hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37 (original) (raw)
Abstract
The 26S proteasome catalyzes the degradation of most proteins in mammalian cells. To better define its composition and associated regulatory proteins, we developed affinity methods to rapidly purify 26S proteasomes from mammalian cells. By this approach, we discovered a novel 46-kDa (407 residues) subunit of its 19S regulatory complex (previously termed ADRM1 or GP110). As its N-terminal half can be incorporated into the 26S proteasome and is homologous to Rpn13, a 156-residue subunit of the 19S complex in budding yeast, we renamed it human Rpn13 (hRpn13). The C-terminal half of hRpn13 binds directly to the proteasome-associated deubiquitinating enzyme, UCH37, and enhances its isopeptidase activity. Knockdown of hRpn13 in 293T cells increases the cellular levels of ubiquitin conjugates and decreases the degradation of short-lived proteins. Surprisingly, an overproduction of hRpn13 also reduced their degradation. Furthermore, transfection of the C-terminal half of hRpn13 slows proteolysis and induces cell death, probably by acting as a dominant-negative form. Thus in human 26S proteasomes, hRpn13 appears to be important for the binding of UCH37 to the 19S complex and for efficient proteolysis.
Keywords: deubiquitination, hRpn13, proteasome, ubiquitin, UCH37
Introduction
Degradation of proteins by the ubiquitin-proteasome pathway is critical in regulating the levels of most cellular proteins and in the rapid elimination of misfolded proteins (Hershko and Ciechanover, 1998; Glickman and Ciechanover, 2002; Goldberg, 2003). The conjugation of a polyubiquitin chain to a protein substrate leads to its rapid binding and ATP-dependent hydrolysis by the 26S proteasome. This 2.4-MDa particle consists of two subcomplexes, the 20S core particle, within which proteolysis takes place, and the 19S regulatory particle, which binds the ubiquitinated substrate (Voges et al, 1999). The 20S proteasome is composed of two outer α-rings and two inner β-rings, each containing seven different, but homologous, subunits. The 19S complex, which binds to one or both ends of the 20S particle, is composed of a base and a lid (Glickman et al, 1998; Leggett et al, 2002). The base contains a ring of six homologous ATPases, which mediate the unfolding and translocation of substrates into the 20S (Smith et al, 2005). The lid contains at least eight subunits, including multiple isopeptidases that catalyze the rapid disassembly of the ubiquitin (Ub) chain (Glickman and Ciechanover, 2002).
Our current knowledge about the composition of the mammalian 26S proteasome has emerged primarily from studies in which this particle was purified by multiple chromatographic steps (Voges et al, 1999). These studies have led to the identification of a set of subunits, whose presence resists relatively stringent purification steps (e.g., high salt concentrations). Recent studies using affinity purification methods in yeast indicate that a number of additional proteins are loosely or transiently bound to these particles and can be dissociated by high salt concentration or upon ATP hydrolysis (Verma et al, 2000; Leggett et al, 2002; Sone et al, 2004; Leggett et al, 2005; Guerrero et al, 2006). One aim of the present study was to develop similar gentle techniques for rapid isolation of 26S proteasomes from mammalian tissues. Such an approach could help identify critical cofactors and regulators of protein degradation, or even novel subunits.
Although proteasome structure and properties are highly conserved in eukaryotes (Smith et al, 2005), a number of differences exist between proteasomes of higher and lower eukaryotes, and several specializations in mammalian proteasomes have been identified, such as the immunoproteasomes that are important in antigen presentation (Rock et al, 2002). Also, the 26S particles in lower eukaryotes seem to contain unique subunits, such as Daq1/Rpn13, a 156 amino-acid subunit of the 19S in budding yeast that appears to be important in the degradation of certain model substrates (Verma et al, 2000). No similar subunit has been reported in mammalian proteasomes, although interestingly, Rpn13 is highly homologous to the N-terminal half of Xoom, a 404 amino-acid protein required for the embryonic development of frogs (Hasegawa et al, 2001).
In the present study, we describe an affinity method for purifying mammalian 26S proteasomes, which is based upon the approach of Leggett et al (2005) for isolating yeast proteasomes. Using this approach, we have discovered a novel human 19S subunit, a 407-residue protein that corresponds to gene products previously termed ADRM1 or ARM1 for ‘adhesion regulating molecule 1' (Simins et al, 1999) or GP110 (Shimada et al, 1991) for a putative surface glycoprotein. However, as we show, this protein is primarily a subunit of the 26S proteasome, and as this paper was submitted, three other groups (Hamazaki et al, 2006; Jorgensen et al, 2006; Yao et al, 2006) have also demonstrated, by other approaches, its presence within the 26S proteasome. As its N-terminal region shares 28% identity with yeast Daq1/Rpn13, we renamed it human Rpn13 (hRpn13). We also show here that it associates with the deubiquitinating enzyme UCH37 and anchors this enzyme to the proteasome. Disassembly of the polyubiquitin chain is an essential step in protein degradation by the 26S particle (Glickman and Ciechanover, 2002). Among the 60 putative isopeptidases in the human genome, three are associated with the 19S complex, POH1/Rpn11, UCH2/UCH37, and Ubp6/Usp14 (Holzl et al, 2000; Leggett et al, 2002; Verma et al, 2002). Interestingly, the isopeptidase UCH37 that binds to hRpn13 is not found in budding yeast (Guterman and Glickman, 2004). When the C-terminal half of hRpn13 that binds to UCH37 is expressed, it can reduce proteolysis and induce cell death, probably by acting as a dominant-negative form of hRpn13. Thus, this novel subunit and its interaction with UCH37 appear to play an important role in proteasomal function.
Results
Affinity purification of human proteasomes
In yeast, attachment of a FLAG-tag to the β4 subunit of the 20S particle or a protein A domain to the RPN11 subunit in the 19S particle has been employed successfully for the affinity purification of 26S proteasomes (Verma et al, 2000; Leggett et al, 2002). To rapidly purify mammalian proteasomes, we made three different constructs encoding the C-terminally tagged mammalian β4 or RPN11, that is, mRPN11-protein A, mβ4-protein A, and mβ4-FLAG. A Tev protease cleavage site was inserted between protein A and mRPN11 (or mβ4). Following transient transfection of human embryonic kidney 293T cells with these constructs, tagged 26S proteasomes were immunoprecipitated with either an IgG or an anti-FLAG antibody (Figure 1A). In extracts of the cells transfected with mRPN11-protein A, the IgG pulled down the 20S subunit, α2, as shown by Western blot, suggesting that the 26S proteasomes were immunoprecipitated. When the cells were transfected with either mβ4-protein A or mβ4-FLAG, α2 was also immunoprecipitated. In order to determine whether FLAG-tagged mβ4 allows the isolation of the 26S proteasome, the cells were cotransfected with mβ4-FLAG and mRPN11-protein A. As expected, mRPN11-protein A was co-immunoprecipitated with the anti-FLAG antibody. These results suggest that all three constructs can be used for the affinity purification of mammalian 26S particles.
Figure 1.
