The chromatoid body of male germ cells: Similarity with processing bodies and presence of Dicer and microRNA pathway components (original) (raw)
Abstract
The chromatoid body is a perinuclear, cytoplasmic cloud-like structure in male germ cells whose function has remained elusive. Here we show that the chromatoid body is related to the RNA-processing body of somatic cells. Dicer and components of microRNP complexes (including Ago proteins and microRNAs) are highly concentrated in chromatoid bodies. Furthermore, we show that Dicer interacts with a germ cell-specific chromatoid body component, the RNA helicase MVH (mouse VASA homolog). Thus, chromatoid bodies seem to operate as intracellular nerve centers of the microRNA pathway. Our findings underscore the importance of posttranscriptional gene regulation and of the microRNA pathway in the control of postmeiotic male germ cell differentiation.
Keywords: Argonaute, microRNA, spermatogenesis, RNA processing
The complex and highly elaborated differentiation program of male germ cells involves multiple and finely tuned levels of gene control (1). During late steps of spermiogenesis there is a striking cessation of transcription, occuring in concert with drastic epigenetic modifications that result in chromatin compaction (2). Concomitantly there are extensive posttranscriptional storing and processing of mRNAs (3).
RNA interference (RNAi) and microRNA (miRNA) pathways are evolutionarily conserved control mechanisms that use RNA molecules to inhibit gene expression at the level of mRNA degradation, translational repression, or chromatin modification and silencing (4–6). RNAi has been shown to be present throughout spermatogenesis in mice (7), but its function and cellular control during germ cell development remain uncertain.
Two classes of 21- to 25-nt small RNAs, small interfering RNAs (siRNAs) and miRNAs, act as sequence-specific regulators of gene expression (5). siRNAs mediate degradation of mRNAs having sequences fully complementary to their sequence, whereas miRNAs are proposed to regulate gene expression by inhibiting protein synthesis through imperfect base-pairing to the 3′ UTR of target mRNAs (4, 5). Both siRNA and miRNA precursors are processed to mature small RNAs in the cytoplasm of cells by the large endonuclease Dicer (4, 8). Mature miRNAs and siRNAs are assembled into miRISC and siRISC (miRNA- and siRNA-induced silencing complexes, respectively), which subsequently act on their targets by translational repression or mRNA cleavage. Essential components of RISC complexes are the members of the Argonaute family of proteins (9). These proteins share the so-called PAZ and PIWI domains and are classified into two subfamilies depending on sequence similarity to either Arabidopsis Argonaute1 or Drosophila Piwi (9, 10). In mammals, Argonaute1 subfamily members, Ago1 to Ago4, have been shown to be involved in the RNAi/miRNA pathway (11, 12). All four members of the PIWI subfamily are expressed mainly in testis (10), and two of them, MIWI and MILI, are crucial for progression through spermatogenesis in mouse (13, 14).
In many organisms, including Drosophila, germ cells are characterized by the accumulation of dense fibrous material into a cytoplasmic structure, called germplasm or nuage (15). The chromatoid body is suggested to be a mammalian counterpart of nuage on the basis of its structural features and protein composition (16). It is a finely filamentous, lobulated perinuclear granule located in the cytoplasm of mammalian postmeiotic round spermatids. Both Drosophila nuage and mouse chromatoid body contain an ATP-dependent DEAD-box RNA helicase, VASA [in the mouse MVH (mouse VASA homolog)] (17–19). Drosophila nuage is thought to function as a site for translational regulation of many important mRNAs, the VASA protein seemingly playing a central role in this process (20). The role of the chromatoid body in the mouse is still elusive, although it was proposed to be involved in RNA storing and processing. In addition to MVH, some other proteins have been reported to localize in the chromatoid body, most of which are known to be involved in RNA metabolism (16). Although there is no DNA in the chromatoid body (21), the presence of RNA has been suggested (22–25).
Here we demonstrate the physical relationship between the chromatoid body and the miRNA machinery in haploid germ cells. Dicer and components of miRISC (also referred to as microRNP), such as various members of the Argonaute family and several miRNA species, concentrate in the chromatoid body. The presence of Dcp1a and other specific proteins in the chromatoid body provides evidence of a functional analogy with the processing bodies (P-bodies), cytoplasmic structures found in somatic cells (ref. 26 and references therein). We have also found that Dicer interacts with the VASA homolog MVH. These findings shed new light on the function of the chromatoid body, a structure whose existence has been known for decades (16) but whose role has remained unknown. In addition, our results underscore the importance of the miRNA pathway in the control of postmeiotic male germ cell differentiation.
Results
Colocalization of the VASA Homolog MVH and MIWI in Chromatoid Body.
