cis- and trans-Acting Determinants of Transcription Termination by Yeast RNA Polymerase II (original) (raw)
Abstract
Most eukaryotic genes are transcribed by RNA polymerase II (Pol II), including those that produce mRNAs and many noncoding functional RNAs. Proper expression of these genes requires efficient termination by Pol II to avoid transcriptional interference and synthesis of extended, nonfunctional RNAs. We previously described a pathway for yeast Pol II termination that involves recognition of an element in the nascent transcript by the essential RNA-binding protein Nrd1. The Nrd1-dependent pathway appears to be used primarily for nonpolyadenylated transcripts, such as the small nuclear and small nucleolar RNAs (snoRNAs). mRNAs are thought to use a distinct pathway that is coupled to cleavage and polyadenylation of the transcript. Here we show that the terminator elements for two yeast snoRNA genes also direct polyadenylated 3′-end formation in the context of an mRNA 3′ untranslated region. A selection for _cis_-acting terminator readthrough mutations identified conserved features of these elements, some of which are similar to cleavage and polyadenylation signals. A selection for _trans_-acting mutations that induce readthrough of both a snoRNA and an mRNA terminator yielded mutations in the Rpb3 and Rpb11 subunits of Pol II that define a remarkably discrete surface on the trailing end of the enzyme. Our results suggest that, at least in budding yeast, protein-coding and noncoding Pol II-transcribed genes use similar mechanisms to direct termination and that the termination signal is transduced through the Rpb3/Rpb11 heterodimer.
A typical eukaryotic chromosome contains hundreds or thousands of genes arrayed along both strands of the DNA. Most of these genes are monocistronic, i.e., correspond to a transcription unit, so each must include transcription start (promoter) and stop (terminator) signals. In the yeast Saccharomyces cerevisiae, the distance between transcription units is often only a few hundred nucleotides or less, so, depending on gene orientation, promoters and terminators are adjacent or even overlapping. Such a genomic arrangement puts stringent demands on the efficiency of transcription termination, since readthrough is expected to result in occlusion of the downstream promoter if the genes are in the same orientation and formation of antisense RNA and/or colliding polymerases if the genes are in the opposite orientation. Either outcome would likely result in aberrant gene expression (36).
Most eukaryotic genes are transcribed by RNA polymerase II (Pol II) (23). Termination of transcription by Pol II is coupled to formation of the 3′ end of the transcript (4, 35), which occurs by different mechanisms for protein-coding and noncoding RNAs. The 3′ ends of mRNAs are formed by cotranscriptional endonucleolytic cleavage, followed by polyadenylation of the 3′ end thus generated (49). The _cis_-acting elements that direct cleavage and polyadenylation in yeast are quite degenerate (22), but statistical analysis of a large number of known and putative processing sites has resulted in algorithms for the probabilistic prediction of mRNA 3′ ends (20). Soon after transcript cleavage, termination of transcription and degradation of the 3′ fragment of the cleaved transcript occurs, although the temporal order and interdependence of these two events are uncertain.
Formation of the 3′ ends of small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs) differs between vertebrates and yeast. In vertebrates, most snoRNAs are processed from pre-mRNA introns (18), while snRNA genes utilize specialized promoters and 3′-end formation elements distinct from those that direct mRNA synthesis (24, 32). In contrast, all yeast snRNAs and most yeast snoRNAs are synthesized autonomously from promoters that appear similar to mRNA promoters. The 3′ ends of these RNAs are generated by either endonucleolytic cleavage or termination, followed by trimming by the nuclear exosome, a complex of 3′ exonucleases (6). In cases where endonucleolytic cleavage of snRNAs or snoRNAs is known to occur, it is not clear if this cleavage is coupled to termination.
We recently reported a mutational analysis of the terminator for a yeast snoRNA gene, SNR13 (39). The SNR13 terminator consists of at least two elements (regions I and II), which together span about 100 nucleotides. Here we place the SNR13 terminator in the 3′ untranslated region (UTR) of a protein-coding gene and find that, surprisingly, it directs polyadenylation of the mRNA. The polyadenylation sites map immediately downstream of region II, which contains sequences similar to mRNA cleavage and polyadenylation elements. A parallel analysis of the SNR65 snoRNA terminator yields similar results, suggesting that bipartite terminators containing cleavage and polyadenylation signals may be a common characteristic of yeast snoRNA genes. Using both the SNR13 snoRNA terminator and the CYC1 mRNA terminator, we selected for _trans_-acting mutations that result in terminator readthrough and obtained substitutions in the Rpb3 and Rpb11 subunits of Pol II. These subunits form a heterodimer that is homologous to the α subunit homodimer of bacterial RNA polymerase and is stabilized by a long coiled-coil interaction. The readthrough mutations cluster at the extreme C terminus of the Rpb11 dimerization helix and an interacting residue in Rpb3. We propose that this surface is a contact point for factors that transmit the termination signal to Pol II.
MATERIALS AND METHODS
Plasmids.
pGAC24 contains the ACT/CUP fusion gene, including CUP1 3′ UTR sequences, between the TDH3 promoter and the PGK1 3′ UTR and polyadenylation site (31). pGAC24ΔUTR lacks the CUP1 3′ UTR but retains the PGK1 3′ UTR and was created by replacing a 751-bp BamHI-SalI fragment of pGAC24 with a 594-bp BamHI-SalI fragment of a PCR product made with the 5′-CUP and CUP1-3′-SalI primers. The 217-bp SNR65/RPS14A intergenic region was amplified by PCR with flanking XhoI sites with primers SNR65-XHO-101F and SNR65-XHO-317R and inserted into the intronic XhoI site of pGAC24 to create pGAC24-SNR65F and pGAC24-SNR65R or into the SalI site of pGAC24-ΔUTR to create pGAC24-65UTR-F and pGAC24-65UTR-R. A single-nucleotide substitution of C for T is encoded within the SNR65-XHO-101F primer relative to the sequence of the SNR65/RPS14A intergenic region currently deposited in the Saccharomyces Genome Database (www.yeastgenome.org/) due to an update in the database occurring after the primers were created. Mutational analysis and mapping of polyadenylation sites were carried out with constructs having this variant sequence. A primer with a sequence matching the current database entry, SNR65-XHO-101F-T, was subsequently used to create versions of pGAC24-SNR65F and pGAC24-SNR65R that perfectly match the SGD sequence. No difference in copper sensitivity was observed with these corrected constructs compared to the variants. The corrected SNR65-XHO101F-T primer was used with SNR65-XHO-236R to create pGAC24-SNR65-101-236.
RNA analysis.
