 11**,** 1217-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe, Y., Fujiyama, A., Ichiba, Y., Hattori, M., Yada, T., Sakaki, Y. & Ikemura, T. (2002) Hum. Mol. Genet. 11**,** 13-21. [DOI] [PubMed] [Google Scholar]
- 26.Eyre-Walker, A. (1992) Nucleic Acids Res. 20**,** 1497-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eyre-Walker, A. & Hurst, L. D. (2001) Nat. Rev. Genet. 2**,** 549-555. [DOI] [PubMed] [Google Scholar]
- 28.Aladjem, M. I., Rodewald, L. W., Lin, C. M., Bowman, S., Cimbora, D. M., Brody, L. L., Epner, E. M., Groudine, M. & Wahl, G. M. (2002) Mol. Cell. Biol. 22**,** 442-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panning, M. M. & Gilbert, D. M. (2005) J. Cell. Biochem., in press. [DOI] [PubMed]
- 30.Dimitrova, D. S. & Gilbert, D. M. (1999) Mol. Cell 4**,** 983-993. [DOI] [PubMed] [Google Scholar]
- 31.Gasser, S. M. (2002) Science 296**,** 1412-1416. [DOI] [PubMed] [Google Scholar]
- 32.Eisenberg, E. & Levanon, E. Y. (2003) Trends Genet. 19**,** 362-365. [DOI] [PubMed] [Google Scholar]
- 33.Shapiro, M. D., Marks, M. E., Peichel, C. L., Blackman, B. K., Nereng, K. S., Jonsson, B., Schluter, D. & Kingsley, D. M. (2004) Nature 428**,** 717-723. [DOI] [PubMed] [Google Scholar]
- 34.Shubin, N. H. & Dahn, R. D. (2004) Nature 428**,** 703-704. [DOI] [PubMed] [Google Scholar]
- 35.Moran, J. & Gilbert, N. (2002) in Mobile DNA II, eds. Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. (Am. Soc. Microbiol. Press, Washington, D.C.), pp. 836-869.
- 36.Han, J. S., Szak, S. T. & Boeke, J. D. (2004) Nature 429**,** 268-274. [DOI] [PubMed] [Google Scholar]
- 37.Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. (2002) Nature 420**,** 520-562. [DOI] [PubMed] [Google Scholar]
- 38.Allen, E., Horvath, S., Tong, F., Kraft, P., Spiteri, E., Riggs, A. D. & Marahrens, Y. (2003) Proc. Natl. Acad. Sci. USA 100**,** 9940-9945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lynch, M. & Conery, J. S. (2003) Science 302**,** 1401-1404. [DOI] [PubMed] [Google Scholar]
- 40.Feng, Q., Moran, J. V., Kazazian, H. H., Jr., & Boeke, J. D. (1996) Cell 87**,** 905-916. [DOI] [PubMed] [Google Scholar]
- 41.Schramke, V. & Allshire, R. (2004) Curr. Opin. Genet. Dev. 14**,** 174-180. [DOI] [PubMed] [Google Scholar]
- 42.Xie, Z., Johansen, L. K., Gustafson, A. M., Kasschau, K. D., Lellis, A. D., Zilberman, D., Jacobsen, S. E. & Carrington, J. C. (2004) PLoS Biol. 2**,** E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakimoto, B. T. (1998) Cell 93**,** 321-324. [DOI] [PubMed] [Google Scholar]
- 44.Smit, A. F., Toth, G., Riggs, A. D. & Jurka, J. (1995) J. Mol. Biol. 246**,** 401-417. [DOI] [PubMed] [Google Scholar]
- 45.Bailey, J. A., Carrel, L., Chakravarti, A. & Eichler, E. E. (2000) Proc. Natl. Acad. Sci. USA 97**,** 6634-6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jaillon, O., Aury, J. M., Brunet, F., Petit, J. L., Stange-Thomann, N., Mauceli, E., Bouneau, L., Fischer, C., Ozouf-Costaz, C., Bernot, A., et al. (2004) Nature 431**,** 946-957. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information