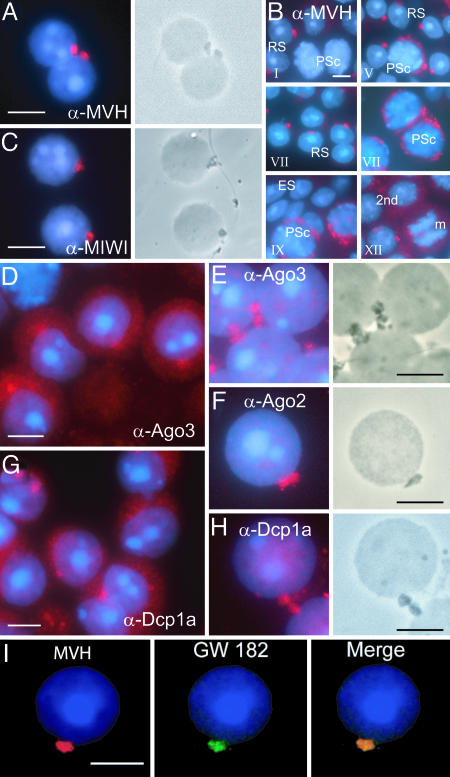
The highly restricted localization of MVH in the chromatoid body has been used as specific marker for this structure (18). We confirmed this observation by performing immunostaining with anti-MVH antibodies on male germ cells isolated from stages II–V of seminiferous epithelial cycle (Fig. 1A). Chromatoid bodies are clearly visible in parallel phase contrast microscopy as electron-dense areas (Fig. 1A). Using the drying-down technique to prepare the cell slides, the cytoplasm is partially lost to expose the chromatoid body, which usually stays in contact with the nucleus.
Fig. 1.
Localization of MVH and Ago proteins in chromatoid body. (A) Drying-down slides containing germ cells from stages II–V were labeled with anti-MVH antibody (red). The localization in chromatoid body was confirmed by parallel phase contrast microscopy. (B) Expression of MVH in chromatoid bodies during spermatogenesis. Squash preparations from stages I, V, VII, IX, and XII were stained with anti-MVH antibody (red) and studied by fluorescence microscopy. RS, round spermatid; PSc, pachytene spermatocyte; 2nd, secondary spermatocyte; m, meiotic division. (C) Localization of MIWI in chromatoid body. Germ cells from stages II–V were labeled with anti-MIWI antibody. (D and E) Ago3 is concentrated in chromatoid bodies. Germ cell squash preparations (D) at stage IV or drying-down slides containing germ cells from stages II–V (E) were immunolabeled by polyclonal anti-Ago3 antibody (red). (F) Ago2 was also shown to be concentrated in chromatoid bodies by using polyclonal anti-Ago2 antibody. (G and H) Localization of the P-body marker, the decapping enzyme Dcp1a, in chromatoid bodies. Germ cell squash preparations at stages III–IV (G) or drying-down slides from stages II–V (H) were immunostained by polyclonal anti-Dcp1a antibody. (I) Chromatoid body localization of a GW body marker protein. Patient sera against human GW182 protein (18033; a gift from Marvin J. Fritzler, University of Calgary, Calgary, Canada) were used for localization studies. For MVH detection, rabbit polyclonal anti-MVH antibody was used as a primary antibody. Alexa Fluor 594 anti-rabbit IgG or Alexa Fluor 488 anti-human IgG were used as secondary antibodies, and DAPI was used to stain nuclei (blue). (Scale bars: 5 μm.)
We analyzed the formation of the chromatoid body along the differentiation of postmeiotic germ cells. Stage-specific sequential squash preparations show MVH expression in chromatoid bodies throughout the development of round spermatids (Fig. 1B). Expression is detected in the cytoplasm of pachytene spermatocytes at stages IV–V within filamentous–granular structures. In round spermatids, the chromatoid body becomes finely compacted, and usually there is only one per cell. The RNA-binding protein MIWI has been shown to have a granular distribution in the cytoplasm of haploid round spermatids (14), but it was unclear whether these granules corresponded to chromatoid bodies. We demonstrate that MIWI is indeed concentrated in chromatoid bodies, colocalizing with MVH (Fig. 1C).
Argonaute Proteins and P-body Components in the Chromatoid Body.
The presence of MIWI in chromatoid bodies prompted us to analyze the localization of Ago subfamily members to establish a possible relationship with the miRNA pathway. The function of Ago proteins in male germ cells is presently unknown. Squash preparations at stage IV showed a granular cytoplasmic localization of Ago3 (Fig. 1D). Although Ago3 was diffused throughout the cytoplasm, the signal was concentrated on one cytoplasmic spot in close contact with the nucleus (Fig. 1D). This granule corresponds to the chromatoid body, as revealed by immunofluorescence of germ cell slides and parallel phase contrast (Fig. 1E). Ago2 was also concentrated in the chromatoid body (Fig. 1F). It has been recently shown that in somatic cells Ago proteins localize in cytoplasmic P-bodies (27–29). We therefore analyzed the localization of a P-body marker, the decapping enzyme component Dcp1a (30), in male germ cells. Strikingly, Dcp1a was also concentrated in chromatoid bodies (Fig. 1 G and H). The distribution of Dcp1a along spermatogenesis is dynamic. Although it has a granular cytoplasmic localization in both meiotic and postmeiotic germ cells, in late pachytene spermatocytes there seem to be several Dcp1a granules (data not shown), which then concentrate postmeiotically in the chromatoid body (Fig. 1H). Two other P-body markers, the 5′-to-3′ exonuclease Xrn1 (data not shown) and the RNA-binding protein GW182 (Fig. 1I) (31), were also found in chromatoid bodies.
