The APC tumor suppressor counteracts β-catenin activation and H3K4 methylation at Wnt target genes (original) (raw)
Abstract
The APC tumor suppressor controls the stability and nuclear export of β-catenin (β-cat), a transcriptional coactivator of LEF-1/TCF HMG proteins in the Wnt/Wg signaling pathway. We show here that β-cat and APC have opposing actions at Wnt target genes in vivo. The β-cat C-terminal activation domain associates with TRRAP/TIP60 and mixed-lineage-leukemia (MLL1/MLL2) SET1-type chromatin-modifying complexes in vitro, and we show that β-cat promotes H3K4 trimethylation at the c-Myc gene in vivo. H3K4 trimethylation in vivo requires prior ubiquitination of H2B, and we find that ubiquitin is necessary for transcription initiation on chromatin but not nonchromatin templates in vitro. Chromatin immunoprecipitation experiments reveal that β-cat recruits Pygopus, Bcl-9/Legless, and MLL/SET1-type complexes to the c-Myc enhancer together with the negative Wnt regulators, APC, and βTrCP. Interestingly, APC-mediated repression of c-Myc transcription in HT29-APC colorectal cancer cells is initiated by the transient binding of APC, βTrCP, and the CtBP corepressor to the c-Myc enhancer, followed by stable binding of the TLE-1 and HDAC1 corepressors. Moreover, nuclear CtBP physically associates with full-length APC, but not with mutant SW480 or HT29 APC proteins. We conclude that, in addition to regulating the stability of β-cat, APC facilitates CtBP-mediated repression of Wnt target genes in normal, but not in colorectal cancer cells.
Keywords: Wnt signaling, β-catenin, MLL1/MLL2/SET1, H3K4 methylation, APC tumor suppressor, CtBP, βTrCP
Wnt/Wg signaling events control body axis formation, cell proliferation, and organogenesis in many organisms (Cadigan 2002; Bienz and Clevers 2003; Gregorieff and Clevers 2005) and are important for self-renewal of intestinal epithelial and hematopoietic stem cells (Radtke and Clevers 2005; Reya and Clevers 2005). In the canonical Wnt/Wg signaling pathway, β-catenin (β-cat) functions as a dedicated transcriptional coactivator of LEF-1/TCF HMG proteins (Cadigan 2002; Bienz and Clevers 2003). In the absence of Wnt signaling, newly synthesized β-cat is targeted to a cytoplasmic “destruction complex” that contains Axin, protein phosphatase 2A, and the adenomatous polyposis coli (APC) tumor suppressor (Xing et al. 2003, 2004; Ha et al. 2004), where it undergoes sequential phosphorylation by casein kinase Iα (CKIα) and glycogen synthase kinase-3β (GSK-3β). Subsequent phosphorylation of APC by these protein kinases induces high-affinity binding to β-cat and disassembly of the Axin complex (Xing et al. 2004). The phosphorylated β-cat is then ubiquitinated by βTrCP and destroyed by proteasome-mediated proteolysis. Wnt ligands initiate a signaling cascade through Frizzled and LDL cell-surface receptors and the Dishevelled protein to ultimately inactivate GSK-3β and break the destruction cycle. The unphosphorylated β-cat then enters the nucleus, binds LEF-1/TCF proteins, and activates Wnt target genes (Bienz and Clevers 2003; Gregorieff and Clevers 2005).
Misregulation of the Wnt signaling pathway is a hallmark of many aggressive human cancers, including colon carcinomas and melanomas (Moon et al. 2004; Reya and Clevers 2005). The vast majority of colorectal adenomas and carcinomas contain sporadic or inherited truncations of the APC tumor suppressor (Kinzler and Vogelstein 1996) or oncogenic stabilizing mutations in β-cat, resulting in the chronic induction of c-Myc and other Wnt target genes. The most frequent cancer-causing mutations of APC are truncations that remove the C-terminal half of the protein, including binding sites for Axin and nuclear export signals, which render the mutant APC proteins unable to target β-cat to the destruction complex and unable to function as a tumor suppressor (for reviews, see Bienz 2002; Henderson and Fagotto 2002). Because Class I mutant APC proteins accumulate in the nucleus, they may also disrupt Wnt signaling by other means. For example, nuclear APC has been suggested to bind and sequester excess β-cat in the nucleus, preventing it from associating with the DNA-bound LEF-1/TCF proteins and dampening transcription in signaling cells (Neufeld et al. 2000a, b; Rosin-Arbesfeld et al. 2000, 2003). Wild-type and mutant APC proteins were also recently discovered to bind to the nuclear transcriptional corepressor, CtBP, potentially to redirect β-cat away from Wnt target genes in vivo (Hamada and Bienz 2004).
Emerging biochemical and genetic studies have elaborated important aspects of the mechanism of Wnt transcriptional regulation. The central armadillo (ARM) repeats of β-cat interact with the Bcl-9/Legless coactivator, which serves to connect β-cat to the Pygopus (Pygo) PhD finger protein (for review, see Bienz and Clevers 2003; Gregorieff and Clevers 2005). Bcl-9/Lgs and Pygo have also been shown to be required for β-cat to accumulate within the nucleus (Townsley et al. 2004). Although it is not clear whether Bcl-9/Lgs and Pygo can interact with DNA-bound β-cat:LEF-1 complexes, genetic studies strongly implicate a role for these coactivators in transcription (Hoffmans and Basler 2004; Hoffmans et al. 2005). Other studies have shown that β-cat is transported from the nucleus by APC, which contains a central nuclear export sequence that is critical for its function (Rosin-Arbesfeld et al. 2003). APC and LEF-1 compete for overlapping binding sites within the β-cat armadillo (ARM) repeats, suggesting that β-cat must switch between different complexes containing APC, Bcl-9/Lgs:Pygo, and LEF-1 before and after transcription. These steps are not well defined biochemically, and various models have been proposed to explain how β-cat exchanges between APC and LEF-1 in the nucleus (for review, see Tolwinski and Wieschaus 2004).
In addition to APC and LEF-1, the β-cat ARM repeats interact with the DNA helicases TIP49a/Pontin52 and TIP49b/TIP48/Reptin52 (Bauer et al. 1998, 2000), which are subunits in the TRRAP/TIP60 histone acetyltransferase (HAT) and mammalian INO80 and SWRCAP/SWR1 chromatin-remodeling complexes (Cai et al. 2003, 2005; Jin et al. 2005). Both TIP60 and TIP49 directly regulate Wnt target genes in vivo (Bauer et al. 2000; Feng et al. 2003; Kim et al. 2005b). Other β-cat-interacting proteins required for Wnt signaling include Brg1-containing chromatin remodeling complexes (Barker et al. 2001) and CBP/p300 (Hecht et al. 2000; Miyagishi et al. 2000). The region C-terminal to the β-cat ARM repeats contains a very strong activation domain; however, the factors that bind this domain are unknown. In vivo, the LEF-1/TCF proteins bind constitutively to Wnt target genes and repress transcription in the absence of Wnt signaling in conjunction with Gro/TLE-1, CtBP, HDAC1, and other corepressors (for review, see Courey and Jia 2001; Chinnadurai 2002).
Here, we report that the β-cat C-terminal activation domain and adjacent ARM 11/12 repeats (CTARM) selectively associate with nuclear chromatin remodeling subunits of TRRAP/TIP60 histone acetyltransferase (HAT), ISW1, and mixed-lineage-leukemia (MLL1/MLL2) SET1-type histone methyltransferase (HMT) complexes, and that β-cat promotes H3K4 trimethylation (H3K4Me3) at the c-Myc gene in vivo. SET1 proteins mediate H3K4Me3 in a manner that depends upon prior ubiquitination of H2B by mammalian BRE1:Rad6 ubiquitin ligase complexes (Kim et al. 2005a; Zhu et al. 2005). The mammalian SET1-related HMT proteins (including hSET1, ALL-1/MLL1, MLL2, ALR-1, and HALR) reside in complexes with other conserved SET1 subunits (including Ash2, menin, HCF, WRD5, and RbBP5), and are frequent direct targets of transcriptional activators (Nakamura et al. 2002; Yokoyama et al. 2004; Dou et al. 2005; Guenther et al. 2005; Milne et al. 2005; Schneider et al. 2005; Wysocka et al. 2005b). We show here that H3K4Me3 levels increase upon induction of c-Myc transcription by β-cat, and decline when transcription is shut off in cells expressing the APC tumor suppressor. Interestingly, repression of c-Myc transcription is accompanied by transient recruitment of APC, βTrCP, and CtBP. Moreover, nuclear CtBP coimmunoprecipitates with full-length APC, but not with Class I or Class II mutant APC proteins. We conclude that β-cat regulates H3K4 methylation and is counteracted directly by APC at Wnt target genes in vivo.
