A cell cycle regulatory network controlling NF-κB subunit activity and function (original) (raw)
Abstract
Aberrantly active NF-κB complexes can contribute to tumorigenesis by regulating genes that promote the growth and survival of cancer cells. We have investigated NF-κB during the cell cycle and find that its ability to regulate the G1-phase expression of key proto-oncogenes is subject to regulation by the integrated activity of IκB kinase (IKK)α, IKKβ, Akt and Chk1. The coordinated binding of NF-κB subunits to the Cyclin D1, c-Myc and Skp2 promoters is dynamic with distinct changes in promoter occupancy and RelA(p65) phosphorylation occurring through G1, S and G2 phases, concomitant with a switch from coactivator to corepressor recruitment. Akt activity is required for IKK-dependent phosphorylation of NF-κB subunits in G1 and G2 phases, where Chk1 is inactive. However, in S-phase, Akt is inactivated, while Chk1 phosphorylates RelA and associates with IKKα, inhibiting the processing of the p100 (NF-κB2) subunit, which also plays a critical role in the regulation of these genes. These data reveal a complex regulatory network integrating NF-κB with the DNA-replication checkpoint and the expression of critical regulators of cell proliferation.
Keywords: cell cycle, IKK, NF-κB, transcription
Introduction
The tumor-promoting role of the classical NF-κB pathway, through IκB kinase (IKK)β-dependent phosphorylation of IκBα and activation of p50/RelA (p65) complexes, is now well established (Kim et al, 2006). However, a role for the noncanonical, p52/p100 (NF-κB2)-dependent pathway in the regulation of cell proliferation and tumorigenesis is also apparent. In mice, for example, overexpression of p52 resulting from deletion of the C-terminus of p100 leads to gastric hyperplasia and increased lymphocyte proliferation (Ishikawa et al, 1997). Moreover, there are tumor-associated truncations of p100 found in human B- and T-cell lymphomas (Rayet and Gelinas, 1999), some of which have transforming effects in murine fibroblasts (Ciana et al, 1997). Furthermore, deregulated p100 processing is associated with T-cell transformation (Xiao et al, 2001) and constitutive activation of p52 with breast tumors (Cogswell et al, 2000). These observations suggest that the p52 NF-κB subunit can function as a regulator of cell proliferation and its aberrant activity could promote cell cycle progression and ultimately tumorigenesis.
Cyclin D1 expression and assembly with CDK4/6 represents one of the key mitogen-regulated events during the G1 phase of the cell cycle. Cyclin D1 represents a unique component of the cell cycle apparatus, with a peak of expression in mid G1 phase (Tashiro et al, 2007). Unlike other Cyclins that are periodically induced during cell cycle progression, the level of Cyclin D1 is controlled by the extracellular mitogenic environment. For this reason, Cyclin D1 is believed to serve as link between extracellular environment and the core cell cycle machinery. Moreover, Cyclin D1 overexpression is frequently associated with human malignancy (Tashiro et al, 2007). Many reports have described a role for NF-κB subunits and IκB kinases (IKK) in the control of Cyclin D1 expression (Guttridge et al, 1999; Hinz et al, 1999; Joyce et al, 2001; Rocha et al, 2003; Demicco et al, 2005; Eddy et al, 2005; Ouyang et al, 2005; Schumm et al, 2006), although NF-κB-independent mechanisms also exist (Bromberg et al, 1998; Watanabe et al, 1998; Albanese et al, 1999; Sabbah et al, 1999). These have described multiple effects, although since many earlier reports were based on transiently transfected reporter plasmids and overexpression, the exact role of each NF-κB subunit in different cell types remains uncertain. However, it has become clear that in some cell types, the p52 (NF-κB2) NF-κB subunit is an important regulator of Cyclin D1 expression (Guttridge et al, 1999; Hinz et al, 1999; Westerheide et al, 2001; Rocha et al, 2003; Schumm et al, 2006). In particular, our laboratory has recently shown that in U2OS human osteosarcoma cells, Cyclin D1 expression is p52 dependent and that endogenous p52 binds the Cyclin D1 promoter (Schumm et al, 2006). We have also found that p53 activation can result in a change in p52 function, where its association with the Bcl-3 coactivator is replaced by an interaction with HDAC1, resulting in repression of the Cyclin D1 promoter (Rocha et al, 2003). In addition, while p52/Bcl-3 complexes can directly bind and activate the Cyclin D1 promoter in keratinocytes (Massoumi et al, 2006; Zhang et al, 2007), p52 also represses the Cyclin D1 promoter in murine thymocytes (Vacca et al, 2006).
Other important regulators of cell proliferation are also targets of NF-κB. A RelB/p52 complex has recently been shown to control transcription of the skp2 gene (S-phase kinase-associated protein 2) (Schneider et al, 2006). Skp2 promotes the degradation of CDKI p27, thus promoting Cyclin E/CDK2 activation and cell cycle progression. In this study, Skp2 expression was shown to depend on IKKα, which regulates the noncanonical NF-κB pathway by phosphorylating the p100 NF-κB subunit and inducing its processing to p52. The RelB/p52 complex has also been described as an activator of c-Myc expression during mammary gland development and carcinogenesis (Demicco et al, 2005). Two functional NF-κB elements have been identified on the promoter of c-Myc and it has been shown that p50, p52 and RelA can also bind this promoter to regulate its expression (La Rosa et al, 1994; Kanda et al, 2000). Regulation of Cyclin D1, c-Myc and Skp2 likely contribute to the overall role of NF-κB as a regulator of cell survival, angiogenesis, metastasis and proliferation in cancer cells (Kim et al, 2006).
In this report, we provide a detailed analysis of cell cycle regulation of p52 and the other NF-κB subunits. These studies reveal that in some cell types, there is dynamic and coordinated recruitment of NF-κB subunits to the Cyclin D1, c-Myc and Skp2 promoters at different stages of the cell cycle, with distinct G1-phase ‘activator' and G2-phase ‘repressor' complexes being observed. Moreover, we demonstrate that the integrated activity of the Akt and Chk1 kinases control NF-κB subunit phosphorylation and function across the cell cycle, suggesting a link between NF-κB activity and the DNA-replication checkpoint.
Results
p100 processing to p52 is inhibited in S phase
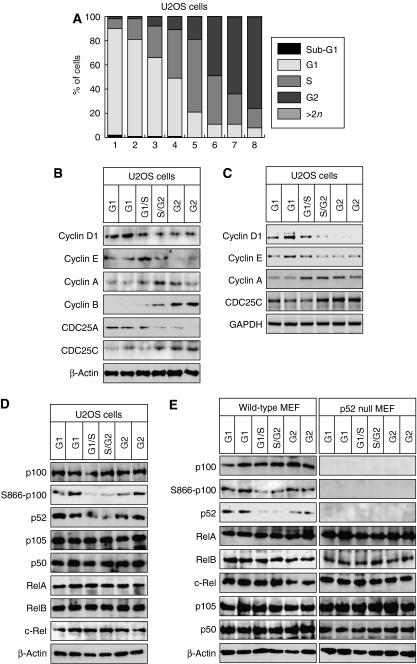
Previously we have shown that the p52 NF-κB subunit regulates cell proliferation in U2OS human osteosarcoma and other cell lines (Schumm et al, 2006). These data suggested that p100/p52 and other NF-κB subunits might themselves be subject to regulation across the cell cycle. To investigate this hypothesis, we used centrifugal elutriation, a drug and starvation-stress free system, to separate cells on the basis of their differential sedimentation rate, allowing enrichment of cell populations at each cell cycle stage. Relative to fluorescent-activated cell sorting (FACS), elutriation allows significant quantities of cells to be isolated, allowing more in-depth analysis.
