Different regulation of wild-type and mutant Cu,Zn superoxide dismutase localization in mammalian mitochondria (original) (raw)
Abstract
The antioxidant enzyme Cu,Zn superoxide dismutase (SOD1) is predominantly localized in the cytosol, but it is also found in mitochondria. Studies in yeast suggest that apoSOD1 is imported into mitochondria and trapped inside by folding and maturation, which is facilitated by its copper chaperone for SOD1 (CCS). Here, we show that in mammalian cells, SOD1 mitochondrial localization is dictated by its folding state, which is modulated by several interconnected factors. First, the intracellular distribution of CCS determines SOD1 partitioning in cytosol and mitochondria: CCS localization in the cytosol prevents SOD1 mitochondrial import, whereas CCS in mitochondria increases it. Second, the Mia40/Erv1 pathway for import of small intermembrane space proteins participates in CCS mitochondrial import in a respiratory chain-dependent manner. Third, CCS mitochondrial import is regulated by oxygen concentration: high (20%) oxygen prevents import, whereas physiological (6%) oxygen promotes it. Therefore, SOD1 localization responds to changes in environmental conditions following redistribution of CCS, which operates as an oxygen sensor. Fourth, all of the cysteine residues in human SOD1 are critical for its retention in mitochondria due to their involvement in intramolecular disulfide bonds and in the interaction with CCS. Mutations in SOD1 are associated with autosomal dominant familial amyotrophic lateral sclerosis. Like the wild-type protein, mutant SOD1 localizes to mitochondria, where it induces bioenergetic defects. We find that the physiological regulation of mitochondrial localization is either inefficient or absent in SOD1 pathogenic mutants. We propose misfolding and aggregation of these mutants that trap them inside mitochondria.
INTRODUCTION
Cu,Zn superoxide dismutase (SOD1) is an abundant protein that catalyzes the dismutation of superoxides to hydrogen peroxide. SOD1 is localized predominantly in the cytosol, but it is also found in other cellular compartments, including nucleus (1,2), endoplasmic reticulum (3) and mitochondria. In the latter, SOD1 is concentrated in the intermembrane space (IMS) (4–7), but it can also be detected in the matrix (8) and on the outer membrane (OM) (9–11). Wild-type (WT) SOD1 in mitochondria is thought to provide protection from superoxide produced by the respiratory chain on the outer side of the inner membrane (IM), which is inaccessible to the matrix-residing MnSOD.
To date, over one hundred mutations in the gene encoding for SOD1 have been linked to familial forms of amyotrophic lateral sclerosis (fALS), a progressive motor neuron disease, which results in selective degeneration of upper and lower motor neurons causing paralysis and death. In SOD1-fALS, a toxic gain of function of the mutant protein is responsible for disease pathogenesis, because several pathogenic mutants retain SOD1 activity and mutant SOD1 does not impair the function of WT SOD1 (12). The toxic mechanism of mutant SOD1 that causes motor neuron death is still unclear, but numerous pathogenic pathways have been postulated, including aberrant production of free radical species, neurofilament dysregulation, impaired axonal transport, glutamate excitotoxicity, induction of apoptosis and proteasome and mitochondrial dysfunction (13). Mutant SOD1 in the mitochondria is thought to contribute to the wide range of abnormalities observed in SOD1-fALS: mitochondrial morphology is affected from presymptomatic stages in mouse models of fALS; swollen mitochondria, filled with vacuoles resulting from the expansion of the IMS, contain mutant SOD1 in large proteinaceous aggregates (6,14,15). Furthermore, oxidative phosphorylation, respiratory chain enzyme activity, calcium handling and mitochondrial transport defects have been described in mutant SOD1 transgenic mice (7,15–20). Although a direct contribution of mutant SOD1 to mitochondrial damage is widely accepted, the mechanisms whereby SOD1 localizes in mitochondria under physiological or pathological conditions in mammalian cells are largely unknown.
The mechanisms of WT and mutant SOD1 import from the cytosol, where it is synthesized, into the various mitochondrial compartments need to be investigated in the context of the more general mechanisms that regulate the composition of the mitochondrial proteome. Mitochondria contain their own DNA, which encodes for 13 polypeptides of the respiratory chain. However, these represent only ∼1% of mitochondrial proteins, whereas the rest is encoded by nuclear DNA, synthesized by cytosolic ribosomes and imported in mitochondria by complex translocation machineries. Proteins destined for the matrix, IMS, IM or OM, are imported by diverse mechanisms that are dictated by their mitochondrial targeting signals, typically N-terminal or internal amino acid motifs. However, some mitochondrial proteins, such as apocytochrome c, are devoid of recognizable targeting signals (21–23). Similarly, SOD1 lacks any recognizable mitochondrial targeting signal. Studies in yeast demonstrated that the apoprotein is able to cross the OM only in its non-metallated and disulfide reduced state, and that the maturation of SOD1, facilitated by mitochondrially localized copper chaperone for SOD1 (CCS), traps the protein in the IMS (24). Recent studies in SOD1-expressing NSC34 (motor neuron-like) cells and transgenic mice have focused on disulfide-linked oligomeric forms of SOD1 trapped in mitochondria (25,26). Based on these observations, the cysteine residues of SOD1 appear to play an essential role in mitochondrial localization of the protein.
The interplay between SOD1 folding and the various components of the mitochondrial import machinery regulates SOD1 localization in mitochondria. In this study, we have investigated this interplay, in both WT and mutant forms of human SOD1 expressed in mammalian cells. We show that protein folding is the major determinant of SOD1 mitochondrial localization and it is physiologically modulated by multiple factors, including the partitioning of CCS between cytosol and mitochondria, oxygen concentration and respiratory chain function. Importantly for fALS, we find that SOD1 pathogenic mutants lose the physiological regulation of mitochondrial import and retention and that their localization in mitochondria depends largely on aberrant folding and aggregation.
RESULTS
WT SOD1 mitochondrial localization in mammalian cells
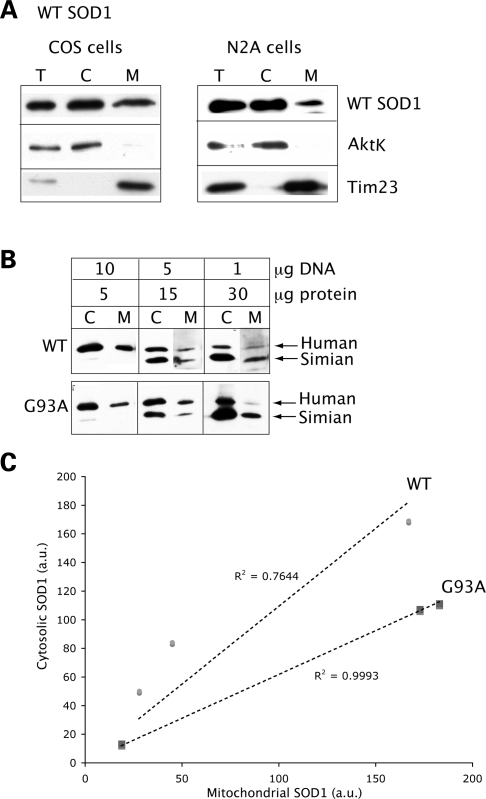
WT SOD1 localization was examined in cytosolic and mitochondrial fractions of transfected COS cells. In all fractionation/localization experiments, SOD1 was detected by western blot using a sheep polyclonal anti-SOD1 antibody. Equal amounts of proteins (5 µg) were loaded in each lane. A proportion of the expressed protein is found in the mitochondrial fraction (Fig. 1A, left panels). AktK, used to assess cytosolic protein contamination in mitochondrial fractions, was undetectable. Tim23, the translocase of the IM, was used as a loading marker for mitochondrial proteins. To confirm WT SOD1 mitochondrial localization in neurons, we expressed the protein in N2A neuroblastoma cells. WT SOD1 is localized to the N2A mitochondrial fraction, in a proportion similar to that found in COS cells (Fig. 1A, right panels).
Figure 1.
WT SOD1 localization in mammalian mitochondria. (A) Western blot of total lysate (T), cytosolic (C) and mitochondrial (M) fractions from COS or N2A neuroblastoma cells transfected with WT SOD1. Equal amounts of proteins (5 µg) were loaded in each lane, and SOD1 was detected by the SOD1 polyclonal antibody. A proportion of the WT SOD1 localizes to mitochondria in both cell types. The same blots were probed with AktK and Tim23 to exclude cytosolic protein contamination and to estimate mitochondrial loading, respectively. (B) COS cells were transfected with WT or G93A mutant SOD1 using different amounts of plasmid DNA (1, 5 and 10 µg). Proteins were loaded for the cytosolic and mitochondrial fractions as indicated (5, 15 and 30 µg), and SOD1 was detected with the SOD1 polyclonal antibody. Human SOD1 migrates higher in the gel than the endogenous simian SOD1. (C) SOD1 levels in cytosolic and mitochondrial fractions were determined by densitometry of western blot immunoreactive bands. SOD1 contents in cytosol and mitochondria were normalized to AktK and Tim23 (data not shown), respectively. The ratio between cytosolic and mitochondrial SOD1 content follows an approximately linear correlation for both WT and mutant G93A SOD1 (the _R_2 values are indicated for both WT and G93A SOD1).
To determine that SOD1 mitochondrial localization is not an artifact due to protein overexpression (27), we transfected COS cells with increasing amounts of plasmid DNA (1, 5 and 10 µg) encoding either WT or G93A mutant SOD1, and compared the ratio between cytosolic and mitochondrial SOD1 content (Fig. 1B). The ratio follows an approximately linear correlation, both for mutant and WT SOD1 (Fig. 1C), suggesting that the amount of SOD1 localized to mitochondria is proportional to the expression levels of the protein, and that mitochondrial localization occurs also when transgenic SOD1 is expressed at levels comparable to or below the endogenous ones.