Northern blots were prepared as described previously (39), with 20 μg of total cellular RNA per lane, and probed with 32P-labeled oligonucleotide 3′CUP-2 or ACT1-probe. To map sites of polyadenylation, 0.1 to 1 μg of total cellular RNA prepared by the glass bead-guanidinium isothiocyanate-hot phenol method (45) was combined with 250 pmol of T16-BSG1 primer in 26.5 μl of H2O, incubated at 95°C for 10 min and 65°C for 10 min, and cooled on ice. Reaction mixtures were brought to 50 μl by the addition of 5 μl of 100 mM dithiothreitol, 5 μl of deoxynucleoside triphosphates (10 mM each), 1 μl of RNasin (Promega), 10 μl of 5× avian myeloblastosis virus reverse transcriptase buffer, and 2.5 μl of avian myeloblastosis virus reverse transcriptase (USB) and incubated at 42°C for 1 h. One-fifth of the reverse transcription (RT) reaction mixture was used directly in PCRs with the T16Bsg1 and 5′-CUP primers and Tfl DNA polymerase (Epicenter Technologies). Products were digested with BamHI and SalI, cloned into pUC118, and sequenced with M13 universal primers.
Isolation of _cis_-acting readthrough mutations.
Error-prone PCR of the ACT-CUP intron region of pGAC24-SNR65F, cotransformation of PCR products with gapped pGAC24, and selection of copper-resistant colonies were carried out as described previously (39).
Isolation of _trans_-acting readthrough mutations.
The genome-wide selection for spontaneous SNR13 terminator readthrough mutations was described previously (39). One mutant (from the 46a background; MATa _cup1_Δ ura3 his3 trp1 lys2 ade2 leu2) having a cold-sensitive growth phenotype was transformed with a YCp50-based yeast genomic library to identify clones that enable growth at 16°C. A single complementing clone with a genomic insert of ∼14.4 kb containing RPB11 and several other genes was identified, and subcloning revealed that the complementing activity was contained within a 1.55-kb EcoRI-SacI fragment containing only RPB11 and the extreme 5′ end of SIN3. Sequencing of the genomic RPB11 locus in the mutant strain revealed a GAG-to-GGG mutation in codon 108.
DNA fragments containing wild-type RPB3, RPB7, and RPB11 alleles were amplified from yeast strain 46α (isogenic with 46a except _MAT_α) with KOD Hi-Fi DNA polymerase (Novagen) and cloned into _URA3_-marked centromere plasmid pRS316. Mutagenesis was carried out by PCR of these plasmid clones with Tfl DNA polymerase (Epicenter) with universal M13 forward and reverse primers and deoxynucleoside triphosphates at 200 μM each. The resultant PCR products were cotransformed with EcoRI/BamHI-digested pRS316 plasmid DNA into yeast strain 46α harboring _LEU2_-marked reporter plasmid pGAC24-SNR13 (40) or pGAC24-CYC83F (39). Transformants were selected on medium lacking uracil and leucine, and colonies were then replica plated onto medium lacking leucine and containing 0.15 or 0.25 mM CuSO4 to select for terminator readthrough. To test for linkage of the copper-resistant growth phenotype to the mutagenized plasmid, strains were streaked onto plates containing 5-fluoroorotic acid and lacking leucine to select for loss of the _URA3_-marked plasmid and then scored for loss of copper resistance.
Plasmid-borne Pol II subunit genes from copper-resistant strains were amplified with KOD Hi-Fi DNA polymerase with M13 universal primers and sequenced with a combination of M13 and gene-specific primers. PCR products from selected RPB11 mutants (see Table 2) were cloned into TRP1_-marked centromere plasmid pRS314 to confirm linkage of the copper resistance phenotype and to test for function in the absence of wild-type RPB11. The genomic RPB11 locus of 46α bearing a second copy of RPB11 on URA3_-marked centromere plasmid pRS316 was disrupted by transformation with an rpb11_Δ::KanMX4 fragment amplified from an rpb11 disruption strain obtained from Research Genetics and selection for G418 resistance. Integration at the RPB11 locus was confirmed by PCR and by dependence on a plasmid-borne RPB11 allele for viability. Mutant rpb11 alleles in pRS314 were transformed into 46α Δ_rpb11, tested for terminator readthrough activity by plating on medium containing copper, and tested for function in the absence of RPB11 by plating on medium containing 5-fluoroorotic acid. All of the mutant alleles tested (see Table 2) conferred resistance to at least 0.15 mM copper.
TABLE 2.
Dominant terminator readthrough mutations in RPB11 and RPB3
| Locus and substitution(s)a | No. of strains | Recessive 30°C growthb |
|---|---|---|
| RPB11 | ||
| E108G | 13 | Normal |
| E108K | 2 | Dead |
| E108V | 1 | NTh |
| L111P | 7c | NT |
| Q112X | 2d | Slow |
| L114P | 2e | Normal |
| A27V, W109X | 1 | Dead |
| T113P, L114X | 1f | NT |
| RPB3: K9E | 12g | NT |
Oligodeoxynucleotides.
The oligodeoxynucleotides used were as follows: 5′-CUP, 5′-CAAGAACTTAGTTTCGACGG-3′; CUP1-3′-SalI, 5′-GACTAGTCGACTTATTTCCCAGAGCAGCA-3′; 3′CUP-2, 5′-ACTCATGACCTTCATTTTGGAAC-3′; T16BSG1, 5′-GCGTCGACGAATTCGTGCAGTTTTTTTTTTTTTTTTV-3′ (V = A-C-G mixture); SNR65-XHO-101F, 5′-TCCGCTCGAGCGATTTTGCAACTCTTGGTA-3′; SNR65-XHO-101F-T, 5′-TCCGCTCGAGCGATTTTGTAACTCTTGGTA-3′; SNR65-XHO-317R, 5′-TCCGCTCGAGGTTATGCATGTATTGTACTT-3′; SNR65-XHO-236R, 5′-TCCGCTCGAGATCAAATCAGCTCATACGTC-3′; ACT1-probe, 5′-TCAGTAAATTTTCGATCTTGGGAAG-3′. Underlining indicates the single-nucleotide substitution of C for T that is encoded within the SNR65-XHO-101F primer relative to the sequence of the SNR65/RPS14A intergenic region currently deposited in the Saccharomyces Genome Database (www.yeastgenome.org/).
RESULTS
Characterization of the SNR65 terminator.
We previously mapped the SNR13 snoRNA gene terminator by inserting PCR-mutagenized SNR13 downstream sequences into the intron of the ACT/CUP fusion gene (31) and selecting for mutations that allow readthrough of the terminator by demanding growth in the presence of >0.1 mM copper (39) (Fig. 1). The SNR13 terminator was found to be bipartite, consisting of two ∼50-bp sequences designated region I and region II. Region I contains sequences expected to bind Nrd1, while region II contains features similar to pre-mRNA cleavage-polyadenylation sites. Each region has weak terminator activity, but both are required for efficient termination and point mutations in both are required for strong readthrough (39).