Fractions of U2OS cells and immortalized mouse embryonic fibroblasts (MEFs) that had been subjected to elutriation were analyzed either by propidium iodide staining and FACS analysis or for enrichment of key cell cycle indicators, such as Cyclins (Figure 1A–C, Supplementary Figure 1A–C). Highly enriched fractions of cells in all phases of the cell cycle could be readily obtained, although S-phase-enriched fractions were less pure than those containing G1- and G2-phase cells. Notably, the expression of Cyclin D1 and Cyclin B1 showed contrasting expression profiles, being preferentially found in G1- and G2-phase-enriched fractions, respectively. Surprisingly, western blotting of whole-cell extracts from each cell cycle stage revealed a significant decrease in p100 processing to p52 in the S-phase-enriched fractions of both U2OS cells and MEFs (Figure 1D and E, Supplementary Figure 1D). This correlated with an inhibition of p100 phosphorylation at serine 866, one of the sites known to induce this processing event. By contrast, no significant changes could be seen in the expression levels of any other NF-κB subunits (Figure 1D and E) or the p52 coactivator Bcl-3 (Supplementary Figure 1E). This effect was also observed in MCF7 cells but not in HUT78 cells, where the NFKB2 (p100) gene is truncated, leading to constitutive p52 production (Thakur et al, 1994) (Supplementary Figure 2).
Figure 1.
p100 processing to p52 is inhibited in S phase. (A) U2OS cells were fractionated by centrifugal elutriation and stained with propidium iodide for analysis by FACS. A representative distribution of cells at different cell cycle stages is shown. Fractions designated G2 will contain some cells undergoing mitosis. However, since only adherent cells were harvested, most mitotic cells are lost. The enrichment of S-phase fractions may be underestimated as cells in early and late S phase may be scored as G1 or G2 phases, respectively. Cell cycle analysis of elutriated fraction used in all experiments can be found in Supplementary Figure 9. (B) Fractionated U2OS whole-cell protein extracts were pooled according to cell cycle stage and subjected to western blot analysis using antibodies to Cyclin D1, E, A and B1, CDC25A and CDC25C. (C) RT–PCR analysis was performed using primers specific to Cyclin D1, E, A, CDC25C and GAPDH control, using total RNA prepared from U2OS cells following centrifugal elutriation. (D, E) Whole-cell extracts prepared from U2OS (D) or w/t and p52/100-null MEF (E) from the indicated cell cycle stages were subjected to western blot analysis using antibodies to the indicated NF-κB subunits.
NF-_κ_B subunits regulate the expression of the Cyclin D1, c-Myc and Skp2 genes
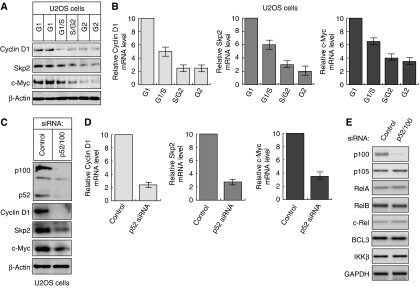
Interestingly, western blot and RNA analysis revealed that in addition to Cyclin D1, c-Myc and Skp2 are also significantly more highly expressed in the G1 phase of U2OS cells (Figure 2A and B) and MEFs (Supplementary Figure 1C). Significantly, knockdown of p52 by siRNA confirmed that p52 is required for high-level expression of Cyclin D1, c-Myc and Skp2 in U2OS cells but does not affect the expression of other NF-κB subunits (Figure 2C–E). This observation is consistent with our previous data concerning p52 regulation of Cyclin D1 in these cells (Rocha et al, 2003; Schumm et al, 2006). Similar results were obtained upon knockdown of the p52 coactivator Bcl3 (Supplementary Figure 1F). Other NF-κB target genes, such as those encoding Bcl-xL, Bcl-2 and A20, were expressed throughout the cell cycle, with increased levels being seen in G2 phase (Supplementary Figure 1E).
Figure 2.
Cell cycle expression of Cyclin D1, c-Myc and Skp2. (A) U2OS whole-cell extracts, from each cell cycle stage, were subjected to western blot analysis using antibodies to Cyclin D1, Skp2 and c-Myc. (B) Cyclin D1, Skp2 and c-Myc are preferentially expressed in G1 phase. Quantitative PCR analysis was performed using primers specific to Cyclin D1, Skp2 and c-Myc and total RNA prepared from U2OS cells following centrifugal elutriation. (C–E) p52 regulates Cyclin D1, Skp2 and c-Myc expression. U2OS cells were transfected with an siRNA targeting p52/p100. Protein (C) and mRNA (D, E) expression were analyzed as shown. In (D), analysis was performed using quantitative, real-time PCR. In these and subsequent experiments, cells were transfected with siRNA on day 1 and harvested on day 4.
This expression profile of c-Myc, Cyclin D1 and Skp2 contrasts with previous data on these factors. Some of these differences might be explained by other studies being performed in highly transformed cells or where techniques such as serum stimulation or chemical synchronization of cells were employed. However, earlier experiments that also analyzed c-Myc expression using elutriation had reported that although serum stimulation of cells did produce a significant increase in c-Myc expression in G1 phase, in an asynchronous population of cycling cells c-Myc levels were evenly distributed across the cell cycle (Hann et al, 1985; Rabbitts et al, 1985; Thompson et al, 1985). To resolve this discrepancy, we first confirmed our original observation using FACS analysis of an asynchronous population of U2OS cells. Again, c-Myc and Cyclin D1 expression was found to be significantly higher in G1-phase cells (Supplementary Figure 2). Moreover, further analysis showed minimal co-expression of Cyclin D1 and c-Myc with Cyclin B1 (not shown). Since we had earlier observed that best results were obtained with early passage U2OS cells, we next analyzed the effect of continuous growth in culture of these cells. Importantly, we found that older U2OS cells showed significantly increased c-Myc, Cyclin D1 and Skp2 expression in later stages of the cell cycle, as judged by both FACS analysis and elutriation (Supplementary Figure 2). To investigate this, we analyzed further cell types. In MCF7 human breast cancer cells, c-Myc expression was found to be similar to that in early passage U2OS cells, while Cyclin D1, and to a lesser extent Skp2, expression was found in all cell cycle stages (Supplementary Figure 2). Cyclin D1 expression in MCF7 cells is not regulated by p52 (Park et al, 2005). By contrast, HUT78 cells showed almost equal expression of c-Myc and Skp2 at all cell cycle stages, but no detectable expression of Cyclin D1 mRNA or protein (Supplementary Figure 2). We therefore conclude that the cell cycle profile of these genes is more variable than previously thought and that other factors, such as cell type, growth conditions and oncogenic mutations, can affect their expression. Nonetheless, early passage U2OS cells and MEFs provide a suitable model system to analyze NF-κB function during the cell cycle.
