The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression (original) (raw)
Abstract
Histone monoubiquitylation is implicated in critical regulatory processes. We explored the roles of histone H2B ubiquitylation in human cells by reducing the expression of hBRE1/RNF20, the major H2B-specific E3 ubiquitin ligase. While H2B ubiquitylation is broadly associated with transcribed genes, only a subset of genes was transcriptionally affected by RNF20 depletion and abrogation of H2B ubiquitylation. Gene expression dependent on RNF20 includes histones H2A and H2B and the p53 tumor suppressor. In contrast, RNF20 suppresses the expression of several proto-oncogenes, which reside preferentially in closed chromatin and are modestly transcribed despite bearing marks usually associated with high transcription rates. Remarkably, RNF20 depletion augmented the transcriptional effects of epidermal growth factor (EGF), increased cell migration, and elicited transformation and tumorigenesis. Furthermore, frequent RNF20 promoter hypermethylation was observed in tumors. RNF20 may thus be a putative tumor suppressor, acting through selective regulation of a distinct subset of genes.
Keywords: RNF20, BRE1, H2B ubiquitylation, tumor suppressor, transcription
Eukaryotic DNA is packaged into a chromatin structure of repeating nucleosomes consisting of DNA wrapped around an octamer of core histone proteins (H2A, H2B, H3, and H4). The histone tails, which protrude from the nucleosome, are subjected to a multitude of covalent modifications believed to play a vital role in chromatin remodeling and transcriptional regulation (Jenuwein and Allis 2001; Berger 2007; Weake and Workman 2008). One such modification is histone H2B monoubiquitylation. In the yeast S. cerevisiae this process is mediated by the E3 ligase BRE1 (Hwang et al. 2003). In mammals, the hBRE1(RNF20)/RNF40 complex was shown to function as the relevant E3 ligase (Kim et al. 2005; Zhu et al. 2005). In yeast, transcription of several inducible genes is impaired in the absence of ubiquitylated H2B (H2Bub) (Kao et al. 2004). Increased levels of H2Bub occur on the GAL1 core promoter and throughout the transcribed region upon transcriptional activation, with both ubiquitylation and deubiquitylation being required for optimal transcription (Henry et al. 2003; Xiao et al. 2005). Moreover, H2B monoubiquitylation was shown to lead to H3 methylation on Lys 4 and Lys 79, considered marks of actively transcribed genes (Briggs et al. 2002; Sun and Allis 2002). Yet, a recent study suggests that H2B ubiquitylation in S. pombe controls transcriptional elongation by RNA polymerase II (Pol II) independently of H3 methylation (Tanny et al. 2007).
Along with the studies linking H2Bub positively with active transcription, other reports suggest a link between H2B ubiquitylation and gene repression in yeast. Thus, H2Bub is involved in subtelomeric gene silencing (Huang et al. 1997; Zhang 2003), likely via indirect effects (van Leeuwen and Gottschling 2002), and ablation of H2Bub up-regulates some genes in addition to down-regulating others (Mutiu et al. 2007; Tanny et al. 2007). Indeed, H2Bub may act as a barrier for transcriptional elongation by blocking the recruitment of the Ctk1 kinase (Wyce et al. 2007).
In mammalian cells, H2Bub was found to associate preferentially with the transcribed region of highly expressed genes, suggesting a positive role in transcription (Minsky et al. 2008). Moreover, RNF20/hBRE1 acts as a transcriptional coactivator of the p53 tumor suppressor protein (Kim et al. 2005), and in vitro transcription elongation assays established a role for H2Bub in facilitating elongation by Pol II (Pavri et al. 2006). Yet, two recent studies suggest that H2Bub may repress transcription and contribute to heterochromatin silencing in mammalian cells (Zhang et al. 2008; Zhao et al. 2008). These studies showed that the deubiquitylating activity of USP22, an H2Bub-specific hydrolase and a subunit of the human SAGA complex, is necessary for activation of SAGA-dependent genes. SAGA-mediated H2B deubiquitylation was also reported to control the development of neuronal connectivity in the Drosophila visual system (Weake et al. 2008). Furthermore, USP22 is required for cell cycle progression (Glinsky et al. 2005; Widschwendter et al. 2007; Zhang et al. 2008).
In this study we analyzed the genome-wide localization of H2Bub and the global transcriptional effects of extensive reduction of H2Bub via RNF20 depletion. The results indicate that RNF20, probably through H2Bub, activates or suppresses distinct gene classes. The sum of these effects on growth promoting and growth restrictive pathways, as well as RNF20-related alterations in human cancer, suggest a key function of RNF20 as a tumor suppressor.
