Cell- and gene-specific regulation of primary target genes by the androgen receptor (original) (raw)
Abstract
The androgen receptor (AR) mediates the physiologic and pathophysiologic effects of androgens including sexual differentiation, prostate development, and cancer progression by binding to genomic androgen response elements (AREs), which influence transcription of AR target genes. The composition and context of AREs differ between genes, thus enabling AR to confer multiple regulatory functions within a single nucleus. We used expression profiling of an immortalized human prostate epithelial cell line to identify 205 androgen-responsive genes (ARGs), most of them novel. In addition, we performed chromatin immunoprecipitation to identify 524 AR binding regions and validated in reporter assays the ARE activities of several such regions. Interestingly, 67% of our AREs resided within ∼50 kb of the transcription start sites of 84% of our ARGs. Indeed, most ARGs were associated with two or more AREs, and ARGs were sometimes themselves linked in gene clusters containing up to 13 AREs and 12 ARGs. AREs appeared typically to be composite elements, containing AR binding sequences adjacent to binding motifs for other transcriptional regulators. Functionally, ARGs were commonly involved in prostate cell proliferation, communication, differentiation, and possibly cancer progression. Our results provide new insights into cell- and gene-specific mechanisms of transcriptional regulation of androgen-responsive gene networks.
Keywords: Androgen receptor (AR), androgen response element (ARE), transcription, steroid receptor, chromatin immunoprecipitation (ChIP), prostate cancer
Androgens are steroid hormones that control male development and reproductive function and are key regulators of prostate cancer cell growth and proliferation. They bind to the androgen receptor (AR), a member of the intracellular receptor family of transcriptional regulators (Chang et al. 1988; Lubahn et al. 1988), which in turn associates with genomic androgen response elements (AREs); in particular, AR can specifically recognize a 15-bp partially palindromic AR binding sequence (ARBS) motif (Claessens et al. 2001). However, localization of AR to specific genomic sites is clearly more complex than simple sequence recognition. Similarly, AR can regulate transcription by distinct mechanisms; depending on cell or gene context, AR can activate or repress transcription, can functionally interact with other transcriptional regulators (Murtha et al. 1997; Sato et al. 1997) or with other intracellular receptors (Chen et al. 1997; Lee et al. 1999; Panet-Raymond et al. 2000; Chuang et al. 2005). Hormone response element (HRE) organization is likely a major determinant of receptor specificity and coregulator recruitment in a given chromosomal context (Lefstin and Yamamoto 1998). Thus, a detailed understanding of androgen action will require identification of genes bound and regulated by AR, identification of coregulators that interact with AR and characterization of the loci where these interactions occur.
RNA expression profiling of LNCaP prostate cancer cells has described ∼500 transcripts whose levels are altered by androgen treatment (DePrimo et al. 2002; Nelson et al. 2002). However, the mechanisms of regulation are nearly unexplored because the primary androgen responsive genes (ARGs), i.e., the subset regulated directly by AR-occupied AREs, have not been defined. The primary ARGs may in turn produce effects on secondary target genes. Thus, identifying primary ARGs would provide insight into signal transduction pathways and transcriptional regulatory networks modulated by AR. Moreover, comparison of AR target genes in two androgen-responsive cell lines, one originating from normal prostate epithelium and the other from prostate carcinoma, might define cell-specific ARGs and suggest possible disease mechanisms.
We shall operationally define AREs as chromosomal regions that recruit AR in a native chromosomal context and modulate transcription in a reporter gene context in response to androgens. Traditionally, experimental searches for AREs have focused on short genomic segments just upstream of the transcription start sites of individual genes (Loreni et al. 1988; Crossley et al. 1992; Murtha et al. 1993; Rennie et al. 1993; Cleutjens et al. 1996); computational approaches have used predefined ARBS motifs to query these same regions, albeit with only modest success (Nelson et al. 2002; Steketee et al. 2004; Magee et al. 2006).
Here we have used RNA expression profiling to identify ARGs, and chromatin immunoprecipitation together with microarray analysis (ChIP–chip) to define a subset of primary ARGs, thus localizing putative AREs to be validated in reporter assays. Rather than focusing primarily on promoter proximal regions (Phuc Le et al. 2005) or CpG islands (Weinmann et al. 2002) or individual chromosomes (Carroll et al. 2005), we developed tiled-oligonucleotide microarrays spanning ∼104 kb centered on the predicted transcription start site of each of several hundred candidate hormone-responsive genes. We assessed the relationship between receptor occupancy and hormone responsive expression, characterized the genomic organization of AR-regulated genes and AREs, surveyed the complexity and composition of the AREs, and examined the physiologic roles of the primary ARGs.
Results
Identification of androgen-responsive genes
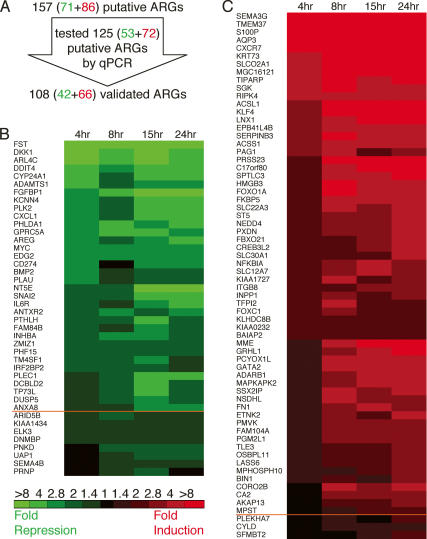
The HPr-1AR cell line, derived from immortalized normal human prostate epithelium, is androgen-responsive and expresses wild-type AR; androgen promotes differentiation of these cells, reminiscent of the normal actions of androgen on the prostate (Ling et al. 2001). We treated HPr-1AR cultures with synthetic androgen R1881, or vehicle as a control, for 4, 8, 15, and 24 h, and identified androgen-regulated transcripts on spotted cDNA microarrays. Of 157 putative ARGs identified by this analysis, 71 were repressed and 86 were induced (Fig. 1A; Supplemental Fig. 1). To validate the expression microarray data, primers were designed for 53 putatively repressed and 72 putatively induced genes, and real-time quantitative polymerase chain reaction (qPCR) subsequently established that 42 were repressed (Fig. 1B) and 66 were induced (Fig. 1C). These 108 differentially expressed genes included 13 (12%) previously identified ARGs, such as SGK (DePrimo et al. 2002; Nelson et al. 2002), as well as 95 (88%) ARGs not previously implicated as androgen targets.
Figure 1.
qPCR validation of the expression of ARGs identified by microarray. (A) Most of the 157 putative ARGs identified by expression microarray analysis were subsequently validated using qPCR. The colorimetric representation shows genes, indicated by HUGO Gene Nomenclature Committee gene symbols, whose transcripts were repressed (B, green) and induced (C, red) by androgen. The color intensity reflects the relative fold change in transcript level for androgen- versus vehicle-treated cells. The time course spanned 4, 8, 15, and 24 h. The mean change in expression level for genes above the orange line was greater than twofold across the time course, whereas the mean change in expression for those genes below the line was greater than 1.5 fold.
Identification of AR binding regions
To identify genomic regions occupied by AR, HPr-1AR cells were treated with R1881 or vehicle for 4 h, a time by which AR occupancy was significantly enriched at target genes such as SGK and FKBP5 (E.C. Bolton and K.R. Yamamoto, unpubl.). Following AR ChIP, immunoprecipitated DNA fragments were amplified, labeled, and hybridized to custom NimbleGen tiled-oligonucleotide microarrays. We interrogated ∼104 kb genomic regions centered on the transcription start sites of 548 candidate hormone-responsive genes to identify AREs and to determine the distribution of AREs at promoter proximal, distal, and intragenic regions. These human candidate genes included our 157 putative and validated ARGs identified in HPr-1AR cells, 241 putative and validated glucocorticoid receptor- (GR) responsive genes identified in A549 lung adenocarcinoma cells and U2OS-GR osteosarcoma cells (Rogatsky et al. 2003; Wang et al. 2004; So et al. 2007), and 150 genes potentially hormone-responsive in other cell types (e.g., 35 ARGs from LNCaP cells; DePrimo et al. 2002; Nelson et al. 2002). GR, which is closely related to AR, binds in vitro with similar affinity as AR to consensus sequences in the elementary half-sites GGTACAnnnTGTTCT (Claessens et al. 2001). Across the ∼57 Mb of DNA interrogated, 593 additional “bystander” genes were incidentally included, giving a total of 1141 genes examined in our tiled microarray (genes assigned by NCBI Build 35, hg17).
The AR ChIP–chip hybridizations directly compared DNA enriched in androgen-treated and control samples. To reduce the rate of stochastic false positives, we intersected putative AR binding peak data for duplicate independent experiments, in which 778 and 714 peaks were detected, with 509 AR binding peaks identified in both replicates. Primers for qPCR were designed for 67 AR binding peaks, and conventional AR ChIP was performed in quadruplicate experiments to establish the remaining false-positive fraction (1 of 64 or <2%), resulting in 508 androgen-stimulated AR binding regions (ARBRs); <10 of these 508 ARBRs may be false positives. Sixteen additional peaks identified in only one of the AR ChIP–chip replicates were also included following validation in quadruplicate experiments by conventional AR ChIP, resulting in 524 ARBRs (Supplemental Table 1).
