Tlx1 and Tlx3 Coordinate Specification of Dorsal Horn Pain-Modulatory Peptidergic Neurons (original) (raw)
Abstract
The dorsal spinal cord synthesizes a variety of neuropeptides that modulate the transmission of nociceptive sensory information. Here, we used genetic fate mapping to show that Tlx3+ spinal cord neurons and their derivatives represent a heterogeneous population of neurons, marked by partially overlapping expression of a set of neuropeptide genes, including those encoding the anti-opioid peptide cholecystokinin, pronociceptive Substance P (SP), Neurokinin B, and a late wave of somatostatin. Mutations of Tlx3 and Tlx1 result in a loss of expression of these peptide genes. Brn3a, a homeobox transcription factor, the expression of which is partly dependent on Tlx3, is required specifically for the early wave of SP expression. These studies suggest that Tlx1 and Tlx3 operate high in the regulatory hierarchy that coordinates specification of dorsal horn pain-modulatory peptidergic neurons.
Keywords: dorsal spinal cord, peptidergic neurons, Tlx3, cell fate specification, transcriptional regulation, pain
Introduction
The dorsal horn of the spinal cord is an integrative center that processes and transmits somatic sensory information. Morphological and functional studies have revealed a tremendous diversity of dorsal horn neurons (Christensen and Perl, 1970; Lima and Coimbra, 1986; Todd and Spike, 1993; Han et al., 1998; Grudt and Perl, 2002; Todd and Koerber, 2006). Diversity of the dorsal horn neurons is further suggested by the expression of neuropeptides, including the opioid-like peptides Dynorphin (DYN) and enkephalin (ENK), the anti-opioid peptide cholecystokinin (CCK), the tachykinin peptides Substance P (SP) and Neurokinin B (NKB), somatostatin (SOM), and others (Marti et al., 1987; Todd and Spike, 1993; Todd and Koerber, 2006; Polgar et al., 2006). Functionally, neuropeptides modulate the transmission of somatic sensory information, particularly those involved with pain perception (Kajander et al., 1990; Xu et al., 1993; Wang et al., 2001; Wiesenfeld-Hallin et al., 2002).
The past decade has seen important progress in understanding dorsal horn neuron development (Caspary and Anderson, 2003; Helms and Johnson, 2003; Fitzgerald, 2005; Ma, 2006). Signals derived from the roof plate pattern the dorsal neural tube, such that precursors are divided into distinct compartments along the dorsoventral axis (Caspary and Anderson, 2003; Helms and Johnson, 2003). Early born neurons dorsal horn interneurons 1–6 (DI1–DI6) migrate ventrally and settle in deep dorsal horn laminas, whereas late born neurons (DILA and DILB) settle in superficial dorsal horn laminas (Caspary and Anderson, 2003; Helms and Johnson, 2003). With the exception of DI1–DI3 neurons, most dorsal horn neurons express Lbx1 at embryonic day 11.5 (E11.5) to E13.5 (Gross et al., 2002; Müller et al., 2002). Lbx1+ neurons are divided into two populations, based on their nonoverlapping expression of the homeobox proteins Pax2 (DI4, DI6, and DILA) versus Tlx3 plus Lmx1b (DI5 and DILB) (Gross et al., 2002; Müller et al., 2002; Cheng et al., 2004). A set of transcription factors (TFs) acts to specify the excitatory versus the inhibitory neuron cell fates (Cheng et al., 2004, 2005; Glasgow et al., 2005; Mizuguchi et al., 2006; Hori et al., 2008). Lbx1 determines a basal GABAergic inhibitory neuron cell fate (Cheng et al., 2005). The homeobox proteins GSH1 and GSH2 control the expression of Tlx3, which in turn antagonizes Lbx1 to determine the glutamatergic excitatory neuron cell fate (Cheng et al., 2004, 2005; Mizuguchi et al., 2006). Ptf1a acts in combination with RBPjk (recombination signal binding protein for IgκJ region) to suppress Tlx3 expression and to promote GABAergic differentiation (Glasgow et al., 2005; Mizuguchi et al., 2006; Hori et al., 2008).
Despite this progress, transcriptional regulation of neuropeptides in the developing spinal cord is poorly understood. In this study, we used genetic fate mapping to show that Tlx3+ neurons or their derivatives express a set of neuropeptides, including SP, CCK, NKB, and a late wave of SOM. Accordingly, expression of these peptide genes is eliminated in mice that lack Tlx3 and Tlx1. Mechanistically, Tlx1 and Tlx3 activate a variety of downstream transcription factors, including the homeobox protein Brn3a, which controls the expression of an early wave of SP expression. Tlx1/3 therefore act as master regulators that coordinate the development of dorsal horn excitatory peptidergic neurons.
Materials and Methods
Animals.
The generation of Tlx1 and Tlx3 mutant mice, Lbx1 mutant mice, FLPe deleter mice, and Brn3a mutant mice has been described previously (Roberts et al., 1994; Rodriguez et al., 2000; Shirasawa et al., 2000; Gross et al., 2002; Quina et al., 2005). The generation of Tlx3 Cre knock-in mice is described in supplemental Figure 1 (available at www.jneurosci.org as supplemental material). To fate map Tlx3-expressing neurons, the Tlx3 Cre knock-in mice were then crossed with Cre-dependent Rosa26-lacZ reporter mice (Soriano, 1999), as described in supplemental Figure 2 (available at www.jneurosci.org as supplemental material). Tlx3 Cre mice were also crossed with another Cre-dependent reporter line, Tau-nLacZ (Hippenmeyer et al., 2005), as described in Figure 1. In all timed matings, the morning that vaginal plugs were observed was considered to be E0.5. All animal procedures are contained in protocols reviewed and approved by the Animal Care Committees at the Dana-Faber Cancer Institute, Harvard Medical School.
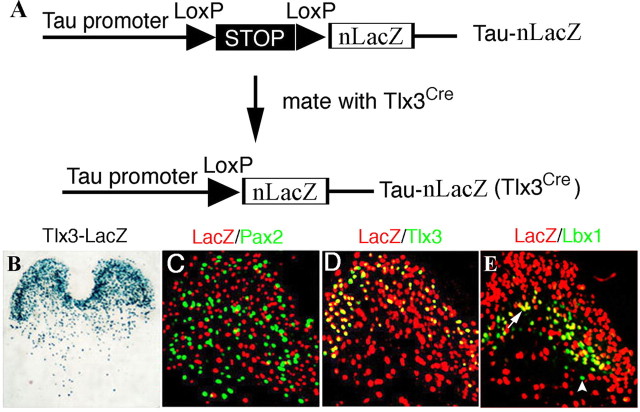
Figure 1.
Persistent and transient Tlx3 expression revealed by fate mapping. A, Tau-nLacZ reporter mice were crossed with Tlx3 Cre mice, allowing the removal of the “_STOP_” cassette, flanked with loxP sites (triangles), by Cre-mediated recombination. Subsequently, the expression of nLacZ protein is driven from the pan-neuronal Tau promoter. B–E, Transverse sections through thoracic P7 spinal cord of Tau-nLacZ(Tlx3Cre) mice. B, X-gal staining marking the distribution of Tlx3+ neurons and their derivatives. C–E, Double staining of the nLacZ protein product (C–E, red) plus Pax2 (C, green), Tlx3 (D, green), or Lbx1 (E, green, arrowhead) proteins. Neurons coexpressing nLacZ and Tlx3 (D) or Lbx1 (E, arrow) appear yellow. All Tlx3+ neurons showed either strong or weak expression of nLacZ (D) (data not shown).
In situ hybridization and immunostaining.
Detailed methods for single- and double-color in situ hybridization (supplemental Fig. 4, available at www.jneurosci.org as supplemental material) have been described previously (Qian et al., 2001). The following mouse in situ probes were amplified with gene-specific sets of PCR primers and cDNA templates prepared from postnatal day 0 (P0) or P7 mouse brain/spinal cords, including Sst (NM_009215; 0.5 kb), Tac1 (NM_009311; 0.85 kb), CCK (NM_031161; 0.34 kb), Pdyn (NM_018863; 0.7 kb), Penk1 (NM_001002927; 0.63 kb), and Tac2 (D14423; 0.44 kb). Chick Tac1 (BI395005; 0.4 kb) was amplified from cDNA from E10 chick spinal cord. In situ probes for dorsal horn functional genes (supplemental Fig. 7, available at www.jneurosci.org as supplemental material) were described previously (Qian et al., 2002; Cheng et al., 2004). To produce double-color in situ hybridization (supplemental Fig. 4, available at www.jneurosci.org as supplemental material), the first in situ hybridization signal [purple, with nitroblue tetrazolium (NBT)/5-bromo-4-chlor-indolyl-phosphate (BCIP) substrates] was photographed, followed by the development of the second signal [brown, with INT (iodonitro tetrazolium)/BCIP as substrates]. This sequential photographic process is helpful in determining whether a cell expresses a single gene or two genes.
