Study of an RNA helicase implicates small RNA–noncoding RNA interactions in programmed DNA elimination in Tetrahymena (original) (raw)
Abstract
Tetrahymena eliminates micronuclear-limited sequences from the developing macronucleus during sexual reproduction. Homology between the sequences to be eliminated and ∼28-nucleotide small RNAs (scnRNAs) associated with an Argonaute family protein Twi1p likely underlies this elimination process. However, the mechanism by which Twi1p–scnRNA complexes identify micronuclear-limited sequences is not well understood. We show that a Twi1p-associated putative RNA helicase Ema1p is required for the interaction between Twi1p and chromatin. This requirement explains the phenotypes of EMA1 KO strains, including loss of selective down-regulation of scnRNAs homologous to macronuclear-destined sequences, loss of H3K9 and K27 methylation in the developing new macronucleus, and failure to eliminate DNA. We further demonstrate that Twi1p interacts with noncoding transcripts derived from parental and developing macronuclei and this interaction is greatly reduced in the absence of Ema1p. We propose that Ema1p functions in DNA elimination by stimulating base-pairing interactions between scnRNAs and noncoding transcripts in both parental and developing new macronuclei.
Keywords: RNA, heterochromatin, small RNA, noncoding RNA, Tetrahymena
Heterochromatin functions in various chromosomal processes, including regulation of gene expression, chromosome segregation, and nuclear organization (for review, see Grewal and Jia 2007). In diverse eukaryotes, RNAi-related mechanisms involving small RNAs complexed with Argonaute family proteins mediate heterochromatin formation (for review, see Martienssen and Moazed 2006; Grewal and Jia 2007). However, the mechanism by which small RNAs target heterochromatin formation is not completely understood. In ciliated protozoans, heterochromatin formation is also induced by an RNAi-related mechanism, followed by programmed DNA elimination of germline-specific sequences from the developing somatic nucleus (for review, see Meyer and Chalker 2006). Thus, programmed DNA elimination in ciliates serves as a model to study small RNA-mediated heterochromatin formation.
Like most ciliated protozoans, Tetrahymena thermophila exhibits nuclear dimorphism. Each cell contains a germline micronucleus (Mic) and a somatic macronucleus (Mac). It is likely that only the Mac contributes to gene expression. In vegetative growth, the Mic and Mac replicate/divide, and sister nuclei are segregated to daughter cells. In the sexual process of conjugation (Fig. 1A; see also Supplemental Fig. S1), the Mic undergoes meiosis to form two haploid pronuclei, one of which is reciprocally exchanged between the two conjugating cells. The migratory and stationary pronuclei then fuse to create a zygotic nucleus that divides mitotically twice to produce the next generation of new Macs and Mics. Then, paired cells separate, one of the two new Mics and the parental Mac are destroyed and, if fed, they resume vegetative growth.
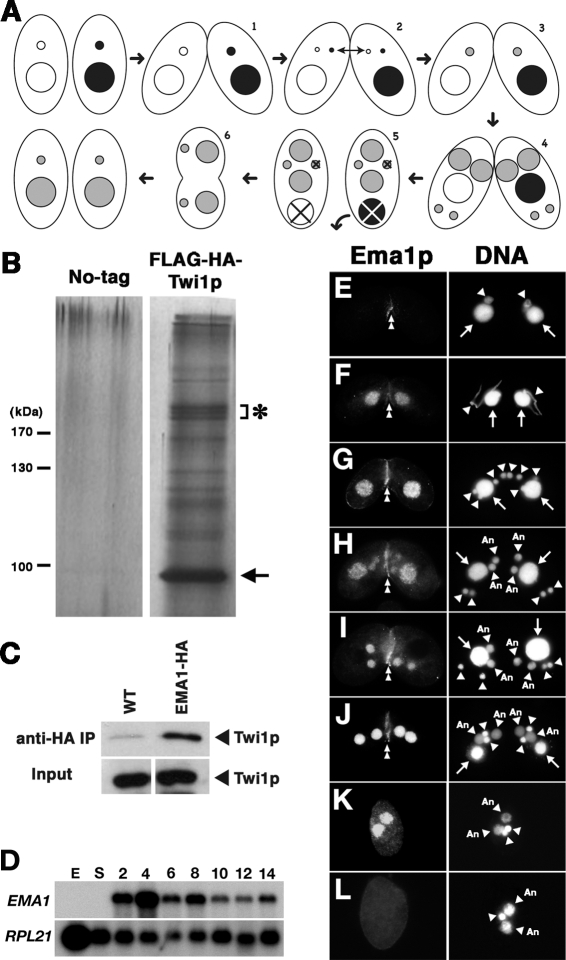
Figure 1.
Identification and characterization of EMA1/Ema1p. (A) Conjugation. In the sexual process of conjugation, two cells mate (1); the Mic undergoes meiosis to form two haploid pronuclei, one of which is reciprocally exchanged between the two conjugating cells (2); the migratory and stationary pronuclei then fuse to create a zygotic nucleus (3); the zygotic nucleus divides mitotically twice to produce the next generation of new Macs and Mics (4); paired cells separate and one of the two new Mics and the parental Mac are destroyed (5); and cells resume vegetative growth with Mic mitosis, followed by cytokinesis (6). (B) Copurification of Ema1p with Flag-HA-Twi1p. Two wild-type strains (No-tag) or two Flag-HA-TWI1 strains were mated and were harvested at 9 h post-mixing. Flag-HA-Twi1p-containing complexes were enriched by gel filtration and immuno-affinity purified first with anti-Flag antibody and then with anti-HA antibody. The purified proteins were separated by SDS-PAGE and analyzed by silver staining. The arrow and the asterisk indicate the positions of Flag-HA-Twi1p and the Twi1p-associated proteins identified by mass spectrometry analysis, respectively. (C) Twi1p coimmunoprecipitates with Ema1p. An EMA1_-HA strain and a wild-type strain were mated and lysed at 6 h post-mixing. (Right) The Ema1p-HA-containing complex was pulled down with an anti-HA antibody. (Left) As a control, two wild-type strains were crossed and processed similarly. Coimmunoprecipitated proteins (anti-HA IP) or total proteins (Input) used for immunoprecipitation were analyzed on Western blot using anti-Twi1p antiserum. (D) Expression of EMA1 mRNA. Total RNAs from exponentially growing (E), starved (S), and conjugating (2, 4, 6, 8, 10, 12, and 14 h post-mixing) wild-type cells were analyzed by Northern hybridization. RPL21 was used as a loading control. (E–L) Localization of Ema1p. Mating pairs of wild-type cells in the early (E) and late (F) premeiosis, pronuclear exchange (G), Mac Anlagen (H,I), Nuclear Alignment (J), Pair Separation (K), and Mic Elimination (L) stages were processed for immunostaining. See Supplemental Figure S1 for the conjugation stages. Ema1p was localized using anti-Ema1p antiserum (left) and DNA was stained by DAPI (right). Arrows, arrowheads, and arrowheads marked with “An” indicate Macs, Mics, and developing new Macs, respectively. The staining detected at the junction of cells (double arrowheads) was also observed in Δ_EMA1 strains (see Fig. 7O,P) and thus represents cross-reaction of the antiserum with other proteins.
After Pair Separation, ∼6000 internal eliminated sequences (IESs) are deleted from the new Mac, and the flanking sequences are ligated (for review, see Yao et al. 2002). The Mic sequences that remain in the future Mac are called Mac-destined sequences (MDSs). IESs in Tetrahymena vary in size (∼0.5–20 kb) and account for ∼15% of the Mic genome. Most IESs are moderately repeated in the Mic genome and many are related to transposable elements. IESs have not been found in coding sequences of Tetrahymena, although some are located in introns. Excision of IESs can occur reproducibly at a specific site or with a limited number of alternative boundaries.
IES elimination is epigenetically regulated by a mechanism that prevents sequences present in the old Mac from being eliminated in the new Mac (Chalker and Yao 1996). Small (∼28 nucleotide [nt]) scan RNAs (scnRNAs) expressed during conjugation (Mochizuki et al. 2002; Lee and Collins 2006) are involved in epigenetic regulation of IES elimination (Mochizuki et al. 2002; Yao et al. 2003). A Dicer-like protein, Dcl1p, is required for IES elimination and for processing Mic noncoding RNA (ncRNA, also called nongenic RNA) transcripts derived from both strands of the Mic sequences (Chalker and Yao 2001) to scnRNAs (Malone et al. 2005; Mochizuki and Gorovsky 2005). An Argonaute protein Twi1p is complexed with scnRNAs (Mochizuki and Gorovsky 2004b) and is also required for IES elimination (Mochizuki et al. 2002).