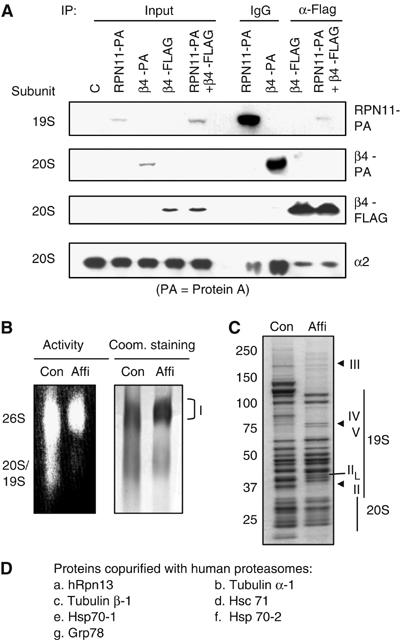
Affinity purification of mammalian proteasomes. (A) Immunoprecipitation of the affinity-tagged human proteasomes. 293T cells were transfected with mRPN11-protein-A, mβ4-protein A, mβ4-FLAG, or both mRPN11-protein-A and mβ4-FLAG. Following immunoprecipitation using an IgG or an anti-FLAG antibody, the subunits of both the 19S and 20S particles were detected by Western blot. Input represents 1% of the cell lysates for immunoprecipitation. (B) In-gel peptidase activity of the affinity purified proteasomes. Both the conventionally-purified (Con) and the affinity-purified (Affi) proteasomes were separated by native PAGE. Left, the peptidase activities of the proteasomes were assayed in the gel using suc-LLVY-amc as a substrate. Right, proteasomes were stained with Coomassie blue. (C) Analysis of affinity-purified proteasomes by SDS–PAGE. Both the conventionally purified (Con) and the affinity-purified (Affi) proteasomes were separated by SDS–PAGE and stained with Coomassie blue. (D) Mass spectrometric analysis of the proteins copurified with mammalian proteasomes. Protein samples for mass spectrometry were analyzed by MALDI-TOF using an Applied Biosystems Voyager DE-STR.
To further characterize the affinity-purified proteasomes, we transfected 293T cells with mRPN11-protein A, isolated the affinity-tagged proteasomes using IgG beads, and released the proteasomes from the beads by incubating them with the Tev protease. Then, we analyzed the peptidase activity of the affinity-purified proteasomes following native PAGE. As in the case of the proteasomes purified by conventional procedures, the affinity-purified proteasomes were able to cleave the standard fluorogenic substrate of the 20S's chymotrypsin-like activity, suc-LLVY-7-amino-4-methylcoumarin (amc) (Figure 1B, left). As shown by Coomassie blue staining, the affinity-purified proteasomes were predominantly in the form of 26S particles (band I, Figure 1B). Upon SDS–PAGE, the affinity-purified proteasomes displayed almost all the known subunit bands found in 19S and 20S, purified by conventional methods (Figure 1C). One clear difference was that the band II migrated more slowly, probably because of the insertion of the Tev cleavage site (15 amino acids) between mRPN11 and protein A. It is noteworthy that there were two unexpected lightly stained bands at about 200 kDa (band III) and 75 kDa (band IV) in both preparations and a novel band (band V) in the affinity-purified proteasomes. These results suggested that certain unidentified proteins were copurified with the affinity-purified proteasomes.
Mass spectrometric analysis of affinity-purified proteasomes
To identify these additional proteins, we excised the affinity-purified 26S proteasomes from the native gel (band I, Figure 1B) and performed mass spectrometric (MALD1-TOF) analysis. All the known proteasome subunits (including UCH37) were identified in the sample (Supplementary Table I) (as shown by repeated identification of tryptic peptides). In addition, two microtubule components, tubulin α-1 and tubulin β-1, were detected in these samples (Figure 1D and Supplementary Table II), as well as a protein of 407 amino acids of unknown function (Figure 1D and Supplementary Table II), which we named hRpn13 as discussed below. None of the additional proteins corresponded in molecular weight to the bands III–V in Figure 1C. Therefore, we analyzed these bands individually by mass spectrometry and found that band III represented the recently discovered proteasome activator, PA200 (Leggett et al, 2002; Ustrell et al, 2002), and bands IV and V presented a mixture of four heat-shock proteins, Hsc71, Hsp70-1, Hsp70-2, and Grp78 (Figure 1D and Supplementary Table II), all of which tend to bind to unfolded proteins and may be nonspecific contaminants, although an Hsp70 family member has been identified as a component interacting with the yeast 26S particles (Guerrero et al, 2006).
The N-terminal half of hRpn13 is conserved in eukaryotes
The novel proteasome-associated protein, which we termed hRpn13, contains a Ser/Thr-rich region and surprisingly, a putative transmembrane domain in its C-terminal half. This polypeptide was first reported to be an interferon-_γ_-inducible 110-kDa cell surface glycoprotein, GP110 (Shimada et al, 1991, 1994), and was also proposed to be an adhesion-related protein, ADRM1/ARM-1 (Simins et al, 1999). However, recent work has suggested that it is an intracellular protein (Lamerant and Kieda, 2005). This sequence is highly conserved in the multicellular organisms (Figure 2A). For example, its homolog in frogs, Xoom, is a 404-amino-acid protein with an 87% identity to hRpn13. A less conserved homolog is found in single-cell eukaryotes. Fission yeast contains a protein with 24% identity and homologies to both the N- and C-terminal halves of hRpn13, whereas budding yeast contains a homologous proteasomal subunit, Daq1/Rpn13, of only 156 amino acids. However, it is homologous only to the N-terminal half of hRpn13 (with 28% identity). Because of this homology to a known 19S subunit and the functional similarities shown below, we termed this polypeptide hRpn13.
Figure 2.
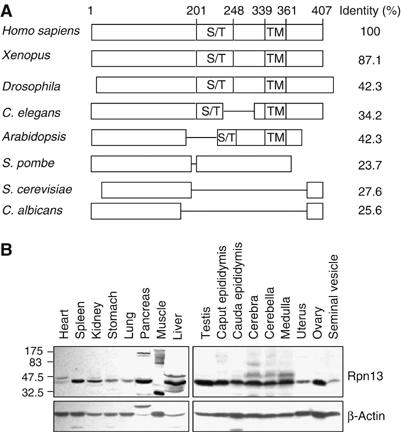
Rpn13 is evolutionally conserved in eukaryotes and is expressed in various mouse tissues. (A) hRpn13, especially its N-terminal half, is highly conserved through evolution. hRpn13 (GenBank accession no. NM 007002) and its homologs in several different organisms were aligned, and identities were calculated as indicated. A Ser/Thr-rich region (S/T) and a putative transmembrane (TM) domain in the C-terminal half are conserved in Rpn13 from multicellular organisms. (B) hRpn13 is expressed in various mouse tissues. Whole-tissue extracts were prepared and analyzed by Western blots.
To detect the protein, we generated a polyclonal antiserum against the recombinant protein expressed in insect cells, which on Western blot reacted with endogenous hRpn13 from 293T cells, as well as the transfected protein (Supplementary Figure 1A). Using this antiserum, we showed that hRpn13 is expressed in all mouse tissues examined (Figure 2B) and in a variety of mammalian cell lines, including ones derived from skeletal muscle (C2C12), kidney (293, 293T, and Cos-7), breast (MCF-7, MDA-MB-453, MDA-MB-468, and A431), adrenal (SW13), lymphocytes (Jurket), colon (HCT116), and embryo (NIH/3T3) (Supplementary Figure 1B). These findings confirm the earlier report that RNA transcripts of this sequence were present in all tissues of rats (Nakane et al, 2000). Interestingly, higher molecular weight species were detected in several mouse tissues, including heart, skeletal muscle, pancreas, liver, and brain, probably because of the post-translational modifications of Rpn13 (Figure 2B).
As per the initial report that the levels of the 110-kDa membrane protein corresponding to hRpn13 are elevated by treatment with interferon-γ (Shimada et al, 1994), and because interferon-γ induces alternative β-subunits of the 20S proteasome (Rock et al, 2002), we repeatedly treated different cell lines with this cytokine, and assayed for its induction. However, the levels of hRpn13 in HeLa cells did not change after treatment with interferon-γ at concentrations that did cause induction of the immunoproteasome subunit, Lmp7/β5i, in these cells (Supplementary Figure 1C). Also, no induction was observed when we used the same antibody as used in the prior study (Shimada et al, 1994), which was kindly provided by J Greiner. Further, that antibody failed to react with the overexpressed or purified hRpn13/GP110. These findings (and those shown below) argue strongly that this sequence does not correspond to an interferon-induced glycoprotein.
hRpn13 is a subunit of the 19S particle
To confirm the association of hRpn13 with the proteasome, lysates of 293T cells were ultracentrifuged through a 10–40% glycerol density gradient. The presence of proteasomes in each fraction was monitored using suc-LLVY-amc, a standard proteasome substrate (Kisselev et al, 2002). hRpn13, like S5a (a 19S subunit), appeared in two overlapping peaks centered at fractions 11 and 17, respectively (Figure 3A, left), but the peak of 26S activity was in fraction 17. The lower molecular weight shoulder on this peak, which contained Rpn13 and some S5a, probably represented free 19S particles. Thus, hRpn13 cosedimented with the 26S proteasome, but also appeared to be present within free 19S particle in these crude extracts. However, in lysates of C2C12 cells, almost all Rpn13 and S5a cosedimented with the 26S particles (Figure 3A, right).