The following antibodies were used for single or double immunostaining: rabbit anti-Pax2 antibody (Zymed Laboratories, South San Francisco, CA), rabbit anti-Brn3a antibody (E. Turner, University of California, San Diego, La Jolla, CA), and guinea pig anti-Lbx1, rabbit anti-Tlx3, and guinea pig anti-Tlx3 antibodies (T. Müller and C. Birchmeier, Max-Delbrück-Center for Molecular Medicine, Berlin, Germany).
For in situ hybridization combined with fluorescent immunostaining, in situ hybridization was first performed without proteinase K treatment. After posthybridization washing, Tlx3, Pax2, Lbx1, or Brn3a proteins were detected by incubation with appropriate antibodies and then with Alexa-488-conjugated secondary antibody (1:200; Invitrogen, Carlsbad, CA) in PBS plus 0.1% Tween 20 solution. After the fluorescent signals were photographed, sections were incubated with alkaline phosphatase-conjugated anti-digoxigenin antibody, followed by development of the in situ hybridization signal with NBT/BCIP substrates. The bright-field views of the in situ hybridization images were inverted and then merged with the fluorescent images. This process avoids the masking of low-level fluorescent signals by nonfluorescent in situ signals.
Immunostaining combined with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) staining was also performed sequentially, with the immunostaining done first. The bright-field images of X-gal staining were inverted and then merged with the immunofluorescence images, thus avoiding the masking of low-level fluorescent signals by β-galactosidase (lacZ) staining signals. In situ hybridization combined with X-gal staining was performed by a similar sequential process, with the X-gal staining performed and photographed first.
In ovo electroporation.
For electroporation studies in chick embryos, a cDNA fragment encoding a Myc-tagged mouse Brn3a fusion protein was cloned to the RCASBP chick viral expression vector (Morgan and Fekete, 1996) to produce the construct RCAS-Brn3a. The purified plasmid DNA was resuspended at concentrations of 5 μg/μl. RCAS-Brn3a plus a GFP expression vector, pCAX-IRES-GFP or pCAX-GFP (Gross et al., 2002), were coinjected into the spinal neural tubes of E2 chick embryos. After electroporation, the embryos were incubated at 39°C for an additional 72–120 h (E5–E7). Embryos with a high level of green fluorescent protein (GFP) fluorescence were fixed, and changes in the expression of genes of interest in the spinal cord were analyzed.
Cell counting.
To count neurons that express Tac1, Sst, Tac2, and Pemk1 per thoracic spinal section of E14.5 or E18.75 wild-type and Tlx1/3 double mutants, three sets of thoracic transverse sections from three pairs of wild-type and mutant embryos (14 μm thickness) were hybridized with probes derived from the cDNAs for each peptide. Positive cells with clear nuclear morphology in the dorsal spinal cord were counted. Values were presented as mean ± SD. The differences in values were considered to be significant at p < 0.05 by Student's t test. To determine the percentage of Tlx3+, Pax2+, or Lbx1+ neurons that express a peptide gene, we again only counted those isolated cells with clear nuclear morphology.
Results
Generation of Tlx3 Cre knock-in mice
Tlx3 exhibits dynamic expression in the developing spinal cord (Qian et al., 2002). To follow the fate of those neurons derived from Tlx3+ cells, we generated a Tlx3 Cre knock-in mouse line, in which the Cre recombinase gene was inserted into the first coding exon of the Tlx3 locus (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). To determine whether Tlx3 Cre expression faithfully reflects in vivo Tlx3 expression, we crossed Tlx3 Cre mice with a Cre-dependent lacZ reporter line, ROSA26-LacZ (supplemental Fig. 2, available at www.jneurosci.org as supplemental material) (Soriano, 1999). In ROSA26-LacZ(Tlx3Cre) mice, Cre-mediated removal of a transcriptional termination cassette allows a constitutive expression of the lacZ protein product, β-galactosidase (Soriano, 1999). Consequently, all derivatives that undergo successful Cre-mediated DNA recombination are labeled by X-gal staining (also called lacZ staining). Examined at E11.5, ROSA26-LacZ(Tlx3Crei) embryos exhibited a lacZ staining pattern that matched endogenous Tlx3 expression revealed by whole-mount in situ hybridization (supplemental Fig. 2, available at www.jneurosci.org as supplemental material), demonstrating that Tlx3 Cre mice are an effective tool for fate-mapping experiments.
Persistent and transient Tlx3 expression in different dorsal horn laminas
To facilitate fate-mapping experiments, we next crossed Tlx3 Cre mice with another Cre-dependent reporter mouse line, Tau-nLacZ, with the resulting double heterozygous mice referred to as Tau-nLacZ(Tlx3Cre) mice. After Cre-mediated removal of a transcriptional termination cassette, this reporter gene encodes β-galactosidase linked to a nuclear localization signal (nLacZ) and is driven from the pan-neuronal Tau promoter (Fig. 1A) (Hippenmeyer et al., 2005). In Tau-nLacZ(Tlx3Cre) mice at P7, X-gal staining showed that nLacZ+ neurons were enriched in the dorsal spinal cord but also present in small numbers in the ventral spinal cord (Fig. 1B). Tlx3+ cells normally give rise to glutamatergic neurons that are intermingled with inhibitory interneurons marked by the expression of Pax2 (Cheng et al., 2004). Consistent with this, virtually no nLacZ+ neurons coexpressed Pax2 (Fig. 1C), providing a key validation of the fidelity of nLacZ expression in Tau-nLacZ(Tlx3Cre) mice. Double staining of Tlx3 protein and nLacZ showed that neurons with persistent Tlx3 expression (nLacZ+;Tlx3+) were enriched in the superficial dorsal horn, whereas neurons with transient Tlx3 expression (nLacZ+;Tlx3−) were distributed throughout the spinal cord but are enriched in areas from deep dorsal horn laminas to the ventral spinal cord (Fig. 1D). Transient Tlx3 expression in a subset of dorsal horn neurons is consistent with the previous finding that Tlx3 expression is switched off in DI3 and a portion of DI5 neurons that settle in deep dorsal horn laminas (Qian et al., 2002).
Most dorsal horn neurons, including Tlx3+ excitatory neurons and Pax2+ inhibitory neurons, develop from Lbx1+ cells (Gross et al., 2002; Müller et al., 2002). In P7 spinal cord of Tau-nLacZ(Tlx3Cre) mice, Lbx1 protein, however, was virtually absent in a majority of nLacZ+ neurons (Fig. 1E), as well as in most Pax2+ neurons (data not shown), implying a transient Lbx1 expression in most dorsal horn neurons. Residual Lbx1+ neurons were located in an intermediate dorsal horn lamina, most of which derive from Tlx3+ neurons, as indicated by the coexpression of Lbx1 and nLacZ (Fig. 1E, arrows). In summary, both Tlx3 and Lbx1 exhibit dynamic expression in the developing spinal cord.
Tlx3+ neurons/derivatives and Pax2+ neurons express two distinct sets of peptide genes
We next examined the expression of the following six peptide genes: Tachykinin 1 (Tac1) encoding the precursor for the SP and Neurokinin A, Tachykinin 2 (Tac2) encoding the precursor for the NKB, CCK encoding the precursor for CCK peptides, Somatostatin (Sst) encoding the precursor for SOM, Prodynorphin (Pdyn) encoding the precursor for DYN, and Preproenkephalin 1 (Penk1) encoding the precursor for ENK.
Figure 2 shows the spatial and temporal expression patterns of these peptide genes. Several features are noteworthy. First, expression of different peptide genes is established at distinct developmental stages. In the dorsal spinal cord, Sst expression starts at E11.5, followed by Tac1 expression at E12.5, CCK expression at E14.5, Pdyn and Penk1 expression at E16.5–P0, and finally Tac2 expression at P5–P7 (Fig. 2) (data not shown). Second, as reported previously (Todd and Spike, 1993), each peptide gene exhibits a unique lamina-specific expression pattern (Fig. 2). Specifically, in the P7 spinal cord, Sst expression is enriched in superficial laminas but is also widely distributed, Tac2 and CCK expression is confined to the intermediate laminas, Tac1 expression is enriched in the deep laminas, Pdyn expression is enriched in superficial laminas, and Penk1 expression is widely distributed (Fig. 2).
Figure 2.