Results
hBRE1/RNF20 regulates selectively the expression of a subset of genes in human cells
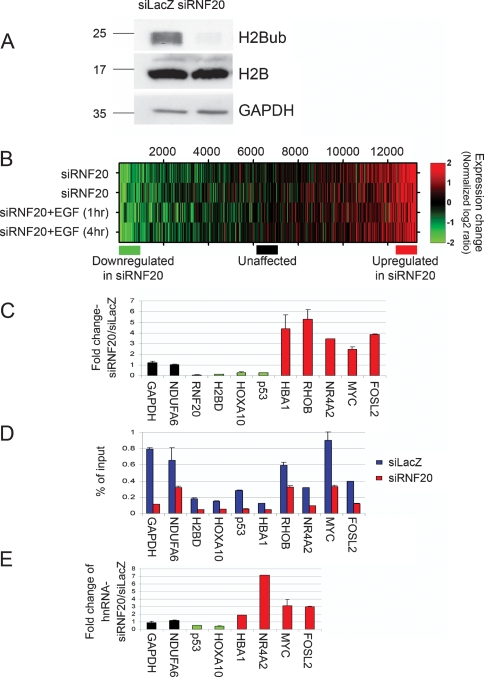
To address the possible involvement of hBRE1/RNF20 in the regulation of mammalian gene expression, we performed expression microarray analysis on HeLa cells after siRNA-mediated knockdown of endogenous RNF20. Consistent with its role as the major mammalian E3 ubiquitin ligase for histone H2B (Kim et al. 2005; Zhu et al. 2005), RNF20 knockdown led to near complete ablation of global ubiquitylated H2B (H2Bub) (Fig. 1A) visualized by Western blotting with H2Bub-specific monoclonal antibodies (Minsky et al. 2008), while not similarly affecting total H2B levels.
Figure 1.
RNF20 depletion decreases H2B ubiquitylation and causes large-scale expression changes. (A) Western blot analysis of H2Bub and total H2B 48 h after transient transfection of HeLa cells with RNF20 siRNA (siRNF20) or LacZ siRNA (siLacZ). GAPDH served as loading control. (B) Expression microarray analysis of the impact of RNF20 knockdown. RNA was extracted from HeLa cells 48 h after transfection with either RNF20 siRNA or LacZ siRNA, with or without EGF treatment (20 ng/mL), and hybridized to Affymetrix HG-U133A oligonucleotide arrays (see Materials and Methods). Colors indicate the normalized log2 expression ratio (siRNF20/siLacZ, with or without EGF as indicated) for each gene. The two top rows represent two independent biological repeats. (Bottom) Colored horizontal bars indicate the three groups of genes (∼1000 each) selected for ChIP-Seq (Fig. 2A) and ChIP/chip (Fig. 2B). (C) qRT–PCR validation of selected genes from B. RNF20-dependent genes (down-regulated in siRNF20), unaffected genes, and genes suppressed by RNF20 (up-regulated in siRNF20) are in green, black, and red, respectively. The Y axis shows fold change in siRNF20 relative to siLacZ. Prior to fold change calculation, values were normalized to GAPDH mRNA in the same sample. The GAPDH bar indicates the ratio between the nonnormalized amounts of GAPDH mRNA in the siRNF20 and siLacZ samples, respectively. (D) ChIP analysis of relative H2Bub abundance on the genes indicated in C, in siLacZ (blue) and siRNF20 (red) HeLa cells. ChIP was done with anti-H2Bub antibodies (Minsky et al. 2008). Immunoprecipitated DNA and input DNA were quantified by qPCR with primers specific for the transcribed regions of the indicated genes. (E) qRT–PCR analysis of precursor RNA. The RNA samples in C were subjected to qRT–PCR analysis with primer pairs derived from intronic sequences of the indicated genes, to quantify nonspliced primary transcripts and splicing intermediates. The Y axis represents fold change in siRNF20 relative to siLacZ, normalized to mature GAPDH mRNA in the same sample.
The results of the expression microarray analysis are shown in Figure 1B, which portrays the ratio between the expression level of each gene following RNF20 knockdown relative to its level in cells transfected with control siRNA. RNF20 depletion affected a similar set of genes in untreated HeLa cells (Fig. 1B, top two rows) and in cells treated with epidermal growth factor (EGF) for 1 or 4 h (Fig. 1B, bottom two rows) (EGF effects are further discussed below).
Previous genome-wide analysis revealed a general positive correlation between gene expression levels and steady-state H2Bub levels (Minsky et al. 2008). Nevertheless, the majority of transcribed genes in HeLa cells were not significantly affected by RNF20 knockdown. The overall correlation between the microarray data from the RNF20 depleted cells and the control cells was ∼0.94, suggesting that H2B ubiquitylation is not rate limiting for global transcription in these cells. Yet, two subgroups of transcripts diverged significantly from the bulk transcriptome. One subgroup was down-regulated upon RNF20 depletion (Fig. 1B, green), implying that H2Bub may be preferentially required for their efficient transcription. Interestingly, a comparable number of transcripts were up-regulated in RNF20-depleted cells (Fig. 1B, red), suggesting that their transcription is typically suppressed, directly or indirectly, by RNF20.
Supplemental Table 1 lists the most affected genes. Figure 1C depicts validation by quantitative RT–PCR (qRT–PCR) of the microarray data for representative genes from each group, as well as for some genes whose expression was not affected by RNF20 depletion. Supplemental Table 1 reveals that the gene subset down-regulated by RNF20 depletion was strongly enriched for genes encoding histones H2A and H2B (P values for H2A and H2B: 1.2 × 10−5 and 5.3 × 10−3, respectively), but not H1, H3, and H4 (P > 0.05). RNF20 knockdown by itself had no visible effect on cell cycle distribution of HeLa cells (Supplemental Fig. 1), and particularly did not alter the percentage of cells in S phase, when histone gene transcription preferentially occurs. Hence, the changes in H2A and H2B mRNA levels following RNF20 depletion are not secondary consequences of cell cycle changes. Rather, this may reflect direct transcriptional regulation by RNF20 activity.
