A Novel Cyclic AMP-Dependent Epac-Rit Signaling Pathway Contributes to PACAP38-Mediated Neuronal Differentiation (original) (raw)
Abstract
Pituitary adenylate cyclase-activating polypeptide (PACAP38) stimulation results in the activation of Gsα protein-coupled receptors to regulate neuronal differentiation in a cyclic AMP (cAMP)-dependent manner. These pathways involve protein kinase A (PKA)-dependent processes, but a growing body of evidence indicates that cAMP also regulates cellular functions through PKA-independent signaling cascades. Here we show that the Rit small GTPase is regulated by PACAP38 in a cAMP-dependent but PKA-independent fashion. Rit activation results from stimulation of the cAMP-activated guanine nucleotide exchange factor Epac but does not appear to rely upon the activation of Rap GTPases, the accepted cellular Epac substrates. Although RNA interference studies demonstrated that Epac is required for PACAP38-mediated Rit activation, neither Epac1 nor Epac2 activates Rit directly, indicating that Epac signals to Rit through a novel mechanism in which Rap signaling is not essential. Loss-of-function analysis demonstrated that Rit makes an important contribution to PACAP38-mediated neuronal differentiation. Surprisingly, although Rit is required for sustained extracellular signal-regulated kinase (ERK) and p38 mitogen-activated protein kinase signaling following nerve growth factor stimulation of pheochromocytoma 6 (PC6) cells, Rit silencing selectively suppressed PACAP38-elicited activation of p38, without obvious effects on ERK signaling in the same cells. Moreover, the ability of PACAP38 to stimulate CREB-dependent transcription and to promote neurite outgrowth was inhibited by Rit knockdown. Together, these studies identify an unsuspected connection between cAMP and Rit signaling pathways and imply that Rit can function downstream of Gsα/cAMP/Epac in a novel signal transduction pathway necessary for PACAP38-mediated neuronal differentiation and CREB signaling.
Neurotrophic signaling pathways regulate a wide range of nerve cell functions, including differentiation, axonal and dendritic growth, survival, and various aspects of learning and memory. Although signaling through the Trk receptor tyrosine kinase family is often associated with these biological processes (19), other cellular factors contribute prominently to the regulation of these diverse biological effects. These include the neuropeptide pituitary adenylate cyclase-activating polypeptide (PACAP38), a member of the vasoactive intestinal peptide/secretin/glucagon family of peptides, which is expressed throughout the nervous system and binds to G protein-coupled receptor family members to promote both neuronal differentiation and survival (47, 53).
In pheochromocytoma 12 (PC12) cells, PACAP38 exposure has been shown to stimulate differentiation to result in a sympathetic response-like neuronal cell phenotype characterized by neurite elongation (11). Although a complete description of the cellular pathways utilized by PACAP38 receptors to promote neuronal differentiation is lacking, activation of adenylate cyclase and regulation of Ras/Raf/extracellular signal-regulated kinase (ERK) mitogen-activated protein kinase (MAPK) signaling have been reported to be key signaling pathways (2, 27). While many of the cellular actions of cyclic AMP (cAMP) have been associated with direct activation of protein kinase A (PKA) and while cAMP analogues are sufficient to induce neurite outgrowth in PC12 cells (7, 24), the neurotrophic effect of PACAP38 does not rely solely upon PKA signaling (27). Instead, recent work has identified the cAMP-activated guanine nucleotide exchange factors (GEFs) Epac1 and Epac2, which regulate the activity of the Rap GTPases, as key mediators of PKA-independent signaling (5, 42). Interestingly, Epac proteins have been reported to couple Gsα activation to ERK activation in neuronal cells (29) and are essential for driving cAMP-mediated differentiation signaling (24). These studies have led to the notion that the cell type-specific actions of cAMP on neuronal differentiation result from both PKA-dependent and PKA-independent signaling, with the coordinated stimulation of Ras and Rap GTPases and their differential effects on the ERK MAP kinase cascade playing particularly important roles (15). Thus, PKA and Epac signaling pathways appear to function together to mediate the cellular effects of cAMP (5, 42).
The recently characterized small GTP-binding protein Rit has emerged as an important mediator of neurotrophin signaling (28, 44, 45, 48). Although Rit shares significant sequence identity with Ras and is expressed in a majority of adult and embryonic tissues, including a variety of primary neurons and the developing brain (28, 48), it shares a unique effector domain with the closely related Rin and Drosophila Ric proteins and lacks a known C-terminal lipidation sequence required for membrane localization of the majority of Ras-like GTPases (18, 44). We have previously demonstrated that Rit stimulates a variety of Ras-responsive promoter elements (39), activates Ral GTPase signal transduction (43), and regulates both ERK and p38 MAP kinase signaling pathways (45, 48). Expression of constitutively active Rit also inhibits growth factor withdrawal-mediated apoptosis and promotes neuritogenesis in pheochromocytoma cells (20, 45, 48). Rit is activated following nerve growth factor (NGF) stimulation, and RNA interference (RNAi)-mediated silencing of endogenous Rit inhibits NGF-mediated neuritogenesis (45). These studies have led to the proposal that Rit signaling plays a critical role in regulating neurotrophic signaling pathways, including roles in regulating aspects of neuronal differentiation, neurite elongation, and survival.
For this report, we investigated the contribution of Rit signal transduction to the PACAP38-Gsα protein-coupled receptor neurotrophic signaling cascade. Our results show that Rit is rapidly stimulated after exposure of pheochromocytoma cells to PACAP38. Rit activation occurs in a cAMP-dependent fashion that is PKA independent. Instead, PACAP38-mediated Rit activation involves a novel cAMP-Epac signaling cascade. Although Epac proteins do not serve as direct Rit GEFs, Epac1 function is necessary for PACAP38-mediated Rit activation, and Rap signaling does not appear to be required, suggesting that an additional regulatory protein(s) is necessary to couple Epac activation to Rit signaling. To directly assess the contribution of Rit to PACAP38 signal transduction, we selectively knocked down Rit, using small interfering RNA (siRNA)-mediated RNA interference. Inhibiting Rit expression in pheochromocytoma cells inhibited both PACAP38-induced p38 MAP kinase activation and CREB activation and disrupted PACAP38-dependent neurite differentiation. Taken together, these data indicate that Rit represents a unique cellular target of PACAP38/cAMP/Epac signaling with crucial roles in neuronal signal transduction and suggest that Rit signaling may represent a general component of cAMP-mediated signal transduction in a variety of biological processes.
MATERIALS AND METHODS
Plasmids and reagents.
Flag-tagged wild-type and mutant human Rit and Ras in the p3xFLAG-CMV-10 vector were described previously (45). Expression vectors for GsαQ227L (QL), Gβ1, and Gγ2, pFA2-CREB, pFR-luciferase reporter, and pRSV-β-gal were kindly provided by J. Kehrl (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD). PACAP38 was purchased from BACHEM California (Torrance, CA), dissolved in water (5 μM), and stored at −20°C before use. Constructs expressing wild-type Epac1 and Epac2 and constitutively active Epac2 and wild-type RapGAP were provided by L. Quilliam (Indiana University School of Medicine, Indianapolis, IN). The cDNA clones for wild-type Rap1A and constitutively active Rap1A (Rap 1A-G12V) were purchased from the University of Missouri-Rolla cDNA Resource Center and subsequently subcloned into pCMV-Myc (BD-Clontech, Palo Alto, CA). ddA, 8-bromo-adenosine-3′,5′-cyclic monophosphate (8-Br-cAMP), 8-(_p_-chlorophenylthio)-2′-_O_-methyl-adenosine-3′,5′-cyclic monophosphate (8-CPT-2′-_O_-Me-cAMP), PD98059, H89, and cholera toxin (CTX) were purchased from Calbiochem (La Jolla, CA), while SB203580 was purchased from Tocris (Ellisville, MO). The following commercial antibodies were used: Flag antibody (Sigma, St. Louis, MO), phospho-specific ERK1/2 mouse monoclonal antibody, phospho-specific p38 MAPK mouse monoclonal antibody, p38 MAPK rabbit polyclonal antibody (Cell Signaling, Beverly, MA), ERK1/2 rabbit polyclonal antibody, and actin antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Biotinylated anti-Flag or antihemagglutinin (anti-HA) monoclonal antibodies were generated using an Amersham protein biotinylation system (Amersham Biosciences, Uppsala, Sweden). GST-RGL3-RBD glutathione-agarose beads were prepared from bacterially expressed GST-RGL3-RBD as described previously (37, 45).
Cell lines, cell culture, and transfection.
PC6 is a subline of PC12 cells that produces neurites in response to NGF but also grows as isolated cells rather than in clumps (the generous gift of T. Vanaman, University of Kentucky, Lexington, KY). The cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) containing 10% (vol/vol) heat-inactivated fetal bovine serum (HyClone, Salt Lake City, UT), 5% (vol/vol) heat-inactivated horse donor serum (Life Technologies), 100 μg/ml streptomycin, and 100 units/ml penicillin at 37°C in a humidified atmosphere of 5% CO2. COS cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and cultured in DMEM supplemented with 10% (vol/vol) fetal bovine serum, 100 μg/ml streptomycin, and 100 U/ml penicillin at 37°C in a humidified atmosphere of 5% CO2. PC6 cells were transfected using Effectene, while COS cells were transfected using Superfect (QIAGEN) as described previously (46).
RNA interference.
The knockdown of rat Rit GTPase in PC6 cells, using shRit208 in the pSUPER-GFP/Neo (OligoEngine) vector, was performed as described previously (45). The vector allows direct synthesis of small hairpin RNA (shRNA) transcripts using the polymerase H1-RNA gene promoter and coexpresses green fluorescent protein (GFP) to allow detection of transfected cells. A siRNA with no predicted target site in the rat genome (shCTR) was inserted into pSUPER-GFP/Neo and served as a negative control. To reconstitute the Rit deficiency in shRit208-treated PC6 cells, cells were cotransfected with 0.2 μg of 3× Flag-tagged human Rit (3×Flag-hRit-WT) and 1.5 μg of shRit208 or shCTR by using Effectene and then subjected to G418 selection (400 μg/ml for 48 h). The expression of wild-type human Rit (hRit-WT) was monitored by immunoblotting.