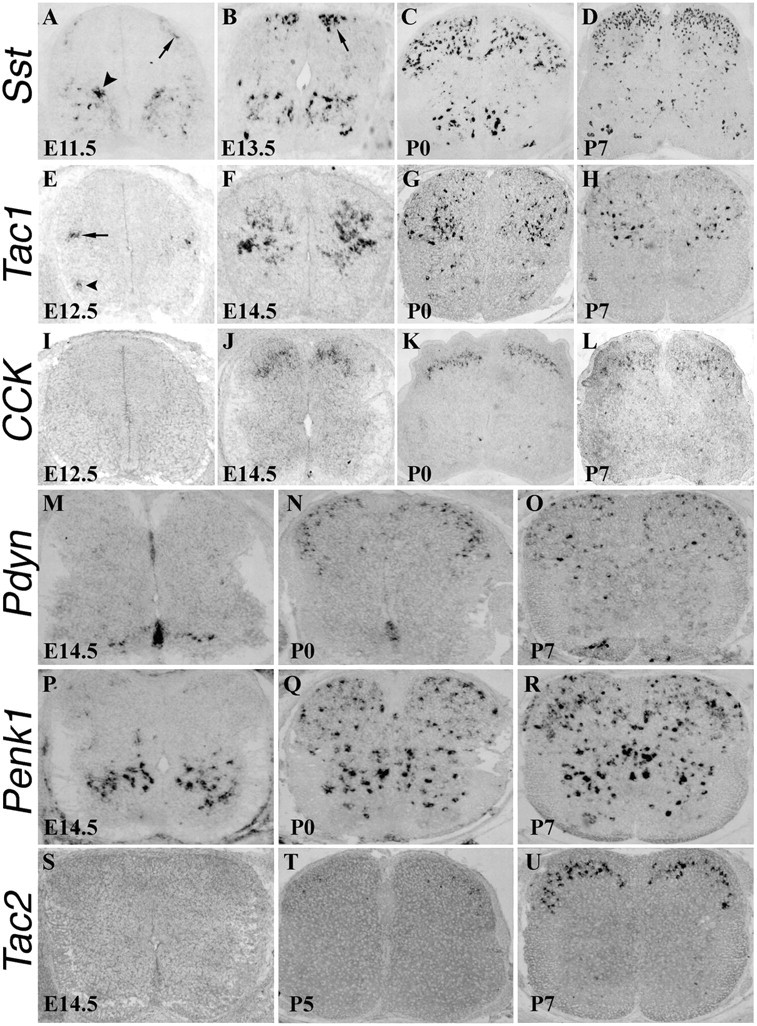
Expression of peptide precursor genes in the developing spinal cord. A–U, In situ hybridization was performed on sections through thoracic spinal cord at various developmental stages using various peptide genes as the probes. A–D, At E11.5, Sst expression was initiated in the dorsal spinal cord (A, arrow) but was already well established in the ventral spinal cord (A, arrowhead). By E13.5, a patch of _Sst_-expressing cells emerged in areas lateral to the dorsal midline (B, arrow). By P0, numerous _Sst_-expressing cells were present throughout the dorsal horn (C), and by P7, Sst expression was enriched in the superficial laminas (D). E–H, At E12.5, Tac1 expression was observed in the intermediate spinal cord area (E, arrow) and in the ventral horn at forelimb levels (E, arrowhead). By E14.5, Tac1 expression was greatly expanded (F). From P0 to P7, most _Tac1_-expressing cells still occupied deep laminas, with the remaining few scattering through superficial laminas (G, H). I–L, _CCK_-expressing cells first emerged at E14.5 in intermediate dorsal horn laminas, and the expression persists at P0–P7. M–R, _Pdyn_-expressing and _Penk1_-expressing cells emerged in the ventral spinal cord at E14.5 (M, P) and then expanded to the dorsal horn from E16.5 to P0 (N, Q) (data not shown). From P0 and on, _Pdyn_-expressing cells were enriched in superficial laminas (N, O), and _Penk1_-expressing cells were scattered throughout the spinal cord (Q, R). S–U, _Tac2_-expressing neurons emerged from P5 to P7 in an intermediate layer.
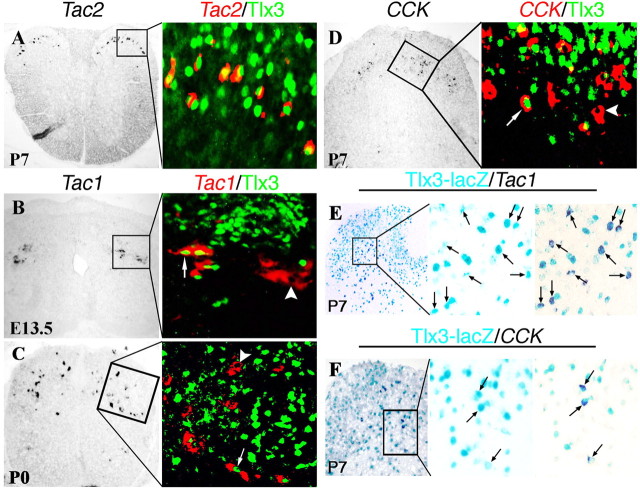
To better understand the relationship between transcriptional regulators and neuropeptide phenotype in the dorsal spinal cord, we undertook a series of double-staining experiments that combined in situ hybridization with peptide cDNAs as the probes and immunostaining with Tlx3 or Pax2 antibodies (Figs. 3, 4). Tac2 expression was confined to a subset of Tlx3+ neurons in intermediate laminas of P7 dorsal spinal cord (Fig. 3A). Only a portion of _Tac1_-expressing neurons expressed Tlx3 at E13.5 (Fig. 3B). At P0 or P7, ∼22.5% (124 of 551) of _Tac1_-expressing neurons and 21.2% (95 of 442) of _CCK_-expressing neurons coexpressed Tlx3 (Fig. 3C,D). However, in P7 Tau-nLacZ(Tlx3Cre) fate-mapping mice, Tac1 and CCK expression was confined exclusively to nLacZ+ neurons (Fig. 3E,F), implying that all _Tac1_-expressing and _CCK_-expressing neurons are derived from Tlx3+ neurons, but Tlx3 expression is transient in most of these peptidergic neurons.
Figure 3.
Expression of Tac2, Tac1, and CCK in Tlx3+ neurons or their derivatives. A–D, Double staining of Tlx3 protein (green) with Tac2, Tac1, or CCK mRNAs (red) on thoracic spinal sections at indicated stages. Note the consistent coexpression of Tac2 with Tlx3 (A), whereas Tac1 and CCK were expressed in both Tlx3+ (arrows) and Tlx3− neurons (arrowheads). Only cells with clear nuclear morphology were analyzed. E, F, A combination of lacZ staining and in situ hybridization. Thoracic spinal sections of P7 Tau-nLacZ(Tlx3Cre) mice were first subjected to lacZ staining (blue). After photographing, the sections were then subjected to in situ hybridization (purple staining). Note a coexpression of lacZ with Tac1 (E, arrows) or CCK (F, arrows).
Figure 4.
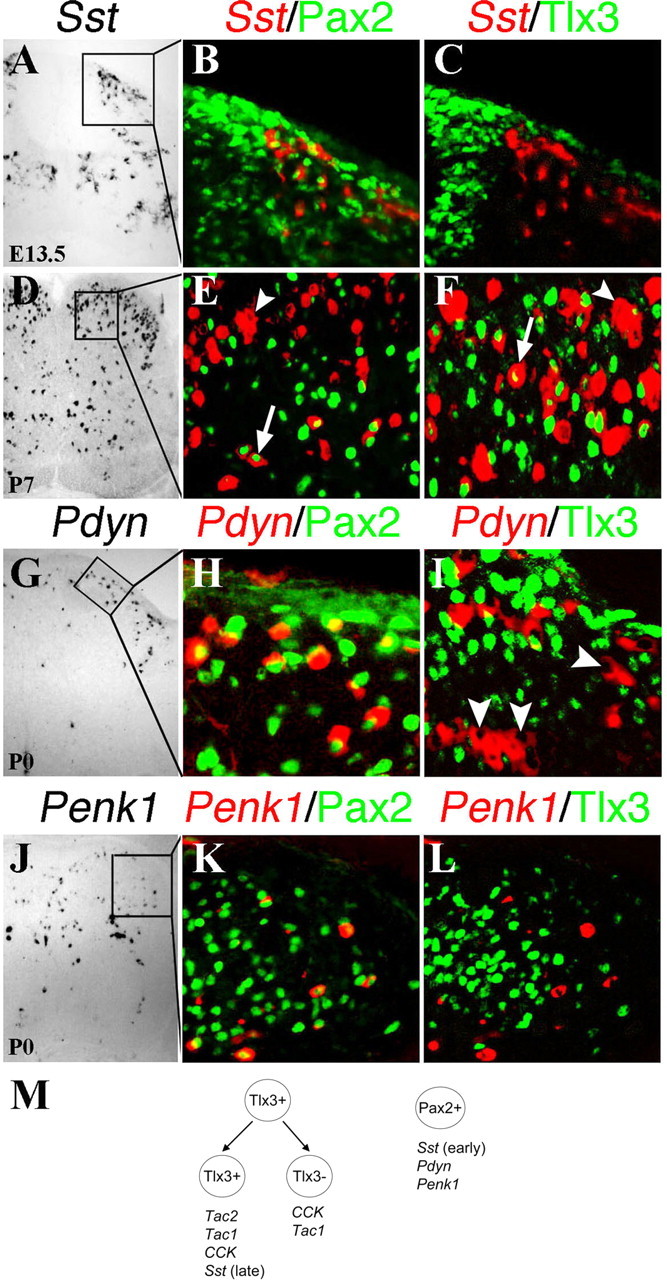
Expression of Sst, Pdyn, and Penk1 in Pax2+ or Tlx3+ neurons. A–L, Double staining of Pax2 protein (B, E, H, K, green) or Tlx3 protein (C, F, I, L, green) with Sst (A–F, red), Pdyn (G–I, red), or Penk1 (J–L, red) mRNA on spinal sections at indicated stages. Bright-field in situ hybridization signals were converted into red pseudo-fluorescent signals. A–C, Note a colocalization of Sst with Pax2 (B) but not with Tlx3 (C) in E13.5 lumbar dorsal horn. D–F, In P7 dorsal horn, Pax2 was expressed in deep lamina _Sst_-expressing neurons (E, arrow) but not in superficial _Sst_-expressing neurons (E, arrowhead). At this stage, a portion of _Sst_-expressing neurons in superficial laminas coexpressed Tlx3 (arrow). Fate mapping showed that the remaining Tlx3-negative _Sst_-expressing neurons in superficial laminas (F, arrowhead) were derived primarily from Tlx3+ neurons (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). G–L, Colocalization of Pdyn and Penk1 with Pax2 (H, K) but not Tlx3 (I, L). Arrows in I indicate _Pdyn_-expressing cells with clear nuclear morphology that lack Tlx3 expression. M, Schematics show two distinct set of peptides that are associated with Tlx3+ neurons (and their derivatives) and Pax2+ neurons, respectively.
