Phosphorylation of spinophilin by ERK and cyclin-dependent PK 5 (Cdk5) (original) (raw)
Abstract
Spinophilin is a protein that binds to protein phosphatase-1 and actin and modulates excitatory synaptic transmission and dendritic spine morphology. We have identified three sites phosphorylated by ERK2 (Ser-15 and Ser-205) and cyclin-dependent PK 5 (Cdk5) (Ser-17), within the actin-binding domain of spinophilin. Cdk5 and ERK2 both phosphorylated spinophilin in intact cells. However, in vitro, phosphorylation by ERK2, but not by Cdk5, was able to modulate the ability of spinophilin to bind to and bundle actin filaments. In neurons and HEK293 cells expressing GFP-tagged variants of spinophilin, imaging studies demonstrated that introduction of a phospho-site mimic (Ser-15 to glutamate) was associated with increased filopodial density. These results support a role for spinophilin phosphorylation by ERK2 in the regulation of spine morphogenesis.
Keywords: actin, dendritic spines, protein phosphatase
Spines are specialized, micrometer-sized protrusions studding the dendritic shafts of neurons. Most excitatory input in the central nervous system is transmitted through synapses that occur on spines (axospinous synapses). Alterations in this structural circuitry by dynamic changes in spine morphology and density may underlie much of the neuronal plasticity in response to experience and learning (1, 2).
Actin is the major cytoskeletal component of dendritic spines. Rearrangements of the actin cytoskeleton are thought to power spine motility. Time-lapse studies of neurons in culture labeled with GFP or GFP-actin reveal highly motile spines whose dynamic nature is inhibited by cytochalasin D, an actin filament-disrupting agent (3, 4). The motility of spines is regulated by extracellular signals, such as glutamate (5, 6) and Ephrin B (7, 8). Moreover, an array of intracellular proteins has been identified that regulate spine morphology and density. Many of these molecules converge on the regulation of the actin cytoskeleton. However, it is unclear how these proteins are activated by signaling at the synapse, how these signals regulate the actin cytoskeleton, and the mechanisms by which this regulation is transduced into changes in spine morphology and density.
Spinophilin is a protein phosphatase-1 (PP1)- and actin-binding protein that is enriched in dendritic spines (9, 10). Its localization and functional properties suggest that spinophilin may serve as a link between synaptic transmission and changes in spine morphology. Spinophilin can regulate synaptic transmission by virtue of its ability to bind PP1, localizing it close to the postsynaptic density, where PP1 can dephosphorylate glutamate receptors. Also, spinophilin knockout mice exhibit reduced synaptic transmission and a deficit in long-term depression. Moreover, spinophilin is involved in spine dynamics. Young spinophilin knockout mice have a higher density of spines than their WT littermates, and hippocampal cultures made from these knockout mice have more numerous filopodia than WT cultures (11). The actin-binding domain of spinophilin is sufficient to localize spinophilin to dendritic spines (12), and spinophilin is able to bind to and bundle F-actin (10, 13).
We have shown (13, 14) that phosphorylation of spinophilin by PKA or Ca2+/calmodulin-dependent PK II (CaMKII) disrupts the ability of spinophilin to bind F-actin. Spinophilin extracted from striatal slices is phosphorylated at sites in addition to those phosphorylated by PKA or CaMKII. Spinophilin contains consensus sequences for other PKs, including ERK1/2 and cyclin-dependent PK 5 (Cdk5). ERK1/2 and Cdk5 are highly expressed in brain, and both have been implicated in the regulation of neuronal plasticity and spine morphology. ERK1/2 is involved in synaptic plasticity (15–18), and biochemical activation of ERK1/2 by spaced membrane depolarizations was shown to be critical for protrusion of new dendritic filopodia (19). Cdk5 has been implicated in adaptive changes related to cocaine addiction (20) and in regulation of spine density induced by cocaine (21).
In this study, we have identified ERK2 and Cdk5 phosphorylation sites within the actin-binding domain of spinophilin, and we show that phosphorylation by ERK2 but not Cdk5 reduces the ability of spinophilin to bind to and bundle F-actin. Last, we determined that ERK2 mediated phosphorylation of spinophilin may be involved in regulation of spine morphogenesis.
Methods
Preparation of Spinophilin and Neurabin Fusion Proteins. Spinophilin and neurabin fusion protein were prepared as described in Supporting Methods, which is published as supporting information on the PNAS web site.
In Vitro Phosphorylation. Phosphorylation reactions were performed by using the protein of interest (2.5 μM spinophilin and 10 μM neurabin) and Cdk5/p35 (22) in 50 mM Mops, pH 7.4/50 mM MgCl2/10 mM DTT or ERK2 (NEB) in 50 mM Tris·HCl, pH 7.5/10 mM MgCl2/1 mM EGTA/2 mM DTT/0.01% Brij 35 at 30°C. Reactions were initiated by adding 100 μM [γ-32P]ATP. Samples were taken from the reaction mixture, and reactions were terminated by dilution in SDS sample buffer and subjected to SDS/PAGE alongside a standard protein curve of β-gal (high-molecular-weight standards, Amersham Biosciences). The stoichiometry of phosphorylation (mol of Pi per mol of protein) was calculated by measuring 32P incorporation into spinophilin and normalizing to the amount of spinophilin protein.
Two-Dimensional Phospho-Peptide Mapping. WT and mutant spinophilin proteins were phosphorylated for 1 h, as described above. Trypsin digestion, peptide extraction, and two-dimensional phospho-peptide mapping were performed as described (14).