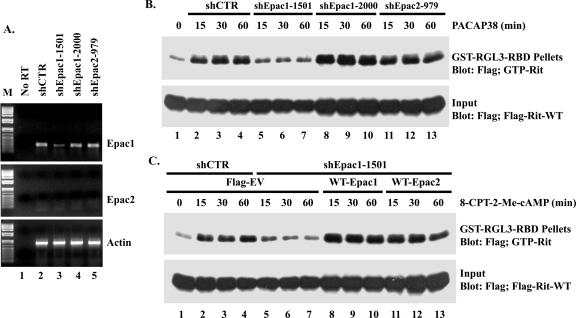
To knock down endogenous Epac proteins in PC6 cells, the gene-specific insert sequences TTCCTAACCATGAGGAACC (target sequence [sense] shEpac1-1501), TTCATGCGACGCTTCAAC (shEpac1-2000), and ACATGAGAATGATCCTGCG (shEpac2-979), which were separated by a nine-nucleotide noncomplementary spacer (TCTCTTGAA) from the reverse complement of the same Epac-specific nucleotide sequences, were synthesized and subsequently subcloned into the BglII and HindIII sites of the pSuper-GFP/Neo vector to generate pSuper-Epac RNA interference constructs (as described in reference 45). Since commercial antibodies against Epac1 and Epac2 failed to detect the endogenous proteins expressed in PC6 cells (data not shown), reverse transcription-PCR (RT-PCR) was used to determine the efficiency of Epac shRNA silencing. Total RNA was isolated with TRIzol reagent (Invitrogen) from PC6 cells transfected with 1.5 μg of pSuper-shCTR, pSuper-shEpac1-1501, pSuper-shEpac1-2000, or pSuper-shEpac2-979, followed by G418 (400 μg/ml) selection for 48 h. Total RNA (2 μg) was subsequently used for reverse transcription (Omniscript; QIAGEN). The following PCR primers were used for PCR: for Epac1, TCTATCACCACTCAGAAGC and TTGAGTCCGCTCTGTGGA; for Epac2, TTCCTGAGGAGATTTAATG and AGTTTAGCTGCTGTCAGC; and for β-actin, GTTTGAGACCTTCAACACCC and ATACTCCTGCTTGCTGATCC.
Neurite outgrowth.
PC6 cells were exposed to DNA-Effectene complexes for 9 to 16 h after pretreatment with a kinase inhibitor (SB203580 or PD98059), as indicated. The cells were then replated after dilution (1:4 dilution), and neurite outgrowth was induced by the addition of 5 to 10 nM PACAP38 and subjected to 400-μg/ml G418 selection (Life Technologies, San Diego, CA). On days 3 and 7 after the initiation of neurite induction, cells were fixed with methanol-acetone (3:1), and images were captured with an Axiovert 200 M phase-contrast microscope (Zeiss) with a 20× objective, using OpenLab 3.1.4 imaging software. The percentage of neurite-bearing cells, the neurite number per cell body, the neurite length, and the number of branch points per neurite were determined in two to four separate experiments as described previously (20, 45).
MAP kinase assay and immunoblotting.
To examine ERK and p38 MAP kinase activation, PC6 cells seeded in six-well plates were transfected with shRit208 or shCTR as a control and then subjected to G418 selection for 48 h. Cells were then serum starved for 5 h and stimulated with PACAP38 (as indicated) prior to the preparation of whole-cell lysates. The phosphorylation status of ERK1/2 and p38 MAP kinases was determined by immunoblotting with phospho-specific antibodies.
Immunoblots were blocked with 1% casein (Sigma) in phosphate-buffered saline (PBS) plus 0.1% Tween 20 (PBST) for 1 h at 25°C and incubated with an appropriate dilution of the primary antibody in 1% casein or 5% bovine serum albumin in PBST for 1 to 2 h. The immunoblots were washed three times with PBST before the addition of horseradish peroxidase-conjugated secondary antibodies or horseradish peroxidase-conjugated streptavidin (Zymed Laboratories Inc., San Francisco, CA), diluted 1:20,000 or 1:40,000 in 1% casein in PBST, respectively. The signal was detected by chemiluminescence (SuperSignal West Pico system; Pierce, Rockford, IL) as described previously (45). Stripping and reprobing of immunoblots to ensure equal expression of recombinant proteins were performed as described previously (45).
p38 MAP kinase activity assay.
To perform p38 MAP kinase assays, PC6 cells were transfected with Flag-tagged p38α, p38γ, or p38δ in the presence of empty 3× HA vector, 3× HA-Rit-Q79L vector, or 3× HA-H-Ras-Q61L vector, and the kinase assay was carried out using a p38 MAP kinase assay kit (Cell Signaling) as described previously (46). The level of phosphorylated glutathione _S_-transferase (GST)-ATF2 was determined by immunoblotting with an anti-phospho-specific ATF2 antibody and used to monitor p38 MAP kinase activity.
Epac GEF assay.
His-Rit (amino acids 1 to 205) was loaded with the fluorescent analogue 2′,3′-_O_-(_N-_methylanthraniloyl)-GDP (mGDP; BioLog Life Science, Germany) and used in nucleotide exchange assays with Epac as described previously (35). In brief, the fluorescence properties of mGDP depend upon the local chemical environment. Thus, the intensity of the emitted fluorescence is approximately twice as high when mGDP is bound in the hydrophobic environment of a G protein. By incubating Rit loaded with mGDP in the presence of an excess of normal GDP, the exchange of mGDP for GDP can be monitored as a decrease in fluorescence intensity. The decay in the fluorescence signal is equal to the rate of nucleotide dissociation, and nucleotide exchange should be accelerated in the presence of an active GEF.
Luciferase reporter gene assay.
CREB transcription activity was analyzed using a luciferase gene reporter assay of PC6 cells. In brief, PC6 cells were transfected with the plasmid pFA2-CREB, pFR-Luci, or pRSV-β-gal in the presence or absence of 3× Flag-Rit-S35N, pSuper-Neo/GFP-shRit208, 3× Flag-Rit-Q79L, 3× Flag-hRit-WT, or empty 3× Flag vector as a negative control. Transfected cells were starved with serum-free DMEM for 5 h, pretreated with or without PD98059 (10 μM) or SB203580 (10 μM) for 30 min, and then stimulated with PACAP38 (10 nM) for 4 h as indicated. The cells were harvested with ligand lysis buffer {25 mM Tris phosphate dibasic, 15% glycerol, 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 1% l-α-lecithin, 1% bovine serum albumin, 4.0 mM EGTA (pH 8.0), 8 mM MgCl2, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride}, and the lysates were incubated at room temperature for 20 min and centrifuged at 13,000 rpm for 5 min. Cleared supernatant (30 μl) was added to a 96-well MicroLite Optiplate (Parkard) containing phosphate-buffered ATP solution (100 mM potassium phosphate-buffered solution [pH 7.8], 3.65 mM ATP, 20 mM MgCl2) (100 μl per well), and the luciferase activities were analyzed with a luminometer (Lmax; Molecular Devices Co., Sunnyvale, California), using d-luciferin (1 mM in 0.1 M potassium phosphate-buffered solution) (Promega, Madison, WI) as the substrate. Supernatants (15 μl) were also transferred to a 96-well clear plate before the addition of β-galactosidase (β-Gal) assay buffer (200 μl; 100 mM _O_-nitrophenyl-β-d-galactopyranoside [ONPG] in sodium phosphate-buffered solution and 50 mM β-mercaptoethanol) and were incubated at 37°C, and β-Gal activity was determined by the absorbance value at 415 nm, using a microplate reader (Bio-Rad).
Rit-GTP and Rap1-GTP precipitation assays.
GST fusion proteins containing the Rit binding domain of RGL3 (residues 610 to 709) were expressed and purified, and Rit activation was assessed essentially as described previously (45). In brief, PC6 or COS cells seeded in six-well plates were transfected with 3× Flag-hRit-WT and incubated for an additional 36 h to allow maximal gene expression. Cells were then starved in serum-free DMEM for an additional 5 h. Cell monolayers were washed once in ice-cold PBS and lysed in GST pull-down assay buffer (20 mM HEPES [pH 7.4], 250 mM NaCl, 50 mM KF, 50 mM β-glycerolphosphate, 1% Triton X-100, 10% glycerol, and 1× protease inhibitor cocktail) by sonication on ice. GST resin (10 μg of the appropriate fusion protein/20 μl glutathione beads) was added to Rit (200 μg)-expressing cell lysates in a total volume of 1 ml and incubated with rotation for 1 h at 4°C, and the resin was recovered in a 4°C microcentrifuge (5 min at 10,000 rpm). The pelleted GST-RGL3-RBD beads were washed once with ice-cold GST pull-down buffer, twice with ice-cold GST pull-down buffer supplemented with 500 mM NaCl, and twice with ice-cold GST pull-down buffer. Bound GTP-Rit was detected by immunoblot analysis using an anti-Flag monoclonal antibody. To determine the effects of PACAP38 on Rit activation, PC6 cells expressing 3× Flag-hRit-WT were stimulated with 10 nM PACAP38 for various times or with increasing amounts of PACAP38 for 15 min following preincubation in serum-free medium for 5 h. To determine the requirement for Gα and Gβγ subunits in Rit activation, PC6 cells were cotransfected with 3× Flag-hRit-WT and either a vector expressing GsαQL or vectors encoding Gβ1 and Gγ2, as indicated, or were stimulated with CTX (5 μg/ml) for the indicated time after pretreatment, with or without ddA (50 μM), for 5 min. To determine the requirement for PKA in PACAP38-mediated Rit activation, PC6 cells transfected with 3× Flag-hRit-WT were pretreated with H89 (10 μM) for 30 min before stimulation with PACAP38, as indicated. To examine the role of cAMP signaling in Rit activation, PC6 cells expressing Flag-Rit-WT were stimulated with 8-Br-cAMP. To determine the requirement for Epac, Rit-transfected PC6 cells were stimulated with 8-CPT-2′-_O_-Me-cAMP (25 μM) or cotransfected with constitutively active Epac2 (CA-Epac2), as indicated. To analyze the role of Rap GTPase function on Rit activity, cells were cotransfected with 3× Flag-hRit-WT and either WT Rap1A, constitutively active Rap1A (Rap1A-G12V), CA-Epac2 and WT Rap1A, or RapGAP. To determine the effect of Epac silencing on Rit activation, PC6 cells were cotransfected with 3× Flag-hRit-WT and shCTR, shEpac1-1501, shEpac1-2000, or shEpac2-979 and then stimulated with PACAP38 (10 nM) or 8-CPT-2′-_O_-Me-cAMP (25 μM). To determine the effect of cAMP signaling on Rit activation in COS cells, cells were transfected with 3× Flag-hRit-WT and either empty 3× Flag vector or CA-Epac2 and stimulated as described above.