ARBR validation and detection of additional ARGs
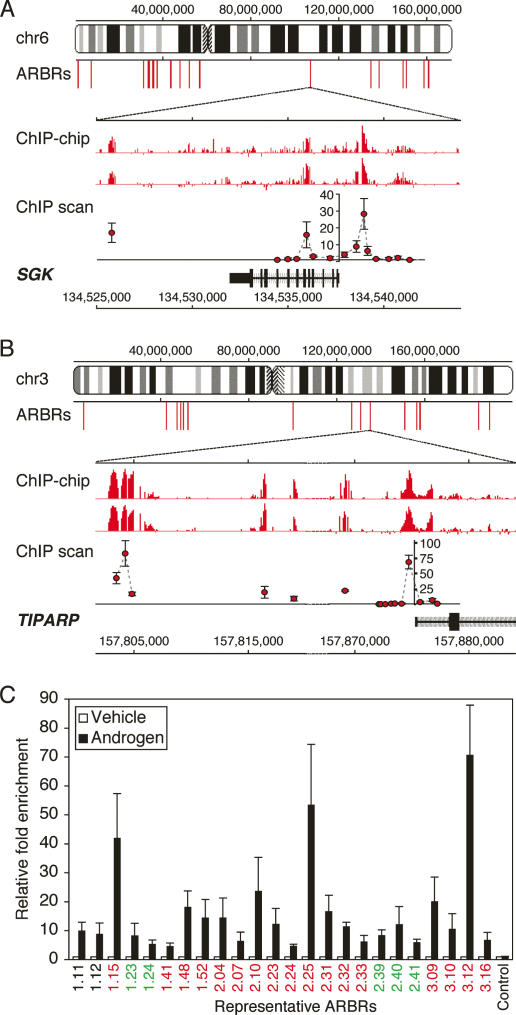
To illustrate our validation process for ChIP–chip data, we describe our analysis of an ARBR 1.2 kb upstream of the SGK gene. In this case, a functional GRE was previously known to reside in this area (Itani et al. 2002). In addition, we mapped an androgen-stimulated DNase I hypersensitive site to this region (Supplemental Fig. 2). Finally, our ChIP–chip and conventional ChIP experiments (Fig. 2A) revealed AR occupancy in this same region (ARBR6.35) as well as in regions 1.6 kb (intron 5) and 11.9 kb (3′ of SGK gene) downstream from the SGK transcription initiation site, defining these regions as ARBRs.
Figure 2.
ARBRs: identification by ChIP–chip and validation by conventional ChIP. The visual representation of ARBRs on chromosomes 6 (A) and 3 (B) are shown. ARBRs are shown as equilateral red bars immediately below the chromosomes. Expanded views of (A) SGK and (B) TIPARP gene loci are shown based on the May 2004 human genome freeze hg17 in the UCSC browser using GenBank RefSeq positions. For SGK (3′–5′ orientation) and TIPARP (5′–3′ orientation), the ChIP–chip signals (red bars) are shown for duplicate experiments and indicate the relative fold enrichment of AR-immunoprecipitated DNA fragments for androgen- versus vehicle-treated cells. The ChIP scanning (Wang et al. 2004) signals (red circles) demonstrate validation of the ChIP–chip data and show the relative fold enrichment of AR ChIP for androgen- versus vehicle-treated cells. Data represent the mean ± SD, n = 4. (C) Additional ARBRs (numbered) identified by ChIP–chip were validated using conventional AR ChIP. The relative fold enrichment of AR ChIP is shown for androgen- versus vehicle-treated cells; data represent the mean ± SD, n = 4. The ARBRs found near or within genes that are repressed (green), induced (red), or unresponsive (black) following androgen treatment are indicated. A region near the HSPA1A gene, which is not occupied by AR, is shown as a control. Compared to the control, all ARBRs demonstrate significant AR occupancy, P < 0.05.
As with SGK, we observed ARBRs near ARGs (within ∼50 kb upstream of or downstream from the transcription start site), including FKBP5, ELL2, TMEPAI, UAP1, and FN1, all of which are androgen-responsive in LNCaP cells (DePrimo et al. 2002; Nelson et al. 2002). ARBRs were also observed near genes not previously implicated as androgen targets, including TIPARP (Fig. 2B). Conventional AR ChIP was performed on a representative subset of ARBRs (Fig. 2C), and >98% were confirmed in this more rigorous test. Taken together, these findings validated the approach used in this study to identify authentic ARBRs and AR regulated genes and confirmed these regions as bona fide in vivo AR targets.
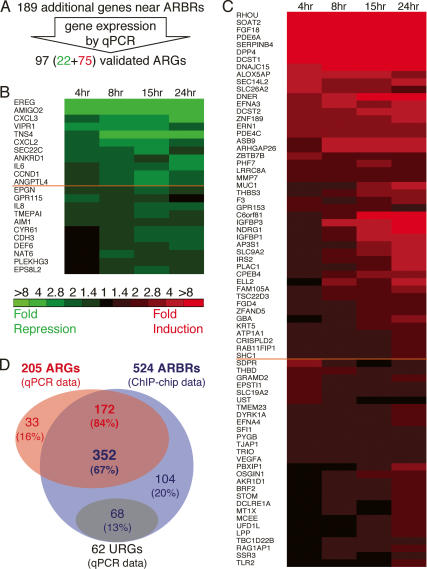
Of the 524 ARBRs we identified, 206 ARBRs (39% of ARBRs) were within ∼50 kb of the transcription start site of a validated ARG identified by our expression microarray analysis, whereas most of the remaining 318 ARBRs (61% of ARBRs) were located near genes absent from the expression microarray. We tested by qPCR 189 of these novel ARBR-proximal genes and found that 22 were androgen-repressed and 75 were androgen-induced (Fig. 3A–C). Of these 97 additional differentially expressed genes, only 10 (10%) were known ARGs (DePrimo et al. 2002; Nelson et al. 2002).
Figure 3.
ARBRs identified by AR ChIP-chip lead to the discovery of novel primary ARGs. (A) Many of the 189 ARBR-proximal genes that were not detected by microarray analysis were, in fact, primary ARGs. The colorimetric representation shows genes, near identified ARBRs, whose transcripts were repressed (B, green) or induced (C, red) by androgen treatment. See Figure 1 for colorimetric and time course details. (D) Of the 524 ARBRs (blue) and 205 (108 initial and 97 additional ARGs, red) differentially expressed ARGs, we found 352 ARBRs near 172 ARGs, which represent direct, functional targets of AR. The response of 62 genes containing in total 68 intragenic ARBRs (gray) was independent of androgen (androgen-unresponsive genes, URGs).
We then sought to determine which of the 108 validated ARGs are also putative primary ARGs by analyzing the overlap between the ARBR and ARG data sets; we found that 75 of the 108 ARGs were near ARBRs. From the 524 ARBRs and all 205 differentially expressed ARGs, we identified 352 ARBRs (67% of ARBRs) near 172 primary ARGs (84% of ARGs) (Fig. 3D). Thus, AR occupancy of ARBRs correlates closely with androgen-responsive expression of linked genes; indeed, we suggest that ChIP–chip is a powerful tool for target gene discovery that is distinct from and complementary to expression microarrary analysis. For the remaining 33 ARGs (16% of ARGs), our failure to detect nearby ARBRs may reflect (1) peaks below the threshold set in the AR ChIP–chip analysis; (2) ARBRs outside of the ∼104-kb windows surveyed here; or (3) ARGs that are secondary rather than primary targets of AR regulation.
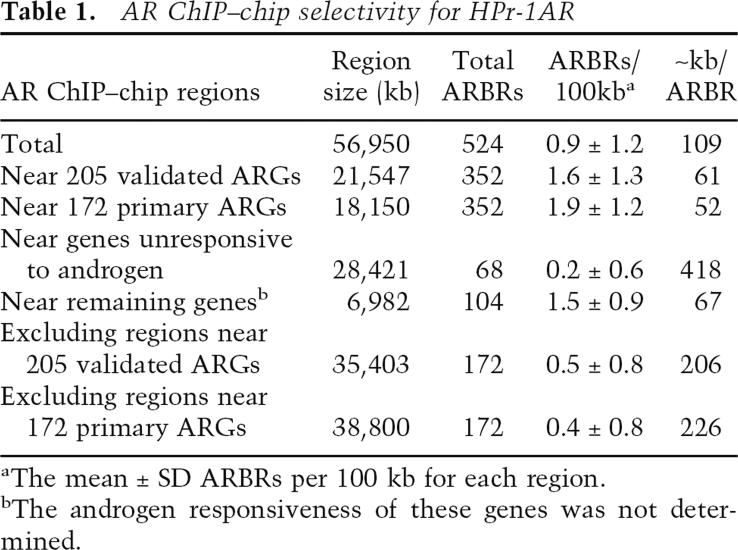
To test whether ARBRs are enriched near ARGs, and to assess AR ChIP–chip selectivity, we determined the mean frequency of ARBR occurrence (Table 1). For the 56,950 kb of DNA scanned, we identified 524 ARBRs, or one ARBR per 109 kb of genomic DNA. Of the 18,150 kb of DNA containing the 172 primary ARGs, we found 352 (67%) ARBRs, i.e., one ARBR per 52 kb of genomic DNA. For the remaining 38,800 kb of DNA scanned, even if all of the remaining 172 (33%) ARBRs were adjacent to genes whose expression is androgen-independent, a maximum of one ARBR per 226 kb was detected. We conclude that, relative to the microarray gene population, biased as it was toward hormone-responsive genes, ARBRs were enriched near ARGs (Wilcoxon rank sum test, P < 10−10), consistent with the ChIP–chip data and the notion that AR occupancy strongly correlates with AR-regulated expression.
Table 1.
AR ChIP–chip selectivity for HPr-1AR
Primary ARG clusters
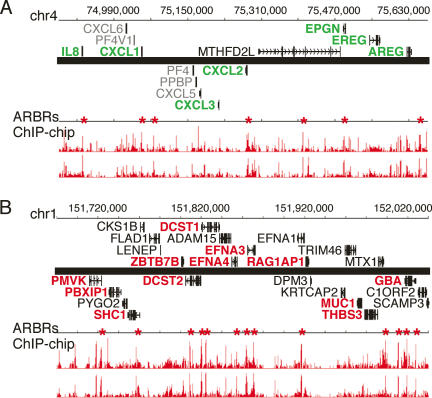
Intersection analysis of the ARG and ARBR data revealed two AR-regulated gene clusters consisting of multiple ARBRs and ARGs. An 800-kb region on chromosome 4 contained seven validated ARBRs, seven repressed ARGs and one expressed but androgen-unresponsive gene (Fig. 4A). Similarly, a 370-kb region on chromosome 1 contained 13 validated ARBRs, 12 induced ARGs, and 12 androgen-unresponsive genes (Fig. 4B). We are intrigued that AR regulation within these ARG clusters is negative or positive but not both, implying functional coordination resulting in part from genomic organization.
Figure 4.
Intersection of ARBR and ARG data reveals primary ARG clusters. (A) An 800-kb region on chromosome 4 containing seven validated ARBRs (red asterisks) was found to contain genes that were repressed (green) or unresponsive (black) following treatment with androgen. (B) A 370-kb region on chromosome 1 containing 13 validated ARBRs (red asterisks) was found to contain genes that were induced (red) or unresponsive (black) following treatment with androgen. Gene loci are shown based on the May 2004 human genome freeze hg17 in the UCSC browser using GenBank RefSeq positions. The androgen responsiveness of some genes was not determined (gray). Genes above the heavy black line are transcribed from left to right whereas those below the line are transcribed from right to left. The longest transcriptional variants for each gene are represented. The ChIP–chip signals (red bars) are shown for duplicate experiments and indicate the relative fold enrichment of AR-immunoprecipitated DNA fragments for androgen- versus vehicle-treated cells.