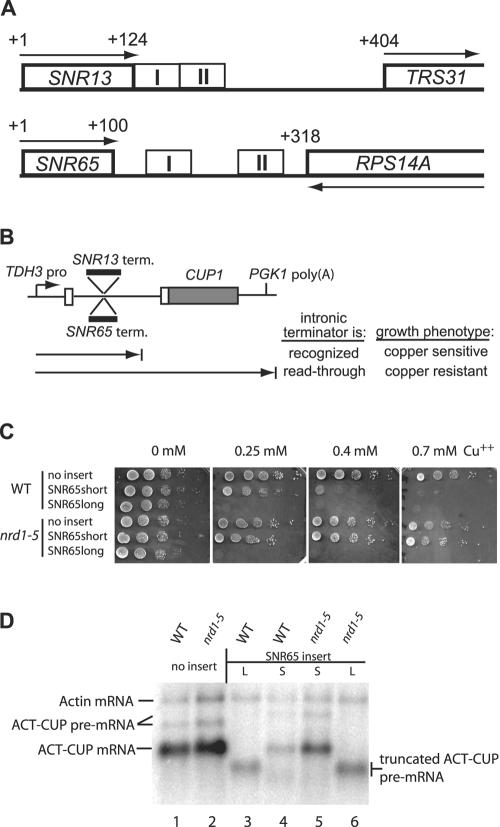
FIG. 1.
(A) Locations of terminator elements downstream of the SNR13 and SNR65 snoRNA genes. Numbers indicate position in base pairs from the transcription start site of the snoRNA genes. Boxes with heavy outlines indicate either the coding sequence for the mature snoRNAs snR13 and snR65 or the open reading frames for the downstream protein-coding genes. Arrows indicate the directions of transcription. Boxes with thin outlines demarcate the region I and region II terminator elements of SNR13 (39) or SNR65 (this study). (B) Scheme for selection of _cis_-acting mutations in snoRNA terminators (term.). See text for details. (C) Copper plate assay for SNR65 terminator function. Tenfold serial dilutions of cultures of the 46α _cup1_Δ (WT [wild type]) and _nrd1_-5 mutant strains harboring ACT/CUP reporter plasmids with no insert or with the indicated SNR65 3′-flanking sequences (SNR65long, bp 101 to 317; SNR65short, bp 101 to 236) inserted into the intron were spotted onto medium lacking leucine and containing the indicated concentrations of copper and incubated for 2 to 3 days at 30°C. (D) Northern analysis of ACT/CUP RNA. Total cellular RNA was prepared from the 46α (WT) and _nrd1_-5 mutant strains harboring ACT/CUP reporter plasmids with no insert or with SNR65long (L) or SNR65short (S) sequences inserted into the intron. The blot was probed with 32P-labeled oligonucleotide ACT1-probe, which is complementary to the ACT1 mRNA 5′ UTR and so detects ACT/CUP mRNA, as well as endogenous ACT1 mRNA.
To search for elements common to snoRNA terminators, we subjected the SNR65 downstream region to the same analysis. We chose SNR65 both because it has a small intergenic interval of only 217 bp and, unlike SNR13, it is convergent with the downstream gene (RPS14A, Fig. 1A). Thus, the SNR65 terminator is adjacent to or interdigitated with a cleavage-and-polyadenylation site on the opposite strand. This arrangement may demand a terminator structure different from that of SNR13, which is transcribed in the same direction as its downstream gene.
It was anticipated that insertion of the 217-bp SNR65/RPS14A intergenic fragment into the ACT/CUP intron in either orientation would result in decreased expression of the ACT/CUP gene, due to either the SNR65 terminator in the forward orientation or the RPS14A cleavage-and-polyadenylation site in the reverse orientation. Surprisingly, while the forward orientation conferred severe copper sensitivity (SNR65long, Fig. 1C), the reverse orientation did not (data not shown). Thus, the RPS14A cleavage-and-polyadenylation site does not appear to function in the context of the ACT/CUP intron.
We examined the role of the Nrd1 protein in SNR65 terminator function by testing copper sensitivity in a _nrd1_-5 mutant strain (Fig. 1C). No appreciable increase in copper resistance was imparted by the _nrd1_-5 mutation with the SNR65long insertion, suggesting that Nrd1 may not be required for SNR65 terminator function. However, sequence alignment of the SNR65 3′-flanking region revealed two blocks of similarity with the SNR13 terminator (Fig. 2), corresponding roughly to regions I and II but separated by a spacer region of about 50 nucleotides. Furthermore, region I of SNR65 is highly similar to a region of the NRD1 5′ UTR that is required for negative autoregulation of Nrd1 level by premature transcription termination (40) (Fig. 2). Therefore, we tested whether a smaller segment of SNR65 3′-flanking sequence containing the region I-like and spacer sequences but not region II (nucleotides 101 to 236) directs Nrd1-dependent termination. Indeed, this smaller region (SNR65short) causes moderate copper sensitivity that is relieved by the _nrd1_-5 mutation (Fig. 1C).
FIG. 2.
Termination-polyadenylation elements of snR13 and snR65 pre-snoRNAs. Nucleotide positions relative to the transcription start site and mature 5′ end are shown above and below the sequences for snR13 and snR65, respectively. Lowercase letters indicate nucleotides contained within mature snR13, while uppercase letters indicate nucleotides downstream of the mature 3′ end of either RNA. Also shown is a portion of the NRD1 5′ UTR (nucleotides −229 to −173 relative to the start codon) that contains a Nrd1-responsive element (40). Sequences that are identical in two or more of the genes are in boldface. Residues at which mutations decrease terminator function in the context of the ACT/CUP fusion gene intron, either singly or in combination (see Table 1), are boxed. SNR13 and SNR65 polyadenylation sites in the context of the ACT/CUP fusion gene 3′ UTR are indicated by filled stars (major sites) or open stars (additional sites).
Northern analysis of RNA from ACT/CUP reporter genes with SNR65 terminator sequences in the intron (Fig. 1D) corroborates the results of the copper growth assay. The SNR65short insertion causes four- to fivefold reduced accumulation of full-length ACT/CUP mRNA relative to endogenous actin mRNA in a wild-type strain, while in the _nrd1_-5 strain the insertion causes only an approximately twofold reduction (compare lanes 1, 4, and 5). The SNR65long insertion results in complete loss of full-length mRNA and the accumulation, instead, of a truncated transcript, and this pattern does not change in a _nrd1_-5 mutant strain (compare lanes 1, 3, and 6). We conclude that the SNR65 terminator consists of proximal elements that are relatively Nrd1 dependent (region I) and distal elements that are relatively Nrd1 independent (region II).
To identify nucleotides critical for SNR65 terminator function, we generated a pool of mutated DNA fragments containing the entire SNR65/RPS14A intergenic region by error-prone PCR and introduced them into the ACT/CUP fusion gene intron in the forward orientation by in vivo homologous recombination. Yeast transformants that survived on plates containing 0.15 mM copper, a concentration restrictive for the wild-type insert, were selected. The ACT/CUP plasmid was rescued from 17 copper-resistant transformants, plasmid linkage of copper resistance was confirmed by retransformation, and the insert from each plasmid was sequenced. The results are displayed in Table 1 and Fig. 2. Interestingly, 14 of the 17 plasmids have mutations affecting the UA dinucleotide at positions 260 and 261, and all 17 have at least one mutation in region II. Mutation of A251, U260, or A261 alone is sufficient to allow growth on 0.15 mM copper. Growth on 0.7 mM copper was observed only when additional mutations are present, either another mutation in region II (A283G) or several mutations in region I (U166A, C172A) and the spacer (U198Δ). The failure to obtain mutations in region I alone suggests that region II is the major determinant for termination on SNR65 and/or that region I of SNR65 has redundant elements not easily disrupted by random mutagenesis.