WT SOD1 mitochondrial content is modulated by CCS
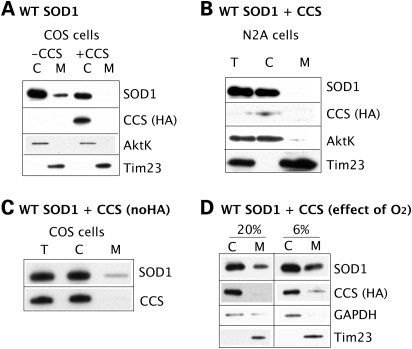
In yeast, mitochondrial SOD1 content depends on the levels of mitochondrial CCS (24). To test whether CCS modulates SOD1 mitochondrial localization in a mammalian system, we co-transfected WT SOD1 and CCS-HA in COS and N2A cells. In both cell types, looking at CCS localization with an antibody against the HA tag, we find that CCS localizes in the cytosol, whereas it is undetectable in mitochondria (Fig. 2A), suggesting that under our experimental conditions, CCS is not efficiently imported in mitochondria. WT SOD1 mitochondrial content is markedly decreased in cells co-expressing CCS when compared with cells transfected with WT SOD1 alone (Fig. 2A). In COS cells expressing CCS-HA, SOD1 mitochondrial content is reduced by 98% when compared with cells expressing WT SOD1 alone. In N2A cells expressing CCS-HA, mitochondrial SOD1 is undetectable (Fig. 2B). This suggests that increased cytosolic CCS may accelerate apoSOD1 folding and maturation, thus preventing its mitochondrial import. To exclude that the HA tag added to the C-terminus of CCS does not interfere with mitochondrial localization and interaction with SOD1, we co-expressed WT SOD1 and a CCS construct without the HA tag in COS cells. We confirm that CCS is not detectable in mitochondria and that WT SOD1 mitochondrial content is decreased also under these conditions (Fig. 2C). The absence of transgenic CCS from mitochondria under our experimental conditions raises a question, as it was shown that mitochondria from the nervous tissue of transgenic mice expressing CCS contain significant amounts of the protein (15). We hypothesized that the difference in CCS mitochondrial import may be due to the oxygen concentration in cultured cells and tissue (∼20 and 6%, respectively). Therefore, we examined CCS localization in cultured cells exposed to 6% oxygen for 48 h after transfection. Under these conditions, we detect the presence of CCS in mitochondria of COS cells and a parallel 60% increase in mitochondrial SOD1 (Fig. 2D). These results suggest that high oxygen concentration promotes oxidative folding of CCS in the cytosol, thus preventing its mitochondrial import, while physiological tissue oxygen concentrations favor CCS mitochondrial localization.
Figure 2.
WT SOD1 in mitochondria is modulated by CCS localization, which depends on oxygen levels. (A) SOD1 mitochondrial localization was assessed in COS cells transfected with WT SOD1 with or without CCS-HA. CCS is detected by the anti-HA antibody in the cytosolic fraction, but not in mitochondria. SOD1 mitochondrial content is decreased by CCS co-expression by 98% when compared with cells expressing WT SOD1 alone. (B) SOD1 mitochondrial localization assessed in N2A cells co-transfected with WT SOD1 and CCS-HA. CCS is detected in the cytosol only. SOD1 is undetectable in N2A mitochondria. (C) To exclude an interference by the HA tag, COS cells were transfected with WT SOD1 and a CCS construct without the C-terminal HA. Again, using an antiserum against CCS, the protein is detected in the cytosol but not in mitochondria. Consistently, mitochondrial WT SOD1 content is reduced in COS cells co-expressing CCS when compared with cells expressing WT SOD1 alone. (D) To assess the effect of oxygen concentration on CCS localization, transfected cells were exposed either to ambient oxygen (20%) or to physiological oxygen (6%) for 48 h. Under 20% oxygen, CCS-HA is undetectable in mitochondria, but it becomes detectable in 6% oxygen. The mitochondrial content of SOD1 is increased in 6% oxygen by 60% when compared with 20% oxygen.
Role of Mia40-Erv1 disulfide relay system in SOD1 mitochondrial import
IMS proteins containing twin CX9C or CX3C cysteine motifs are imported into mitochondria in a unique manner. The import of these IMS proteins, such as COX17 and the small Tim proteins, involves a series of redox reactions with the receptor protein Mia40 and its partner Erv1 (28). These reactions result in the formation of intramolecular disulfide bonds in the imported proteins that contribute to their retention in the IMS.
Although human SOD1 does not contain the typical cysteine repeat motifs, it has four cysteines, two of which, C57 and C146, form an intramolecular disulfide bridge. CCS folding also involves intramolecular disulfide bonds. Furthermore, the interaction between CCS and SOD1 occurs through a transitory intermolecular disulfide bond between C57 of SOD1 and C229 of CCS (29). These chemical properties of the CCS/SOD1 interactions are reminiscent of the Mia40 pathway. It has been suggested that CCS acts as a receptor protein, similar to Mia40, in the import of SOD1 into mitochondria (28), but so far the only evidence supporting this hypothesis is that in Erv1-depleted yeast, the amount of SOD1 in mitochondria was reduced (28).
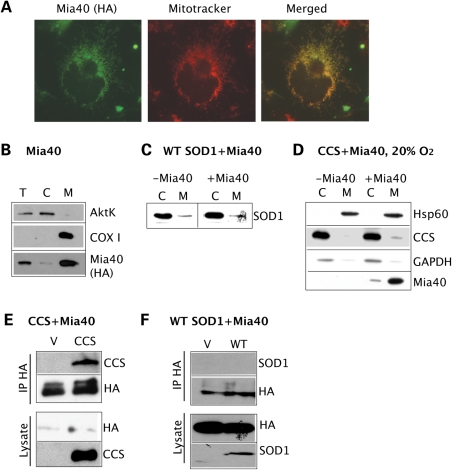
To determine if Mia40 pathway is involved in CCS/SOD1 mitochondrial import, we constructed a Mia40 expression plasmid with a C-terminal HA tag. As expected, Mia40 localizes to mitochondria in COS cells by immuno-colocalization with Mitotracker (Fig. 3A) and by cell fractionation (Fig. 3B). When we co-transfect WT SOD1 with Mia40, we find no change in SOD1 mitochondrial content when compared with cells co-transfected with an empty vector (Fig. 3C). However, when CCS is co-expressed with Mia40, a portion of CCS, which normally is undetectable in the mitochondrial fraction, is now found in mitochondria (Fig. 3D). To establish if there is a physical interaction between Mia40 and either SOD1 or CCS, we performed co-immunoprecipitation experiments of crosslinked proteins using the HA antibody for Mia40 pull-down. We find that CCS (Fig. 3E), but not WT SOD1 (Fig. 3F), co-immunoprecipitates with Mia40. Taken together, these results suggest that CCS, but not SOD1, is a direct substrate for the Mia40/Erv1 import pathway.
Figure 3.
Role of Mia40-Erv1 disulfide relay system in SOD1 mitochondrial import. (A) By immunocytochemistry, Mia40 (in green) co-localizes with mitochondria labeled with Mitotracker (in red), as shown by the yellow color in the merged image. (B) Mia40 expression and localization were determined in COS cells transfected with Mia40-HA. Cellular fractionation shows Mia40 localized almost exclusively in the mitochondrial fraction. Cytochrome oxidase subunit I (COXI) was used as a mitochondrial marker and AktK as a cytosolic marker. (C) SOD1 mitochondrial content is unaffected by co-expression with Mia40 when compared with empty vector control (−Mia40). (D) In cells transfected with CCS with or without Mia40 under 20% oxygen, a small proportion of CCS localizes to mitochondria only when co-expressed with Mia40. In this case, Hsp60 was used as a mitochondrial marker. (E) In cells co-expressing CCS and Mia40(HA) or empty vector, immunoprecipitation of Mia40 using the HA antibody shows that CCS co-immunoprecipitates with Mia40. (F) In cells co-expressing SOD1 and Mia40(HA) or empty vector, immunoprecipitation using the HA antibody does not show WT SOD1 co-immunoprecipitation with Mia40.
Regulation of mitochondrial CCS and SOD1 content by the respiratory chain
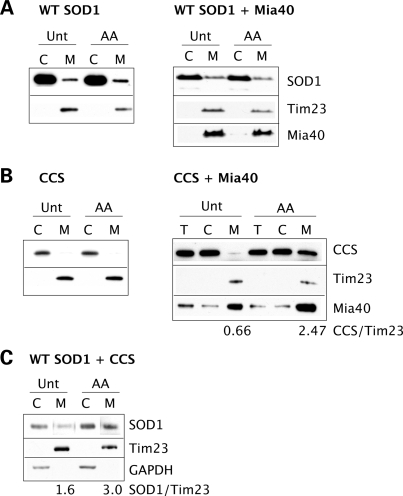
In yeast, the Mia40/Erv1 disulfide relay system is linked to and regulated by the mitochondrial respiratory chain (30). Erv1 is oxidized by donating electrons to cytochrome c, which in turn is reduced by complex III and oxidized by complex IV. Blocking complex III with specific inhibitors, such as antimycin A, results in an increase of oxidized cytochrome c and a sequential buildup of oxidized Erv1 and Mia40, leading to increased efficiency of the import system. To test whether the respiratory chain modulates SOD1 and CCS content in mammalian mitochondria in a Mia40-dependent manner, we co-transfected Mia40 and either CCS or WT SOD1 in 20% oxygen, and treated cells with antimycin A for 16 h prior to fractionation. Although complex III inhibition has virtually no effect on SOD1 (Fig. 4A), the relative mitochondrial CCS content normalized by Tim23 shows approximately a 4-fold increase when the protein is co-expressed with Mia40 (Fig. 4B, right panel), further suggesting that CCS is an Mia40/Erv1 substrate. This increase in CCS mitochondrial content by antimycin A treatment was also found under 6% oxygen (data not shown). To test whether the presence of CCS in mitochondria affects SOD1 localization, we treated cells co-expressing WT SOD1 and CCS with antimycin A, resulting in a 2-fold increase of mitochondrial SOD1 content (Fig. 4C). As antimycin A had no effect on SOD1 without the co-expression of CCS (Fig. 4A), we conclude that blockage of complex III and increased cytochrome c oxidation affect SOD1 import indirectly through Mia40/Erv1 modulation of CCS import.