Interestingly, each ARG cluster contains genes of related function: The androgen-repressed ARGs on chromosome 4 include IL8, CXCL1, CXCL2, and CXCL3, which encode CXC chemokines involved in angiogenesis (Belperio et al. 2000). Also on chromosome 4, EPGN, EREG, and AREG encode epithelial growth factor (EGF)-like mitogens that stimulate MAP kinase signaling (Kochupurakkal et al. 2005). Within the induced cluster on chromosome 1, two ephrin genes, EFNA4 and EFNA3, are AR-regulated. Ephrin receptors are receptor-protein tyrosine kinases involved in cell migration and angiogenesis (Holder and Klein 1999). Thus, the AR-regulated genes within these clusters are involved in key cellular processes, dysregulation of which may influence cancer progression.
ARBRs display ARE activities
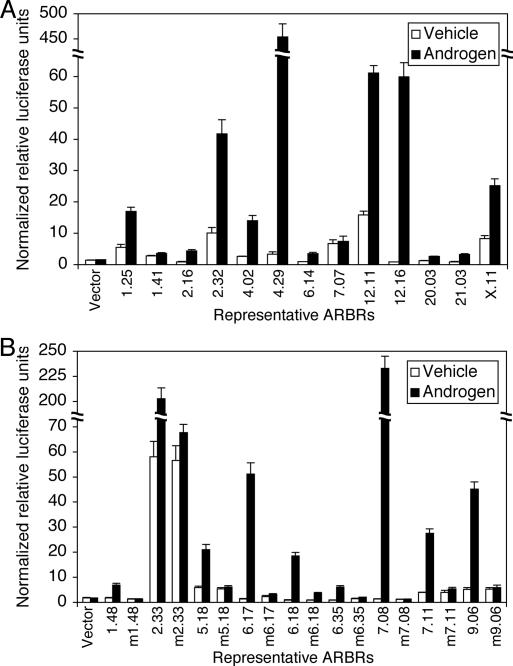
To initially assess whether our ARBRs are indeed functional AREs, we cloned 22 ∼500-bp fragments, each containing an ARBR found within or near an androgen-induced gene, into a reporter plasmid, upstream of a minimal E4 TATA sequence (Lin et al. 1988) driving the expression of luciferase. HPr-1AR cells transfected with individual ARBR reporter constructs were treated with R1881 or vehicle control for ∼20 h, which is sufficient for robust hormone-dependent changes in luciferase activity for constructs containing AREs. Twenty (91%) of the 22 tested ARBRs displayed significant androgen-induced luciferase activity (Fig. 5A,B). Moreover, mutation of the putative ARBSs contained within the ARBRs abolished nearly all of the androgen-induced luciferase activity (Fig. 5B). These data demonstrate that AR occupies ARBRs by ARBS recognition, and that the vast majority of the ARBRs tested conferred AR regulation in a reporter context. We conclude that ARBRs correspond in general to functional AREs at primary ARGs, and from hereon we shall denote such ARBRs as AREs.
Figure 5.
ARBRs display androgen response element (ARE) activities. (A) Thirteen sequences of ∼500 bp, each containing individual ARBRs (numbered), were cloned upstream of a minimal promoter sequence driving the expression of luciferase. (B) Nine additional sequences of ∼500 bp, each containing individual ARBRs (numbered), were also cloned as described (see Materials and Methods). These nine ARBRs and the corresponding ARBR mutants, which contain mutated ARBSs (indicated by the letter m preceding the ARBR number), were tested in cell-based reporter assays. HPr-1AR cells transfected with each of the ARBR reporter constructs were treated with vehicle (open bars) or androgen R1881 (solid bars) and subsequently were assayed for androgen-stimulated luciferase activity. The ARBRs shown correspond to sequences upstream of, within, and downstream from ARGs. Data represent the mean ± SD, n = 3.
ARE distribution within or near ARGs
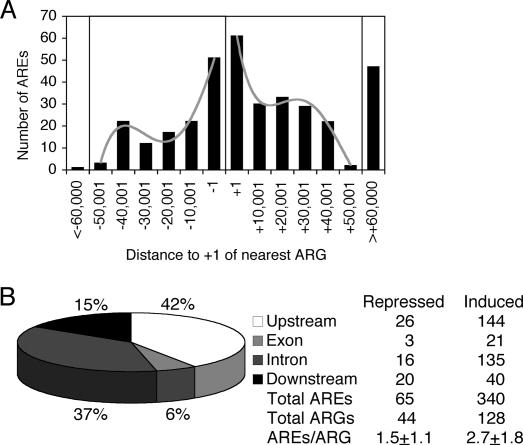
We sought to determine the location of AREs relative to the transcription start site of the closest ARGs; as 6% of our ARGs are annotated with two or more start sites, we independently analyzed the distribution of the AREs relative to the longest (Fig. 6A) and shortest (Supplemental Fig. 3) transcripts. For the 172 ARGs near AREs, only ∼33% of AREs (32% relative to the longest variants and 33% relative to the shortest variants) resided within 10 kb of the predicted transcription start site; most were located downstream from the ARG transcription start site. Similarly, for the 67% of AREs found >10 kb from the ARG transcription start site, most were situated downstream. Relative to the substructure of the primary ARGs, 43% of AREs were intragenic (exonic and intronic), 15% were downstream (Fig. 6B), and 42% were upstream; among ARGs with upstream AREs, only 28% were within 5 kb of the transcription start site. We examined 154 of the 185 genes containing intragenic AREs and found that 92 (60%) were ARGs, whereas the expression of 62 (40%) of these genes was independent of androgen in HPr-1AR. Taken together, our data indicate that most ARGs identified by expression profiling are primary ARGs, but that most ARGs lack promoter-proximal AREs.
Figure 6.
Distribution analysis of AREs located near ARGs. (A) The distribution of AREs identified by ChIP–chip was plotted relative to the predicted transcription start site (+1) of the longest transcriptional variant of the nearest ARG. AREs reside great distances upstream and downstream, as well as near the promoter. Notably, fewer AREs were found upstream of than downstream from ARG transcription start sites of the longest transcriptional variants. Relative distances upstream of (negative) and downstream (positive) from the transcription start sites (vertical black line) were assigned to 10 kb bins. AREs were generally found beyond 10 kb of the transcription start site. (B) The location of AREs is shown relative to the predicted gene substructure of the longest transcriptional variant of the nearby ARGs. When the distribution of all AREs near all ARGs was assessed, most AREs were situated downstream from the transcription start site. These distributions were subdivided based on whether the ARG was repressed or induced, suggesting biases in terms of ARE location and the mean AREs/ARG ratios.
Interestingly, ARE density was significantly lower for androgen-repressed genes (1.5 ± 1.1 AREs/ARG, mean ± SD) than for induced genes (2.7 ± 1.8 AREs/ARG, Wilcoxon rank sum test, P < 10−5). In addition, repressed ARGs contained intragenic AREs (19 of 65 AREs, 29%) at a lower frequency than did induced genes (156 of 340 AREs, 46%), whereas a larger fraction of AREs resided downstream from repressed ARGs (20 of 65 AREs, 31%) compared to induced genes (40 of 340 AREs, 12%) (Fig. 6B). Conceivably, then, hormonal activation or repression of an ARG may relate in some way to ARE position and the number of AREs per ARG, implying that it may be important for mechanistic studies to identify multiple AREs near primary AR target genes. We speculate that androgen-mediated transcriptional regulation may commonly employ combinatorial coordination of multiple AREs located upstream of, within, and downstream from an ARG, e.g., SGK (Fig. 2A) and TIPARP (Fig. 2B).
Potential composite AREs
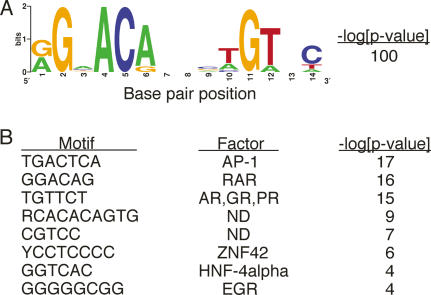
Although relatively few native HREs have been examined, many of these appear to be composite elements, with binding sequences for two or more factors defining platforms for regulatory complex assembly (Lefstin and Yamamoto 1998; Carroll et al. 2005). We employed unbiased searches using BioProspector (Liu et al. 2002) or MobyDick (Bussemaker et al. 2000; So et al. 2007) to identify recurring sequence motifs among our 524 AREs (500-bp sequences centered at the AR binding peak) (Fig. 7). As expected, the most prominent motifs were the partially palindromic ARBS itself, occurring in 69% of AREs (Fig. 7A), and the TGTTCT half-site of the ARBS (Fig. 7B). In addition, we identified several other recurring motifs, with Bonferroni corrected P < 10−3; by comparing these sequence motifs with matrices in TRANSFAC (Matys et al. 2003), we concluded that potential binding sequences for AP-1, retinoic acid receptor (RAR), zinc finger protein 42 (ZNF42), hepatocyte nuclear factor 4 receptor α (HNF-4α), and early growth response (EGR) factor (Fig. 7B) were detected in >142 (27%) of our 524 AREs.
Figure 7.
ARBSs and other _cis_-regulatory motifs are enriched within AREs. Unbiased motif searches of all 524 AREs revealed the presence of significantly recurring motifs. (A) BioProspector analysis identified a consensus ARBS, which is visually represented in WebLogo (http://weblogo.berkeley.edu). (B) MobyDick analysis detected a consensus AR half-site motif as well as putative motifs for AP-1, RAR, ZNF42, HNF-4α, and EGR. These enriched sequence motifs are represented using IUPAC symbols (http://www.iupac.org). Motifs where the putative regulatory factors were not determined (ND) are indicated. _P_-value calculations are described in Materials and Methods.
Detection of additional motifs within AREs suggested potential points of regulatory cross-talk and may facilitate analysis of combinatorial regulation at the molecular level. Indeed, several of the identified sequence motifs appear from other studies to be functional. For example, our computational analysis revealed an AP-1 binding motif at ARE4.13 of the interleukin-8 (IL8) gene, which Freund et al. (2004) showed to be important for AP-1-mediated transcriptional regulation of IL8 promoter expression. We suggest that many AREs are likely composite elements, and that regulatory cross-talk at AREs may typically modulate transcription of ARGs.