TABLE 1.
_cis_-acting readthrough mutations in the SNR65 terminator
| Substitution(s)a | No. of strains | Copper resistant to concn (mM) of: |
|---|---|---|
| A251G | 1 | 0.15 |
| U260C | 4 | 0.15 |
| A261C | 1 | 0.15 |
| A261G | 7 | 0.15 |
| U250C, U268G | 1 | 0.15 |
| A251G, A281G | 1 | 0.15 |
| A261G, A283G | 1 | 0.7 |
| U166A, C172A, U198Δ, U260C | 1 | 0.7 |
Polyadenylation of mRNA directed by snoRNA terminators.
To test if a snoRNA terminator can function from within an mRNA 3′ UTR, the SNR13 and SNR65 terminators were placed individually, in both orientations, between the stop codon and polyadenylation site of the ACT/CUP fusion gene in place of deleted CUP1 3′ UTR sequences (Fig. 3A). In no case did the insertion confer strong copper sensitivity on the _cup1_Δ strain, indicating that functional ACT/CUP mRNA production was not significantly impaired. However, the SNR13 forward insert did result in slower growth on 0.7 to 1.0 mM copper plates, which could be rescued by a mutation in the gene for Nrd1 (data not shown). Total cellular RNA from the transformants was analyzed by Northern blotting with a probe complementary to CUP1 (Fig. 3B). In the reverse orientation, the SNR13 insert increases the length of the ACT/CUP mRNA but apparently does not result in any new 3′ ends (Fig. 3B, compare lanes 1 and 3). This result was expected since the reverse orientation should encode no 3′-end formation elements. There does appear to be a decrease in the steady-state level of the mRNA, perhaps because the altered 3′ UTR sequence destabilizes the transcript.
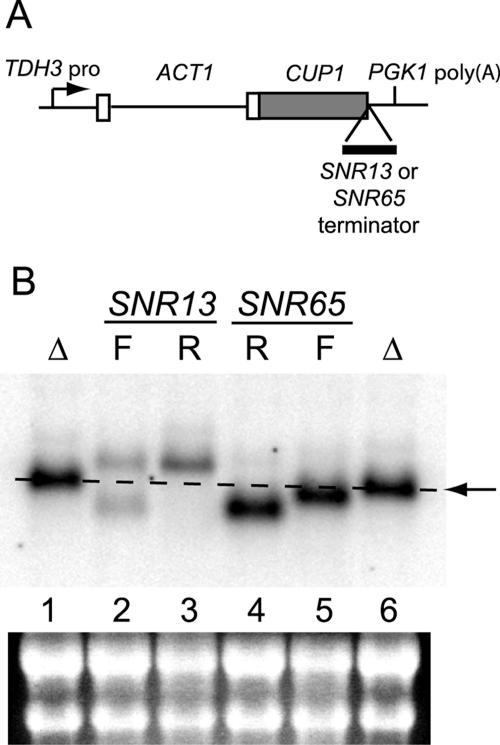
FIG. 3.
(A) Scheme for mapping of functional polyadenylation sites in snoRNA terminators. The CUP1 3′ UTR (157 nucleotides) was deleted prior to insertion of either the SNR13 or the SNR65 terminator at the site of the deletion. (B) Northern analysis of transcripts from ACT/CUP fusion gene alleles. Total cellular RNA was extracted from the 46α _cup1_Δ strain carrying plasmid-borne ACT/CUP alleles with the following modifications: lanes 1 and 6, deletion of 157 nucleotides of the CUP1 3′ UTR (Δ); lanes 2 and 3, insertion of SNR13 nucleotides +125 to +232 in the forward (F; lane 2) and reverse (R; lane 3) orientations; lanes 4 and 5, insertion of SNR65 nucleotides +101 to +317 in the reverse (lane 4) and forward (lane 5) orientations. The arrow and dashed line mark the position of mRNA with only the CUP1 3′ UTR deletion. The blot was probed with 32P-labeled oligonucleotide 3′CUP-2, complementary to the CUP1 coding region. Ethidium bromide staining of the portion of the filter containing the large and small rRNAs is shown below the lane numbers, for normalization.
In the forward orientation, the SNR13 insert results in the formation of two sets of transcripts in roughly equal amounts, one of the size expected for use of the PGK1 polyadenylation site present in the ACT/CUP fusion gene and a shorter set of the size expected if the snoRNA terminator was utilized (Fig. 3B, lane 2). To determine if the shorter transcripts are polyadenylated, and to map the sites of polyadenylation, cDNA was prepared from the RNA with reverse transcriptase and an oligo(dT) primer. The resulting cDNA was amplified by PCR with the oligo(dT) primer and a _CUP1_-specific reverse primer, cloned, and sequenced. Strikingly, the polyadenylation sites map to the downstream end of region II of the SNR13 insert (Fig. 2). Of 19 clones sequenced, 11 had +211 and two had +210 as the last nucleotide before the poly(A) tail. The remaining six clones correspond to six different poly(A) sites from position +218 to position +277. All but two of the sites are within SNR13 sequences. Curiously, none of the clones correspond to mRNAs cleaved and polyadenylated at the PGK1 poly(A) site, even though such transcripts appear to be similar in abundance to transcripts ending in SNR13 sequences (Fig. 3B, lane 2). The RT-PCR also lacked products of the length expected for the PGK1 poly(A) site, which suggests that the RT-PCR conditions we used favored synthesis of the shorter cDNAs. Our results indicate that the SNR13 terminator can direct polyadenylation when present in the 3′ UTR of an mRNA-encoding gene.
The SNR65 terminator also elicited polyadenylation in the ACT/CUP 3′ UTR. The 217-bp fragment bearing the SNR65 terminator contains the RPS14A cleavage-and-polyadenylation site in the opposite orientation, so it was expected to direct 3′-end formation in either orientation. Indeed, both the forward and reverse orientations of the insert direct the formation of shorter transcripts than are seen in the absence of an insert (Fig. 3B, compare lanes 4 and 5 with lane 6), indicative of cleavage and/or termination within the insert. Both the SNR65 and RPS14A terminator elements appear to be more efficient than the SNR13 terminator in the context of the ACT/CUP 3′ UTR, as little or no utilization of the PGK1 cleavage-and-polyadenylation site is apparent. The fact that the transcripts produced from the reverse orientation of the SNR65 insert are approximately the same length as those produced from the forward orientation of the SNR13 insert (Fig. 3B, compare lanes 2 and 4) indicates that the RPS14A polyadenylation site is about 100 nucleotides downstream of the stop codon, between regions I and II of the SNR65 terminator (Fig. 1A). Indeed, of seven cDNAs analyzed for the reverse SNR65 insert, six have poly(A) sites between +205 and +220 (Fig. 2, in the opposite strand of that shown). The seventh clone has a poly(A) site at position +181. Thus, the SNR65 and RPS14A terminators are indeed interdigitated on opposite strands.