Figure 4.
Regulation of mitochondrial CCS and SOD1 content by the respiratory chain. Mitochondrial CCS and WT SOD1 contents were assessed in cells treated with the complex III inhibitor, antimycin A (AA) and compared with untreated cells (Unt). (A) AA has minimal effects on SOD1 mitochondrial content in cells transfected with WT SOD1 alone (left panel) or with Mia40 (right panel). (B) AA treatment results in approximately a 4-fold increase of mitochondrial CCS content when the protein is expressed together with Mia40 (right panel), but not when it is expressed by itself (left panel). The relative CCS mitochondrial content is shown at the bottom. (C) In cells co-expressing CCS and SOD1, AA results in an increase in the mitochondrial content of SOD1. The relative SOD1 mitochondrial content is shown at the bottom.
Human SOD1 mutagenesis
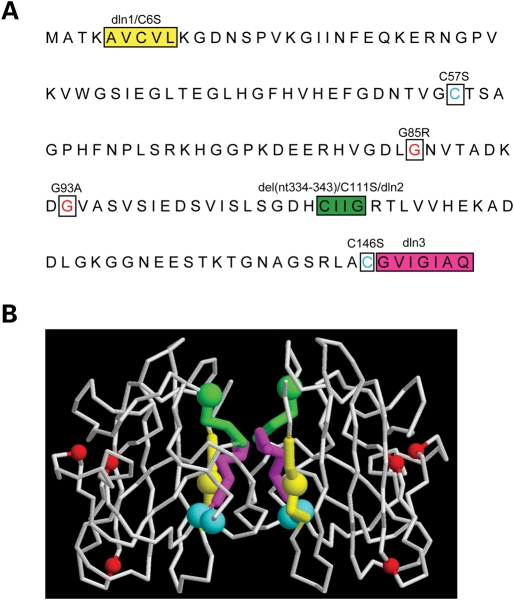
Although most nuclear encoded mitochondrial proteins contain mitochondrial targeting signals or sequence motifs, such as cysteine repeats, that guide their translocation across the mitochondrial membranes and favor their retention inside mitochondria, these elements are not obvious in SOD1. To determine whether there are regions of SOD1 critical for its mitochondrial import and/or retention, we created various SOD1 mutants by site-directed mutagenesis (Fig. 5A). To assess the maturation (folding, metallation and dimerization) state of each SOD1 mutant, we performed superoxide dimutase in-gel activity assays and determined the sensitivity to proteinase K digestion (Table 1). As expected, SOD1 mutants with preserved enzymatic activity are also resistant to proteinase K digestion (31,32).
Figure 5.
Human SOD1 mutagenesis. (A) The SOD1 residues that were changed by in vitro mutagenesis to create the mutants used in the study are highlighted. Hydrophobic stretches were removed to create deletion 1 (dln1, yellow), deletion 2 (dln2, green) and deletion 3 (dln3, magenta) mutants. Four cysteine residues (C6, C57, C111, C146) were changed to serines. C6 and C111 are located within the regions encompassing dln1 and dln2, respectively. The cysteines residues forming the intramolecular disulfide bridge, C57 and C146, are indicated in blue. Pathogenic mutants G85R and G93A SOD1 are shown in red. The Frameshift SOD1 mutant has a 35 amino acid-altered C-terminus with an early truncation at amino acid 145. (B) Tridimensional representation of dimerized SOD1. The mutated residues are indicated by colors as in (A). The hydrophobic stretches of SOD1 are located at the dimer interface. C57 and C146 (in blue), which form the intramolecular disulfide bond, are faced opposing each other.
Table 1.
Characterization of the SOD1 mutants used in this study
| Type | Mutation | Description | SOD1 activity | PK sensitivity |
|---|---|---|---|---|
| Cysteines | C6S | + | − | |
| C57S | − | + | ||
| C111S | + | − | ||
| C146S | − | + | ||
| Hydophobic Stretches | Deletion 1 | Δ5–7 | − | + |
| Deletion 2 | Δ111–114 | − | + | |
| Deletion 3 | Δ147–153 | − | + | |
| Pathogenic | G93A | + | − | |
| G93A/C111S | + | − | ||
| G85R | − | + | ||
| G85R/C111S | − | + | ||
| Frameshift | del(nt334–343) | Stop at 145 | − | + |
Effects of cysteine residue substitutions on SOD1 mitochondrial localization
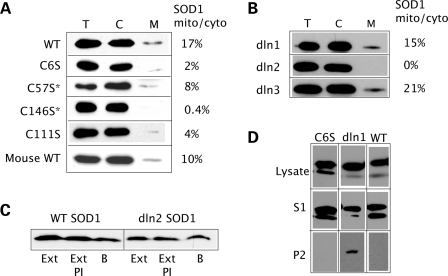
Because cysteine residues are critical for the mitochondrial localization of small IMS proteins that are imported through the Mia40/Erv1 pathway, we investigated the involvement of each of the four cysteines residues of SOD1 (C6, C57, C111 and C146) in its mitochondrial localization. Each cysteine residue was mutagenized to serine and mitochondrial content of SOD1 was assessed. We find that all four cysteine mutants are poorly localized to mitochondria (Fig. 6A). C57 and C146 are involved in the intramolecular disulfide bond; thus C57S and C146S mutants lack this critical step in SOD1 folding. In addition, elimination of the disulfide bond alters loop IV of SOD1 and disrupts dimer interface interactions (33). Failure to fold and dimerize is likely to allow these mutants to slip out of the mitochondrial OM import pore, as shown for other small IMS proteins such as Tim13 (34).
Figure 6.
Cysteine substitutions and ablation of hydrophobic stretch 2 reduced SOD1 mitochondrial localization. (A) All cysteine to serine mutations in human SOD1 result in significantly reduced mitochondrial localization when compared with WT. Asterisk indicates the cysteines involved in the intramolecular disulfide bond formation. Mouse SOD1, which naturally contains a serine at position 111, localizes to mitochondria much more abundantly than human C111S SOD1. The proportion of mitochondrial/cytosolic SOD1 is shown next to each panel. (B) Deletion of hydrophobic stretches in dln1 and dln3 mutants does not impair SOD1 mitochondrial localization, whereas the dln2 mutant is undetectable in the mitochondrial fraction. The proportion of mitochondrial/cytosolic SOD1 is shown. (C) Cytosolic fractions containing WT and dln2 SOD1 were treated with mitochondrial extracts (Ext) from untransfected COS cells. As controls, samples were incubated with ME plus protease inhibitors (Ext PI) or mitochondrial resuspension buffer alone (B). Neither WT nor dln2 SOD1 are degraded by proteases contained in the ME. (D) Detergent solubility assay to detect SOD1 aggregates in total lysates from cells expressing C6S, dln1 and WT SOD1. Only the dln1 mutant protein forms detergent-insoluble aggregates detected in the P2 fraction, whereas WT and C6S SOD1 are only found in the soluble (S1) fraction.
Unlike C57S and C146S, C6S and C111S mutants are able to fold and retain enzymatic activity (Table 1), yet they show reduced mitochondrial localization. Neither C6 nor C111 are involved in intramolecular disulfide bonds and they must affect SOD1 mitochondrial localization by a different mechanism. Both residues have been implicated in the formation of intermolecular disulfide bonds (35). It could be speculated that protein–protein interactions involving C6 and C111 are required for maturation and retention of SOD1 in mitochondria. Recently, C111 was found to be critical for mitochondrial import in NSC34 cells (25). Removal of the sulfhydryl group of C111 does not affect dismutase activity and proteinase K resistance (Table 1, Supplementary Material, Fig. S1). Thus, C111S SOD1 is much like the WT protein, but localizes poorly to mitochondria (Fig. 6A). The role of C111 in mitochondrial import is particularly puzzling because it is not a conserved residue. Only in humans, chimpanzee and chickens, residue 111 is a cysteine, whereas most other species, including mice and rats, have a serine at position 111. Mouse SOD1 expressed in COS cells localizes to mitochondria (Fig. 6A), despite lacking the sulfhydryl group of C111. In fact, there are reports of rodent SOD1 in mitochondria (5). This indicates that C111 in non-human mammals is not necessary for import/retention of SOD1, suggesting that the mechanisms of mitochondrial localization may differ between human and non-human SOD1.