Figure 3.
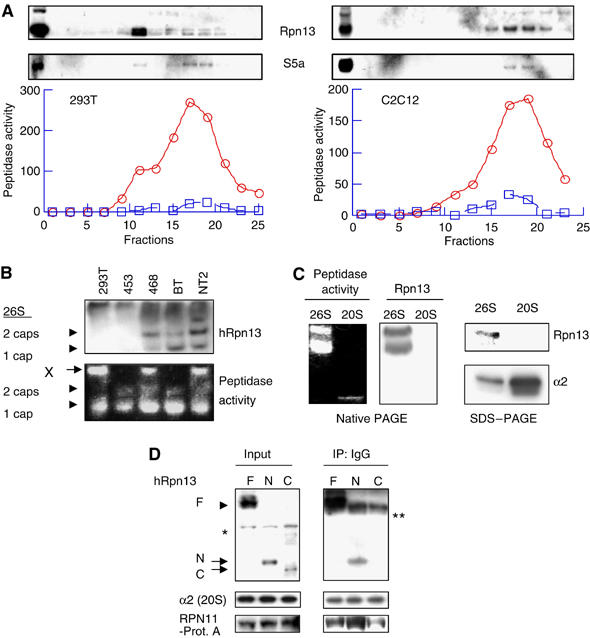
hRpn13 is a subunit of the 19S particle that associates with it through its N-terminal region. (A) Cosedimentation of hRpn13 with proteasomes during glycerol-gradient centrifugation. The lysates (3 mg proteins in 1 ml buffer) from 293T (left) or C2C12 (right) cells were loaded on a 10–40% of glycerol gradient, and centrifuged at 100 000 g (24 000 r.p.m., Beckman Rotor AH628) for 12 h. Peptidase activities from 26S (circle) and 20S (square) in each fraction were monitored by measuring the production of amc from the substrate suc-LLVY-amc in the absence and presence of 0.02% SDS, respectively. Distribution of S5a/Rpn10 (a 19S subunit) and hRpn13 was assayed in each fraction by Western blot. (B) hRpn13 associates with certain forms of proteasomes. Cell extracts (40 μg protein/well) were separated by native PAGE. In-gel peptidase activity was determined by incubating the gel with suc-LLVY-amc and then visualizing the active proteasomes under UV lights. (C) hRpn13 is present in the 19S particle. Left, the proteasomes purified by conventional methods were separated by native PAGE. In-gel peptidase activity was determined as in (B). Right, the proteasomes were separated by SDS–PAGE, and the α2 subunit of the 20S and hRpn13 were assayed by Western blot. (D) hRpn13 binds to proteasomes through its N-terminal region. 293T cells were cotransfected with mRPN11-protein-A and Myc/His6-tagged hRpn13 or its truncated forms. The cell lysates were immunoprecipitated using IgG beads. *stands for nonspecific bands ** heavy chain of IgG.
To further test whether hRpn13 is associated with proteasomes in different cell types, crude lysates from five different cell lines were subjected to native PAGE. The proteasomes' peptidase activities from the extracts of 293 T, MDA-MB-468 (human breast cancer), and NT2 (human teratocarcinoma) cells migrated together with hRpn13. However, surprisingly in these extracts, but not those from human breast cancer MDA-MB-453 and BT474 cells, the peptidase activity was present primarily in very large structures (larger than double-capped 26S proteasomes) that hardly entered the SDS gel (structure ‘X' in Figure 3B). hRpn13 was also found in such large structures from all five cell lines (Figure 3B). Presumably these very large structures represent 26S particles that in vivo are associated with other components and therefore fail to migrate into the gel under these lysis conditions. Although active single-capped 26S proteasomes were detected in the lysates from all five cell lines, hRpn13 was surprisingly only detectable in the single-capped 26S proteasomes from three cell lines (MDA-MB-468, BT474, and NT2 cells). These results suggest that hRpn13 associates only with certain forms of proteasomes or that in certain complexes its accessibility by antibodies (and therefore its detection) may be limited.
To learn if hRpn13 was present in standard preparations of the 26S particle, studied previously, we attempted to detect it in the 26S proteasomes purified from rabbit muscle by the conventional multistep chromatographic approaches. Following native PAGE, Rpn13 was detected in the 26S, but not in the 20S proteasome (Figure 3C, left). Similarly, after SDS–PAGE, Rpn13 was also only detected in the 26S proteasome (Figure 3C, right). Thus Rpn13 in mammals, like its yeast homolog, is a subunit of the 19S particle.
To determine which region of hRpn13 associates with the proteasome, we constructed the following three plasmids encoding full-length or truncated hRpn13 with C-terminal Myc/His6 double tags: the full-length protein, the N-terminal region (hRpn13-Δ201–407), and the C-terminal region (hRpn13-Δ1–200). Following cotransfection of 293T cells with the plasmid for mRPN11-protein A and each of these plasmids, proteasomes were immunoprecipitated from the cell lysates with IgG beads (Figure 3D). Both the full-length and the N-terminal half of hRpn13, but not its C-terminal half, were co-immunoprecipitated with mRPN11-protein A (Figure 3D). Thus hRpn13 binds to the 19S subcomplex through its N-terminal region.
The C-terminal half of hRpn13 associates directly with that of UCH37
In order to define the possible functions of hRpn13, we studied whether it associates with other proteins in the cell in addition to being a part of the 26S proteasome. We transfected 293T cells with Myc/His6-tagged hRpn13 and then carried out immunoprecipitation experiments using an anti-Myc antibody. Following SDS–PAGE of the immunoprecipitate, the Coomassie staining revealed about 11 bands (Figure 4A). As the distribution pattern of these bands was quite different from that of proteasomes, hRpn13 probably forms additional complexes following overexpression. Mass spectrometric analysis showed that the isopeptidase UCH37, tubulin α-1, and tubulin β-1 were among the hRpn13-associated proteins. As UCH37 was the only known proteasome component in this complex, we examined whether hRpn13 could directly bind to UCH37. We expressed His-tagged hRpn13 in insect cells and GST-UCH37 in Escherichia coli and purified the recombinant proteins. After they were incubated together for 1 h, the His6-hRpn13 could be pulled down together with GST-UCH37 (Figure 4B). In contrast, when RPN12 (another 19S subunit) was incubated with hRpn13 in place of UCH37, hRpn13 could not be pulled down with Rpn12.
Figure 4.