Results
The β-cat activation domain associates with nuclear TRRAP/TIP60, ISW1, and MLL1/MLL2 SET1-type complexes
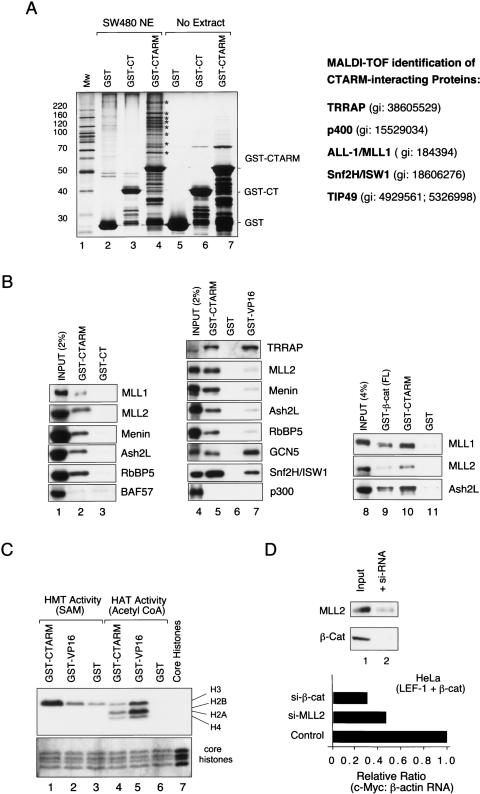
We showed previously that β-cat:LEF-1 transcription on chromatin in vitro can be selectively inhibited, or “squelched,” by a fragment of β-cat that spans the C terminus through ARM repeats 11 and 12 (GST-CTARM) (Tutter et al. 2001). The inhibition was specific because the β-cat CTARM fragment did not block the transcriptional activity of the Notch enhancer complex in vitro or disrupt the cooperative binding of β-catenin and LEF-1 to chromatin. Shorter fragments of β-cat containing either the C-terminal domain (GST-CT) or ARM repeats 11 and 12 alone (GST-ARM11/12) had no effect on β-cat activity in vitro. To characterize the proteins that bind this region of β-cat, the GST-CTARM protein was coupled to glutathione-S-Sepharose beads and incubated with nuclear extracts from SW480 colorectal cancer (CRC) cells in GST-pulldown experiments. The nuclear proteins that remained bound to the GST-CTARM beads after stringent washing were visualized by SDS-PAGE and silver staining (Fig. 1A). Multiple nuclear proteins were found to bind GST-CTARM (Fig. 1A, lane 4, asterisks), but not GST (Fig. 1A, lane 2), or GST-CT (Fig. 1A, lane 3) beads. These factors were not present in the purified GST-CTARM preparation (Fig. 1A, lane 7), indicating that they derive from the SW480 nuclear extract (Fig. 1A, lane 7).
Figure 1.
Identification of proteins that bind to the β-cat CTARM domain in GST-pulldown experiments carried out using SW480CRC nuclear extracts. (A) SDS-PAGE analysis of the proteins bound to GST-CTARM (lane 4), GST-CT (lane 3), and GST (lane 2) after incubation with SW480 nuclear extract, or of GST-CTARM (lane 7), GST-CT (lane 6), or GST (lane 5) proteins in the absence of extract; proteins were visualized by silver stain. The GI protein accession numbers of the CTARM-interacting proteins identified by MALDI-TOF are listed at the right. (B, left) Immunoblot analysis of HeLa nuclear proteins bound to either GST-CTARM (lane 2) or GST-CT (control; lane 3) beads. For comparison, 2% of each input extract is shown in lane 1. (Middle) Immunoblot comparison of SW480 nuclear proteins bound to GST-CTARM (lane 5), GST (control, lane 6), or GST-VP16 activation domain (lane 7). The input SW480 nuclear extract is shown in lane 4. (Right) Analysis of MLL1, MLL2, and Ash2L in SW40 nuclear extract (input, lane 8) or bound to GST-β-cat-FL (lane 9), GST-CTARM (lane 10) or GST (lane 11) beads. (C) Analysis of HMT (lanes 1–3) or HAT (lanes 4–6) activities present in the GST-CTARM (lanes 1,4), GST (lanes 3,6), or GST-VP16 (lanes 2,5) pulldown fractions. The input core histones are shown in lane 7. (Bottom) The core histones in each reaction were visualized by Coomassie stain. (D) Analysis of exogenous β-cat:LEF-1 activation of the endogenous c-Myc gene in HeLa cells transfected with siRNAs against β-cat or MLL2. The inset shows an immunoblot of MLL2 and β-cat protein levels in control cells (lane 1), or cells expressing the siRNA targeted to each protein (lane 2).
To identify the β-cat-interacting proteins, the proteins marked by asterisks in lane 4 of Figure 1A were excised in sections from the gel, eluted by trypsin proteolysis, and analyzed by MALDI-TOF mass-spectrometry. The major CTARM-interacting proteins identified in these experiments were subunits from three different chromatin remodeling complexes. These include the transcription/transformation domain-associated protein TRRAP, the SNF2-related helicase p400, and the bacterial RuvB-related single-stranded DNA-dependent AAA+ ATPases TIP49a/Pontin52 and TIP49b/TIP48/Reptin, all of which are subunits of TRRAP/TIP60 HAT complexes (Cai et al. 2005). Interestingly, TIP49a was previously shown to interact directly with β-cat ARM repeats 2–5 (Bauer et al. 1998; Bauer et al. 2000), and both TIP49a and TIP60 have been shown to be important coactivators for β-cat in vivo (Feng et al. 2003; Kim et al. 2005b).
The CTARM pulldown fraction also included subunits from two other chromatin complexes, specifically the Imitation Switch nucleosome-remodeling ATPase, ISW1, which is found in several distinct remodeling complexes, and the SET1-type complex protein, ALL-1/MLL1. Immunoblots confirmed the identification of TRRAP, MLL1, and ISW1 in the CTARM pulldown fractions from HeLa (Fig. 1B, lanes 1–3) and SW480 (Fig. 1B, lanes 4–7) nuclear extracts, and further revealed the presence of the MLL2 HMT (Fig. 1B, lanes 2,5) as well as the common SET1 subunits, menin (MEN-1 tumor suppressor), the Rb-interacting protein, RbBP5, and Ash2. Additional pulldown experiments indicated that GST-β-cat-FL (full-length β-catenin) also associates with nuclear MLL1 and MLL2 proteins (Fig. 1B, lanes 8–11). As expected, none of the GST-CTARM-interacting factors bound to either GST-CT or GST-beads. Moreover, the GST-CTARM protein did not interact with other nuclear proteins required for H2B ubiquitination, such as Paf1 or Rad6, nor did it bind the 19S proteasomal subunit Rpt6/SUG1, or the BAF57 or Brm chromatin remodeling subunits (Fig. 1B; data not shown). Although it has been reported that the β-cat CTARM domain interacts with the p300/CBP histone acetyltransferase (Hecht et al. 2000; Miyagishi et al. 2000), this association must be relatively weak, because it was not detected in our experiments (Fig. 1B, lanes 4–7).
To evaluate the relative strengths of these interactions, immunoblots were also carried out with parallel fractions obtained using the GST-VP16 activation domain, which is known to bind MLL1 and SAGA/GCN5 complexes. As shown in Figure 1B, the β-cat CTARM protein binds MLL1/MLL2/SET1 complex subunits more strongly than does the VP16 activation domain, whereas the SAGA GCN5 subunit binds more avidly to the VP16 activation domain (cf. lanes 5 and 7). The TRRAP protein associates very tightly with both activation domains, whereas the ISW1 subunit preferentially binds the CTARM protein. Because the TRRAP, TIP49, and ISW1 proteins are each present in multiple remodeling complexes, more detailed studies will be needed to further characterize these CTARM-interacting complexes, and in particular, to identify which subunit(s) interact directly with the β-cat activation domain.
To assess whether the CTARM protein associates with a functional HMT complex, we analyzed the ability of the GST-CTARM and GST-VP16 pulldown fractions to methylate and acetylate purified core histones in vitro. As shown in Figure 1C, the CTARM pulldown fraction contained high levels of a histone H3-specific methyltransferase activity (lane 1), but relatively weak HAT activity (lane 4), whereas the GST-VP16 fraction displayed weak HMT activity (lane 2), but strong HAT activity (lane 5), consistent with the enrichment of GCN5 in this fraction. To assess whether the SET1-type complexes are required for β-cat activity in vivo, levels of MLL2 in HeLa cells were reduced by a targeted small interfering RNA (siRNA), and the effect on c-Myc mRNA was examined in the presence of transfected β-cat and LEF-1 expression vectors. As shown in Figure 1D, endogenous c-Myc mRNA levels declined modestly in the presence of the MLL2-specific siRNA, and more substantially with an siRNA directed against β-cat. In all, these data suggest that the β-cat activation domain associates with active histone H3 methylation complexes, and that MLL2 might contribute to β-cat-mediated induction of c-Myc transcription in vivo.
Ubiquitin is required for β-cat _trans_-activation of chromatin pBRE templates in vitro
These findings raised the question of whether the H3K4 methylation or H2B ubiquitination steps might be important for β-cat activity in vitro. The in vitro chromatin-based transcription assays measure RNA initiation by primer extension, and thus do not require RNA polymerase II (RNAPII) Ser-2 phosphorylation or elongation factors. Although H3K4Me3 has been strongly linked to transcription elongation, MLL1/Set1 is present at most active RNAPII promoters in mammalian cells (Guenther et al. 2005), and thus could be required generally for RNA initiation or promoter clearance. Indeed, recent studies indicate that MLL1 complexes strongly stimulate transcription initiation on chromatin templates in vitro (Dou et al. 2005).
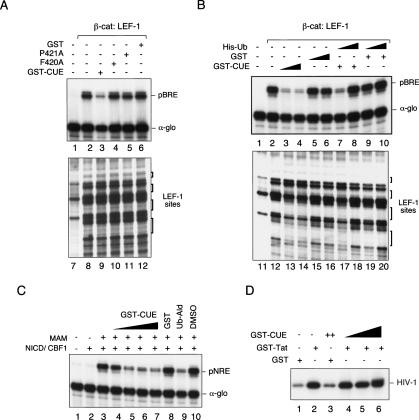
These observations lead us to ask whether the endogenous ubiquitin present in the Drosophila embryo chromatin-assembly extract might be necessary for β-cat: LEF-1 transcription in vitro. To address this question, chromatin transcription reactions were incubated with a protein containing the Vsp9p CUE domain, which binds tightly to monoubiquitin (Lima 2003). Interestingly, the GST-CUE protein strongly inhibited β-cat:LEF-1 activation of pBRE chromatin templates in vitro (Fig. 2A, lane 3). Mutant GST-CUE proteins that are unable to bind ubiquitin failed to block transcription (F420A [Fig. 2A, lane 4] or P421A [Fig. 2A, lane 5]), and GST alone had no effect (Fig. 2A, lane 6). The GST-CUE protein did not block the cooperative binding of β-cat and LEF-1 to chromatin (Fig. 2A, cf. lanes 9–12 and 8), nor did it interfere with nucleosome assembly (data not shown). Importantly, GST-CUE inhibition could be overcome completely by adding exogenous ubiquitin, either as His-tagged ubiquitin, GST-tagged ubiquitin, or a chain-terminating form of ubiquitin (Fig. 2B, cf. lanes 3 and 8; data not shown). The chromatin footprint experiments revealed a slight but reproducible change in DNase I hypersensitivity at one of the β-cat:LEF-1-binding sites (Fig. 2A), which might result from modification of the pBRE template. Ubiquitination of β-cat itself is unlikely, because the ΔN β-cat protein used in these experiments lacks the N-terminal residues for phosphorylation and ubiquitination.