Production and Purification of Phospho-Specific Spinophilin Antibodies. Phospho-specific antibodies were produced essentially as described (13). Phospho-peptides (incorporating a cysteine residue at the C terminus) contained the following sequences: GPLR(pS)ASPHC (residues 11–19, _phospho_-Ser-15) and LRSA(pS)PHRSC (residues 13–21, _phospho_-Ser-17). Rabbits were immunized with phospho-peptides conjugated to hemocyanin. Antibodies were affinity-purified by using protein A linked to Sepharose, as well as dephospho- and phospho-peptide linked to Sulfolink gel (Pierce).
Cell Culture and Transfection. HEK293 cells were plated on poly-d-lysine-coated coverslips at a density of 1.5 × 105 cells. HEK293 and COS-7 cells were transiently transfected with the GFP-tagged actin binding domain of spinophilin or Flag-tagged spinophilin cDNA (23), respectively, by using Lipofectamine 2000 (Invitrogen). Primary hippocampal cultures from embryonic day 18–19 Sprague–Dawley rats were prepared and transfected as described (12). Neurons were transfected on DIV6 and fixed for immunocytochemistry on DIV13. CD8 was coexpressed as a cell-morphology marker where indicated. A ratio of 20:1 spinophilin construct cDNA/CD8 cDNA was used.
Preparation and Incubation of Neostriatal Slices. Neostriatal slices were prepared from male C57BL/6 mice at 6–8 weeks of age, as described (24). Slices were treated with drugs as specified in each experiment. Drugs were obtained from the following sources: okadaic acid and cyclosporin A were obtained from LC Laboratories (Woburn, MA); roscovitine was obtained from Calbiochem; butyrolactone was obtained from Biomol (Plymouth Meeting, PA); and purvanalol and allosterpaullone were obtained from Sigma. After drug treatment, slices were frozen on dry ice and stored at -80°C until assayed.
Lysate Preparation and Immunoblotting. Neostriatal slice lysates were immunoblotted as described (24). After 48 h, transfected COS-7 cells were serum starved overnight, before treatments. After preincubation with 10 μM U0126 for 30 min and/or stimulation with EGF (times and doses as indicated), COS-7 cells were lysed by scraping into NuPAGE SDS/PAGE sample buffer (Invitrogen), heating to 70°C for 10 min, and sonicating. Lysates were run on 3–8% Tris·acetate gels or 4–12% Bis-Tris gels (Invitrogen). Blots were probed with _phospho_-Ser-17 spinophilin antibody (1:1,000) or _phospho_-ERK1/2 antibody (NEB; 1:2,000), and they were then stripped and reprobed with spinophilin antibody (9) or ERK1/2 antibody (NEB; 1:2,000). Primary antibody binding was revealed by using a horseradish peroxidase-linked goat-anti rabbit secondary antibody and the ECL detection system (Amersham Biosciences). Chemiluminescence was detected by autoradiography using autoradiography film (Kodak), and protein bands were quantified by densitometry using National Institutes of Health image 1.61 software.
Actin-Binding Assay. Phosphorylated spinophilin was prepared for actin binding as described (14) by using Cdk5 and ERK2 in the buffers described in detail above. Under these conditions, the stoichiometry of phosphorylation was ≈0.3 (Cdk5) or 1 (ERK2) mol of Pi per mol of protein. Protein concentrations were determined by using the bicinchoninic acid (BCA) protein-assay method, with BSA as a standard. Actin polymerization, binding to spinophilin, and quantification of sedimented spinophilin were performed as described (14).
Actin-Bundling Assay. The actin binding domain of spinophilin was phosphorylated, and actin bundling was performed as described for the actin-binding assay. To selectively sediment bundled actin, reaction mixtures were subjected to a low-speed centrifugation of 10,000 × g at room temperature. The supernatant and pellets were subjected to SDS/PAGE, and the amount of bundled actin was calculated as a percentage of the total actin input.
Immunocytochemistry and Microscopy. HEK293 cells and neurons were fixed with 4% paraformaldehyde/4% sucrose (wt/vol) in 0.01 M PBS (138 mM NaCl/2.7 mM KCl) for 10 min. Nonspecific staining was blocked by 30 min of incubation with 10% BSA in PBS. CD8 was visualized by incubation with rat anti-mouse CD8 antibody (Caltag, 1:500 in 10% BSA for 1 h), followed by washing and incubation in Alexa Fluor 568-labeled goat anti-rat secondary antibody (Invitrogen, 1:500 in 10% BSA for 1 h). After washing in PBS, coverslips were mounted on slides by using Gel/Mount (Biomeda, Foster City, CA). Images of fluorescent proteins were acquired by using a Zeiss 510 laser-scanning microscope. Spine density and length were analyzed by using metamorph software. In control experiments using HEK293 cells, we analyzed the number of filopodia by using the fluorescent signals of the WT and mutant GFP-tagged actin binding domain of spinophilin. We observed no detectable difference in the localization of spinophilin after mutation. WT spinophilin and mutants were found throughout the cell, including the filopodia, as observed by double-labeling of actin with Alexa phalloidin 568. However, in HEK293 cells we observed that GFP itself did not localize to the filopodia, and therefore, for this condition we used CD8 as a cell volume marker to count filopodia. Expression of CD8 did not change the density of filopodia, as observed by comparison of HEK293 cells expressing the GFP-tagged actin binding domain of spinophilin with or without CD8. In neurons coexpressing CD8 and GFP-tagged full-length spinophilin, filopodial density and length were analyzed by using the CD8 immunofluorescent signal. We observed no detectable difference in the localization of spinophilin in neurons after mutation.