ARGs modulate diverse biological processes
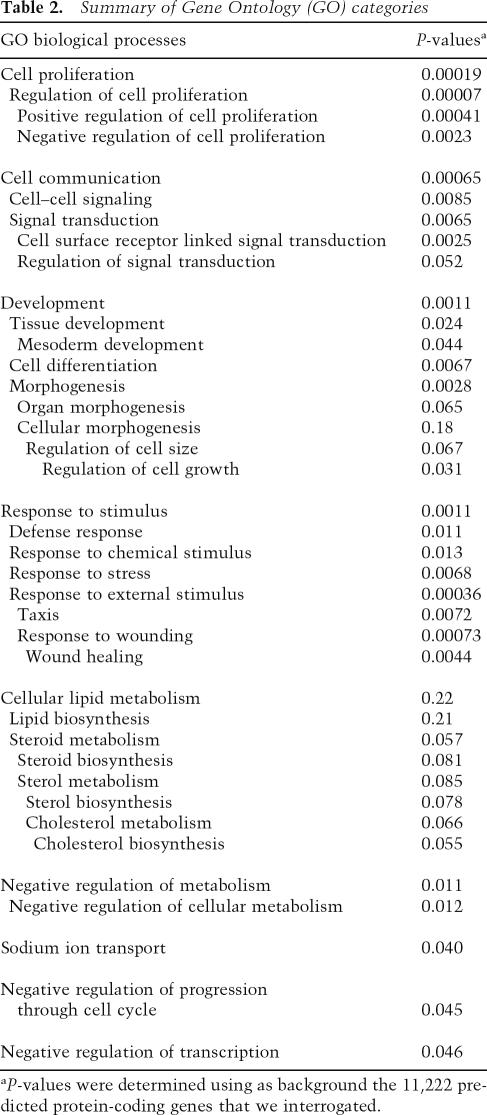
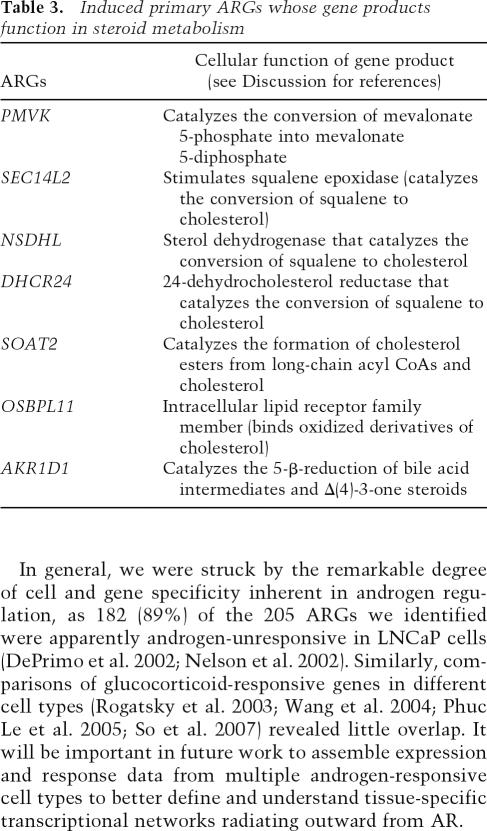
To infer biological processes regulated by androgens and AR, the 205 validated ARGs were surveyed by shared Gene Ontology (GO) Biological Processes data using the Database for Annotation, Visualization and Integrated Discovery (DAVID) 2006 (Dennis et al. 2003). The enriched GO Biological Process categories (Table 2) were dominated by ARGs whose products control cell proliferation and differentiation, cell–cell signaling, and morphogenesis, consistent with the known physiologic roles of androgens as modulators of cellular proliferation, communication, and development. We found it striking that seven ARGs whose products function in steroid metabolism were induced by androgens (Table 3), suggesting that some of the resulting lipid products may modulate cellular responses to steroids, or exert other signaling effects.
Table 2.
Summary of Gene Ontology (GO) categories
Table 3.
Induced primary ARGs whose gene products function in steroid metabolism
Comparing the HPr-1AR ARGs to those previously reported in LNCaP prostate carcinoma cells, we found that only 23 (11%) of the 205 HPr-1AR ARGs were also androgen-responsive in LNCaP cells (Table 4; DePrimo et al. 2002; Nelson et al. 2002). Remarkably, nine (39%) of these 23 ARGs were differentially regulated, i.e., androgen-induced in one cell line and androgen-repressed in the other. Moreover, 17 (74%) of these 23 ARGs were found to contain nearby AREs, and therefore are primary AR targets.
Table 4.
Intersection of ARGs for HPr-1AR and LNCaP cells
These results underscore the strong cell specificity of AR function and suggest that, in principle, extending these analyses of primary AR-regulated genes could reveal the androgen-responsive gene networks that modulate prostate cell proliferation, communication, differentiation, and possibly cancer progression.
Discussion
We found that multiple genes involved in prostate cell growth, proliferation, and perhaps cancer progression are regulated directly by AR. AREs were detected near 84% of ARGs, suggesting that, during our relatively short times of hormone exposure, most ARGs in HPr-1AR cells are primary targets of AR rather than secondary consequences of hormone exposure, and that AR occupancy is a major determinant of AR responsiveness. It will be interesting to extend our cell culture analysis to assess the relative roles for primary and secondary ARGs in developing and adult prostate tissue, and during prostate cancer progression.
Roles of ARGs in biological processes
Consistent with the physiologic roles of androgens as modulators of cellular proliferation, signaling, and development, genes associated with those processes were enriched among our ARGs and primary AR target genes (Table 2). In principle, genes whose expression differs between normal and tumor cells or undergoes change during tumor progression may serve as biomarkers of different cancer stages. Of the 20 ARGs that modulate cell proliferation in HPr-1AR, 15 were not reported to be androgen-responsive in LNCaP cells, and two genes (CYR61 and ADAMTS1) that were repressed in HPr-1AR are induced in LNCaP cells (Table 4). Along with AR itself, eight primary ARGs (SNAI2, KRT5, INHBA, IL6, IL6R, ARL4C, F3, and HMGB3) and one ARG (FOXO1A) in HPr-1AR are commonly expressed aberrantly during prostate carcinogenesis (Giri et al. 2001; LaTulippe et al. 2002; Lapointe et al. 2004; Dong et al. 2006). Moreover, ARL4C, HMGB3, and AR expression are elevated strikingly in metastatic prostate cancers compared to nonrecurrent primary prostate cancers, whereas expression of F3 is decreased (LaTulippe et al. 2002). It will be interesting in future studies to assess whether these and other ARG products that regulate cell proliferation emerge as therapeutic targets or diagnostic markers.
An intriguing subset of primary ARGs involved in cellular lipid metabolism includes seven steroid metabolism genes, all of which are androgen-induced. These include four ARGs (Table 3; PMVK, SEC14L2, NSDHL, and DHCR24, an ARG in LNCaP cells and an unresponsive AR-occupied gene in HPr-1AR) whose products function in de novo cholesterol biosynthesis (Olivier et al. 1999; Shibata et al. 2001; Waterham 2006) and three whose products either process sterols (SOAT2 and AKR1D1) or function as a receptor for oxidized sterols (OSBPL11) (Kondo et al. 1994; Chang et al. 2001; Lehto and Olkkonen 2003). Another induced ARG is long-chain acyl CoA synthetase (ACSL1), which activates long-chain fatty acids for β-oxidation or acylation by cholesterol acyl transferase (SOAT2); SOAT2 in turn converts cholesterol, from which steroids are derived, into cholesterol esters for storage. Together, these primary ARGs expand the AR signaling network to include specific lipid metabolism pathways; some of the resulting lipid products may modulate cellular responses to steroids or exert other signaling effects.
In general, we were struck by the remarkable degree of cell and gene specificity inherent in androgen regulation, as 182 (89%) of the 205 ARGs we identified were apparently androgen-unresponsive in LNCaP cells (DePrimo et al. 2002; Nelson et al. 2002). Similarly, comparisons of glucocorticoid-responsive genes in different cell types (Rogatsky et al. 2003; Wang et al. 2004; Phuc Le et al. 2005; So et al. 2007) revealed little overlap. It will be important in future work to assemble expression and response data from multiple androgen-responsive cell types to better define and understand tissue-specific transcriptional networks radiating outward from AR.
Potential composite AREs
In 69% of AREs (Fig. 7A), we readily identified a partially palindromic ARBS motif, and mutation of the motif virtually abolished the ARE activities (Fig. 5B). The remaining AREs lacked a statistically significant consensus ARBS motif and might be “tethering elements”, at which AR associates through protein–protein interactions with DNA-bound nonreceptor regulatory factors, rather than itself binding to DNA (Murtha et al. 1997; Sato et al. 1997; Lefstin and Yamamoto 1998). In any case, our approach was broadly effective in identifying functional AREs that contain or lack consensus ARBSs, and that either promote or repress AR-regulated transcription.
Combinations of _cis_-regulatory motifs can function as composite response elements to integrate multiple signals and to elicit distinct regulatory effects in different cellular or physiologic contexts. These biological nodes present opportunities to define regulatory interactions at the molecular and functional levels. As a first step, we scanned for additional sequence motifs within the AREs and identified several significantly recurring motifs, including putative sites for AP-1, RAR, ZNF42, HNF-4α, and EGR. We note that AP-1 motifs are known to operate at several ARGs (Murtha et al. 1997; Sato et al. 1997; Chen et al. 2006), and that increases in AR and the AP-1 factor c-Jun correlate with prostate cancer progression (LaTulippe et al. 2002; Tiniakos et al. 2006). Assessing regulatory cross-talk between AR and AP-1 signaling at AREs will thus be informative but technically challenging as the dimeric AP-1 factors are drawn from a superfamily of genes (c-Jun, JunB, and JunD) that can homo- or heterdimerize with c-Fos, FosB, Fra-1, and Fra-2.