Rap1A GTP loading levels were analyzed using a Rap1 activation assay kit (Upstate, Temecula, CA) following the manufacturer's instructions, with minor modifications. In brief, PC6 cells were transfected with Myc-Rap1A-WT together with an expression vector for Flag-RapGAP (25 to 200 ng, as indicated) or CA-Epac2 (50 ng) or an empty vector control. After 36 h, the cells were starved with serum-free DMEM for 5 h before being stimulated with or without PACAP38 (10 nM, 10 min) or 8-CPT-2-_O_-Me-cAMP (25 μM, 10 min), and whole-cell lysates were prepared using assay buffer (20 mM HEPES [pH 7.4], 150 mM NaCl, 50 mM KF, 50 mM β-glycerolphosphate, 5 mM MgCl2, 1 mM Na3VO4, 1% Triton X-100, 10% glycerol plus 1× protease inhibitor cocktail). Total cell lysates (400 μg) were incubated with 25 μl of recombinant GST-RalGDS-RBD agarose (650 μg protein per ml of resin) by end-over-end rotation at 4°C for 1 h. The resins were recovered and washed as described above for the Rit precipitation assay. The resin-bound fractions were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and cellular GTP-Rap1A levels were analyzed by immunoblotting with anti-Myc monoclonal antibody. Total lysates (10 μg) were immunoblotted to ensure equal protein expression.
RESULTS
Role for Rit in PACAP38-induced neurite outgrowth.
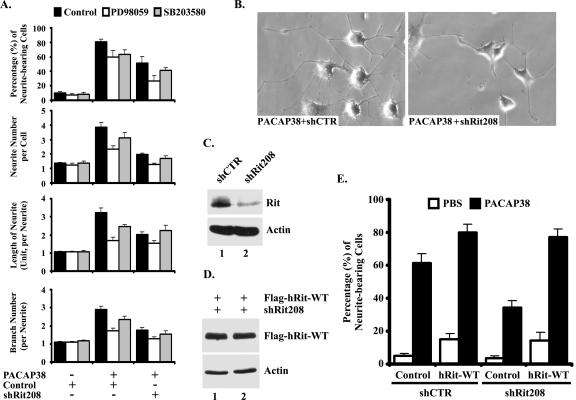
PACAP38 potently stimulates neurite outgrowth in PC12 cells, acting via the PAC1 receptor to activate a diverse array of canonical signaling cascades, including the ERK MAP kinase cascade (53). Rit is rapidly activated following NGF stimulation in PC6 cells (a PC12 subline), and we have recently shown that Rit silencing inhibits NGF-mediated neurite elongation by attenuating both ERK and p38 MAP kinase activation (45). Since PACAP38-induced differentiation depends upon ERK MAP kinase activation (2, 27), we examined the role of Rit signaling in this process. To directly assess whether Rit contributes to PACAP38-mediated differentiation, we selectively inhibited the expression of Rit by using siRNA-mediated RNA interference (16). We have previously demonstrated that the small hairpin interfering RNA shRit208 potently and specifically reduces endogenous rat Rit protein expression in PC6 cells by >80%, whereas a control shRNA with no predicted target in the rat genome (shCTR) has no effect on Rit expression (45) (Fig. 1C). As shown in Fig. 1, shRit208-mediated silencing significantly inhibited the formation of PACAP38-dependent neurite extensions. Quantification of transfected cells showed that Rit depletion resulted in a >36% reduction in the number of neurite-bearing cells following PACAP38 stimulation and resulted in decreased neurite length (37%), neurite branching (39%), and neurite number per cell body (48%) (Fig. 1A and B). This inhibition was approximately equivalent to that induced by pharmacological blockade of either MEK/ERK kinase activity (10 μM PD98059) or p38 MAP kinase activity (10 μM SB203580) (Fig. 1A). Treatment of shRit208-transfected cells with either PD98059 or SB203580 alone resulted in a further slight reduction in neurite outgrowth (Fig. 1A), although treatment with the combination of PD98059 and SB203580 did not further reduce neuritogenesis (data not shown). Similar results were seen with PACAP38-stimulated, shCTR-transfected PC6 cells, indicating that both MEK/ERK and p38 MAP kinase signaling cascades play central roles in PACAP38-mediated neurite outgrowth. Because shRit208 is specific for rat Rit, we reconstituted the Rit deficiency by cotransfecting PC6 cells with wild-type human Rit and shRNA expression vectors. As expected, human Rit escaped shRit208-mediated gene silencing (Fig. 1D) and fully restored PACAP38-mediated PC6 cell differentiation (Fig. 1E), demonstrating the specificity of the shRNA-mediated knockdown. Overexpression of the wild-type protein alone was sufficient to induce modest neurite outgrowth (Fig. 1E), underscoring the ability of Rit to contribute to neuronal differentiation signaling (45, 48). Taken together, these data suggest that Rit plays an important role in PACAP38-dependent neuronal differentiation.
FIG. 1.
Rit is required for PACAP38-mediated neuronal differentiation. (A and B) Rit knockdown attenuates PACAP-induced neurite outgrowth. PC6 cells were transfected with either a control shRNA vector (shCTR) or a Rit-selective shRNA (shRit208), neurite formation was induced with PACAP38 (5 nM), and transformed cells were enriched by drug selection (G418). On days 3 and 7 after PACAP38 stimulation, the percentages of neurite-bearing cells, neurite numbers, neurite lengths, and neurite branches were counted or calculated as described in Materials and Methods. The results are shown for day 7 as means ± standard deviations (SD [error bars]) for three independent experiments. Pretreatment of control cells with either a MEK/ERK (10 μM PD98059) or p38 (10 μM SB203580) kinase inhibitor was used as a control. Representative micrographs from 7-day cultures are shown (B). (C) shRit208 mediates Rit silencing. PC6 cells were transfected with 1.5 μg of shRit208 and subjected to G418 selection (400 μg/ml, 48 h). Endogenous Rit protein levels were determined by immunoblot analysis with an anti-Rit monoclonal antibody. (D) Human Rit is not subject to shRit208-mediated silencing. PC6 cells were transfected with 0.2 μg of 3× Flag-hRit-WT in the presence of 1.5 μg of shRit208 and subjected to G418 (400 μg/ml) selection for 48 h. The expression levels of 3× Flag-hRit-WT were determined by anti-Flag immunoblotting, and actin expression served as a loading control. (E) Human Rit (hRit) restores PACAP38-induced neurite outgrowth to shRit208-expressing PC6 cells. PC6 cells were transfected as described for panel D, and differentiation was stimulated with PACAP38 as described for panel A. Neurite outgrowth was analyzed on day 3 after PACAP38 stimulation, and the percentages of neurite-bearing cells are shown as means ± SD for three independent experiments.
Rit is required for PACAP38-mediated p38, but not ERK, activation.
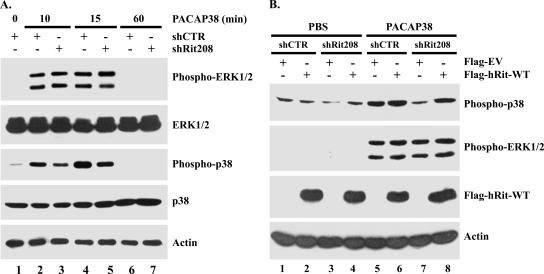
Since PACAP38-mediated neuritogenesis is associated with sustained activation of both cAMP/PKA and MAP kinase cascades (2, 24, 27), we next examined whether Rit silencing altered PACAP38-induced MAP kinase signaling. Kinase activation in shRNA-transfected PC6 cells was monitored by immunoblotting with phospho-specific MAP kinase antibodies following PACAP38 stimulation. As opposed to the case for cells expressing control shRNAs, in which PACAP38-induced ERK and p38 MAP kinase activation was detected within 10 min and remained elevated at 15 min but returned to basal levels within 1 h following stimulation (Fig. 2A, lanes 2, 4, and 6), Rit silencing inhibited p38 activation in response to PACAP38 treatment (Fig. 2A, lanes 3, 5, and 7). Quantification of these data indicated that the loss of Rit inhibits p38 activation approximately 50% (data not shown). Surprisingly, ERK activation was not affected by Rit depletion at any time (Fig. 2A), consistent with a role for Rit signaling in p38, but not ERK, MAP kinase activation following PACAP38 stimulation. These alterations were not a consequence of changes in protein expression, since both ERK and p38 MAP kinase protein levels remained constant during these studies (Fig. 2A). As expected, expression of the human Rit protein rescued PACAP38-mediated p38 activation, demonstrating the specificity of shRit208-mediated knockdown (Fig. 2B). Taken together, these data suggest that Rit signaling contributes to PACAP38-mediated neurite outgrowth, in part through regulation of p38 kinase signaling.
FIG. 2.