Sst exhibited a more complex expression pattern. At E13.5, ∼65.3% (68 of 104) of _Sst_-expressing cells in the dorsal spinal cord expressed Pax2 (Fig. 4B), but none of them expressed Tlx3 (Fig. 4C). At P7, cells coexpressing Pax2 and Sst were confined to deep laminas (Fig. 4E). At this stage, a new population of _Sst_-expressing neurons was detected in superficial laminas that coexpressed Tlx3 (Fig. 4F). Further examination of Sst expression in Tau-nLacZ(Tlx3Cre) mice showed that a majority of _Sst_-expressing neurons in superficial laminas were nLacZ+ (supplemental Fig. 3, available at www.jneurosci.org as supplemental material) and were thus derived from Tlx3+ neurons. Therefore, early and late waves of _Sst_-expressing neurons are primarily associated with Pax2+ and Tlx3+ neurons (and their derivatives), respectively, although some _Sst_-expressing neurons may develop from cells that do not express Pax2 or Tlx3. Pdyn and Penk1 were expressed in neurons that coexpressed Pax2 (Fig. 4H,K) but not Tlx3 (Fig. 4I,L).
In summary, Tlx3+ neurons or their derivatives express Tac1, Tac2, CCK, and a late wave of Sst, whereas Pax2+ neurons express a different set of peptide genes, including Pdyn, Penk1, and an early wave of Sst (summarized in Fig. 4M). Double-color in situ hybridizations further showed that Tac1, Tac2, CCK, and Sst exhibited partially overlapping expression patterns (supplemental Fig. 4, available at www.jneurosci.org as supplemental material), thereby revealing a tremendous diversity of dorsal horn peptidergic neurons.
Tlx1 and Tlx3 are required for peptide gene expression
We next analyzed peptide gene expression in mice that lacked both Tlx3 and its related gene Tlx1, because Tlx3 and Tlx1 exhibit a partial redundancy in cervical and thoracic spinal cord (Cheng et al., 2004). Expression of Tac1 and CCK in the dorsal spinal cord was virtually eliminated in _Tlx1/3_−/− mice, from E13.5 to E18.75 (Figs. 5A–D, 6). Because increased cell death has not been observed in _Tlx1/3_−/− spinal cords during embryonic development (Qian et al., 2002), the simplest interpretation of these results is that Tlx1/3 are required to establish these peptidergic transmitter phenotypes.
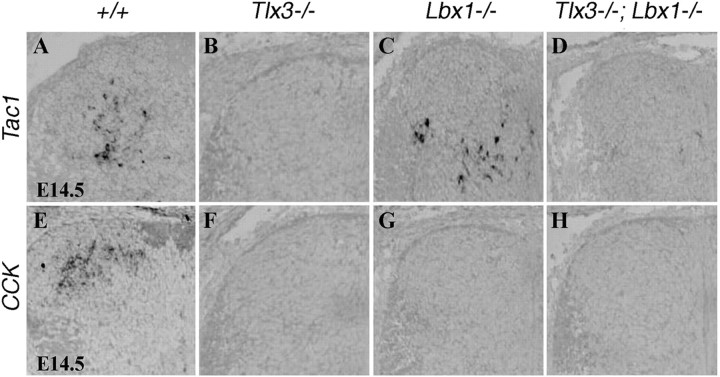
Figure 5.
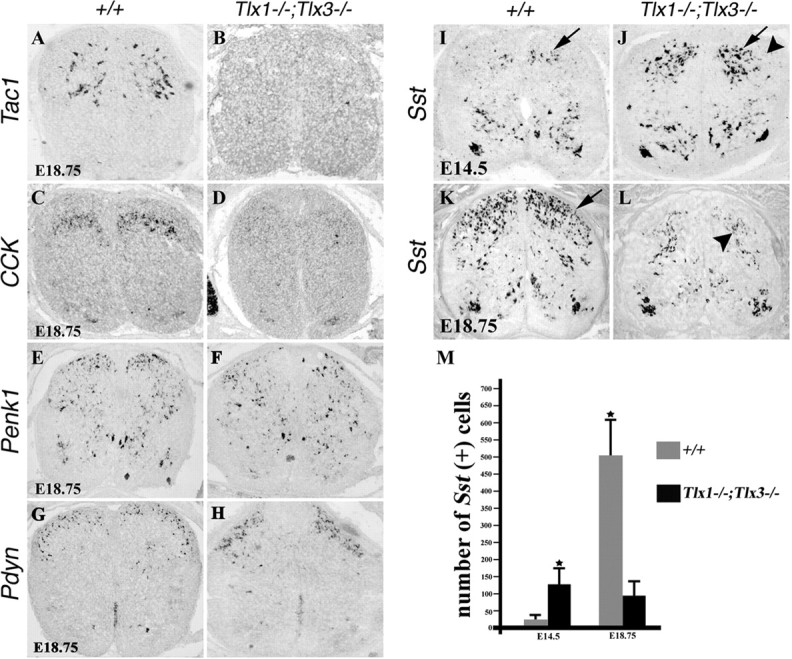
Loss of peptide gene expression in _Tlx1/3_−/− mice. A–L, In situ hybridization was performed on sections through wild-type or _Tlx1/3_−/− thoracic spinal cords at E14.5 or E18.75. A–D, Note a loss of Tac1 and CCK expression in mutants. E–H, Penk1 and Pdyn expression was not reduced (as indicated by quantitative data; see Results). I, J, Sst expression was expanded in the deep dorsal horn of E14.5 _Tlx1/3_−/− embryos (arrows). However, no Sst expression was detected in the most superficial dorsal horn (J, arrowhead). K, L, Sst expression was lost in the superficial dorsal horn of E18.75 _Tlx1/3_−/− mice (arrow), whereas expression in deep dorsal horn laminas persisted (L, arrowhead). M, Quantitative data showed the numbers of _Sst_-expressing neurons per wild-type or _Tlx1/3_−/− dorsal spinal cord section.
Figure 6.
Lbx1 is required for CCK expression. A–H, In situ hybridization was performed on sections of E14.5 lumbar spinal cords with indicated genotypes, with Tac1 and CCK as the probes.
Tlx1/3, however, exerted both negative and positive effects on Sst expression. At E14.5, the number of _Sst_-expressing neurons in dorsal thoracic spinal cord increased by fivefold in _Tlx1/3_−/− embryos compared with wild-type embryos (Fig. 5I,J,M), and most of these _Sst_-expressing neurons were confined to the intermediate and deep dorsal laminas (Fig. 5J). We previously reported that there is a marked increase of Pax2+ neurons in _Tlx1/3_−/− spinal cord (Cheng et al., 2004). Surprisingly, a double staining of Sst and Pax2 showed that only 28.2% of _Sst_-expressing neurons in E14.5 _Tlx1/3_−/− dorsal horn coexpressed Pax2 (supplemental Fig. 5, available at www.jneurosci.org as supplemental material), implying that most of these ectopic _Sst_-expressing neurons were derived from _Tlx1/3_−/− cells that are incapable of switching on Pax2 expression. A potential source could be DI3 interneurons that express Tlx3, but not Lbx1, which is required for Pax2 expression (Helms and Johnson, 2003; Fitzgerald, 2005).