Figure 2.
Ubiquitin is required for β-cat:LEF-1 transcription of chromatin templates in vitro. (A) The CUE domain competes for transcription on chromatin in vitro. Primer-extension analysis of RNA from a chromatin-assembled LEF-1 reporter gene (pBRE) in the presence (lanes 2–6) or absence (lane 1) of 120 nM recombinant ΔN β-cat protein. Recombinant LEF-1 (ΔAD; 120 nM) was present in all reactions, and the α-globin (α-glo) gene was added as a control for nonchromatin (DNA) transcription. Where indicated, reactions contained 15 μg (2.5 μM) each of GST-CUE (lane 3), GST-CUE-F420A (lane 4), GST-CUE-P421A (lane 5), or GST alone (lane 6). Lanes 7–12 show DNase I footprint analyses of the binding of β-cat:LEF-1 in reactions 1–6, respectively. (B) Ubiquitin rescues the inhibitory effect of GST-CUE on β-cat-regulated transcription in vitro. (Top) Primer extension analysis of pBRE and α-glo (control) RNA in reactions containing (lanes 2–10) or lacking (lane 1) recombinant β-cat and LEF-1. Where indicated, reactions contained low (2.5 μM, lanes 3,7,8) or high (10 μM, lane 4) levels of GST-CUE, or low (2.5 μM, lanes 5,9,10) or high (10 μM, lane 6) GST control, in either the absence of exogenous ubiquitin (lanes 1–6) or in the presence of either 1 μM (lanes 7,9) or 10 μM (lanes 8,10) of His-tagged ubiquitin. (C) The GST-CUE domain blocks Notch enhancer activation of chromatin templates in vitro. Where indicated, reactions contained (lanes 2–10) or lacked (lane 1) the recombinant Notch intracellular domain (NICD) and CBF1 DNA-binding protein. Reactions 3–10 also contained the Notch coactivator Mastermind (MAM). GST-CUE levels were 170 nM (lane 4), 830 nM (lane 5), 1.7 μM (lane 6), and 3.3 μM (lane 7), and the GST control was added at 5.3 μM (lane 8). Ubiquitin aldehyde (1.8 nM) and MG132 (4 nM) in DMSO were present in lane 9, and the DMSO alone was tested as a control in lane 10. RNA synthesized from the CBF1-dependent promoter was analyzed by primer extension; α-glo was added as a control for nonchromatin transcription. (D) The CUE domain does not block HIV-1 Tat-regulated transcription initiation or elongation on the HIV-1 promoter DNA in vitro. HIV-1 RNA was analyzed by the run-off elongation assay in the presence of GST-Tat (lanes 2,4–6), GST-CUE (lanes 3–6), or GST alone (lanes 1,3).
Interestingly, the GST-CUE protein also blocked transcription of pNRE chromatin templates by a recombinant Notch enhancer complex (containing CBF1, the Notch intracellular domain, and Mastermind) (Fig. 2C, lanes 4–7), indicating that the requirement for ubiquitin may be general. Both Notch and β-cat-regulated transcription were also blocked by ubiquitin-aldehyde (Fig. 2C, lane 9; data not shown), which inhibits deubiquitinating enzymes. In contrast, the Vsp9p CUE domain did not block transcription from α-globin (α-glo) promoter on a nonchromatin (DNA) template (Fig. 2A–C), nor did it interfere with basal- or Tat-activated HIV-1 transcription elongation on naked DNA in vitro (Fig. 2D). These data suggest that ubiquitination, like acetylation, may be generally required for enhancer- and chromatin-dependent initiation of transcription in vitro, and that further studies in this system should be useful in defining the enzymes and mechanism involved.
β-cat and other Wnt pathway-specific regulators cycle on and off the c-Myc enhancer in LiCl-treated cells in vivo
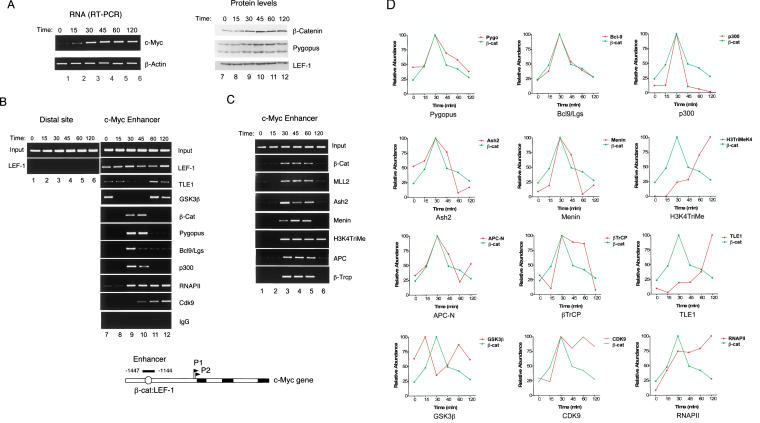
We next used chromatin immunoprecipitation (ChIP) experiments to assess whether β-cat regulates H3K4 trimethylation at Wnt target genes in vivo. C2C12 (mouse myoblast) cells were synchronized and β-cat was induced by treatment with lithium, a GSK-3β inhibitor, under conditions shown previously to induce β-catenin (Baek et al. 2003). As shown in Figure 3A, c-Myc RNA levels increased rapidly upon the addition of LiCl (lanes 1–6), accompanied by an increase in nuclear β-cat protein (lanes 7–12). Although the C2C12 cells were incubated continuously with lithium, β-cat and associated coactivators appeared transiently at either 30–45 min (Fig. 3B; lanes 9,10), or 30–60 min (Fig. 3C; lanes 3–5), and then were replaced by the TLE1 corepressor. In contrast, LEF-1 bound constitutively to the c-Myc enhancer (Fig. 3B, lanes 7–12). The ChIP conditions used here were specific, because LEF-1 was not detected at a distal upstream region of the c-Myc gene (Fig. 3B, lanes 1–6), nor was c-Myc enhancer DNA recovered in immunoprecipitates with control IgG serum (Fig. 3B, lanes 7–12).
Figure 3.
ChIP analysis of β-cat induction of murine c-Myc transcription in lithium-treated C2C12 cells. (A, left, lanes 1–6) RT–PCR analysis of c-Myc RNA in C2C12 cells treated with 10 mM LiCl for the times indicated above each lane; β-actin mRNA levels were monitored as a control. (Right, lanes 7–12) The levels of β-cat, Pygo, and LEF-1 in the lithium-treated C2C12 cells were monitored by immunoblot. (B) ChIP analysis of the c-Myc enhancer at different times following induction of β-cat by exposure of cells to 10 mM LiCl. The primers used to monitor the upstream LEF-1 sites in the mouse c-Myc gene enhancer are indicated in the schematic diagram relative to the P1 RNA start site (+1). (Lanes 1–6) A distal (10 Kb) upstream region was monitored as a control. Lanes 7–12 show the transcription factor binding to the Wnt-responsive c-Myc enhancer (ref). (C) ChIP analysis of MLL2 and SET1-type complex subunits at the c-Myc enhancer following exposure to 10 mM LiCl for the times indicated above each lane. (D) Quantitative PCR analysis of some of the ChIP samples shown in B and C. In each graph, the kinetics of β-cat localization to the c-Myc enhancer (shown in green) is compared with the specific transcription factor indicated at the bottom of each panel.
β-Catenin was recruited to the c-Myc enhancer together with Pygopus and Bcl-9/Lgs (Fig. 3B, lanes 9,10), which are Wnt pathway-specific coregulators that appear to be required for both transcription and nuclear transport of β-cat. MLL2 and the SET1-type subunits Ash2 and menin were also present with β-cat (Fig. 3C, lanes 3–5), and H3K4Me3 increased strongly at the c-Myc gene. Because the GSK-3β kinase, which is the direct target of LiCl, has also been detected in the nucleus, we asked whether it might also be present at the c-Myc enhancer. As shown in Figure 3B, GSK-3β was present at the enhancer with kinetics similar to those of the TLE-1 corepressor (Fig. 3B, lanes 7,11,12), and opposite of that of β-cat. The rapid disappearance of the GSK-3β kinase upon treatment of cells with lithium raises the possibility that it functions as part of the corepressor complex at inactive Wnt target genes. More detailed kinetic ChIP analyses indicate that this cyclic pattern of alternating coactivator and corepressor complexes repeats for multiple cycles in cells that are persistently exposed to LiCl (S. Wang and K.A. Jones, unpubl.). Although the enhancer factors cycle on and off of the enhancer, levels of RNAPII and the CDK9 transcription elongation factor steadily accumulate at the gene (Fig. 1B), in parallel with the rise in steady-state c-Myc mRNA levels.
Given that GSK3β was present at the c-Myc enhancer, we decided to test for the presence of other negative Wnt regulators, in particular the APC tumor suppressor and β-cat ubiquitin ligase, βTrCP. Interestingly, both factors were detected at the c-Myc enhancer; however, they did not colocalize with TLE1, but rather appeared together with β-cat (Fig. 3C, lanes 3–5). Although proteins involved in degradation of other short-lived activators have been localized to enhancer elements of target genes (Muratani and Tansey 2003), it was unexpected to find APC with β-cat at the c-Myc gene, because the two proteins interact in a mutually exclusive manner in vitro. Consequently, it has been assumed that β-cat must transfer between APC-containing nuclear import/export complexes and DNA-bound LEF-1/TCF enhancer complexes. The ChIP data suggest an alternate scenario in which APC, functioning as a nuclear chaperone, accompanies β-cat both on and off the DNA. A more quantitative assessment of the exchange kinetics for some of these transcription factors was provided by real-time PCR analysis of chromatin complexes (Fig. 3D), which supports the conclusion that APC associates with the c-Myc enhancer with kinetics similar to those observed for β-cat, Pygo, and Bcl-9/Lgs.