Because P-body components localize to the chromatoid body, we questioned whether the male germ cell-specific MVH could potentially be targeted to P-bodies in somatic cells. To this end, FLAG-tagged MVH and HA-Ago2 or HA-Ago3 were ectopically expressed in HeLa cells. Indeed, MVH colocalized to the same cytoplasmic granules with Ago2 and Ago3 (Fig. 6, which is published as supporting information on the PNAS web site). MVH mutants containing only the N terminus [FLAG-MVH(1–199)] or lacking the central ATPase domain (FLAG-MVHΔ283–503) (see Fig. 5) failed to colocalize with Ago proteins (Fig. 6). MVH-positive granules were shown to correspond to P-bodies by coimmunostaining of endogenous Dcp1a (Fig. 6). Thus, the P-bodies of somatic cells and the chromatoid body of male germ cells share a number of significant similarities.
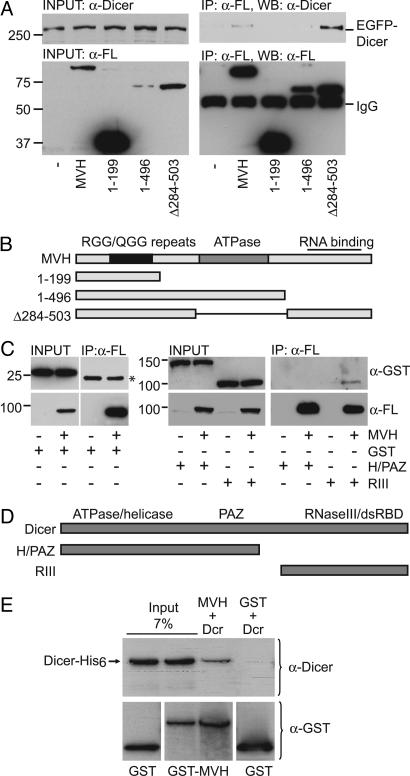
Fig. 5.
Dicer interacts with MVH. (A) Interaction of the full-length EGFP-Dicer with MVH. IP was performed from COS-1 lysates overexpressing EGFP-Dicer and FLAG-MVH, FLAG-MVH(1–199), FLAG-MVH(1–496), or FLAG-MVHΔ284–503 with anti-FLAG antibody (α-FL), and the immunoblotting was performed by using either anti-Dicer antibody to detect coimmunoprecipitated Dicer or anti-FLAG antibody to detect MVH. (B) Schematic diagram of the MVH constructs used in this study. (C) The RNaseIII region of Dicer mediates the interaction with MVH. GST, GST-Helicase/PAZ containing the N-terminal helicase domain and the central PAZ domain of Dicer (H/PAZ), or GST-RNaseIII containing the C-terminal RNaseIII region and dsRNA binding domain of Dicer (RIII) was coexpressed with FLAG-MVH. After immunoprecipitation with anti-FLAG antibody, immunoblotting was done with anti-GST or anti-FLAG antibody. An asterisk indicates the IgG light chain. (D) Schematic representation of the Dicer mutants used in this study. (E) MVH and Dicer interact in vitro. Recombinant GST-MVH and Dicer-6xHis were incubated with glutathione Sepharose. After binding and washes, beads were run into a polyacrylamide gel, and immunoblotting was performed with anti-GST or anti-Dicer antibody.
miRNAs Are Concentrated in Chromatoid Bodies.