By E18.75, Sst expression in the superficial dorsal horn, which is primarily derived from Tlx3+ neurons, was eliminated in _Tlx1/3_−/− mice (Fig. 5K,L), whereas Sst expression in deep laminas was not affected (Fig. 5L). As a result of this, there is a fivefold reduction in the number of _Sst_-expressing neurons in the dorsal spinal cord of E18.75 _Tlx1/3_−/− mice compared with wild-type mice (Fig. 5M). These data suggest a dual function of Tlx1/3: activating and repressing Sst expression in superficial and deep dorsal horn laminas, respectively.
Expression of Pdyn and Penk1, which are confined to Pax2+ cells in wild-type embryos, did not exhibit obvious changes in E18.75 _Tlx1/3_−/− mice (Fig. 5 E–H). The number of _Penk1_-expressing cells per dorsal horn section at thoracic axial levels was 115 ± 12 in wild-type mice and 123 ± 17 in _Tlx1/3_−/− mice (p > 0.5). The numbers of _Pdyn_-expressing cells in E18.75 dorsal spinal cord were also comparable, 65 ± 6 in wild-type mice versus 69 ± 4 in _Tlx1/3_−/− mice. However, the distribution of _Pdyn_-expressing cells may have been slightly affected, with an apparent increase of the density of _Pdyn_-expressing cells in the superficial laminas (Fig. 5, compare H and G). We previously showed that mutations of Tlx1 and Tlx3 result in a transformation of glutamatergic neurons into Pax2+ GABAergic neurons (Cheng et al., 2004). The lack of a significant increase of _Pdyn_-expressing and _Penk1_-expressing neurons suggests an incomplete switch in cell fate.
In summary, Tlx1/3 are required to establish the expression of a set of peptide genes, including Tac1, CCK, and Sst.
Tlx1/3 use distinct pathways to control peptide and glutamate transmitters: a requirement of Lbx1 for CCK expression
Tlx1/3 acts to antagonize Lbx1 to specify the glutamatergic transmitter phenotype in dorsal horn excitatory neurons (Cheng et al., 2005). Accordingly, a loss of the expression of Slc17a6, which encodes the vesicular glutamate transporter 2 (VGLUT2) and the specific marker for dorsal horn glutamatergic neurons, in _Tlx3_−/− embryos is restored in _Tlx3_−/−;_Lbx1_−/− embryos (Cheng et al., 2005). To determine whether peptide transmitter phenotypes are established in a similar way, we analyzed peptide gene expression in _Tlx3_−/− and _Lbx1_−/− single knock-out mice and _Tlx3_−/−;_Lbx1_−/− double knock-out mice at E14.5. E14.5 was chosen, because cell death occurs after E14.5 in the caudal spinal cord of Lbx1 mutant mice (Gross et al., 2002; Cheng et al., 2005). In addition, lumbar spinal cords were analyzed because Tlx3, but not Tlx1, operates at this axial level (Cheng et al., 2005).
Expression of Tac1 was eliminated in E14.5 _Tlx3_−/− mice (Fig. 6, compare A and B) but not affected in _Lbx1_−/− mice (Fig. 6, compare A and C). Furthermore, unlike a restoration of VGLUT2 expression (Cheng et al., 2005), Tac1 expression was not recovered in _Tlx3_−/−;_Lbx1_−/− mice (Fig. 6D), suggesting that Tlx3 controls Tac1 expression through an _Lbx1_-independent pathway. More surprisingly, CCK expression was eliminated in _Tlx3_−/− and _Lbx1_−/− single knock-out mice and in Tlx3_−/−;Lbx1_−/− double knock-out mice (Fig. 6 E–H), suggesting that both Lbx1 and Tlx3 are required for the expression of CCK. Tlx3 therefore uses distinct pathways to specify glutamate and peptide transmitters.
Brn3a is required for the early wave of Tac1 expression
Tlx1/3 are required for the expression of a set of transcription factors in the dorsal spinal cord (Qian et al., 2002). We hypothesized that Tlx1/3 might use these downstream transcription factors to control the expression of peptide genes. To test this hypothesis, we examined the expression of neuropeptide genes in mice with a null mutation of the Pou4f1 gene, encoding the Brn3a homeobox transcription factor (Quina et al., 2005).
Brn3a was expressed primarily in deep dorsal horn laminas and to a lesser extent in the most superficial laminas in P0 spinal cord (Fig. 7A,B). Double immunostaining showed that only a portion of Brn3a+ neurons coexpressed Tlx3 (Fig. 7A, arrow). This is consistent with previously demonstrated Brn3a expression in early born DI1 and DI2 interneurons that lack Tlx3 expression (Gowan et al., 2001; Qian et al., 2002; Helms and Johnson, 2003). Accordingly, Brn3a expression was essentially, but not completely, eliminated in _Tlx1/3_−/− mice (Fig. 7B).
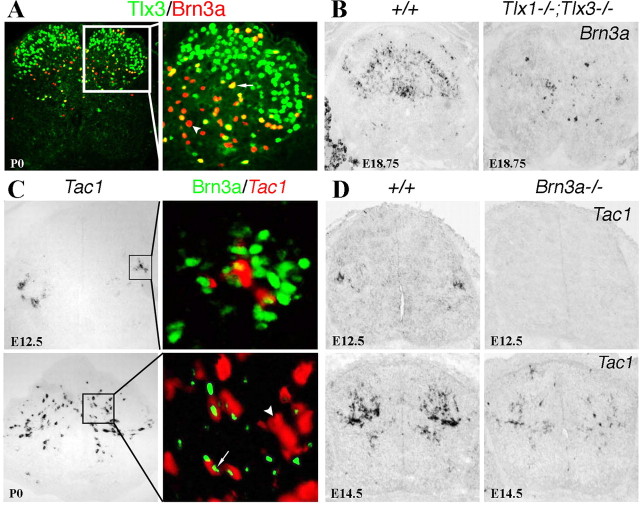
Figure 7.
Brn3a controls an early wave of Tac1 expression. A, Double staining of Tlx3 (green) and Brn3a (red, arrowhead) protein in P0 thoracic spinal cord. Coexpressing cells appear in yellow (arrow). B, In situ hybridization with the Brn3a probe on sections of the thoracic spinal cords of E18.75 wild-type and _Tlx1/3_−/− mice. Note a reduction of Brn3a expression in the mutant spinal cord. C, Double staining of Brn3a protein (green) and Tac1 mRNA (red) on sections of lumbar E12.5 and P0 wild-type spinal cords. Note that Tac1 was expressed exclusively in Brn3a+ neurons at E12.5 but in both Brn3a+ (arrow) and Brn3a− (arrowhead) cells at P0. D, In situ hybridization of Tac1 on sections through lumbar wild-type or _Brn3a_−/− spinal cords at indicated developmental stages.
Because Tac1 is also expressed predominantly in deep dorsal horn laminas, we performed a double staining of Brn3a protein and Tac1 mRNA in the developing spinal cord. We found that at E12.5, virtually all _Tac1_-expressing neurons in the intermediate level of the spinal cord coexpressed Brn3a, but only a fraction of Brn3a+ neurons exhibited Tac1 expression (Fig. 7C). From E12.5 to P0, a new population of _Tac1_-expressing neurons that did not express Brn3a emerged (Fig. 7C).
Consistent with this expression pattern, Tac1 expression was virtually eliminated in the caudal spinal cord of E12.5 _Brn3a_−/− embryos (Fig. 7D), but only reduced in E14.5 _Brn3a_−/− spinal cord (Fig. 7D), suggesting a specific role of Brn3a in controlling the early wave of Tac1 expression.
Because of incomplete loss of Brn3a expression in _Tlx1/3_−/− mice (Fig. 7B), two distinct models may explain a loss of Tac1 expression in both _Tlx1/3_−/− and _Brn3a_−/− mice. First, Tlx1/3 and Brn3a act in a cascade to control Tac1 expression (in other words, Tac1 is established in cells in which Brn3a expression is dependent on Tlx1/3). Second, Tlx1/3 and Brn3a act in combination, meaning that Tac1 expression is established in Tlx3+;Brn3a+ neurons in which Brn3a expression is independent of Tlx3. To distinguish these models, we analyzed Tac1 and Brn3a expression in E12.5 wild-type and _Tlx1/3_−/− embryos. At this stage, Tac1 expression was confined to a lateral region in the middle of the wild-type spinal cord (supplemental Fig. 6, available at www.jneurosci.org as supplemental material). In _Tlx1/3_−/− embryos, expression of both Tac1 and Brn3a was eliminated from this lateral region, whereas _Tlx3_-independent Brn3a expression was located in a dorsomedial area (supplemental Fig. 6, available at www.jneurosci.org as supplemental material). These data are more consistent with the first model that Tlx3 and Brn3a act sequentially to control Tac1 expression.