Of particular note, lowering RNF20 had pronounced effects on the expression of key growth-regulating genes. Thus, RNF20 depletion strongly decreased the expression of the p53 tumor suppressor gene. Conversely, RNF20 depletion elevated the expression of several proto-oncogenes, most notably c-myc and c-fos.
The expression of many genes was only mildly affected by RNF20 depletion and consequent reduction of H2B ubiquitylation. It could be argued that high H2Bub levels are nevertheless maintained on those genes, despite the drop in overall H2B ubiquitylation. Chromatin immunoprecipitation (ChIP) with H2Bub-specific antibodies was therefore performed (Fig. 1D). All genes examined, irrespective of transcriptional response to RNF20 depletion, exhibited a significant reduction in associated H2Bub upon RNF20 knockdown. Further ChIP analysis confirmed the presence of associated RNF20 protein (Supplemental Fig. 2A). Moreover, ChIP analysis of the RNF20-dependent p53 gene (Supplemental Fig. 2B) revealed that RNF20 knockdown augmented the retention of RNA Pol II in the region adjacent to the TSS (1 Kb), while reducing Pol II occupancy near the 3′ end of the gene (18 Kb). As expected, the 1-Kb region was populated primarily with initiating Pol II (P-Ser5), whereas the 18-Kb region was associated with elongating Pol II (P-Ser2) (Supplemental Fig. 2C,D).
Changes in relative mRNA abundance do not necessarily imply altered transcription rates. To address this issue more definitively, we assessed the impact of RNF20 knockdown on the relative amounts of the primary transcripts corresponding to the mRNAs in Figure 1C. To that end, qRT–PCR was performed as in Figure 1C, but using primers derived from intronic sequences, which allow quantification of heterogeneous nuclear RNA. This method provides a good approximation of relative transcription rates (Kuroda et al. 2005; Phelps et al. 2006). As seen in Figure 1E, the distinct RNF20 responsiveness patterns displayed by the mature transcripts (Fig. 1C) were shared by their primary transcripts, strongly arguing that RNF20 depletion indeed affects the transcription rates of those genes.
Together, these results suggest that RNF20 mediates H2B monoubiquitylation on all or most genes, but reduction of H2Bub correlates with strong up- or down-regulation of only certain subsets of genes.
Genes suppressed by RNF20 are associated with higher levels of H2Bub
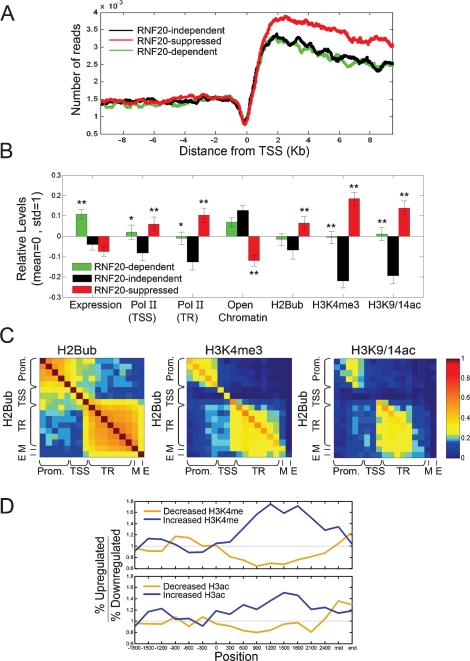
In order to explore the molecular basis for the selective regulation of gene expression by RNF20, we generated a high-resolution map of H2Bub distribution across the human genome. ChIP with H2Bub-specific antibodies (Minsky et al. 2008) was performed on HeLa cells transfected with LacZ siRNA, and the precipitated DNA was subjected to high-throughput sequencing using the Solexa 1G technology (“ChIP-Seq”). In agreement with our earlier results (Minsky et al. 2008), H2Bub was preferentially associated with the transcribed region while being excluded from the upstream region (Fig. 2A). H2Bub levels increased sharply downstream from the transcriptional start site, and declined gradually thereafter. Surprisingly, the subgroup of RNF20-suppressed genes (up-regulated in siRNF20, red) exhibited significantly higher association with H2Bub along the transcribed region when compared with the other two subgroups. Of note, analysis of published ChIP-Seq data, obtained with T cells (Schones et al. 2008), indicates that the three gene subgroups do not differ intrinsically in their overall patterns of nucleosome occupancy (Supplemental Fig. 3). Hence, both the paucity of H2Bub in the promoter region and the differential association of H2Bub with the transcribed regions of the different subgroups are specific features of this particular histone modification.
Figure 2.