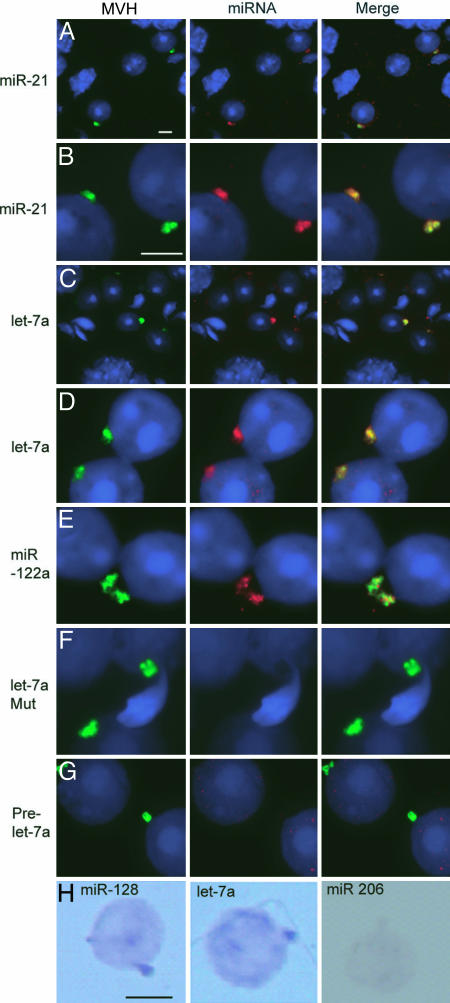
We performed in situ hybridizations using probes specific for various miRNAs known to be expressed in testis to study whether their localization overlaps with Ago proteins in chromatoid bodies. The analysis on squash preparation of stages V–VI, or drying-down slides of germ cells from stages II–VI, revealed high concentration of miR-21 and let-7a in perinuclear granules in round spermatids (Fig. 2A–D). The signal overlapped with MVH signal, clearly demonstrating that miRNAs accumulated in chromatoid bodies. miR-122a also colocalized with MVH in chromatoid body (Fig. 2E). A mutant let-7a probe, used as a negative control, gave no signal (Fig. 2F). Interestingly, no enrichment of signal was detected with the prelet7a RNA probe, indicating that it is primarily the processed form of let7a that localizes to chromatoid bodies (Fig. 2G). Chromatoid body localization of miR-128 and let-7a, but not of nontestis-expressed control miRNA miR-206, was also demonstrated by in situ hybridization of drying-down slides by using the colorimetric detection method (Fig. 2H).
Fig. 2.
Localization of miRNA in chromatoid bodies. (A–G) In situ hybridization was performed with DIG-labeled locked nucleotide probes specific for different miRNAs. Except in A and C, samples fixed by the drying-down method were used for analysis. In A and C, squashed samples from stage II–VI seminiferous tubules were used. The miRNA-specific probes used in each panel are indicated. The prelet-7a probe was designed to detect the prelet-7a but not the mature let-7a. let-7a Mut is a mutant version of let-7a-specific probe. After in situ hybridization, miRNA signals were detected with the Roche fluorescent antibody enhancer set for DIG detection (red). MVH signals are in green, and the nucleus was stained with DAPI (blue). The merged pictures are also shown. (H) detection of miRNA by the colorimetric method. miR-206-specific probe is used as a negative control. (Scale bars: 5 μm.)
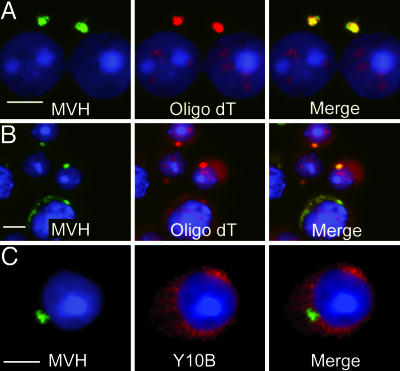
Localization of the RISC components and miRNAs in chromatoid bodies motivated us to study whether mRNA molecules, the natural targets for miRNA-mediated regulation, also concentrate in chromatoid bodies. In situ hybridization using oligo(dT) probe clearly demonstrated accumulation of mRNAs in chromatoid bodies (Fig. 3A and B). Interestingly, antibodies specifically recognizing rRNA did not stain chromatoid body (Fig. 3C); likewise, antibodies recognizing the small ribosomal subunit protein S6 showed no signal overlapping with the MVH-positive chromatoid bodies (data not shown), indicating that active translation at this specific stage of spermatogenesis occurs in locations distinct from chromatoid bodies.
Fig. 3.
Poly(A)+ RNA is concentrated in chromatoid bodies. A Cy3-labeled oligo(dT) probe was used to detect mRNAs with a polyA tail in cells fixed by the drying-down method (A) and also in cells of stage V–VII squash preparations (B). After in situ hybridization with Cy3-labeled oligo(dT), MVH signals were revealed by immunofluorescence (green). DAPI was used to stain the nucleus. (C) Ribosomal RNA localization in round spermatids. Ribosomes localized to cytoplasmic regions (red) distinct from the chromatoid body positive for MVH (green). (Scale bars: 5 μm.)
Dicer Is Concentrated in the Chromatoid Body.