Expression of other _Tlx3_-dependent genes, including CCK, Sst, the Gria2 glutamate receptor gene (Cheng et al., 2004), and the TRPC3 transient receptor potential channel gene (Li et al., 2006), was not grossly affected in Brn3a mutants (supplemental Fig. 7, available at www.jneurosci.org as supplemental material), suggesting a specific role of Brn3a in controlling the early wave of Tac1 expression.
To determine whether Brn3a is sufficient to promote Tac1 expression, we performed gain-of-function analyses by using chick electroporation technique (Itasaki et al., 1999). Electroporation of a Brn3a expression vector, RCAS-Brn3a, in E2 chick neural tubes resulted in an induction of Tac1 expression at E5 (Fig. 8B) and even more at E7 (Fig. 8D). Electroporation with control vectors did not affect Tac1 expression (Fig. 8E,F), suggesting that Tac1 induction by RCAS-Brn3a electroporation was not caused by side effects associated with proviral vector electroporation (Hermann and Logan, 2003). As in the case with wild-type mouse spinal cord (Fig. 7C), only a portion of Brn3a+ neurons coexpressed Tac1 (data not shown), suggesting that Brn3a needs a specific cellular context to activate Tac1.
Figure 8.
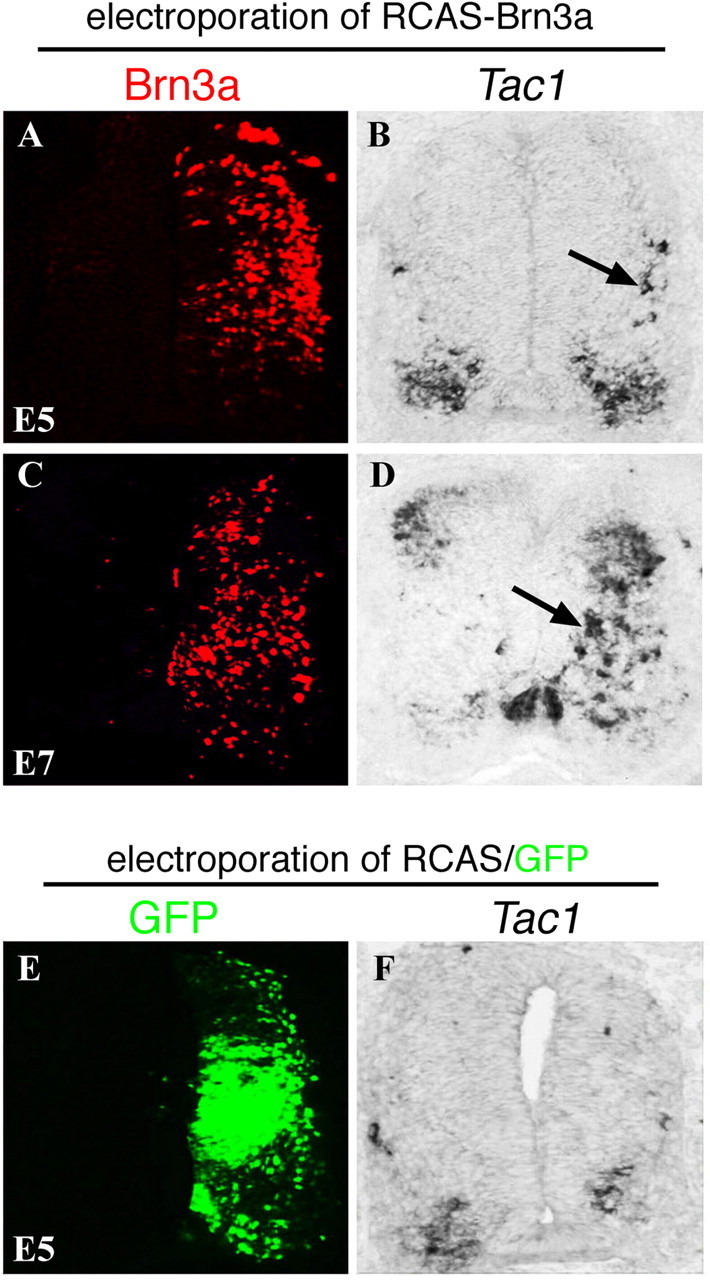
Brn3a induces Tac1 expression in the chick neural tube. A–D, Electroporation of RCAS-Brn3a (plus pCAX-GFP, not shown) into the right side of chick neural tubes at E2, followed by analysis of the expression of exogenous Brn3a protein by immunostaining (A, C) and Tac1 expression by in situ hybridization at E5 (A, B) or E7 (C, D). Arrows indicate ectopic Tac1 expression. E, F, Electroporation with control vectors (RCAS plus pCAX-GFP, referred to as RCAS/GFP). GFP expression (E) was used to monitor electroporation efficacy. Note a lack of SP induction (F).
Discussion
Ontogeny of dorsal horn peptidergic neurons
This study suggests that dorsal horn peptidergic neurons emerge from distinct neuronal populations. Expression of Tac1, Tac2, CCK, and the late wave of Sst is confined to Tlx3+ neurons or their derivatives, and the development of these peptidergic neurons is compromised in mice that lack Tlx3 and Tlx1. Expression of Pdyn, Penk1, and a portion of early wave Sst is restricted to Pax2+ neurons, and their development is independent of Tlx1 or Tlx3. Our data also suggest that some _Sst_-expressing neurons may develop from cells that do not express Tlx3 or Pax2. Tlx3 and Pax2 are associated with excitatory and inhibitory neurons, respectively, at least at embryonic stages (Cheng et al., 2004). Consistently, neurons that produce Neurokinin B (the product of Tac2), Substance P (the product of Tac1), and a late wave of SOM (the product of Sst) belong to glutamatergic excitatory neurons (Proudlock et al., 1993; Todd et al., 2003; Todd and Koerber, 2006; Polgar et al., 2006). Also consistent with an association with Pax2+ neurons, a small number of SOM+ neurons located in the deep dorsal horn are inhibitory neurons (Proudlock et al., 1993).
Separate genetic controls of glutamate and peptide transmitter phenotypes
Tlx1/3 are known to antagonize Lbx1 to control the expression of VGLUT2, the vesicular glutamate transporter and the specific marker for dorsal horn glutamatergic neurons (Todd et al., 2003; Cheng et al., 2004; Fremeau et al., 2004). Loss of VGLUT2 expression in Tlx3 mutant mice is restored in Tlx3_−/−;Lbx1_−/− double mutants (Cheng et al., 2005). However, expression of the _Tlx1/3_-dependent peptide genes is not restored in Tlx3_−/−;Lbx1_−/− double mutants, implying that Tlx1/3 use distinct pathways to coordinate glutamate and peptide transmitters. A separate control of these transmitters is supported by the fact that all dorsal horn excitatory neurons use glutamate as a fast transmitter, whereas individual peptide transmitters are confined to a small subset of dorsal horn neurons.
The development of _Tlx1/3_-dependent peptidergic neurons is subject to complex genetic control. The early, but not late, Tac1 expression is dependent on Brn3a. Moreover, Tlx1/3 can exert both positive and negative effects on Sst expression in different dorsal horn lamina. A surprising result is that CCK expression is dependent on both Lbx1 and Tlx3, despite that Tlx3 antagonizes Lbx1 to control VGLUT2 expression. One potential solution for these seemly conflicting Tlx3 activities is that glutamate and CCK transmitter phenotypes are established at distinct stages. VGLUT2 expression is established soon after cells exit from the cell cycle, and at this stage Tlx3 acts to remove an inhibitory effect of Lbx1 on VGLUT2 expression (Cheng et al., 2005). CCK expression, however, is established at E14.5 (Fig. 2), when Tlx3 protein has been extinguished in most _CCK_-expressing neurons (supplemental Fig. 8, available at www.jneurosci.org as supplemental material). Our hypothesis is that Tlx3 extinguishment allows Lbx1 to escape a Tlx3-mediated inhibition, and Lbx1 might in turn act together with an unknown _Tlx3_-dependent event (established at earlier stages) to control CCK expression.