Heterochromatin is involved in IES elimination. Methylation of histone H3 on Lys 9 (H3K9me) and/or Lys 27 (H3K27me), as well as chromodomain proteins that can bind to these methylated histones, are hallmarks of heterochromatin in diverse eukaryotes. In Tetrahymena, both H3K9me and H3K27me occur in the developing Mac and are specifically associated with eliminated IES sequences (Taverna et al. 2002; Liu et al. 2007). Both H3K9me and K27me in the developing new Mac are essential for IES elimination and depend on Ezl1p, an E(z)-related histone methyltransferase (Liu et al. 2007). The chromodomain protein Pdd1p is also essential for IES elimination (Coyne et al. 1999), can bind to both H3K9me and K27me, and could act as a “reader” of these modifications (Taverna et al. 2002; Liu et al. 2007). IES elimination is also sensitive to treatment with a histone deacetylase inhibitor (Duharcourt and Yao 2002). Together, these results indicate involvement of heterochromatin in DNA elimination. Dcl1p and Twi1p, required for production and accumulation of scnRNAs, respectively, are required for accumulation and/or targeting of H3K9/K27me and for DNA elimination (Liu et al. 2004, 2007; Malone et al. 2005; Mochizuki and Gorovsky 2005). Thus, heterochromatin formation occurs downstream from the RNAi-related mechanism in the IES elimination pathway.
scnRNAs homologous to IES sequences became enriched as conjugation proceeded (Mochizuki and Gorovsky 2004b) and most of the scnRNAs cloned from the late stage of conjugation are complementary to IES sequences (Lee and Collins 2006). Thus, scnRNAs are selected for IES specificity after they are diced. Based on these and other observations, we proposed the scnRNA model (Mochizuki et al. 2002; Mochizuki and Gorovsky 2004a), which postulates that although scnRNAs are derived from both MDSs and IESs of the Mic genome, scnRNAs having homology with any Mac DNA sequence (=MDSs) are degraded in the parental Mac (“scanning” for selection of scnRNA). As a result, scnRNAs become enriched in IES sequences. We further proposed that these IES-specific scnRNAs, complexed to Twi1p, move to the developing Mac, where they identify homologous sequences as IESs and target them for heterochromatin formation via accumulation of H3K9/K27me and Pdd1p, followed by DNA elimination. In the model, a given scnRNA should interact with genomic sequences either in the parental Mac to induce its own degradation or in the developing Mac to induce H3K9/K27me.
Here we report that a Twi1p-associated RNA helicase, Ema1p, plays an important role in the interactions between Twi1p and Mac chromatins and between Twi1p and Mac ncRNAs. We further show that Ema1p is required for IES elimination of some loci, for selective down-regulation of scnRNAs and for accumulation of H3K9me and H3K27me in the developing new Mac. These data suggest that Ema1p facilitates the interaction between nascent ncRNAs from Mac chromatins and the Twi1p–scnRNA complex, and this interaction plays important roles in DNA elimination.
Results
Ema1p interacts with Twi1p
Twi1p, a Tetrahymena Argonaute protein is required for scnRNA accumulation and for IES elimination (Mochizuki et al. 2002) and is physically associated with scnRNAs (Mochizuki and Gorovsky 2004b). To further understand the Twi1p–scnRNA complex, we sought to identify Twi1p-associated proteins. A complex containing N-terminally tagged Flag-HA-Twi1p was purified from conjugating cells at 9 h post-mixing when Mac development is occurring and analyzed by SDS-PAGE and silver staining (Fig. 1B). Two closely migrating proteins copurified with Flag-HA-Twi1p (asterisk in Fig. 1B) were identified by mass spectrometry analysis (Supplemental Table S1).
One of the copurifying proteins was encoded by TTHERM_00088150 (http://www.ciliate.org), a gene we named EMA1. Ema1p, the protein predicted from the EMA1 cDNA sequence (AB292216 in DDBJ/EMBL/GenBank), is a DExH box RNA helicase. Although many RNA helicases are involved in RNAi-related pathways in a variety of eukaryotes, none of them are close relatives of Ema1p. The proteins closely related to Ema1p are XP_001458153 and XP_001423262 in Paramecium, DHX57 and DHX36 in human, AT2G35920 in Arabidopsis, and GH07148p in the fruit fly. DHX36 was recently identified as an AGO1- and AGO2-binding protein (Höck et al. 2007). Thus, these proteins may have a conserved role in RNAi-related mechanisms by modulating the functions of Argonaute proteins.
We confirmed the Twi1p–Ema1p interaction by coimmunoprecipitation using a strain expressing _EMA1_-HA and an anti-HA antibody. Twi1p was highly enriched in the immunoprecipitate from mating _EMA1_-HA cells (Fig. 1C). Thus, Ema1p reciprocally associates with Twi1p.
Ema1p localizes in old and new Macs during conjugation
EMA1 mRNA was detected from early (2 h; meiotic prophase) to late (14 h; when DNA elimination occurs) conjugation stages, but not in growing cells or in starved cells (Fig. 1D). EMA1 mRNA is therefore expressed exclusively during conjugation.
Ema1p was localized by immunostaining using anti-Ema1p antiserum. Ema1p was not detected in vegetative cells (data not shown). During conjugation, Ema1p was first observed in the parental Mac when Mics were in meiotic prophase (Fig. 1A [stage 1],E). Ema1p localized in the parental Mac until the new Macs developed (Fig. 1A [until stage 4], F,G), after which, staining in the parental (old) Mac disappeared and became localized in the new Macs (Fig. 1H,I). Ema1p remained localized in the new Macs until the Pair Separation (Fig. 1A [stage 5], J,K). Then, it disappeared before DNA elimination occurred (∼14 h post-mixing) (Fig. 1L). Similar transitions of localization from the old Mac to the new Mac were also observed for Twi1p, Ezl1p, Pdd1p, and Pdd2p, all of which are required for DNA elimination (Coyne et al. 1999; Nikiforov et al. 1999; Mochizuki et al. 2002; Liu et al. 2007).
Parental EMA1 is required for producing viable progeny
To determine the function(s) of EMA1, EMA1 knockout (KO, Δ_EMA1_) strains were constructed. Part of the coding sequence, including the conserved helicase domain, was replaced by a drug resistance marker in all EMA1 loci in the polyploid Mac (Fig. 2A). By Southern hybridization, a strong band with the expected size of the KO locus, and a faint band with the expected size of the endogenous EMA1 locus, were observed (Fig. 2B). The faint band was ∼30 times less intense than the KO band, reflecting the relative amounts of DNA in Mic and Mac (Woodard et al. 1972), suggesting that complete replacement of the Mac EMA1 gene had occurred. Consistent with this, Ema1p was not detected in Δ_EMA1_ cells during the early to mid stages of conjugation (Fig. 2C). Small amounts of Ema1p were detected in Δ_EMA1_ cells in late stages of conjugation (Fig. 2C, at 12 h post-mixing), indicating that in Δ_EMA1_ cells the intact EMA1 loci in the new Macs could be expressed (zygotic expression). In Δ_EMA1_ cells, Ema1p was never detected in the parental Mac, but first appeared in the new Mac at Nuclear Alignment stage at 12 h post-mixing (Fig. 7O,P, below; Supplemental Fig. S2), although it was detected in the new Mac as early as 6 h post-mixing in the wild-type cells (Fig. 1H). These results argue that, in Δ_EMA1_ strains, Ema1p in the parental Mac is completely eliminated and its accumulation in the developing new Mac is severely delayed.
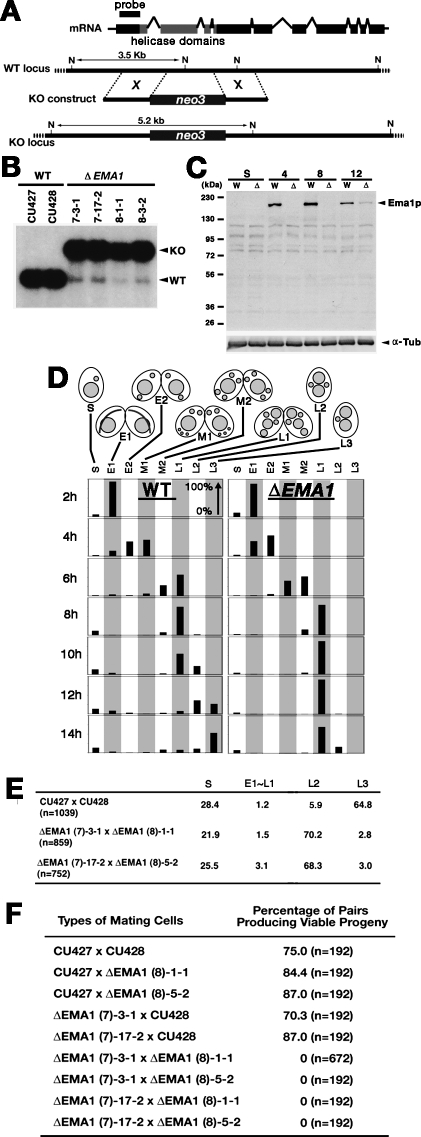
Figure 2.
Characterization of Δ_EMA1_ cells. (A) EMA1 locus and KO construct. A part of the EMA1 coding sequence, including the conserved helicase domains (shown in gray) was replaced by the drug-resistance marker neo3. Upon transformation, the KO construct was introduced into the EMA1 locus by homologous recombination. (B) Southern hybridization of Δ_EMA1_ strains. Total DNA isolated from wild-type (WT) or Δ_EMA1_ strains was digested with NdeI (N in A), and the blot was hybridized with the probe shown in A. Positions of the bands for wild-type and KO loci are indicated with arrowheads. (C) Ema1p expression in Δ_EMA1_ strains. (Top panel) Ema1p expression in the wild-type (W) and Δ_EMA1_ (Δ) strains in starved (S) and mating (4, 8, and 12 h post-mixing) cells was analyzed by Western blot using anti-Ema1p antiserum. (Bottom panel) For a loading control, the amount of α-tubulin was analyzed. (D) Developmental profiles of conjugation in wild-type and Δ_EMA1_ strains. Conjugation stage wild-type (CU427 × CU428) and Δ_EMA1_ [(7)-3-1 X (8)-1-1] cells were observed by DAPI staining. The stages categorized were single unmated cells (S), premeiosis (E1), meiosis (E2), prezygotic (M1), post-zygotic (M2), Mac development (L1), Pair Separation (2 Mics) (L2), and Mic elimination (L3). See Supplemental Figure S1 for the developmental stages. (E) Δ_EMA1_ causes arrest at Pair Separation stage. At 36 h post-mixing, the progeny of wild-type (CU427 and CU428) or Δ_EMA1_ cells were fixed and nuclei were observed by DAPI staining. The stages categorized were the same as in D except stages E1∼L1 were combined. (F) Δ_EMA1_ cells fail to produce viable progeny. At 6∼8 h post-mixing, single mating pairs were placed into drops of medium and incubated for ∼60 h at 30°C. Completion of conjugation was confirmed by testing for expression of the marker specific for newly developed Macs.