Repression of c-Myc transcription in HT29-APC cells is accompanied by binding of APC, βTrCP_,_ and CtBP to the enhancer
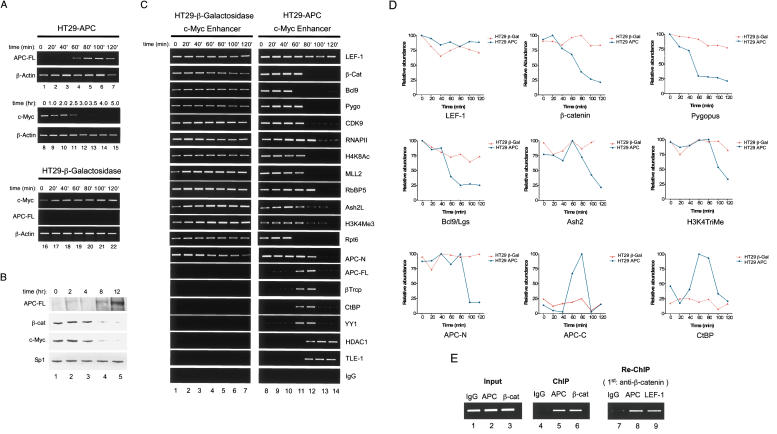
These observations suggested to us that destruction complex subunits like APC and βTrCP might participate in transcription, for example, to disassemble the Wnt enhancer complex or mediate the exchange of coactivators and corepressor complexes between transcription cycles. This possibility was difficult to assess in the lithium-treated C2C12 cells, which continuously induce β-cat, but we reasoned that it might be useful to examine by ChIP the shut-off of c-Myc transcription upon expression of APC in colon cancer cells. For these experiments, we used an engineered HT29-APC colorectal cancer cell line (Morin et al. 1996), which contains a truncated Class II mutant APC that is unable to degrade the endogenous β-cat, as well as an integrated full-length APC transgene under the control of a zinc-inducible metallothionein (MT) promoter. The induction of wild-type APC by zinc in these cells promotes the degradation of β-cat and represses Wnt target gene expression. As a control, ChIP experiments were carried out in HT29-β-Gal cells, which contain a β-Galactosidase transgene expressed from the identical zinc-inducible MT promoter (Morin et al. 1996).
As shown in Figure 4A, exposure of HT29-APC cells to zinc caused a rapid induction of APC RNA (lanes 1–7), whereas steady-state c-Myc RNA levels noticably declined between 2 and 3 h later (lanes 8–15). In contrast, β-actin mRNA levels were unaffected by zinc in either cell line, and zinc had no effect on c-Myc mRNA levels in the control HT29-β-Gal cells (Fig. 4A, lanes 16–22). Immunoblotting of full-length APC protein was relatively insensitive, but showed a gradual accumulation of the protein at 8–12 h after induction with zinc, whereas steady-state levels of both β-cat and c-Myc protein significantly declined between 4 and 8 h (Fig. 4B, lanes 1–5). No changes were observed in the level of the Sp1 transcription factor in these extracts, which was monitored as a control. ChIP analysis revealed that LEF-1, β-cat, Pygo, Bcl-9/Lgs, and the MLL2/SET1-type complex subunits were present at the active c-Myc enhancer in both cell lines in the absence of zinc (Fig. 4C, lane 1 for HT29-β-Gal cells, and lane 8 for the HT29-APC cells). As expected, RNAPII, CDK9, acetylated H4K8, and trimethylated H3K4 were also present at high levels at the active c-Myc gene (Fig. 4C, lanes 1–7). Interestingly, the endogenous Class II mutant HT29 APC protein was also recruited to the active c-Myc enhancer in vivo. However, unlike the situation in LiCl-treated C2C12 cells, β-cat and the associated coactivators did not cycle on and off of the enhancer, but rather remained stably bound, like LEF-1, to the c-Myc gene.
Figure 4.
Analysis of APC-induced shut-off of human c-Myc gene transcription in the HT29-APC CRC cell line. (A, top, lanes 1–7) RT–PCR analysis of the induction of APC-FL mRNA in HT29-APC cells after exposure to ZnCl2 for different times (in minutes), as indicated above each lane. RT–PCR analysis of β-actin mRNA is shown below as a control. (Middle, lanes 8–15) RT–PCR analysis of the decline in c-Myc mRNA levels in the ZnCl2-treated HT29-APC cells, with times of exposure to ZnCl2 (in hours) indicated above each lane. RT–PCR analysis of β-actin mRNA is shown as a control. (Bottom, lanes 16–22) RT–PCR analysis of c-Myc, APC-FL, and β-actin mRNA levels in the control HT29-β-Gal cells, which were exposed to ZnCl2 for the times indicated above each lane. (B, lanes 1–5) Immunoblot analysis of steady-state levels of APC-FL, β-cat, and c-Myc proteins in extracts from HT29-APC cells treated with ZnCl2 for the different times (in hours) indicated above each lane. Levels of the Sp1 transcription factor are included as a control. (C) ChIP analysis of transcription factors bound to the c-Myc gene in HT29-β-Gal cells (lanes 1–7) or in HT29-APC cells (lanes 8–14) by exposure to ZnCl2 for the different amounts of time (in minutes) indicated above each lane. (D) ChIP analysis by RT–PCR for some of the same factors analyzed in C. Each panel analyzes transcription factor occupancy at the c-Myc enhancer in HT29-APC cells (blue) vs. the control HT29-β-Gal cells (red). FL-APC was detected with antisera specific to a C-terminal domain (APC-C), whereas the mutant APC protein was detected using an antisera specific to the N terminus of the protein (APC-N). (E) Re-ChIP analysis to test whether the mutant HT29 APC protein and endogenous β-catenin co-occupy the c-Myc enhancer in HT29 CRC cells. ChIP in HT29-β-Gal cells (without zinc) was first carried out using the α-β-cat antibody, and the immunocomplex was diluted in 10 mM dithiothreitol. Aliquots were then immunoprecipitated with α-APC (N terminus-specific) or α-LEF-1 antisera, and the precipitated chromatin was amplified by PCR using primers specific for the c-Myc enhancer.
Although exposure to zinc had no effect on any of these transcription factor interactions in the control HT29-β-Gal cells (Fig. 4C, lanes 1–7), induction of the full-length APC protein caused a rapid dissociation of the Wnt enhancer proteins (β-cat, Bcl-9/Lgs, and Pygo) and associated coactivators (MLL2, menin, RbBP5, Ash2) from the c-Myc gene in HT29-APC cells (Fig. 4C, lanes 8–14). RNAPII and H3K4Me3 levels declined gradually in these experiments, whereas LEF-1 levels remained unchanged. Most interestingly, the full-length APC protein transiently appeared at the c-Myc enhancer during the loss of the β-cat and coactivator proteins (Fig. 3C,D), in conjunction with βTrCP, CtBP, and YY1 proteins. At later times, only the TLE-1 corepressor and HDAC1 corepressors were detected with LEF-1 at the repressed gene (Fig. 4C, lanes 8–14). These data suggest that the APC-mediated shut-off of c-Myc transcription proceeds in two steps, initiated by the transient binding of full-length APC and the CtBP corepressor to the enhancer, and followed by the stable binding of TLE-1 and HDAC1.
To examine the exchange of transcription coactivator and corepressor complexes in more detail, the association of LEF-1, β-cat, Pygo, Bcl-9/Lgs, APC, and CtBP with the c-Myc enhancer in HT29-APC and HT29-β-Gal cells was analyzed by RT–PCR (Fig. 4D). These results indicate that β-cat, Pygo, and Bcl-9/Lgs exit the promoter with similar kinetics, whereas Ash2 and H3K4Me3 levels decline more slowly. Both APC-FL and CtBP were detected at the c-Myc enhancer in HT29-APC cells at times overlapping the loss of β-cat and coactivator subunits. To further assess whether the truncated mutant APC resides in a stable multiprotein complex with β-cat in HT29 cells, chromatin complexes from the HT29-β-Gal cell line were isolated with β-cat antibody and subjected to reChIP with antisera to APC and LEF-1. As shown in Figure 4E, both APC and LEF-1 could be immunoprecipitated from the initial β-cat-containing chromatin complex (lanes 8,9), indicating that the three factors are closely associated in vivo. Most importantly, we note that the exchange of coactivator and corepressor complexes at the c-Myc enhancer significantly preceeds the decline in β-cat protein levels that result from APC-mediated proteolytic degradation, strongly suggesting that APC acts directly and immediately to facilitate the repression of Wnt target genes.
Binding of β-cat to CKIδ-phosphorylated APC blocks binding to LEF-1 and inhibits LEF-1:β-cat transcription in vitro
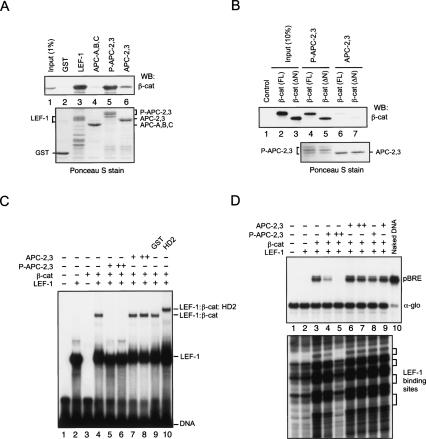
These observations led us to re-examine the question of whether β-cat can interact simultaneously with either Pygo and Bcl-9/Lgs, or APC, when bound to LEF-1 on DNA. We initially assessed whether GST-LEF-1, or different domains of APC, could bind the endogenous β-cat protein in pulldown experiments carried with SW480 nuclear extracts (Fig. 5A). For these studies, we used human APC proteins GST-APC-A,B,C, which contains all three 15-amino-acid repeats; or GST-APC-2,3, which contains the second and third 20-amino-acid repeat (Xing et al. 2003). The native β-cat in the extract bound with high affinity to GST-LEF-1 (Fig. 5A, lane 3), weakly to the APC-2,3 fragment (Fig. 5A, lane 6), and not at all to the APC-A,B,C protein (Fig. 5A, lane 4). Phosphorylation of the APC-2,3 by CK1δ (P-APC) enhances its affinity for recombinant β-cat by several hundredfold in vitro (Xing et al. 2004), and strongly increased binding to β-cat in our experiments (Fig. 5A, lane 5). CK1δ-phosphorylation of GST-APC-2,3 also strongly enhanced binding to both full-length and N-terminal-truncated (ΔN β-cat) recombinant β-cat in vitro (Fig. 5B, cf. lanes 4,5 and 6,7).