RNF20-suppressed genes are associated with higher levels of H2Bub, H3K3me3, and H3K9/14Ac, higher Pol II occupancy, and compact chromatin. (A) Pattern of H2Bub distribution in HeLa cells on the three subgroups indicated by horizontal bars in Figure 1B, determined from high-throughput sequencing. For each gene, the number of sequence reads mapped to the vicinity of the TSS (10 kb upstream or downstream) was smoothed with a moving average of 300 bp. The smoothed data was averaged over RNF20-independent genes (black), RNF20-dependent genes (down-regulated in siRNF20, green) and RNF20-suppressed genes (up-regulated in siRNF20, red). Position 0 denotes the TSS. (B) Average values of expression and chromatin features in HeLa cells (siLacZ) for the subgroups indicated by horizontal bars in Figure 1B. Subgroups are designated as in A. Average expression in HeLa cells was calculated for each individual gene on the basis of the siLacZ microarrays in Figure 1A. The open chromatin score of each gene was defined by the closest genomic position assayed in the microarrays of Gilbert et al. (2004). All other chromatin features, including Pol II occupancy at the TSS and in the promoter-proximal part of the transcribed region (TR), as well as extent of histone H2B ubiquitylation (H2Bub), histone H3 Lys 4 trimethylation (H3K4me3), and acetylation of histone H3 Lys 9 and Lys 14 (H3K9/14ac) were determined by ChIP/chip analysis with appropriate antibodies and a custom tiling array (see Materials and Methods). Only probes overlapping the TSS and the transcribed region were included in the analysis. H2Bub signals were normalized to total H2B, H3 modifications were normalized to total H3, and Pol II signals were normalized to input DNA. All properties were further normalized to zero mean and unit standard deviation (Std), by subtracting their means and dividing by their Std. Error bars were calculated by bootstrapping. (*) _P_-value <0.05; (**) _P_-value <0.01 of regulated genes (red, green) relative to nonregulated genes (black). See Supplemental Table 3 for a complete list of _P_-values. (C) Correlation between modifications at different positions, calculated over 3000 genes represented in the custom array. For each pair of positions (relative to the TSS), we calculated the correlation between H2Bub at one position and H2Bub, H3K4me3, or H3K9/14ac at the other position. Positions are grouped into promoter (Prom), TSS, promoter-proximal part of the transcribed region (TR), middle (M) and end (E) of the transcribed region. (D) Correlation between differential expression changes and changes in histone H3 Lys 4 trimethylation (top) or histone H3 acetylation (bottom) upon RNF20 depletion. Gene group assignments (up-regulated or down-regulated by siRNF20) were as in Figure 1B. Histone modification data was obtained through ChIP/chip analysis using the custom array described in B probed with chromatin prepared form HeLa cells (either siRNF20 or siLacZ). For each position relative to the TSS, the 750 genes with the largest increase or decrease in the indicated modification at that position upon RNF20 knockdown were selected, and the ratio between the number of siRNF20 up-regulated genes and siRNF20 down-regulated genes within this set was calculated. Within the first 2.5 Kb of the transcribed region, increases in either modification were preferentially associated with up-regulation by siRNF20 and decreases in either modification were preferentially associated with down-regulation by siRNF20, while such associations were not observed at the promoter region or at the middle (mid) or end of the coding region.
Genes suppressed by RNF20 are associated with closed chromatin but higher constitutive RNA Pol II occupancy
Expression microarray analysis, performed on HeLa cells transfected with control LacZ siRNA, revealed that the genes dependent on RNF20 for optimal transcription (i.e., down-regulated by RNF20 knockdown) (Fig. 1B, green) tended to be expressed constitutively at higher than average levels (Fig. 2B, green; statistical analysis in Supplemental Table 3), whereas genes suppressed by RNF20 (i.e., up-regulated by RNF20 knockdown) (Fig. 2B, red) were expressed at levels slightly lower than those of typical RNF20-independent genes (Fig. 2B, black).
Next, ChIP plus microarray analysis (ChIP/chip) was performed in order to investigate the chromatin status of the different gene groups. To that end, we used a custom DNA microarray representing the transcription start site (TSS) as well as the TSS-proximal (900–2400 nucleotides downstream from the TSS), central and 3′-proximal regions of the transcribed region of the 3000 genes indicated by the bars in Figure 1A (see the Materials and Methods). Unexpectedly, despite being transcribed at rather modest rates, the RNF20-suppressed genes displayed exceptionally high Pol II occupancy at the TSS and within the TSS-proximal part of the transcribed region, as well as highly elevated constitutive levels of H3K4me3 and H3K9/14Ac. Furthermore, these genes also exhibited higher H2Bub levels, consistent with the ChIP-Seq (Solexa) data in Figure 2A. Analysis of the entire set of 3000 genes indicated that H2B ubiquitylation was significantly correlated with H3K4 trimethylation throughout the TSS-proximal part of the transcribed region (Fig. 2C), similar to what has been found for H3K4 dimethylation (Minsky et al. 2008). Interestingly, histone acetylation was significantly correlated with H2Bub only in the most 5′ part of that region.
Notably, when we analyzed the data obtained with T cells (Barski et al. 2007), using the same three groups of genes as in Figure 2B, the group defined by us as RNF20-suppressed was again found to exhibit higher PolII occupancy despite lower constitutive expression levels (data not shown). The display of a similar behavior in very dissimilar cell types argues that it represents an inherent, distinct feature of this gene set.
An additional distinct feature of these genes became apparent when we compared the chromatin state of the three groups of genes using data from a genome-wide low-resolution analysis of higher order chromatin structure (Gilbert et al. 2004). Notably, when compared with either of the two other groups, RNF20-suppressed genes had a remarkably lower open chromatin score (Fig. 2B), implying that they reside preferentially within compact chromatin regions. Together, these data suggest that genes suppressed by RNF20, presumably through H2B ubiquitylation, may be in a transcriptionally “on” state, as indicated by the presence of transcription-associated histone marks, but their actual transcription is disproportionately inefficient, perhaps being hindered by an unfavorable locally closed chromatin context. Alternatively, particular gene modifications such as H3K4me3 may actually recruit transcriptional repressor complexes rather than activators (Berger 2007).