The polyadenylation sites for the SNR65 terminator were mapped by the same method as that described for the SNR13 terminator, and the results are displayed in Fig. 2. Eight cDNA clones were analyzed; four had poly(A) sites at position +303, and the remainder had sites between +287 and +297. Thus, as for SNR13, the SNR65 poly(A) sites are at the downstream end of region II. It therefore appears that at least two snoRNA terminators contain cleavage and polyadenylation signals at their downstream ends, although we cannot exclude the possibility that the poly(A) tail derives from termination followed by cleavage-independent polyadenylation by Trf4 (see Discussion).
_trans_-acting terminator readthrough mutations in Pol II subunits.
We have also used the ACT-CUP reporter to select for mutations in _trans_-acting factors that induce readthrough of the SNR13 terminator (38-40). The factors thus identified include two RNA-binding proteins, Nrd1 and Nab3, a helicase, Sen1, and a protein phosphatase, Ssu72. Nrd1 and Sen1 bind to the C-terminal domain of the largest subunit of Pol II, Rpb1 (42, 48), while Ssu72 binds to the second largest Pol II subunit, Rpb2 (16), and utilizes the Pol II C-terminal domain as a substrate (29). Nab3 binds to Nrd1 (10). A plausible model for the function of these four factors is that Nrd1 and Nab3 recognize terminator sequences in the nascent transcript, possibly assisted by Sen1, and transmit the termination signal to Pol II, perhaps via Ssu72. The mechanism by which Pol II receives the termination signal is unknown, and no mutations in Pol II subunits had been obtained in the readthrough selection.
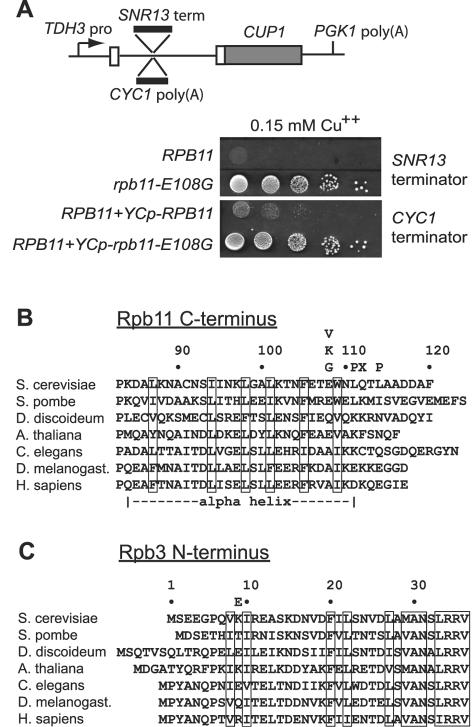
Here we report the identification of terminator readthrough mutations in two subunits of Pol II. Selection for spontaneous copper-resistant mutants with a haploid _cup1_Δ yeast strain bearing the ACT/CUP fusion gene with the SNR13 terminator inserted in the intron yielded a cold-sensitive mutation that was complemented by a plasmid bearing RPB11, the gene encoding the second smallest subunit of Pol II (46). The RPB11 locus was amplified from the cold-sensitive strain by PCR and sequenced, revealing a missense mutation that substitutes glycine for glutamate at residue 108 of the 120-amino-acid Rpb11 protein (_rpb11_-E108G). When cloned into a low-copy-number plasmid and transformed into a wild-type RPB11 strain, the _rpb11_-E108G allele dominantly confers readthrough of the SNR13 terminator (data not shown), as well as the poly(A) site/terminator from the protein-coding CYC1 gene (Fig. 4A). These results confirm that the _rpb11_-E108G mutation is solely responsible for the copper-resistant phenotype and demonstrate that the substitution in Rpb11 affects recognition of both a snoRNA and an mRNA terminator.
FIG. 4.
Terminator readthrough mutations in the Rpb11 and Rpb3 subunits of Pol II. (A) Scheme for selecting _trans_-acting mutations that induce readthrough of either the snR13 snoRNA gene terminator or the Cyc1 protein-coding gene terminator. In the examples shown, the _rpb11_-E108G mutation induces readthrough of the SNR13 terminator when present in the genome and dominantly induces readthrough of the CYC1 terminator when present on a low-copy-number plasmid. (B) Readthrough substitutions isolated in Rpb11 are located at the C terminus and are shown above an alignment of Rpb11 orthologs. Only single-residue substitutions are shown (see Table 2). Numbers refer only to residues in the S. cerevisiae protein. Conserved hydrophobic residues in the α-helical segment are boxed. Humans have several isoforms of Rpb11 that differ at the C terminus (21); isoform a is shown. (C) The readthrough substitution isolated in Rpb3 is located at the N terminus and is shown above an alignment of Rpb3 orthologs. The most-conserved residues are boxed.
The Rpb11-E108G substitution lies near the C-terminal end of a seven-turn α-helix that forms a coiled-coil interaction with the third largest subunit of Pol II, Rpb3 (12). To search for other substitutions in the Rpb3/11 heterodimer that interfere with termination, RPB3 and RPB11 were randomly mutated by error-prone PCR and introduced into the reporter strain by transformation. Transformants were selected for dominant induction of readthrough of either the SNR13 or the CYC1 terminator. As a control, randomly mutated RPB7 was put through the same selection. The Rpb7 subunit maps to a different face of Pol II than the Rpb3/11 heterodimer (2, 5) (Fig. 5). Very few copper-resistant colonies were obtained after transformation with mutated RPB7, and these strains retained copper resistance after selection against the _URA3_-marked RPB7 plasmid, indicating that readthrough was due to spontaneous mutations in the genome or ACT/CUP reporter. In contrast, hundreds of copper-resistant colonies were obtained after transformation with mutated RPB11. When 15 of these candidate RPB11 mutant strains were subjected to selection against the _URA3_-marked plasmid, all 15 lost copper resistance, indicating that RPB11 plasmid-linked mutations could readily be selected. Copper-resistant strains were also readily obtained with mutated RPB3. The RPB3 mutant candidates were not subjected to the plasmid linkage test, but sequencing of the plasmid-borne RPB3 gene from 12 candidates revealed that they all contain the same substitution (see below).