Figure 5.
Analysis of the interactions between recombinant human β-cat and human LEF-1, human Bcl-9 (HD2 domain; amino acids 288–419), APC-A,B,C, or CKI-phosphorylated or unphosphorylated APC-2,3 proteins in vitro. The (human) APC fragments used here are APC-A,B,C, which contains all three 15-amino-acid repeats (amino acids 1100–1189), and APC-2,3, which contains the second and third 20-amino-acid repeat (amino acids 1362–1540; Xing et al. 2003). (A) CKI-phosphorylation of APC-2,3 enhances binding to β-cat in GST pulldown experiments carried out using SW480 nuclear extracts. SW480 nuclear extract was incubated with Glutathione-S-Sepharose 4B beads coupled to the following GST fusion proteins: GST-LEF-1, GST-APC-A,B,C, CKI-phosphorylated (P-APC) or unphosphorylated GST-APC-2,3, or GST (control), and examined for associated nuclear β-cat by Western blot after stringent washing of the beads. The β-cat in the SW480 extract (input, 1%) is shown in lane 1. The bead-coupled GST fusion proteins are visualized by Ponceau S stain as a loading control. (B) FL and ΔN β-catenin were produced by in vitro transcription and translation in a coupled reticulocyte lysate (Promega) and tested for binding to unphosphorylated (lanes 6,7) or CK1δ-phosphorylated (lanes 4,5) GST-APC-2,3. The proteins were visualized by Ponceau S stain in the bottom panel. (C) Phosphorylated APC-2,3 blocks binding of β-cat to LEF-1 in EMSA experiments in vitro. Recombinant LEF-1 (ΔAD; 1 μM) was present in lanes 2, and 4–10, and β-cat (ΔN; 1 μM) was present in lanes 3–10. Where indicated, reactions also contained 2 μM (lane 5) or 12 μM (lane 6) of CK1δ-phosphorylated GST-APC-2,3, or 2 μM (lane 7) or 12 μM (lane 8) GST-APC-2,3 (unphosphorylated), or 25 μM GST (lane 9) or GST-HD2 (lane 10). (D) CK1δ-phosphorylated APC blocks β-cat:LEF-1 transcription of the chromatin pBRE template in vitro. Reactions contained 100 or 600 nM of CK1δ-phosphorylated GST-APC-2,3 (lanes 4,5), or GST-APC-2,3 (unphosphorylated; lanes 6,7). The phosphorylated and unphosphorylated GST-APC-2,3 (100 nM) proteins were added after chromatin assembly was completed in the reactions shown in lanes 8 and 9, respectively.
To evaluate the DNA-binding activity of these complexes, EMSA experiments were carried out with recombinant LEF-1, β-cat, Bcl-9 (HD2 domain), and fragments of the APC β-cat-interacting regions in vitro. As shown in Figure 5C, β-cat readily forms a ternary complex with LEF-1 on DNA (lane 4); however, the migration of this complex is unaffected by high levels of GST-APC-2,3 or GST (lanes 7,8,9, respectively). In contrast, the CK1δ-phosphorylated GST-APC-2,3 competed for the formation of the β-cat:LEF-1:DNA complex (Fig. 5C, lanes 5,6), indicating that β-cat is unable to bind simultaneously to P-APC-2,3 and LEF-1. The opposite result was obtained with the Bcl-9 HD2 domain, which readily bound β-cat:LEF-1 on DNA (Fig. 5C, lane 10), suggesting that β-cat can simultaneously bind LEF-1 and this region of Bcl-9. As expected from these results, the CK1δ-phosphorylated GST-APC-2,3 protein strongly blocked β-cat-regulated transcription in vitro (Fig. 5D, cf. lanes 3 and 4,5) and also inhibited the cooperative binding of β-cat and LEF-1 to chromatin in DNase I footprint experiments (Fig. 5D, bottom, cf. lanes 3 and 5). We conclude from these experiments that β-cat cannot efficiently bind unphosphorylated APC, and that CK1 phosphorylation of APC might induce high-affinity binding to β-cat and trigger its dissociation from LEF-1.
Wild-type, but not mutant, APC proteins associate with the CtBP corepressor in extracts
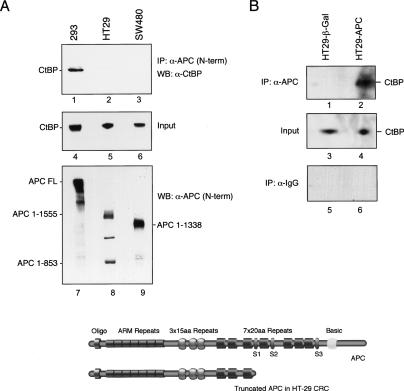
The colocalization of APC with CtBP at the c-Myc enhancer during the shut-off of c-Myc transcription was particularly interesting in light of a recent report that the two proteins interact directly, both in vivo and in vitro (Hamada and Bienz 2004). However, we noted that CtBP was not present with the truncated APC at the active c-Myc in HT29-β-Gal cells, but rather appeared only with the full-length APC protein, suggesting that CtBP does not associate with truncated APC proteins on DNA in vivo. Consequently, coimmunoprecipitation experiments were carried out to examine the binding of CtBP to native full-length and mutant APC proteins in nuclear extracts derived from 293 colorectal cancer cell lines. As shown in Figure 6A, CtBP was readily detected in immunoprecipitates of full-length APC from nuclear extracts of 293 cells (lane 1), but did not associate with the truncated APC proteins present in nuclear extracts of HT29 (lane 2) or SW480 (lane 3) CRC cells, despite equivalent levels of CtBP in each extract (lanes 4–6). We conclude that, in nuclear extracts, CtBP preferentially interacts with the full-length APC protein.
Figure 6.
CtBP associates with full-length APC, but not with cancer-associated truncated APC proteins. (A) Coimmunoprecipitation of CtBP with full-length or mutant APC proteins was analyzed by immunoblot using nuclear extracts derived from 293T cells, SW480, or HT29 CRC cells, as indicated above each lane. (Lanes 1–3) APC was immunoprecipitated using an antibody specific to the N-terminal half of the protein, and any associated CtBP was detected by immunoblot. The input CtBP is shown (by immunoblot) in lanes 4–6, and the input APC proteins are shown (by immunoblot) in lanes 7–9. (B) Newly expressed full-length APC in HT29-APC cells associates with CtBP. HT29-APC and HT29-β-Gal cells were treated with 100 μM ZnCl2 to induce full-length APC expression, and association with CtBP was determined by immunoblot of anti-APC immunoprecipitates (lanes 1,2) or with control anti-IgG serum (lanes 5,6). The input CtBP protein in each extract is shown in lanes 3 and 4. A schematic of wild-type APC and the mutant HT29 APC is shown at the bottom. (S1–S3) Axin-binding sites.
To confirm this observation, we tested whether CtBP associates with full-length APC protein induced in zinc-treated HT29 cells. As shown in Figure 6B (lane 4), CtBP was readily detected in immunoprecipitates of APC-FL from the HT29-APC cell lines, but did not associate with the mutant APC protein immunoprecipitated from the control zinc-treated HT29-β-Galactosidase cells (lane 3), despite equal levels of CtBP in the extracts (lanes 1,2). No CtBP was detected with control IgG antiserum (Fig. 6Blanes 5,6). These data confirm the strong interaction between APC and CtBP reported previously by Hamada and Bienz (2004) and extend these findings by showing that, in nuclear extracts, CtBP does not physically associate with Class I or Class II mutant APC proteins. We were unable to confirm the previous mapping of the CtBP-interacting site to the APC 15-amino-acid repeat domain, and also failed to detect CtBP in GST-pulldown experiments with fragments of APC that contain all three 15-amino-acid repeats (data not shown). The cause of this apparent discrepancy may lie in the stringent binding conditions of our assays and our use of nuclear, rather than whole-cell, extracts. Taken together, these experiments strongly suggest that the full-length APC protein preferentially interacts with nuclear CtBP. These data suggest an important role for nuclear APC in the regulation of Wnt transcription, as outlined schematically in Figure 7 and discussed further below.
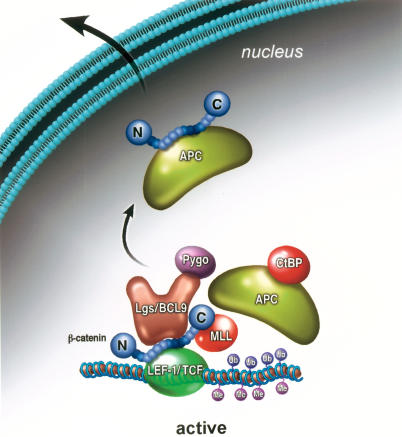
Figure 7.
Model for the role of APC in the exchange of coactivator and corepressor complexes at Wnt target genes. At the active gene, β-cat interacts with Bcl-9:Pygo and with MLL1/MLL2 complexes to promote H3K4Me3. The APC tumor suppressor controls the switch between transcriptional coactivator and corepressor complexes, mediated through its ability to interact with CtBP. Phosphorylation of APC by GSK-3β and CK1 may induce APC to bind β-cat, and dislodge it from LEF-1. β-cat may then be exported from the nucleus as part of an APC complex, or, alternatively, it might be ubiquitinated by βTrCP for degradation at the DNA or elsewhere in the cell.
Discussion
A role for APC in CtBP-mediated repression of Wnt transcription in vivo.