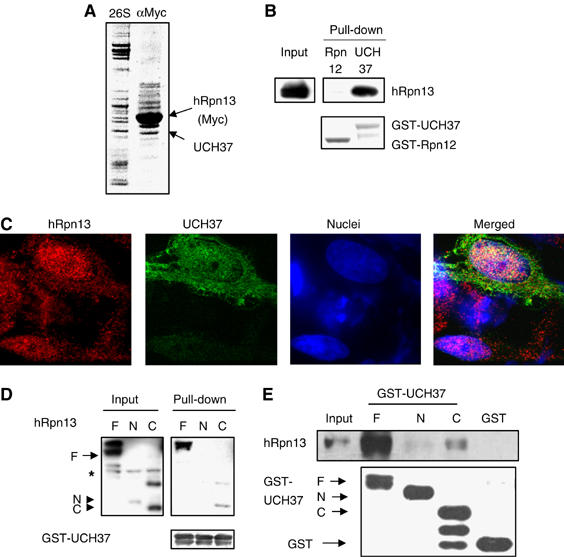
hRpn13 directly associates with UCH37. (A) hRpn13 co-immunoprecipitates with UCH37. 293T cells were transfected with hRpn13-Myc/His6, and incubated for 2 days. Following immunoprecipitation with an anti-Myc antibody, hRpn13-Myc and associated proteins were separated by SDS–PAGE, and stained with Coomassie blue. Conventionally purified 26S proteasomes were included in the gel as a reference. (B) hRpn13 directly binds to UCH37. Purified His-tagged hRpn13 and GST-fused UCH37 or Rpn12 were incubated for 1 h, and a pull-down assay was performed using GSH-beads. (C) UCH37 colocalizes with hRpn13. HeLa cells were transfected with Myc/His6-tagged UCH37, and the localization of the tagged UCH37 and endogenous hRpn13 was determined by immunostaining. (D) hRpn13 binds to UCH37 through its C-terminal region. 293T cells were either transfected with Myc-tagged hRpn13 (labeled as F) or with a similar construct encoding its N-terminal (N) or C-terminal half (C), and incubated for 2 days. Purified GST-UCH37 was incubated with the three cell lysates, and a pull-down assay was carried out using GSH-beads. (E) UCH37 binds to hRpn13 in vitro through its C-terminal half. Purified GST or GST-fused to the full-length (F), the N-terminal (N) or the C-terminal half (C) of UCH37 was incubated with purified recombinant His-tagged hRpn13. Pull-down assays using GSH-beads were then performed, and proteins were analyzed by Western blot.
hRpn13 possesses a putative transmembrane domain, and conflicting conclusions about its subcellular localization have been published (Shimada et al, 1994; Lamerant and Kieda, 2005). To determine the localization of hRpn13, we performed immunostaining of HeLa cells. As observed under a confocal microscope, hRpn13 was present in punctuated particles in both the nucleus and the cytosol, but its levels in the nucleus were higher than those in the cytosol (Figure 4C). Transiently transfected UCH37 partially colocalized with the endogenous hRpn13 in both the cytosol and the nucleus (Figure 4C). Thus, hRpn13 can directly and specifically associate with UCH37 in vivo and after purification in vitro.
To define further this association of hRpn13 and UCH37, we attempted to determine which parts of these proteins bind to each other. As noted previously, Rpn13 in Saccharomyces cerevisiae is homologous to the N-terminal half of hRpn13, which binds to the proteasome. Its C-terminal half, which contains both a Ser/Thr-rich region and a putative transmembrane motif, is not conserved in budding yeast, which also does not contain a homolog of UCH37. To test the possibility that the C-terminal half of hRpn13 binds to UCH37, we incubated GST-UCH37 with the lysates from cells that were transfected with either the full-length, N-terminal half (Δ201–407) or C-terminal half (Δ1–200) of hRpn13. The GSH-beads could pull down the GST-UCH37 together with the full-length and the C-terminal half of hRpn13, but not its N-terminal half (Figure 4D). Thus, hRpn13 binds to UCH37 through its C-terminal half and probably thereby anchors UCH37 to the proteasome.
The N-terminal half of UCH37 harbors its ubiquitin (Ub) hydrolase domain, but the role of its C-terminal half still remains unknown. To examine whether the C-terminal half of UCH37 may bind to hRpn13, we constructed and expressed in E. coli two truncated GST-fusion proteins: GST-UCH37-N-terminal half (Δ227–329) and GST-UCH37-C-terminal half (Δ1–226). When purified, His6-hRpn13 was incubated with either truncated or the full-length GST-UCH37; both the full-length and the C-terminal half, but not the N-terminal half of UCH37, could pull down hRpn13 (Figure 4E). Thus UCH37, through its C-terminal half, binds to the C-terminal half of hRpn13.
hRpn13 increases the isopeptidase activity of UCH37
As shown above, UCH37 binds to a site in the C-terminal region of hRpn13, although UCH37 may also bind to other proteasome subunits. In fact, the homolog of UCH37 in fission yeast, UCH2, can bind to Rpn10/S5a, another 19S subunit (Stone et al, 2004). To determine whether UCH37 also binds to S5a in mammalian proteasomes, purified GST-UCH37 was incubated with purified His-tagged S5a. As shown in Figure 5A, S5a could be pulled down by GST-UCH37, but not GST, suggesting that UCH37 binds to S5a. Thus, there are two, probably complementary, mechanisms by which UCH37 can be recruited to the proteasome.
Figure 5.
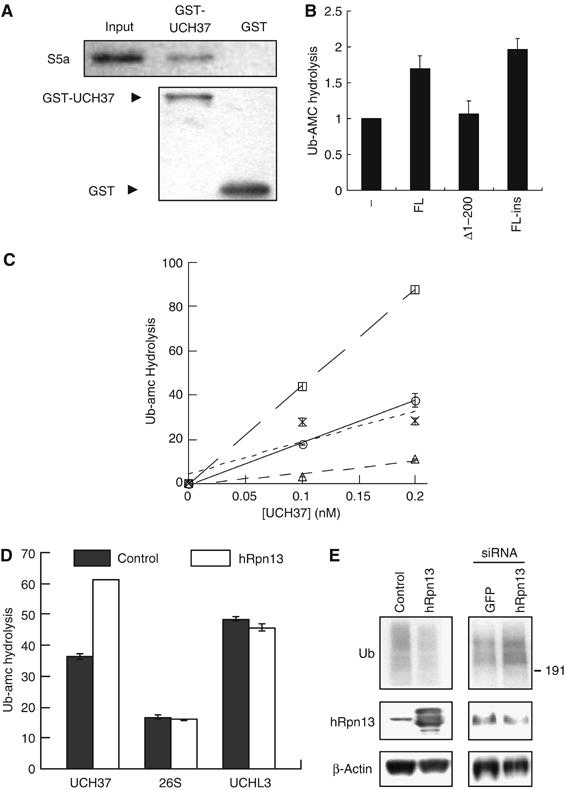
hRpn13 promotes isopeptidase activity of UCH37. (A) UCH37 binds to S5a in vitro. Purified GST or GST-fused UCH37 was incubated with purified recombinant His-tagged S5a for 1 h. Pull-down assays using GSH beads were then performed. The protein levels were analyzed by Western blot. (B) Recombinant hRpn13 stimulates the ubiquitin-amc (Ub-amc) hydrolysis by UCH37. In the presence of 0.5 μM of Ub-amc, 0.1 nM of UCH37 was incubated with 0 or 10 nM of the full-length hRpn13 (FL, from bacteria), the C-terminal half of hRpn13 (Δ1–200, from bacteria), or the full-length hRpn13 expressed in insect cells (FL-ins). Ub-amc hydrolysis (in arbitrary units) was determined during incubation for 30 min by monitoring the release of amc. (C) Unlike hRpn13, pure S5a/Rpn10 cannot promote the isopeptidase activity of UCH37 in vitro. UCH37 at indicated concentrations, was incubated with 0 (circle) or 10 nM of hRpn13 (square, expressed in insect cells), S5a (cross), or hRpn13 plus 10 nM of Ub-aldehyde (triangle). Ub-amc hydrolysis (in arbitrary units) was determined as in (B). (D) hRpn13 does not increase the isopeptidase activity of the 26S proteasome or UCHL3. Ub-amc was incubated (as in 5B) without (control) or with 10 nM of purified hRpn13 in the presence of UCH37 (0.1 nM), the 26S proteasome (0.24 μg/ml, about 0.1 nM), or UCHL3 (0.01 nM). Ub-amc hydrolysis (in arbitrary units) was determined as in (B). (E) hRpn13 decreases the levels of ubiquitin conjugates in cells. Left, 293T cells were transfected with an empty vector or Myc/His6-tagged hRpn13. Right, 293T cells were transfected with pBS/U6/GFP (GFP siRNA) or pBS/U6/hRpn13 (hRpn13 siRNA). The cells were incubated for 2 days after transfection, and the contents of hRpn13, β-actin, and large ubiquitin conjugates (molecular weights >191 kDa) were assayed by Western blot.