Histone H3 methylation is only partially associated with H2B ubiquitylation and is insufficient to account for the selective impact of RNF20 on gene expression
H2B monoubiquitylation was previously reported to regulate histone H3 Lys 4 (H3K4) methylation in both yeast and mammals (Dover et al. 2002; Sun and Allis 2002; Kim et al. 2005). This would appear consistent with the positive correlation between the distribution patterns of H2Bub and H3K4me3 (Fig. 2C).
However, although RNF20 knockdown led to a dramatic decrease in global H2B ubiquitylation in HeLa cells (Fig. 1A), the overall amount of trimethylated histone H3K4 (H3K4me3) was affected only mildly (<1.5-fold) (Supplemental Fig. 4); this differs from the situation reported for HEK-293T cells, where a similar knockdown of RNF20 elicited a pronounced reduction in H3K4me3 (Kim et al. 2005). Hence, at least in HeLa cells, H3K4 trimethylation is either only partially dependent on H2Bub or else is very stable (note that the ChIP analysis was performed 48 h after RNF20 knockdown). Moreover, ChIP/chip analysis with anti-H3K4me3 antibodies did not reveal a significant change in the extent of this modification at most of the RNF20-regulated genes following RNF20 knockdown, despite their altered transcription rates (data not shown). Thus, for most RNF20-regulated genes, knockdown of RNF20 and loss of H2B ubiquitylation modulate transcription through a mechanism that apparently does not require changes in H3K4 trimethylation.
Yet, a subset of RNF20-regulated genes did exhibit a modest change in the pattern of H3K4 trimethylation upon RNF20 knockdown. In those genes, most of which were up-regulated upon RNF20 knockdown, increased H3K4me3 abundance was observed at the promoter proximal part of the transcribed region (Fig. 2D, top panel). Conversely, decreased H3K4me3 abundance within the same region was preferentially seen in genes whose expression was down-modulated by RNF20 knockdown. A similar effect, albeit more modest, was observed for histone H3 acetylation (Fig. 2D, bottom panel). In conclusion, while altered H3K4 methylation may contribute to the effects of RNF20 knockdown on the transcription rates of some genes, it does not appear to account for the majority of transcriptome changes that occur within 2 d of RNF20 depletion.
RNF20 preferentially modulates the expression of EGF-inducible genes
The results described thus far establish a gene-selective role for RNF20 in the maintenance of basal transcription patterns in mammalian cells. We next sought to determine the impact of RNF20 on induced gene expression. EGF elicits a transcriptional program in which distinct clusters of genes are induced at different time points after addition of the growth factor (Amit et al. 2007). Expression microarray analysis was therefore performed on control and RNF20-depleted HeLa cells 0, 1, or 4 h after EGF stimulation. Overall, the majority of the genes defined as RNF20 dependent under basal conditions displayed a very similar pattern of RNF20 dependence also in the presence of EGF (Fig. 1B), validating the robustness of this subgroup definition and the reproducibility of the expression microarray analysis. Surprisingly, a substantial overlap (P = 5 × 10−6) was observed between the group of genes induced by EGF alone (Supplemental Table 2) and those induced by RNF20 knockdown alone (Supplemental Table 1). Similar chromatin features may thus be shared between genes that are constitutively suppressed through the activity of RNF20 and genes poised for rapid activation by environmental cues.
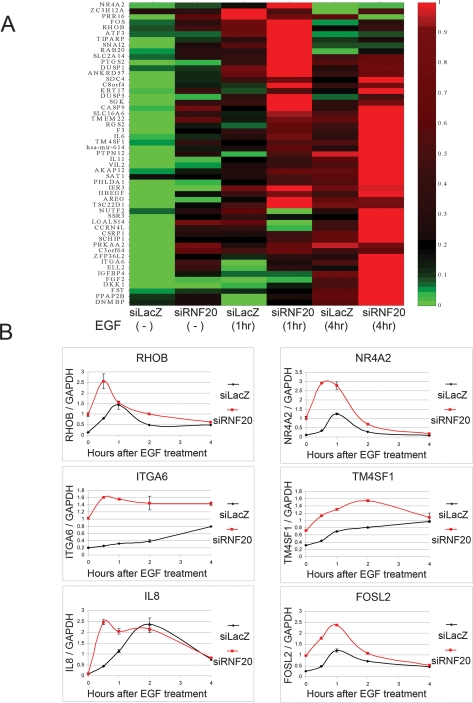
Figure 3A depicts the corresponding expression microarray analysis, focusing on the genes that responded positively to both EGF exposure and RNF20 knockdown. As reported (Amit et al. 2007), some genes were strongly induced by EGF within 1 h, whereas others were induced only after longer treatment. Regardless of EGF induction kinetics, these genes were also up-regulated by RNF20 knockdown. Moreover, the combination of EGF treatment and RNF20 knockdown often had a stronger inductive effect than either condition alone. Hence, RNF20 depletion potentiates the transcriptional response to EGF, at least with regard to many of the genes participating in this response.
Figure 3.