Results
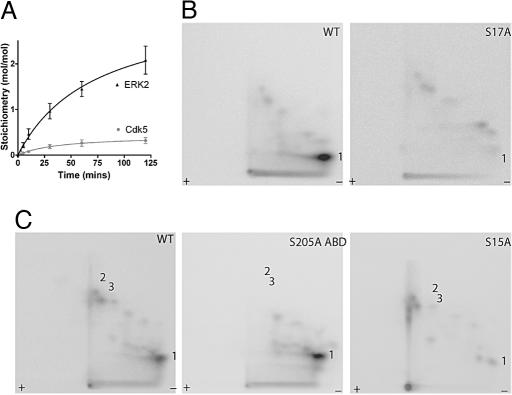
Phosphorylation of Spinophilin by Cdk5 and ERK2 in Vitro. Cdk5 or ERK2 phosphorylated spinophilin in a time-dependent manner to a maximal stoichiometry of 0.3 and 2.1 mol of Pi per mol of spinophilin, respectively (Fig. 1_A_). The two-dimensional phosphopeptide maps of spinophilin revealed one major peptide phosphorylated by Cdk5 (peptide 1, Fig. 1_B_) and one major and two minor peptides phosphorylated by ERK2 (peptides 1–3, Fig. 1_C_).
Fig. 1.
Spinophilin is phosphorylated in vitro by Cdk5 and ERK2. (A) Time course of phosphorylation of full-length spinophilin by Cdk5 and ERK2. Spinophilin was phosphorylated in the presence of [γ-32P]ATP and the stoichiometry of phosphorylation was assessed after SDS/PAGE and measurement of incorporation of [γ-32P] phosphate into spinophilin. (B) Two-dimensional peptide maps of full-length WT or Ser-17-to-alanine mutant spinophilin (S17A) phosphorylated in vitro by Cdk5. (C) Two-dimensional peptide maps of full-length spinophilin (WT), a Ser-205-to-alanine mutant of the actin-binding domain (residues 1–221) (S205A ABD), or a Ser-15-to-alanine mutant full-length spinophilin (S15A) each phosphorylated in vitro by ERK2. Phosphopeptides are marked numerically.
To determine the residues that were phosphorylated by Cdk5 and ERK2, we used two-dimensional peptide mapping to analyze a number of spinophilin mutants in which consensus phosphorylation sites were changed to alanine (Fig. 1 B and C). Mutation of Ser-17 (an excellent Cdk5 consensus site) to alanine resulted in the loss of peptide 1 after incubation with Cdk5. Spinophilin has five consensus sites for phosphorylation by ERK2, two of which are found in the actin-binding domain (residues 1–221). After incubation of spinophilin (1–221) with ERK2, two-dimensional peptide maps revealed the same pattern of phosphopeptides as full-length spinophilin, indicating that all ERK2 phosphorylation sites are located within residues 1–221 (Fig. 7, which is published as supporting information on the PNAS web site). Mutation of Ser-205 to alanine led to the disappearance of the two minor phosphopeptides (2 and 3) (Fig. 1_C_). Two-dimensional peptide mapping suggested that the major peptide 1 phosphorylated by ERK2 was likely to be the same as peptide 1 phosphorylated by Cdk5. However, mutation of Ser-17 to alanine did not effect phosphorylation of spinophilin by ERK2 (Fig. 7). Mutation of Ser-15 to alanine led to the disappearance of peptide 1, after incubation with ERK2 (Fig. 1_C_).
To confirm Ser-15 as the major site phosphorylated by ERK2 and Ser-17 as the major site for Cdk5, and for use in other studies, we developed phospho-specific antibodies against _phospho_-Ser-15- and _phospho_-Ser-17-spinophilin. The _phospho_-Ser-15 antibodies only recognized spinophilin that had been phosphorylated by ERK2 and the _phospho_-Ser-17 antibody only recognized spinophilin that had been phosphorylated by Cdk5. No detectable binding to dephospho_-spinophilin was observed. (Fig. 8_A, which is published as supporting information on the PNAS web site). These antibodies were also site-specific. The _phospho_-Ser-15 antibody did not recognize the Ser-15-to-alanine mutant phosphorylated by ERK2, and the phospho_-Ser-17 antibody did not recognize the Ser-17-to-alanine mutant phosphorylated by Cdk5 (Fig. 8_B).
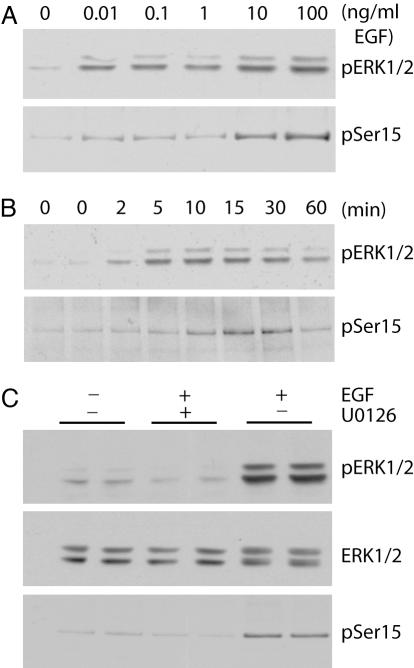
EGF Stimulates Phosphorylation of Spinophilin in an ERK1/2-Dependent Manner in COS-7 Cells. The activation of ERK1/2 by growth factors has been extensively characterized in cell lines, particularly in COS-7 cells. Flag-tagged spinophilin was transiently transfected into COS-7 cells and cells were treated with increasing doses of EGF for 10 min or for increasing time periods with a fixed dose of EGF (100 ng/ml). By using phospho-specific antibody for _phospho_-ERK1/2 or _phospho_-Ser-15 spinophilin, we found ERK1/2 and spinophilin to be phosphorylated in a similar dose- and time-dependent manner (Fig. 2 A and B). Activation of ERK2 did not lead to phosphorylation at Ser-17 (data not shown). The peak of ERK1/2 and spinophilin phosphorylation occurred after 15 min of treatment with 100 ng/ml EGF. By using these conditions, incubation with U0126 (a specific inhibitor of ERK activation) before EGF stimulation abolished both ERK1/2 phosphorylation and spinophilin phosphorylation at Ser-15 (Fig. 2_C_).