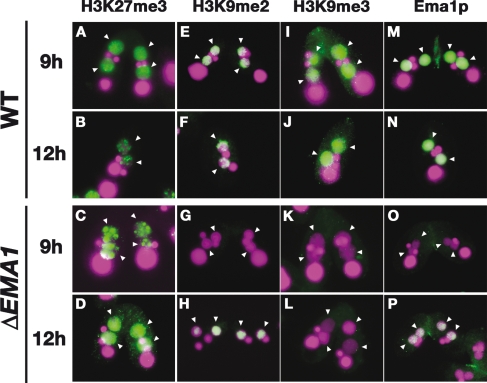
Figure 7.
H3K9/K27me in Δ_EMA1_ cells. Wild-type (WT, top) and Δ_EMA1_ (bottom) conjugating cells at 9 or 12 h post-mixing were processed for immunofluorescent staining (green) using anti-H3K27me3 (A–D), anti-H3K9me2 (E–H), anti-H3K9me3 (I–L), or anti-Ema1p (M–P) Abs. DNA was stained with DAPI (purple). Arrowheads indicate developing new Macs.
The Δ_EMA1_ strains showed no obvious defects during vegetative growth (data not shown). They mate normally but meiosis (between stages 1 and 2 in Fig. 1A; stage E2 in Supplemental Fig. S1) and Pair Separation (stages 4 and 5 in Fig. 1A; stages L1–L2 in Supplemental Fig. S1) were delayed (Fig. 2D). The significance of the meiotic delay is not clear. Delayed Pair Separation in Δ_EMA1_ strains is possibly related to defective heterochromatin formation (see below). Nonetheless, exconjugants (progeny) containing new Macs and Mics were produced in the Δ_EMA1_ cells (L2 in Fig. 2D). Thus, at the cytological level, nuclear differentiation appeared to occur normally without parental EMA1.
The exconjugants from wild-type cells eliminate one of the two Mics (stage 5 in Fig. 1A; stage L3 in Supplemental Fig. S1) and, if the cells are fed, the remaining Mic divides, followed by resumption of vegetative growth. However, the majority of the exconjugants from Δ_EMA1_ cells retained two Mics (arrested in stage L2 in Supplemental Fig. S1), even at 36 h post-mixing (Fig. 2E). Therefore, the progeny of Δ_EMA1_ cells were expected not to resume vegetative growth. Indeed, all single pairs of Δ_EMA1_ cells placed into nutrient medium did not grow and eventually died (Fig. 2F). Thus, parental EMA1 is essential for the formation of viable conjugation progeny.
Elimination of a subset of IESs is inhibited in ΔEMA1
Lack of viable conjugation progeny and retention of the two Mics observed in the Δ_EMA1_ strains are phenotypes reported in mutant strains in which genes affecting IES elimination (DCL1, EZL1, PDD1, PDD2, and TWI1) were disrupted (Coyne et al. 1999; Nikiforov et al. 1999; Mochizuki et al. 2002; Malone et al. 2005; Mochizuki and Gorovsky 2005; Liu et al. 2007). To determine whether EMA1 is required for IES elimination, we analyzed the elimination of four different IESs: M, R, Cal, and Tlr1 elements (Fig. 3). Pairs of mating Δ_EMA1_ strains were isolated at ∼8 h post-mixing and allowed to complete conjugation. At 36 h post-mixing, one of the two separated exconjugants was analyzed by nested PCR to assess DNA elimination. Because IES elimination occurs at around 14 h post-mixing in wild-type cells, we reasoned that 36 h post-mixing should allow observation of DNA elimination in mutant cells, even if it was delayed. As negative controls for DNA elimination, pairs of Δ_TWI1_ and Δ_EZL1_ (encoding H3K9/K27 methyltransferase) strains (Supplemental Fig. S3) were also analyzed.
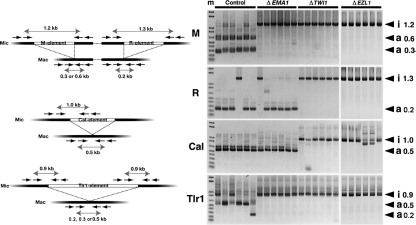
Figure 3.
DNA elimination in the progeny of ΔEMA1 cells. (Left) Schematic drawings of the IES elimination assays. Solid horizontal lines indicate MDSs and the open boxes indicate IESs. The M and R elements are ∼2.5 kb apart on the same Mic chromosome. Four primers (arrows) that flank each IES were used for nested PCR. For the assays of Cal and Tlr-1 elements, primers complementary to IESs were also used for the same PCR. (Right) Results of the IES elimination assays. The sizes of the unprocessed (Mic form) and the processed (Mac form) products are marked by arrowheads with “i” and “a”, respectively. (m) Molecular weight marker.
As expected, PCR products from the rearranged Mac forms of DNA were detected for all of the IES sites tested in the control wild-type strains (Fig. 3, control lanes) and only unrearranged, Mic forms of DNA were observed in the Δ_TWI1_ and Δ_EZL1_ strains (Fig. 3), arguing that the RNAi machinery and heterochromatin were required for DNA elimination of all of the IESs tested here. Surprisingly, disruption of parental EMA1 had different effects on different IESs: elimination of the M and Tlr1 elements was inhibited in the progeny of Δ_EMA1_ strains, while elimination of the R and Cal elements was not (Fig. 3). Thus, parental EMA1 is required for the elimination of some loci, but not of others. As far as we know, Δ_EMA1_ is the first Tetrahymena mutant reported to be defective in the DNA elimination of a subset of IESs .
The elimination of the M element was also inhibited in an EMA1 point mutant K188A (Walker-type ATP-binding motif substitution) (Supplemental Fig. S4), arguing that Ema1p was likely to have ATP-dependent RNA helicase activity that was involved in DNA elimination.
Ema1p is not required for scnRNA loading and passenger strand removal
RNA helicase A is required for the loading of siRNA to Argonaute proteins in human cells (Robb and Rana 2007). To determine whether Ema1p is required for the loading of scnRNAs to Twi1p, a Twi1p-containing complex was immunoprecipitated from wild-type and Δ_EMA1_ strains at 4 h post-mixing using an anti-Twi1p antibody, and the presence of coimmunoprecipitated scnRNAs was analyzed. Denaturing gel analysis (Supplemental Fig. S5A) revealed that the amount of scnRNAs complexed to Twi1p was similar in wild-type and in Δ_EMA1_ strains, suggesting that Ema1p was not required for scnRNA loading onto Twi1p.
scnRNAs are made from double-stranded Mic ncRNAs by the Dicer-like protein Dcl1p (Malone et al. 2005; Mochizuki and Gorovsky 2005). There should therefore be a mechanism to remove one of two scnRNA strands (the passenger strand); otherwise, the scnRNA would not be able to recognize its complementary sequences. Native gel analysis of the scnRNAs prepared above indicated that the amount of double-stranded scnRNAs associated with Twi1p in the Δ_EMA1_ strains was similar to that in the wild-type strains (Supplemental Fig. S5B). Thus, Ema1p is not required for the passenger strand removal of scnRNAs.
Ema1p is required for Twi1p–chromatin interaction
Next, we compared the localization of Twi1p in the presence and absence of Ema1p using an anti-Twi1p antibody. Both in wild-type and Δ_EMA1_ cells, Twi1p was localized in the parental Mac in early to mid-stages (Supplemental Fig. S6A,B,D,E) and in the developing new Mac in late stages (Supplemental Fig. S6C,F) of conjugation. Thus, Twi1p localized in Macs normally in the absence of Ema1p.
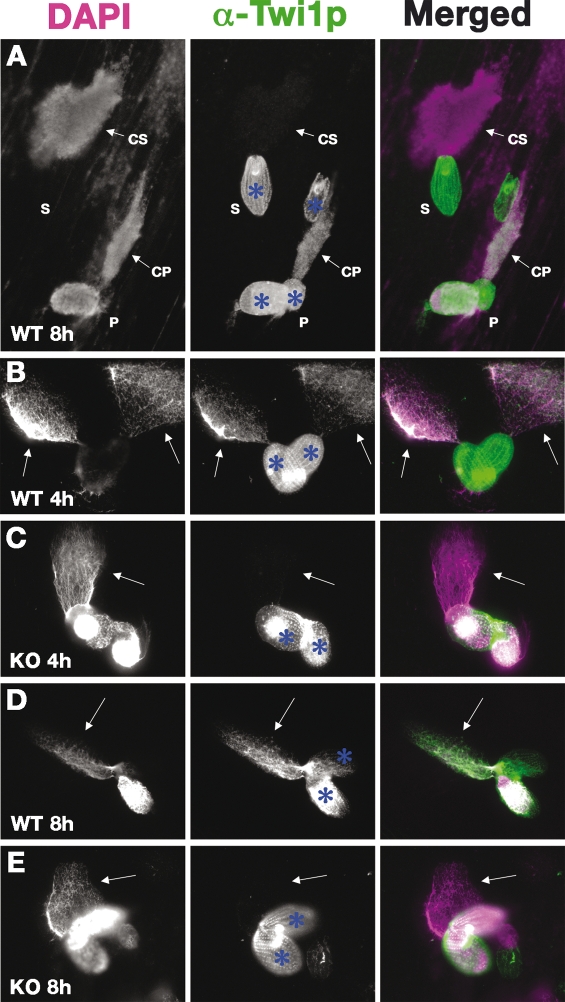
We previously hypothesized that the Twi1p–scnRNA complex interacts with chromatin to induce scnRNA degradation in the parental Mac, followed by H3K9me and DNA elimination in the new Mac (Mochizuki and Gorovsky 2004c). Therefore, we analyzed the association between Twi1p and chromatin. We prepared chromatin spreads from wild-type and Δ_EMA1_ strains and analyzed the localization of Twi1p on the chromatin. In wild-type cells, Twi1p colocalized with DNA spread out from the mating cells (CP in Fig. 4A), but not with that from nonmating cells (CS in Fig. 4A). As Twi1p is expressed only in mating cells (Mochizuki et al. 2002), this method could specifically detect Twi1p on chromatin.