RNF20 modulates the expression of genes induced by EGF. (A) Clustering analysis of genes up-regulated by both RNF20 knockdown and EGF (20 ng/mL). Genes were included on the basis of the microarray data in Figure 1B (Supplemental Tables 1, 2). Each column represents expression values following the treatment indicated at the bottom. Log2 expression values of each gene were normalized into [0, 1], where 0 and 1 indicate the lowest and highest expression value of that gene in the six treatments, respectively. (B) qRT–PCR analysis of changes in the expression levels of the indicated representative genes at various times after exposure to EGF of HeLa cells transiently transfected with RNF20 siRNA (red) or LacZ siRNA (black). All values are normalized to GAPDH mRNA.
Detailed kinetic analysis by qRT–PCR was performed on representative genes. As seen in Figure 3B, all genes tested were indeed positively affected by the combination of RNF20 knockdown and EGF treatment. In most cases (with the exception of IL8), basal expression levels were significantly up-regulated by RNF20 knockdown. In all cases, the combination of both treatments led to overall higher transcript levels, and in some cases, induction following EGF exposure was more rapid (e.g., RHOB, ITGA6). Acceleration, but without a change in extent of induction, was observed for IL8. Hence, in agreement with the data in Figure 3A, RNF20 knockdown facilitates the EGF-induced transition of numerous genes into an efficiently transcribed state.
RNF20 knockdown attenuates p53 activity
RNF20 knockdown led to a pronounced decrease in p53 mRNA in HeLa cells (Fig. 1C). Similar results were obtained in MCF10A cells, derived from normal human mammary epithelium. RNF20 knockdown in MCF10A cells resulted in near-complete ablation of H2B ubiquitylation as well as prominent down-regulation of p53 protein (Supplemental Fig. 5) and RNA (data not shown). Furthermore, RNF20 interacts physically with p53 and is recruited by it to chromatin, where its serves as a transcriptional coactivator for p53 (Kim et al. 2005).
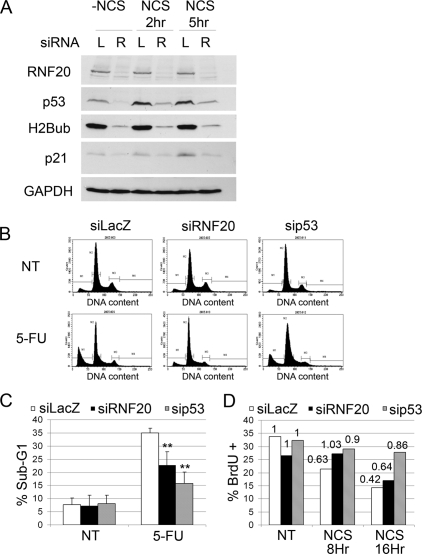
To study the impact of RNF20 on p53 activity, we subjected MCF10A to the genotoxic agent Neocarzinostatin (NCS). As seen in Figure 4, NCS induced a severalfold increase in p53 levels, which was markedly dampened by RNF20 knockdown. Concomitantly, both basal and NCS-induced levels of p21, encoded by a p53 target gene, were attenuated by RNF20 depletion.
Figure 4.
Knockdown of RNF20 compromises p53 activation. (A) MCF10A cells were transiently transfected with RNF20 siRNA (siRNF20) or LacZ siRNA (siLacZ) as control. Forty-eight hours later, cells were treated with NCS (200 ng/μL) for 2 and 5 h, and harvested. Equal amounts of protein from each sample were resolved by SDS-PAGE, followed by Western blot analysis with antibodies against p53 (DO1+PAb1801), RNF20, p21, H2Bub, and GAPDH. (B) HCT116 cells were transiently transfected with RNF20 siRNA, p53 siRNA, or LacZ siRNA as control. Forty-eight hours later cells were treated with 5-FU (50 μg/mL) for 24 h, harvested, fixed, and subjected to FACS-assisted DNA content analysis. (NT) Nontreated. (C) Quantification of the percentage of cells with sub-G1 DNA content, indicative of apoptosis, in HCT116 cells treated or not treated with 5-FU. Values were averaged from three independent experiments as in B. (**) _P_-value <0.01. (D) U2OS cells were transfected as in B. Forty-eight hours later, cells were treated with NCS (200 ng/μL) for 8 or 16 h, and BrdU incorporation was analyzed by FACS. The _Y_-axis indicates the percentage of BrdU-positive cells. (NT) Nontreated. Numerical values above each bar denote the extent of BrdU incorporation relative to the nontreated sample of the corresponding siRNA-transfected population.
We next asked whether RNF20 depletion affected p53-dependent biological responses to stress. Indeed, RNF20 knockdown reduced the extent of 5-Fluorouracil (5-FU)-induced apoptosis in p53-positive HCT116 cells, albeit somewhat less effectively than p53 knockdown (Fig. 4B,C). Moreover, it also attenuated and delayed NCS-induced growth arrest, assayed by BrdU incorporation in U2OS cells (Fig. 4D); the effect became less pronounced after extended exposure to genotoxic stress, consistent with the gradual accumulation of p53 under such conditions (data not shown). These findings argue that RNF20 is required for maintaining maximal p53 functionality in a variety of cell types.