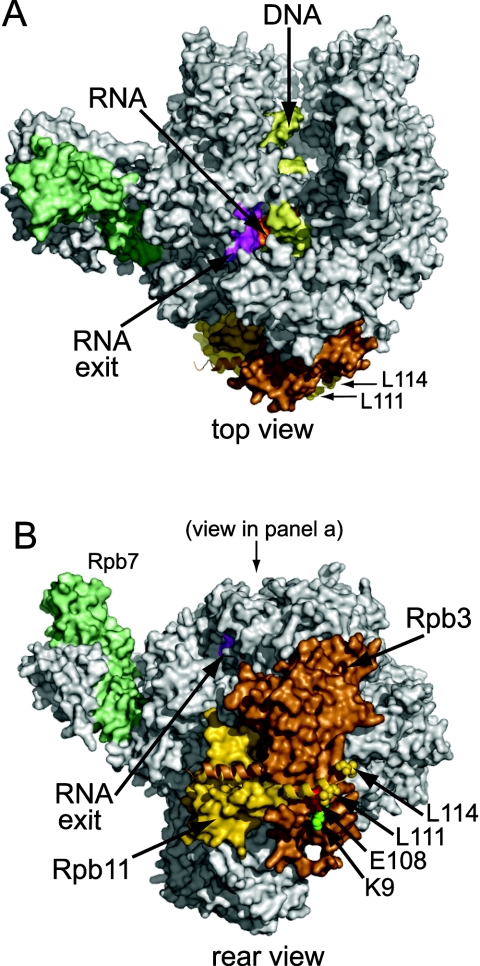
FIG. 5.
Shown are top (A) and rear (B) orthogonal views of 12-subunit yeast Pol II with a transcription bubble mimic in the active site (27). DNA (pale yellow) enters from the top in panel A and exits toward the viewer. RNA (orange) exits through a pore that is covered in part by the lid (magenta). Rpb3 (copper) and Rpb11 (gold) are at the back of Pol II, while Rpb7 (pale green) extends from the side. Rpb11 residue E108 is red, while L111 and L114 are gold. (Residues 115 to 120 are not resolved in the crystal structure.) Rpb3 residue K9 is bright green. Rpb11-E108 is close to Rpb3-K9 in the back view. The C termini of Rpb3 and Rpb11 are shown as ribbons to emphasize the coiled-coil structure. This figure was created with PyMOL (DeLano Scientific) by using pdb file 1Y1W (www.rcsb.org/pdb).
Sequencing of 29 RPB11 and 12 RPB3 readthrough alleles identified the substitutions listed in Table 2. By far the most common mutation selected in RPB11 is the E108G substitution that was also obtained in the genome-wide spontaneous mutation selection, suggesting that this substitution provides an optimal balance between readthrough of the intronic terminator and recognition of native terminators. Two other substitutions were recovered at position 108, E108K and E108V. The rarity of these substitutions relative to E108G could be due to a semidominant detrimental effect on cell growth. Consistent with this notion, a test of growth phenotypes in the absence of wild-type RPB11 revealed that an _rpb11_-E108G mutant strain grows normally at 30°C, but the _rpb11_-E108K mutant strain is not viable (Table 2), despite the dominant effect of the _rpb11_-E108K allele on terminator recognition. The difference in function of Rpb11-E108G and Rpb11-E108K suggests that the substitution of glycine for glutamate simply results in loss of a favorable contact, whereas substitution of lysine for glutamate results in a steric and/or ionic clash in addition to the loss of a specific contact.
Strikingly, all other mutations selected in RPB11 result in changes C terminal to E108 (Fig. 4B). The most common of these were the substitutions L111P and L114P. In addition, nonsense mutations were obtained at codons 109, 112, and 114, either alone or in combination with a substitution (Table 2). Thus, alterations in the structure of the extreme C-terminal segment of Rpb11 appear to result in inefficient recognition of terminators by Pol II. The fact that the readthrough mutations are dominant indicates that the alterations in Rpb11 do not prevent assembly into Pol II, nor do they strongly inhibit initiation or elongation by Pol II.
All 12 Rpb3 mutants identified in the selection encode a substitution of glutamate for lysine 9 (K9E, Fig. 4C). Remarkably, K9 of Rpb3 lies within a few angstroms of Rpb11-E108 in crystal structures of yeast Pol II (Fig. 5). Rpb3-K9 is not part of the C-terminal α-helix that forms a coiled coil with Rpb11; rather, it is in a β-sheet that cradles the C terminus of the Rpb11 α-helix. The asymmetry of the selected substitutions in Rpb11 and Rpb3 suggests that it is not simply disruption of the coiled coil that results in readthrough, but rather alterations in the surface at one end of the coiled coil. A different domain of Rpb3 was previously implicated in the activation of transcription (41); our mutations are the first evidence for a function of the Rpb3/11 heterodimer in termination.
The basic side chain of Rpb3-K9 and the acidic side chain of Rpb11-E108 have the potential of forming an ionic interaction. However, this potential is not conserved in several of the known orthologs of yeast Rpb3/11 (Fig. 4). Furthermore, the recessive lethal growth defect of an _rpb11_-E108K mutant strain is not rescued by transformation with _rpb3_-K9E on a low-copy-number plasmid (data not shown), suggesting that an ionic interaction between these two residues is not sufficient for normal Rpb3/11 activity.
DISCUSSION
While it has been known for some time that termination of transcription by Pol II on protein-coding genes is coupled to cleavage and polyadenylation, the elements that direct termination of Pol II on noncoding genes are poorly characterized. Furthermore, the mechanism by which the termination signal of any gene is transduced from factors that recognize the terminator elements to Pol II is obscure. This study illuminates both the nature of _cis_-acting elements that induce termination on yeast snoRNA genes and the way in which Pol II receives the signal that such elements are present.
The SNR13 and SNR65 terminators contain polyadenylation signals.
The terminators from two yeast snoRNA genes, SNR13 (7, 33, 39) and SNR47 (7), have previously been studied in some detail. Both terminators are bipartite in structure, with functional elements centered roughly 45 bp apart. We have now characterized a third snoRNA terminator, that of SNR65. Interestingly, it too is bipartite, but the region I and region II elements are centered about 95 bp apart, due to the presence of a 50-bp spacer sequence. Within this spacer lies the cleavage-and-polyadenylation site for the convergently transcribed gene RPS14A. Thus, separation of regions I and II may be an adaptation to allow interdigitated terminators on opposite strands. Such flexibility in terminator architecture contributes to the difficulty in identifying terminators by sequence alone. Interestingly, regions I of the SNR13 and SNR65 terminators, and the NRD1 autoregulatory element, share fairly extensive sequence similarity (Fig. 2), which suggests that natural terminators are more than just a collection of low-affinity Nrd1 (GUAA) and Nab3 (UCUU) binding sites (7).