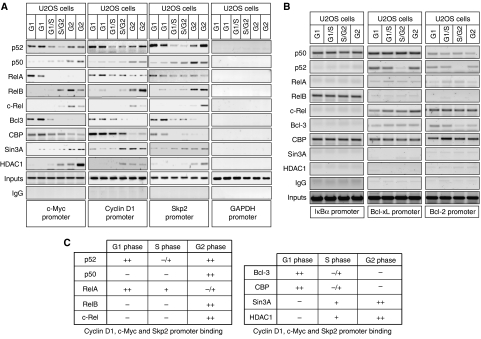
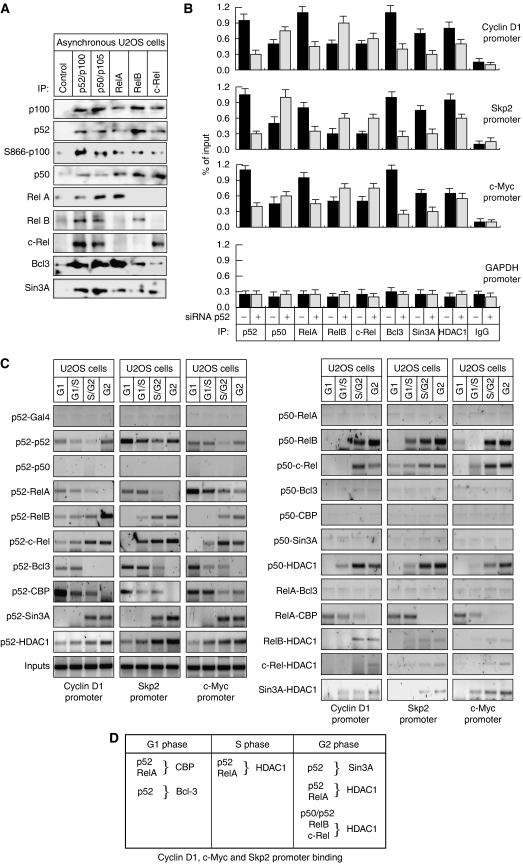
Therefore, to learn more about the recruitment of p52 and other NF-κB subunits to these promoters during the cell cycle of U2OS cells, chromatin immunoprecipitation (ChIP) assays were performed (Figure 3A). These demonstrated a surprisingly complex but similar pattern of NF-κB subunit binding to the Cyclin D1, c-Myc and Skp2 promoters. Consistent with the decrease in p100 processing observed in S-phase (Figure 1D and E), p52 associated most strongly with these promoters in G1 and G2 phases. RelA binding, although present to some extent throughout the cell cycle, was significantly higher during G1 phase (Figure 3A and C, see also Figure 5B and Supplementary Figure 3). By contrast, p50, RelB and c-Rel only significantly interacted with the Cyclin D1, c-Myc and Skp2 promoters in G2 phase, when expression of these genes is reduced (Figure 3A) (Supplementary Figure 3). Also correlating with the expression patterns of these genes, the coactivator proteins Bcl-3 and CBP bound these promoters in G1 phase, while the corepressor proteins Sin3A and HDAC1 bound during G2 phase. Analysis of these ChIP samples using quantitative PCR is shown in Supplementary Figure 3. The Cyclin D1 and c-Myc promoters contain multiple κB elements (see Supplementary Figure 10). However, the same NF-κB recruitment pattern is observed with primers covering any of these NF-κB-binding sites (not shown), although possible DNA-looping effects and the limits of ChIP assay resolution mean that it should not be concluded that all κB elements are binding the same combinations of NF-κB subunits.
Figure 3.
Cell cycle-dependent binding of NF-κB subunits to the Cyclin D1, c-Myc and Skp2 promoters. (A, B) Analysis of NF-κB subunit, coactivator and corepressor recruitment to the promoters of NF-κB target genes in U2OS cells. ChIP analysis of the Cyclin D1, Skp2, c-Myc and GAPDH control promoters (A), or IκBα, Bcl-xL and Bcl-2 promoters (B) was performed using antibodies to the indicated proteins in U2OS cells following centrifugal elutriation. Further details on ChIP primers used in U2OS cell experiments can be found in Supplementary Figure 10. (C) Table summarizing ChIP assay data from Figure 3A and Supplementary Figure 3 describing NF-κB subunit and coactivator/corepressor binding to the Cyclin D1, c-Myc and Skp2 promoters.
Figure 5.
RelA phosphorylation changes during the cell cycle. (A) Whole-cell lysates were prepared from U2OS cells following centrifugal elutriation and immunoblotted for RelA S468 and S536 phosphorylation and β-actin. RelA was immunoprecipitated before western blot analysis for RelA T505 phosphorylation. (B) ChIP analysis of S468-, T505- and S536-phosphorylated RelA recruitment to the Cyclin D1, Skp, c-Myc and GAPDH promoters in each cell cycle stage of U2OS cells. (C, D) S468-, T505- and S536-phosphorylated RelA differentially associate with coactivators and corepressors at different stages of the cell cycle. ReChIP analysis of the Cyclin D1, Skp2 and c-Myc gene promoters using elutriated U2OS cells was performed using the indicated combinations of antibodies. (E) Table summarizing ChIP and ReChIP assay data from (C) and (D) describing the co-occupancy of the Cyclin D1, c-Myc and Skp2 promoters by differentially phosphorylated forms of RelA.
To determine whether this pattern of NF-κB promoter occupancy is gene specific, we analyzed the IκBα, Bcl-xL and Bcl-2 promoters (Figure 3B). ChIP analysis demonstrated that the pattern of NF-κB binding in these promoters were not the same as with the G1-regulated promoters. Continuous NF-κB subunit binding was observed across the cell cycle, apart from a dip in p52 recruitment in S phase, correlating with the decrease in p100 processing seen before. These results demonstrated that the dynamic recruitment of NF-κB subunits is specific to cell cycle promoters and does not result from changes in nuclear localization (also data not shown).
Different NF-_κ_B complexes are present on the Cyclin D1, c-Myc and Skp2 promoters in G1 and G2 phases
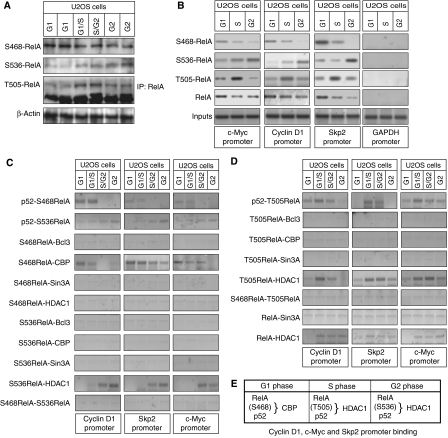
To determine the relationship between the different NF-κB subunits, co-immunoprecipitation assays were performed using extracts prepared from asynchronous U2OS cells. Interestingly, all NF-κB subunits were seen to interact, with the exception of RelA with RelB, RelA with c-Rel and RelB with c-Rel (Figure 4A). Moreover, c-Rel and RelB complexes only weakly interact with Bcl-3. We have previously observed that p52 can interact with HDAC1 (Rocha et al, 2003) and found that at least in part this is mediated by an interaction with the Sin3A corepressor (S Rocha and N Perkins, unpublished). Here, we found that all the NF-κB subunits can associate, either directly or indirectly, with Sin3A (Figure 4A). We next investigated the effect of depleting p52 on recruitment of the other NF-κB subunits to the Cyclin D1, c-Myc and Skp2 promoters (Figure 4B). ChIP analysis demonstrated that following knockdown of p52/100 by siRNA in U2OS cells, the recruitment of p52, RelA, Bcl-3 and Sin3A was significantly reduced, almost to background levels. By contrast, the recruitment of p50, RelB and c-Rel increased. A slight decrease in HDAC1 recruitment was observed suggesting that this is not totally p52 dependent. These results were consistent with ChIP analysis performed in p52/p100-null (_nfkb2_−/−) MEF cells, where the recruitment of RelA, Bcl-3 and Sin3A on the Cyclin D1, c-Myc and Skp2 promoters was also not seen (Supplementary Figure 4). Similarly, these cells displayed increased levels of p50, RelB and c-Rel binding to these G1-regulated promoters, while HDAC1 recruitment was diminished but not abolished (Supplementary Figure 4).
Figure 4.
Different NF-κB homodimer and heterodimer complexes are present on Cyclin D1, c-Myc and Skp2 promoters in G1 and G2 phases. (A) NF-κB subunits were immunoprecipitated from whole-cell lysates prepared from asynchronous cells and analyzed by western blotting as indicated. (B) The effect of depleting p52 on protein binding to the Cyclin D1, Skp2 and c-Myc promoters. U2OS cells were transfected with an siRNA targeting p52/p100. Cyclin D1, Skp2, c-Myc and GAPDH promoters were analyzed by using ChIP and Q-PCR as shown. (C) Distinct NF-κB subunit complexes bind the Cyclin D1, Skp2 and c-Myc promoters. ReChIP analysis of the Cyclin D1, Skp2 and c-Myc promoters was performed using the indicated antibodies. (D) Table summarizing ChIP and ReChIP assay data from (B) and (C) and Supplementary Figure 4 describing the co-occupancy of the Cyclin D1, c-Myc and Skp2 promoters by different NF-κB subunits and coactivator/corepressors.