Figure 4.
Association of Twi1p with chromatin in Δ_EMA1_ cells. (A–E) Cells with partially spread chromatin from wild-type (A,B,D) or Δ_EMA1_ (C,E) strains were fixed at the indicated time points post-mixing (mating). The localization of Twi1p (middle column; green in the right column) was analyzed by immunostaining using an anti-Twi1p antibody. DNA was counterstained with DAPI (left column; purple in the right column). (S) Single, nonmating cell; (P) paired mating cells; (CS) chromatin from a single cell; (CP) chromatin from paired cells. Arrows in B–E indicate chromatin from mating cells. In this experiment, the cell cortex (blue asterisks) has background staining for anti-Twi1p.
Twi1p was localized on the chromatin both at early (4 h post-mixing) (Fig. 4B) and late (8 h post-mixing) (Fig. 4D) conjugation stages in the wild-type cells. In contrast, no Twi1p was detected on the chromatins from Δ_EMA1_ cells at either time point (Fig. 4C,E). These results argue that Twi1p is associated with chromatin both in the parental and the new Macs in an Ema1p-dependent manner.
ncRNA is transcribed both in parental and in newly developed Macs
Recent studies in Schizosaccharomyces pombe suggest that an Argonaute protein Ago1 interacts with chromatin using base pairing between small RNAs and nascent transcripts to induce heterochromatin formation (for review, see Martienssen and Moazed 2006; Grewal and Jia 2007). Because Ema1p is a putative RNA helicase, Ema1p could act in the Twi1p–chromatin interaction by unwinding target RNAs to promote their interactions with scnRNAs. This hypothesis predicts the existence of nascent ncRNAs, transcribed from both the parental and the developing new Mac, which interact with scnRNAs to mediate Twi1p–chromatin interactions.
In Tetrahymena, ncRNAs heterogeneous in size (∼0.2–1 kb) that hybridized to the M element IES sequence and the repetitive IES sequence pTt2512 were detected (Chalker and Yao 2001). These ncRNAs have heterogeneous 3′ (Chalker and Yao 2001) and 5′ (U. Schöberl and K. Mochizuki, unpubl.) termini. Because the probes used in these studies came from IESs, and the ncRNAs first appeared early in conjugation, it was assumed that these transcripts were derived from the Mic. In addition, ncRNA complementary to an IES sequence was increased during conjugation when the IES was ectopically introduced into the parental Mac (Chalker and Yao 2001; Chalker et al. 2005). Thus, the parental Mac could also express ncRNA from exogenous DNA. However, it was not clear whether endogenous sequences of the parental Mac also make ncRNAs.
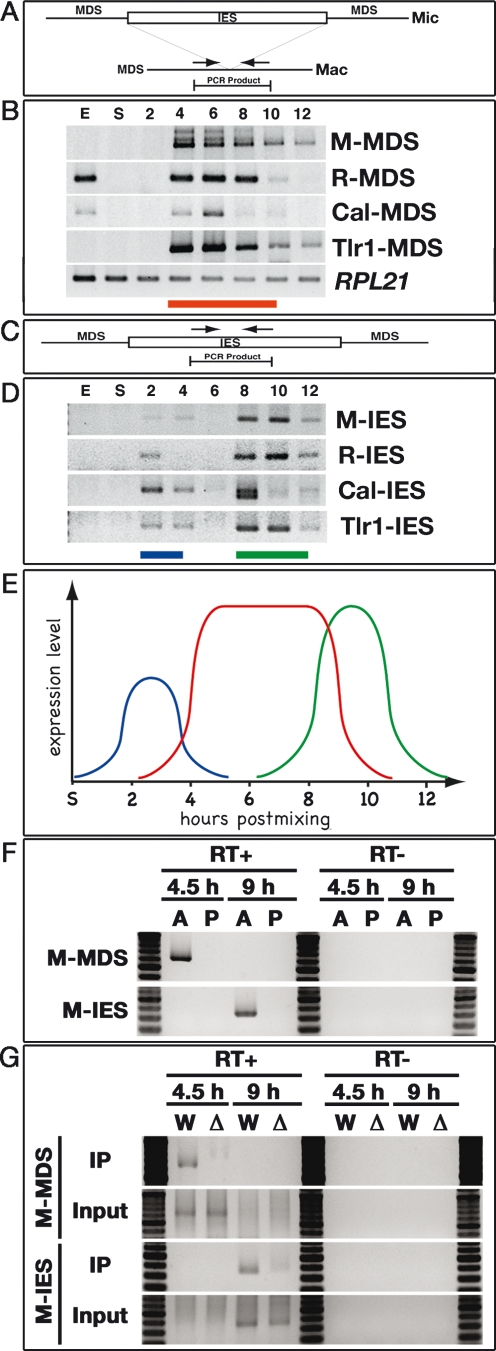
To observe ncRNAs from the parental Mac, we used a set of primers flanking the M, R, Cal, or Tlr1 elements for RT–PCR (Fig. 5A). We believe these regions do not include any coding sequences, because in Tetrahymena removal of IESs is often imprecise at the nucleotide level (Austerberry et al. 1989) and such variability would result in frame shifts if it occurred within genes. As shown in Figure 5B, transcripts were detected for the all IES-flanking Mac loci tested. These parental Mac ncRNAs were mainly detected in mid-conjugation stages (4–8 h post-mixing, corresponding to stages 2–4 in Fig. 1A), but some were also detected in exponentially growing cells. The biological significance of the vegetatively expressed ncRNAs is not clear. In mid-conjugation stages, most, if not all, of the Twi1p–scnRNA complexes are localized in the parental Mac (Mochizuki et al. 2002) where they could associate with these Mac transcripts. This interaction could play an essential role in the post-transcriptional selection of scnRNA.
Figure 5.
Three types ncRNA. (A–D) Exponentially growing (E), starved (S), and conjugating (2, 4, 6, 8, 10, and 12 h post-mixing) wild-type cells were analyzed by RT–PCR. (A) Schematic drawing of the analysis of ncRNA transcription from the parental Mac. Sets of primers (arrows) that flank four different IESs were used to amplify the cDNAs. (B) Results of RT–PCR for Mac loci flanking the indicated elements. The red horizontal bar indicates stages in which the ncRNAs are up-regulated. Constitutively expressed RPL21 mRNA was amplified as the positive control for the RT reaction. (C) Schematic drawing of ncRNA analysis from IESs. Sets of primers (arrows) in four different IESs were used to amplify the cDNAs. (D) The results of RT–PCR for the indicated IES elements. Blue and green horizontal bars indicate stages in which the ncRNAs are up-regulated. (E) Schematic summary of ncRNA transcription analyses. Relative expression levels (vertical axis) and expression timing (horizontal axis) of the ncRNAs from the Mic (blue), the parental Mac (red), and the developing Mac (green) are represented. (F) Coimmunoprecipitation of ncRNAs with Twi1p. (Lanes labeled as A) Conjugating wild-type strains at 4.5 or 9 h post-mixing were lysed and the Twi1p complex was immunoprecipitated with anti-Twi1p antibody. Coimmunoprecipitated RNA was analyzed by RT–PCR (RT+) to detect M-MDS (from parental Mac) or M-IES (from developing new Mac) transcripts as above. As a control, a similar experiment was performed with preimmune serum (lanes labeled as P). In parallel, a similar experiment was performed without the reverse transcription reaction (RT−). (G) Coimmunoprecipitation assay of ncRNAs with Twi1p in the wild-type (W) or Δ_EMA1_ strains (Δ). Conjugating cells were lysed at 4.5 or 9 h post-mixing and the Twi1p complex was immunoprecipitated with anti-Twi1p antibody (IP). Coimmunoprecipitated RNA was analyzed by RT–PCR (RT+) to detect M-MDS or M-IES transcripts. RNA was also extracted from part of the lysate and analyzed by RT–PCR (Input). As a negative control, a similar experiment was performed without the reverse transcription reaction (RT−).
Next, we sought to observe ncRNAs from IESs. Transcription from IESs could occur from the Mic and from the developing new Mac that has not yet undergone DNA elimination. The same cDNAs described above were used to detect ncRNAs from IES sequences using primer sets specific for M, R, Cal, and Tlr1 element IESs (Fig. 5C,D). As expected of Mic transcripts, ncRNAs from all of these IESs were detected in early conjugation stages (during Mic meiosis, 2 and 4 h post-mixing) (before stage 2 in Fig. 1A; stages E1 and E2 in Supplemental Fig. S1) and fell to low or undetectable levels at mid-conjugation (6 h post-mixing) (right before stage 4 in Fig. 1A; stage M2 in Supplemental Fig. S1). Importantly, the level of these transcripts increased again at 8–10 h post-mixing (stage 4 in Fig. 1A; stage L1 in Supplemental Fig. S1), when new Mac development started. This biphasic pattern suggests that ncRNA transcription from IES sequences occurs twice, first at meiotic prophase, the only stage when RNA synthesis and the transcription apparatus are detected in the Mic (Sugai and Hiwatashi 1974; Stargell and Gorovsky 1994; Mochizuki and Gorovsky 2004c), and then when new Macs develop.