ARE distribution, combinatorial regulation, and ARG clusters
Studies of promoter proximal AREs have yielded an emerging view of coordinated recruitment of AR, nuclear coregulators, chromatin remodeling factors, and the transcriptional machinery (Shang et al. 2002; Metzger et al. 2005; Wang et al. 2005; Yamane et al. 2006). Whether such findings present a complete picture, however, is a question brought into focus by results showing that most AREs are quite remote from ARGs. We suggest that some remote AREs may act analogously to the β-globin locus control region (LCR), which exerts temporal control over expression of genes within the β-globin gene cluster from 60 kb away (Lawrence 2003); Takayama et al. (2007) hypothesize that distal intergenic as well as intragenic AREs function to regulate primary ARGs.
We identified 172 primary AR target genes and 524 AREs. In agreement with a recent ChIP–chip study of 10 androgen-responsive genes in LNCaP (Takayama et al. 2007), we found that most AREs were >10 kb upstream of and, or more typically, downstream from the ARG transcription start sites. As functional enhancers, AREs can operate relatively remote from promoters, and thus provide little selective pressure to assume more promoter proximal positions. Our results parallel those from ChIP–chip studies of other transcriptional regulators (Martone et al. 2003; Euskirchen et al. 2004; So et al. 2007). We also commonly observed multiple AREs per ARG (Figs. 2, 4, 5), suggesting combinatorial regulation of individual primary ARGs.
Interestingly, 31 (15%) ARGs were themselves linked in gene clusters, consisting of ARBRs and multiple ARGs. In addition to the two large ARG clusters described here (Fig. 4A,B), we also identified five small ARG clusters consisting of at least one intergenic ARE flanked by two ARGs (E.C. Bolton and K.R. Yamamoto, unpubl.). Strikingly, all of the ARGs within any given cluster were similarly regulated, i.e., either induced or repressed by androgens, but not a mixture of both. However, some genes within a given cluster nonetheless escape AR-mediated regulation in HPr-1AR cells. Conceivably, AR-mediated regulation within ARG clusters may progress through discrete stages during male development, analogous to the temporal regulation of the β-globin locus (Lawrence 2003).
The challenges associated with testing the coordination of several AREs spanning ∼100 kb for even a single ARG are daunting. One possible approach for future work is to use integrated bacterial artificial chromosome-based reporter genes in mouse genetic studies (DiLeone et al. 2000; Dodou et al. 2004). In any case, it seems likely that intricate collaborations of multiple AREs may orchestrate recruitment of coregulatory complexes that dictate the spatiotemporal expression of AR target genes.
The present study advances our understanding of the organization and function of AREs and nearby ARGs, and also illuminates new unknowns, such as the mechanisms that restrict AR occupancy to a subset of potential AREs in a given cell context, or the contributions of individual AREs to ARG regulation, especially in the context of ARG clusters. Addressing these and other challenges raised by our study will continue to provide new insights into the mechanisms of transcriptional regulation of genes and gene networks by AR and other transcriptional regulators.
Materials and methods
Cell culture, plasmids, transfections, and reporter assays
HPr-1AR cells, which express wild-type AR, were grown as described (Ling et al. 2001) in keratinocyte serum free medium (Cat. # 17005–042, Invitrogen) and a 5% CO2 atmosphere. Plasmid pGL4.10-E4TATA (provided by Yuriy Shostak, University of California at San Francisco, San Francisco, CA) and pBEC22 (Renilla luciferase vector) were created by insertion of a 50-bp minimal E4 TATA promoter sequence (Lin et al. 1988), driving luciferase expression, into the Bgl II to Hind III sites of vectors pGL4.10 and pGL4.70 (Promega), respectively. Individual ∼500-bp ARBR fragments were amplified from human genomic DNA by PCR (primer sequences in Supplemental Table 2) and inserted into the Kpn I to Xho I sites upstream of the E4 TATA sequence. The QuikChange kit (Stratagene) was used to mutate two to four of the underlined positions (GGAACAnnnT GTNCN) of putative ARBSs contained within the ARBRs (primer sequences in Supplemental Table 2). Mutation of either of two consensus ARBSs within ARBR7.08 reduced androgen-induced luciferase activity (E.C. Bolton and K.R. Yamamoto, unpubl.), and mutation of both ARBSs (mARBR7.08) abolished it. For ARBR6.35 and mARBR6.35, DNA linkers containing only the 15-bp ARBS and mutated ARBS, respectively, were inserted into the Kpn I to Xho I sites of pGL4.10-E4TATA. HPr-1AR cells were grown in 48-well plates, cotransfected with 150 ng of reporter constructs and 150 ng of pBEC22 (Renilla luciferase vector) using Gene Juice (Novagen). After overnight transfection, cells were treated with 0.1% ethanol vehicle or 1 nM R1881 (New England Nuclear) for ∼20 h, and luciferase activities were measured using the Dual-Luciferase Reporter Assay (Promega) and an Ultra Evolution plate reader (Tecan).
RNA isolation, reverse transcription, and qPCR
Total RNA was isolated from HPr-1AR cells using QIAshredder and RNeasy Mini kit (Qiagen). Random-primed cDNA was prepared from total RNA using the ProtoScript First Strand cDNA kit (New England Biolabs). cDNA was used in 50-μL qPCR reactions containing 1 unit of AmpliTaq Gold DNA polymerase (Applied Biosystems), 2.5 mM MgCl2, 500 nM of each primer (primer sequences in Supplemental Table 3), 150 μM dNTP mix, 0.2× SYBR green I dye (Invitrogen), and 1× ROX reference dye (Invitrogen) in 1× PCR Buffer II (Applied Biosystems). Primers were designed using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi), and those that efficiently amplified (Ginzinger 2002) single products of the expected sizes were used for qPCR. We performed qPCR with a 7300 Real Time PCR System (Applied Biosystems). Data were analyzed using the ∂∂Ct method (Applied Biosystems) and normalized to the expression of the RPL19 gene, which is not regulated by androgen or AR.
ChIP and ChIP–chip design and analysis
ChIP and ChIP scanning assays were performed as described (Wang et al. 2004; So et al. 2007) with the following modifications. HPr-1AR cells in medium were treated with 0.1% ethanol vehicle or 1 nM R1881 for 4 h at 37°C, 1% formaldehyde for 3 min as the dishes cooled from 37°C to 22°C, and 125 mM glycine for 10 min as the dishes cooled from 22°C to 4°C. Cells were rinsed with phosphate-buffered saline at 4°C and lysed in ice-cold IP lysis buffer (50 mM HEPES-KOH, pH 7.4, 1 mM EDTA, 150 mM NaCl, 10% glycerol, 0.5% Triton X-100, supplemented with protease inhibitors), and harvested by scraping; cell lysis continued as cells were mixed at 4°C for 10 min. Nuclei were collected by centrifugation (500_g_ for 5 min at 4°C) and resuspended in 2 mL of ice-cold RIPA buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 150 mM NaCl, 5% glycerol, 0.1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, supplemented with protease inhibitors). Nuclei were sonicated until an average DNA fragment size of 100–500 bp was achieved (assessed by agarose gel electrophoresis). For ∼107 HPr-1AR nuclei, 6 μg of anti-AR antibody (PG-21, Upstate) was used for immunoprecipitation. Immunoprecipitated material was washed 5× at 4°C with RIPA buffer containing 300 mM NaCl and 100 μg/mL yeast tRNA, and resuspended in 80 μL of proteinase K solution (TE, pH 8.0, 0.7% SDS, 200 μg/mL proteinase K). After reversing cross-links, DNA fragments were purified using QIAquick PCR Purification kit (Qiagen).
Ligation-mediated PCR (LMPCR) amplification was performed as described (So et al. 2007) to amplify AR ChIP samples. Using qPCR of numerous control amplicons, we validated that the LMPCR-amplified products yielded levels of AR occupancy similar to unamplified AR ChIP samples. For conventional AR ChIP, qPCR data were normalized to a region 140 bp upstream of the HSPA1A gene, which is not occupied by AR (primer sequences in Supplemental Table 4). Primers for ChIP qPCR were designed as described above.
For the ChIP–chip microarray, ∼104 kb human genomic regions centered on the transcription start sites of 548 candidate hormone-responsive genes were tiled with isothermal 50-mer oligos that were spaced ∼54 bp apart on repeat-masked DNA. Where ∼104-kb regions overlapped, the surrounding genomic regions were tiled further, bidirectionally. Human DNA sequences from NCBI Build 35 (hg17) were retrieved using the UCSC Genome Browser (http://genome.ucsc.edu). NimbleGen Systems (http://www.nimblegen.com) labeled the LMPCR-amplified AR ChIP samples from vehicle and R1881-treated HPr-1AR cells with Cy3 and Cy5, respectively, hybridized labeled samples to the custom microarrays, and measured relative signal intensities. Androgen-enriched peaks were detected by Mpeak (http://www.stat.ucla.edu/~zmdl/mpeak; Kim et al. 2005) using default parameters with the following modifications: threshold prefilter = 2(mean) + 4(SD); local maximum = 350 bp.
Computational analysis
For analysis of enriched motifs, human repeat-masked DNA sequences from NCBI Build 35 (hg17) were retrieved using the UCSC Genome Browser (http://genome.ucsc.edu). BioProspector analysis (Liu et al. 2002) was initially performed using nucleotide widths (w) 12, 14, 16, and 18 on ARBRs to identify ARBSs, and the top motifs were masked to identify other motifs. For MobyDick analysis (Bussemaker et al. 2000), human and human/mouse aligned sequences (UCSC Genome Browser) were used as inputs to identify enriched motifs (So et al. 2007). Similar motifs were clustered and Bonferroni corrected _P_-values of enrichment were determined as described (So et al. 2007) (see Supplemental Materials and Methods). The top BioProspector w14 position weight matrix (PWM) was used to score AREs for putative ARBSs with a false-positive rate of <10%. This upper bound was calculated from randomly sampling unbound regions (Supplemental Fig. 4).
For other motifs with P < 10−3, PWMs were constructed and compared to known binding sites in TRANSFAC professional v.9.3 (Matys et al. 2003). To associate motifs with putative regulatory factor binding sites, we measured the distance between the PWMs and those representing binding sites for known regulatory factors using relative entropy (Kullback-Liebler divergence) with a cutoff <6.0.