Tlx1 and Tlx3 orchestrate a set of downstream transcription factors to specify dorsal horn neuron subtypes
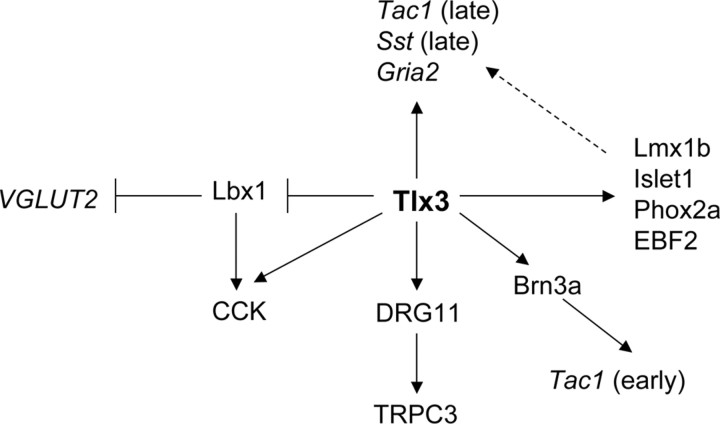
Our findings argue that Tlx1/3 act as “master regulators” in coordinating dorsal horn excitatory neuron development. Virtually all known functional genes that are preferentially expressed in glutamatergic neurons within the dorsal spinal cord are eliminated in _Tlx1/3_−/− mice, including VGLUT2 (Cheng et al., 2004), the glutamate receptor gene Gria2 (Kerr and Todd, 1998; Cheng et al., 2004), the channel gene TRPC3 (Li et al., 2006), and a set of peptide genes described in this study. Tlx1/3 activate a set of downstream transcription factors, some of which appears to control a portion of _Tlx1/3_-dependent differentiation programs (Fig. 9). For example, Brn3a is required for the early wave of Tac1 expression but is dispensable for the expression of other _Tlx1/3_-dependent genes (supplemental Fig. 6, available at www.jneurosci.org as supplemental material). DRG11, encoded by the homeobox gene Prrxl1 (Saito et al., 1995; Chen et al., 2001), is required for the expression of TRPC3 (Li et al., 2006) but not Gria2 or any category I peptide genes (C. Lopes and D. Lima, unpublished data). Finally, other _Tlx1/3_-dependent transcription factors such as Islet1, Phox2a, and early B-cell factor 2 (EBF2) are all expressed in a fraction of dorsal horn neurons (Tiveron et al., 1996; Qian et al., 2002; Li et al., 2006), and therefore they likely contribute to specification of other specialized dorsal horn neuron subtypes. One challenging unsolved question is to understand how Tlx1/3 are able to activate distinct downstream differentiation programs in distinct dorsal horn neuron contexts.
Figure 9.
Tlx1/3 coordinate dorsal horn neuron development. Tlx3 acts to antagonize Lbx1 to control VGLUT2 expression and the glutamatergic transmitter phenotype. Both Tlx3 and Lbx1 are required for CCK expression, and transient Tlx3 expression may allow Lbx1 to work together with a putative _Tlx3_-dependent downstream event to control CCK expression (see Discussion). Tlx3 acts via Brn3a to control the early wave of Tac1 expression, but Brn3a is not required for the expression of other _Tlx3_-dependent genes such as CCK, Sst, TRPC3, or Gria2. Tlx3 acts via DRG11 to control the expression of TRPC3 (Li et al., 2006), but DRG11 is not required for the expression of any _Tlx3_-dependent peptide genes or Gria2 (C. Lopes and D. Lima, unpublished data). Tlx3 is required for the expression of many other dorsal horn transcription factors such as Islet1, Phox2a, EBF2 [wrongly called EBF3 in our previous paper (Qian et al., 2002)], and Lmx1b (Qian et al., 2002). These downstream factors may control other unknown downstream events or control the expression of those _Tlx3_-dependent but _Brn3a/DRG11_-independent target genes such as late-wave Tac1, late-wave Sst, or Gria2 (dashed arrow).
Modular control of the development of the mammalian nervous system
One important concept in developmental biology is that specification of individual neuronal cell types is controlled by a unique combination of transcription factors (TFs), or combinatorial TF codes (Shirasaki and Pfaff, 2002; Thor and Thomas, 2002). However, after late neuronal phenotypes are analyzed, it becomes increasingly evident that a TF code established in newly born neurons specifies more than one neuronal cell type. As aforementioned, a set of peptidergic neurons and other excitatory neurons in the superficial dorsal horn develop from DI5/DILB neurons that share the same TF code, by coexpressing Tlx3 and Lmx1b (Gross et al., 2002; Müller et al., 2002; Cheng et al., 2004), and Tlx3 coordinates the development of these neurons. In the ventral spinal cord, Engrailed1+ V1 interneurons are composed of multiple neuron subtypes involved with locomotion controls (Sapir et al., 2004; Alvarez et al., 2005). In dorsal root ganglia, the Runx1 runt domain transcription factor is required for the development of a variety of nociceptive sensory neurons (Chen et al., 2006; Ibanez and Ernfors, 2007; Marmigere and Ernfors, 2007; Woolf and Ma, 2007). The emerging theme is that a TF code established in newly formed neurons coordinates specification of a heterogeneous group of neurons that perform related physiological functions, thereby implying a modular control of the development of the mammalian nervous system.
Footnotes
This work was supported by National Institutes of Health Grants R01NS47710 and P01NS047572 from the National Institute of Neurological Disorders and Stroke. C.L. is supported by a visiting student scholarship from Portugal, and Q.M. is a Claudia Adams Barr Scholar. We thank Drs. Senji Shirasawa and Stan Korsmeyer for Tlx1 and Tlx3 null mice, Silvia Arber for Tau-nLacZ reporter mice, Susan Dymecki for FLPe deleter mice, Philippe Soriano for ROSA26-lacZ reporter mice, Mengqing Xiang for early phase analysis of Brn3a mutant phenotype, and Thomas Muller and Carmen Birchmeier for Lbx1 and Tlx3 antibodies. We also thank Drs. Jane Johnson and Chuck Stiles for critical reading of this manuscript.
References
- Alvarez FJ, Jonas PC, Sapir T, Hartley R, Berrocal MC, Geiman EJ, Todd AJ, Goulding M. Postnatal phenotype and localization of spinal cord V1 derived interneurons. J Comp Neurol. 2005;493:177–192. doi: 10.1002/cne.20711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Anderson KV. Patterning cell types in the dorsal spinal cord: what the mouse mutants say. Nat Rev Neurosci. 2003;4:289–297. doi: 10.1038/nrn1073. [DOI] [PubMed] [Google Scholar]
- Chen CL, Broom DC, Liu Y, de Nooij JC, Li Z, Cen C, Samad OA, Jessell TM, Woolf CJ, Ma Q. Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron. 2006;49:365–377. doi: 10.1016/j.neuron.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Chen Z, Rebelo A, White F, Malmberg AB, Baba H, Lima D, Woolf CJ, Basbaum AI, Anderson DJ. The paired homeodomain protein DRG11 is required for the projection of cutaneous sensory afferent fibers to the dorsal spinal cord. Neuron. 2001;31:59–73. doi: 10.1016/s0896-6273(01)00341-5. [DOI] [PubMed] [Google Scholar]
- Cheng L, Arata A, Mizuguchi R, Qian Y, Karunaratne A, Gray PA, Arata S, Shirasawa S, Bouchard M, Luo P, Chen CL, Busslinger M, Goulding M, Onimaru H, Ma Q. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat Neurosci. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- Cheng L, Samad OA, Xu Y, Mizuguchi R, Luo P, Shirasawa S, Goulding M, Ma Q. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat Neurosci. 2005;8:1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- Christensen BN, Perl ER. Spinal neurons specifically excited by noxious or thermal stimuli: marginal zone of the dorsal horn. J Neurophysiol. 1970;33:293–307. doi: 10.1152/jn.1970.33.2.293. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M. The development of nociceptive circuits. Nat Rev Neurosci. 2005;6:507–520. doi: 10.1038/nrn1701. [DOI] [PubMed] [Google Scholar]
- Fremeau RTJ, Voglmaier S, Seal RP, Edwards RH. VGLUTs define subsets of excitatory neurons and suggest novel roles for glutamate. Trends Neurosci. 2004;27:98–103. doi: 10.1016/j.tins.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Glasgow SM, Henke RM, Macdonald RJ, Wright CV, Johnson JE. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development. 2005;132:5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- Gowan K, Helms AW, Hunsaker TL, Collisson T, Ebert PJ, Odom R, Johnson JE. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/s0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Gross MK, Dottori M, Goulding M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron. 2002;34:535–549. doi: 10.1016/s0896-6273(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Grudt TJ, Perl ER. Correlations between neuronal morphology and electrophysiological features in the rodent superficial dorsal horn. J Physiol (Lond) 2002;540:189–207. doi: 10.1113/jphysiol.2001.012890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han ZS, Zhang ET, Craig AD. Nociceptive and thermoreceptive lamina I neurons are anatomically distinct. Nat Neurosci. 1998;1:218–225. doi: 10.1038/665. [DOI] [PubMed] [Google Scholar]