Fig. 2.
ERK1/2-dependent phosphorylation of spinophilin at _phospho_-Ser-15 in response to EGF stimulation. COS-7 cells were transiently transfected with Flag-tagged spinophilin. Cells were also serum-starved overnight to reduce the basal phosphorylation of spinophilin. (A) Cells were treated with increasing concentrations of EGF for 10 min as indicated. (B) Cells were treated with EGF (100 ng/ml) for increasing time as indicated. (C) Cells were pretreated without (-) or with (+) U0126 (10 μM) for 30 min as indicated and then treated without (-) or with (+) 100 ng/ml EGF for 15 min as indicated. Cell lysates were analyzed by SDS/PAGE and immunoblotting using antibodies to total ERK1/2, _phospho_-ERK1/2 (pERK1/2), and _phospho_-Ser-15 of spinophilin (pSer15). The immunoblots are representative of three experiments. Total levels of ERK1/2 and spinophilin remained the same for all treatments (data not shown).
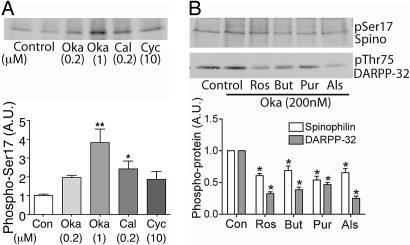
Cdk5-Dependent Phosphorylation of Spinophilin in Neostriatal Neurons. Cdk5 and its activators p35 and p39 are expressed in neurons, but mechanisms involved in their regulation remain poorly characterized. In neostriatal slices, spinophilin exhibited a low basal level of phosphorylation at Ser-17, as determined by using the _phospho_-Ser-17 antibody. Treatment of neostriatal slices with okadaic acid, a PP1/PP2A inhibitor with higher selectivity for PP2A, increased phosphorylation at Ser-17, the increase in phosphorylation being greater at 1 μM than at 200 nM (Fig. 3_A_). However, the effect of calyculin A, an inhibitor with similar selectivity for PP1 and PP2A, was similar to okadaic acid (200 nM). Cyclosporin A, an inhibitor of protein phosphatase 2B, had an effect similar to okadaic acid (200 nM) and calyculin A. These results suggest that multiple protein phosphatases may act on Ser-17, with PP2A likely playing a prominent role (see ref. 25 for discussion of the use of phosphatase inhibitors in striatal slices). Application of various Cdk5 inhibitors (roscovitine, butyrolactone, purvanalol, or allosterpaullone) in the presence of okadaic acid decreased phosphorylation at Ser-17 by ≈40–50% (Fig. 3_B_). In the same samples, comparable decreases were seen in phosphorylation at Thr-75 of DARPP-32, a physiological substrate for Cdk5 in neostriatal neurons (26).
Fig. 3.
Cdk5 inhibitors reduce phosphorylation of spinophilin at Ser-17 in neurons. (A) Neostriatal slices were treated with the following phosphatase inhibitors: okadaic acid (Oka, 0.2 or 1 μM), calyculin A (Cal, 0.2 μM), or cyclosporin A (Cyc, 10 μM). (B) Slices were treated with the following different Cdk5 inhibitors: roscovitine (Ros, 50 μM), butyrolactone (But, 20 μM), purvanalol (Pur, 50 μM), or allosterpaullone (Als, 50 μM) for 60 min in the presence of okadaic acid (0.2 μM). Cell lysates were analyzed by SDS/PAGE and immunoblotting using antibodies to spinophilin (pSer17 Spino) or DARPP-32 (pThr75 DARPP-32) antibody as indicated. Cumulative data from at least four experiments are shown in the bar graphs (means ± SEM). *, P < 0.01 (one-way ANOVA with post hoc Newman–Keuls multiple-comparison test).
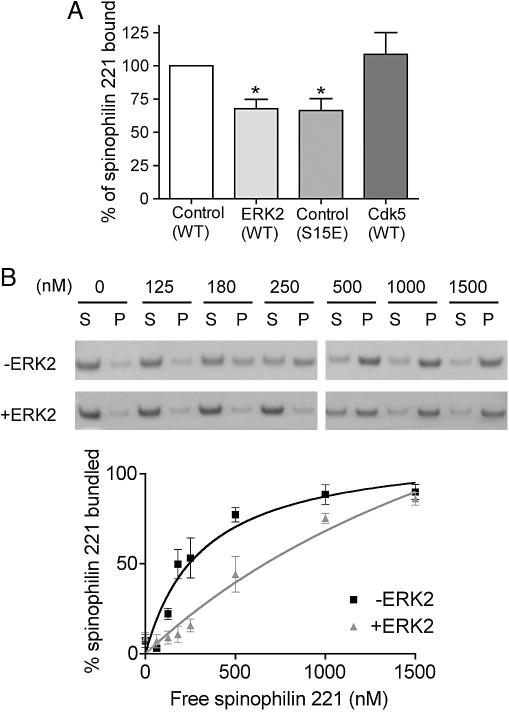
Phosphorylation of Spinophilin by ERK2 but Not Cdk5 Regulates Its Interaction with F-Actin. The minimal actin binding domain of spinophilin has been localized to residues 1–154, and it likely extends to include amino acids 221 (10, 12, 13, 27). The major sites phosphorylated by ERK2 and Cdk5 are located within this domain. Therefore, we tested whether phosphorylation by ERK2 or Cdk5 could regulate the interaction of spinophilin (1–221) with F-actin. Based on our previous studies (14), we chose a fixed concentration of spinophilin (50 nM) that was within the dynamic range of the binding curve for F-actin. Phosphorylation by ERK2 reduced spinophilin binding to F-actin by 32% (Fig. 4_A_). Furthermore, mutation of Ser-15 to glutamate to produce a phosphorylation-state mimic, reduced the amount of spinophilin bound to actin by a similar magnitude of 34% (Fig. 4_A_). However, phosphorylation by Cdk5 had no effect on spinophilin binding to F-actin (Fig. 4_A_). Mutation of Ser-17 to glutamate also had no effect on binding to F-actin (data not shown).