Combining the data above, we conclude that three types of ncRNA transcription occur during conjugation (Fig. 5E). The first is transcription in the Mic at meiotic prophase (Fig. 5E, blue line). These Mic ncRNAs are probably processed to scnRNAs (Malone et al. 2005; Mochizuki and Gorovsky 2005). The second is transcription from the parental Mac at mid-conjugation stages (Fig. 5E, red line). The last is the transcription from IESs in the developing Mac at late stages of conjugation (Fig. 5E, green line). Since ncRNAs from the intergenic sequence between HHF2 and HHT2 were also detected from early to late conjugation stages (Supplemental Fig. S7), ncRNA transcriptions are not limited from IES-associated sequences, but probably occur genome-wide.
Similar RT–PCR studies using Δ_EMA1_ strains showed that the expression of all three classes of ncRNAs occurred normally (Supplemental Fig. S8), suggesting that the defects observed in Δ_EMA1_ strains are not caused by abnormal ncRNA transcription.
Ema1p is required for efficient Twi1p–ncRNA interaction
Twi1p associated with chromatin in an Ema1p-dependent manner in both the parental and developing new Macs (Fig. 4), and ncRNA was detected in both types of Mac (Fig. 5). These results led us to test the possibility that the interaction between Twi1p and chromatin is mediated by nascent ncRNA.
Cell lysates were prepared from mating wild-type cells at 4.5 h (around stage 2 in Fig. 1A) and 9 h post-mixing (stage 4 in Fig. 1A; stage L1 in Supplemental Fig. S1), when Twi1p is present in (Mochizuki et al. 2002) and the ncRNAs are expressed from (Fig. 5) the parental and developing new Macs, respectively. Twi1p was immunoprecipitated with an anti-Twi1p antibody and the coimmunoprecipitated RNA was used for cDNA production. Immunoprecipitation with preimmune serum was used as a control. ncRNAs from the parental Mac and the developing new Mac were observed by RT–PCR using primers flanking the M elements described above and with primers in the M element IES, respectively. As shown in Figure 5F, the parental Mac ncRNAs were specifically coimmunoprecipted with Twi1p at 4.5 h post-mixing (Fig. 5F). Similarly, the ncRNAs from the new Mac were detected specifically in the RNA coimmunoprecipted with Twi1p at 9 h post-mixing (Fig. 5F). Thus, Twi1p interacts with the ncRNA transcribed in both the parental and the developing new Mac.
Next, to determine whether these Twi1p–ncRNA interactions were dependent on Ema1p, a similar experiment was performed using Δ_EMA1_ strains. The ncRNAs coimmunoprecipitated with Twi1p were greatly reduced in the absence of EMA1 (Fig. 5G), indicating that Ema1p facilitates interaction between Twi1p and the ncRNAs. Because Ema1p is also required for the interaction between Twi1p and Mac chromatin (Fig. 4), these findings suggest that Ema1p mediates Twi1p association with the chromatin-associated nascent ncRNAs.
EMA1 is required for selective down-regulation of scnRNAs homologous to a repeated MDS
Because Twi1p associates with scnRNAs and is required for their accumulation (Mochizuki and Gorovsky 2004b), it was important to analyze the expression of scnRNA in Δ_EMA1_ strains. Because specific scnRNAs themselves have not been well characterized, we first studied the expression patterns of scnRNAs in wild-type cells.
We demonstrated previously that the total amount of scnRNAs remained relatively constant during conjugation (Fig. 6A; Mochizuki et al. 2002). In addition, we used endogenous scnRNAs as probes for hybridization to Southern blots containing Mac and Mic DNA to demonstrate that scnRNAs homologous to IES sequences became enriched as conjugation proceeds (Mochizuki and Gorovsky 2004b). These observations suggested that scnRNAs derived from IESs should persist in the later stages of conjugation, while those derived from MDSs should disappear. However, Chalker et al. (2005) reported that scnRNAs homologous to the M element IES detected by Northern hybridization were present in large amounts in very early conjugation stages, but were reduced to low levels in mid to late conjugation.
Figure 6.
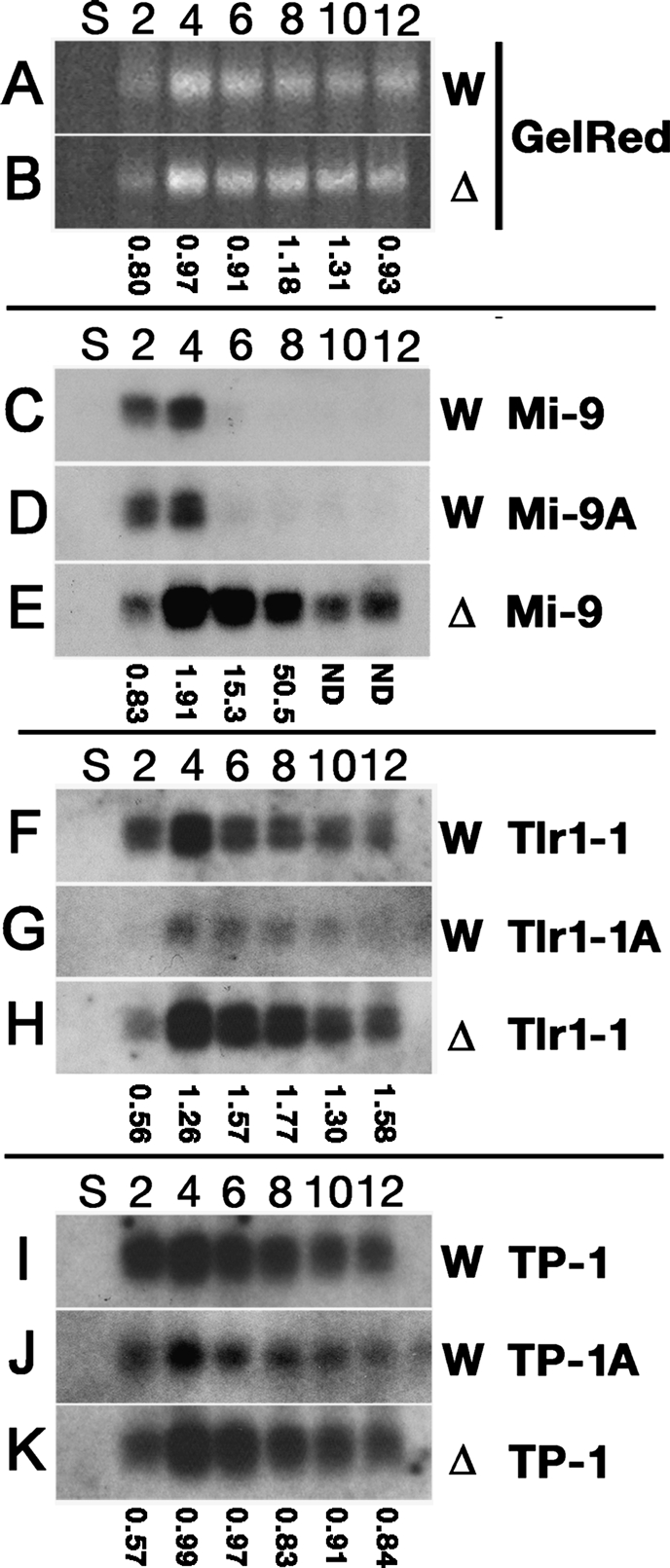
Expression of scnRNAs in Δ_EMA1_ cells. Total RNA was extracted from starved (S) or conjugating (2, 4, 6, 8, 10, and 12 h post-mixing) wild-type (W) or Δ_EMA1_ (Δ) strains and separated in sequencing gels. (A,B) Bulk scnRNAs (∼28 nt) were visualized by staining the gels with GelRed. (C–K) Blots were hybridized with the probes indicated (right). Quantitation of the signals are shown as ratios of the signals (Δ_EMA1_/wild-type) obtained from gel staining and Northern blots.
In an attempt to reconcile these observations, we examined the expression of scnRNAs homologous to the M element IES. Six 50-b DNA probes homologous to different regions of the M element IES (see Supplemental Fig. S9) were used to detect scnRNAs on Northern blots of RNA from different stages of conjugation. Five of the six probes tested failed to detect any scnRNAs, while one of them, the Mi-9 probe, detected scnRNAs (Fig. 6C) expressed in conjugating cells 2–4 h post-mixing (before stage 2 in Fig. 1A), but not thereafter. Similar results were obtained using the probe Mi-9A, which was complementary to Mi-9 (Fig. 6D), demonstrating the specificity of the hybridization.