To further investigate the different NF-κB complexes present at these promoters during the cell cycle, serial chromatin immunoprecipitations (ReChIP) on the Cyclin D1, c-Myc and Skp2 promoters were performed (Figure 4C and D). Interestingly, p52 and p50 binding to all promoters appears mutually exclusive. Moreover, while p50 could be readily detected binding with c-Rel or RelB in G2 phase, no detectable signal was seen with p50 and either RelA, CBP or Bcl-3 at any point of the cell cycle. p50 was found associated with HDAC1 but interestingly not with Sin3A, consistent with the effects observed upon p52/100 siRNA treatment and in p52/100-null (_nfkb2_−/−) MEF cells (Figure 4B and Supplementary Figure 4). These results indicate that p50 binds these genes when they show reduced levels of expression and suggest that it functions as a repressor of Cyclin D1, c-Myc and Skp2 in the G2 phase of the cell cycle. However, there is also clear evidence for functional redundancy, since p52 and RelA complexes also recruit corepressors to these promoters (Figure 4B and Supplementary Figure 4). Interestingly, RelA does not bind the promoter at the same time as Bcl-3, suggesting that p52 homodimer complexes with Bcl-3 bind mutually exclusively with p52/RelA heterodimers, which likely recruit CBP. Importantly, although U2OS cells possess many NF-κB complexes, many of these, such as the common p50/RelA heterodimer, appear to be excluded from the Cyclin D1, c-Myc and Skp2 promoters (Figure 4A and C).
RelA phosphorylation changes during the cell cycle
Although western blot analysis indicated no major changes in RelA levels during the cell cycle, we were interested in whether other, more subtle, effects might be occurring, such as phosphorylation. There is a low basal level of nuclear RelA and S468 and S536 RelA phosphorylation in U2OS cells. Interestingly, these show a distinct and opposing cell cycle distribution: RelA is predominantly S468 phosphorylated in G1 phase, while S536 phosphorylation is low in G1 phase, becoming apparent in S phase and peaking in G2 phase (Figure 5A, Supplementary Figure 5A). We also observed RelA phosphorylation at T505, which has previously been shown to be phosphorylated by Chk1 in response to cisplatin treatment and induction of the ARF tumor suppressor (Rocha et al, 2005; Campbell et al, 2006). This peaked in S phase and continued at a lower level in G2-phase enriched samples, consistent with Chk1 activation by the DNA replication checkpoint. Immunoprecipitation analysis confirmed that p52/100 is associated with T505-phosphorylated RelA in S and G2 phases, with S468-phosphorylated RelA in G1 phase and with S536-phosphorylated RelA in S and G2 phase (data not shown).
To determine whether the different phosphorylated forms of RelA were actively involved in gene regulation, we analyzed their recruitment to the Cyclin D1, Skp2 and c-Myc promoters (Figure 5B). This confirmed that phosphorylated RelA is bound to the promoters of these genes and that this is cell cycle dependent: S468-phosphorylated RelA is recruited in G1 phase, T505-phosphorylated RelA binding peaks in S phase, while S536-phosphorylated RelA binding is highest in G2 phase (Figure 5B). Significantly, this latter effect occurs despite the overall level of RelA associated with the promoters decreasing in G2 phase. To investigate the possible role of these RelA phosphorylations in the transcriptional activity of NF-κB complexes, ReChIP analysis was performed. These experiments revealed that RelA S468 phosphorylation correlates with CBP recruitment but not with Bcl-3 and could not be detected with HDAC1. By contrast, both T505 and S536 phosphorylations were not observed with CBP or Bcl-3 but were found associated with HDAC1 (Figure 5C–E). Interestingly, T505 phosphorylated RelA/HDAC1 binding starts in S phase and subsequently declines, while S536 phosphorylated RelA/HDAC1 comes up in G2 phase. No association was seen with Sin3A. Taken together with earlier ReChIP data (Figure 4C), this suggests recruitment of this corepressor occurs through p52 complexes. Moreover, the association of T505-phosphorylated RelA with HDAC1 is consistent with previous observations, where induction of ARF or cisplatin treatment were found to induce T505 phosphorylation and RelA association with HDAC1, concomitant with the inhibition of NF-κB transcriptional activity (Rocha et al, 2005; Campbell et al, 2006).
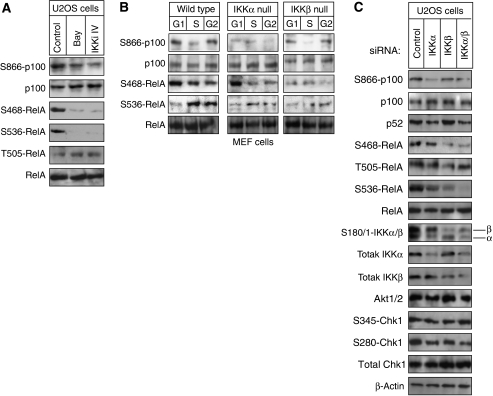
Differential RelA phosphorylation in the G1 and G2 phases of the cell cycle is IKK dependent
Consistent with previous reports (Perkins, 2006), IKKβ inhibitor treatment of asynchronous U2OS cells revealed that both RelA S468 and S536 phosphorylations are dependent on IKK activity (Figure 6A). Interestingly, in MEF cells, G2-phase S536 phosphorylation was dependent on both IKKα and IKKβ, consistent with reports that both kinases can phosphorylate this site (Perkins, 2006) (Figure 6B). By contrast, G1-phase phosphorylation of Ser468 is IKKβ dependent only, which suggests separation of IKK functions between the different cell cycle stages. These results were confirmed by siRNA depletion analysis of both IKKα and β (Figure 6C) and an in vitro kinase assay using immunoprecipitated IKKβ (Supplementary Figure 5B).
Figure 6.
RelA phosphorylation in G1 and G2 phases of the cell cycle is IKK dependent. (A) Asynchronous U2OS cells were treated with the Bay11-7082 and IKKiIV ([5-(p-Fluorophenyl)-2-ureido]thiophene-3-carboxamide) IKK inhibitors, for the indicated times. Whole-cell lysates were prepared and immunoblotted for S468- and S536-phosphorylated RelA. (B) Whole-cell extracts from wild-type and IKKα- or IKKβ-null MEFs from each cell cycle stages were subjected to western blot analysis using antibodies to phosphorylated RelA and p100. (C) U2OS cells were transfected with siRNAs targeting IKKα or IKKβ whole-cell lysates were prepared and analyzed by western blotting as shown.
Analysis of wild-type, IKKα- and IKKβ-null MEF cells demonstrated that S866 phosphorylation of p100 was also due to IKKα in the G1 and G2 phases of the cell cycle (Figure 6B). Furthermore, knockdown of IKKα and IKKβ by siRNA in U2OS cells confirmed that p100 processing to p52 is also IKKα dependent in this cell type (Figure 6C). Moreover, immunoprecipitation analysis followed by a kinase assay further confirmed that IKKα from G1- and G2-phase cells, but not S-phase cells, could phosphorylate p100 (Supplementary Figure 5B). Together these results confirm that the basal level of NF-κB activity we see in U2OS and immortalized MEFS results from constitutive IKKα and -β activity.