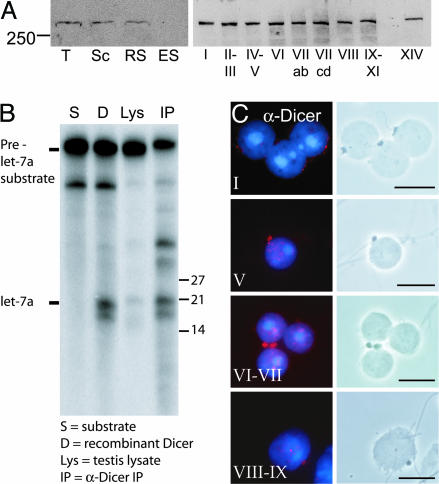
The presence of Ago proteins and miRNAs in the chromatoid body directed us to study Dicer expression during spermatogenesis. Dicer is present in both meiotic spermatocytes and postmeiotic round spermatids but not in elongated spermatids (Fig. 4A). Expression is constant along the stages of the seminiferous epithelial cycle (Fig. 4A). We have also established that the Dicer protein in testis is enzymatically active (Fig. 4B). Immunostaining of germ cell slides from specific stages revealed that the expression of Dicer is indeed concentrated in chromatoid bodies (Fig. 4C). Finally, a detailed analysis along the differentiation of male germ cells shows a stage-dependent distribution of Dicer in the chromatoid body: In step 1 round spermatids, Dicer expression is still very low, but it increases at stages VI–VII. At stage VIII, Dicer levels in the chromatoid body decrease significantly. Thus, Dicer localization to the chromatoid body is highly dynamic, being at its peak coordinately with the other elements of the miRNA machinery.
Fig. 4.
Localization of Dicer in chromatoid body. (A) Expression of Dicer protein in testis. Western blot analysis of the protein extracts of rat total testis cells (T), pachytene spermatocytes (Sc), round spermatids (RS), and elongated spermatids (ES), as well as of different stages of the rat seminiferous epithelial cycle (indicated as Roman numerals I–XIV), was performed by using anti-Dicer antibody. (B) premiRNA-processing activity in testis. prelet-7a substrate was processed when testis extract (Lys) or anti-Dicer immunoprecipitate (IP) was added to the reaction. Reaction with recombinant Dicer (D) was used as a positive control. (C) Dicer is expressed in the chromatoid body of late round spermatids. The drying-down slides were immunostained with the anti-Dicer 349 antibody and studied by both fluorescence microscopy (red, Dicer; blue, DNA) and phase contrast microscopy to confirm the chromatoid body localization. Similar staining was obtained with anti-Dicer 350 antibody. (Scale bars: 10 μm.)
Dicer Interacts with the VASA Homolog MVH.
Although Dicer is known to interact with other proteins involved in RNA silencing (refs. 32 and 33 and references therein), its presence in the chromatoid body raised the possibility that it could interact with germ cell-specific elements, in particular MVH. FLAG-tagged MVH and EGFP-tagged Dicer were ectopically coexpressed in COS-1 cells and then coimmunoprecipitated (Fig. 5A). We found that MVH and Dicer interact. We have used a number of MVH deletions to establish the protein domain involved in Dicer interaction (Fig. 5 A and B). The mutants MVH(1–199) and MVH(1–496) did not interact with Dicer, despite containing the central ATPase motif and the N-terminal RGG/QGG repeats known to be involved in protein/RNA interactions (17). This result shows that the C terminus of MVH, which contains a RNA-binding domain (17), is required for Dicer interaction (Fig. 5A). Interestingly, the MVHΔ284–503 deletion, which lacks the central ATPase motif, interacts with Dicer more efficiently than the full-length MVH protein. This finding may suggest a negative regulatory role of this region in controlling the interaction with Dicer.
Dicer is a multidomain protein that includes a RNA helicase/ATPase domain, the DUF283 and PAZ signatures, two neighboring RNase III-like domains, and a dsRNA binding domain (34). Our coimmunoprecipitation experiments demonstrate that the C-terminal RNaseIII region is sufficient for interaction with MVH (Fig. 5C). In support of this observation, a large portion of the Dicer protein containing the N-terminal ATPase/helicase domains and the central PAZ signatures did not interact with MVH (Fig. 5 C and D). Interestingly, HIWI, the human homolog of mouse MIWI, has also been shown to interact with the C terminus of Dicer (32). This interaction was used as a positive control (data not shown). Finally, the direct interaction between MVH and Dicer was confirmed by in vitro interaction assays by using bacterially generated recombinant GST-MVH and Dicer-6xHis (Fig. 5E). Thus, MVH appears to be a male germ cell-specific component of the Dicer complex.
Discussion
The molecular nature and function of the chromatoid body in germ cells have been elusive and debated for many years (16). Our findings support a scenario where the chromatoid body would function as an RNA storing and processing structure. Specifically, the presence of Dicer/Argonaute and miRNAs reveals that the chromatoid body occupies a privileged position in posttranscriptional control of gene expression through the small RNAs pathway.