Effect of hydophobic stretches on SOD1 aggregation and mitochondrial import
Strings of hydrophobic amino acids serve as mitochondrial internal targeting signal for various IM proteins (22). SOD1 contains three such strings, specifically residues 5–9, 112–115 and 148–153, located at the protein dimer interface (Fig. 5B). We created deletion mutants of SOD1 (named deletion 1–3), each with an ablation of one of three hydrophobic domains. SOD1 localization experiments revealed that deletions 1 and 3 are found in mitochondrial fractions, whereas deletion 2 does not localize to mitochondria (Fig. 6B). Taken together, these results suggest that the hydrophobic stretches 1 and 3 of SOD1 are not necessary determinants for mitochondrial localization. The absence of deletion 2 from mitochondria could be explained by lack of import, lack of retention or protein degradation in mitochondria. To exclude that deletion 2 is degraded by mitochondrial proteases, we treated the cytosolic fractions from COS cells expressing WT or deletion 2 SOD1 with concentrated mitochondrial extracts. Western blot shows that there is no degradation of either protein, indicating that the absence of deletion 2 in mitochondria is due to either lack of import or retention (Fig. 6C). The residues ablated in deletion 2 include C111, which is involved in the mitochondrial localization of SOD1 (Fig. 6A), suggesting that loss of C111 is responsible for the lack of mitochondrial localization of deletion 2. On the other hand, deletion 1 is found in mitochondria despite lacking C6, which is also involved in SOD1 mitochondrial localization (Fig. 6A). In the case of this mutant, there must be other determinants that overcome the loss of C6.
We hypothesized that a discriminating factor between deletion 1 and C6S mutants could be their tendency to misfold and form aggregates, which may contribute to mitochondrial retention of SOD1, even in the absence of proper folding. Thus, we performed detergent solubility assays of cell homogenates for detection of aggregated protein precipitates containing SOD1. Upon solubilization with NP40 and high-speed centrifugation, deletion 1 is detected both in the detergent soluble fraction, containing monomeric or oligomeric species, and in the insoluble fraction, containing precipitated high molecular weight multimeric species, suggesting that this mutant aggregates. C6S is only found in the soluble fraction, similar to WT SOD1, indicating that this mutant does not form high molecular weight multimeric aggregates (Fig. 6D). Table 2 summarizes the results of the protein solubility assays in the remaining mutants. Deletion 3 displays abundant aggregates and deletion 2 a small amount of aggregation, whereas C57S, C111S and C146S mutants do not show aggregation. Therefore, aggregate formation in the mitochondria is one likely mechanism for mutant SOD1 mitochondrial retention that may supersede the loss of cysteine residues.
Table 2.
Correlation between the folding and aggregation of SOD1 mutants and their mitochondrial localization
| SOD1 | Folding | Aggregation | Mitochondrial localization |
|---|---|---|---|
| WT | + | − | + |
| Mouse WT | + | − | + |
| C6S | + | − | − |
| C57S | − | − | − |
| C111S | + | − | − |
| C146S | − | − | − |
| Deletion 1 | − | + | + |
| Deletion 2 | − | + | − |
| Deletion 3 | − | + | + |
| G93A | + | + | + |
| G93A/C111S | + | + <G93A | + |
| G85R | − | + | + |
| G85R/C111S | − | + <G85R | + |
| Frameshift | − | + | +a |
Mitochondrial localization of SOD1 mutants associated with fALS
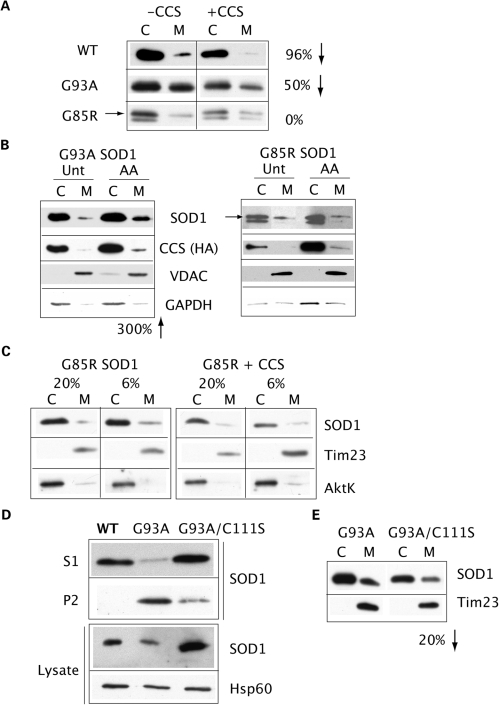
We studied the mitochondrial localization and modulation of import/retention in the SOD1 mutants G93A and G85R, which are associated with human fALS. We find that both mutants localize to mitochondria (Fig. 7A, left panels). However, the modulation of SOD1 mitochondrial localization by CCS co-expression is different than in WT SOD1 (Fig. 7A, right panels). The relative mitochondrial content of G93A mutant SOD1 is only reduced by ∼50%, whereas in WT SOD1 cells the decrease is much greater (96%). The apparent discrepancy with the increase in mitochondrial content of G93A SOD1 observed in CCS/G93A double transgenic mice (15) is explained by tissue culture conditions, where COS cells are exposed to high oxygen, which promotes the oxidative folding of CCS in the cytoplasm and prevents SOD1 import (Fig. 2). The relative mitochondrial content of G85R mutant SOD1 is completely unaffected by CCS co-expression. G93A mutant content in mitochondria is increased ∼3-fold by complex III inhibition with antimycin A, via enhanced CCS mitochondrial localization (Fig. 7B, left panels), similarly to WT SOD1 (Fig. 4C). However, G85R mutant mitochondrial localization is completely unchanged by antimycin A, despite an expected increase in CCS mitochondrial content (Fig. 7B, right panels). These results show that the two SOD1 mutants display different responses to CCS localization. This could be explained by the fact that, unlike the highly unstable G85R SOD1 that presumably interacts poorly with CCS, G93A SOD1 retains WT-like properties involving CCS, such as the capacity to become enzymatically active (Table 1), despite a tendency to misfold and aggregate (Table 2). Finally, G85R mutant content in mitochondria is completely unchanged by lowering the oxygen concentration to 6%, with or without the co-expression of CCS (Fig. 7C). Taken together, these results indicate that pathogenic SOD1 mutants are either partially (WT-like mutants such as G93A) or not at all (unstable mutants such as G85R) subjected to the physiological regulation of mitochondrial import. Hence, the mechanisms of mitochondrial localization of these mutants are likely to involve misfolding and aggregation.
Figure 7.
Mitochondrial localization of pathogenic SOD1 mutants. (A) The mitochondrial content of WT, G93A, and G85R SOD1 was assessed with and without CCS co-expression. The co-expression of CCS reduces WT SOD1 mitochondrial localization by 96% (downward arrow), as compared to WT SOD1 expressed alone. Co-expression of CCS reduces G93A mitochondrial content by 50%, as compared to G93A SOD1 expressed alone. CCS has no effect on relative G85R SOD1 mitochondrial localization. The arrow denotes the band corresponding to human G85R SOD1, which migrates slightly above simian endogenous SOD1. (B) Cells co-transfected with G93A or G85R SOD1 and CCS, treated with (AA) or without (Unt) antimycin A. G93A SOD1 mitochondrial content is increased 300% by AA, as compared to Unt (left panels), reflecting the increase in mitochondrial CCS. G85R SOD1 is not affected by changes in CCS mitochondrial localization induced by AA (right panels). (C) Mitochondrial localization of G85R SOD1 is not affected by changes in oxygen concentration, regardless of CCS expression. (D) Mitochondria from WT, G93A and G93A/C111S SOD1-expressing cells were separated into NP40 detergent soluble and insoluble fractions. WT SOD1 is found exclusively in the detergent soluble fraction (S1), whereas the G93A mutant is largely contained in the detergent-insoluble fraction (P2). G93A/C111S mutant has a reduced detergent-insoluble component when compared with the G93A mutant. Expression of SOD1 and the levels of the mitochondrial matrix protein Hsp60 are shown by western blot of cell lysates (bottom panels). (E) Mitochondrial localization of G93A/C111S SOD1 is decreased by 20% when compared with G93A SOD1.
Because C111S substitution prevents mitochondrial localization of WT SOD1 (Fig. 6A), and C111S is also known to reduce aggregation of mutant SOD1 (35–37), we tested the effects of C111S on G93A mutant aggregation and mitochondrial localization. We first compared the formation of high molecular weight aggregates in G93A mutant and G93A/C111S double mutant by detergent solubility assays. When compared with the G93A mutant, G93A/C111S SOD1 contains ∼90% less protein in the insoluble fraction in mitochondria (Fig. 7D) and in whole cells (data not shown), relative to the amount of protein found in the soluble fraction. However, this 90% reduction in high molecular weight aggregates only results in a 20% decrease in mitochondrial localization of the G93A/C111S protein relative to the G93A mutant (Fig. 7E). Similarly, a G85R/C111S mutant displayed reduced detergent-insoluble aggregates with no change in the levels of mitochondrial localization of the protein (data not shown). These results indicate that the misfolding properties of the pathogenic mutants override the effect of C111S on SOD1 mitochondrial localization. Therefore, mutant SOD1 misfolding, perhaps resulting in intramitochondrial hetero- or homo-oligomers of SOD1 with itself or other proteins, is sufficient to trap the protein in mitochondria, without requiring the formation of high molecular weight aggregates.