- Helms AW, Johnson JE. Specification of dorsal spinal cord interneurons. Curr Opin Neurobiol. 2003;13:42–49. doi: 10.1016/s0959-4388(03)00010-2. [DOI] [PubMed] [Google Scholar]
- Hermann PM, Logan CC. Electroporation of proviral RCAS DNA alters gene expression in the embryonic chick hindbrain. Biotechniques. 2003;35:942–946. doi: 10.2144/03355st01. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Cholewa-Waclaw J, Nakada Y, Glasgow SM, Masui T, Henke RM, Wildner H, Martarelli B, Beres TM, Epstein JA, Magnuson MA, Macdonald RJ, Birchmeier C, Johnson JE. A nonclassical bHLH Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev. 2008;22:166–178. doi: 10.1101/gad.1628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez CF, Ernfors P. Hierarchical control of sensory neuron development by neurotrophic factors. Neuron. 2007;54:673–675. doi: 10.1016/j.neuron.2007.05.023. [DOI] [PubMed] [Google Scholar]
- Itasaki N, Bel-Vialar S, Krumlauf R. “Shocking” developments in chick embryology: electroporation and in ovo gene expression. Nat Cell Biol. 1999;1:E203–E207. doi: 10.1038/70231. [DOI] [PubMed] [Google Scholar]
- Kajander KC, Sahara Y, Iadarola MJ, Bennett GJ. Dynorphin increases in the dorsal spinal cord in rats with a painful peripheral neuropathy. Peptides. 1990;11:719–728. doi: 10.1016/0196-9781(90)90187-a. [DOI] [PubMed] [Google Scholar]
- Kerr RC, MD, Todd AJ. GluR1 and GluR2/3 subunits of the AMPA-type glutamate receptor are associated with particular types of neurone in laminae I-III of the spinal dorsal horn of the rat. Eur J Neurosci. 1998;10:324–333. doi: 10.1046/j.1460-9568.1998.00048.x. [DOI] [PubMed] [Google Scholar]
- Li MZ, Wang JS, Jiang DJ, Xiang CX, Wang FY, Zhang KH, Williams PR, Chen ZF. Molecular mapping of developing dorsal horn-enriched genes by microarray and dorsal/ventral subtractive screening. Dev Biol. 2006;292:555–564. doi: 10.1016/j.ydbio.2006.01.033. [DOI] [PubMed] [Google Scholar]
- Lima D, Coimbra A. A Golgi study of the neuronal population of the marginal zone (lamina I) of the rat spinal cord. J Comp Neurol. 1986;244:53–71. doi: 10.1002/cne.902440105. [DOI] [PubMed] [Google Scholar]
- Ma Q. Transcriptional regulation of neuronal phenotype in mammals. J Physiol (Lond) 2006;575:379–387. doi: 10.1113/jphysiol.2006.113449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmigere F, Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat Rev Neurosci. 2007;8:114–127. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- Marti E, Gibson SJ, Polak JM, Facer P, Springall DR, Van Aswegen G, Aitchison M, Koltzenburg M. Ontogeny of peptide- and amine-containing neurones in motor, sensory, and autonomic regions of rat and human spinal cord, dorsal root ganglia, and rat skin. J Comp Neurol. 1987;266:332–359. doi: 10.1002/cne.902660304. [DOI] [PubMed] [Google Scholar]
- Mizuguchi R, Kriks S, Cordes R, Gossler A, Ma Q, Goulding M. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat Neurosci. 2006;9:770–778. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- Morgan BA, Fekete DM. Manipulating gene expression with replication-competent retroviruses. Methods Cell Biol. 1996;51:185–218. doi: 10.1016/s0091-679x(08)60629-9. [DOI] [PubMed] [Google Scholar]
- Müller T, Brohmann H, Pierani A, Heppenstall PA, Lewin GR, Jessell TM, Birchmeier C. The homeodomain factor Lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron. 2002;34:551–562. doi: 10.1016/s0896-6273(02)00689-x. [DOI] [PubMed] [Google Scholar]
- Polgar E, Furuta T, Kaneko T, Todd A. ) Characterization of neurons that express preprotachykinin B in the dorsal horn of the rat spinal cord. Neuroscience. 2006;139:687–697. doi: 10.1016/j.neuroscience.2005.12.021. [DOI] [PubMed] [Google Scholar]
- Proudlock F, Spike RC, Todd AJ. Immunocytochemical study of somatostatin, neurotensin, GABA, and glycine in rat spinal dorsal horn. J Comp Neurol. 1993;327:289–297. doi: 10.1002/cne.903270210. [DOI] [PubMed] [Google Scholar]
- Qian Y, Fritzsch B, Shirasawa S, Chen CL, Choi Y, Ma Q. Formation of brainstem (nor)adrenergic centers and first-order relay visceral sensory neurons is dependent on homeodomain protein Rnx/Tlx3. Genes Dev. 2001;15:2533–2545. doi: 10.1101/gad.921501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Shirasawa S, Chen C, Cheng L, Ma Q. Proper development of relay somatic sensory neurons and D2/D4 interneurons requires homeobox genes Rnx/Tlx3 and Tlx1. Genes Dev. 2002;16:1220–1233. doi: 10.1101/gad.982802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quina LA, Pak W, Lanier J, Banwait P, Gratwick K, Liu Y, Velasquez T, O'Leary DD, Goulding M, Turner EE. Brn3a-expressing retinal ganglion cells project specifically to thalamocortical and collicular visual pathways. J Neurosci. 2005;25:11595–11604. doi: 10.1523/JNEUROSCI.2837-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CW, Shutter JR, Korsmeyer SJ. Hox11 controls the genesis of the spleen. Nature. 1994;368:747–749. doi: 10.1038/368747a0. [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Saito T, Greenwood A, Sun Q, Anderson DJ. Identification by differential RT-PCR of a novel paired homeodomain protein specifically expressed in sensory neurons and a subset of their CNS targets. Mol Cell Neurosci. 1995;6:280–292. doi: 10.1006/mcne.1995.1022. [DOI] [PubMed] [Google Scholar]
- Sapir T, Geiman EJ, Wang Z, Velasquez T, Mitsui S, Yoshihara Y, Frank E, Alvarez FJ, Goulding M. Pax6 and engrailed 1 regulate two distinct aspects of renshaw cell development. J Neurosci. 2004;24:1255–1264. doi: 10.1523/JNEUROSCI.3187-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Shirasawa S, Arata A, Onimaru H, Roth KA, Brown GA, Horning S, Arata S, Okumura K, Sasazuki T, Korsmeyer SJ. Rnx deficiency results in congenital central hypoventilation. Nat Genet. 2000;24:287–290. doi: 10.1038/73516. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Thor S, Thomas J. Motor neuron specification in worms, flies and mice: conserved and “lost” mechanisms. Curr Opin Genet Dev. 2002;12:558–564. doi: 10.1016/s0959-437x(02)00340-4. [DOI] [PubMed] [Google Scholar]
- Tiveron MC, Hirsch MR, Brunet JF. The expression pattern of the transcription factor Phox2 delineates synaptic pathways of the autonomic nervous system. J Neurosci. 1996;16:7649–7660. doi: 10.1523/JNEUROSCI.16-23-07649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd AJ, Koerber HR. Neurochemical substrates of spinal nociception. In: McMahon SB, Koltzenburg M, editors. Wall and Melzack's textbook of pain. Ed 5. Edinburgh, UK: Churchill Livingstone; 2006. pp. 73–90. [Google Scholar]
- Todd AJ, Spike RC. The localization of classical transmitters and neuropeptides within neurons in laminae I-III of the mammalian spinal dorsal horn. Prog Neurobiol. 1993;41:609–645. doi: 10.1016/0301-0082(93)90045-t. [DOI] [PubMed] [Google Scholar]
- Todd AJ, Hughes DI, Polgár E, Nagy GG, Mackie M, Ottersen OP, Maxwell DJ. The expression of vesicular glutamate transporters VGLUT1 and VGLUT2 in neurochemically defined axonal populations in the rat spinal cord with emphasis on the dorsal horn. Eur J Neurosci. 2003;17:13–27. doi: 10.1046/j.1460-9568.2003.02406.x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gardell LR, Ossipov MH, Vanderah TW, Brennan MB, Hochgeschwender U, Hruby VJ, Malan TPJ, Lai J, Porreca F. Pronociceptive actions of dynorphin maintain chronic neuropathic pain. J Neurosci. 2001;21:1779–1786. doi: 10.1523/JNEUROSCI.21-05-01779.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenfeld-Hallin Z, Xu XJ, Hökfelt T. The role of spinal cholecystokinin in chronic pain states. Pharmacol Toxicol. 2002;91:398–403. doi: 10.1034/j.1600-0773.2002.910619.x. [DOI] [PubMed] [Google Scholar]
- Woolf CJ, Ma Q. Nociceptors-noxious stimulus detectors. Neuron. 2007;55:353–364. doi: 10.1016/j.neuron.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Xu XJ, Puke MJ, Verge VM, Wiesenfeld-Hallin Z, Hughes J, Hokfelt T. Up-regulation of cholecystokinin in primary sensory neurons is associated with morphine insensitivity in experimental neuropathic pain in the rat. Neurosci Lett. 1993;152:129–132. doi: 10.1016/0304-3940(93)90500-k. [DOI] [PubMed] [Google Scholar]