Acknowledgments
We thank the members of the Yamamoto lab for discussions and reagents, and Hee Jung Chung, Mark Diamond, Miles Pufall, Nirao Shah, Sebastiaan Meijsing, and Tony Gerber for insightful comments on the manuscript. Cell line HPr-1AR and plasmid pGL4.10-E4TATA were generous gifts from Patrick Ming-Tat Ling (University of Hong Kong) and Yuriy Shostak, respectively. This work was supported by NRSA postdoctoral fellowship F32DK65402 to E.C.B.; predoctoral fellowship 10GB-0104 of the University of California Breast Cancer Research Program to A.Y.S.; NSF predoctoral fellowship to C.C.; Packard Fellowship in Science and Engineering to H.L.; and research grants from NIH (CA101042 to C.M.H., GM070808 to H.L., CA020535 to K.R.Y.).
Footnotes
References
- Belperio J.A., Keane M.P., Arenberg D.A., Addison C.L., Ehlert J.E., Burdick M.D., Strieter R.M., Keane M.P., Arenberg D.A., Addison C.L., Ehlert J.E., Burdick M.D., Strieter R.M., Arenberg D.A., Addison C.L., Ehlert J.E., Burdick M.D., Strieter R.M., Addison C.L., Ehlert J.E., Burdick M.D., Strieter R.M., Ehlert J.E., Burdick M.D., Strieter R.M., Burdick M.D., Strieter R.M., Strieter R.M. CXC chemokines in angiogenesis. J. Leukoc. Biol. 2000;68:1–8. [PubMed] [Google Scholar]
- Bussemaker H.J., Li H., Siggia E.D., Li H., Siggia E.D., Siggia E.D. Building a dictionary for genomes: Identification of presumptive regulatory sites by statistical analysis. Proc. Natl. Acad. Sci. 2000;97:10096–10100. doi: 10.1073/pnas.180265397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J.S., Liu X.S., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Liu X.S., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Shao W., Hestermann E.V., Geistlinger T.R., Hestermann E.V., Geistlinger T.R., Geistlinger T.R., et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Chang C.S., Kokontis J., Liao S.T., Kokontis J., Liao S.T., Liao S.T. Molecular cloning of human and rat complementary DNA encoding androgen receptors. Science. 1988;240:324–326. doi: 10.1126/science.3353726. [DOI] [PubMed] [Google Scholar]
- Chang T.Y., Chang C.C., Lin S., Yu C., Li B.L., Miyazaki A., Chang C.C., Lin S., Yu C., Li B.L., Miyazaki A., Lin S., Yu C., Li B.L., Miyazaki A., Yu C., Li B.L., Miyazaki A., Li B.L., Miyazaki A., Miyazaki A. Roles of acyl-coenzyme A:cholesterol acyltransferase-1 and -2. Curr. Opin. Lipidol. 2001;12:289–296. doi: 10.1097/00041433-200106000-00008. [DOI] [PubMed] [Google Scholar]
- Chen S., Wang J., Yu G., Liu W., Pearce D., Wang J., Yu G., Liu W., Pearce D., Yu G., Liu W., Pearce D., Liu W., Pearce D., Pearce D. Androgen and glucocorticoid receptor heterodimer formation. A possible mechanism for mutual inhibition of transcriptional activity. J. Biol. Chem. 1997;272:14087–14092. doi: 10.1074/jbc.272.22.14087. [DOI] [PubMed] [Google Scholar]
- Chen S.Y., Cai C., Fisher C.J., Zheng Z., Omwancha J., Hsieh C.L., Shemshedini L., Cai C., Fisher C.J., Zheng Z., Omwancha J., Hsieh C.L., Shemshedini L., Fisher C.J., Zheng Z., Omwancha J., Hsieh C.L., Shemshedini L., Zheng Z., Omwancha J., Hsieh C.L., Shemshedini L., Omwancha J., Hsieh C.L., Shemshedini L., Hsieh C.L., Shemshedini L., Shemshedini L. c-Jun enhancement of androgen receptor transactivation is associated with prostate cancer cell proliferation. Oncogene. 2006;25:7212–7223. doi: 10.1038/sj.onc.1209705. [DOI] [PubMed] [Google Scholar]
- Chuang K.H., Lee Y.F., Lin W.J., Chu C.Y., Altuwaijri S., Wan Y.J., Chang C., Lee Y.F., Lin W.J., Chu C.Y., Altuwaijri S., Wan Y.J., Chang C., Lin W.J., Chu C.Y., Altuwaijri S., Wan Y.J., Chang C., Chu C.Y., Altuwaijri S., Wan Y.J., Chang C., Altuwaijri S., Wan Y.J., Chang C., Wan Y.J., Chang C., Chang C. 9-cis-retinoic acid inhibits androgen receptor activity through activation of retinoid X receptor. Mol. Endocrinol. 2005;19:1200–1212. doi: 10.1210/me.2004-0181. [DOI] [PubMed] [Google Scholar]
- Claessens F., Verrijdt G., Schoenmakers E., Haelens A., Peeters B., Verhoeven G., Rombauts W., Verrijdt G., Schoenmakers E., Haelens A., Peeters B., Verhoeven G., Rombauts W., Schoenmakers E., Haelens A., Peeters B., Verhoeven G., Rombauts W., Haelens A., Peeters B., Verhoeven G., Rombauts W., Peeters B., Verhoeven G., Rombauts W., Verhoeven G., Rombauts W., Rombauts W. Selective DNA binding by the androgen receptor as a mechanism for hormone-specific gene regulation. J. Steroid Biochem. Mol. Biol. 2001;76:23–30. doi: 10.1016/s0960-0760(00)00154-0. [DOI] [PubMed] [Google Scholar]
- Cleutjens K.B., van Eekelen C.C., van der Korput H.A., Brinkmann A.O., Trapman J., van Eekelen C.C., van der Korput H.A., Brinkmann A.O., Trapman J., van der Korput H.A., Brinkmann A.O., Trapman J., Brinkmann A.O., Trapman J., Trapman J. Two androgen response regions cooperate in steroid hormone regulated activity of the prostate-specific antigen promoter. J. Biol. Chem. 1996;271:6379–6388. doi: 10.1074/jbc.271.11.6379. [DOI] [PubMed] [Google Scholar]
- Crossley M., Ludwig M., Stowell K.M., De Vos P., Olek K., Brownlee G.G., Ludwig M., Stowell K.M., De Vos P., Olek K., Brownlee G.G., Stowell K.M., De Vos P., Olek K., Brownlee G.G., De Vos P., Olek K., Brownlee G.G., Olek K., Brownlee G.G., Brownlee G.G. Recovery from hemophilia B Leyden: An androgen-responsive element in the factor IX promoter. Science. 1992;257:377–379. doi: 10.1126/science.1631558. [DOI] [PubMed] [Google Scholar]
- Dennis G., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Gao W., Lane H.C., Lempicki R.A., Lane H.C., Lempicki R.A., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. doi: 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- DePrimo S.E., Diehn M., Nelson J.B., Reiter R.E., Matese J., Fero M., Tibshirani R., Brown P.O., Brooks J.D., Diehn M., Nelson J.B., Reiter R.E., Matese J., Fero M., Tibshirani R., Brown P.O., Brooks J.D., Nelson J.B., Reiter R.E., Matese J., Fero M., Tibshirani R., Brown P.O., Brooks J.D., Reiter R.E., Matese J., Fero M., Tibshirani R., Brown P.O., Brooks J.D., Matese J., Fero M., Tibshirani R., Brown P.O., Brooks J.D., Fero M., Tibshirani R., Brown P.O., Brooks J.D., Tibshirani R., Brown P.O., Brooks J.D., Brown P.O., Brooks J.D., Brooks J.D. Transcriptional programs activated by exposure of human prostate cancer cells to androgen. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0032 . RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLeone R.J., Marcus G.A., Johnson M.D., Kingsley D.M., Marcus G.A., Johnson M.D., Kingsley D.M., Johnson M.D., Kingsley D.M., Kingsley D.M. Efficient studies of long-distance Bmp5 gene regulation using bacterial artificial chromosomes. Proc. Natl. Acad. Sci. 2000;97:1612–1617. doi: 10.1073/pnas.97.4.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E., Verzi M.P., Anderson J.P., Xu S.M., Black B.L., Verzi M.P., Anderson J.P., Xu S.M., Black B.L., Anderson J.P., Xu S.M., Black B.L., Xu S.M., Black B.L., Black B.L. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- Dong X.Y., Chen C., Sun X., Guo P., Vessella R.L., Wang R.X., Chung L.W., Zhou W., Dong J.T., Chen C., Sun X., Guo P., Vessella R.L., Wang R.X., Chung L.W., Zhou W., Dong J.T., Sun X., Guo P., Vessella R.L., Wang R.X., Chung L.W., Zhou W., Dong J.T., Guo P., Vessella R.L., Wang R.X., Chung L.W., Zhou W., Dong J.T., Vessella R.L., Wang R.X., Chung L.W., Zhou W., Dong J.T., Wang R.X., Chung L.W., Zhou W., Dong J.T., Chung L.W., Zhou W., Dong J.T., Zhou W., Dong J.T., Dong J.T. FOXO1A is a candidate for the 13q14 tumor suppressor gene inhibiting androgen receptor signaling in prostate cancer. Cancer Res. 2006;66:6998–7006. doi: 10.1158/0008-5472.CAN-06-0411. [DOI] [PubMed] [Google Scholar]
- Euskirchen G., Royce T.E., Bertone P., Martone R., Rinn J.L., Nelson F.K., Sayward F., Luscombe N.M., Miller P., Gerstein M., Royce T.E., Bertone P., Martone R., Rinn J.L., Nelson F.K., Sayward F., Luscombe N.M., Miller P., Gerstein M., Bertone P., Martone R., Rinn J.L., Nelson F.K., Sayward F., Luscombe N.M., Miller P., Gerstein M., Martone R., Rinn J.L., Nelson F.K., Sayward F., Luscombe N.M., Miller P., Gerstein M., Rinn J.L., Nelson F.K., Sayward F., Luscombe N.M., Miller P., Gerstein M., Nelson F.K., Sayward F., Luscombe N.M., Miller P., Gerstein M., Sayward F., Luscombe N.M., Miller P., Gerstein M., Luscombe N.M., Miller P., Gerstein M., Miller P., Gerstein M., Gerstein M., et al. CREB binds to multiple loci on human chromosome 22. Mol. Cell. Biol. 2004;24:3804–3814. doi: 10.1128/MCB.24.9.3804-3814.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund A., Jolivel V., Durand S., Kersual N., Chalbos D., Chavey C., Vignon F., Lazennec G., Jolivel V., Durand S., Kersual N., Chalbos D., Chavey C., Vignon F., Lazennec G., Durand S., Kersual N., Chalbos D., Chavey C., Vignon F., Lazennec G., Kersual N., Chalbos D., Chavey C., Vignon F., Lazennec G., Chalbos D., Chavey C., Vignon F., Lazennec G., Chavey C., Vignon F., Lazennec G., Vignon F., Lazennec G., Lazennec G. Mechanisms underlying differential expression of interleukin-8 in breast cancer cells. Oncogene. 2004;23:6105–6114. doi: 10.1038/sj.onc.1207815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzinger D.G. Gene quantification using real-time quantitative PCR: An emerging technology hits the mainstream. Exp. Hematol. 2002;30:503–512. doi: 10.1016/s0301-472x(02)00806-8. [DOI] [PubMed] [Google Scholar]