A frameshift mutation induces SOD1 misfolding and mitochondrial accumulation
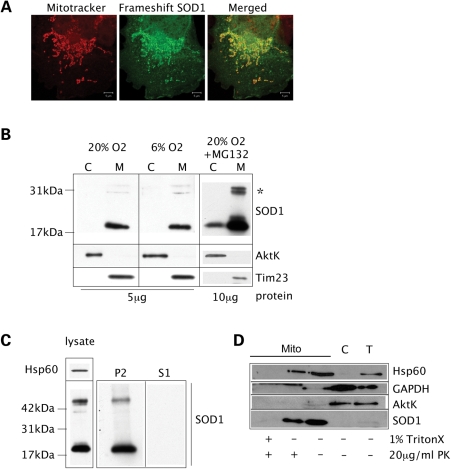
To further test the hypothesis that misfolding and aggregation of mutant SOD1 determines mitochondrial localization, we generated a mutant, where a 10 base-pair deletion (nt334–343) results in a frameshift after amino acid position 110. Between amino acid 110 and the premature stop at position 145, this mutant SOD1 (named Frameshift SOD1) contains a 35 amino acid modified C-terminus. Surprisingly, Frameshift SOD1 is found predominantly in mitochondria, as shown by co-localization of SOD1 and mitochondria labeled with Mitotracker in COS cells (Fig. 8A). Cell fractionation revealed that Frameshift SOD1 mutant is not detectable in the cytosol and is exclusively localized to mitochondria, both under 20 and 6% oxygen (Fig. 8B, left panels). The absence of this mutant in the cytosolic fraction is presumably due to proteasomal degradation, because after complete proteasome inhibition with 20 µm MG132 overnight, the protein becomes detectable also in the cytosol (Fig. 8B, right panel). Another, structurally different proteasome inhibitor, epoxomycin (50 nm) had exactly the same effect (data not shown). Therefore, the Frameshift SOD1 mutant accumulates within the mitochondrial membranes, where it is protected from proteasome degradation. Frameshift SOD1 mutant lacks both C111 and C146, which are necessary for WT SOD1 mitochondrial localization. Therefore, mitochondrial localization of Frameshift SOD1 must be mediated through misfolding and aggregation. In fact, this mutant is found exclusively in the form of detergent-insoluble aggregates in mitochondria (Fig. 8C). To confirm that these aggregates are indeed localized within the outer mitochondrial membrane and not on its external surface, we performed proteinase K treatment on mitochondrial fractions. In intact mitochondria, Frameshift SOD1 is protected from degradation, but the protein is fully degraded once the membranes are solubilized with detergents (Fig. 8D). Taken together, these results indicate that misfolding and aggregation of mutant SOD1 is a crucial mechanism leading to the accumulation of unstable SOD1 mutants in mitochondria.
Figure 8.
A highly aggregatable Frameshift SOD1 mutant accumulates in mitochondria. (A) COS cells transfected with Frameshift SOD1 were immunostained with the monoclonal anti-human SOD1 antibody. This mutant (in green) co-localizes with mitochondria labeled with Mitotracker (in red), as shown by the merged image. (B) Fractionation studies of Frameshift SOD1-expressing cells, cultured in 20 or 6% oxygen, show that this mutant is undetectable in the cytosol and it accumulates exclusively in mitochondria (left panels). Upon proteasome inhibition with 20 µm MG132, Frameshift SOD1 becomes detectable also in the cytosolic fraction (right panel). The amounts of protein loaded are shown at the bottom of each panel. The asterisk denotes mutant SOD1 oligomers resistant to SDS and β-mercaptoethanol. (C) In mitochondria, Frameshift SOD1 is only found in NP-40-insoluble aggregates (P2), as determined by a detergent solubility assay. (D) Intact mitochondria (Mito) from Frameshift SOD1-expressing cells were treated with (+) or without (−) proteinase K (PK). Most of the Frameshift SOD1 associated with mitochondria is still detectable after PK treatment, but is degraded when mitochondrial membranes are solubilized with Triton X, suggesting that Frameshift SOD1 is localized inside mitochondria. T, total cell lysate; C, cytosolic fraction. Hsp60 is used as mitochondrial matrix marker. GAPDH and AktK are used as cytosolic markers.
DISCUSSION
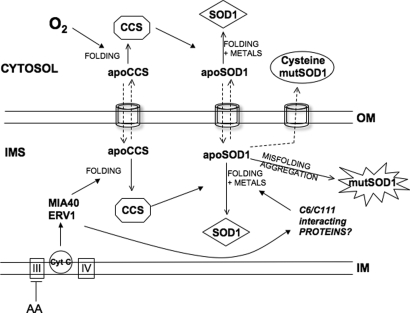
Our studies reveal multiple factors affecting the mitochondrial localization of SOD1 in mammalian cells. The physiological regulation of WT SOD1 mitochondrial localization involves its interactions with CCS and the proper folding and maturation of SOD1. A different set of rules applies to mutant forms of SOD1 that fail to fold correctly, because these mutants are largely unresponsive to the physiological control, and their mitochondrial localization depends mostly on misfolding and aggregation. Figure 9 summarizes our understanding of the regulation of SOD1 mitochondrial localization. ApoSOD1 synthesized by cytosolic ribosomes may follow different fates: (i) its folding and maturation involving the acquisition of Cu and Zn and dimerization keeps the protein localized in the cytosol. CCS in the cytosol promotes intramolecular disulfide bond formation and insertion of Cu, thus accelerating SOD1 maturation and preventing its translocation into mitochondria. (ii) A proportion of apoSOD1 finds its way into the mitochondrial translocation machinery and is imported across the OM. It is possible that this event is more frequent when SOD1 is synthesized by perimitochondrial ribosomes, allowing apoSOD1 to engage in the mitochondrial general import pore before it can fold. Similar to many other IMS proteins, SOD1 does not contain an N-terminal cleavable signal typical of matrix proteins; therefore, it does not require membrane potential or ATP for its mitochondrial import. Once inside the IMS, apoSOD1 is subjected to folding and maturation into the enzymatically active protein. The folding process is necessary for mitochondrial retention, because otherwise apoSOD1 escapes from the IMS back into the cytosol.
Figure 9.
Model of regulation of SOD1 mitochondrial localization. OD1 and CCS can enter mitochondria through the general import pore of the TOM complex only in their apo-forms. The rate of folding and maturation of these proteins in the cytosol (i) determine the probability of their import in mitochondria (ii). Oxygen accelerates cytosolic maturation of CCS and consequently of SOD1, preventing mitochondrial import of both proteins. Conversely, retention in mitochondria of CCS and SOD1 is dependent upon their folding inside, which is regulated by the respiratory chain via the disulfide relay system, Mia40/Erv1, and by other unknown proteins that interact with SOD1 cysteine residues. Non-aggregatable SOD1 mutants that fail to fold due to the lack of critical cysteine residues involved in inter- or intramolecular disulfide bonds are not retained in mitochondria. On the other hand, misfolding and aggregation override the physiological regulation and determine the mitochondrial localization of pathogenic mutant SOD1. IM, mitochondrial inner membrane; IMS, intermembrane space; OM, outer membrane; AA, antimycin A; cyt c, cytochrome c.
In yeast, mechanisms of mitochondrial protein retention, involving folding mediated by disulfide bonds and metal acquisition, have been described for some resident IMS proteins such as the small Tim proteins (34) and SOD1 (24). Thus, the probability that SOD1 becomes terminally localized in mitochondria depends on the efficacy of its maturation process in a CCS-dependent manner. Our results indicate that in mammalian cells, mitochondrial localization of CCS is regulated by the respiratory chain activity through the Mia40/Erv1 system, because mitochondrial localization of CCS becomes detectable when Mia40 is co-expressed (Figs 3D and 4B), and is enhanced by complex III inhibition with antimycin A (Fig. 4B). Furthermore, CCS mitochondrial content is affected by oxygen concentration, because CCS becomes detectable in mitochondria when oxygen concentration is reduced from 20 (high oxygen) to 6% (physiological oxygen, Fig. 2D). As mitochondrial respiration is unchanged in 20 and 6% oxygen (38,39), we conclude that the absence of detectable CCS in mitochondria under 20% oxygen does not depend on respiratory chain activity, but is due primarily to the propensity of CCS to fold in the cytosol under high oxygen conditions. It is known that for some IMS proteins imported by the Mia40/Erv1 system, such as Tim9 and Tim10, the rate of oxidative folding in the cytosol is faster than the rate of mitochondrial import in ambient oxygen (40). The different intracellular CCS distribution between high and physiological oxygen conditions can explain why the protein localizes abundantly in mitochondria from tissues of the CCS transgenic mouse (15), whereas it is undetectable in mitochondria when expressed in cultured cells exposed to air oxygen (Fig. 2). The physiological meaning of the enhanced CCS and SOD1 localization in mitochondria under lower oxygen remains to be determined, but it could be speculated that cells sense the shift from 20–6% oxygen as a condition of ‘relative hypoxia’ and that by increasing SOD1 in the IMS, they prepare for the boost in mitochondrial ROS production that follows the return to normoxia (41). Alternatively, accelerated oxidative maturation of CCS, and secondarily of SOD1, in the cytosol may represent a response to enhanced superoxide production from non-mitochondrial sources, such as NAPDH oxidase (42) occurring under high oxygen (20%) conditions.
The lack of mitochondrial localization of the C6S and C111S SOD1 mutants suggests that SOD1 establishes interactions with mitochondrial proteins other than CCS. These two mutants have WT-like activity and do not aggregate (Tables 1 and 2); thus, they are expected to interact normally with CCS. Furthermore, the C6S and C111S mutations do not affect the intramolecular disulfide bond. However, these two cysteines have the potential to form intermolecular disulfide bonds and contribute to protein aggregation of pathogenic mutants of SOD1 (25,26,35,43), although the latter may not involve necessarily disulfide bridging (37). Thus, it can be hypothesized that the lack of mitochondrial localization of the C6S and C111S SOD1 is due to their involvement in conformational modifications and/or interactions with IMS chaperones that participate in SOD1 maturation. For example, C111 is implicated in the initial transfer of Cu to SOD1 (44). In the case of human SOD1, protein interactions involving C6 and/or C111 must occur at the initial phase of import, preceding the subsequent steps of folding and maturation that retain SOD1 in mitochondria; in the absence of these interactions, SOD1 escapes from the IMS (Fig. 9). The requirement of C111 for mitochondrial localization appears to be species-specific, because mouse WT SOD1, which contains a serine at position 111, localizes to mitochondria. This indicates that the amino acid differences between the human and mouse SOD1 (27 in total) affect the structure of mouse SOD1 in a way that allows for mitochondrial retention in the absence of C111.