To determine whether this association with hRpn13 or S5a may influence the activity of UCH37, we first expressed recombinant His-tagged hRpn13 in bacteria. When the purified full-length hRpn13 was incubated with purified UCH37, the hydrolysis of Ub-amc (a model fluorogenic substrate for isopeptidases) by UCH37 increased by 69% (Figure 5B). In contrast, under similar conditions, the C-terminal half of hRpn13 had no effect on the activity of UCH37. To confirm this result, we expressed His-tagged hRpn13 in insect cells and found that hRpn13 from insect cells could also significantly promote the activity of UCH37 (Figure 5B and C). The detected Ub-amc hydrolysis was due to the isopeptidase activity of UCH37, as the inhibitor of isopeptidases, Ub-aldehyde, greatly reduced this activity (Figure 5C). Also in similar experiments, hRpn13 was not found to affect the activity of a typical deubiquitinating enzyme, UCHL3 (Figure 5D). On the other hand, although S5a also bound to UCH37, it did not increase its capacity to digest Ub-amc (Figure 5C). Also, when hRpn13 was incubated with 26S proteasomes, no change in their isopeptidase activity was observed, presumably because proteasomes already contain sufficient endogenous Rpn13 to stimulate Ub-amc hydrolysis by UCH37 maximally (Figure 5D). Thus, the novel 19S subunit hRpn13, perhaps together with Rpn10/S5a, appears to recruit UCH37 to the proteasome by binding to its C-terminal domain and stimulates its deubiquitinating activity.
hRpn13 influences the degradation of short-lived proteins in cells
To examine the role of hRpn13 in cells, we transfected 293T cells with Myc-tagged hRpn13 and found that overexpression of hRpn13 decreased the cell content of Ub conjugates (Figure 5E). To examine the effects of reducing hRpn13 expression, a RNA interference (RNAi) vector was prepared. It reduced, though only partially, the endogenous content of hRpn13 but nevertheless increased the total levels of Ub conjugates in the cells. No such effect was seen with a control vector expressing siRNA for GFP. Thus, hRpn13 can regulate the cellular levels of high molecular weight Ub conjugates, presumably by promoting protein deubiquitination.
Changes in the overall content of ubiquitinated proteins would be expected to alter rates of protein degradation by the proteasome. To determine whether rates of protein degradation are altered, we transfected 293T cells with hRpn13 or its C-terminal half (hRpn13-Δ1–200), and then analyzed the degradation of proteins in these cells by a pulse-chase protocol. As shown in Figure 6A, overexpression of hRpn13 decreased the degradation of the most short-lived proteins (those hydrolyzed in 20 min), but subsequently degradative rates were similar to those in control cells. These effects are consistent with the reduction in the content of Ub conjugates. Interestingly, transfection of hRpn13-Δ1–200, which can bind to UCH37, but not the proteasome, also reduced degradation of these short-lived cell components (Figure 6A). This reduction in proteolysis by the C-terminal half of hRpn13 is most likely because it acts as a dominant-negative form of hRpn13 that reduces the binding of UCH37 to the proteasome (also see below).
Figure 6.
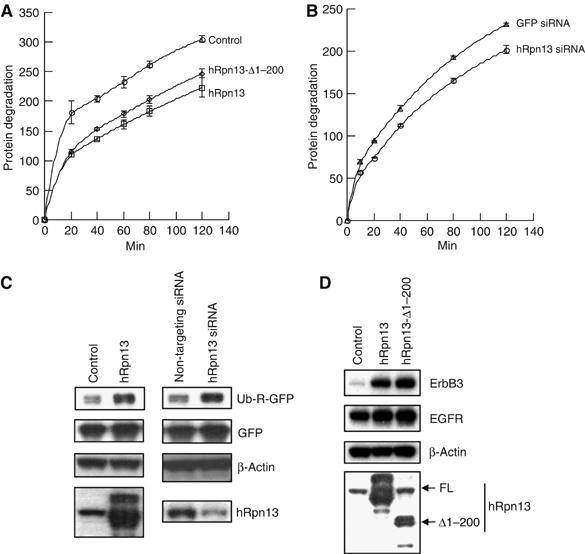
hRpn13 content influences rates of degradation of short-lived proteins in 293T cells. (A) Overexpression of hRpn13 or its C-terminal region reduces the degradation of short-lived proteins. 293T cells were transfected with an empty vector, hRpn13 or its C-terminal region (hRpn13-Δ1–200). After 2 days, the rate of degradation of cellular proteins was determined by pulse-chase analysis using [3H]tyrosine. A similar small reduction in degradation was seen in at least three experiments. (B) Decreasing the levels of hRpn13 also slows the degradation of short-lived proteins. 293T cells were transfected with pBS/U6/GFP (GFP siRNA) or pBS/U6/hRpn13 (hRpn13 siRNA). Then, degradation of short-lived proteins was determined as in (A). (C) Overexpression or knockdown of hRpn13 reduces the degradation of the model N-end rule substrate, Ub-R-GFP. Left, 293T cells were cotransfected with Ub-R-GFP, GFP, and hRpn13 (or empty vector). Right, 293T cells were cotransfected with Ub-R-GFP, GFP, and siRNA oligos (100 nM) for hRpn13 (or nontargeting siRNA oligos). The cells were incubated for 3 days after transfection, and the protein levels were analyzed by Western blot. (D) Degradation of the natural proteasomal substrate, ErbB3, is reduced by overexpression of hRpn13 or its C-terminal half. 293T cells were cotransfected with ErbB3, EGFR, and hRpn13 (or hRpn13-Δ1–200 or empty vector), and incubated for 2 days after transfection. Protein levels were analyzed by Western blot.
In an attempt to further clarify the role of hRpn13, we employed RNAi to knockdown hRpn13 levels. Transfection of the plasmids expressing siRNA for hRpn13 in 293T cells slightly (but consistently) reduced the degradation of short-lived proteins (Figure 6B). No such effect was seen with a control vector expressing siRNA against GFP. This result and those shown below are consistent with a defect in the proteasome's capacity to degrade Ub conjugates (Figure 5E), but is surprising as it suggests that both increasing and decreasing the levels of hRpn13 reduce proteolysis.
To further confirm the above results, we employed a model proteasome substrate, Ub-R-GFP. Ub-R-GFP is rapidly degraded through the N-end rule pathway in which proteins are selectively ubiquitinated and then degraded by the proteasome, depending on the identity of their N-terminal residuals (Varshavsky, 1997; Dantuma et al, 2000). As shown in Figure 6C, overexpression of hRpn13 had little effect on the levels of control protein GFP, but markedly increased the levels of Ub-R-GFP, apparently by inhibiting the proteasomal degradation. Similarly, transfection of the siRNA oligos for hRpn13 also greatly enhanced the levels of Ub-R-GFP, but not those of GFP (Figure 6C).
To further verify these results, we used another proteasomal substrate, ErbB3 (a member of the epidermal growth factor receptor family), which is known to be rapidly degraded by the proteasome after its Nrdp1-mediated ubiquitination (Diamonti et al, 2002; Qiu and Goldberg, 2002). Transfection of either hRpn13 or its dominant-negative form (the C-terminal half of hRpn13) dramatically increased the levels of cotransfected ErbB3, but not EGFR/ErbB1 (Figure 6D), a known substrate of lysosomes (Waterman et al, 1999). Taken together, these results argue that the levels of hRpn13 have to be precisely regulated in vivo to maintain the rate of protein degradation by the proteasome.