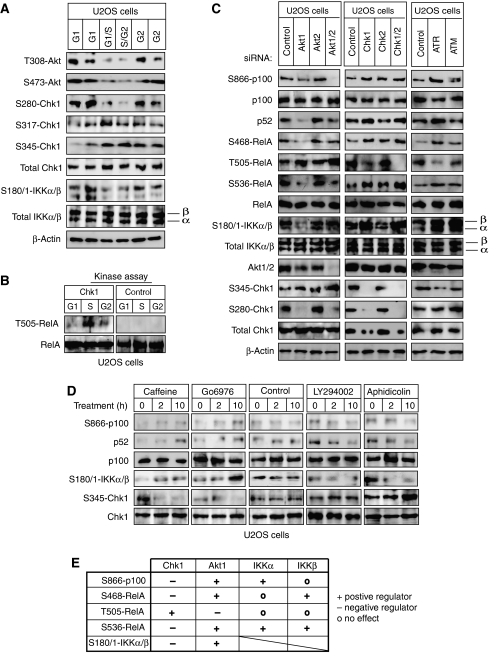
Active levels of Akt and Chk1 fluctuate during the cell cycle and regulate IKK/NF-_κ_B activity
Taken together, these experiments revealed a previously unknown and dynamic regulation of NF-κB activity across the cell cycle, involving changes in promoter binding, p100 processing and subunit phosphorylation. Although many of these activities depend at some level on IKKα or β, our observations suggested that other signaling pathways must coordinate these events and integrate with NF-κB pathway signaling in a cell cycle-dependent manner. Since we had previously determined that Chk1 can regulate RelA function (Rocha et al, 2005; Campbell et al, 2006), we first examined its role as a more global regulator of NF-κB activity.
Further analysis of U2OS cells following centrifugal elutriation demonstrated that active Chk1 was significantly enriched in S-phase fractions, consistent with activation of the ATR-dependent, DNA replication checkpoint (Figure 7A) (Petermann and Caldecott, 2006). Chk1 has previously been shown to be inactivated by Akt (PKB) phosphorylation at S280 (Puc et al, 2005). Interestingly, we also observed S280 phosphorylated Chk1 in both G1 and G2 phases of the cell cycle and this correlated with Akt phosphorylation at T308 and S473, both markers for its activation (Figure 7A) and exclusion of Chk1 from the nucleus (Supplementary Figure 5C). Confirming the biological activity of Chk1 in these S-phase fractions and consistent with previous reports from our laboratory (Rocha et al, 2005; Campbell et al, 2006), the RelA T505 residue was phosphorylated by Chk1 immunoprecipitated from S-phase U2OS cell extracts but not from G1- or G2-phase extracts (Figure 7B).
Figure 7.
Antagonistic regulation of NF-κB by Akt1 and Chk1. (A) Akt and Chk1 activity fluctuates during the cell cycle. Whole cells extracts from elutriated U2OS cells were subjected to western blot analysis using antibodies to phosphorylated Akt, Chk1 and IKKα/β. (B) Chk1 was immunoprecipitated from whole-cell extracts prepared from elutriated U2OS cells. The immunoprecipitate was incubated with whole-cell lysate from serum-starved U2OS cells. The lysates were analyzed by western blotting with antibodies to T505-phosphorylated RelA and total RelA. (C) U2OS cells were transfected with siRNAs targeting Akt1, Akt2, Chk1, Chk2, ATR and ATM in U2OS cells. Whole-cell lysates were prepared and analyzed by western blotting with antibodies to the indicated phosphorylated NF-κB and IKK subunits. Note, the basal level of some bands can be seen varying between panels. This is due to selection of different gel exposures to best allow visualization of induction or repression of signal strength. All experiments shown were derived from the same set of protein extracts, although separate gels were required to generate the complete data set. (D) Asynchronous U2OS cells were treated with caffeine and Gö6976, which inhibit ATR and Chk1, respectively, aphidicolin that causes S-phase arrest leading to an activation of Chk1, or LY294002, an inhibitor of PI-3 kinase activity and the Akt pathway, for the indicated times. Whole-cell lysates were prepared and immunoblotted for S866-phosphorylated p100, S180/1-phosphorylated IKKα/β, S345-phosphorylated Chk1 and total p100. (E) Table summarizing the regulation of different markers of NF-κB pathway activity by the Akt1, Chk1 and IKK kinases.
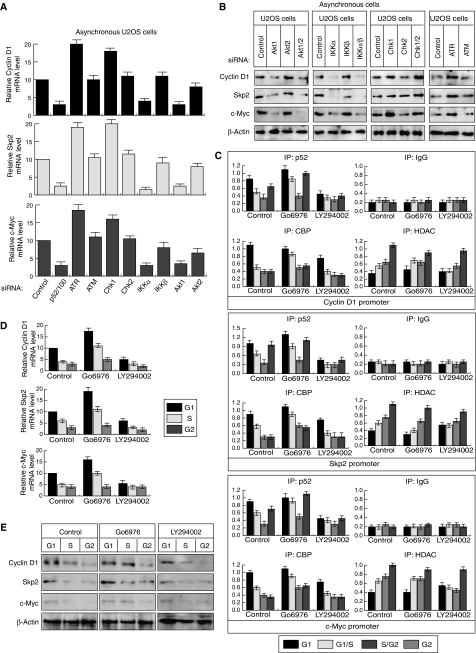
These observations suggested an antagonistic relationship between Chk1 and Akt activity during the cell cycle that could provide an explanation for some of the cell cycle regulation of NF-κB we had observed. Therefore, using siRNAs targeting each pathway, we explored this hypothesis further, together with its impact on NF-κB and IKK activity. Significantly, as judged by RelA S468, S536 and T505 phosphorylation, p100 phosphorylation and processing to p52 together with IKKα and β phosphorylation, entirely opposing effects were seen upon inhibition of Akt1 and Chk1: while Akt1 inhibition downregulates markers of NF-κB activation, Chk1 inhibition has the opposite effect (Figure 7C). We also observed opposing effects on RelA T505 phosphorylation but here Akt1 inhibition stimulated its levels. Inhibition of ATR had essentially the same effect as Chk1 inhibition. By contrast, inhibition of Akt2, Chk2 or ATM had minimal effects, the exception being a significant increase in IKKβ phosphorylation upon ATM inhibition. This latter effect, while potentially interesting, was not pursued further. A combination of western blotting (Figure 7C) and real-time PCR analysis (Supplementary Figure 6) confirmed the specificity of the siRNAs used. The effects on p100 phosphorylation and processing were further confirmed by treatment of U2OS cells with small molecule inhibitors of these pathways: treatment with caffeine, an ATR/ATM inhibitor (Rocha et al, 2005) or Gö6976, which inhibits the Chk1 pathway (Rocha et al, 2005; Bain et al, 2007), increased p100 processing to p52 (Figure 7D). Also consistent with the siRNA results, aphidicolin treatment, which causes arrest in S phase and Chk1 activation, or LY294002, which inhibits the PI-3 kinase pathway and Akt activation, resulted in inhibition of processing (Figure 7D).
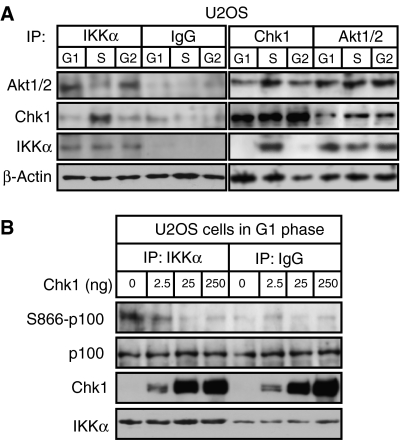
Chk1, Akt and IKK_α_ differentially interact during the cell cycle
To learn more about the relationship between Akt, Chk1 and IKK activity, we investigated their association by co-immunoprecipitation analysis. Surprisingly, we found a dramatic switch in IKKα interaction with Akt and Chk1 during S phase. While IKKα and Akt are seen to interact in G1- and G2-phase-enriched protein extracts, this association is reduced in S-phase-enriched samples and is replaced by an IKKα/Chk1 complex (Figure 8A). By contrast, Akt and Chk1 associate throughout the cell cycle (Figure 8A), despite Akt phosphorylation of Chk1 at S280 being inhibited in S phase (Figure 7A). This result is consistent with Akt being required for IKKα activity in G1 and G2 (Figure 7). Moreover, it suggested that Chk1 might directly inhibit IKKα activity in S phase. In agreement with this hypothesis, we found that purified recombinant Chk1 potently inhibited the ability of immunoprecipitated IKKα from G1-phase extracts to phosphorylate p100 in vitro (Figure 8B). The effect was lost upon either heat inactivation of Chk1 or inclusion of the Chk1 inhibitor Gö6976, demonstrating that Chk1 kinase activity is required for this effect (Supplementary Figure 7).