Although our experimental mutant C6S SOD1 is not found in mitochondria, a pathogenic C6F mutant localizes to mitochondria and causes respiratory chain dysfunction in NSC34 cells (25). Therefore, there must be a difference between the substitution of C6 with serine, a polar uncharged amino acid, and phenylalanine, a nonpolar, hydrophobic amino acid. In fact, C6S SOD1 is enzymatically active, proteinase K insensitive and detergent soluble (Fig. 6, Tables 1 and 2), whereas C6F is incapable of proper folding, is proteinase K sensitive, cannot establish the intramolecular disulfide bond (32) and forms detergent-insoluble aggregates (37). This suggests that misfolding and aggregation of the C6F pathogenic mutant is the principal determinant for its mitochondrial localization, superseding the lack of C6.
Our studies of pathogenic and experimental SOD1 mutants with propensity to form insoluble high molecular weight aggregates, such as G85R, G93A, deletion 1 and deletion 3 (Table 2), show that aggregation is the principal determinant for their mitochondrial localization. In these mutants, all of the physiological modulating factors are either inefficient, in the case of G93A, or completely inactive, in the case of G85R. It was shown that a proportion of mutant SOD1 localizes to the cytoplasmic surface of the OM (11), presumably because aggregates form prior or during mitochondrial import. Our experimental frameshift mutant Frameshift SOD1 displays extremely high propensity to aggregate (Fig. 8C), but the aggregates are entirely localized within the boundaries of the mitochondrial membranes (Fig. 8D). Therefore, even an aggregation-prone SOD1 can enter the IMS, where it is trapped by its misfolding. In the case of unstable proteins, such as the Frameshift SOD1 and the pathogenic G127X (9,11), mitochondria may provide a protective compartment inaccessible to the ubiquitin-proteasome system. Hence, these mutants are rapidly degraded in the cytosol, but accumulate in mitochondria.
We have shown that aggregation is sufficient to trap mutant SOD1 in mitochondria. However, the formation of high molecular weight aggregates is not a necessary requirement for mitochondrial localization, because decreasing the amount of such aggregates by introducing a C111S substitution does result in a proportional decrease of mutant SOD1 mitochondrial localization (Fig. 7E). As pathogenic SOD1 mutants can misfold and form low molecular weight oligomers, even in the absence of C111 (37), oligomerization or even just misfolding of the monomer must be sufficient to trap SOD1 in mitochondria.
In summary, we propose that WT and mutant SOD1 mitochondrial localization respond to different regulatory mechanisms. Mutant SOD1 tends to lose the physiological regulation and accumulates in mitochondria due to misfolding, whereas WT SOD1 responds to environmental cues through the interaction with CCS. As the content of CCS in mammalian cells is 15–30-fold less than that of SOD1 (45), it is likely that the localization of CCS is the limiting factor, which dictates the subcellular distribution of enzymatically active SOD1. When the cell is exposed to an increased oxidative environment, such as in the presence of high oxygen, CCS localizes primarily in the cytosol and retains SOD1 in this compartment. A shift towards a more oxidative environment in the IMS, for example during mitochondrial oxidative stress, will result in a more efficient disulfide relay import system, enhancing CCS and SOD1 import to scavenge mitochondrial superoxide. Therefore, CCS may function as a redox sensor that determines the localization of enzymatically active SOD1 in the cell compartment where it is most needed.
MATERIALS AND METHODS
Expression plasmids
Human SOD1 cDNAs (WT, G93A and G85R mutants) were cloned into pcDNA3.0 (Invitrogen, Carlsbad, CA, USA). Mouse CCS, mouse WT SOD1 and human Mia40 cDNAs cloned in pSPORT6 were obtained from the ATCC mammalian gene Image collection. Mouse CCS and human Mia40 were amplified by PCR with primer extension to add a C-terminal HA tag. The PCR products were cloned into pCR2.1-TOPO. The inserts were then excised and cloned into pcDNA3.0 at the _Not_I and _Hin_dIII sites for mammalian cell expression.
Human SOD1 site-directed mutagenesis
All mutant SOD1 constructs were made with WT SOD1 as template using the QuikChange II XL Site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) with appropriate oligonucleotides, following the manufacturer's instructions. For G93A/C111S and G85R/C111S constructs, G93A and G85R mutant SOD1 plasmids served as templates. All mutations were confirmed by DNA sequencing.
Cell culture
COS-7 (hereto referred to as COS cells) and N2A neuroblastoma cells (ATCC, Manassas, VA, USA) were cultured in 5% CO2 at 37°C. Media containing Advanced DMEM supplemented with 2% fetal bovine serum (FBS) and Gluta-max was used for COS cells. DMEM:Opti-MEM (1:1) supplemented with 5% FBS was used for N2A cells. Media and supplements were purchased from Invitrogen. Cells grown in 6% oxygen were placed in a Modulator Incubator Chamber (Billups-Rothernberg, Inc., Del Mar, CA, USA) filled with a gas mixture containing 5% CO2, 6% O2 and 89% nitrogen.
Transfection and cell fractionation
COS and N2A cells were transfected with expression plasmids using the FuGene6 transfection reagent (Roche, Indianapolis, IN, USA) or Lipofectamine 2000 (Invitrogen), respectively, according to the manufacturers' instructions. In double transfection experiments, the total amount of DNA (10 µg for a 15 cm plate of cells) was equally split between the two plasmids.
For mitochondrial localization experiments, transfections were performed in 15 cm culture plates, which were further expanded into three 15 cm plates on the day following transfection. Forty-eight h after transfection, cells were harvested by trypsinization, rinsed in sterile phosphate buffered saline (PBS), resuspended in sucrose isolation buffer and fractionated by differential centrifugation into cytosolic and enriched mitochondrial fractions, according to established protocols (46). Mitochondria-rich fractions were washed with isolation buffer containing 150 mm KCl to remove proteins peripherally associated with mitochondrial membranes by electrostatic interactions.
In a subset of experiments, 32 h after transfection, cells were treated with proteasome or respiratory chain inhibitors for 16 h prior to cell fractionation. Respiratory chain complex III was inhibited with antimycin A (1 µm). For proteasomal inhibition, MG132 (20 µm) or epoxomycin (50 nm) were used. All inhibitors were from Sigma-Aldrich (St Louis, MO, USA).
Western blot analyses
SDS–polyacrylamide gel electrophoresis (PAGE) was performed in 12% gels. Protein concentrations were determined by a colorimetric assay (BioRad, Hercules, CA, USA). For SOD1 localization experiments, fractions containing 5 µg of proteins were loaded per well. Samples were heated at 75°C for 10 min prior to electrophoresis. After SDS–PAGE, proteins were transferred onto polyvinylidene difluoride membranes (Bio-Rad) at 75 V for 75 min and the membranes were blocked in 5% milk made in Tris-buffered saline with 0.1% Tween-20 (TBS-T) for 1 h at room temperature. Primary antibodies were applied overnight at 4°C. After washing in TBS-T, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Membranes were washed in TBS-T and immunoreactive bands were revealed with enhanced chemiluminescence reagent (Pierce, Rockford, IL, USA).
Western blot quantification
The relative amount of SOD1 localized to mitochondria was estimated by densitometry of the western blot bands using NIH image software. To correct for potential differences in gene expression, mitochondrial SOD1 was normalized by the mitochondrial marker Tim23, whereas cytosolic SOD1 was normalized by AktK or GAPDH. For each experiment, the relative cytosolic content of SOD1 was set at 1 and the relative mitochondrial content was expressed as a percentage of cytosolic SOD1. Changes in mitochondrial SOD1 content between test and control conditions were quantified by comparing relative mitochondrial contents determined as described earlier.
Antibodies for western blot
There are several commercially available antibodies with variable affinities for WT and mutant SOD1 from different species. We tested three antibodies, a rabbit polyclonal (Stressgen, Ann Arbor, MI, USA), a mouse monoclonal (Santa Cruz Biotechnology, Santa Cruz, CA) and a sheep polyclonal (Calbiochem, La Jolla, CA, USA) to select the one that detects both WT and mutant SOD1 with comparable affinities. We found that the polyclonal sheep antibody detects the human SOD1 with similar affinity for both the mutant and WT proteins (Supplementary Material, Fig. S2). Therefore, unless otherwise specified, the polyclonal sheep antibody was used in all experiments to detect transfected SOD1.
Other antibodies used in localization experiments included: GAPDH (Abcam, Cambridge, MA, USA), Tim23 (BD Biosciences, San Jose, CA, USA), cytochrome c (Cell Signaling, Danvers, MA, USA), COXI, VDAC (Invitrogen), AktK (Santa Cruz Biotechnology), polyclonal HA (Abcam) and CCS (gift from Dr J. Rothstein, Johns Hopkins University, Baltimore, MD, USA).
Immunocytochemistry
COS cells were grown on glass coverslips placed in 24-well plates. Forty-eight h after transfection, mitochondria were stained with 250 nm Mitotracker Red (Invitrogen) for 30 min at 37°C. Then, cells were fixed in 4% paraformaldehyde, followed by permeabilization with 0.1% Triton X and blocking in PBS containing 1% bovine serum albumin and 10% normal goat serum. Cells were incubated with primary antibodies against HA or monoclonal anti-human SOD1 diluted in blocking buffer for 2 h with gentle shaking. Fluorescently labeled secondary antibodies were diluted in blocking buffer and applied to cells for 1 h. After three washes in PBS, the coverslips were mounted onto glass slides and dried. All steps were performed at room temperature. Immunostained cells were imaged with a Zeiss LSM 510 laser scanning confocal microscope with a 63× Plan Apochromat oil immersion lens with aperture 1.4 using a photomultiplier (Carl Zeiss MicroImaging, Inc., Germany). A series of z-sections were taken spanning the thickness of the cell, with intervals between sections set at 0.5 µm. Z-stack images were projected onto a single plane using the LSM Image Browser software (Carl Zeiss MicroImaging, Inc.) and digital magnification was 2× (total magnification was 126×).