We previously noted that region II of the SNR13 terminator has sequence similarity to yeast cleavage-and-polyadenylation sites (39). Here we show that both the SNR13 and SNR65 terminators direct polyadenylation when placed in the 3′ UTR of a reporter gene. In either case, polyadenylation occurs at the downstream boundary of region II, within a few nucleotides of sites predicted by the algorithm of Graber et al. (20), which was developed to identify cleavage-and-polyadenylation sites in protein-coding genes. We therefore presume that these polyadenylated 3′ ends are produced by the cleavage and polyadenylation factors that act on mRNAs. While we cannot rule out the possibility that these 3′ ends are generated by termination and are subsequently polyadenylated by Trf4, which marks nuclear RNAs for exosomal degradation (25, 30, 43, 47), we consider this scenario unlikely since the transcripts accumulate to high levels and are translated efficiently, judging by their ability to confer copper resistance. Furthermore, our observations are consistent with the fact that several cleavage and polyadenylation factors have been implicated in termination on snoRNA genes. These include CFIA subunits Rna14 and Rna15 (17, 33) and holo-CPF subunits Pti1, Ref2, Ssu72, and Swd2 (9, 14-16, 19, 34, 39).
Earlier studies detected polyadenylated forms of two noncoding RNAs, telomerase RNA (8) and U2 snRNA (1). Approximately 5 to 10% of telomerase RNA is polyadenylated in wild-type yeast cells, whereas polyadenylation of U2 RNA is observed only when processing of precursor U2 RNA by Rnt1 endonuclease is inhibited. Several snoRNA genes produce extended and polyadenylated transcripts when the nuclear exosome is inhibited by deletion of the gene for the Rrp6 subunit, and this polyadenylation is dependent on Pap1, the poly(A) polymerase that acts on mRNAs (44). It is not clear if the polyadenylation sites of these noncoding RNAs map to the 3′ ends of their terminator sequences and thus are mechanistically similar to cleavage and polyadenylation directed by the SNR13 and SNR65 terminators.
It is intriguing that the _SNR65_-RPS14A intergenic region did not confer copper sensitivity when inserted into the ACT/CUP fusion gene intron in the RPS14A orientation, yet it efficiently directed cleavage and polyadenylation when inserted into the ACT/CUP 3′ UTR. One interpretation of this result is that the RPS14A cleavage-and -polyadenylation signal can only be recognized after Pol II has transcribed several hundred nucleotides of RNA. In contrast, the CYC1 cleavage-and-polyadenylation signal strongly decreases ACT/CUP gene expression when inserted into the intron (Fig. 4A). The CYC1 coding region is only 330 bp long, while the RPS14A coding region (with intron) is 721 bp long. Thus, the dichotomous behavior of the terminator elements of these two genes may reflect their adaptation to different gene lengths. This model is consistent with the finding that Nrd1, Nab3 (34), and Sen1 (E.J.S. and D.A.B., unpublished data) exhibit high occupancy over both the 5′ and 3′ ends of tested protein-coding regions, suggestive of recruitment upon initiation of transcription, while cleavage and polyadenylation factors peak primarily over the 3′ end of coding regions, implying recruitment only after significant elongation (28, 34). We propose that the Nrd1-dependent termination pathway is most active within a few hundred base pairs of the transcription start site, while the cleavage-and-polyadenylation-dependent termination pathway operates primarily at sites several hundred base pairs downstream of the start site. Intermediate sites, such as the CYC1 poly(A) site, might utilize components of both pathways.
Substitutions in the Rpb3/11 heterodimer cause terminator readthrough.
How might the selected substitutions in the Rpb3/11 heterodimer induce terminator readthrough? The location of the substitutions on the rear surface of Pol II suggests that they may interfere with the interaction of Pol II with an extrinsic termination factor. Termination of Pol II transcription appears to require the binding of protein factors to terminator elements in the nascent transcript (3, 37). Although the Rpb3/11 mutations are distant from the RNA exit pore (Fig. 5), a basic channel that may provide a path for the nascent transcript (“groove 2” of reference 11) leads from the RNA exit pore to the vicinity of Rpb3/11. Therefore, a termination factor that binds to the nascent transcript may contact Pol II at the Rpb3/11 heterodimer. Such a factor may induce termination by causing an allosteric change in the elongating polymerase, or it may reach around the back of the enzyme and interact more directly with macromolecules in or near the active site. It is interesting that the Rpb3/11 heterodimer plays a central role in the interaction of Pol II with the 21-subunit Mediator protein complex (13). While Mediator functions in initiation of transcription and its involvement in termination has not been proposed, one can imagine that binding of Mediator to Pol II at the end of a gene may serve to couple termination to reinitiation.
There are several other possible candidates for a factor that interacts with Rpb3/11 to promote termination. Because our mutations cause readthrough of both a snoRNA and an mRNA terminator, factors known to be common to both pathways are of particular interest. Such factors currently include the cleavage and polyadenylation factors mentioned above, as well as the helicase Sen1 (38, 40; E.J.S. and D.A.B., unpublished). Given that the Rpb3/11 heterodimer is homologous to the N-terminal domains of the α subunit homodimer in bacterial RNA polymerase, it is intriguing that mutations in the Escherichia coli α subunit C-terminal domain have been shown to cause readthrough of terminators that are dependent on the helicase rho (26). These observations raise the possibility that the signaling pathway for transcription termination in eukaryotes and prokaryotes may employ analogous interactions between a transcript-associated helicase and the upstream end of RNA polymerase.
Acknowledgments
We thank E. Settles for technical assistance; J. Keck, M. Lopper, M. Killoran, and S. Butcher for help in creating Fig. 5; and R. Gourse, W. Ross, R. Landick, J. Dahlberg, and J. S. Butler for discussions and comments on the manuscript.
This work was supported by Public Health Service grant GM44665.