The data presented here support a model in which the APC tumor suppressor functions directly to counteract β-cat-mediated transcription at Wnt target genes in vivo. This possibility was first suggested by the finding that full-length APC cycles on and off the c-Myc enhancer in conjunction with β-cat and associated coactivators in LiCl-treated C2C12 cells (Fig. 3). In contrast, the enhancer complex appears to be stable and does not cycle in HT29 CRC cells, which contain a Class II APC mutant protein that is unable to degrade β-cat (Fig. 4). Most strikingly, the binding of the full-length APC protein to the c-Myc gene in HT29-APC cells correlates with the rapid disassembly of the Wnt enhancer complex in vivo and the subsequent decline in steady-state c-Myc mRNA levels, both of which significantly precede the drop in β-cat protein levels that occurs as a result of proteolytic degradation in the cytoplasm (Fig. 4). Thus, the effect of APC on c-Myc transcription appears to be immediate and direct, and may serve to coordinate the switch between the β-cat coactivator and TLE1 corepressor complexes (Fig. 7).
The β-cat enhancer complex includes the Wnt coactivators Pygopus and Bcl-9/Lgs, which control the retention of β-cat in the nucleus (Townsley et al. 2004) and may also function directly in transcription (Hoffmans and Basler 2004; Hoffmans et al. 2005). The observation that APC can also regulate nuclear transport of β-cat (Rosin-Arbesfeld et al. 2003) raises the possibility that these factors may reside within a larger regulatory complex that chaperones β-cat in and out of the nucleus and mediates its release from the DNA. Indeed, sequential ChIP (re-ChIP) data indicate that the mutant APC in HT29 colorectal cancer cells exists in a stable complex with β-cat and LEF-1 at the active c-Myc gene (Fig. 4E). This finding is unexpected because β-cat cannot bind simultaneously to APC and LEF-1, and thus, if the full-length APC is part of a larger β-cat:LEF enhancer complex, it may interact with other subunits. Alternatively, the full-length APC and β-cat may exist in different complexes that rapidly exchange at the enhancer. Our current data indicate that targeting is mediated by the N-terminal half of the APC protein, and that CtBP and βTrCP appear only in conjunction with the full-length APC protein. How APC is recruited to Wnt enhancers remains an open and important question.
The ChIP experiments also suggest that APC-mediated inhibition of c-Myc transcription in HT29 cells occurs in two steps, initiated by transient binding of APC, βTrCP, CtBP, and YY1 to the enhancer, and followed by stable binding of the TLE-1 and HDAC1 corepressors. The transient recruitment of APC and CtBP, at the time when β-cat, Bcl-9, Pygo, and other Wnt enhancer factors leave the DNA, strongly suggests a role for these factors in the exchange of Wnt coactivator and corepressor complexes. In this respect it is interesting that CtBP was shown recently to associate with APC, both in vivo and in vitro (Hamada and Bienz 2004). Our results confirm a high-affinity interaction between CtBP and the full-length APC protein induced in HT29-APC cells, as well as with the native (full-length) APC protein in 293 cells (Fig. 6). Consequently, APC may function to recruit CtBP to Wnt enhancers (Fig. 7). Although both CtBP and TLE-1 are well-established corepressors of Wnt target genes, the different functions of the two types of corepressors remain unclear, and the ChIP data suggest that they act at distinct steps. Together, these data suggest that APC counteracts β-cat function in the nucleus, as well as in the cytoplasm, and may facilitate turnover of the enhancer complex at responsive genes by recruiting βTrCP and CtBP.
Cancer-associated mutant APC proteins do not interact with CtBP
Whereas CtBP interacts strongly with full-length APC, it was not detected in immunoprecipitates of the mutant APC proteins in SW480 or HT29 CRC cells (Fig. 6). These findings contradict a recent report that CtBP binds to the APC 15-amino-acid repeat sequences present in both full-length and mutant APC proteins (Hamada and Bienz 2004). We attribute this difference to the stringent binding conditions of our assays and to our use of nuclear rather than whole-cell extracts. The observation that only the full-length APC protein can interact with CtBP may explain why neither CtBP nor βTrCP were present with the mutant APC protein at the active c-Myc gene in vivo (Fig. 4). Importantly, the model outlined in Figure 7 strongly supports the earlier conclusion from Hamada and Bienz (2004) that CtBP is not recruited to Wnt enhancers through binding to the LEF-1/TCF proteins, and we speculate instead that CtBP and βTrCP are brought to the enhancer by APC. Interestingly, CtBP complexes contain a subunit with amine oxidase activity, LSD1, which is able to reverse mono- and di-H3K4 methylation in vitro (Shi et al. 2003; Wysocka et al. 2005a), and therefore can also alter the epigenetic state of the template. Given these observations, it will be particularly interesting to assess whether CtBP and βTrCP are also required for the exchange of coactivator and corepressor complexes at Wnt enhancers in vivo.
A central role for H3K4 trimethylation in β-cat-regulated transcription
To elaborate the mechanism of repression, it is important to understand how β-cat activates target genes. The data presented here indicate that histone H3K4 trimethylation plays a key role in β-cat transactivation. Our biochemical studies indicate that the C-terminal activation domain of β-cat physically associates with chromatin remodeling or modifying subunits from the TRRAP/TIP60, ISW1, and trithorax-related MLL1/MLL2 SET1-type HMT complexes (Fig. 1). SET1-type complexes mediate trimethylation of histone H3K4, a signature chromatin modification of highly active genes that has been strongly correlated with transcription elongation (for review, see Hampsey and Reinberg 2003). Trimethylation of H3K4 requires prior ubiquitination of H2B at K120, and is stimulated in vivo by the Bre1:Rad6 ubiquitin ligase and the 19S proteasome ATPase subunits, Rpt4 and Rpt6 (Ezhkova and Tansey 2004). We show that levels of H3K4Me3 increase strongly upon induction of c-Myc transcription by β-cat in lithium-treated C2C12 cells (Fig. 3), and gradually decline when the gene is repressed in HT29-APC cells. The observation that β-cat associates with an ISW1 complex is interesting in light of recent data indicating that yeast Isw1p ATPases appear to remodel chromatin in a manner that attenuates RNAPII transcription and facilitates elongation in response to H3K4 trimethylation (Morillon et al. 2003, 2005; Santos-Rosa et al. 2003). By inference, these studies suggest that β-cat regulates early stages of transcription elongation at target genes by recruiting chromatin complexes that direct H3K4 trimethylation.
Multiple roles for ubiquitin in transcription
The observation that the β-cat activation domain, which is required for transcription in vitro (Tutter et al. 2001), associates with H3K4 methylation factors lead us to ask whether ubiquitin is required for β-cat activity in vitro. We find that β-cat transcription of chromatin pBRE templates in vitro is specifically blocked by the monoubiquitin-binding Vsp9 CUE domain and can be rescued by exogenous ubiquitin. The conclusion that transcription initiation on chromatin requires ubiquitin supports recent reports that purified MLL1/SET1 complex proteins strongly enhance transcription initiation on chromatin templates in vitro (Dou et al. 2005). In contrast, ubiquitin was not required for either initiation or elongation of transcription on naked DNA. Although ubiquitination of short-lived acidic activators has also been strongly linked with transcription elongation in vivo (Muratani and Tansey 2003), our experiments were carried out with a truncated β-cat protein lacking the N-terminal regulatory domain, suggesting that β-cat itself is unlikely to be the target of ubiquination in these experiments. The finding that the transcription by the recombinant Notch enhancer complex also requires ubiquitin further suggests that this is a general phenomenon of transcription on chromatin, consistent with recent studies showing a widespread requirement for this step in regulated RNAPII transcription activation (Dou et al. 2005; Guenther et al. 2005; Milne et al. 2005; Wysocka et al. 2005b). These findings warrant additional mechanistic studies to establish whether ubiquitination of H2B enhances transcription initiation in vitro, and to better elucidate how ubiquitination and methylation steps are coordinated during the early stages of transcription.
Because both GSK-3β and βTrCP are present with APC at the c-Myc enhancer, it is possible that phosphorylation or ubiquitination of β-cat and phosphorylation of APC may be an important trigger for the disassembly of the enhancer complex in vivo. Phosphorylation of the APC 20-amino-acid repeats by CKIδ greatly enhances binding to β-cat and disrupts its interaction with LEF-1 (Ha et al. 2004), and blocks binding and transcription in vitro (Fig. 5). One possibility is that APC may bind and transport β-cat out of the nucleus for proteolytic degradation in the cytoplasm. Alternatively, it is possible that ubiquitinated β-cat may undergo proteolytic degradation at the DNA, as proteolysis was recently shown to stimulate the activity of the yeast GCN4 and GAL4 activators in vivo (Lipford et al. 2005). In HT29 cells, the mutant APC protein is present at the active gene; however, βTrCP is not recruited and the enhancer complex does not turn over, which suggests that ubiquitination of β-cat may not be required for transcription activation, but instead may serve to promote the exchange of coactivator and corepressor complexes, and facilitate repression of transcription at Wnt target genes. Collectively, these findings suggest that there may be multiple ubiquin-dependent steps that regulate in the process of activation and turnover at Wnt target genes in vivo.
Lastly, the observation that both APC and β-cat enter the nucleus to regulate Wnt target gene expression further highlights the need to assess whether the Wnt pathway evolved to directly coordinate changes in cell adhesion, morphology, polarity, or migration with changes in gene regulation in the nucleus. Thus, β-cat is not only a nuclear transcriptional coactivator, but also associates with membrane-bound cadherins to control cell adhesion, and with actin microfilaments, to control cell morphology (Nelson and Nusse 2004; Bienz 2005; Harris and Peifer 2005). Similarly, APC controls actin cytoskeletal organization and cell mobility through its interactions with microtubules and F-actin (Bienz and Hamada 2004). Given the many emerging connections between actin signaling and transcription, including the role of histone methyltransferases in actin polymerization (Nolz et al. 2005), these questions have important implications for understanding tissue development and tumor metastasis.