Fig. 4.
Phosphorylation of spinophilin by ERK2 but not Cdk5 decreases spinophilin binding to and bundling of actin filaments in vitro. (A) WT spinophilin (1–221) was phosphorylated with either Cdk5 or ERK2 and then incubated with F-actin. Spinophilin (1–221) in which Ser-15 was mutated to glutamate also was analyzed. The amount of _phospho_-spinophilin or mutant spinophilin (1–221) bound to actin was calculated as a percentage of _dephospho_-spinophilin (1–221). Data represent means ± SEM for two experiments performed in triplicate. *, P < 0.01 (one-way ANOVA with post hoc Dunnet's multiple-comparison test). (B) Spinophilin (1–221) was incubated without or with ERK2 and incubated with F-actin. The immunoblots and graph demonstrate the effect of ERK2 phosphorylation on actin bundling. Actin bundling was calculated as a fraction of the amount of actin in the pellet compared with total actin input. Data represent means ± SEM for three experiments performed in duplicate or triplicate.
Spinophilin has been shown to bundle F-actin (10), a process that possibly involves homodimerization, mediated by its C-terminal coiled-coil domain. However, a recent study has indicated that spinophilin has a second actin-binding domain located between residues 151–282 (27) that may contribute to F-actin bundling. As expected, we found that spinophilin (1–221) was able to bundle actin in a concentration-dependent manner (Fig. 4_B_). Phosphorylation by ERK2 caused a right shift in the bundling curve, demonstrating that the bundling ability of spinophilin was decreased by phosphorylation by ERK2 (Fig. 4_B_).
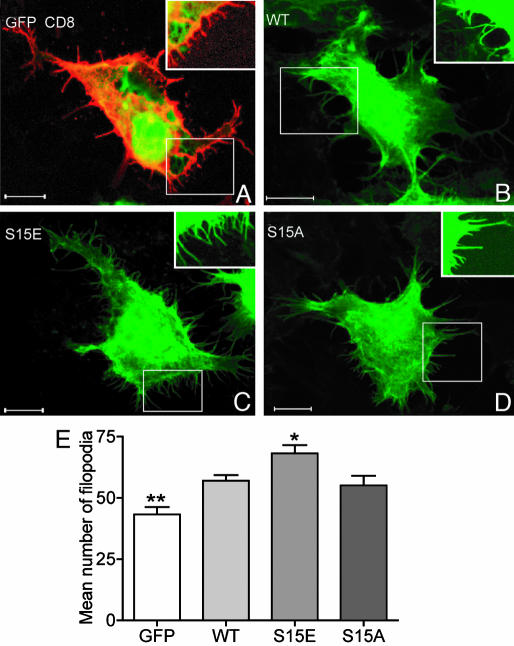
Spinophilin Phosphorylation Increases Protrusion Density in HEK293 Cells and Hippocampal Neurons. The retraction and extension of filopodia are largely determined by dynamics of the underlying actin cytoskeleton. Because spinophilin phosphorylation by ERK2 reduced its ability both to bind and to bundle actin, we next investigated whether spinophilin phosphorylation could alter filopodial actin dynamics. In initial studies, we expressed in HEK293 cells, the actin-binding domain of spinophilin as well as mutants where Ser-15 was replaced with alanine, to prevent phosphorylation, or glutamate, to mimic the phosphorylated condition (Fig. 5 A–D). Cells expressing the actin-binding domain of spinophilin (WT sequence) had a greater number of filopodia than control cells (those expressing GFP alone) (≈30% increase, Fig. 5 Lower). Cells expressing the Ser-15 to glutamate mutant had significantly more filopodia (19% increase compared with WT) than those expressing either WT spinophilin or the Ser-15-to-alanine mutant.
Fig. 5.
A _phospho_-spinophilin mimic increases filopodial density in HEK293 cells. The images show representative HEK293 cells expressing GFP (green) and CD8 (red) (A), WT GFP-tagged spinophilin (1–221) (WT, green) (B), GFP-spinophilin (1–221) with a Ser-15-to-glutamate mutation (S15E) (C), and GFP-spinophilin (1–221) with a Ser-15-to-alanine mutation (S15A) (D). HEK293 cells were serum-starved overnight to reduce the basal phosphorylation of spinophilin. GFP did not localize to the filopodia (A), and therefore, we used CD8 as a marker to analyze filopodia. (B–D) The GFP signal associated with tagged sphinophilin (1–221) was used to analyze filopodia. Insets show sections of filopodia imaged at higher gain intensity. (Scale bar, 10 μm.) (E) The bar graph shows the mean number of filopodia per cell ± SEM (18 cells were counted for each expressed protein). The mean number of filopodia was 43 ± 13 for GFP, 57 ± 2 for WT spinophilin (1–221), 68 ± 3 for the Ser-15-to-glutamate mutant, and 55 ± 4 for the Ser-15-to-alanine mutant. *, P < 0.05; **, P < 0.01 (one-way ANOVA with post hoc Newman–Keuls multiple-comparison test against WT).