Immunoprecipitation
Transfected COS cells were cross-linked with 2 mm dithiobis [succinimidylpropionate] (DSP, Pierce), dissolved in DMSO for 30 min at room temperature, followed by incubation in 20 mm Tris (pH 7.6) for 15 min to stop the reaction. Samples were washed three times in PBS, lysed in RIPA buffer containing 20 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate and a protease inhibitor cocktail and cleared by centrifugation at 10 000_g_ for 5 min at 4°C. The supernatants were incubated overnight at 4°C with protein G-Sepharose beads (Zymed, S. San Francisco, CA, USA), which had been preadsorbed with the rat monoclonal HA antibody (Roche) for 2 h at room temperature and collected by brief centrifugation. The following day, the beads were washed three times in RIPA buffer and boiled for 10 min in Laemmli buffer containing 50 mm DTT prior to electrophoresis. Immunoprecipitated proteins and proteins in cellular lysates were detected by SDS–PAGE and western blots using polyclonal SOD1 and CCS antibodies.
Proteinase K sensitivity assay
For characterization of mutant SOD1 folding, total cell lysates (100 µg) from COS cells expressing SOD1 were incubated with 100 µg/ml proteinase K (Sigma-Aldrich) for 20 min at room temperature, followed by proteolysis termination in 2 mm phenylmethylsulfonyl fluoride (PMSF) for 10 min. For mitochondrial protein localization studies, mitochondrial fractions (10 µg) were incubated with 20 µg/ml proteinase K for 20 min on ice, followed by PMSF inactivation. An aliquot of the mitochondrial fraction was incubated with 1% Triton X to break mitochondrial membranes prior to proteinase K treatment. Proteinase K-treated samples were then added to Laemmli buffer and analyzed by SDS–PAGE/western blotting using SOD1, AktK, GAPDH and Hsp60 (Stressgen) antibodies.
Detergent extraction and high-speed centrifugation for SOD1 aggregate analysis
High molecular weight protein aggregates were analyzed by NP-40 detergent extraction and centrifugation as described before (47), with some modifications. Total cells or enriched mitochondria (100 µg) were pelleted and lysed in 100 µl of lysis buffer (TEN) containing 10 mm Tris–HCl, pH 8.0, 1 mm EDTA, 100 mm NaCl and protease inhibitor cocktail (Roche). The lysate was mixed with 100 µl of TEN + 1% NP-40, sonicated for 30 s and centrifuged at 100 000_g_ for 10 min. The supernatant (S1) was transferred to a new tube and the pellet was resuspended in 200 µl TEN + 0.5% NP-40, sonicated and centrifuged as above. The supernatant was discarded and the pellet (P2) was resuspended in 20 µl of TEN + 0.5% NP-40. Levels of SOD1 in S1, containing detergent soluble proteins, and P2, containing insoluble aggregated proteins, were analyzed by SDS–PAGE and western blotting with SOD1 antibodies.
SOD1 proteolysis by mitochondrial extracts
Mitochondria isolated from untransfected COS cells were sonicated for 15 s to release soluble proteins, including proteases. The cytosolic fractions from COS cells expressing either WT or deletion 2 SOD1 were incubated with the mitochondrial extracts (4:1 by volume) at 37°C for 30 min, with or without the addition of a cocktail of protease inhibitors. Another aliquot of sample was treated with mitochondrial resuspension buffer without mitochondrial proteins, as a control. Samples were added to Laemmli buffer and analyzed by SDS–PAGE/western blotting using the polyclonal SOD1 antibody.
SOD in-gel activity assays
COS cells expressing SOD1 were lysed in PBS containing 0.5% Triton X-100 on ice for 20 min and cleared by centrifugation at 6000_g_ for 5 min at 4°C. Samples were electrophoresed in a 10% native acrylamide gel. To determine SOD activity, the native gel was incubated in the dark with 1.23 mm Nitro blue tetrazolium (Sigma-Aldrich) on an orbital shaker at room temperature for 15 min, followed by a wash in 0.1 M KH2PO4, pH 7.0 (48). The gel was then incubated at room temperature in a buffer with 28 µm riboflavin and 28 mm TEMED in 0.1 M KH2PO4 for 15 min on an orbital shaker in the dark. After washing once in 0.1 M KH2PO4, the gel was exposed to a 13 W Sylvania fluorescent tube for 10 min. Purified human SOD1 (0.5 and 1 unit, Sigma-Aldrich) was used as a positive control. An aliquot of samples was analyzed by SDS–PAGE/western blotting with the polyclonal SOD1 antibody to determine SOD1 expression.
FUNDING
The Robert Packard ALS Research Center ‘The New York Community Trust’; National Institute of Health/National Institute of Neurological Disease and Stroke (P01-NS011766 and R01-NS051419 to G.M., F31 NS054554 to H.K.); Muscular Dystrophy Association.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG Online.
Supplementary Material
[Supplementary Data]
ACKNOWLEDGEMENTS
We thank Dr Jeffrey Rothstein (Johns Hopkins University) for kindly providing the CCS antibody.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Crapo J.D., Oury T., Rabouille C., Slot J.W., Chang L.Y. Copper,zinc superoxide dismutase is primarily a cytosolic protein in human cells. Proc. Natl Acad. Sci. USA. 1992;89:10405–10409. doi: 10.1073/pnas.89.21.10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang L.Y., Slot J.W., Geuze H.J., Crapo J.D. Molecular immunocytochemistry of the CuZn superoxide dismutase in rat hepatocytes. J. Cell Biol. 1988;107:2169–2179. doi: 10.1083/jcb.107.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kikuchi H., Almer G., Yamashita S., Guegan C., Nagai M., Xu Z., Sosunov A.A., McKhann G.M., II, Przedborski S. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc. Natl Acad. Sci. USA. 2006;103:6025–6030. doi: 10.1073/pnas.0509227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaarsma D., Rognoni F., van Duijn W., Verspaget H.W., Haasdijk E.D., Holstege J.C. CuZn superoxide dismutase (SOD1) accumulates in vacuolated mitochondria in transgenic mice expressing amyotrophic lateral sclerosis-linked SOD1 mutations. Acta Neuropathol. (Berl.) 2001;102:293–305. doi: 10.1007/s004010100399. [DOI] [PubMed] [Google Scholar]
- 5.Okado-Matsumoto A., Fridovich I. Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J. Biol. Chem. 2001;276:38388–38393. doi: 10.1074/jbc.M105395200. [DOI] [PubMed] [Google Scholar]
- 6.Higgins C.M., Jung C., Ding H., Xu Z. Mutant Cu, Zn superoxide dismutase that causes motoneuron degeneration is present in mitochondria in the CNS. J. Neurosci. 2002;22:RC215. doi: 10.1523/JNEUROSCI.22-06-j0001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattiazzi M., D'Aurelio M., Gajewski C.D., Martushova K., Kiaei M., Beal M.F., Manfredi G. Mutated human SOD1 causes dysfunction of oxidative phosphorylation in mitochondria of transgenic mice. J. Biol. Chem. 2002;277:29626–29633. doi: 10.1074/jbc.M203065200. [DOI] [PubMed] [Google Scholar]
- 8.Vijayvergiya C., Beal M.F., Buck J., Manfredi G. Mutant superoxide dismutase 1 forms aggregates in the brain mitochondrial matrix of amyotrophic lateral sclerosis mice. J. Neurosci. 2005;25:2463–2470. doi: 10.1523/JNEUROSCI.4385-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J., Lillo C., Jonsson P.A., Velde C.V., Ward C.M., Miller T.M., Subramaniam J.R., Rothstein J.D., Marklund S., Andersen P.M., et al. Toxicity of familial ALS-linked SOD1 mutants from selective recruitment to spinal mitochondria. Neuron. 2004;43:5–17. doi: 10.1016/j.neuron.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 10.Pasinelli P., Belford M.E., Lennon N., Bacskai B.J., Hyman B.T., Trotti D., Brown R.H., Jr Amyotrophic lateral sclerosis-associated SOD1 mutant proteins bind and aggregate with Bcl-2 in spinal cord mitochondria. Neuron. 2004;43:19–30. doi: 10.1016/j.neuron.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 11.Vande Velde C., Miller T.M., Cashman N.R., Cleveland D.W. Selective association of misfolded ALS-linked mutant SOD1 with the cytoplasmic face of mitochondria. Proc. Natl Acad. Sci. USA. 2008;105:4022–4027. doi: 10.1073/pnas.0712209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borchelt D.R., Guarnieri M., Wong P.C., Lee M.K., Slunt H.S., Xu Z.S., Sisodia S.S., Price D.L., Cleveland D.W. Superoxide dismutase 1 subunits with mutations linked to familial amyotrophic lateral sclerosis do not affect wild-type subunit function. J. Biol. Chem. 1995;270:3234–3238. doi: 10.1074/jbc.270.7.3234. [DOI] [PubMed] [Google Scholar]
- 13.Pasinelli P., Brown R.H. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat. Rev. Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 14.Higgins C.M., Jung C., Xu Z. ALS-associated mutant SOD1G93A causes mitochondrial vacuolation by expansion of the intermembrane space and by involvement of SOD1 aggregation and peroxisomes. BMC Neurosci. 2003;4:16. doi: 10.1186/1471-2202-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Son M., Puttaparthi K., Kawamata H., Rajendran B., Boyer P.J., Manfredi G., Elliott J.L. Overexpression of CCS in G93A-SOD1 mice leads to accelerated neurological deficits with severe mitochondrial pathology. Proc. Natl Acad. Sci. USA. 2007;104:6072–6077. doi: 10.1073/pnas.0610923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung C., Higgins C.M.J., Xu Z. Mitochondrial electron transport chain complex dysfunction in a transgenic mouse model for amyotrophic lateral sclerosis. J. Neurochem. 2002;83:535–545. doi: 10.1046/j.1471-4159.2002.01112.x. [DOI] [PubMed] [Google Scholar]