RNF20 knockdown increases cell migration and promotes cell transformation
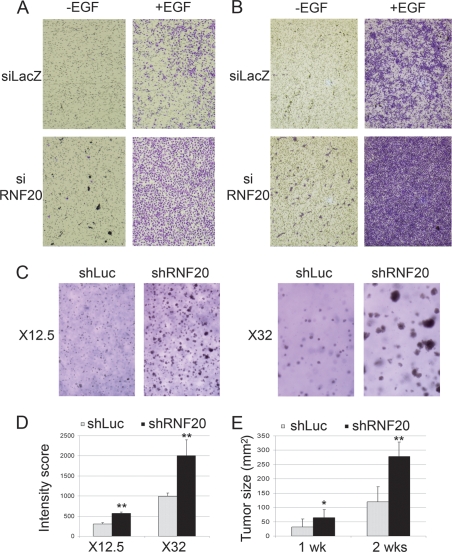
As shown above, RNF20 knockdown alone mimicked some of the transcriptional effects of EGF, and further augmented these effects when applied in combination with EGF. The biological significance of these observations was explored in MCF10A cells. MCF10A cells are usually nonmigratory (Fig. 5A,B, siLacZ − EGF), but can be stimulated to migrate by addition of EGF (Fig. 5A,B, siLacZ + EGF). Remarkably, knockdown of RNF20 alone was sufficient to induce some, albeit very modest, MCF10A cell migration (Fig. 5A,B, siRNF20 − EGF). The most dramatic effect was seen when both treatments were combined (Fig. 5A,B, siRNF20 + EGF), resulting in a robust increase in cell migration. Hence, down-regulation of RNF20 strongly enhances the biological effect of EGF. Since enhanced cell migration is a distinctive feature of advanced tumors, this finding suggests that decreased RNF20 expression may potentially facilitate cancer progression. Such facilitation may also stem from the more general ability of RNF20 down-regulation to augment the transcriptional output of growth stimulatory signal transduction pathways.
Figure 5.
RNF20 knockdown increases EGF-induced cell migration and anchorage independent cell proliferation. (A,B) MCF10A cells (50,000 [_A_] or 80,000 [_B_]) were transiently transfected with RNF20 siRNA (siRNF20) or LacZ siRNA (siLacZ) and plated 48 h later in the upper compartment of a 24-well Transwell tray (Corning). Cells were allowed to migrate for 16 h with or without EGF. Migrating cells were visualized by staining the membrane with methyl violet followed by photography under a binocular. (C) NIH3T3 cells (100,000 per 3.5cm dish), stably expressing lentivirus-encoded RNF20-specific shRNA (shRNF20) or shLuciferase were plated in soft agar. Two weeks later, colonies were photographed under a binocular at the indicated magnification. (D) Quantification of the data in C. See the Materials and Methods for details. (E) NIH 3T3 cells stably expressing RNF20 shRNA (clone #1) or Luciferase shRNA (shLuc) as a control were resuspended in PBS and injected subcutaneously into the left flank of nude mice. Tumor size (in square millimeters) was determined by caliper measurement 1 and 2 wk after injection.
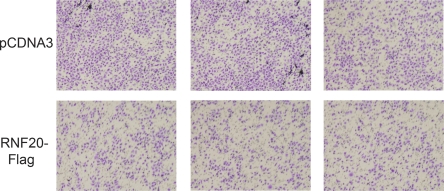
Further support for the potential oncogenic outcome of RNF20 down-regulation was provided through the use of mouse NIH-3T3 cells. Five recombinant lentiviruses expressing shRNA complementary to different sequences within the mouse RNF20 gene were generated; infection with either one of these viruses achieved a substantial reduction in RNF20 and H2Bub (Supplemental Fig. 6). Unlike what we observed in a variety of human cell types, RNF20 depletion did not reduce p53 mRNA levels in NIH-3T3 cells (data not shown). Cells stably expressing RNF20 shRNA were assessed for their ability to form colonies in soft agar—a strong indicator of neoplastic transformation in vitro. Unlike their counterparts expressing control shRNA directed against firefly luciferase, NIH-3T3 cells expressing RNF20 shRNA formed large colonies in soft agar, indicative of anchorage-independent growth (Fig. 5C, quantified in D). Furthermore, RNF20-depleted NIH-3T3 cells were significantly more tumorigenic when assayed in nude mice (Fig. 5E). Conversely, overexpression of RNF20 in HeLa cells caused a substantial reduction in their migration capacity (Fig. 6), suggesting that they have been rendered less malignant.
Figure 6.
RNF20 overexpression reduces HeLa cell migration. HeLa cells were transiently transfected with RNF20 siRNA (siRNF20) or LacZ siRNA (siLacZ) and plated 48 h later in the upper compartment of a 24-well Transwell tray (Corning). Cells were allowed to migrate for 16 h. Migrating cells were visualized by staining the membrane with methyl violet followed by photography under a binocular. Experiments were performed in triplicate. Each panel corresponds to a different transwell.
Together, our data strongly argue that RNF20 activity is inversely correlated with tumorigenic behavior, supporting the possibility that RNF20 possesses features of a tumor suppressor.