- Giri D., Ozen M., Ittmann M., Ozen M., Ittmann M., Ittmann M. Interleukin-6 is an autocrine growth factor in human prostate cancer. Am. J. Pathol. 2001;159:2159–2165. doi: 10.1016/S0002-9440(10)63067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder N., Klein R., Klein R. Eph receptors and ephrins: Effectors of morphogenesis. Development. 1999;126:2033–2044. doi: 10.1242/dev.126.10.2033. [DOI] [PubMed] [Google Scholar]
- Itani O.A., Liu K.Z., Cornish K.L., Campbell J.R., Thomas C.P., Liu K.Z., Cornish K.L., Campbell J.R., Thomas C.P., Cornish K.L., Campbell J.R., Thomas C.P., Campbell J.R., Thomas C.P., Thomas C.P. Glucocorticoids stimulate human sgk1 gene expression by activation of a GRE in its 5′-flanking region. Am. J. Physiol. Endocrinol. Metab. 2002;283:E971–E979. doi: 10.1152/ajpendo.00021.2002.. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Richmond T.A., Wu Y., Green R.D., Ren B., Wu Y., Green R.D., Ren B., Green R.D., Ren B., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochupurakkal B.S., Harari D., Di-Segni A., Maik-Rachline G., Lyass L., Gur G., Kerber G., Citri A., Lavi S., Eilam R., Harari D., Di-Segni A., Maik-Rachline G., Lyass L., Gur G., Kerber G., Citri A., Lavi S., Eilam R., Di-Segni A., Maik-Rachline G., Lyass L., Gur G., Kerber G., Citri A., Lavi S., Eilam R., Maik-Rachline G., Lyass L., Gur G., Kerber G., Citri A., Lavi S., Eilam R., Lyass L., Gur G., Kerber G., Citri A., Lavi S., Eilam R., Gur G., Kerber G., Citri A., Lavi S., Eilam R., Kerber G., Citri A., Lavi S., Eilam R., Citri A., Lavi S., Eilam R., Lavi S., Eilam R., Eilam R., et al. Epigen, the last ligand of ErbB receptors, reveals intricate relationships between affinity and mitogenicity. J. Biol. Chem. 2005;280:8503–8512. doi: 10.1074/jbc.M413919200. [DOI] [PubMed] [Google Scholar]
- Kondo K.H., Kai M.H., Setoguchi Y., Eggertsen G., Sjoblom P., Setoguchi T., Okuda K.I., Bjorkhem I., Kai M.H., Setoguchi Y., Eggertsen G., Sjoblom P., Setoguchi T., Okuda K.I., Bjorkhem I., Setoguchi Y., Eggertsen G., Sjoblom P., Setoguchi T., Okuda K.I., Bjorkhem I., Eggertsen G., Sjoblom P., Setoguchi T., Okuda K.I., Bjorkhem I., Sjoblom P., Setoguchi T., Okuda K.I., Bjorkhem I., Setoguchi T., Okuda K.I., Bjorkhem I., Okuda K.I., Bjorkhem I., Bjorkhem I.1994. . Cloning and expression of cDNA of human Δ 4-3-oxosteroid 5 β-reductase and substrate specificity of the expressed enzyme Eur. J. Biochem. 219357–363. [DOI] [PubMed] [Google Scholar]
- Lapointe J., Li C., Higgins J.P., de van Rijn M., Bair E., Montgomery K., Ferrari M., Egevad L., Rayford W., Bergerheim U., Li C., Higgins J.P., de van Rijn M., Bair E., Montgomery K., Ferrari M., Egevad L., Rayford W., Bergerheim U., Higgins J.P., de van Rijn M., Bair E., Montgomery K., Ferrari M., Egevad L., Rayford W., Bergerheim U., de van Rijn M., Bair E., Montgomery K., Ferrari M., Egevad L., Rayford W., Bergerheim U., Bair E., Montgomery K., Ferrari M., Egevad L., Rayford W., Bergerheim U., Montgomery K., Ferrari M., Egevad L., Rayford W., Bergerheim U., Ferrari M., Egevad L., Rayford W., Bergerheim U., Egevad L., Rayford W., Bergerheim U., Rayford W., Bergerheim U., Bergerheim U., et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc. Natl. Acad. Sci. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaTulippe E., Satagopan J., Smith A., Scher H., Scardino P., Reuter V., Gerald W.L., Satagopan J., Smith A., Scher H., Scardino P., Reuter V., Gerald W.L., Smith A., Scher H., Scardino P., Reuter V., Gerald W.L., Scher H., Scardino P., Reuter V., Gerald W.L., Scardino P., Reuter V., Gerald W.L., Reuter V., Gerald W.L., Gerald W.L. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- Lawrence J.G. Gene organization: Selection, selfishness, and serendipity. Annu. Rev. Microbiol. 2003;57:419–440. doi: 10.1146/annurev.micro.57.030502.090816. [DOI] [PubMed] [Google Scholar]
- Lee Y.F., Shyr C.R., Thin T.H., Lin W.J., Chang C., Shyr C.R., Thin T.H., Lin W.J., Chang C., Thin T.H., Lin W.J., Chang C., Lin W.J., Chang C., Chang C. Convergence of two repressors through heterodimer formation of androgen receptor and testicular orphan receptor-4: A unique signaling pathway in the steroid receptor superfamily. Proc. Natl. Acad. Sci. 1999;96:14724–14729. doi: 10.1073/pnas.96.26.14724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefstin J.A., Yamamoto K.R., Yamamoto K.R. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- Lehto M., Olkkonen V.M., Olkkonen V.M. The OSBP-related proteins: A novel protein family involved in vesicle transport, cellular lipid metabolism, and cell signalling. Biochim. Biophys. Acta. 2003;1631:1–11. doi: 10.1016/s1388-1981(02)00364-5. [DOI] [PubMed] [Google Scholar]
- Lin Y.S., Carey M.F., Ptashne M., Green M.R., Carey M.F., Ptashne M., Green M.R., Ptashne M., Green M.R., Green M.R. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988;54:659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- Ling M.T., Chan K.W., Choo C.K., Chan K.W., Choo C.K., Choo C.K. Androgen induces differentiation of a human papillomavirus 16 E6/E7 immortalized prostate epithelial cell line. J. Endocrinol. 2001;170:287–296. doi: 10.1677/joe.0.1700287. [DOI] [PubMed] [Google Scholar]
- Liu X.S., Brutlag D.L., Liu J.S., Brutlag D.L., Liu J.S., Liu J.S. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- Loreni F., Stavenhagen J., Kalff M., Robins D.M., Stavenhagen J., Kalff M., Robins D.M., Kalff M., Robins D.M., Robins D.M. A complex androgen-responsive enhancer resides 2 kilobases upstream of the mouse Slp gene. Mol. Cell. Biol. 1988;8:2350–2360. doi: 10.1128/mcb.8.6.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubahn D.B., Joseph D.R., Sullivan P.M., Willard H.F., French F.S., Wilson E.M., Joseph D.R., Sullivan P.M., Willard H.F., French F.S., Wilson E.M., Sullivan P.M., Willard H.F., French F.S., Wilson E.M., Willard H.F., French F.S., Wilson E.M., French F.S., Wilson E.M., Wilson E.M. Cloning of human androgen receptor complementary DNA and localization to the X chromosome. Science. 1988;240:327–330. doi: 10.1126/science.3353727. [DOI] [PubMed] [Google Scholar]
- Magee J.A., Chang L.W., Stormo G.D., Milbrandt J., Chang L.W., Stormo G.D., Milbrandt J., Stormo G.D., Milbrandt J., Milbrandt J. Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology. 2006;147:590–598. doi: 10.1210/en.2005-1001. [DOI] [PubMed] [Google Scholar]
- Martone R., Euskirchen G., Bertone P., Hartman S., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Euskirchen G., Bertone P., Hartman S., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Bertone P., Hartman S., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Hartman S., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Royce T.E., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Luscombe N.M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Rinn J.L., Nelson F.K., Miller P., Gerstein M., Nelson F.K., Miller P., Gerstein M., Miller P., Gerstein M., Gerstein M., et al. Distribution of NF-κB-binding sites across human chromosome 22. Proc. Natl. Acad. Sci. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Karas D., Kel A.E., Kel-Margoulis O.V., Kel A.E., Kel-Margoulis O.V., Kel-Margoulis O.V., et al. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E., Wissmann M., Yin N., Muller J.M., Schneider R., Peters A.H., Gunther T., Buettner R., Schule R., Wissmann M., Yin N., Muller J.M., Schneider R., Peters A.H., Gunther T., Buettner R., Schule R., Yin N., Muller J.M., Schneider R., Peters A.H., Gunther T., Buettner R., Schule R., Muller J.M., Schneider R., Peters A.H., Gunther T., Buettner R., Schule R., Schneider R., Peters A.H., Gunther T., Buettner R., Schule R., Peters A.H., Gunther T., Buettner R., Schule R., Gunther T., Buettner R., Schule R., Buettner R., Schule R., Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Murtha P., Tindall D.J., Young C.Y., Tindall D.J., Young C.Y., Young C.Y. Androgen induction of a human prostate-specific kallikrein, hKLK2: Characterization of an androgen response element in the 5′ promoter region of the gene. Biochemistry. 1993;32:6459–6464. doi: 10.1021/bi00076a020. [DOI] [PubMed] [Google Scholar]