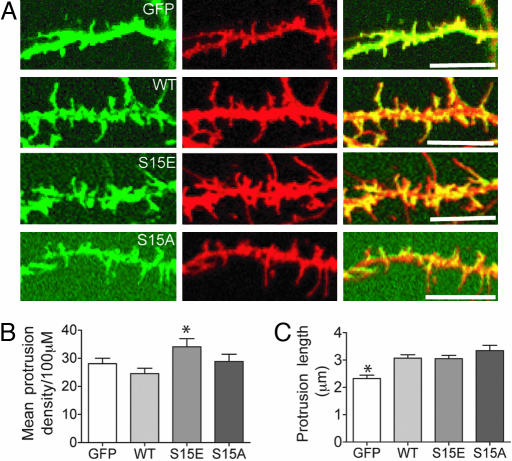
We next assessed the possible relevance of Ser-15 phosphorylation in regulating filopodial morphology in hippocampal neurons. Representative segments of dendrites from neurons coexpressing CD8 (red) and either GFP (green) or GFP-tagged full-length spinophilin are shown in Fig. 6_A_. Expression of WT spinophilin increased the length of protrusions by ≈30% compared with control neurons expressing GFP alone (Fig. 6_B_). However, filopodial density was not significantly increased by spinophilin expression (Fig. 6_C_). Note that expression of spinophilin in neurons in which Ser-15 was mutated to glutamate resulted in an increased density of filopodia (35% increase compared with WT) compared with neurons expressing either the WT spinophilin or Ser-15 to alanine (Fig. 6_C_). Together, these results suggest that phosphorylation at Ser-15 could regulate filopodial initiation, extension, or retraction in cell lines and neurons.
Fig. 6.
A _phospho_-spinophilin mimic increases filopodial density but not filopodial length in hippocampal neurons. (A) Representative segments of dendrites from hippocampal neurons expressing GFP, GFP-tagged WT spinophilin (WT), a Ser-15-to-glutamate mutant (S15E), and a Ser-15-to-alanine mutant (S15A). (Left) GFP fluorescence (green). (Center) CD8 labeling used as a volume marker (red). (Right) Merge of green and red images. (Scale bar, 10 μm.) (B and C) The bar graphs show the mean number of filopodia per neuron ± SEM (B) or mean spine length ± SEM (C). The following numbers of neurons from at least three separate sets of cultures were counted; 7 GFP, 21 WT, 13 Ser-15-to-glutamate, and 9 Ser-15-to-alanine. The mean number of protrusions per 100 μm was 28.0 ± 5.2 for cells expressing GFP, 24.6 ± 1.9 for WT spinophilin, 34.1 ± 2.9 for the Ser-15-to-glutamate mutant, and 28.8 ± 2.6 for the Ser-15-to-alanine mutant. The protrusion length was 2.3 ± 0.1 μm for neurons expressing GFP/CD8, 3.1 ± 0.1 μm for WT spinophilin/CD8, 3.1 ± 0.1 μm for the Ser-15-to-glutamate mutant, and 3.3 ± 0.2 μm for the Ser-15-to-alanine mutant. *, P < 0.05 (one-way ANOVA with post hoc Dunnet's multiple-comparison test against WT).
Discussion
In the present study, we show that spinophilin is phosphorylated in vitro by ERK2 and Cdk5. Mapping of the phosphorylation sites revealed that they were located in close proximity, at Ser-15 and Ser-17, respectively, in the N-terminal actin-binding domain of spinophilin. We have demonstrated that spinophilin can be phosphorylated by PKA and CaMKII within its actin-binding domain (13, 14). The PKA and CaMKII phosphorylation sites are not conserved in neurabin, a homologue of spinophilin, suggesting that these two proteins can be regulated differentially by phosphorylation. However, the ERK2 and Cdk5 phosphorylation sites are conserved in neurabin, suggesting that spinophilin and neurabin also have common mechanisms of regulation. Indeed, neurabin can be phosphorylated, in vitro, at Ser-15 and Ser-17 by ERK2 and Cdk5, respectively (Fig. 9, which is published as supporting information on the PNAS web site). Phosphorylation of spinophilin by ERK2, but not Cdk5, reduces its ability to bind to and bundle actin filaments. The basis for the ability of phosphorylation of Ser-15, but not of Ser-17, to influence actin binding is not known, but it presumably reflects a precise molecular interaction between spinophilin and actin. Despite the close proximity of the Cdk5 and ERK2 phosphorylation sites in spinophilin, it does not appear that phosphorylation at either site affects the kinetics of phosphorylation at the other site in vitro (M.F., A.C.N., and P.G., unpublished data).
We observed phosphorylation at both Ser-15 and Ser-17 in intact cells, suggesting that spinophilin is a physiological target for ERK2 and Cdk5. However, phosphorylation of Ser-15 was detected in nonneuronal cells and was found only at low levels in striatal slices, whereas phosphorylation of Ser-17 was detected in striatal slices but was found only at low levels in nonneuronal cells (Figs. 2 and 3, and data not shown). Spinophilin, although enriched in neurons, is expressed in nonneuronal cells (9), and its phosphorylation by ERK may contribute to regulation of actin in nonneuronal cells. The lack of phosphorylation of Ser-17 in nonneuronal cells is explained by the low levels of Cdk5 activity. However, ERK is readily activated in neurons, including by treatment with okadaic acid (as used in this study). Therefore, the reason for the lack of phosphorylation of Ser-15 in neurons is not clear at this time. Possibly, the interaction of spinophilin with F-actin in neurons precludes phosphorylation of this site. As mentioned above, spinophilin is also phosphorylated by PKA and CaMKII within its actin-binding domain (13, 14). It is possible that multiple synergistic signaling pathways acting on spinophilin are required for Ser-15 phosphorylation in neurons.