Figure 8.
Chk1 is associated with and inhibits IKKα activity. (A) IKKα associates with Chk1 in S phase. IKKα, Akt1/2 and Chk1 were immunoprecipitated from whole-cell extracts prepared from elutriated U2OS cells. The immunoprecipitates were resolved by SDS–PAGE before western blotting with antibodies to IKKα, Chk1 or Akt1/2. A β-actin western blot from the input material is also shown. (B) Chk1 inhibits IKKα activity in vitro. IKKα was immunoprecipitated from G1-phase-enriched extract prepared from elutriated U2OS cells. The immunoprecipitate was incubated with whole-cell lysate from serum-starved U2OS cells with or without the indicated quantities of recombinant Chk1. These lysates were then analyzed by western blotting with antibodies to S866-phosphorylated p100, total Chk1, total p100 and IKKα.
Antagonism between Akt and Chk1 regulates the cell cycle-dependent expression of Cyclin D1, c-Myc and Skp2
We next investigated, using siRNAs targeting each pathway and quantitative PCR or western blot analysis, whether the antagonistic regulation of NF-κB by Akt and Chk1 translated into effects on Cyclin D1, Skp2 and c-Myc expression. As predicted, depletion of Akt1 consistently inhibited Cyclin D1, Skp2 and c-Myc mRNA and protein levels to the same as those seen upon p52/p100 and IKKα inhibition (Figure 9A and B). By contrast, ATR or Chk1 inhibition had the opposite effect. We next examined the effect inhibiting these pathways would have on promoter binding by p52, CBP and HDAC1. Consistent with the effects on expression levels, inhibition of the Akt pathway using LY294002 strongly reduced promoter binding by p52, with significant effects also being seen for CBP and HDAC1 (Figure 9C, see also Supplementary Figure 8). Again, Chk1 inhibition had the opposite effect, with Gö6976 treatment stimulating p52 and CBP recruitment. Notably, the most significant effects in both cases were seen in S-phase-enriched samples. This suggested that Akt or Chk1 inhibition might affect the cell cycle profile of Cyclin D1, c-Myc and Skp2 expression seen previously (Figure 2). Significantly, Chk1 inhibition resulted in enhanced expression of Cyclin D1, Skp2 and c-Myc in S phase, although repression in G2 was still observed, while Akt inhibition had the opposing effect (Figure 9D and E, Supplementary Figure 8B).
Figure 9.
Chk1 and Akt1 antagonistically regulate Cyclin D1, c-Myc and Skp2 activity. (A, B) The expression of Cyclin D1, Skp2 and c-Myc expression is regulated by Akt1 and Chk1. Quantitative PCR analysis (A) or western blot analysis (B) was performed on extracts prepared from asynchronous U2OS cells treated with the indicated siRNAs. (C) Analysis of p52 NF-κB, HDAC1 and CBP recruitment to the promoters of NF-κB target genes in each cell cycle stage following Chk1 and Akt pathway inhibition. ChIP analysis of the Cyclin D1, Skp2, c-Myc and GAPDH promoters was performed in U2OS cells following centrifugal elutriation of U2OS cells with or without Gö6976 or LY294002 treatment. Analysis of the other NF-κB subunits is shown in Supplementary Figure 8. (D, E) RNA and whole-cell protein extracts were prepared from elutriated U2OS cells treated with Gö6976 or LY294002 and subjected to quantitative PCR (D) or western blot (E) analysis.
Discussion
Our results reveal a complex regulatory network coordinating NF-κB subunit promoter binding, p52/100 processing and RelA phosphorylation with the parallel control of important target genes during the cell cycle (Figure 10). Exchanges of NF-κB subunits on promoters, together with associated coactivators/corepressors, has been observed previously, although these examples followed treatment with a specific NF-κB-activating stimulus (Saccani et al, 2003; Hoberg et al, 2004). Here, the exchange of NF-κB complexes on the Cyclin D1, c-Myc and Skp2 promoters, between G1, S and G2 phases of the cell cycle, demonstrates a level of complexity of NF-κB target gene regulation during the cell cycle not previously appreciated. Taken together with our previous report (Schumm et al, 2006) and recent data highlighting the likely frequency of tumor-associated ‘driver mutations' in IKKα (Luo et al, 2007), these results demonstrate the likely importance of the noncanonical pathway as a regulator of cell proliferation in some tumor types.
Figure 10.
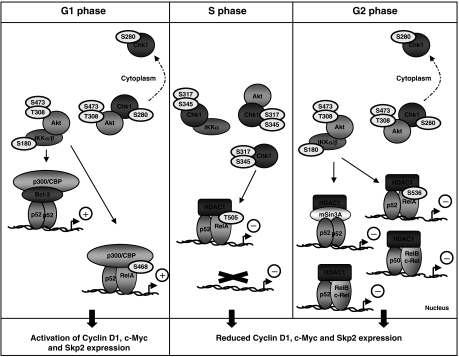
Model depicting the integration of NF-κB activity with the Akt1, IKK and Chk1 pathways during the cell cycle. During G1 and G2 phases, Akt activity is required for IKKα and β activity, resulting in the processing of p100 to p52 and the phosphorylation of RelA at serines 468 or 536. At the same time, Akt acts to inhibit Chk1, by phosphoylating it at serine 280. However, in S phase, Chk1 becomes activated and Akt activity is suppressed. Chk1 becomes directly associated with IKKα, inhibiting its activity. As a consequence, processing of p100 to p52 is inhibited. Furthermore, RelA becomes phosphorylated at Thr505, resulting in recruitment of HDAC1 to the Cyclin D1, c-Myc and Skp2 promoters. In G2 phase, NF-κB-mediated suppression of these genes is maintained, an effect associated with the recruitment of additional NF-κB complexes containing p50, RelB and c-Rel, even in the presence of active Akt and inhibited Chk1, suggesting an additional regulatory mechanism.
We have analyzed these pathways in a tumor cell line that has become dependent upon the p52 NF-κB subunit (Schumm et al, 2006). Significantly, we also detected these regulatory events in immortalized mouse fibroblasts. However, the cell cycle expression patterns of Cyclin D1, c-Myc and Skp2 can vary between cell lines (Supplementary Figure 2). In addition, prolonged growth of U2OS cells in culture results in increased G2 expression of these genes, suggesting that it is loss of a repressive mechanism that accounts for differences in c-Myc, Skp2 and Cyclin D1 expression between some cell types. This suggests that in some tumors and transformed cell lines, the balance between positive and negative regulatory pathways controlling Cyclin D1, c-Myc and Skp2 expression might break down, leading to higher levels of expression throughout the cell cycle. Alternatively, it is possible that the mechanism of controlling the expression of these genes we observe here may be more restricted in vivo, possibly being a feature of specific cell types or only being revealed in certain contexts, in response to specific stimuli. The identity and prevalence of this G2-phase repression mechanism will be the subject of future investigations.
In different contexts, NF-κB subunits may also contribute to regulation of these genes in different ways. For example, consistent with our data, Schneider et al (2006) recently demonstrated that in asynchronous populations of MiaPaCa2 pancreatic cancer cells, IKKα activity is required for Skp2 expression. Moreover, this report also found that knockdown of IKKα resulted in a change in NF-κB subunit binding to the Skp2 promoter, with a switch from p52/RelB to p50/RelB complexes being observed, concomitant with repression of Skp2 expression. This dynamic change in promoter binding of NF-κB subunits is reminiscent of our results, although we suggest a repressor rather than activator role for p52/RelB in U2OS cells. Similarly, the Gerondakis laboratory reported reduced c-Myc levels in B and T cells derived from p50/c-Rel-null mice, implying an activator rather than repressor role for these subunits in these cells (Grumont et al, 2004). We have previously noted (Schumm et al, 2006) that the proliferative role of p52 is cell type specific. Taken together, these results indicate the dangers of overextrapolating from any single cell type. The multiple NF-κB pathways are regulated by a large array of stimuli in many cell contexts, which can result in clear functional differences, such as activation or repression of specific target genes by the same NF-κB subunit (Campbell et al, 2004; Perkins, 2007).