To understand why different probes for the M element IES gave such different results, we examined the Tetrahymena genome database and confirmed that a ∼190-bp sequence (referred to as the MI repeat), including the sequence in the Mi-9 probe, was highly repeated in the sequences found in the Tetrahymena “Mac” genome database, as also recently noted by Kowalczyk et al. (2006). MI repeats were found both in long (≥10 kb) and in short (<10 kb) genomic scaffolds (Supplemental Fig. S9). Short scaffolds are thought to represent contaminants of the Mic genome in the Mac preparations used for genomic library construction, while long scaffolds are thought to represent true assemblies of Mac chromosomes (Eisen et al. 2006). We conclude, therefore, that the scnRNAs detected by the Mi-9 probe and by the M-IES probes studied by Chalker et al. (2005) were derived not only from the M element IES, but also from other MI repeats located in many different MDS and IES loci. Since the other (non-Mi-9) M element probes are probably complementary only to the single-copy sequences of the M element IES, we believe the expression levels of the scnRNAs homologous to them were too low to be detected in our experiments. Because it is actually an MDS sequence, the large amount of scnRNAs homologous to MI repeats in early conjugation and their dramatic reduction in the midstages of conjugation is consistent with our previous observation that scnRNAs homologous to sequences in the parental Mac (=MDSs) are reduced in the midstages of conjugation (Mochizuki and Gorovsky 2004b).
Next, we examined MI repeat scnRNAs in the Δ_EMA1_ strains. In Δ_EMA1_ cells, bulk scnRNA levels were comparable with those in the wild-type cells (Fig. 6A,B). Thus, EMA1 is not required for the accumulation of bulk scnRNAs. Interestingly, in the absence of EMA1 expression, scnRNAs homologous to Mi-9 probes were continuously detected, even in late stages of conjugation where they were not detected in wild-type cells (Fig. 6C,E). This difference was not due to developmental defects in Δ_EMA1_ cells because although they proceed more slowly through the meiotic stage, most Δ_EMA1_ strains exhibit normal initiation of the appearance of scnRNAs between 0 and 2 h (Fig. 6) and cytologically normal new Macs by 8 h post-mixing (Fig. 2D). Thus, we conclude that Ema1p is involved in the selective reduction of scnRNAs homologous to widely dispersed MI repeats and possibly of scnRNAs homologous to other MDSs.
scnRNAs homologous only to IESs are not affected in the absence of EMA1
Next, we analyzed the expression of scnRNA homologous to IES-specific sequences using 50 b DNA probes homologous to moderately repeated sequences found only in IESs. The probe Tlr1-1 is complementary to the Tlr1-element IES (Wells et al. 1994). The probe TP-1 is complementary to the antisense strand of a predicted ORF (TTHERM_01785770). Both are related to transposable elements (Wells et al. 1994; Eisen et al. 2006). Several sequences identical or similar to Tlr1-1 or TP-1 were found only in very short (<4 kb) scaffolds in the Tetrahymena Mac genome database (data not shown) and, therefore, were likely to be in IESs as described above.
In dramatic contrast to the scnRNA homologous to the MI repeat (Fig. 6C), in wild-type cells, the scnRNAs homologous to the Tlr1-1 and TP-1 were detected throughout conjugation (Fig. 6F,I). Similar results were obtained using the probes Tlr1-1A and TP-1A, which were complementary to Tlr1-1 and TP-1, respectively (Fig. 6G,J). These results are consistent with our previous observation that scnRNAs homologous to IES sequences persist to late stages of conjugation (Mochizuki and Gorovsky 2004b).
Next, we examined IES-specific scnRNAs present in the Δ_EMA1_ strains by Northern hybridization using the Tlr1-1 and TP-1 probes and found that there were no obvious differences in their expression between the wild-type and the Δ_EMA1_ strains (Fig. 6, cf. F and and K). Together with above results, we conclude that Ema1p is not required for the production of scnRNAs, but is involved in the selective down-regulation of scnRNAs homologous to MDSs.
H3K9 and K27me in the developing new Mac are affected in ΔEMA1 Cells
H3K9me2 and H3K27me3 accumulate specifically in IES-associated chromatin during DNA elimination in the developing new Mac (Taverna et al. 2002; Liu et al. 2007). H3K9me3 also specifically accumulates in the developing new Mac (Liu et al. 2007). Because Twi1p is associated with Ema1p and is essential for the accumulation of H3K9me2 and H3K27me3 (Liu et al. 2004, 2007) in the developing new Mac, we compared the accumulation of these modifications in wild-type and Δ_EMA1_ strains by immunostaining.
As reported, all of these modifications accumulated in the developing new Macs from the Nuclear Alignment stage (9 h post-mixing) (Figs. 1A, [slightly after stage 4], 7A,E,I; stage L1 in Supplemental Fig. S1) to the Pair Separation stage before DNA elimination started (12 h post-mixing) (Figs. 1A [stage 5], 7B,F,J; stage L2 in Supplemental Fig. S1) in wild-type cells. In Δ_EMA1_ cells, these modifications were differentially affected. H3K27me3 occurred in the developing new Mac at 9 h post-mixing, but it showed punctate localization at the periphery of the nuclei (Fig. 7C) instead of the even distribution observed in the wild-type cells at the same stage (Fig. 7A). In the later stages, H3K27me3 was evenly distributed in the new Mac in the Δ_EMA1_ strains (Fig. 7D), possibly due to the activity of zygotically expressed Ema1p (Fig. 7P).
In Δ_EMA1_ strains, both H3K9me2 and H3K9me3 were not detected in the developing Macs in early developmental stages (9 h post-mixing) (Fig. 7G,K). In most of the Δ_EMA1_ cells in the late stages of conjugation (12 h post-mixing), H3K9me2 accumulation reached a level similar to that of wild-type cells (Fig. 7H). This delayed appearance was well correlated with the accumulation of zygotic Ema1p in Δ_EMA1_ strains (Fig. 7P). H3K9me3 was not detected even in the very late stages of conjugation (Fig. 7L). Thus, Ema1p is essential for the accumulation of H3K9me3. We conclude that Ema1p is required for proper induction of H3K9me2 and H3K9me3, and only H3K9me2 can be partially restored by zygotic Ema1p.
Liu et al. (2007) reported that H3K27me was upstream of H3K9me. Thus, H3K27me3, H3K9me2, and H3K9me3 should occur sequentially, and the different effect of the loss of parental EMA1 on these modifications correlates well with this sequential event. These suggest that Ema1p is likely essential for accumulation of all of these modifications, and the zygotic Ema1p can restore only the earlier steps of the sequential events.
Discussion
We identified a putative RNA helicase, Ema1p, as part of a complex containing the Argonaute protein Twi1p. Ema1p is required for interaction between Twi1p and chromatin in both parental and developing new Macs. We also demonstrated the presence of ncRNAs in both parental and developing new Macs and showed that Ema1p is essential for the efficient interaction between Twi1p and these ncRNAs. In the parental Mac, Ema1p is required for selective down-regulation of scnRNAs homologous to MI repeat MDS sequences. In the new Mac, it is required for the induction of H3K9/K27me, and for the elimination of a subset of IESs. We propose that Ema1p functions similarly in both parental and new Macs to facilitate interaction between the Twi1p– scnRNA complex and nascent ncRNAs to induce these events.
Possible functions of Ema1p in DNA elimination
We propose that Ema1p mediates scnRNA–chromatin interaction by facilitating the interaction between scnRNA and nascent ncRNA because (1) it is required for Twi1p–chromatin interaction in both parental and new Macs (Fig. 4), (2) is required for efficient interaction between Twi1p and the ncRNA transcribed from both parental and new Macs (Fig. 5) and, (3) Twi1p interact with scnRNA (Mochizuki and Gorovsky 2004b). This scnRNA–nascent RNA interaction most likely serves to identify the scnRNAs homologous to MDS sequences that will be down-regulated in the parental Mac (Fig. 6) and those homologous to the IESs that will induce H3K9/K27me (Fig. 7) and IES elimination (Fig. 3) in the developing new Mac. While there is no direct evidence supporting the idea that ncRNAs bridge the Twi1p– chromatin interaction, the observations that the loss of Ema1p disrupts both the binding of Twi1p to chromatin and its association with ncRNA support this hypothesis. Future studies will be designed to understand the biogenesis of ncRNA and to perturb it genetically to clarify the precise role (or roles) of ncRNA in the DNA elimination.
In the fission yeast S. pombe, association of the Argonaute protein Ago1 with chromatin and the initiation and spreading of heterochromatin are proposed to involve base pairing between siRNAs and nascent RNA polymerase II transcripts (Motamedi et al. 2004; Djupedal et al. 2005; Kato et al. 2005; Bühler et al. 2006). A putative RNA helicase Hrr1 in S. pombe associates with Ago1 and is required for its localization to the centromeric repeats whose transcription initiates heterochromatin formation (Motamedi et al. 2004). Although Hrr1 and Ema1p belong to different classes of RNA helicases and their exact roles are unclear, they could still have analogous functions, as they both interact with Argonaute proteins and are essential for making heterochromatin.
scnRNA selection is likely to be achieved by the selective degradation of scnRNA complementary to MDSs
We demonstrated previously that scnRNA complementary to IES sequences were selectively enriched during the course of conjugation (Mochizuki and Gorovsky 2004b). In this study, we show that scnRNAs complementary to the MI repeat, which was found in both IESs and MDSs, were selectively down-regulated in the mid to late stages of conjugation, while scnRNAs complementary to the IES sequences were stably expressed throughout the conjugation. Thus, as suggested (Mochizuki et al. 2002), scnRNA selection is likely to be achieved, at least in part, by the selective down-regulation of scnRNAs complementary to MDSs.
Since the Dicer-like protein Dcl1p, which is responsible for the production of scnRNAs, is detected exclusively in the Mic in early conjugation, and the Mic-derived ncRNA found in early conjugation increases when DCL1 is knocked out (Malone et al. 2005; Mochizuki and Gorovsky 2005), scnRNAs are likely to be derived from Mic ncRNAs in early conjugation stages. The fact that ectopic expression of ncRNAs from parental Mac did not cause up-regulation of scnRNAs from these transcripts suggested that the parental Mac ncRNA does not contribute to produce scnRNAs (Chalker et al. 2005). These results suggest that the down-regulation of scnRNAs complementary to MDSs is likely to occur by their selective degradation.