- Murtha P.E., Zhu W., Zhang J., Zhang S., Young C.Y., Zhu W., Zhang J., Zhang S., Young C.Y., Zhang J., Zhang S., Young C.Y., Zhang S., Young C.Y., Young C.Y. Effects of Ca2+ mobilization on expression of androgen-regulated genes: Interference with androgen receptor-mediated transactivation by AP-I proteins. Prostate. 1997;33:264–270. doi: 10.1002/(sici)1097-0045(19971201)33:4<264::aid-pros7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Nelson P.S., Clegg N., Arnold H., Ferguson C., Bonham M., White J., Hood L., Lin B., Clegg N., Arnold H., Ferguson C., Bonham M., White J., Hood L., Lin B., Arnold H., Ferguson C., Bonham M., White J., Hood L., Lin B., Ferguson C., Bonham M., White J., Hood L., Lin B., Bonham M., White J., Hood L., Lin B., White J., Hood L., Lin B., Hood L., Lin B., Lin B. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc. Natl. Acad. Sci. 2002;99:11890–11895. doi: 10.1073/pnas.182376299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier L.M., Chambliss K.L., Gibson K.M., Krisans S.K., Chambliss K.L., Gibson K.M., Krisans S.K., Gibson K.M., Krisans S.K., Krisans S.K. Characterization of phosphomevalonate kinase: Chromosomal localization, regulation, and subcellular targeting. J. Lipid Res. 1999;40:672–679. [PubMed] [Google Scholar]
- Panet-Raymond V., Gottlieb B., Beitel L.K., Pinsky L., Trifiro M.A., Gottlieb B., Beitel L.K., Pinsky L., Trifiro M.A., Beitel L.K., Pinsky L., Trifiro M.A., Pinsky L., Trifiro M.A., Trifiro M.A. Interactions between androgen and estrogen receptors and the effects on their transactivational properties. Mol. Cell. Endocrinol. 2000;167:139–150. doi: 10.1016/s0303-7207(00)00279-3. [DOI] [PubMed] [Google Scholar]
- Phuc Le P., Friedman J.R., Schug J., Brestelli J.E., Parker J.B., Bochkis I.M., Kaestner K.H., Friedman J.R., Schug J., Brestelli J.E., Parker J.B., Bochkis I.M., Kaestner K.H., Schug J., Brestelli J.E., Parker J.B., Bochkis I.M., Kaestner K.H., Brestelli J.E., Parker J.B., Bochkis I.M., Kaestner K.H., Parker J.B., Bochkis I.M., Kaestner K.H., Bochkis I.M., Kaestner K.H., Kaestner K.H. Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genet. 2005;1:e16. doi: 10.1371/journal.pgen.0010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie P.S., Bruchovsky N., Leco K.J., Sheppard P.C., McQueen S.A., Cheng H., Snoek R., Hamel A., Bock M.E., MacDonald B.S., Bruchovsky N., Leco K.J., Sheppard P.C., McQueen S.A., Cheng H., Snoek R., Hamel A., Bock M.E., MacDonald B.S., Leco K.J., Sheppard P.C., McQueen S.A., Cheng H., Snoek R., Hamel A., Bock M.E., MacDonald B.S., Sheppard P.C., McQueen S.A., Cheng H., Snoek R., Hamel A., Bock M.E., MacDonald B.S., McQueen S.A., Cheng H., Snoek R., Hamel A., Bock M.E., MacDonald B.S., Cheng H., Snoek R., Hamel A., Bock M.E., MacDonald B.S., Snoek R., Hamel A., Bock M.E., MacDonald B.S., Hamel A., Bock M.E., MacDonald B.S., Bock M.E., MacDonald B.S., MacDonald B.S., et al. Characterization of two cis-acting DNA elements involved in the androgen regulation of the probasin gene. Mol. Endocrinol. 1993;7:23–36. doi: 10.1210/mend.7.1.8446105. [DOI] [PubMed] [Google Scholar]
- Rogatsky I., Wang J.C., Derynck M.K., Nonaka D.F., Khodabakhsh D.B., Haqq C.M., Darimont B.D., Garabedian M.J., Yamamoto K.R., Wang J.C., Derynck M.K., Nonaka D.F., Khodabakhsh D.B., Haqq C.M., Darimont B.D., Garabedian M.J., Yamamoto K.R., Derynck M.K., Nonaka D.F., Khodabakhsh D.B., Haqq C.M., Darimont B.D., Garabedian M.J., Yamamoto K.R., Nonaka D.F., Khodabakhsh D.B., Haqq C.M., Darimont B.D., Garabedian M.J., Yamamoto K.R., Khodabakhsh D.B., Haqq C.M., Darimont B.D., Garabedian M.J., Yamamoto K.R., Haqq C.M., Darimont B.D., Garabedian M.J., Yamamoto K.R., Darimont B.D., Garabedian M.J., Yamamoto K.R., Garabedian M.J., Yamamoto K.R., Yamamoto K.R. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc. Natl. Acad. Sci. 2003;100:13845–13850. doi: 10.1073/pnas.2336092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Sadar M.D., Bruchovsky N., Saatcioglu F., Rennie P.S., Sato S., Lange P.H., Gleave M.E., Sadar M.D., Bruchovsky N., Saatcioglu F., Rennie P.S., Sato S., Lange P.H., Gleave M.E., Bruchovsky N., Saatcioglu F., Rennie P.S., Sato S., Lange P.H., Gleave M.E., Saatcioglu F., Rennie P.S., Sato S., Lange P.H., Gleave M.E., Rennie P.S., Sato S., Lange P.H., Gleave M.E., Sato S., Lange P.H., Gleave M.E., Lange P.H., Gleave M.E., Gleave M.E. Androgenic induction of prostate-specific antigen gene is repressed by protein-protein interaction between the androgen receptor and AP-1/c-Jun in the human prostate cancer cell line LNCaP. J. Biol. Chem. 1997;272:17485–17494. doi: 10.1074/jbc.272.28.17485. [DOI] [PubMed] [Google Scholar]
- Shang Y., Myers M., Brown M., Myers M., Brown M., Brown M. Formation of the androgen receptor transcription complex. Mol. Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- Shibata N., Arita M., Misaki Y., Dohmae N., Takio K., Ono T., Inoue K., Arai H., Arita M., Misaki Y., Dohmae N., Takio K., Ono T., Inoue K., Arai H., Misaki Y., Dohmae N., Takio K., Ono T., Inoue K., Arai H., Dohmae N., Takio K., Ono T., Inoue K., Arai H., Takio K., Ono T., Inoue K., Arai H., Ono T., Inoue K., Arai H., Inoue K., Arai H., Arai H. Supernatant protein factor, which stimulates the conversion of squalene to lanosterol, is a cytosolic squalene transfer protein and enhances cholesterol biosynthesis. Proc. Natl. Acad. Sci. 2001;98:2244–2249. doi: 10.1073/pnas.041620398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So A.Y., Chaivorapol C., Bolton E.C., Li H., Yamamoto K.R., Chaivorapol C., Bolton E.C., Li H., Yamamoto K.R., Bolton E.C., Li H., Yamamoto K.R., Li H., Yamamoto K.R., Yamamoto K.R. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steketee K., Ziel-van der Made A.C., van der Korput H.A., Houtsmuller A.B., Trapman J., Ziel-van der Made A.C., van der Korput H.A., Houtsmuller A.B., Trapman J., van der Korput H.A., Houtsmuller A.B., Trapman J., Houtsmuller A.B., Trapman J., Trapman J. A bioinformatics-based functional analysis shows that the specifically androgen-regulated gene SARG contains an active direct repeat androgen response element in the first intron. J. Mol. Endocrinol. 2004;33:477–491. doi: 10.1677/jme.1.01478. [DOI] [PubMed] [Google Scholar]
- Takayama K., Kaneshiro K., Tsutsumi S., Horie-Inoue K., Ikeda K., Urano T., Ijichi N., Ouchi Y., Shirahige K., Aburatani H., Kaneshiro K., Tsutsumi S., Horie-Inoue K., Ikeda K., Urano T., Ijichi N., Ouchi Y., Shirahige K., Aburatani H., Tsutsumi S., Horie-Inoue K., Ikeda K., Urano T., Ijichi N., Ouchi Y., Shirahige K., Aburatani H., Horie-Inoue K., Ikeda K., Urano T., Ijichi N., Ouchi Y., Shirahige K., Aburatani H., Ikeda K., Urano T., Ijichi N., Ouchi Y., Shirahige K., Aburatani H., Urano T., Ijichi N., Ouchi Y., Shirahige K., Aburatani H., Ijichi N., Ouchi Y., Shirahige K., Aburatani H., Ouchi Y., Shirahige K., Aburatani H., Shirahige K., Aburatani H., Aburatani H., et al. Identification of novel androgen response genes in prostate cancer cells by coupling chromatin immunoprecipitation and genomic microarray analysis. Oncogene. 2007;26:4453–4463. doi: 10.1038/sj.onc.1210229. [DOI] [PubMed] [Google Scholar]
- Tiniakos D.G., Mitropoulos D., Kyroudi-Voulgari A., Soura K., Kittas C., Mitropoulos D., Kyroudi-Voulgari A., Soura K., Kittas C., Kyroudi-Voulgari A., Soura K., Kittas C., Soura K., Kittas C., Kittas C. Expression of c-jun oncogene in hyperplastic and carcinomatous human prostate. Urology. 2006;67:204–208. doi: 10.1016/j.urology.2005.07.045. [DOI] [PubMed] [Google Scholar]
- Wang J.C., Derynck M.K., Nonaka D.F., Khodabakhsh D.B., Haqq C., Yamamoto K.R., Derynck M.K., Nonaka D.F., Khodabakhsh D.B., Haqq C., Yamamoto K.R., Nonaka D.F., Khodabakhsh D.B., Haqq C., Yamamoto K.R., Khodabakhsh D.B., Haqq C., Yamamoto K.R., Haqq C., Yamamoto K.R., Yamamoto K.R. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc. Natl. Acad. Sci. 2004;101:15603–15608. doi: 10.1073/pnas.0407008101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Carroll J.S., Brown M., Carroll J.S., Brown M., Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Waterham H.R. Defects of cholesterol biosynthesis. FEBS Lett. 2006;580:5442–5449. doi: 10.1016/j.febslet.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Weinmann A.S., Yan P.S., Oberley M.J., Huang T.H., Farnham P.J., Yan P.S., Oberley M.J., Huang T.H., Farnham P.J., Oberley M.J., Huang T.H., Farnham P.J., Huang T.H., Farnham P.J., Farnham P.J. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes & Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y., Tempst P., Wong J., Zhang Y., Wong J., Zhang Y., Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]