Rit is required for PACAP38-mediated p38 activation. (A) Rit silencing attenuates PACAP38-mediated p38 MAP kinase signaling but is not required for ERK activation. PC6 cells were transfected with shCTR or shRit208 and enriched by G418 (400 μg/ml) selection for 2 days. Cells were then serum starved for 5 h prior to PACAP38 (5 nM) stimulation, and the phosphorylation status of ERK1/2 and p38 MAP kinases was analyzed by immunoblotting with the appropriate phospho-specific antibodies. (B) Human Rit restores PACAP38-mediated p38 signaling to shRit208-expressing PC6 cells. PC6 cells were transfected as described in the legend to Fig. 1D and subjected to G418 (400 μg/ml) selection for 48 h. Prior to the preparation of whole-cell lysates, cells were serum starved for 5 h and stimulated with PACAP38 (10 nM) for 15 min, as indicated. The levels of phosphorylated p38 and ERK1/2 MAP kinases were analyzed by immunoblotting with phospho-specific antibodies. The expression levels of human Rit and actin levels were examined by Western blotting. The results shown are from one experiment that was representative of the four experiments performed.
Isoform-specific p38 regulation by Rit.
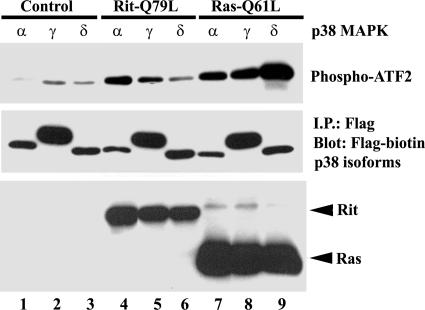
Previous work has suggested that Rit may signal selectively to the p38γ isoform to transform NIH 3T3 cells (40), and we have found that the Rin GTPase displays isoform-selective p38 activation (46). Therefore, we next examined the roles of individual p38 kinase isoforms in Rit-mediated signaling. The specificity of Rit p38 kinase cascade activation in PC6 cells was examined using in vitro kinase assays of PC6 cells coexpressing Flag-tagged p38 isoforms and constitutively active Rit (RitQ79L). As shown in Fig. 3, RitQ79L activated p38α and p38γ but did not noticeably stimulate p38δ kinase activity. As a general positive control, we expressed H-RasQ61L, a constitutively active H-Ras mutant which potently stimulates all three p38 isoforms (Fig. 3). These studies suggest that isoform-specific p38 MAP kinase activation may contribute to Rit-mediated signaling.
FIG. 3.
Rit activates p38α and p38γ MAP kinases. PC6 cells were cotransfected with Flag-tagged p38 MAP kinase isoforms α, γ, and δ in the presence of empty 3× HA vector (control), 3× HA-hRit-Q79L, or 3× HA-H-Ras-Q61L. The transfected p38 isoforms were immunoprecipitated (I.P.) with an anti-Flag monoclonal antibody from 400 μg of total cell lysate, and the immunoprecipitates were subjected to in vitro p38 MAP kinase assays as described in Materials and Methods. The data shown are representative of three independent experiments.
Rit signaling contributes to PACAP38-mediated CREB activation.
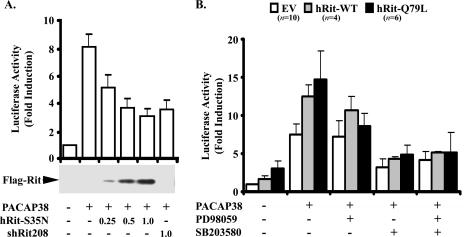
CREB has been identified as an important cAMP-controlled transcriptional regulator in a wide range of neuronal processes (30). Although PKA is an important mediator of CREB phosphorylation, other regulatory kinases in neurons, including MSK1, a kinase activated downstream of both ERK and p38 MAP kinase cascades, have been shown to play a critical role in neurotrophin-mediated CREB activation (1). Therefore, we hypothesized that Rit knockdown might result in a loss of PACAP38-mediated CREB activation. To test this possibility, PC6 cells were cotransfected with a CREB-regulated reporter construct and either a dominant inhibitory Rit mutant (RitS35N) or shRit208 to knock down endogenous Rit expression. Stimulation of mock-transfected PC6 cells with PACAP38 induced expression of the luciferase reporter, with an eightfold maximal induction (Fig. 4A). In contrast, shRNA-mediated Rit knockdown resulted in a potent inhibition of reporter gene activity (Fig. 4A). Increasing levels of RitS35N expression resulted in a dose-dependent inhibition of PACAP38-mediated CREB activation. In contrast, expression of either wild-type Rit or constitutively active Rit (RitQ79L) alone resulted in modest stimulation of the CREB-regulated reporter (2-fold and >3-fold, respectively) but significantly enhanced PACAP38-dependent CREB transcriptional activation, leading to a >12-fold induction of the luciferase reporter (Fig. 4B). Pharmacological blockade of p38 (10 μM SB203580), but not inhibition of MEK/ERK signaling (10 μM PD98059), significantly inhibited PACAP38-dependent CREB activation, suggesting that the p38 kinase cascade is crucial for this signaling event. ERK activation was required for the enhanced CREB activation seen with coexpressed WT Rit and RitQ79L, since PD98059 treatment reduced the CREB activity to that seen with PACAP38-stimulated control cells. However, p38 activity was critical, since SB203580 treatment largely inhibited PACAP38-mediated CREB activation in all cells, and the addition of both PD98059 and SB203580 did not further suppress CREB activity (Fig. 4B). Taken together, these findings suggest that Rit signaling contributes to PACAP38-mediated activation of CREB, possibly through regulation of p38 MAP kinase activity.
FIG. 4.
Rit is involved in PACAP38-mediated CREB activation. (A) PC6 cells were transfected with 100 ng of pFA2-CREB, pFR-Luciferase reporter, and pRSV-β-gal in the absence or presence of 3× Flag-Rit-S35N (0.25, 0.5, and 1.0 μg) or shRit208, as indicated. Cells were serum starved for 5 h prior to PACAP38 (10 nM) stimulation for 4 h where indicated, and the luciferase activities were analyzed as described in Materials and Methods. The results are presented as means ± SD for three experiments performed in duplicate. (B) p38 MAP kinase signaling contributes to PACAP38/Rit signaling to CREB. PC6 cells were transfected with the CREB reporter system as described for panel A in the presence of either empty 3× Flag vector (EV), 3× Flag-RitWT, or 3× Flag-Rit-Q79L. The cells were serum starved for 5 h and pretreated with either PD98059, SB203580, or a combination of both (10 μM each) for 30 min before PACAP38 stimulation (10 nM for 4 h). The results are presented as means ± SD for 4 to 10 experiments performed in triplicate.
PACAP38 activates Rit in a Gsα-mediated and cAMP-dependent manner.
We reasoned that if Rit serves as a key element in PACAP38-mediated signaling, it must be activated following PACAP38 stimulation. To examine the cellular GTP-binding status of Rit, we utilized a GST fusion protein containing the Rit binding domain (RBD) of RGL3 to selectively precipitate GTP-bound Rit as described previously (45). PC6 cells transiently transfected with Flag-tagged Rit were incubated in serum-deficient medium for 5 h prior to treatment with PACAP38, and then pull-down experiments were carried out on cell lysates prepared at various times following PACAP38 stimulation. Although Rit protein levels were constant during the course of the treatment (Fig. 5A) and serum-starved PC6 cells contained low levels of Rit-GTP, PACAP38 stimulation led to a rapid and dose-dependent increase in the levels of GTP-bound Rit (Fig. 5A). Activation of Rit was detected within 1 min following PACAP38 stimulation and remained elevated for at least 1 h (Fig. 5A). Activation was dose dependent, with activation observed with as little as 2.5 nM PACAP38 and reduced Rit activity observed with concentrations above 20 nM. Taken together, these data demonstrate that Rit is activated following PACAP38 stimulation.
FIG. 5.
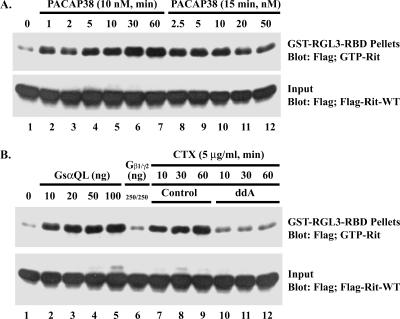
PACAP38 activates Rit in a Gsα-dependent manner. (A) PACAP38 activates Rit in PC6 cells. PC6 cells were transfected with an expression construct for 3× Flag-hRit-WT. Prior to preparation of whole-cell lysates, cells were serum starved for 5 h and then exposed to 10 nM PACAP38 for the indicated time (lanes 2 to 7) or to the indicated concentrations of PACAP38 for 15 min (lanes 8 to 12). Whole-cell lysates (200 μg) were subjected to GST pull-down assay, using GST-RGL3-RBD agarose, and the levels of GTP-bound Rit were determined as described in Materials and Methods. Total lysates (5 μg) were immunoblotted with an anti-Flag monoclonal antibody to ensure equal expression of 3× Flag-hRit-WT in the assay. (B) Gsα and CTX, but not Gβγ, activate Rit. PC6 cells were cotransfected with expression constructs for 3× Flag-hRit-WT and either a constitutively active Gsα (GsαQL) mutant or a combination of Gβ1 and Gγ2 vectors, as indicated, and Rit activation was determined using a GST-RGL3-RBD pull-down assay as described above. To examine the requirement for adenylate cyclase/cAMP in Rit activation, serum-starved (5 h) PC6 cells transfected with 3× Flag-Rit-WT were treated with or without ddA (50 μM) for 5 min and then exposed to CTX (5 μg/ml) for the indicated time periods, and the levels of GTP-bound Rit were determined using GST-RGL3-RBD as described above.