Why is elimination of only a subset of IESs dependent on parental Ema1p?
Parental EMA1 is essential for the elimination of only a subset of IESs (Fig. 3). Because two RNAi machineries, Twi1p and Dcl1p, are required for elimination of the R and Cal elements (Fig. 8; Malone et al. 2005), whose elimination is not dependent on parental EMA1 expression, there may be an IES elimination pathway that is dependent on scnRNA, but not on scnRNA selection and parental Ema1p. Mic ncRNA, which produces scnRNAs, might be biased for IESs at some loci and the derived scnRNAs could possess sufficient specificity to cause IES elimination without the scnRNA selection process. Alternatively, ncRNA in the new Mac might be produced more efficiently from IESs than from their flanking MDSs at some loci and be targeted correctly even by unselected scnRNAs. It is interesting to note in this regard that the DNA elimination of a foreign sequence inserted into the Mic is known to be subject to the position effect (Liu et al. 2005) and thus, ncRNA transcription in the Mic and/or new Mac may occur nonuniformly.
Figure 8.
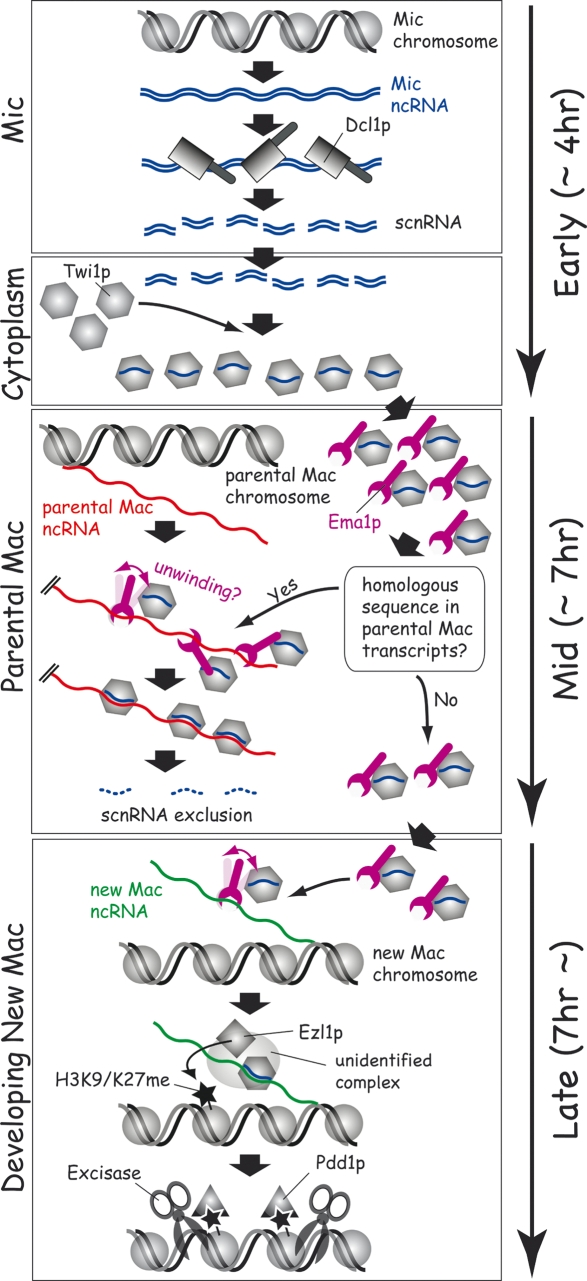
A refined scnRNA model. The sequentially occurring events are drawn from top to bottom. The approximate stages when the events occur are shown on the right with arrows. See the text for details.
It is known that the centromeric histone H3 variant Cna1p plays an essential role in IES elimination (Cui and Gorovsky 2006). Thus, the physical location of IESs in Mic may contribute to the position effect. Differences in sensitivity to homology-dependent maternal effects have also been reported (Fillingham et al. 2001). However, the patterns of dependence on Ema1p do not correlate with either of these processes. The characterization of more than the currently known small number of IESs in Tetrahymena will enable us to better define what causes sensitivity to this protein.
Refining the scnRNA model
The present study enables the inclusion of Ema1p and ncRNAs in a refined version of the scnRNA model (Fig. 8). In the early stages of conjugation (soon after mating, in the first 4 h post-mixing, before stage 2 in Fig. 1A), bidirectional ncRNA transcription occurs in the Mic in meiotic prophase. The resulting double-stranded Mic ncRNAs (shown as blue wavy lines in Fig. 8) are processed to ∼28-nt scnRNAs by Dcl1p. The scnRNAs are then transferred to the cytoplasm, where they form a complex with accumulating Twi1p. Then, the scnRNA–Twi1p complex localizes in the parental Mac in the midstages of conjugation (4 to ∼7 h post-mixing, before stage 4 in Fig. 1A). In parallel, ncRNAs are made from parental Mac chromosomes (red wavy lines in Fig. 8). We propose that scnRNAs homologous to the parental Mac are selectively degraded in a homology-dependent process that is achieved by interactions between scnRNAs and the parental Mac ncRNAs. Ema1p (drawn in purple) likely functions in this selective elimination of scnRNAs either by unwinding the parental Mac ncRNAs to enhance the scnRNA–ncRNAs interaction and/or to enhance its turnover. scnRNAs complexed with ncRNAs could be digested by a specific ribonuclease or the interaction might displace Twi1p from the scnRNA, exposing the scnRNA to nonspecific ribonucleases. Next, we hypothesize that the remaining IES-specific scnRNAs, accompanied by Twi1p and Ema1p, localize to the developing new Mac in the late stages of conjugation (>7 h, after stage 4 in Fig. 1A) and target the methylation of H3K9/K27 to the chromatin transcribing ncRNAs (green wavy lines in Fig. 8) that are complementary with them. We suggest that the interaction between scnRNA–ncRNAs recruits a complex containing a histone methyltransferase, Ezl1p, to induce H3K9/K27me. Again, we propose that Ema1p is involved in the homology-dependent methylation of H3K9/27 by enhancing the interaction between scnRNA and their complementary nascent ncRNAs. Then, H3K9/27me attracts Pdd1p, a chromodomain protein, to establish a heterochromatin-like structure. Finally, we propose that this heterochromatin serves as a platform to attract an unidentified endonuclease, Excisase, which cuts out the IES and rejoins the flanking sequences.
A pleasing aspect of this refined model is that it argues that both of the essential processes of IES elimination in Tetrahymena, scanning and IES targeting, are likely to be mediated by small RNA-nascent RNA interactions. Thus, in both heterochromatin formation in S. pombe and IES elimination in Tetrahymena, epigenetic modifications of chromatin function are likely to have at their core mechanisms that are similar to those in conventional RNAi.
Materials and methods
Strains and culture conditions
Wild-type B2086, CU427, and CU428 strains of T. thermophila were provided by Dr. P.J. Bruns (Cornell University, Ithaca, NY). Flag-HA-TWI1, EMA1 knockout (Δ_ΕΜΑ1_), EMA1_-HA, Δ_EZL1, P1-EMA1 (wild type), and P1-EMA1 (K188A) strains are described in the Supplemental Material. Δ_RPB3_ homozygous heteterokaryon strains and Δ_TWI1_(somatic) strains were described previously (Mochizuki et al. 2002; Mochizuki and Gorovsky 2004b). Cells were grown in SPP medium (Gorovsky et al. 1975) containing 1% or 2% proteose peptone at 30°C. For conjugation, growing cells (∼5 × 105 cells per milliliter) of two different mating types were washed, prestarved (∼12–24 h) and mixed in 10 mM Tris (pH 7.5) at 30°C. In the experiments shown in Figure 6, the culture was refed at 4 h post-mixing by adding 1/3 vol of 4× SPP medium to limit the initiation of mating to that period.
Immunopurifications
Tandem affinity purification and identification of Twi1p-associated proteins, immunoprecipitation of Ema1p-HA, and analysis of RNA associated with Twi1p are described in the Supplemental Material.
Immunostainings
Cells were fixed and processed as described (Mochizuki et al. 2002; Loidl and Scherthan 2004) with a 1:200 dilution of anti-Ema1p serum (see Supplemental Material), 1:500 dilution of anti-Twi1p serum (see Supplemental Material), 1:50 dilution of anti-dimethyl histone H3 Lys 9 (H3K9me2) antibody or 1:250 anti-H3K9me3 antibody or 1:250 anti-H3K27me3 antibody followed by incubation in 1:500 to 1:2000 diluted Alexa488-conjugated anti-rabbit IgG (Invitrogen). Anti-methyl histone antibodies were a gift from T. Jenuwein (IMP, Vienna, Austria).
Progeny viability test
Viability of progeny was analyzed as described (Mochizuki et al. 2002), except the mating pairs were isolated at 6–8 h post-mixing and the growth of cells was examined at ∼60 h after cloning.
DNA elimination assays
Pairs of the RPB3 KO homozygous heterokaryon (control), Δ_EMA1_, Δ_EZL1_, or Δ_TWI1_ strains were isolated into drops of SPP medium at ∼8 h post-mixing and allowed to complete conjugation. One of the two separated exconjugants was analyzed at ∼36 h post-mixing by PCR as described previously (Mochizuki et al. 2002). Details of the DNA elimination assays are in the Supplemental Material.