REFERENCES
- 1.Abou Elela, S., and M. Ares, Jr. 1998. Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 17**:**3738-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armache, K. J., H. Kettenberger, and P. Cramer. 2003. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl. Acad. Sci. USA 100**:**6964-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birse, C. E., L. Minvielle-Sebastia, B. A. Lee, W. Keller, and N. J. Proudfoot. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280**:**298-301. [DOI] [PubMed] [Google Scholar]
- 4.Buratowski, S. 2005. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 17**:**257-261. [DOI] [PubMed] [Google Scholar]
- 5.Bushnell, D. A., and R. D. Kornberg. 2003. Complete, 12-subunit RNA polymerase II at 4.1- Å resolution: implications for the initiation of transcription. Proc. Natl. Acad. Sci. USA 100**:**6969-6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler, J. S. 2002. The yin and yang of the exosome. Trends Cell Biol. 12**:**90-96. [DOI] [PubMed] [Google Scholar]
- 7.Carroll, K. L., D. A. Pradhan, J. A. Granek, N. D. Clarke, and J. L. Corden. 2004. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol. Cell. Biol. 24**:**6241-6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapon, C., T. R. Cech, and A. J. Zaug. 1997. Polyadenylation of telomerase RNA in budding yeast. RNA 3**:**1337-1351. [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, H., X. He, and C. Moore. 2004. The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Mol. Cell. Biol. 24**:**2932-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conrad, N. K., S. M. Wilson, E. J. Steinmetz, M. Patturajan, D. A. Brow, M. S. Swanson, and J. L. Corden. 2000. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154**:**557-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cramer, P., D. A. Bushnell, J. Fu, A. L. Gnatt, B. Maier-Davis, N. E. Thompson, R. R. Burgess, A. M. Edwards, P. R. David, and R. D. Kornberg. 2000. Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288**:**640-649. [DOI] [PubMed] [Google Scholar]
- 12.Cramer, P., D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292**:**1863-1876. [DOI] [PubMed] [Google Scholar]
- 13.Davis, J. A., Y. Takagi, R. D. Kornberg, and F. A. Asturias. 2002. Structure of the yeast RNA polymerase II holoenzyme: mediator conformation and polymerase interaction. Mol. Cell 10**:**409-415. [DOI] [PubMed] [Google Scholar]
- 14.Dheur, S., L. T. A. Vo, F. Voisinet-Hakil, M. Minet, J. M. Schmitter, F. Lacroute, F. Wyers, and L. Minvielle-Sebastia. 2003. Pti1p and Ref2p found in association with the mRNA 3′ end formation complex direct snoRNA maturation. EMBO J. 22**:**2831-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dichtl, B., R. Aasland, and W. Keller. 2004. Functions for S. cerevisiae Swd2p in 3′ end formation of specific mRNAs and snoRNAs and global histone 3 lysine 4 methylation. RNA 10**:**965-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dichtl, B., D. Blank, M. Ohnacker, A. Friedlein, D. Roeder, H. Langen, and W. Keller. 2002. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell 10**:**1139-1150. [DOI] [PubMed] [Google Scholar]
- 17.Fatica, A., M. Morlando, and I. Bozzoni. 2000. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J. 19**:**6218-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filipowicz, W., and V. Pogacic. 2002. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 14**:**319-327. [DOI] [PubMed] [Google Scholar]
- 19.Ganem, C., F. Devaux, C. Torchet, C. Jacq, S. Quevillon-Cheruel, G. Labesse, C. Facca, and G. Faye. 2003. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J. 22**:**1588-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graber, J. H., G. D. McAllister, and T. F. Smith. 2002. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 30**:**1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grandemange, S., S. Schaller, S. Yamano, S. Du Manoir, G. V. Shpakovski, M. G. Mattei, C. Kedinger, and M. Vigneron. 2001. A human RNA polymerase II subunit is encoded by a recently generated multigene family. BMC Mol. Biol. 2**:**14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo, Z., and F. Sherman. 1996. 3′-end-forming signals of yeast mRNA. Trends Biochem. Sci. 21**:**477-481. [DOI] [PubMed] [Google Scholar]
- 23.Hahn, S. 2004. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11**:**394-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez, N. 2001. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 276**:**26733-26736. [DOI] [PubMed] [Google Scholar]
- 25.Kadaba, S., A. Krueger, T. Trice, A. M. Krecic, A. G. Hinnebusch, and J. Anderson. 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 18**:**1227-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kainz, M., and R. L. Gourse. 1998. The C-terminal domain of the alpha subunit of Escherichia coli RNA polymerase is required for efficient rho-dependent transcription termination. J. Mol. Biol. 284**:**1379-1390. [DOI] [PubMed] [Google Scholar]
- 27.Kettenberger, H., K. J. Armache, and P. Cramer. 2004. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 16**:**955-965. [DOI] [PubMed] [Google Scholar]
- 28.Kim, M., S. H. Ahn, N. J. Krogan, J. F. Greenblatt, and S. Buratowski. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23**:**354-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishnamurthy, S., X. He, M. Reyes-Reyes, C. Moore, and M. Hampsey. 2004. Ssu72 is an RNA polymerase II CTD phosphatase. Mol. Cell 14**:**387-394. [DOI] [PubMed] [Google Scholar]
- 30.LaCava, J., J. Houseley, C. Saveanu, E. Petfalski, E. Thompson, A. Jacquier, and D. Tollervey. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121**:**713-724. [DOI] [PubMed] [Google Scholar]
- 31.Lesser, C. F., and C. Guthrie. 1993. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics 133**:**851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medlin, J. E., P. Uguen, A. Taylor, D. L. Bentley, and S. Murphy. 2003. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J. 22**:**925-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morlando, M., P. Greco, B. Dichtl, A. Fatica, W. Keller, and I. Bozzoni. 2002. Functional analysis of yeast snoRNA and snRNA 3′-end formation mediated by uncoupling of cleavage and polyadenylation. Mol. Cell. Biol. 22**:**1379-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nedea, E., X. He, M. Kim, J. Pootoolal, G. Zhong, V. Canadien, T. Hughes, S. Buratowski, C. L. Moore, and J. Greenblatt. 2003. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 278**:**33000-33010. [DOI] [PubMed] [Google Scholar]
- 35.Proudfoot, N. 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16**:**272-278. [DOI] [PubMed] [Google Scholar]
- 36.Shearwin, K. E., B. P. Callen, and J. B. Egan. 2005. Transcriptional interference—a crash course. Trends Genet. 21**:**339-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steinmetz, E. J., and D. A. Brow. 1998. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc. Natl. Acad. Sci. USA 95**:**6699-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinmetz, E. J., and D. A. Brow. 1996. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol. 16**:**6993-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinmetz, E. J., and D. A. Brow. 2003. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol. Cell. Biol. 23**:**6339-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinmetz, E. J., N. K. Conrad, D. A. Brow, and J. L. Corden. 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413**:**327-331. [DOI] [PubMed] [Google Scholar]
- 41.Tan, Q., K. L. Linask, R. H. Ebright, and N. A. Woychik. 2000. Activation mutants in yeast RNA polymerase II subunit RPB3 provide evidence for a structurally conserved surface required for activation in eukaryotes and bacteria. Genes Dev. 14**:**339-348. [PMC free article] [PubMed] [Google Scholar]
- 42.Ursic, D., K. Chinchilla, J. S. Finkel, and M. R. Culbertson. 2004. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res. 32**:**2441-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vanacova, S., J. Wolf, G. Martin, D. Blank, S. Dettwiler, A. Friedlein, H. Langen, G. Keith, and W. Keller. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3**:**e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Hoof, A., P. Lennertz, and R. Parker. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20**:**441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wise, J. A. 1991. Preparation and analysis of low molecular weight RNAs and small ribonucleoproteins. Methods Enzymol. 194**:**405-415. [DOI] [PubMed] [Google Scholar]
- 46.Woychik, N. A., K. McKune, W. S. Lane, and R. A. Young. 1993. Yeast RNA polymerase II subunit RPB11 is related to a subunit shared by RNA polymerase I and III. Gene Expr. 3**:**77-82. [PMC free article] [PubMed] [Google Scholar]
- 47.Wyers, F., M. Rougemaille, G. Badis, J. C. Rousselle, M. E. Dufour, J. Boulay, B. Regnault, F. Devaux, A. Namane, B. Seraphin, D. Libri, and A. Jacquier. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121**:**725-737. [DOI] [PubMed] [Google Scholar]
- 48.Yuryev, A., M. Patturajan, Y. Litingtung, R. V. Joshi, C. Gentile, M. Gebara, and J. L. Corden. 1996. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl. Acad. Sci. USA 93**:**6975-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63**:**405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]