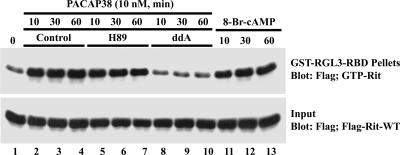
In PC12 cells, PACAP38 signals through the PAC1 Gsα protein-coupled receptor to promote neuronal differentiation (11). While PAC1 can stimulate a variety of signaling cascades, including phospholipase C, phosphatidylinositol 3-kinase, and L-type Ca2+ channel activation (53), it couples predominantly to the Gsα-adenylate cyclase-cAMP signaling pathway. Since we have recently shown that Rit is not activated by elevated levels of intracellular calcium or diacylglycerol in PC6 cells (45), we next examined whether Rit was regulated in a Gsα-cAMP-dependent manner. Consistent with a role for Gsα signaling in PACAP38-mediated Rit activation, cells overexpressing a GTPase-deficient activated mutant of the α subunit Gsα (GsαQL), but not βγ subunits, resulted in potent Rit activation (Fig. 5B). Furthermore, treatment of PC6 cells with CTX, which is known as an activator of Gsα proteins (6), increased GTP-bound Rit in a time-dependent fashion (Fig. 5B). Cholera toxin-mediated Rit activation was potently suppressed by the direct adenylate cyclase inhibitor ddA (50 μM) (Fig. 5B). In keeping with a role for adenylate cyclase/cAMP signaling in Rit activation, ddA also inhibited PACAP38-induced Rit activation (Fig. 6). The same effect was observed upon the addition of 8-Br-cAMP, a nonhydrolyzable cAMP analog, which alone was sufficient to induce potent Rit activation (Fig. 6). Taken together, these data suggest that Rit activation by PACAP38 is dependent, at least in part, on cAMP formation.
FIG. 6.
PACAP38-mediated Rit activation is cAMP dependent but PKA independent. PC6 cells expressing 3× Flag-hRit-WT were preincubated with H89 (10 μM, 60 min) or ddA (50 μM, 5 min) and then stimulated with PACAP38 (10 nM) or exposed to 8-Br-cAMP (50 μM) for the indicated times. Cell lysates (200 μg) were analyzed by GST-RGL3-RBD pull-down assay as described in Materials and Methods, and GTP-bound Rit levels were determined by immunoblot analysis using biotinylated anti-Flag antibody. Equal expression of recombinant Flag-tagged human Rit was controlled by immunoblotting cell lysates (5 μg) with anti-Flag antibody (input).
To determine whether cAMP-dependent Rit activation required PKA signaling, PACAP38-stimulated PC6 cells were pretreated with the PKA inhibitor H89 (10 μM). Whereas preincubation with ddA reduced Rit-GTP levels, H89 had no significant effect on Rit activation (Fig. 6), while it effectively inhibited PACAP38-mediated CREB activation (data not shown). We infer from these results that PACAP38-mediated Rit activation is cAMP dependent but PKA independent.
Involvement of Epac in PACAP38-mediated Rit activation: Rap signaling is not essential.
The inability of H89 to inhibit Rit activation by PACAP38 suggested that the action of cAMP was mediated by another cellular effector. The recently identified cAMP-GEF/Epac proteins exhibit guanine nucleotide exchange factor activities, exert diverse effects on a variety of cellular functions, and are thought to mediate many of the PKA-independent effects of cAMP-regulated signaling (4, 42). Therefore, we next examined the effects of the membrane-permeative Epac-selective cAMP analog 8-CPT-2′-_O_-Me-cAMP on Rit activity in PC6 cells (4, 7). As illustrated in Fig. 7A, 8-CPT-2′-_O_-Me-cAMP treatment resulted in a potent and rapid increase in GTP-bound Rit levels. Indeed, 8-CPT-2′-_O_-Me-cAMP-induced stimulation was as pronounced as 8-Br-cAMP-mediated Rit activation, implicating cAMP-Epac signaling in PACAP38-dependent Rit activation (Fig. 7A).
FIG. 7.
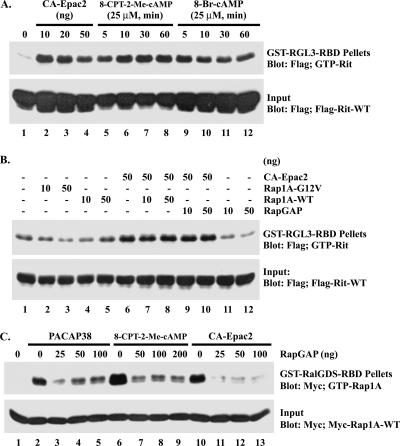
Epac induces the activation of Rit in PC6 cells. (A) Epac2 induces Rit activation in PC6 cells. PC6 cells were transfected with 3× Flag-hRit-WT (1 μg), with or without constitutively active Epac2 (10, 20, or 50 ng). Prior to the preparation of whole-cell lysates, cells were serum starved for 5 h. Rit-GTP was recovered using GST-RGL3-RBD pull-down analysis as described in Materials and Methods. PC6 cells transfected with 3× Flag-hRit-WT were exposed to either 8-Br-cAMP (25 μM) or the Epac-selective cAMP analog 8-CPT-2′-_O_-Me-cAMP (25 μM) for the indicated times, and the levels of GTP-bound Rit were analyzed using a GST-RGL3-RBD pull-down assay. (B) Epac-mediated Rit activation does not appear to require Rap1A. PC6 cells were cotransfected with 3× Flag-hRit-WT and an active form of Rap1A (Rap1A-G12V), wild-type Rap1A (Rap1A-WT), or RapGAP in the presence or absence of constitutively active Epac2 (CA-Epac2), as indicated. Transfected cells were serum starved for 5 h, whole-cell lysates were prepared, and Rit activation was accessed using GST-RGL3-RBD pull-down analysis. The expression level of recombinant Rit was determined by immunoblot analysis. The data shown are from one experiment that is representative of the four experiments performed. (C) RapGAP inhibits Rap1A activation. PC6 cells expressing Myc-Rap1A-WT in the presence of Flag-RapGAP or empty Flag vector were serum starved and then stimulated with PACAP38 (10 nM, 10 min) (lanes 2 to 5) or 8-CPT-2-Me-cAMP (25 μM, 10 min) (lanes 6 to 9) or cotransfected with Flag-CA-Epac2 (50 ng) (lanes 10 to 13). The GTP-loading levels of Rap1A were determined as described in Materials and Methods.
To substantiate the involvement of Epac proteins in PACAP38-mediated Rit activation, PC6 cells were transiently transfected with a constitutively active mutant of Epac2 (CA-Epac2), which lacks the cAMP-binding domain and has been shown to promote cAMP-independent Rap activation (10). As shown in Fig. 7A, expression of CA-Epac2 alone resulted in potent Rit activation.
Since Epac proteins act as GEFs for Rap GTPases but not for H-Ras (4, 10, 36), we next asked whether Epac-mediated Rit activation was indirect, involving downstream signaling following Rap activation. However, overexpression of a constitutively active Rap1A mutant (Rap1A-G12V) failed to promote elevated GTP-Rit levels (Fig. 7B). Furthermore, expression of wild-type Rap1A did not further enhance CA-Epac2-mediated Rit activation (Fig. 7B). Although these data suggest that Rap1 is not required for PACAP38-induced Rit activation, Epac proteins stimulate both Rap1 and Rap2 GTPases, so we next examined whether another Rap isoform might mediate Rit activation. To address this issue, we overexpressed RapGAP, which has previously been shown to inhibit Rap signaling in PC12 cells (12, 38). RapGAP was cotransfected with CA-Epac2 and Flag-tagged Rit, and Rit activation was detected following GST-RGL3-RBD pull-down. As shown in Fig. 7B, overexpression of RapGAP had no effect on CA-Epac2-mediated Rit activation, although RapGAP expression was shown to potently inhibit PACAP38-, 8-CPT-2′-_O_-Me-cAMP-, and CA-Epac2-mediated Rap1 activation (Fig. 7C). Taken together, these data suggest that Epac plays a central role in PACAP38-dependent Rit activation but that Rap GTPase activity is not essential for this signaling cascade. Instead, these data suggest that Epac2 may directly activate Rit.
Epac proteins are not RitGEFs.
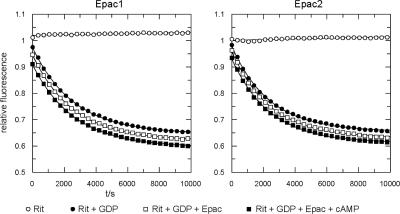
The ability of CA-Epac2 and Epac-selective cAMP analogs to activate Rit suggested that Epac proteins might directly catalyze GDP/GTP exchange on Rit. To determine whether the activation of Rit by Epac results from a direct interaction, we measured Rit activation in vitro. Equal amounts of the recombinant catalytic domains of Epac1 and Epac2 (200 nM) were incubated with fluorescent mGDP-loaded Rit (100 nM) in the presence of excess unlabeled GDP, and the exchange of guanine nucleotides was followed in real time as a decrease in fluorescence (35). As shown in Fig. 8, when these rates were compared with the intrinsic exchange rate for mGDP-Rit measured in the same experiment, no difference was seen. Thus, Epac1 and Epac2 do not exhibit catalytic activity toward Rit in vitro.
FIG. 8.
Epac proteins are not direct RitGEFs. The graphs show the nucleotide exchange of Rit. The dissociation of the Rit-mGDP complex was monitored by the time-dependent decrease in fluorescence intensity. Rit-mGDP (200 nM) was loaded with the fluorescent nucleotide analogue mGDP in the presence (squares) or absence (circles) of inactive (open squares) or activated (500 μM cAMP treated) (closed squares) 100 nM Epac1 (left panel) or 100 nM Epac2 (right panel) after the addition of a 100-fold excess of unlabeled GDP, as indicated. For clarity of presentation, the curves were shifted 0.025 relative units to avoid the overlay of symbols.
To extend the in vitro results, we investigated the ability of cAMP signaling to activate Rit in COS cells, a cell line which does not express endogenous Epac proteins (9). As expected for Epac-dependent signaling cascades, the expression of constitutively active GsαQL or treatment with either 8-Br-cAMP or cholera toxin failed to activate Rit in COS cells (data not shown). In contrast to PC6 cells, COS cells transfected with constitutively active Epac2 together with Flag-Rit-WT or expressing WT-Epac2 and Flag-Rit-WT and stimulated with 8-CPT-2′-_O_-Me-cAMP fail to activate Rit (data not shown). These results suggest that additional cellular factors required to couple Epac to Rit activation are lacking in COS cells.