- 17.Kirkinezos I.G., Bacman S.R., Hernandez D., Oca-Cossio J., Arias L.J., Perez-Pinzon M.A., Bradley W.G., Moraes C.T. Cytochrome c association with the inner mitochondrial membrane is impaired in the CNS of G93A-SOD1 mice. J. Neurosci. 2005;25:164–172. doi: 10.1523/JNEUROSCI.3829-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Damiano M., Starkov A.A., Petri S., Kipiani K., Kiaei M., Mattiazzi M., Flint Beal M., Manfredi G. Neural mitochondrial Ca2+ capacity impairment precedes the onset of motor symptoms in G93A Cu/Zn-superoxide dismutase mutant mice. J. Neurochem. 2006;96:1349–1361. doi: 10.1111/j.1471-4159.2006.03619.x. [DOI] [PubMed] [Google Scholar]
- 19.De Vos K.J., Chapman A.L., Tennant M.E., Manser C., Tudor E.L., Lau K.F., Brownlees J., Ackerley S., Shaw P.J., McLoughlin D.M., et al. Familial amyotrophic lateral sclerosis-linked SOD1 mutants perturb fast axonal transport to reduce axonal mitochondria content. Hum. Mol. Genet. 2007;16:2720–2728. doi: 10.1093/hmg/ddm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Son M., Leary S.C., Romain N., Pierrel F., Winge D.R., Haller R.G., Elliott J.L. Isolated cytochrome c oxidase deficiency in G93A SOD1 mice over-expressing CCS protein. J. Biol. Chem. 2008;283:12267–12275. doi: 10.1074/jbc.M708523200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfanner N., Wiedemann N., Meisinger C., Lithgow T. Assembling the mitochondrial outer membrane. Nat. Struct. Mol. Biol. 2004;11:1044–1048. doi: 10.1038/nsmb852. [DOI] [PubMed] [Google Scholar]
- 22.Neupert W. Protein import into mitochondria. Annu. Rev. Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 23.Herrmann J.M., Hell K. Chopped, trapped or tacked–protein translocation into the IMS of mitochondria. Trends. Biochem. Sci. 2005;30:205–211. doi: 10.1016/j.tibs.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Field L.S., Furukawa Y., O'Halloran T.V., Culotta V.C. Factors controlling the uptake of yeast copper/zinc superoxide dismutase into mitochondria. J. Biol. Chem. 2003;278:28052–28059. doi: 10.1074/jbc.M304296200. [DOI] [PubMed] [Google Scholar]
- 25.Ferri A., Cozzolino M., Crosio C., Nencini M., Casciati A., Gralla E.B., Rotilio G., Valentine J.S., Carri M.T. Familial ALS-superoxide dismutases associate with mitochondria and shift their redox potentials. Proc. Natl Acad. Sci. USA. 2006;103:13860–13865. doi: 10.1073/pnas.0605814103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng H.X., Shi Y., Furukawa Y., Zhai H., Fu R., Liu E., Gorrie G.H., Khan M.S., Hung W.Y., Bigio E.H., et al. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc. Natl Acad. Sci. USA. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergemalm D., Jonsson P.A., Graffmo K.S., Andersen P.M., Brannstrom T., Rehnmark A., Marklund S.L. Overloading of stable and exclusion of unstable human superoxide dismutase-1 variants in mitochondria of murine amyotrophic lateral sclerosis models. J. Neurosci. 2006;26:4147–4154. doi: 10.1523/JNEUROSCI.5461-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., Herrmann J.M. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 29.Lamb A.L., Torres A.S., O'Halloran T.V., Rosenzweig A.C. Heterodimeric structure of superoxide dismutase in complex with its metallochaperone. Nat. Struct. Biol. 2001;8:751–755. doi: 10.1038/nsb0901-751. [DOI] [PubMed] [Google Scholar]
- 30.Bihlmaier K., Mesecke N., Terziyska N., Bien M., Hell K., Herrmann J.M. The disulfide relay system of mitochondria is connected to the respiratory chain. J. Cell Biol. 2007;179:389–395. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratovitski T., Corson L.B., Strain J., Wong P., Cleveland D.W., Culotta V.C., Borchelt D.R. Variation in the biochemical/biophysical properties of mutant superoxide dismutase 1 enzymes and the rate of disease progression in familial amyotrophic lateral sclerosis kindreds. Hum. Mol. Genet. 1999;8:1451–1460. doi: 10.1093/hmg/8.8.1451. [DOI] [PubMed] [Google Scholar]
- 32.Bruns C.K., Kopito R.R. Impaired post-translational folding of familial ALS-linked Cu, Zn superoxide dismutase mutants. EMBO J. 2007;26:855–866. doi: 10.1038/sj.emboj.7601528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hornberg A., Logan D.T., Marklund S.L., Oliveberg M. The coupling between disulphide status, metallation and dimer interface strength in Cu/Zn superoxide dismutase. J. Mol. Biol. 2007;365:333–342. doi: 10.1016/j.jmb.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 34.Lutz T., Neupert W., Herrmann J.M. Import of small Tim proteins into the mitochondrial intermembrane space. EMBO J. 2003;22:4400–4408. doi: 10.1093/emboj/cdg421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niwa J., Yamada S., Ishigaki S., Sone J., Takahashi M., Katsuno M., Tanaka F., Doyu M., Sobue G. Disulfide bond mediates aggregation, toxicity, and ubiquitylation of familial amyotrophic lateral sclerosis-linked mutant SOD1. J. Biol. Chem. 2007;282:28087–28095. doi: 10.1074/jbc.M704465200. [DOI] [PubMed] [Google Scholar]
- 36.Cozzolino M., Amori I., Pesaresi M.G., Ferri A., Nencini M., Carri M.T. Cysteine 111 affects aggregation and cytotoxicity of mutant Cu,Zn-superoxide dismutase associated with familial amyotrophic lateral sclerosis. J. Biol. Chem. 2008;283:866–874. doi: 10.1074/jbc.M705657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karch C.M., Borchelt D.R. A limited role for disulfide cross-linking in the aggregation of mutant SOD1 linked to familial amyotrophic lateral sclerosis. J. Biol. Chem. 2008;283:13528–13537. doi: 10.1074/jbc.M800564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffman D.L., Salter J.D., Brookes P.S. Response of mitochondrial reactive oxygen species generation to steady-state oxygen tension: implications for hypoxic cell signaling. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H101–H108. doi: 10.1152/ajpheart.00699.2006. [DOI] [PubMed] [Google Scholar]
- 39.Jones D.P. Intracellular diffusion gradients of O2 and ATP. Am. J. Physiol. 1986;250:C663–C675. doi: 10.1152/ajpcell.1986.250.5.C663. [DOI] [PubMed] [Google Scholar]
- 40.Morgan B., Lu H. Oxidative folding competes with mitochondrial import of the small Tim proteins. Biochem. J. 2008;411:115–122. doi: 10.1042/BJ20071476. [DOI] [PubMed] [Google Scholar]
- 41.Li C., Jackson R.M. Reactive species mechanisms of cellular hypoxia-reoxygenation injury. Am. J. Physiol. Cell Physiol. 2002;282:C227–C241. doi: 10.1152/ajpcell.00112.2001. [DOI] [PubMed] [Google Scholar]
- 42.Marden J.J., Harraz M.M., Williams A.J., Nelson K., Luo M., Paulson H., Engelhardt J.F. Redox modifier genes in amyotrophic lateral sclerosis in mice. J. Clin. Invest. 2007;117:2913–2919. doi: 10.1172/JCI31265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banci L., Bertini I., Durazo A., Girotto S., Gralla E.B., Martinelli M., Valentine J.S., Vieru M., Whitelegge J.P. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: a possible general mechanism for familial ALS. Proc. Natl Acad. Sci. USA. 2007;104:11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H., Zhu H., Eggers D.K., Nersissian A.M., Faull K.F., Goto J.J., Ai J., Sanders-Loehr J., Gralla E.B., Valentine J.S. Copper(2+) binding to the surface residue cysteine 111 of His46Arg human copper-zinc superoxide dismutase, a familial amyotrophic lateral sclerosis mutant. Biochemistry. 2000;39:8125–8132. doi: 10.1021/bi000846f. [DOI] [PubMed] [Google Scholar]
- 45.Rothstein J.D., Dykes-Hoberg M., Corson L.B., Becker M., Cleveland D.W., Price D.L., Culotta V.C., Wong P.C. The copper chaperone CCS is abundant in neurons and astrocytes in human and rodent brain. J. Neurochem. 1999;72:422–429. doi: 10.1046/j.1471-4159.1999.0720422.x. [DOI] [PubMed] [Google Scholar]
- 46.Pallotti F., Lenaz G. Isolation and subfractionation of mitochondria from animal cells and tissue culture lines. Methods Cell Biol. 2001;65:1–35. doi: 10.1016/s0091-679x(01)65002-7. [DOI] [PubMed] [Google Scholar]
- 47.Wang J., Slunt H., Gonzales V., Fromholt D., Coonfield M., Copeland N.G., Jenkins N.A., Borchelt D.R. Copper-binding-site-null SOD1 causes ALS in transgenic mice: aggregates of non-native SOD1 delineate a common feature. Hum. Mol. Genet. 2003;12:2753–2764. doi: 10.1093/hmg/ddg312. [DOI] [PubMed] [Google Scholar]
- 48.Beauchamp C., Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]