Chromatin spreading
Chromatin spreading was performed (Loidl et al. 1998) with the following modifications. Mating cells (20 μL) concentrated to ∼5 × 106 cells per milliliter in 10 mM Tris (pH7.5) were dropped onto slides and mixed with 100 μL of ice-cold 5% Lipsol detergent (Barloworld Scientific). After ∼5–10 sec incubation, 200 μL of ice-cold fixation solution (4% paraformaldehyde and 3.4% sucrose) were added. The mixture was spread and dried. The slide was washed with PBS twice and used for immunofluorescent staining as described above.
RT–PCR
Total RNA was extracted using TRIzol reagent (Invitrogen) from wild-type (B2086 and CU428) or Δ_EMA1_ [Δ_EMA1_ (7)-3-1 and Δ_EMA1_ (8)-1-1] strains and analyzed by RT–PCR using the primers listed in Supplemental Table S2. Detailed descriptions for RT–PCR can be found in the Supplemental Material.
Northern hybridization
Total RNA extracted using TRIzol reagent (Invitrogen) from 6.7 × 105 starved or mating cells was separated on 15% denaturing polyacrylamide gels, transferred to Hybond N+ membranes (GE Healthcare) by semidry electroblotting (2 mA/cm2, 75 min), fixed by UV (1200 J/cm2), and baked for 30 min at 80°C. Membranes were probed with 10 pmol 5′-32P-labeled DNA oligos (Supplemental Table S2) in Ultra-Hyb Oligo solution (Ambion) for ∼24 h at 37°C. Membranes were washed twice in 2× SSC containing 0.5% SDS for 30 min at 37°C and subjected to autoradiography. The intensity of the signals was quantified by ImageJ version 1.37.
Acknowledgments
We thank Thomas Jenuwein (IMP, Vienna) for the anti H3K9me and H3K27me antibodies, and Jürgen Knoblich and Henriette Kurth (IMBA) for critical reading of the manuscript. Tetrahymena genome sequence data were obtained from the J. Craig Venter Institute (http://www.tigr.org). This work was supported by National Institutes of Health grants GM21793 and GM72752 to M.A.G. and a Canadian Institutes for Health Research grant (MOP13347) to R.E.P. L.A. is supported by the IMBA Ph.D. program (European Research Council). K.W.M.S. obtained infrastructure support from the Ontario Research and Development Challenge Fund and MDS SCIEX. K.M. is a Junior Group Leader at IMBA, funded by the Austrian Academy of Sciences.
Footnotes
References
- Austerberry C.F., Snyder R.O., Yao M.C. Sequence microheterogeneity is generated at junctions of programmed DNA deletions in Tetrahymena thermophila. Nucleic Acids Res. 1989;17:7263–7272. doi: 10.1093/nar/17.18.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler M., Verdel A., Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Chalker D.L., Yao M.C. Non-Mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Mol. Cell Biol. 1996;16:3658–3667. doi: 10.1128/mcb.16.7.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker D.L., Yao M.C. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes & Dev. 2001;15:1287–1298. doi: 10.1101/gad.884601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker D.L., Fuller P., Yao M.C. Communication between parental and developing genomes during Tetrahymena nuclear differentiation is likely mediated by homologous RNAs. Genetics. 2005;169:149–160. doi: 10.1534/genetics.104.032300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne R.S., Nikiforov M.A., Smothers J.F., Allis C.D., Yao M.C. Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Mol. Cell. 1999;4:865–872. doi: 10.1016/s1097-2765(00)80396-2. [DOI] [PubMed] [Google Scholar]
- Cui B., Gorovsky M.A. Centromeric histone H3 is essential for vegetative cell division and for DNA elimination during conjugation in Tetrahymena thermophila. Mol. Cell. Biol. 2006;26:4499–4510. doi: 10.1128/MCB.00079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djupedal I., Portoso M., Spahr H., Bonilla C., Gustafsson C.M., Allshire R.C., Ekwall K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes & Dev. 2005;19:2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duharcourt S., Yao M.C. Role of histone deacetylation in developmentally programmed DNA rearrangements in Tetrahymena thermophila. Eukaryot. Cell. 2002;1:293–303. doi: 10.1128/EC.1.2.293-303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J.A., Coyne R.S., Wu M., Wu D., Thiagarajan M., Wortman J.R., Badger J.H., Ren Q., Amedeo P., Jones K.M., et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J.S., Bruno D., Pearlman R.E. Cis-acting requirements in flanking DNA for the programmed elimination of mse2.9: A common mechanism for deletion of internal eliminated sequences from the developing macronucleus of Tetrahymena thermophila. Nucleic Acids Res. 2001;29:488–498. doi: 10.1093/nar/29.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S.I., Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Gorovsky M.A., Yao M.C., Keevert J.B., Pleger G.L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9:311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Höck J., Weinmann L., Ender C., Rudel S., Kremmer E., Raabe M., Urlaub H., Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Goto D.B., Martienssen R.A., Urano T., Furukawa K., Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- Kowalczyk C.A., Anderson A.M., Arce-Larreta M., Chalker D.L. The germ line limited M element of Tetrahymena is targeted for elimination from the somatic genome by a homology-dependent mechanism. Nucleic Acids Res. 2006;34:5778–5789. doi: 10.1093/nar/gkl699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.R., Collins K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes & Dev. 2006;20:28–33. doi: 10.1101/gad.1377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepère G., Bétermier M., Meyer E., Duharcourt S. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes & Dev. 2008.;22:1501–1512. doi: 10.1101/gad.473008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Mochizuki K., Gorovsky M.A. Histone H lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc. Natl. Acad. Sci. 2004;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Song X., Gorovsky M.A., Karrer K.M. Elimination of foreign DNA during somatic differentiation in Tetrahymena thermophila shows position effect and is dosage dependent. Eukaryot. Cell. 2005;4:421–431. doi: 10.1128/EC.4.2.421-431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Taverna S.D., Muratore T.L., Shabanowitz J., Hunt D.F., Allis C.D. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes & Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J., Scherthan H. Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. J. Cell Sci. 2004;117:5791–5801. doi: 10.1242/jcs.01504. [DOI] [PubMed] [Google Scholar]
- Loidl J., Klein F., Engebrecht J. Genetic and morphological approaches for the analysis of meiotic chromosomes in yeast. Methods Cell Biol. 1998;53:257–285. doi: 10.1016/s0091-679x(08)60882-1. [DOI] [PubMed] [Google Scholar]
- Malone C.D., Anderson A.M., Motl J.A., Rexer C.H., Chalker D.L. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol. Cell. Biol. 2005;25:9151–9164. doi: 10.1128/MCB.25.20.9151-9164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen R., Moazed D. In: RNAi and heterochromatin assembly. Epigenitics. Allis C.D., et al., editors. Cold Spring Habor Laboratory Press; Cold Spring Harbor, New York: 2006. pp. 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E., Chalker D.L. Epigenetics in Ciliates. In: Allis C.D., et al., editors. Epigenetics. Cold Spring Habor Laboratory Press; Cold Spring Harbor, New York: 2006. pp. 127–150. [Google Scholar]
- Mochizuki K., Gorovsky M.A. Small RNAs in genome rearrangement in Tetrahymena. Curr. Opin. Genet. Dev. 2004a;14:181–187. doi: 10.1016/j.gde.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Gorovsky M.A. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes & Dev. 2004b;18:2068–2073. doi: 10.1101/gad.1219904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K., Gorovsky M.A. RNA polymerase II localizes in Tetrahymena thermophila meiotic micronuclei when micronuclear transcription associated with genome rearrangement occurs. Eukaryot. Cell. 2004c;3:1233–1240. doi: 10.1128/EC.3.5.1233-1240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K., Gorovsky M.A. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes & Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K., Fine N.A., Fujisawa T., Gorovsky M.A. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Motamedi M.R., Verdel A., Colmenares S.U., Gerber S.A., Gygi S.P., Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Nikiforov M.A., Smothers J.F., Gorovsky M.A., Allis C.D. Excision of micronuclear-specific DNA requires parental expression of pdd2p and occurs independently from DNA replication in Tetrahymena thermophila. Genes & Dev. 1999;13:2852–2862. doi: 10.1101/gad.13.21.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb G.B., Rana T.M. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol. Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Stargell L.A., Gorovsky M.A. TATA-binding protein and nuclear differentiation in Tetrahymena thermophila. Mol. Cell. Biol. 1994;14:723–734. doi: 10.1128/mcb.14.1.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugai T., Hiwatashi K. Cytologic and autoradiographic studies of the micronucleus at meiotic prophase in Tetrahymena pyriformis. J. Protozool. 1974;21:542–548. doi: 10.1111/j.1550-7408.1974.tb03695.x. [DOI] [PubMed] [Google Scholar]
- Taverna S.D., Coyne R.S., Allis C.D. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- Wells J.M., Ellingson J.L., Catt D.M., Berger P.J., Karrer K.M. A small family of elements with long inverted repeats is located near sites of developmentally regulated DNA rearrangement in Tetrahymena thermophila. Mol. Cell. Biol. 1994;14:5939–5949. doi: 10.1128/mcb.14.9.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard J., Kaneshiro E., Gorovsky M.A. Cytochemical studies on the problem of macronuclear subnuclei in Tetrahymena. Genetics. 1972;70:251–260. doi: 10.1093/genetics/70.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M.C., Duharcourt S., Chalker D.L. Genome-wide rearrangements of DNA in ciliates. In: Craig N., et al., editors. Mobile DNA II. Academic Press; New York: 2002. pp. 730–758. [Google Scholar]
- Yao M.C., Fuller P., Xi X. Programmed DNA deletion as an RNA-guided system of genome defense. Science. 2003;300:1581–1584. doi: 10.1126/science.1084737. [DOI] [PubMed] [Google Scholar]