PACAP38-mediated Rit activation requires Epac1.
Since the data in Fig. 8 indicate that Epac proteins do not function as RitGEFs, we next examined whether PACAP38-dependent Rit activation requires Epac function. Because commercial Epac-selective antibodies were unable to detect endogenous Epac1/2 expression in PC6 cells (data not shown), RT-PCR was used to examine endogenous Epac expression. As shown in Fig. 9A, only Epac1 is expressed in PC6 cells. Two different shRNAs for rat Epac1 (shEpac1-1501 and shEpac1-2000) and a single shRNA targeting Epac2 (shEpac2-979) were generated, and RT-PCR studies found that endogenous Epac1 expression was reduced by shEpac1-1501, while transfection with the remaining Epac shRNAs had no effect on Epac1 mRNA levels (Fig. 9A). We first tested the effect of Epac1 loss on PACAP38-induced Rit activation. As shown in Fig. 9B, shEpac1-1501-mediated silencing significantly diminished the activation of Rit. Neither shEpac1-2000, shEpac2-979, nor shCTR disrupted PACAP38-mediated Rit activation (Fig. 9B), consistent with the inability of these constructs to knock down Epac1 expression (Fig. 9A). Depletion of cellular Epac1 by transient transfection with Epac1-specific shRNA (shEpac1-1501) also inhibited the activation of Rit by the Epac-specific cAMP analog 8-CPT-2′-_O_-Me-cAMP (Fig. 9C). Since the shRNAs used in these experiments are specific for the rat, we reconstituted the Epac deficiency by cotransfecting PC6 cells with either wild-type human Epac1 or wild-type human Epac2 and the shEpac1-1501 expression vector. Both Epac proteins escaped shEpac1-1501-mediated gene silencing (data not shown) and fully restored 8-CPT-2′-_O_-Me-cAMP-mediated Rit activation (Fig. 9C). From these results, we conclude that while Epac does not serve as a direct RitGEF, it is a critical downstream signaling element in PACAP38-dependent Rit activation.
FIG. 9.
Epac1 is required for Rit activation in PC6 cells. (A) Expression of shEpac1-1501 induces Epac1 silencing. PC6 cells were transfected with shCTR (lane 2), shEpac1-1501 (lane 3), shEpac1-2000 (lane 4), or shEpac2-979 (lane 5) and subjected to G418 (400 μg/ml) enrichment for 2 days. Purified RNAs were isolated and subjected to RT-PCR using Epac1-, Epac2-, and actin-specific primers. Non-reverse transcription controls are shown (lane 1). M, 1-kb Plus DNA ladder (Invitrogen). (B) Epac1-1501 silencing attenuates PACAP38-dependent Rit activation. PC6 cells were cotransfected with 3× Flag-Rit-WT and either shCTR, shEpac1-1501, shEpac1-2000, or shEpac2-979. The cells were starved with serum-free DMEM for 5 h and stimulated with 10 nM PACAP38, and the level of GTP-bound Rit was determined as described in Materials and Methods. (C) Epac1 and Epac2 restore cAMP-mediated Rit activation to shEpac1-1501-expressing PC6 cells. PC6 cells were transfected with 3× Flag-Rit-WT in the presence of shCTR or shEpac1-1501, together with an empty 3× Flag, WT Epac1, or WT Epac2 expression vector, and stimulated with 8-CPT-2′-_O_-Me-cAMP (25 μM) for the indicated times. The GTP-Rit levels were analyzed using a GST-RGL3-RBD pull-down assay as described above.
DISCUSSION
PACAP38 and its receptors are critical regulators of neuronal proliferation, differentiation, and survival and play important roles in a variety of neurological processes, including learning and memory formation (47, 53, 54). In this report, we describe a novel PACAP38 receptor signaling pathway in which cAMP-dependent activation of Epac leads to stimulation of a Rit signaling pathway to promote both neurite elongation and the regulation of CREB transcriptional activity. We show that Rit activation is induced by the stimulation of adenylate cyclase (by the expression of activated Gsα or treatment with cholera toxin) and suppressed by direct adenylate cyclase inhibition (ddA), indicating that cAMP is critical for PACAP38-mediated Rit activation. The action of cAMP appears to be independent of PKA, since the inhibition of the kinase pathway (H89) did not disrupt Rit activation. Instead, the cAMP-activated GEF Epac is the critical cAMP effector. Although Epac does not directly activate Rit, Epac is necessary for PACAP38-dependent Rit activation and does not appear to require the actions of the Rap GTPases. Importantly, shRNA-mediated Rit silencing inhibited PACAP38-stimulated neuronal differentiation (Fig. 1). Although the regulation of sustained ERK signaling appears central to the contributions of previously identified Ras family GTPases, including Ras and Rap, to cAMP-mediated neuronal differentiation (5, 15, 49), Rit predominantly regulates p38 MAP kinase signaling (Fig. 2). These data provide the first evidence that Rit can be activated by Gsα protein-coupled receptors and identify an important function for Epac/Rit in the generation of prolonged p38 MAP kinase activation needed for PACAP38-dependent neurite outgrowth and CREB activation.
The persistence of PACAP38-mediated Rit activation in the presence of PKA inhibition (Fig. 6) and the stimulation of Rit by selective Epac pathway activation (Fig. 7) allowed us to identify Rit as a novel target of cAMP/Epac signaling. Since the catalytic GEF domain of Epac has been shown to activate Rap but not Ras, Ral, or R-Ras (10) and since a recent study identified Epac as a key regulator of Rap1-mediated Rac activation (31), we examined whether Epac-mediated Rit regulation was indirect, being controlled in a Rap-dependent fashion. Surprisingly, activated Rap1 failed to promote Rit GTP loading, and the expression of RapGAP, which results in a general blockade of Rap signaling (Fig. 7C), did not alter Epac2-mediated Rit activation (Fig. 7B). Thus, Rap signaling does not appear to play a central role in Epac2-mediated Rit regulation. More importantly, while shRNA studies indicated that Epac function is required for PACAP38-mediated Rit activation (Fig. 9), Epac1 and -2 do not serve to directly regulate Rit (Fig. 8). Indeed, the studies presented here suggest that Rit activation requires additional regulatory molecules. Thus, although the expression of constitutively active Epac2 potently stimulates Rit in PC6 cells, CA-Epac2 failed to elevate GTP-Rit levels in COS cells (Fig. 7 and data not shown). These finding indicate that Epac-dependent Rit activation relies on an additional regulatory protein(s) that is not universally expressed. Identifying the cellular factors involved in cAMP/Epac-regulated Rit signaling remains an important challenge for the future. To date, Epac signaling has been assumed to involve Rap-dependent events (4, 42). Examination of the physiological role of Epac proteins is ongoing but has already established important roles for these regulators in many biological processes (4, 42). As these investigations continue, it will be necessary to consider the contribution of Rit, in addition to that of Rap, to these diverse cellular signaling pathways. In the nervous system, G protein-coupled receptors that couple to Gsα-adenylate cyclase-cAMP signaling are activated by a diverse array of ligands, including biogenic amines and peptide and nonpeptide neurotransmitters (32, 33). The possibility that these signaling cascades may require Epac/Rit signaling is currently under investigation.
Although the mechanism of cAMP-mediated neuronal differentiation remains controversial, cAMP-induced ERK activation that occurs independently of PKA appears to play a central role (15). Recently, Kiermayer et al. (24) reported that cAMP-mediated PKA activation results in ERK kinase signaling that increases PC12 cell proliferation, whereas selective Epac stimulation results in sustained PKA-independent ERK activation and promotes neuronal differentiation. Indeed, several studies have found that selective Epac activation is capable of inducing neurite extension in PC12 cells, although the mechanism by which Epac stimulates differentiation remains unclear (7, 24). Thus, our findings that PACAP38-mediated Rit activation relies upon cAMP-Epac signaling (Fig. 5, 6, and 7) and that Rit knockdown inhibits PACAP38-induced neuritogenesis (Fig. 1) are in keeping with the importance of Epac to differentiation. However, while the regulation of sustained ERK signaling appears central to the role of Ras and Rap signaling in cAMP-mediated neuronal differentiation (5, 15, 49), Rit silencing resulted in the selective loss of p38 signaling without inhibiting PACAP38-mediated ERK activation (Fig. 2). We speculate that Epac may play a dual signaling function in cAMP-stimulated neurons, activating the ERK cascade, perhaps in a Rap-dependent manner (15), while also regulating Rit activity to control p38 kinase cascade signaling. Because ERK and p38 signaling pathways share common cellular targets but also display distinct signaling activities (50, 52), these data suggest that cAMP-Epac pathway signaling requires both MAP kinase cascades to exert its cellular effects. Thus, Epac may allow cross talk between the MAP kinase signaling pathways and serve to modulate the biological outcome of cAMP signaling. These results also indicate that sustained p38 activation plays a critical role in PACAP38-dependent neuronal differentiation and CREB activation. These findings are in agreement with a number of recent studies suggesting that p38 signaling plays a central role in both neuronal differentiation and survival (50) and are consistent with the sustained activation of p38 following PACAP38 stimulation (Fig. 2), with recent studies demonstrating that disruption of p38 signaling inhibits neurite outgrowth in several systems, including PACAP38-stimulated PC12 cells (17, 21, 22, 41) (Fig. 1), and with the importance of p38 signaling to PACAP38/Epac/Rit-dependent CREB activation (Fig. 4).