Expression of the C-terminal half of hRpn13 induces cell death
Many proteasome subunits are essential for cell viability, but most isopeptidases are disposable for cells (Guterman and Glickman, 2004). The homolog of UCH37 in Drosophila, dUCH37, does not seem to be required for proteasome activity in S2 cells (Wojcik and DeMartino, 2002), but the homolog of hRpn13 in frogs, Xoom, is required for embryonic development (Hasegawa et al, 2001). To determine the importance of hRpn13 in survival of mammalian cells, HeLa cells were transiently transfected with the Myc/His6-tagged full-length, the N-terminal half, or the C-terminal half of hRpn13 and were then analyzed by immunostaining (Figure 7A). Unlike endogenous hRpn13, transiently transfected hRpn13 and its fragments were primarily localized in the cytosol. The full-length protein, which was widely distributed in the cytosol, seemed to cause slight aggregation of mitochondria near the nuclei. In contrast, its N-terminal half formed punctuated perinuclear patterns, but had no clear effect on cell morphology, including the shapes of mitochondria and nuclei. However, the expression of the C-terminal half of hRpn13 led to the condensation of nuclei and a marked aggregation of mitochondria, as were also observed during apoptosis induced by TNFα and cycloheximide. Thus, at high levels, the C-terminal half of hRpn13 appears to induce cell death. In order to clarify the mechanism of this effect, we asked whether the C-terminal half of hRpn13 (which itself does not bind to the proteasome) might interact with full-length hRpn13. GST pull-down assays demonstrated that unlike UCH37, hRpn13 did not bind to its C-terminal half in vitro (Figure 7B). Thus, the C-terminal half of hRpn13 induces cell death, probably by preventing UCH37, but not hRpn13, from binding to the proteasome.
Figure 7.
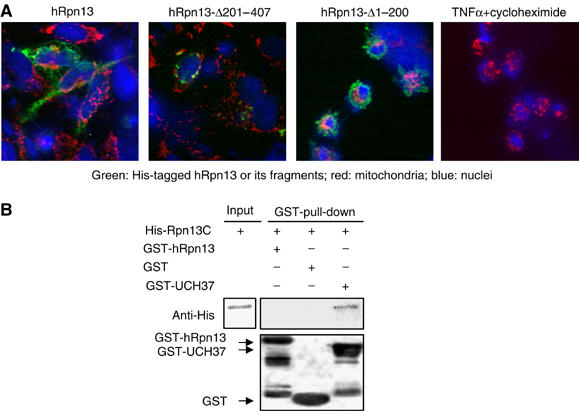
Expression of the C-terminal half (but not the N-terminal half) of hRpn13 induces cell death. (A) HeLa cells were transfected with Myc/His6-tagged hRpn13, hRpn13-Δ201–407, or hRpn13-Δ1–200. After 1 day, hRpn13 and its fragments were visualized by immunofluorescence staining with an anti-His6 antibody, mitochondria with MitroTracker Red, and nuclei with DAPI. The apoptosis-like features, the condensation of nuclei and the severe aggregation of mitochondria, were observed in more than 90% cells (about 50 cells examined) that were transfected with hRpn13-Δ1–200. For comparison with apoptotic cell death, the cells were treated with 20 ng/ml of TNFα and 1 μM of cycloheximide for 4 h. (B) The C-terminal half of hRpn13 does not bind to full-length hRpn13, though it binds to UCH37. The purified His-hRpn13-Δ1–200 was incubated with GST, GST-hRpn13 or GST-UCH37 for 1 h. Pull-down assays were performed using glutathione beads, and the protein levels were determined by Western blot. Thus, the C-terminal half of hRpn13 causes cell death, probably by blocking UCH37 binding to the proteasome.
Discussion
Affinity purification of mammalian proteasomes
The new affinity methods described here allowed rapid and gentle purification of 26S proteasomes from cultured cells. The particles obtained exhibit the characteristic peptidase activities and known subunits of the 26S proteasome, but it remains to be determined whether these gentle isolation methods yield particles that differ in other functional properties from proteasomes isolated by conventional methods. In addition to the known subunits and hRpn13, these preparations contained tubulins-α and -β and several heat-shock proteins of the Hsp70 family, which have not been previously reported as proteins associated with mammalian proteasomes. Homologous heat-shock proteins have been found to be associated with yeast proteasomes purified by analogous methods (Verma et al, 2000; Guerrero et al, 2006), but tubulins-α and -β themselves have not been previously found to be associated with proteasomes, although PAC2, a tubulin-specific chaperone, has been detected in the affinity-purified yeast proteasome (Verma et al, 2000). As tubulins can play a role in protein translocation, their isolation with the proteasomes raises the possibility that they may function in transporting proteasomes or even in substrate delivery.
A number of observations presented here indicate that hRpn13 is in fact an integral subunit of the human 26S proteasome. This sequence initially identified in mammalian cells as GP110 was proposed to be an interferon-γ-regulated surface membrane protein (Shimada et al, 1991) containing a putative transmembrane sequence. However, we showed that like other proteasome subunits, hRpn13 is localized in both the cytosol and the nucleus, that it cosediments with proteasomes upon glycerol-density gradient centrifugation, and that it is also present in the proteasomes purified by conventional chromatographic approaches. Also, hRpn13 directly associates with UCH37, a known proteasome constituent. After our paper was submitted for publication, three other groups have also reported that hRpn13 is a component of the 19S regulatory complex, where it serves to bind UCH37 (Hamazaki et al, 2006; Jorgensen et al, 2006; Yao et al, 2006). Finally, the homolog of hRpn13 in budding yeast, Daq1/Rpn13, has been identified as a subunit of the 19S particle (Verma et al, 2000; Sone et al, 2004).
The role of hRpn13 in deubiquitination
An intriguing finding likely to be of mechanistic importance was that purified hRpn13 binds to UCH37 and enhances its isopeptidase activity. Presumably, this activation is important in the disassembling of Ub conjugates during proteasomal degradation. Two other isopeptidases, the cysteine peptidase, Ubp6/USP14, which is structurally related to UCH37, and the metallopeptidase, POH1/Rpn11, are also located in the 26S proteasome (Leggett et al, 2002; Yao and Cohen, 2002). Rpn11 is a highly conserved Jab1/MPN isopeptidase of the Jabl/MPN family that is important in yeast in cleaving the polyUb chain from the substrate (Verma et al, 2002; Yao and Cohen, 2002). In contrast, UCH37 and USP14/Ubp6 release sequentially the most distal Ubs of a polyUb chain. However, the exact roles of UCH37 and the other proteasome-associated isopeptidases in degrading Ub conjugates are uncertain. Budding yeast do not have a homolog of UCH37, and Ubp6 plays the predominant role in regulating the disassembly of the polyubiquitin chain and protein degradation by their 26S proteasomes (Leggett et al, 2002). Nevertheless, budding yeast lacking Ubp6 are viable. In fission yeast, the homolog of UCH37, UCH2, seems to be responsible for the majority of proteasomal isopeptidase activity (Stone et al, 2004). But, UCH2-deficient fission yeast also survived, and thus, other deubiquitinating enzymes, such as Rpn11, can support proteasomal degradation.
It is noteworthy that the C-terminal region of hRpn13 (residues 201–407), which binds to UCH37, is partially conserved in fission yeast, but not in budding yeast. Interestingly, UCH37 can also bind to the proteasome through Rpn10/S5a (Figure 5B) (Stone et al, 2004), which is located at the hinge between the lid and the base of the 19S complex (Glickman et al, 1998), close to where UCH37 has been localized (Holzl et al, 2000). Although the Rpn10/S5a subunit thus appears to interact with UCH37 in the 19S complex, this interaction fails to activate UCH37, unlike the binding of its C-termini to that of Rpn13. Presumably both interactions function within the 19S complex, but the C-terminal region of hRpn13 evolved specifically to activate UCH37. Although this activation by free hRpn13 was only two-fold against Ub-amc, the stimulation of UCH37 activity against natural polyUb chains or by proteasomally bound hRpn13 are important questions for future study.