Materials and methods
Plasmids, recombinant proteins, and antibodies
His-tagged human Pygopus (amino acids 1–160) and His-tagged human Bcl-9 (amino acids 1181–1426) cDNAs were PCR-amplified and cloned in-frame with a 6-histidine tag between the BamHI and HindIII restriction sites of the pET-28a(+) (Novagen) expression plasmid. The protocol for His-tagged protein purification has been described previously (Tutter et al. 2001), and the antigens were used to raise polyclonal antiserum in rabbits (Pocono Rabbit Farm and Laboratory, Inc.). Commercial antisera used were β-cat, APC, βTrCP, and GSK-3β (Santa Cruz Biotechnology); MLL1, MLL2, RbBP5, Menin, Ash2L (Bethyl Laboratories), and H4K8Ac and H3K4Me3 (Upstate).
Cells
Mouse C2C12 and human SW480 cell lines were obtained from the American Type Culture Collection. C2C12 cells were cultured in DMEM medium supplemented with 10% FBS, and SW480 were cultured in Leibovitz L-15 medium supplemented with 10% FBS. The human HT29-APC and HT-29-β-Galactosidase cell lines, which contain integrated APC and β-Gal transgenes expressed from the MT promoter, were maintained as described previously (Morin et al. 1996) and protein expression was induced by exposure of the cells to 100 mM ZnCl2 (Sigma) in the medium for the different times indicated in Figure 4.
Chromatin immunoprecipitation experiments
Approximately 3 × 107 to 5 × 107 C2C12 cells were plated 48 h prior to formaldehyde cross-linking and syncronized by serum starvation. Two hours before induction, the medium was replaced with complete DMEM. The C2C12 cells were then cultured in 10 mM LiCl (Sigma) for 0–2 h as indicated for each experiment in Figure 2. For ChIP analysis in HT29 cells, full-length APC was induced by the addition of 100 mM ZnCl2 to the medium. The protocol for the ChIP experiments was described previously (Weinmann and Farnham 2002). Qiaprep Spin Miniprep colums (Qiagen) were used for DNA purification, and immunoprecipitated DNA was analyzed using the HotStart Polymerase kit (Qiagen). PCR primers were designed to amplified regions specific to the human (HT29 experiments) or mouse (C2C12 experiments) c-Myc gene. Under these conditions, the PCR product depended linearly on the amount of genomic DNA added to the reaction. PCR products were analyzed on 1.8% agarose/TBE gels with ethidium-bromide stain. PCR of the input DNA prior to immunoprecipitation was used as a control. Primers for the human c-Myc promoter monitored the third LEF-1 site were as described (Sasaki et al. 2003). The c-Myc primers were human forward, 5′-GTGAATACACGTTTGCGG GTTAC-3′; human reverse, 5′-AGAGACCCTTGTGAAAAAA ACCG-3′; mouse forward, 5′-CTAGAACCAATGCACAGAG C-3′; and mouse reverse, 5′-CTCCCAGGACAAACCCAAGC-3′. The sequence for negative control in mouse c-Myc promoter was located 10 kb upstream of the TATA box: forward, 5′-ACA CACCTTGAATCCCGT-3′; and reverse, 5′-CCCAGCTAGAA TGAAGAAG-3′. For the re-ChIP experiments, complexes were eluted by incubation for 30 min at 37°C in 50 μL 10 mM DTT. After centrifugation, the supernatant was diluted 20 times with Dilution Buffer (1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris-Hcl at pH 8.1) and subjected again to the ChIP procedure.
Quantitative PCR experiments were performed in 25-μL reactions using the 96-well micro titer dish format and biological samples were analyzed in triplicate. SYBR GREEN (Applied Biosystems) was used as a marker for DNA amplification on an ABI Prism 7900HT apparatus and SDS 2.1 software for the data analysis (Applied Biosystems). Line graphs represent the relative value of the averaged immunoprecipitation percentage, assigning the value 100% to the maximum percentage of immunoprecipitation in each sample. Primers for murine c-Myc gene in C2C12 cells were Mmyc-QPCR-1F, CACACACATACGAAG GCAAAA; Mmyc-QPCR-1R, AAAAGTCGGCCCTGATCAG; and for the human c-Myc gene in HT29 cells: Hmyc-QPCR-1F, CCCAAAAAAAGGCACGGAA; Hmyc-QPCR-2R, TATTGGA AATGCGGTCATGC.
Coimmunoprecipitation and immunoblot analyses
To determine whether APC proteins associate with CtBP by coimmunoprecipitation, aliquots of 200 μg of precleared nuclear extracts derived from 293, HT29, or SW480 cells were diluted 1:5 in Binding Buffer (20 mM HEPES at pH 7.9, 20% Glycerol, 0.5 mM DTT, 0.5 mM PMSF, 1 μg/mL leupeptin, 1 μg/mL aprotinin, 0.1% NP-40), and incubated at 4°C for 90 min in successive steps with 1 μg of anti-APC (Santa Cruz Biotechnology) and 20 μL of protein-G agarose (Santa Cruz Biotechnology), respectively. The beads were then washed with 3 × 500 μL Wash Buffer (20 mM HEPES at pH 7.9, 75 mM KCl, 2.5 mM MgCl2, 1 mM DTT, 0.1% [v/v] Nonidet P-40, 0.5 mM PMSF, 1 μg/mL leupeptin, 1 μg/mL aprotinin) and eluted by boiling in SDS sample buffer and analyzed by SDS-PAGE.
GST-pulldown, HMT/HAT assays, and in vitro transcription and binding experiments
GST-pulldown and in vitro transcription experiments were carried out using protocols described previously (Tutter et al. 2001). For pulldown experiments from SW480CRC extracts, the nuclear extract was dialyzed against HM 0.1M (20 mM HEPES at pH 7.9, 100 mM KCl, 0.2 mM EDTA, 12.5 mM MgCl2, 10% glycerol) before use. An aliquot of 300 μL (500 μg) of nuclear extract was incubated with 700 μL of HM 0.1M, and 40 μL suspended GST fusion protein-coupled Glutathione Sepharose 4B beads (10 μg, final 100 nM), and incubated for 4 h at 4°C with rocking, washed five times in buffer (50 mM HEPES at pH 7.9, 150 mM NaCl, 0.2 mM EDTA, 12.5 mM MgCl2, 40 μM ZnSO4, 10% Glycerol, and 0.5% Tween20), and analyzed by SDS-PAGE for silver-stain and immunoblot analysis, or for enzymatic activity in the HMT and HAT assays. The HMT assay was carried out in a 50-μL reaction (final volume) in reaction buffer HM 0.06M (NaCl). Half of the sample beads were incubated with [3H]-S-Adenosyl Methionine (SAM) or [3H]Acetyl-CoA in the presence of 10 mM ATP, and 2 μg of purified core histones for 30 min at 30°C. For in vitro phosphorylation of APC, ∼30 μg of purified GST-APC (3 × 20-amino-acid repeat) was incubated with 0.5 μL (500 U) of CKIδ (New England Biolabs) with or without 1 mM ATP in 30 μL reaction in the following final buffer concentration: 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 5 mM DTT, and incubated at 30°C for 30 min prior to analysis by SDS-PAGE. Excised proteins were identified using a Bruker Ultraflex TOF/TOF MALDI mass-spectrometer.
For the siRNA knockdown experiments, LEF-1 and β-cat transient expression vectors were transfected into HeLa cells together with siRNAs for β-cat (5′-UCAUGCACCUUUGC GUGAGTT-3′; 5′-GCUCAUCAUACUG-GCUAGUTT-3′) or MLL2 (5′-GCUGCUAGAAUCUGCGUUCTT-3′; 5′-AUUCUG CCACGUCUGUGGATT-3′) using Oligofectamine (Invitrogen). LiCl was added to a final concentration of 15 mM at 2 h prior to harvesting the total RNA, which was 48 h post-transfection, and β-cat activity was compared relative to an untransfected control.
Acknowledgments
We are grateful to Wolfgang Fischer and Jessica Read of the Peptide Biology Laboratory, The Salk Institute, for mass-spectrometry identification of CTARM-interacting proteins; to Loni Pickle for carrying out the CUE domain inhibition experiment on nonchromatin templates; and to Reiko Landry for expert assistance in the early stages in this project. We also thank Bert Vogelstein (Johns Hopkins University) for the HT29-APC cell line, Wenqing Xu (University of Washington, Seattle) for the GST-APC expression vectors, and Marianne Bienz (MRC, Cambridge, U.K.) and Kurt Basler (University of Zurich, Switzerland) for Pygopus and Bcl-9 cDNAs, respectively, which were used to generate antisera. We are also grateful to M. Bienz for helpful discussions and suggestions throughout the course of this work. This work was funded by NIH grants to K.A.J. (PO1CA054418 and GM067127-03).