To date, very little was known about the function of siRNA and miRNA pathways and the expression and localization of the components of these pathways during spermatogenesis. RNAi has been shown to be active during the whole spermatogenesis program (7), and many miRNAs whose expression is enriched in testis have been identified (35, 36). Here we demonstrate that the main component of the RNA-silencing pathway, Dicer, is expressed in both meiotic and postmeiotic cells at all stages of the seminiferous epithelial cycle.
Dicer processes miRNA precursor molecules folded into dsRNA-like hairpins to mature miRNAs. These are subsequently transferred to the miRISC complex, where the effector phase of the process takes place (4, 8). A direct interaction between Dicer and Ago proteins may be required for the transfer of miRNAs to the effector complex (8). The presence in the chromatoid body of several miRNAs, of the enzyme that processes them, and of proteins essential for the effector phase of mRNA silencing strongly supports the notion that functional miRNA-mediated mRNA control is taking place in this compartment. Interestingly, a potential connection between germplasm (nuage) and miRNA pathway has also been suggested in Drosophila. Indeed, mutation of the nuage protein Maelstrom causes mislocalization of two miRNA pathway proteins, Dicer and Ago2 (37).
MIWI, a member of the PIWI subfamily of Argonaute proteins, localizes to the chromatoid body (Fig. 1). Argonaute1 subfamily members are widely expressed in many tissues. In contrast, all four members of PIWI subfamily are expressed mainly in testis (10), but their role in the RNAi pathway is still obscure (9). MIWI associates with ribohomopolymers, and specifically with transcripts of CREM-target genes, suggesting a functional connection of MIWI to RNA processing (13). Importantly, mice deficient for either MIWI or CREM display a similar block in spermatogenesis (13, 38). Because MIWI and the Argonaute1 subfamily members Ago2 and Ago3 colocalize in the chromatoid body (Fig. 1), it is tempting to speculate that MIWI may operate as a male germ cell-specific miRNA pathway component involved in functional mRNA regulation in testis.
In yeast and mammalian somatic cells, mRNAs targeted to translational repression and degradation accumulate in cytoplasmic P-bodies, where the enzymes involved in the RNA decay pathway are also concentrated (30, 39–41). Recently, Ago proteins, and also miRNAs and miRNA-repressed mRNAs, were demonstrated to localize in P-bodies in mammalian cells (27–29), suggesting that RNA silencing and RNA decay pathways may take place in the same cellular compartments. The current model proposes that mammalian miRNAs inhibit the initiation of translation of mRNAs that subsequently accumulate in P-bodies for storage (29). We have demonstrated that several P-body markers are highly concentrated in chromatoid bodies (Fig. 1). These results suggest that the chromatoid body of male germ cells and P-bodies in somatic cells are functionally related, both acting as a site for mRNA decay and mRNA translational repression by the miRNA pathway. Thus, our findings provide an attractive interpretation of the phenomenon of translational repression that occurs postmeiotically in spermatids (42, 43).
One significant result reported here is that an essential component of the chromatoid body, the RNA helicase/ATPase MVH, interacts with Dicer. MVH may operate as an anchoring element for RNA-silencing components, or it might have a more functional role. Interestingly, the turnover of the RISC cleavage activity depends on ATP, suggesting that release of the cleaved mRNA halves may involve RNA helicases (8, 44). Thus, our finding reveals MVH as a potential germ cell-specific candidate for providing helicase/ATPase activity to the RISC complex.
The chromatoid body is endowed with the remarkable property of moving very actively and three-dimensionally in the cytoplasm of round spermatids. During these movements chromatoid bodies make frequent contacts with the nuclear envelope. Continuity in electron-dense material between the nucleus and the chromatoid body through nuclear pore complexes has been observed (16). One hypothesis suggests that rapidly moving chromatoid bodies collect mRNA getting out from the nucleus. Interestingly, miRNA precursors processed in the nucleus are exported to the cytoplasm through nuclear pore complexes (45). Based on our findings we suggest a model in which premiRNAs transported to the cytoplasm are loaded through nuclear pores to the chromatoid body. Thus, the chromatoid body functions as a subcellular concentration site for components of the miRNA pathway, centralizing the miRNA posttranscriptional control system in the cytoplasm of haploid male germ cells.
Materials and Methods
Immunofluorescence.
Squash preparations or drying-down slides (46) were fixed in 4% paraformaldehyde, and immunofluorescence was performed by using specific polyclonal or monoclonal antibodies and Alexa Fluor 594 and 488 secondary antibodies (Molecular Probes).
miRNA in Situ.
Locked nucleotide probes were labeled with digoxigenin (DIG) by using the terminal transferase 3′ DIG-tailing kit (Roche). Fixed cells were permeabilized in 70% ethanol, rehydrated in 2× SSC for 15 min, and hybridized overnight at 37°C with 15 ng of the probe. After three washes, slides were incubated with anti-DIG alkaline phosphate-conjugated Fab fragment (1:5,000; Roche), and signals were developed with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate in the dark. In colocalization studies the Fluorescent Antibody Enhancer Set for DIG Detection (Roche) was used.