Inhibition of IKK activity in S-phase results from Chk1 activity, which is active during this part of the cell cycle in U2OS cells, presumably as a consequence of the DNA-replication checkpoint. Significantly, Chk1 co-immunoprecipitates with IKKα in S-phase protein extracts and can inhibit IKKα phosphorylation of p100 in vitro. Whether this results from an ability of Chk1 to directly phosphorylate IKKα and inhibit its catalytic activity is not currently known. Nonetheless, this reveals a new regulatory pathway linking NF-κB activity with checkpoint kinase function. This is consistent with another link we have previously found between Chk1 and NF-κB, namely its ability to phosphorylate RelA at T505 (Rocha et al, 2005; Campbell et al, 2006). Here we confirm this observation but also reveal that it is not solely as a consequence of certain forms of extrinsic DNA damage or ARF tumor suppressor induction. Rather, we find RelA T505 phosphorylation occurring during S phase. Moreover, consistent with our previous observation that this modification turns RelA into a repressor of target gene expression, we demonstrate that the switch from S468 to T505 phosphorylation correlates with a switch from coactivator to corepressor recruitment to the Cyclin D1, c-Myc and Skp2 promoters. Chk1 could therefore coordinately regulate NF-κB function to ensure the inhibition of NF-κB-dependent G1-phase gene expression at the start of S phase.
Signaling through the PI-3 kinase/Akt pathway counterbalances the inhibitory activity of Chk1. Previously, Akt has been shown to inhibit Chk1 activity through phosphorylation at serine 280, resulting in the cytoplasmic sequestration of Chk1 (Puc et al, 2005). We also see evidence for this effect and observe oscillations in Akt activity across the cell cycle, with peaks in active, phosphorylated, forms of Akt being seen in G1 and G2 phases. The ability of IKKβ to phosphorylate RelA on S536 has previously been shown to be regulated by Akt (Perkins, 2006). Furthermore, Akt has been shown to regulate basal processing of p100 to p52 (Gustin et al, 2006). Our results are also consistent with these reports. However, here we extend this observation to reveal a remarkable degree of integration between Akt's ability to regulate multiple markers of both classical and noncanonical NF-κB pathway activity, and Chk1 activity during S phase (Figure 7E). Moreover, tipping the balance in favor of either pathway significantly affects both the NF-κB-regulated expression level and cell cycle profile of Cyclin D1, c-Myc and Skp2. In tumor cell lines with high levels of Akt activity, it is possible that many of the more subtle effects we observe here will not be seen to the same extent. Further investigation of these pathways will give mechanistic insight into how NF-κB/IKK function is subverted during tumorigenesis to promote cell growth and survival.
Materials and methods
Cells
Human Osteosarcoma U2OS, MCF7 breast cancer cells and wild-type, IKKα-null, IKKβ-null, p52-null or RelA-null MEF cells were maintained at 5% CO2 in Dulbecco's modified Eagle's medium (Cambrex) supplemented with 10% fetal bovine serum (Invitrogen), 1% penicillin–streptomycin (Cambrex) and 1% L-glutamine (Cambrex). U20S cells were obtained from the European Collection of Cell Cultures (ECACC). IKK-null MEFs were a kind gift from Professor Inder Verma (Salk Institute), while p52-null and RelA-null MEFs were provided by Professor Ron Hay (Dundee). HUT78 cells were obtained from the ECACC and grown in RPMI 1640 media supplemented with 10% fetal bovine serum (Invitrogen), 1% penicillin–streptomycin (Cambrex) and 1% L-glutamine (Cambrex).
Centrifugal elutriation
Centrifugal elutriation was carried out using a Beckman J6-M1 centrifuge equipped with a JE 5.0 rotor and a large chamber (Beckman Coulter) connected to a Pump Masterflex pump (model 7016-20) (Grdina et al, 1984; White et al, 1984). All steps were performed at room temperature in PBS containing 1% FBS and 25 mM EDTA, and centrifugation was carried out at 2000 r.p.m. Cells (2 × 108) were loaded into the chamber, and equilibration was carried out at a flow rate of 85 ml/min for 15 min. The flow rate was increased from 100 to 200 ml/min, successively by 5 ml/min for each 200 ml fraction.
For other Materials and methods, please see online Supplementary Information.
Supplementary Material
Supplementary Figures
Supplementary Figure Legends
Acknowledgments
We thank Inder Verma, Ron Hay and John Rouse for supplying reagents. We also thank the current members of the NDP Laboratory, Katie Schumm, Sonia Rocha, Kirsteen Campbell and the Division of Gene Regulation and Expression at the University of Dundee for their help and support. BB is funded by a project grant from the Association of International Cancer Research.
References
- Albanese C, D'Amico M, Reutens AT, Fu M, Watanabe G, Lee RJ, Kitsis RN, Henglein B, Avantaggiati M, Somasundaram K, Thimmapaya B, Pestell RG (1999) Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J Biol Chem 274: 34186–34195 [DOI] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie J, McLauchlan H, Klevernic I, Arthur S, Alessi D, Cohen P (2007) The selectivity of protein kinase inhibitors; a further update. Biochem J Sep 13 (E-pub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg JF, Fan Z, Brown C, Mendelsohn J, Darnell JE Jr (1998) Epidermal growth factor-induced growth inhibition requires Stat1 activation. Cell Growth Differ 9: 505–512 [PubMed] [Google Scholar]
- Campbell KJ, Rocha S, Perkins ND (2004) Active repression of antiapoptotic gene expression by RelA(p65) NF-κB. Mol Cell 13: 853–865 [DOI] [PubMed] [Google Scholar]
- Campbell KJ, Witty JM, Rocha S, Perkins ND (2006) Cisplatin mimics ARF tumor suppressor regulation of RelA (p65) nuclear factor-κB transactivation. Cancer Res 66: 929–935 [DOI] [PubMed] [Google Scholar]
- Ciana P, Neri A, Cappellini C, Cavallo F, Pomati M, Chang CC, Maiolo AT, Lombardi L (1997) Constitutive expression of lymphoma-associated NFKB-2/Lyt-10 proteins is tumorigenic in murine fibroblasts. Oncogene 14: 1805–1810 [DOI] [PubMed] [Google Scholar]
- Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS Jr (2000) Selective activation of NF-κB subunits in human breast cancer: potential roles for NF-κB2/p52 and for Bcl-3. Oncogene 19: 1123–1131 [DOI] [PubMed] [Google Scholar]
- Demicco EG, Kavanagh KT, Romieu-Mourez R, Wang X, Shin SR, Landesman-Bollag E, Seldin DC, Sonenshein GE (2005) RelB/p52 NF-κB complexes rescue an early delay in mammary gland development in transgenic mice with targeted superrepressor IκB-α expression and promote carcinogenesis of the mammary gland. Mol Cell Biol 25: 10136–10147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SF, Guo S, Demicco EG, Romieu-Mourez R, Landesman-Bollag E, Seldin DC, Sonenshein GE (2005) Inducible IκB kinase/IκB kinase epsilon expression is induced by CK2 and promotes aberrant nuclear factor-κB activation in breast cancer cells. Cancer Res 65: 11375–11383 [DOI] [PubMed] [Google Scholar]
- Grdina DJ, Meistrich ML, Meyn RE, Johnson TS, White RA (1984) Cell synchrony techniques. I. A comparison of methods. Cell Tissue Kinet 17: 223–236 [DOI] [PubMed] [Google Scholar]
- Grumont R, Lock P, Mollinari M, Shannon FM, Moore A, Gerondakis S (2004) The mitogen-induced increase in T cell size involves PKC and NFAT activation of Rel/NF-κB-dependent c-myc expression. Immunity 21: 19–30 [DOI] [PubMed] [Google Scholar]