Phosphorylation-induced release of spinophilin from the actin cytoskeleton could relocalize spinophilin within filopodia and perhaps in more mature spines. We have shown that spinophilin can regulate α-amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA) and NMDA receptors and synaptic plasticity through anchoring PP1 in close proximity to glutamate receptors (11, 28, 29). Such repositioning of the spinophilin-PP1 complex within the spine after phosphorylation by ERK2 might contribute to altered phosphorylation of glutamate receptors.
Filopodia and spines are enriched for actin and these dendritic protrusions undergo changes in number and morphology during development as well as in mature neurons (30). It has been suggested that, during central nervous system development, filopodia dynamically extend and retract and that this motility may contribute to the formation and plasticity of synaptic connections (31). Recent studies have suggested that the actin-binding domain of neurabin may play a role in actin polymerization during spine morphogenesis (32, 33). In this study, we demonstrate that over-expression of spinophilin can also regulate filopodial dynamics in HEK293 cells and in neurons. Furthermore, through the use of a phosphorylation site mimic, our results suggest that phosphorylation of the actin-binding domain of spinophilin can influence the regulation of filopodial morphology perhaps by altering the dynamics of actin polymerization. Spine morphogenesis is regulated by synaptic activity, particularly by glutamate receptors. Signaling pathways in neurons leading to changes in spine shape and density are not clearly defined, and many intracellular molecules have been implicated. ERK2 and Cdk5 are both activated in neurons by glutamate receptors (15, 34–37) and implicated in neuronal plasticity and spine morphogenesis. In this study, we have provided evidence that spinophilin phosphorylation may regulate filopodial morphogenesis. Therefore, phosphorylation of spinophilin may be an important link between synaptic signaling, actin dynamics, and spine morphogenesis.
Supplementary Material
Supporting Information
Acknowledgments
We thank Linda Hsieh-Wilson and Patrick Allen (The Rockefeller University) for generous gifts of spinophilin constructs and Sophorn Chip for assistance with primary hippocampal culture preparation. This work was supported by National Institutes of Health Grants MH40899 and DA1044 (to P.G. and A.C.N.).
Author contributions: M.F., S.A.B., H.C.H., A.N., P.G., and A.C.N. designed research; M.F., K.U., S.A.B., Y.K., and A.N. performed research; A.C.N. contributed new reagents/analytic tools; M.F., K.U., S.A.B., Y.K., H.C.H., A.N., P.G., and A.C.N. analyzed data; and M.F., S.A.B., H.C.H., A.N., P.G., and A.C.N. wrote the paper.
Abbreviations: Cdk5, cyclin-dependent PK 5; PP1, protein phosphatase-1; CaMKII, calmodulin-dependent PK II.
References
- 1.Lendvai, B., Stern, E. A., Chen, B. & Svoboda, K. (2000) Nature 404**,** 876-881. [DOI] [PubMed] [Google Scholar]
- 2.Trachtenberg, J. T., Chen, B. E., Knott, G. W., Feng, G., Sanes, J. R., Welker, E. & Svoboda, K. (2002) Nature 420**,** 788-994. [DOI] [PubMed] [Google Scholar]
- 3.Dunaevsky, A., Tashiro, A., Majewska, A., Mason, C. & Yuste, R. (1999) Proc. Natl. Acad. Sci. USA 96**,** 13438-13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer, M., Kaech, S., Knutti, D. & Matus, A. (1998) Neuron 20**,** 847-854. [DOI] [PubMed] [Google Scholar]
- 5.Fischer, M., Kaech, S., Wagner, U., Brinkhaus, H. & Matus, A. (2000) Nat. Neurosci. 3**,** 887-894. [DOI] [PubMed] [Google Scholar]
- 6.Halpain, S., Hipolito, A. & Saffer, L. (1998) J. Neurosci. 18**,** 9835-9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henkemeyer, M., Itkis, O. S., Ngo, M., Hickmott, P. W. & Ethell, I. M. (2003) J. Cell Biol. 163**,** 1313-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penzes, P., Beeser, A., Chernoff, J., Schiller, M. R., Eipper, B. A., Mains, R. E. & Huganir, R. L. (2003) Neuron 37**,** 263-274. [DOI] [PubMed] [Google Scholar]
- 9.Allen, P. B., Ouimet, C. C. & Greengard, P. (1997) Proc. Natl. Acad. Sci. USA 94**,** 9956-9961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satoh, A., Nakanishi, H., Obaishi, H., Wada, M., Takahashi, K., Satoh, K., Hirao, K., Nishioka, H., Hata, Y., Mizoguchi, A. & Takai, Y. (1998) J. Biol. Chem. 273**,** 3470-3475. [DOI] [PubMed] [Google Scholar]