RNA Processing Assays.
The internally32P-labeled prelet-7 RNA was dissolved in water and renaturated by incubation at 90°C for 1 min. Processing assays (50 μl) were carried out as previously described in refs. 34 and 47. 32P-labeled substrate (3–5 fmol) was incubated with a testis extract or anti-Dicer immunoprecipitate in buffer containing 20 mM Tris·HCl (pH 7.5), 2 mM MgCl2, 75 mM NaCl, and 10% glycerol at 37°C. RNA was extracted with phenol/chloroform and analyzed by 10% denaturing PAGE.
Immunoprecipitation.
COS-1 cells transfected with indicated plasmids were collected and lysed in a buffer containing 50 mM Tris·HCl (pH 8.0), 170 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, 1 mM DTT, and 1:1,000 protease inhibitor mixture. Immunoprecipitation was performed with anti-FLAG M2 (Sigma) monoclonal antibody and protein G Sepharose (Amersham Pharmacia Biosciences). After washes, proteins were released in 2× Laemmli sample buffer. Immunoblotting was performed by anti-GST (1:1,000), anti-FLAG M2 (1:1,000), or anti-Dicer 349 (1:1,000) antibody.
Recombinant Proteins and in Vitro Interaction.
Recombinant GST-MVH was purified from the BL21 Escherichia coli strain with glutathione Sepharose (Amersham Pharmacia) and eluted from the beads with 20 mM glutathione. Recombinant Dicer was purified as described (47). In vitro protein interactions with recombinant proteins were analyzed in buffer containing 30 mM Tris·HCl (pH 7.5), 1 mM MgCl2, 100 mM NaCl, and 0.5% Nonidet P-40 by using standard methods.
Specific reagents and detailed additional procedures are fully described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Supporting Information
Acknowledgments
We thank H. Lin (Duke University Medical Center, Durham, NC), T. Noce (Mitsubishi Kagaku Institute of Life Sciences, Tokyo), T. Hobman (University of Alberta, Edmonton, AB, Canada), J. Lykke-Andersen (University of Colorado, Boulder, CO), M. J. Fritzler (University of Calgary, Calgary, AB, Canada), M. Kiledjian (Rutgers, The State University of New Jersey, Piscataway, NJ), J. Steitz (Howard Hughes Medical Institute, New Haven, CT), and M. Drozdz and T. Zoller (both from Freidrich Miescher Institute for Biomedical Research, Basel) for antibodies, and we thank M. Carmell (Cold Spring Laboratory, Cold Spring Harbor, NY) and all members of the P.S.-C. and W.F. laboratories for help and stimulating discussions. N.K. was supported by the European Molecular Biology Organization, and S.K. was supported by a fellowship from the Fondation pour la Recherche Médicale. S.N.B. is the recipient of a fellowship from the Human Frontiers Science Program. Our studies are supported by grants from the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Fondation de la Recherche Médicale, and Université Louis Pasteur. The P.S.-C. laboratory is an “Equipe Labelisée” of La Ligue Contre le Cancer. Research in the Friedrich Miescher Institute, a part of the Novartis Research Foundation, was partially supported by EC FP6 STREP Program LSHG-CT-2004.
Abbreviations
siRNA
small interfering RNA
miRNA
microRNA
RNAi
RNA interference
RISC
RNA-induced silencing complex
MVH
mouse VASA homolog
P-body
processing body
DIG
digoxigenin.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sassone-Corsi P. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- 2.Kimmins S., Sassone-Corsi P. Nature. 2005;434:583–589. doi: 10.1038/nature03368. [DOI] [PubMed] [Google Scholar]
- 3.Steger K. Anat. Embryol. 2001;203:323–334. doi: 10.1007/s004290100176. [DOI] [PubMed] [Google Scholar]
- 4.Filipowicz W., Jaskiewicz L., Kolb F. A., Pillai R. S. Curr. Opin. Struct. Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Zamore P. D., Haley B. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein E., Allis C. D. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 7.Shoji M., Chuma S., Yoshida K., Morita T., Natatsuji N. Dev. Biol. 2005;282:524–534. doi: 10.1016/j.ydbio.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 8.Sontheimer E. J. Nat. Rev. Mol. Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- 9.Carmell M. A., Xuan Z., Zhang M. Q., Hannon G. J. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- 10.Sasaki T., Shiohama A., Minoshima S., Shimizu N. Genomics. 2003;82:323–330. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 11.Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Liu J., Carmell M. A., Rivas F. V., Marsden C. G., Thomson J. M., Song J. J., Hammond S. M., Joshua-Tor L., Hannon G. J. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 13.Deng W., Lin H. Dev. Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 14.Kuramochi-Miyagawa S., Kimura T., Ijiri T. W., Isobe T., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W., et al. Development (Cambridge, U.K.) 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 15.Ikenishi K. Dev. Growth Differ. 1998;40:1–10. doi: 10.1046/j.1440-169x.1998.t01-4-00001.x. [DOI] [PubMed] [Google Scholar]