- Gustin JA, Korgaonkar CK, Pincheira R, Li Q, Donner DB (2006) Akt regulates basal and induced processing of NF-κB2 (p100) to p52. J Biol Chem 281: 16473–16481 [DOI] [PubMed] [Google Scholar]
- Guttridge DC, Albanese C, Reuther JY, Pestell RG, Baldwin AS Jr (1999) NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol 19: 5785–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann SR, Thompson CB, Eisenman RN (1985) c-myc oncogene protein synthesis is independent of the cell cycle in human and avian cells. Nature 314: 366–369 [DOI] [PubMed] [Google Scholar]
- Hinz M, Krappmann D, Eichten A, Heder A, Scheidereit C, Strauss M (1999) NF-κB function in growth control: regulation of cyclin D1 expression and G0/G1-to-S-phase transition. Mol Cell Biol 19: 2690–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoberg JE, Yeung F, Mayo MW (2004) SMRT derepression by the IκB kinase α: a prerequisite to NF-κB transcription and survival. Mol Cell 16: 245–255 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Carrasco D, Claudio E, Ryseck RP, Bravo R (1997) Gastric hyperplasia and increased proliferative responses of lymphocytes in mice lacking the COOH-terminal ankyrin domain of NF-κB2. J Exp Med 186: 999–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce D, Albanese C, Steer J, Fu M, Bouzahzah B, Pestell RG (2001) NF-κB and cell-cycle regulation: the cyclin connection. Cytokine Growth Factor Rev 12: 73–90 [DOI] [PubMed] [Google Scholar]
- Kanda K, Hu HM, Zhang L, Grandchamps J, Boxer LM (2000) NF-κB activity is required for the deregulation of c-myc expression by the immunoglobulin heavy chain enhancer. J Biol Chem 275: 32338–32346 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Hawke N, Baldwin AS (2006) NF-κB and IKK as therapeutic targets in cancer. Cell Death Differ 13: 738–747 [DOI] [PubMed] [Google Scholar]
- La Rosa FA, Pierce JW, Sonenshein GE (1994) Differential regulation of the c-myc oncogene promoter by the NF-κB rel family of transcription factors. Mol Cell Biol 14: 1039–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JL, Tan W, Ricono JM, Korchynskyi O, Zhang M, Gonias SL, Cheresh DA, Karin M (2007) Nuclear cytokine-activated IKKα controls prostate cancer metastasis by repressing Maspin. Nature 446: 690–694 [DOI] [PubMed] [Google Scholar]
- Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R (2006) Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-κB signaling. Cell 125: 665–677 [DOI] [PubMed] [Google Scholar]
- Ouyang W, Ma Q, Li J, Zhang D, Liu ZG, Rustgi AK, Huang C (2005) Cyclin D1 induction through IκB kinase β/nuclear factor-κB pathway is responsible for arsenite-induced increased cell cycle G1–S phase transition in human keratinocytes. Cancer Res 65: 9287–9293 [DOI] [PubMed] [Google Scholar]
- Park KJ, Krishnan V, O'Malley BW, Yamamoto Y, Gaynor RB (2005) Formation of an IKKα-dependent transcription complex is required for estrogen receptor-mediated gene activation. Mol Cell 18: 71–82 [DOI] [PubMed] [Google Scholar]
- Perkins ND (2006) Post-translational modifications regulating the activity and function of the nuclear factor κB pathway. Oncogene 25: 6717–6730 [DOI] [PubMed] [Google Scholar]
- Perkins ND (2007) Integrating cell-signalling pathways with NF-κB and IKK function. Nat Rev Mol Cell Biol 8: 49–62 [DOI] [PubMed] [Google Scholar]
- Petermann E, Caldecott KW (2006) Evidence that the ATR/Chk1 pathway maintains normal replication fork progression during unperturbed S phase. Cell Cycle 5: 2203–2209 [DOI] [PubMed] [Google Scholar]
- Puc J, Keniry M, Li HS, Pandita TK, Choudhury AD, Memeo L, Mansukhani M, Murty VV, Gaciong Z, Meek SE, Piwnica-Worms H, Hibshoosh H, Parsons R (2005) Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell 7: 193–204 [DOI] [PubMed] [Google Scholar]
- Rabbitts PH, Watson JV, Lamond A, Forster A, Stinson MA, Evan G, Fischer W, Atherton E, Sheppard R, Rabbitts TH (1985) Metabolism of c-myc gene products: c-myc mRNA and protein expression in the cell cycle. EMBO J 4: 2009–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayet B, Gelinas C (1999) Aberrant rel/nfkb genes and activity in human cancer. Oncogene 18: 6938–6947 [DOI] [PubMed] [Google Scholar]
- Rocha S, Garrett MD, Campbell KJ, Schumm K, Perkins ND (2005) Regulation of NF-κB and p53 through activation of ATR and Chk1 by the ARF tumour suppressor. EMBO J 24: 1157–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha S, Martin AM, Meek DW, Perkins ND (2003) p53 represses cyclin D1 transcription through down regulation of Bcl-3 and inducing increased association of the p52 NF-κB subunit with histone deacetylase 1. Mol Cell Biol 23: 4713–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah M, Courilleau D, Mester J, Redeuilh G (1999) Estrogen induction of the cyclin D1 promoter: involvement of a cAMP response-like element. Proc Natl Acad Sci USA 96: 11217–11222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G (2003) Modulation of NF-κB activity by exchange of dimers. Mol Cell 11: 1563–1574 [DOI] [PubMed] [Google Scholar]
- Schneider G, Saur D, Siveke JT, Fritsch R, Greten FR, Schmid RM (2006) IKKα controls p52/RelB at the skp2 gene promoter to regulate G1- to S-phase progression. EMBO J 25: 3801–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumm K, Rocha S, Caamano J, Perkins ND (2006) Regulation of p53 tumour suppressor target gene expression by the p52 NF-κB subunit. EMBO J 25: 4820–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro E, Tsuchiya A, Imoto M (2007) Functions of cyclin D1 as an oncogene and regulation of cyclin D1 expression. Cancer Sci 98: 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur S, Lin HC, Tseng WT, Kumar S, Bravo R, Foss F, Gelinas C, Rabson AB (1994) Rearrangement and altered expression of the NFKB-2 gene in human cutaneous T-lymphoma cells. Oncogene 9: 2335–2344 [PubMed] [Google Scholar]
- Thompson CB, Challoner PB, Neiman PE, Groudine M (1985) Levels of c-myc oncogene mRNA are invariant throughout the cell cycle. Nature 314: 363–366 [DOI] [PubMed] [Google Scholar]
- Vacca A, Felli MP, Palermo R, Di Mario G, Calce A, Di Giovine M, Frati L, Gulino A, Screpanti I (2006) Notch3 and pre-TCR interaction unveils distinct NF-κB pathways in T-cell development and leukemia. EMBO J 25: 1000–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe G, Albanese C, Lee RJ, Reutens A, Vairo G, Henglein B, Pestell RG (1998) Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol Cell Biol 18: 3212–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerheide SD, Mayo MW, Anest V, Hanson JL, Baldwin AS Jr (2001) The putative oncoprotein Bcl-3 induces cyclin D1 to stimulate G(1) transition. Mol Cell Biol 21: 8428–8436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RA, Grdina DJ, Meistrich ML, Meyn RE, Johnson TS (1984) Cell synchrony techniques. II. Analysis of cell progression data. Cell Tissue Kinet 17: 237–245 [DOI] [PubMed] [Google Scholar]
- Xiao G, Harhaj EW, Sun SC (2001) NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol Cell 7: 401–409 [DOI] [PubMed] [Google Scholar]
- Zhang J, Warren MA, Shoemaker SF, Ip MM (2007) NFκB1/p50 is not required for tumor necrosis factor-stimulated growth of primary mammary epithelial cells: implications for NFκB2/p52 and RelB. Endocrinology 148: 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Figure Legends