- 11.Feng, J., Yan, Z., Ferreira, A., Tomizawa, K., Liauw, J. A., Zhuo, M., Allen, P. B., Ouimet, C. C. & Greengard, P. (2000) Proc. Natl. Acad. Sci. USA 97**,** 9287-9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grossman, S. D., Hsieh-Wilson, L. C., Allen, P. B., Nairn, A. C. & Greengard, P. (2002) Neuromolecular Med. 2**,** 61-69. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh-Wilson, L. C., Benfenati, F., Snyder, G. L., Allen, P. B., Nairn, A. C. & Greengard, P. (2003) J. Biol. Chem. 278**,** 1186-1194. [DOI] [PubMed] [Google Scholar]
- 14.Grossman, S. D., Futter, M., Snyder, G. L., Allen, P. B., Nairn, A. C., Greengard, P. & Hsieh-Wilson, L. C. (2004) J. Neurochem. 90**,** 317-324. [DOI] [PubMed] [Google Scholar]
- 15.Bailey, C. H., Kaang, B. K., Chen, M., Martin, K. C., Lim, C. S., Casadio, A. & Kandel, E. R. (1997) Neuron 18**,** 913-924. [DOI] [PubMed] [Google Scholar]
- 16.Atkins, C. M., Selcher, J. C., Petraitis, J. J., Trzaskos, J. M. & Sweatt, J. D. (1998) Nat. Neurosci. 1**,** 602-609. [DOI] [PubMed] [Google Scholar]
- 17.Berman, D. E., Hazvi, S., Rosenblum, K., Seger, R. & Dudai, Y. (1998) J. Neurosci. 18**,** 10037-10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crow, T., Xue-Bian, J. J., Siddiqi, V., Kang, Y. & Neary, J. T. (1998) J. Neurosci. 18**,** 3480-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu, G. Y., Deisseroth, K. & Tsien, R. W. (2001) Nat. Neurosci. 4**,** 151-158. [DOI] [PubMed] [Google Scholar]
- 20.Bibb, J. A., Chen, J., Taylor, J. R., Svenningsson, P., Nishi, A., Snyder, G. L., Yan, Z., Sagawa, Z. K., Ouimet, C. C., Nairn, A. C., Nestler, E. J. & Greengard, P. (2001) Nature 410**,** 376-380. [DOI] [PubMed] [Google Scholar]
- 21.Norrholm, S. D., Bibb, J. A., Nestler, E. J., Ouimet, C. C., Taylor, J. R. & Greengard, P. (2003) Neuroscience 116**,** 19-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito, T., Onuki, R., Fujita, Y., Kusakawa, G., Ishiguro, K., Bibb, J. A., Kishimoto, T. & Hisanaga, S. (2003) J. Neurosci. 23**,** 1189-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh-Wilson, L. C., Allen, P. B., Watanabe, T., Nairn, A. C. & Greengard, P. (1999) Biochemistry 38**,** 4365-4373. [DOI] [PubMed] [Google Scholar]
- 24.Hamada, M., Higashi, H., Nairn, A. C., Greengard, P. & Nishi, A. (2004) J. Neurochem. 90**,** 1094-1103. [DOI] [PubMed] [Google Scholar]
- 25.Nishi, A., Snyder, G. L., Nairn, A. C. & Greengard, P. (1999) J. Neurochem. 72**,** 2015-2021. [DOI] [PubMed] [Google Scholar]
- 26.Bibb, J. A., Snyder, G. L., Nishi, A., Yan, Z., Meijer, L., Fienberg, A. A., Tsai, L. H., Kwon, Y. T., Girault, J. A., Czernik, A. J., et al. (1999) Nature 402**,** 669-671. [DOI] [PubMed] [Google Scholar]
- 27.Barnes, A. P., Smith, F. D., III, VanDongen, H. M., VanDongen, A. M. & Milgram, S. L. (2004) Brain Res. Mol. Brain Res. 124**,** 105-113. [DOI] [PubMed] [Google Scholar]
- 28.Stafstrom-Davis, C. A., Ouimet, C. C., Feng, J., Allen, P. B., Greengard, P. & Houpt, T. A. (2001) Learn. Mem. 8**,** 272-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan, Z., Hsieh-Wilson, L., Feng, J., Tomizawa, K., Allen, P. B., Fienberg, A. A., Nairn, A. C. & Greengard, P. (1999) Nat. Neurosci. 2**,** 13-17. [DOI] [PubMed] [Google Scholar]
- 30.Rao, A. & Craig, A. M. (2000) Hippocampus 10**,** 527-541. [DOI] [PubMed] [Google Scholar]
- 31.Dailey, M. E. & Smith, S. J. (1996) J. Neurosci. 16**,** 2983-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliver, C.J., Terry-Lorenzo, R.T., Elliott, E., Christensen-Bloomer, W.A., Li, S., Brautigan, D.L., Colbran, R.J. & Shenolikar, S. (2002) Mol. Cell. Biol. 22**,** 4690-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zito, K., Knott, G., Shepherd, G. M., Shenolikar, S. & Svoboda, K. (2004) Neuron 44**,** 321-334. [DOI] [PubMed] [Google Scholar]
- 34.Liu, F., Ma, X. H., Ule, J., Bibb, J. A., Nishi, A., DeMaggio, A. J., Yan, Z., Nairn, A. C. & Greengard, P. (2001) Proc. Natl. Acad. Sci. USA 98**,** 11062-11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurino, M., Fukunaga, K., Ushio, Y. & Miyamoto, E. (1995) J. Neurochem. 65**,** 1282-1289. [DOI] [PubMed] [Google Scholar]
- 36.Fiore, R. S., Murphy, T. H., Sanghera, J. S., Pelech, S. L. & Baraban, J. M. (1993) J. Neurochem. 61**,** 1626-1633. [DOI] [PubMed] [Google Scholar]
- 37.Xia, Z., Dudek, H., Miranti, C. K. & Greenberg, M. E. (1996) J. Neurosci. 16**,** 5425-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information