The finding that Rit knockdown had no effect on PACAP38-dependent ERK signaling was unexpected, because earlier studies have shown that Rit function is required to couple NGF stimulation to B-Raf/ERK and p38 MAP kinase signaling in PC6 cells (45). The organization of higher-order molecular complexes by scaffolding proteins is one mechanism known to confer specificity to MAP kinase signaling (25, 34) and allows the distinct regulation of Ras and Rap GTPases following NGF activation in PC12 cells (19). The ability of Rit to promote distinct MAP kinase pathway activation, even within the same cell, reinforces the notion that unique signaling complexes may be involved in the activation of Rit following NGF (45) and PACAP38 stimulation. Analogous to their ability to promote selective MAP kinase signaling pathways, distinct scaffolding proteins may also mediate the observed ERK and p38 kinase signaling specificity following Rit activation (34). Recent work has found Epac1 associated with an A kinase-anchoring protein (mAKAP) signaling complex. mAKAP is expressed in both striated myocytes and neurons, and the scaffolded mAKAP/Epac1 complex appears to integrate cAMP, calcium, and MAP kinase signals to allow differential Gsα protein-coupled receptor signaling (13, 14). Additional regulatory factors, such as the expression of specific p38 phosphatases or the expression of proteins directing the activation of specific kinase modules, may also help explain the isoform-selective activation of p38α and p38γ by Rit (Fig. 3). Why Rit stimulates only a subset of p38 kinases is not clear (26), but these data are consistent with a recent study demonstrating isoform-specific p38γ activation by Rit in NIH 3T3 cells (40). The nature of the molecular machinery that links Rit and p38 in PACAP38-stimulated neurons remains to be characterized.
RNAi-knockdown studies were also used to demonstrate a role for Rit in PACAP38-mediated CREB activation. cAMP response element (CRE)-mediated transcription is a convergence point for multiple pathways initiated by G protein-coupled receptor and receptor tyrosine kinase activation in PC12 cells through the regulation of both CRE-binding protein (CREB) and CREB-binding protein (CBP) (30). Transcription of CRE-dependent genes is required for PC12 cell differentiation, and the suppression of CREB activity following Rit silencing might explain the requirement for Rit activity in differentiation following PACAP38 treatment of PC6 cells (Fig. 4). How might the loss of Rit result in a selective loss of CREB transcription? The expression of CRE-regulated genes can be controlled by a variety of signaling cascades that include PKA, calmodulin kinase, and MAP kinase pathways (30). Induction of these pathways leads to the phosphorylation and activation of CREB. The present study suggests that Rit plays a prominent role in PACAP38-induced p38 MAP kinase activation (Fig. 2), and recent work has established p38 MAP kinase signaling as a key regulator of CREB. p38 does not appear to directly activate CREB; instead, other protein kinases that are activated by p38 signaling are involved in CREB phosphorylation (30). Among these are the MSK1 and MSK2 kinases, which are activated downstream of both ERK and p38 MAP kinase pathways in vivo (1, 8, 51, 55). The possibility that these kinases contribute to Rit-mediated CREB regulation is currently under investigation. In addition, recent studies have identified a role for CREB in the control of activity-dependent neuronal survival (3, 23). Thus, Rit signaling might also contribute to PACAP38-dependent neuronal survival, consistent with earlier studies suggesting a role for Rit in NGF-mediated neuronal survival (48). Future studies will be needed to examine this possibility in neurons.
In summary, through RNAi-mediated knockdown of Rit expression in pheochromocytoma cells, we have discovered a novel in vivo requirement for Rit in PACAP38 receptor-specific signaling. The marked effect of Rit silencing on PACAP38-mediated p38 activation, but not on the activation of ERK, suggests that Rit provides a function in PACAP38 signaling that cannot be compensated for by other small GTPases. Our data support a model in which Rit is activated by cAMP through a PKA-independent pathway. Instead, activation of the adenylyl cyclase-coupled PACAP38 receptor stimulates Epac to promote Rit activation, in a manner that does not appear to rely upon Rap signaling (Fig. 7B) or the direct regulation of Rit by Epac (Fig. 8). These data indicate that Epac-regulated Rit activation is important for coupling cAMP to both CREB transcription and p38 MAP kinase activation and that Rit signaling contributes to PACAP38-induced neuronal differentiation. Thus, the cAMP-Epac-Rit signaling module may be an important general regulator of neuronal signaling. How widely this system operates downstream of other Gsα- and adenylyl cyclase-dependent signaling cascades and the role of p38 MAP kinase signaling in neuronal function are important issues that remain to be addressed. The identification of additional molecules that link cAMP-Rit to the p38 cascade should provide insight into the role of Rit signaling in neuronal regulation and development.
Acknowledgments
The cDNAs encoding GsαQ227L (QL), Gβ1, and Gγ2 and the plasmids needed to analyze CREB transcription were kindly provided by J. Kehrl (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD). The cDNAs encoding WT Epac1/2, constitutively active Epac2, and RapGAP were a gift from L. Quilliam (Indiana University School of Medicine, Indianapolis, IN).
This work was supported by Public Health Service grant NS045103 (to D.A.A.) from the National Institute of Neurological Disorders and Stroke and by grant P20RR20171 from the COBRE program of the National Center for Research Resources, a component of the National Institutes of Health (NIH). H.R. was supported by the Chemical Sciences of The Netherlands Organization for Scientific Research (NWO-CW).
The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
Footnotes
▿
Published ahead of print on 25 September 2006.
REFERENCES
- 1.Arthur, J. S., A. L. Fong, J. M. Dwyer, M. Davare, E. Reese, K. Obrietan, and S. Impey. 2004. Mitogen- and stress-activated protein kinase 1 mediates cAMP response element-binding protein phosphorylation and activation by neurotrophins. J. Neurosci. 24**:**4324-4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrie, A. P., A. M. Clohessy, C. S. Buensuceso, M. V. Rogers, and J. M. Allen. 1997. Pituitary adenylyl cyclase-activating peptide stimulates extracellular signal-regulated kinase 1 or 2 (ERK1/2) activity in a Ras-independent, mitogen-activated protein kinase/ERK kinase 1 or 2-dependent manner in PC12 cells. J. Biol. Chem. 272**:**19666-19671. [DOI] [PubMed] [Google Scholar]
- 3.Bito, H., and S. Takemoto-Kimura. 2003. Ca(2+)/CREB/CBP-dependent gene regulation: a shared mechanism critical in long-term synaptic plasticity and neuronal survival. Cell Calcium 34**:**425-430. [DOI] [PubMed] [Google Scholar]
- 4.Bos, J. L. 2003. Epac: a new cAMP target and new avenues in cAMP research. Nat. Rev. Mol. Cell Biol. 4**:**733-738. [DOI] [PubMed] [Google Scholar]
- 5.Bos, J. L., J. de Rooij, and K. A. Reedquist. 2001. Rap1 signalling: adhering to new models. Nat. Rev. Mol. Cell Biol. 2**:**369-377. [DOI] [PubMed] [Google Scholar]
- 6.Cassel, D., and Z. Selinger. 1977. Mechanism of adenylate cyclase activation by cholera toxin: inhibition of GTP hydrolysis at the regulatory site. Proc. Natl. Acad. Sci. USA 74**:**3307-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen, A. E., F. Selheim, J. de Rooij, S. Dremier, F. Schwede, K. K. Dao, A. Martinez, C. Maenhaut, J. L. Bos, H. G. Genieser, and S. O. Doskeland. 2003. cAMP analog mapping of Epac1 and cAMP kinase. Discriminating analogs demonstrate that Epac and cAMP kinase act synergistically to promote PC-12 cell neurite extension. J. Biol. Chem. 278**:**35394-35402. [DOI] [PubMed] [Google Scholar]
- 8.Deak, M., A. D. Clifton, L. M. Lucocq, and D. R. Alessi. 1998. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 17**:**4426-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Rooij, J., N. M. Boenink, M. van Triest, R. H. Cool, A. Wittinghofer, and J. L. Bos. 1999. PDZ-GEF1, a guanine nucleotide exchange factor specific for Rap1 and Rap2. J. Biol. Chem. 274**:**38125-38130. [DOI] [PubMed] [Google Scholar]
- 10.de Rooij, J., F. J. Zwartkruis, M. H. Verheijen, R. H. Cool, S. M. Nijman, A. Wittinghofer, and J. L. Bos. 1998. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature 396**:**474-477. [DOI] [PubMed] [Google Scholar]
- 11.Deutsch, P. J., and Y. Sun. 1992. The 38-amino acid form of pituitary adenylate cyclase-activating polypeptide stimulates dual signaling cascades in PC12 cells and promotes neurite outgrowth. J. Biol. Chem. 267**:**5108-5113. [PubMed] [Google Scholar]
- 12.de Vries-Smits, A. M., L. van der Voorn, J. Downward, and J. L. Bos. 1995. Measurements of GTP/GDP exchange in permeabilized fibroblasts. Methods Enzymol. 255**:**156-161. [DOI] [PubMed] [Google Scholar]
- 13.Dodge-Kafka, K. L., and M. S. Kapiloff. 2006. The mAKAP signaling complex: integration of cAMP, calcium, and MAP kinase signaling pathways. Eur. J. Cell Biol. 85**:**593-602. [DOI] [PubMed] [Google Scholar]