The role of hRpn13 in addition to regulation of deubiquitination
Although there is no homolog of UCH37/UCH2 in budding yeast, Daq1/Rpn13 is still required for degradation of certain ubiquitinated proteins (e.g., substrates of the UFD pathway) (Verma et al, 2000). Therefore, hRpn13 also seems to be important in assembly of the 19S complex or its function at some UCH37/UCH2-independent steps. In frogs, the homolog of hRpn13, Xoom, is required for embryonic development (Hasegawa et al, 2001), which presumably reflects a critical role for protein degradation or for UCH37 during frog development. Knockdown of hRpn13 in 293T cells, though incomplete, reduced overall degradation of short-lived proteins reproducibly, and this two-fold inhibition caused a large accumulation of the rapidly degraded model substrate Ub-R-GFP, which is degraded by the proteasome after ubiquitination by the ‘N-end rule' ligase E3α/Ubr1 (Varshavsky, 1997). Furthermore, similar decreases in proteolysis were seen upon expression of high amounts of the C-terminal half of hRpn13, which probably acts as a dominant-negative form that inhibits UCH37 binding, but allows 19S assembly. As this construct eventually induced cell death, hRpn13 appears to be critical for proteolysis. Moreover, because overproduction of hRpn13 also reduced degradation and Ub conjugate accumulation, the precise level of hRpn13 appears important, as would be expected for a key subunit of this critical structure.
Materials and methods
Cell culture and transfections
293T adenovirus-transformed human embryonic kidney cells and HeLa human epitheloid carcinoma cells, and all other cell lines were maintained in DMEM (Cellgro), supplemented with 10% fetal bovine serum and antibiotics in 5% CO2. All cell lines and serum were obtained from the American Type Culture Collection. Except where noted, transfection was carried out using Trans-IT transfection kit (Mirus Inc.).
DNA or RNA oligos and expression vectors
For mammalian expression, the C-terminus of mouse Rpn11 or β4 was fused with protein A (kindly provided by Dr D Finley) or FLAG tag (DYDDDDK), and a Tev protease cleavage site was inserted between the proteasome subunit and protein A. These expression cassettes were subcloned at _Nhe_I/_Xho_I sites of pcDNA3.1 (Invitrogen). UCH37, UCHL3, and the full-length (residues 1–407), the N-terminal region (Δ201–407), or the C-terminal region (Δ1–200) of hRpn13 with Myc-His6 tags at the C-termini were subcloned at the _Hin_dIII/_Eco_RI sites of pcDNA6 (Invitrogen). For expression in insect cells, hRpn13 with a C-terminal His6 tag was subcloned into the baculoviral vector pAcHLT-A (BD Biosciences) at _Eco_RI/_Not_I sites. For bacterial expression, GST-fused UCH37 and RPN12 were constructed at the _Eco_RI/_Xho_I sites of pGEX-4T-2 (Pharmacia), and the full-length or the C-terminal half (Δ1–200) of hRpn13 was subcloned into the _Eco_R1/_Xho_I sites of pET28b (Novagen). The plasmid for Ub-R-GFP was kindly provided by Dr MG Masucci. For siRNA expression, BS/U6/hRpn13 was constructed using specific DNA oligos based on the 21-nt sequence in the coding region of the hRpn13 cDNA (5′-GGGTCTACGTGCTGAAGTTCA-3′), and BS/U6/GFP was obtained as described previously (Qiu et al, 2004). The sequences of all constructs were verified by DNA sequencing. In addition, a pool of four siRNA oligos specific for hRpn13 (Smartpool) and a nontargeting siRNA oligo were purchased (Dharmacon Inc.) and transfected using trans-IT-TKO transfection kit (Mirus Inc.).
Co-immunoprecipitation and Western blot analysis
Proteins were extracted and immunoprecipitated in the buffer containing 20 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM ATP, 1 mM EDTA, 1 mM DTT, and 0.1 mM Phenylmethylsulfonyl fluoride (PMSF). Except where noted, the levels of proteins were analyzed by Western blot using horseradish peroxidase-conjugated or alkaline-phosphatase-conjugated secondary antibodies. Antibodies against α2 and RPN12 were purchased from Affiniti, β-actin and FLAG from Sigma, Myc from Oncogene, and IgG-resin from ICN. The polyclonal antiserum against hRpn13 was raised from rabbits using full-length hRpn13 as an antigen (Rockland).
Peptidase and isopeptidase assays
The proteasome's chymotrypsin-like activity was assayed using the fluorogenic substrate succinyl LLVY-7amc as described (Holzl et al, 2000; Kisselev et al, 2002). To stimulate the 20S peptidase activity, 0.02% SDS was added. The Ub C-terminal hydrolase activity of the purified isopeptidases and the 26S proteasome were assayed using Ub-amc as a substrate. The release of amc from the substrate was monitored continuously at 380/460 nm (excitation/emission).
Affinity purification of mammalian proteasomes
Each of the 20 plates (100 mm) of 293T cells was transfected with 6 μg pcDNA3.1-mRPN11-protein-A, and incubated for 24 h. Then, the cells were split onto 20 large plates (150 mm), and incubated for 2 more days. The cells were harvested by scrapping, suspended in the lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10% glycerol, 5 mM MgCl2, 1 mM PMSF, and 5 mM ATP), and broken by sonication. The 16 000 g supernatants of the cell lysates were incubated with 1 ml of IgG resins (ICN) for 1 h. The resins were washed three times with 2 ml of lysis buffer, and once with Tev buffer (50 mM Tris, pH 8.0, 1 mM EDTA, and 1 mM DTT). The proteasomes were released from the resins by incubating with 100 units of His-tagged Tev protease (Gibco BRL) in 1.5 ml of the Tev buffer for 1 h. The His-tagged Tev protease in the proteasome solution was removed by incubating with 50 μl of Ni-resins (Qiagen) for 15 min. To concentrate the proteasome and to remove the free form of RPN11, an ultrafiltration through a 5 kDa filter (Bio-Rad) was performed.
Purification of proteasomes from rabbit skeletal muscle by conventional methods
Purification of 20S and 26S proteasomes was carried out as described by Kisselev et al (2002) with a few modifications. Briefly, the 100 000 g supernatant from homogenized rabbit muscle (100 g) was incubated with DE52 DEAE cellulose (100 ml). The resin was then washed on a glass filter with buffer A (20 mM Tris–HCl, pH7.5, 10% glycerol, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, and 2 mM ATP) containing 50 mM NaCl, and then packed in a column. Proteasomes were eluted with 250 ml of buffer A with a 50–300 mM NaCl gradient. The pooled fractions with proteasome peptidase activity were loaded on a Uno Q column (6 ml, Bio-Rad), and then eluted with 25 ml of buffer A, with a 100–350 mM NaCl gradient. 20S and 26S proteasomes were separated into two pools. The 20S pool was further dialyzed against buffer B (20 mM Na-HEPES, pH 7.5, 0.5 mM EDTA, 1 mM DTT, and 10% glycerol), loaded on a Heparin HiTrap column (5 ml, Amersham/Pharmacia), and eluted with 50 ml of buffer B containing a 0–250 mM NaCl gradient. The 26S proteasome pool from the Uno Q step was finally purified by ultracentrifugation (100 000 g for 24 h) on a 20–40% glycerol gradient.