Footnotes
References
- Baek S.H, Kioussi C, Briata P, Wang D, Nguyen H.D, Ohgi K.A, Glass C.K, Wynshaw-Boris A, Rose D.W, Rosenfeld M.G. Regulated subset of G1 growth-control genes in response to derepression by the Wnt pathway. Proc. Natl. Acad. Sci. 2003;100:3245–3250. doi: 10.1073/pnas.0330217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H. The chromatin remodelling factor Brg-1 interacts with β-catenin to promote target gene activation. EMBO J. 2001;20:4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A, Huber O, Kemler R. Pontin52, an interaction partner of β-catenin, binds to the TATA box binding protein. Proc. Natl. Acad. Sci. 1998;95:14787–14792. doi: 10.1073/pnas.95.25.14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A, Chauvet S, Huber O, Usseglio F, Rothbacher U, Aragnol D, Kemler R, Pradel J. Pontin52 and reptin52 function as antagonistic regulators of β-catenin signalling activity. EMBO J. 2000;19:6121–6130. doi: 10.1093/emboj/19.22.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M. The subcellular destinations of APC proteins. Nat. Rev. Mol. Cell Biol. 2002;3:328–338. doi: 10.1038/nrm806. [DOI] [PubMed] [Google Scholar]
- Bienz M. β-Catenin: A pivot between cell adhesion and Wnt signalling. Curr. Biol. 2005;15:R64–R67. doi: 10.1016/j.cub.2004.12.058. [DOI] [PubMed] [Google Scholar]
- Bienz M, Clevers H. Armadillo/β-catenin signals in the nucleus—Proof beyond a reasonable doubt? Nat. Cell Biol. 2003;5:179–182. doi: 10.1038/ncb0303-179. [DOI] [PubMed] [Google Scholar]
- Bienz M, Hamada F. Adenomatous polyposis coli proteins and cell adhesion. Curr. Opin. Cell Biol. 2004;16:528–535. doi: 10.1016/j.ceb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Cadigan K.M. Wnt signaling—20 years and counting. Trends Genet. 2002;18:340–342. doi: 10.1016/s0168-9525(02)02707-5. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Tomonori-Sato C, Sato S, Sorokina I, Parmely T.J, Conaway R.C, Conaway J.L. Identification of new subunits of the multiprotein mammalian TRRAP/Tip60-containing histone acetyltransferase complex. J. Biol. Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Florens L, Swanson S.K, Kusch T, Li B, Workman J.L, Washburn M.P, Conaway R.C, Conaway J.L. The mammalian YL1 protein is a shared subunit of the TRRAP/Tip60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 2005;280:13665–13670. doi: 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell. 2002;9:213–224. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- Courey A.J, Jia S. Transcriptional repression: The long and the short of it. Genes & Dev. 2001;15:2786–2796. doi: 10.1101/gad.939601. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne T.A, Tackett A.J, Smith E.R, Fukuda A, Wysocka J, Allis C.D, Chait B.T, Hess J.L, Roeder R.G. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Tansey W.P. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol. Cell. 2004;13:435–442. doi: 10.1016/s1097-2765(04)00026-7. [DOI] [PubMed] [Google Scholar]
- Feng Y, Lee N, Fearon E.R. TIP49 regulates β-catenin-mediated neoplastic transformation and T-cell factor target gene induction via effects on chromatin remodeling. Cancer Res. 2003;63:8726–8734. [PubMed] [Google Scholar]
- Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: From endoderm to cancer. Genes & Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- Guenther M.G, Jenner R.G, Chevalier B, Nakamura T, Croce C.M, Canaani E, Young R.A. Global and Hox-specific roles for the MLL1 methyltransferase. Proc. Natl. Acad. Sci. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha N.C, Tonozuka T, Stamos J.L, Choi H.J, Weis W.I. Mechanism of phosphorylation-dependent binding of APC to β-catenin and its role in β-catenin degradation. Mol. Cell. 2004;15:511–521. doi: 10.1016/j.molcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Hamada F, Bienz M. The APC tumor suppressor binds to C-terminal binding protein to divert nuclear β-catenin from TCF. Dev. Cell. 2004;7:677–685. doi: 10.1016/j.devcel.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. Tails of intrigue: Phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- Harris T.J, Peifer M. Decisions, decisions: β-Catenin chooses between adhesion and transcription. Trends Cell Biol. 2005;15:234–237. doi: 10.1016/j.tcb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Hecht A, Vleminckx K, Stemmler M.P, van Roy F, Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of β-catenin in vertebrates. EMBO J. 2000;19:1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B.R, Fagotto F. The ins and outs of APC and β-catenin nuclear transport. EMBO Rep. 2002;3:834–839. doi: 10.1093/embo-reports/kvf181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmans R, Basler K. Identification and in vivo role of the Armadillo-Legless interaction. Development. 2004;131:4393–4400. doi: 10.1242/dev.01296. [DOI] [PubMed] [Google Scholar]
- Hoffmans R, Stadeli R, Basler K. Pygopus and legless provide essential transcriptional coactivator functions to armadillo/β-catenin. Curr. Biol. 2005;15:1207–1211. doi: 10.1016/j.cub.2005.05.054. [DOI] [PubMed] [Google Scholar]
- Jin J, Cai Y, Yao T, Gottschalk A.J, Florens L, Swanson S.K, Gutierrez J.L, Coleman M.K, Workman J.L, Mushegian A, et al. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J. Biol. Chem. 2005;280:41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- Kim J, Hake S.B, Roeder R.G. The human homologue of yeast BRE1 functions as a transcriptional coactivation through direct activator interactions. Mol. Cell. 2005a;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Kim J.H, Kim B, Cai L, Choi H.J, Ohgi K.A, Tran C, Chen C, Chung C.H, Huber O, Rose D.W, et al. Transcriptional regulation of metastasis suppressor gene by Tip60 and β-catenin complexes. Nature. 2005b;434:921–926. doi: 10.1038/nature03452. [DOI] [PubMed] [Google Scholar]
- Kinzler K.W, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- Lima C.D. CUE’d up for monoubiquitin. Cell. 2003;113:554–556. doi: 10.1016/s0092-8674(03)00398-2. [DOI] [PubMed] [Google Scholar]
- Lipford J.R, Smith G.T, Chi Y, Deshaies R.J. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–116. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- Milne T.A, Dou Y, Martin M.E, Brock H.W, Roeder R.G, Hess J.L. MLL associates specifically with a subset of transcriptionally active target genes. Proc. Natl. Acad. Sci. 2005;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishi M, Fujii R, Hatta M, Yoshida E, Araya N, Nagafuchi A, Ishihara S, Nakajima T, Fukamizu A. Regulation of Lef-mediated transcription and p53-dependent pathway by associating β-catenin with CBP/p300. J. Biol. Chem. 2000;275:35170–35175. doi: 10.1074/jbc.C000258200. [DOI] [PubMed] [Google Scholar]
- Moon R.T, Kohn A.D, De Ferrari G.V, Kaykas A. WNT and β-catenin signalling: Diseases and therapies. Nat. Rev. Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, O'Sullivan J, Kent N, Proudfoot N, Mellor J. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell. 2003;115:425–435. doi: 10.1016/s0092-8674(03)00880-8. [DOI] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic histone methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Morin P.J, Vogelstein B, Kinzler K.W. Apoptosis and APC in colorectal tumorigenesis. Proc. Natl. Acad. Sci. 1996;93:7950–7954. doi: 10.1073/pnas.93.15.7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M, Tansey W.P. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce C.M, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- Nelson W.J, Nusse R. Convergence of Wnt, β-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld K.L, Nix D.A, Bogerd H, Kang Y, Beckerle M.C, Cullen B.R, White R.L. Adenomatous polyposis coli protein contains two nuclear export signals and shuttles between the nucleus and cytoplasm. Proc. Natl. Acad. Sci. 2000a;97:12085–12090. doi: 10.1073/pnas.220401797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld K.L, Zhang F, Cullen B.R, White R.L. APC-mediated downregulation of β-catenin activity involves nuclear sequestration and nuclear export. EMBO Rep. 2000b;1:519–523. doi: 10.1093/embo-reports/kvd117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolz J.C, Gomez T.S, Billadeau D.D. The Ezh2 methyltransferase complex: Actin up in the cytosol. Trends Cell Biol. 2005;15:514–517. doi: 10.1016/j.tcb.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Radtke F, Clevers H. Self-renewal and cancer of the gut: Two sides of a coin. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Rosin-Arbesfeld R, Townsley F, Bienz M. The APC tumour suppressor has a nuclear export function. Nature. 2000;406:1009–1012. doi: 10.1038/35023016. [DOI] [PubMed] [Google Scholar]
- Rosin-Arbesfeld R, Cliffe A, Brabletz T, Bienz M. Nuclear export of the APC tumour suppressor controls β-catenin function in transcription. EMBO J. 2003;22:1101–1113. doi: 10.1093/emboj/cdg105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bernstein B.E, Karabetsou N, Morillon A, Weise C, Schreiber S.L, Mellor J, Kouzarides T. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell. 2003;12:1325–1332. doi: 10.1016/s1097-2765(03)00438-6. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Suzuki H, Yagi K, Furuhashi M, Yao R, Susa S, Noda T, Arai Y, Miyazono K, Kato M. Lymphoid enhancer factor 1 makes cells resistant to transforming growth factor β-induced repression of c-myc. Cancer Res. 2003;63:801–806. [PubMed] [Google Scholar]
- Schneider J, Wood A, Lee J.S, Schuster R, Dueker J, Maguire C, Swanson S.K, Florens L, Washburn M.P, Shilatifard A. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol. Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine J.R, Lan F, Ogawa H, Luke M.P, Nakatani Y, Shi Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Tolwinski N.S, Wieschaus E. A nuclear escort for β-catenin. Nat. Cell Biol. 2004;6:579–580. doi: 10.1038/ncb0704-579. [DOI] [PubMed] [Google Scholar]
- Townsley F.M, Cliffe A, Bienz M. Pygopus and Legless target Armadillo/β-catenin to the nucleus to enable its transcriptional co-activator function. Nat. Cell Biol. 2004;6:626–633. doi: 10.1038/ncb1141. [DOI] [PubMed] [Google Scholar]
- Tutter A.V, Fryer C.J, Jones K.A. Chromatin-specific regulation of LEF-1-β-catenin transcription activation and inhibition in vitro. Genes & Dev. 2001;15:3342–3354. doi: 10.1101/gad.946501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann A.S, Farnham P.J. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26:37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Milne T.A, Allis C.D. Taking LSD 1 to a new high. Cell. 2005a;122:654–658. doi: 10.1016/j.cell.2005.08.022. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne T.A, Dou Y, Zhang X, Burlingame A.L, Roeder R.G, Brivanlou A.H, Allis C.D. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005b;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Xing Y, Clements W.K, Kimelman D, Xu W. Crystal structure of a β-catenin/axin complex suggests a mechanism for the β-catenin destruction complex. Genes & Dev. 2003;17:2753–2764. doi: 10.1101/gad.1142603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Clements W.K, Le Trong I, Hinds T.R, Stenkamp R, Kimelman D, Xu W. Crystal structure of a β-catenin/APC complex reveals a critical role for APC phosphorylation in APC function. Mol. Cell. 2004;15:523–533. doi: 10.1016/j.molcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero D.J, Kitabayashi I, Herr W, Cleary M.L. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham A.-D, Mandal S, Erdjument-Bromage H, Temptst P, Reinberg D. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol. Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]