- 16.Parvinen M. Int. J. Androl. 2005;28:189–201. doi: 10.1111/j.1365-2605.2005.00542.x. [DOI] [PubMed] [Google Scholar]
- 17.Fujiwara Y., Komiya T., Kawabata H., Sato M., Fujimoto H., Furusawa M., Noce T. Proc. Natl. Acad. Sci. USA. 1994;91:12258–12262. doi: 10.1073/pnas.91.25.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toyooka Y., Tsunekawa N., Takahashi Y., Matsui Y., Satoh M., Noce T. Mech. Dev. 2000;93:139–149. doi: 10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka S. S., Toyooka Y., Akasu R., Katoh-Fukui Y., Nakahara Y., Suzuki R., Yokoyama M., Noce T. Genes Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- 20.Styhler S., Nakamura A., Swan A., Suter B., Lasko P. Development (Cambridge, U.K.) 1998;125:1569–1578. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- 21.Biggiogera M., Fakan S., Leser G., Martin T. E., Gordon J. Mol. Reprod. Dev. 1990;26:150–158. doi: 10.1002/mrd.1080260209. [DOI] [PubMed] [Google Scholar]
- 22.Söderström K. O., Parvinen M. J. Cell Biol. 1976;70:239–246. doi: 10.1083/jcb.70.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walt H., Armbruster B. L. Cell Tissue Res. 1984;236:487–490. doi: 10.1007/BF00214254. [DOI] [PubMed] [Google Scholar]
- 24.Figueroa J., Burzio L. O. Cell Tissue Res. 1998;291:575–579. doi: 10.1007/s004410051027. [DOI] [PubMed] [Google Scholar]
- 25.Saunders P. T., Millar M. R., Maguire S. M., Sharpe R. M. Mol. Reprod. Dev. 1992;33:385–391. doi: 10.1002/mrd.1080330404. [DOI] [PubMed] [Google Scholar]
- 26.Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fitzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen G. L., Blau H. M. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 28.Liu L., Valencia-Sanchez M. A., Hannon G. J., Parker R. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pillai R. S., Bhattacharyya S. N., Artus C. G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 30.Cougot N., Babajko S., Séraphin B. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z., Jakymiw A., Wood M. R., Eystathioy T., Rubin R. L., Fritzler M. J., Chan E. K. J. Cell Sci. 2004;117:5567–5578. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- 32.Tahbaz N., Kolb F. A., Zhang H., Jaronczyk K., Filipowicz W., Hobman T. C. EMBO Rep. 2004;5:189–194. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haase A. D., Jaskiewicz L., Zhang H., Laine S., Sack R., Gatignol A., Filipowicz W. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H., Kolb F. A., Jaskiewicz L., Westhof E., Filipowicz W. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 35.Barad O., Meiri E., Avniel A., Aharonov R., Barzilai A., Bentwich I., Einav U., Gilad S., Hurban P., Karov Y., et al. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Z., Raabe T., Hecht N. B. Biol. Reprod. 2005;73:427–433. doi: 10.1095/biolreprod.105.040998. [DOI] [PubMed] [Google Scholar]
- 37.Findley S. D., Tamanaha M., Clegg N. J., Ruohola-Baker H. Development (Cambridge, U.K.) 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- 38.Nantel F., Monaco L., Foulkes N. S., Masquilier D., LeMeur M., Henriksen K., Dierich A., Parvinen M., Sassone-Corsi P. Nature. 1996;380:159–162. doi: 10.1038/380159a0. [DOI] [PubMed] [Google Scholar]
- 39.Sheth U., Parker R. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brengues M., Teixeira D., Parker R. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrei M. A., Ingelfinger D., Heintzmann R., Achsel T., Rivera-Pomar R., Luhrmann R. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleene K. C. Dev. Biol. 1993;159:720–731. doi: 10.1006/dbio.1993.1277. [DOI] [PubMed] [Google Scholar]
- 43.Sassone-Corsi P. Cell. 1997;88:163–166. doi: 10.1016/s0092-8674(00)81834-6. [DOI] [PubMed] [Google Scholar]
- 44.Haley B., Zamore P. D. Nat. Struct. Mol. Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 45.Cullen B. R. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Kotaja N., Kimmins S., Brancorsini S., Hentsch D., Vonesch J. L., Davidson I., Parvinen M., Sassone-Corsi P. Nat. Methods. 2004;1:249–254. doi: 10.1038/nmeth1204-249. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H., Kolb F. A., Brondani V., Billy E., Filipowicz W. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information