Pulse-chase analysis of protein degradation
Degradation of short-lived proteins was detected by pulse-chase analysis as described previously (Gronostajski et al, 1985) with a few modifications. Briefly, 293T cells were incubated with DMEM containing 10% dialysed FBS and 5 μCi ml−1 of [3H]tyrosine for 2 h, and then chased in the medium with 10% FBS and 2 mM of unlabeled tyrosine for 0–120 min. 200 μl of the medium were sampled every time, and mixed with 20 μl of 100% trichloroacetic acid. Following centrifugation, the radioactivities in supernatants were determined by scintillation. After the final chase, the cells were lysed in 0.2 N of NaOH, and subjected for measurement of total radioactivity in the cells. Protein degradation in arbitrary units was derived from the radioactivity released in the medium normalized to the total radioactivity incorporated in cells.
Mass spectrometry
Protein samples were analyzed by MALDI-TOF using an Applied Biosystems Voyager-DE-STR at the Tappan Mass Spectrometry Facility (Harvard Medical School).
Supplementary Material
Legend for Supplementary Figure
Supplementary Figure 1
Supplementary Table 1
Supplementary Table 2
Acknowledgments
We are indebted to Drs D Leggett and D Finley for their suggestions and protein A cDNA, Dr M G Masucci for Ub-R-GFP plasmid, Dr H-T Kim for pET-S5a plasmid, G-P Li and S-C Chang for technical assistance, and the members of the Goldberg Lab for valuable discussions. This work was supported by grants from the NIGMS (5 R01 GM46147-10) and Ellison Foundation to ALG, and grants for Distinguished Young Scholars (No. 30525033) and for the Creative Research Group (No. 30421003) from the National Natural Science Foundation of China, and the National Basic Research Program of China (No. 2006CB504001, No. 2006CBOD0606 & No. 2006CBOF1002) from the Ministry of Science and Technology of China.
References
- Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG (2000) Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol 18: 538–543 [DOI] [PubMed] [Google Scholar]
- Diamonti AJ, Guy PM, Ivanof C, Wong K, Sweeney C, Carraway KL III (2002) An RBCC protein implicated in maintenance of steady-state neuregulin receptor levels. Proc Natl Acad Sci USA 99: 2866–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94: 615–623 [DOI] [PubMed] [Google Scholar]
- Goldberg AL (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426: 895–899 [DOI] [PubMed] [Google Scholar]
- Gronostajski RM, Pardee AB, Goldberg AL (1985) The ATP dependence of the degradation of short- and long-lived proteins in growing fibroblasts. J Biol Chem 260: 3344–3349 [PubMed] [Google Scholar]
- Guerrero C, Tagwerker C, Kaiser P, Huang L (2006) An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol Cell Proteomics 5: 366–378 [DOI] [PubMed] [Google Scholar]
- Guterman A, Glickman MH (2004) Deubiquitinating enzymes are IN/(trinsic to proteasome function). Curr Protein Pept Sci 5: 201–211 [DOI] [PubMed] [Google Scholar]
- Hamazaki J, Iemura SI, Natsume T, Yashiroda H, Tanaka K, Murata S (2006) A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J 25: 4524–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, Sakurai N, Kinoshita T (2001) Xoom is maternally stored and functions as a transmembrane protein for gastrulation movement in Xenopus embryos. Dev Growth Differ 43: 25–31 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Holzl H, Kapelari B, Kellermann J, Seemuller E, Sumegi M, Udvardy A, Medalia O, Sperling J, Muller SA, Engel A, Baumeister W (2000) The regulatory complex of Drosophila melanogaster 26S proteasomes. Subunit composition and localization of a deubiquitylating enzyme. J Cell Biol 150: 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen JP, Lauridsen AM, Kristensen P, Dissing K, Johnsen AH, Hendil KB, Hartmann-Petersen R (2006) Adrm1, a putative cell adhesion regulating protein, is a novel proteasome-associated factor. J Mol Biol 360: 1043–1052 [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Kaganovich D, Goldberg AL (2002) Binding of hydrophobic peptides to several non-catalytic sites promotes peptide hydrolysis by all active sites of 20 S proteasomes. Evidence for peptide-induced channel opening in the alpha-rings. J Biol Chem 277: 22260–22270 [DOI] [PubMed] [Google Scholar]
- Lamerant N, Kieda C (2005) Adhesion properties of adhesion-regulating molecule 1 protein on endothelial cells. Febs J 272: 1833–1844 [DOI] [PubMed] [Google Scholar]
- Leggett DS, Glickman MH, Finley D (2005) Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol Biol 301: 57–70 [DOI] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D (2002) Multiple associated proteins regulate proteasome structure and function. Mol Cell 10: 495–507 [DOI] [PubMed] [Google Scholar]
- Nakane T, Inada Y, Itoh F, Chiba S (2000) Rat homologue of the human M(r) 110 000 antigen is the protein that expresses widely in various tissues. Biochim Biophys Acta 1493: 378–382 [DOI] [PubMed] [Google Scholar]
- Qiu XB, Goldberg AL (2002) Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc Natl Acad Sci USA 99: 14843–14848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu XB, Markant SL, Yuan J, Goldberg AL (2004) Nrdp1-mediated degradation of the gigantic IAP, BRUCE, is a novel pathway for triggering apoptosis. EMBO J 23: 800–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, York IA, Saric T, Goldberg AL (2002) Protein degradation and the generation of MHC class I-presented peptides. Adv Immunol 80: 1–70 [DOI] [PubMed] [Google Scholar]
- Shimada S, Ogawa M, Schlom J, Greiner JW (1991) Identification of a novel tumor-associated Mr 110 000 gene product in human gastric carcinoma cells that is immunologically related to carcinoembryonic antigen. Cancer Res 51: 5694–5703 [PubMed] [Google Scholar]
- Shimada S, Ogawa M, Takahashi M, Schlom J, Greiner JW (1994) Molecular cloning and characterization of the complementary DNA of an M(r) 110 000 antigen expressed by human gastric carcinoma cells and upregulated by gamma-interferon. Cancer Res 54: 3831–3836 [PubMed] [Google Scholar]
- Simins AB, Weighardt H, Weidner KM, Weidle UH, Holzmann B (1999) Functional cloning of ARM-1, an adhesion-regulating molecule upregulated in metastatic tumor cells. Clin Exp Metastasis 17: 641–648 [DOI] [PubMed] [Google Scholar]
- Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL (2005) ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell 20: 687–698 [DOI] [PubMed] [Google Scholar]
- Sone T, Saeki Y, Toh-e A, Yokosawa H (2004) Sem1p is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. J Biol Chem 279: 28807–28816 [DOI] [PubMed] [Google Scholar]
- Stone M, Hartmann-Petersen R, Seeger M, Bech-Otschir D, Wallace M, Gordon C (2004) Uch2/Uch37 is the major deubiquitinating enzyme associated with the 26S proteasome in fission yeast. J Mol Biol 344: 697–706 [DOI] [PubMed] [Google Scholar]
- Ustrell V, Hoffman L, Pratt G, Rechsteiner M (2002) PA200, a nuclear proteasome activator involved in DNA repair. EMBO J 21: 3516–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A (1997) The ubiquitin system. Trends Biochem Sci 22: 383–387 [DOI] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR III, Koonin EV, Deshaies RJ (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298: 611–615 [DOI] [PubMed] [Google Scholar]
- Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ (2000) Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell 11: 3425–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 68: 1015–1068 [DOI] [PubMed] [Google Scholar]
- Waterman H, Levkowitz G, Alroy I, Yarden Y (1999) The RING finger of c-Cbl mediates desensitization of the epidermal growth factor receptor. J Biol Chem 274: 22151–22154 [DOI] [PubMed] [Google Scholar]
- Wojcik C, DeMartino GN (2002) Analysis of Drosophila 26 S proteasome using RNA interference. J Biol Chem 277: 6188–6197 [DOI] [PubMed] [Google Scholar]
- Yao T, Cohen RE (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419: 403–407 [DOI] [PubMed] [Google Scholar]
- Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE (2006) Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol 8: 994–1002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legend for Supplementary Figure
Supplementary Figure 1
Supplementary Table 1
Supplementary Table 2