- 14.Dodge-Kafka, K. L., J. Soughayer, G. C. Pare, J. J. Carlisle Michel, L. K. Langeberg, M. S. Kapiloff, and J. D. Scott. 2005. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature 437**:**574-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumaz, N., and R. Marais. 2005. Integrating signals between cAMP and the RAS/RAF/MEK/ERK signalling pathways. Based on the anniversary prize of the Gesellschaft fur Biochemie und Molekularbiologie Lecture delivered on 5 July 2003 at the Special FEBS Meeting in Brussels. FEBS J. 272**:**3491-3504. [DOI] [PubMed] [Google Scholar]
- 16.Elbashir, S. M., J. Harborth, K. Weber, and T. Tuschl. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26**:**199-213. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, T. O., J. F. Rehfeld, and F. C. Nielsen. 2000. Cyclic AMP-induced neuronal differentiation via activation of p38 mitogen-activated protein kinase. J. Neurochem. 75**:**1870-1877. [DOI] [PubMed] [Google Scholar]
- 18.Harrison, S. M., J. L. Rudolph, M. L. Spencer, P. D. Wes, C. Montell, D. A. Andres, and D. A. Harrison. 2005. Activated RIC, a small GTPase, genetically interacts with the Ras pathway and calmodulin during Drosophila development. Dev. Dyn. 232**:**817-826. [DOI] [PubMed] [Google Scholar]
- 19.Huang, E. J., and L. F. Reichardt. 2003. Trk receptors: roles in neuronal signal transduction. Annu. Rev. Biochem. 72**:**609-642. [DOI] [PubMed] [Google Scholar]
- 20.Hynds, D. L., M. L. Spencer, D. A. Andres, and D. M. Snow. 2003. Rit promotes MEK-independent neurite branching in human neuroblastoma cells. J. Cell Sci. 116**:**1925-1935. [DOI] [PubMed] [Google Scholar]
- 21.Ishii, T., E. Satoh, and M. Nishimura. 2001. Integrin-linked kinase controls neurite outgrowth in N1E-115 neuroblastoma cells. J. Biol. Chem. 276**:**42994-43003. [DOI] [PubMed] [Google Scholar]
- 22.Iwasaki, S., M. Iguchi, K. Watanabe, R. Hoshino, M. Tsujimoto, and M. Kohno. 1999. Specific activation of the p38 mitogen-activated protein kinase signaling pathway and induction of neurite outgrowth in PC12 cells by bone morphogenetic protein-2. J. Biol. Chem. 274**:**26503-26510. [DOI] [PubMed] [Google Scholar]
- 23.Kandel, E. R. 2001. The molecular biology of memory storage: a dialogue between genes and synapses. Science 294**:**1030-1038. [DOI] [PubMed] [Google Scholar]
- 24.Kiermayer, S., R. M. Biondi, J. Imig, G. Plotz, J. Haupenthal, S. Zeuzem, and A. Piiper. 2005. Epac activation converts cAMP from a proliferative into a differentiation signal in PC12 cells. Mol. Biol. Cell 16**:**5639-5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolch, W. 2005. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 6**:**827-837. [DOI] [PubMed] [Google Scholar]
- 26.Kyriakis, J. M., and J. Avruch. 2001. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81**:**807-869. [DOI] [PubMed] [Google Scholar]
- 27.Lazarovici, P., H. Jiang, and D. Fink, Jr. 1998. The 38-amino-acid form of pituitary adenylate cyclase-activating polypeptide induces neurite outgrowth in PC12 cells that is dependent on protein kinase C and extracellular signal-regulated kinase but not on protein kinase A, nerve growth factor receptor tyrosine kinase, p21(ras) G protein, and pp60(c-src) cytoplasmic tyrosine kinase. Mol. Pharmacol. 54**:**547-558. [DOI] [PubMed] [Google Scholar]
- 28.Lee, C. H. J., N. G. Della, C. E. Chew, and D. J. Zack. 1996. Rin, a neuron-specific and calmodulin-binding small G-protein, and Rit define a novel subfamily of Ras proteins. J. Neurosci. 16**:**6784-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, S. L., N. N. Johnson-Farley, D. R. Lubinsky, and D. S. Cowen. 2003. Coupling of neuronal 5-HT7 receptors to activation of extracellular-regulated kinase through a protein kinase A-independent pathway that can utilize Epac. J. Neurochem. 87**:**1076-1085. [DOI] [PubMed] [Google Scholar]
- 30.Lonze, B. E., and D. D. Ginty. 2002. Function and regulation of CREB family transcription factors in the nervous system. Neuron 35**:**605-623. [DOI] [PubMed] [Google Scholar]
- 31.Maillet, M., S. J. Robert, M. Cacquevel, M. Gastineau, D. Vivien, J. Bertoglio, J. L. Zugaza, R. Fischmeister, and F. Lezoualc'h. 2003. Crosstalk between Rap1 and Rac regulates secretion of sAPPalpha. Nat. Cell Biol. 5**:**633-639. [DOI] [PubMed] [Google Scholar]
- 32.Marinissen, M. J., and J. S. Gutkind. 2001. G-protein-coupled receptors and signaling networks: emerging paradigms. Trends Pharmacol. Sci. 22**:**368-376. [DOI] [PubMed] [Google Scholar]
- 33.Martin, B., R. L. de Maturana, R. Brenneman, T. Walent, M. P. Mattson, and S. Maudsley. 2005. Class II G protein-coupled receptors and their ligands in neuronal function and protection. Neuromol. Med. 7**:**3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrison, D. K., and R. J. Davis. 2003. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 19**:**91-118. [DOI] [PubMed] [Google Scholar]
- 35.Rehmann, H. 2005. Characterization of the activation of the Rap-specific exchange factor epac by cyclic nucleotides. Methods Enzymol. 407**:**159-173. [DOI] [PubMed] [Google Scholar]
- 36.Rehmann, H., J. Das, P. Knipscheer, A. Wittinghofer, and J. L. Bos. 2006. Structure of the cyclic-AMP-responsive exchange factor Epac2 in its auto-inhibited state. Nature 439**:**625-628. [DOI] [PubMed] [Google Scholar]
- 37.Rosario, M., H. F. Paterson, and C. J. Marshall. 2001. Activation of the Ral and phosphatidylinositol 3′ kinase signaling pathways by the Ras-related protein TC21. Mol. Cell. Biol. 21**:**3750-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubinfeld, B., S. Munemitsu, R. Clark, L. Conroy, K. Watt, W. J. Crosier, F. McCormick, and P. Polakis. 1991. Molecular cloning of a GTPase activating protein specific for the Krev-1 protein p21rap1. Cell 65**:**1033-1042. [DOI] [PubMed] [Google Scholar]
- 39.Rusyn, E. V., E. R. Reynolds, H. Shao, T. M. Grana, T. O. Chan, D. A. Andres, and A. D. Cox. 2000. Rit, a non-lipid-modified Ras-related protein, transforms NIH 3T3 cells without activating the ERK, JNK, p38 MAPK or PI3K/Akt pathways. Oncogene 19**:**4685-4694. [DOI] [PubMed] [Google Scholar]
- 40.Sakabe, K., H. Teramoto, M. Zohar, B. Behbahani, H. Miyazaki, H. Chikumi, and J. S. Gutkind. 2002. Potent transforming activity of the small GTP-binding protein Rit in NIH 3T3 cells: evidence for a role of a p38gamma-dependent signaling pathway. FEBS Lett. 511**:**15-20. [DOI] [PubMed] [Google Scholar]
- 41.Sakai, Y., H. Hashimoto, N. Shintani, S. Tomimoto, K. Tanaka, A. Ichibori, M. Hirose, and A. Baba. 2001. Involvement of p38 MAP kinase pathway in the synergistic activation of PACAP mRNA expression by NGF and PACAP in PC12h cells. Biochem. Biophys. Res. Commun. 285**:**656-661. [DOI] [PubMed] [Google Scholar]
- 42.Seino, S., and T. Shibasaki. 2005. PKA-dependent and PKA-independent pathways for cAMP-regulated exocytosis. Physiol. Rev. 85**:**1303-1342. [DOI] [PubMed] [Google Scholar]
- 43.Shao, H., and D. A. Andres. 2000. A novel RalGEF-like protein, RGL3, as a candidate effector for Rit and Ras. J. Biol. Chem. 275**:**26914-26924. [DOI] [PubMed] [Google Scholar]
- 44.Shao, H., K. Kadono-Okuda, B. S. Finlin, and D. A. Andres. 1999. Biochemical characterization of the Ras-related GTPases Rit and Rin. Arch. Biochem. Biophys. 371**:**207-219. [DOI] [PubMed] [Google Scholar]
- 45.Shi, G. X., and D. A. Andres. 2005. Rit contributes to nerve growth factor-induced neuronal differentiation via activation of b-Raf-extracellular signal-regulated kinase and p38 mitogen-activated protein kinase cascades. Mol. Cell. Biol. 25**:**830-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi, G. X., J. Han, and D. A. Andres. 2005. Rin GTPase couples nerve growth factor signaling to p38 and b-Raf/ERK pathways to promote neuronal differentiation. J. Biol. Chem. 280**:**37599-37609. [DOI] [PubMed] [Google Scholar]
- 47.Somogyvari-Vigh, A., and D. Reglodi. 2004. Pituitary adenylate cyclase activating polypeptide: a potential neuroprotective peptide. Curr. Pharm. Des. 10**:**2861-2889. [DOI] [PubMed] [Google Scholar]
- 48.Spencer, M. L., H. Shao, and D. A. Andres. 2002. Induction of neurite extension and survival in pheochromocytoma cells by the Rit GTPase. J. Biol. Chem. 277**:**20160-20168. [DOI] [PubMed] [Google Scholar]
- 49.Stork, P. J., and J. M. Schmitt. 2002. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 12**:**258-266. [DOI] [PubMed] [Google Scholar]
- 50.Takeda, K., and H. Ichijo. 2002. Neuronal p38 MAPK signalling: an emerging regulator of cell fate and function in the nervous system. Genes Cells 7**:**1099-1111. [DOI] [PubMed] [Google Scholar]
- 51.Tan, Y., J. Rouse, A. Zhang, S. Cariati, P. Cohen, and M. J. Comb. 1996. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 15**:**4629-4642. [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas, G. M., and R. L. Huganir. 2004. MAPK cascade signalling and synaptic plasticity. Nat. Rev. Neurosci. 5**:**173-183. [DOI] [PubMed] [Google Scholar]
- 53.Vaudry, D., B. J. Gonzalez, M. Basille, L. Yon, A. Fournier, and H. Vaudry. 2000. Pituitary adenylate cyclase-activating polypeptide and its receptors: from structure to functions. Pharmacol. Rev. 52**:**269-324. [PubMed] [Google Scholar]
- 54.Waschek, J. A. 2002. Multiple actions of pituitary adenylyl cyclase activating peptide in nervous system development and regeneration. Dev. Neurosci. 24**:**14-23. [DOI] [PubMed] [Google Scholar]
- 55.Wiggin, G. R., A. Soloaga, J. M. Foster, V. Murray-Tait, P. Cohen, and J. S. Arthur. 2002. MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol. Cell. Biol. 22**:**2871-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]