The RNF20 promoter is hypermethylated in breast cancer tumors
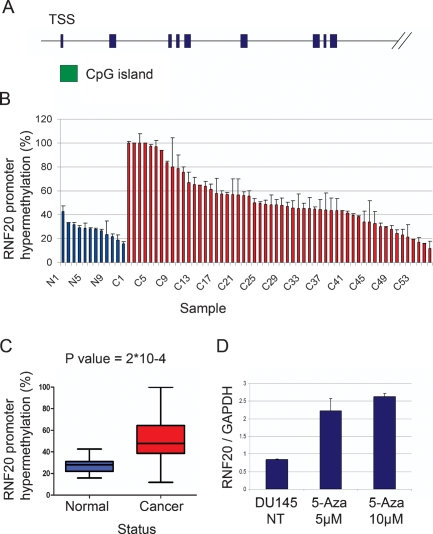
Tumor suppressors are often silenced in the course of tumor development via promoter hypermethylation, occurring selectively within CpG islands (Fraga and Esteller 2005). The RNF20 gene contains a prominent CpG island (Fig. 7A). We therefore performed methylation-specific qPCR to compare the extent of RNF20 promoter hypermethylation in normal breast tissue and in breast cancer tissue. Analysis of 56 cancer DNA samples (C1–C56) and 12 normal DNA samples (N1–N12) revealed RNF20 promoter hypermethylation in a substantial number of tumors when compared with normal tissue (Fig. 7B). This difference was statistically significant (Fig. 7C). The observed RNF20 promoter hypermethylation was not due to a nonspecific increase in overall CpG island methylation in those individual tumors as other CpG islands corresponding to the Lats2 and miR-34a genes displayed distinct and nonoverlapping methylation patterns in the same set of tumors (Supplemental Fig. 7).
Figure 7.
The RNF20 promoter is hypermethylated in breast cancer tumors. (A) Schematic map of the human RNF20 locus. Green box indicates the CpG island. Only the first nine exons are shown (blue boxes). (B) Genomic DNA from 56 breast cancer tumors (C) and 12 normal breast tissue specimens (N) was subjected to methylation-specific qPCR. Each bar depicts the relative methylation level of the CpG island located upstream of the RNF20 gene in an individual DNA sample, relative to a universal methylated human DNA standard, set as 100%. Alu primers were used in order to normalize for the total amount of genomic DNA in each sample. (C) Statistical analysis of the data in B. Boxes represent the values of 50% of the samples of each group. The horizontal bar within each box represents the mean of all samples, and vertical lines show the lowest and highest value. (D) DU145 cells were treated with 5 μM or 10 μM of 5-Aza-2′-Deoxycytidine (5-Aza), or left untreated (NT). Four days later, cells were harvested, and RNA was extracted. RNF20 mRNA was quantified by qRT–PCR and normalized to GAPDH.
A recent study (Varambally et al. 2005) revealed that RNF20 is often underexpressed in metastatic prostate cancer (http://www.ncbi.nlm.nih.gov/sites/entrez?db=geo&term="GDS1439"[ACCN]+RNF20). To examine a possible link between RNF20 promoter hypermethylation and RNF20 expression, we therefore used DU-145 cells derived from a prostate cancer brain metastasis. These cells exhibit substantial RNF20 promoter hypermethylation (>40%; data not shown). Exposure of DU-145 cells to 5′-deoxy-azacytidine (5-Aza), an inhibitor of DNA methylation, indeed led to a substantial increase in RNF20 mRNA (Fig. 7D), coupled with increased cell death (data not shown). Thus, at least in some tumors, RNF20 promoter hypermethylation may serve to dampen RNF20 expression.
Together, our data suggest that RNF20 possesses tumor suppressor features, and that during tumor progression these effects of RNF20 may be overcome by promoter hypermethylation.
Discussion
H2B monoubiquitylation is widely associated with actively transcribed genes in mammalian cells (Minsky et al. 2008). Nevertheless, extending earlier observations in yeast (Mutiu et al. 2007; Tanny et al. 2007), we found that only a subset of genes is significantly dependent on RNF20 for basal expression.
The exact contribution of H2B ubiquitylation to the transcription of this subset of genes remains to be established. Recent evidence suggests that H2Bub affects preferentially transcriptional elongation (Xiao et al. 2005; Pavri et al. 2006; Laribee et al. 2007; Shukla and Bhaumik 2007; Tanny et al. 2007; Wyce et al. 2007). Remarkably, many genes encoding histones H2A and H2B, but not H3 or H4, are regulated by RNF20, suggesting that the status of H2B ubiquitylation may control overall rates of de novo H2B synthesis. At present we cannot rule out that some of the effects of RNF20 manipulation may involve other, unknown targets of its E3 activity, or may represent secondary effects of changes in a more limited subset of direct target genes.
Although a dependence of H3K4 methylation on prior H2B ubiquitylation has been convincingly documented (Dover et al. 2002; Kim et al. 2005; Osley 2006; Lee et al. 2007), near-complete ablation of H2Bub by RNF20 knockdown affected global H3K4me3 levels only mildly (Supplemental Fig. 4). Whereas H2B ubiquitylation is very dynamic and reversible (Minsky et al. 2008), histone methylation is rather stable (Shilatifard 2006). It is thus possible that 48 h of RNF20 knockdown was not long enough to impose significant changes in global H3K4me3 levels. Nevertheless, our data implies that, at least within the subgroups of genes defined as RNF20 regulated, modulation of transcription by RNF20 is not strictly dependent on changes in H3K4 trimethylation, in line with yeast data (Tanny et al. 2007).
Why is only a subset of genes modulated by RNF20 knockdown? Conceivably, BRE1/RNF20 may affect the activity of only some transcriptional regulators. However, we could not identify a significant enrichment for specific transcription factor motifs among the RNF20-regulated genes (data not shown). Alternatively, only genes located in particular chromatin domains may be affected by H2Bub. Indeed, we found that some of the genes responding similarly to RNF20 knockdown reside in close proximity to each other, within a distance of two to five genes (data not shown); this trend was more significant among RNF20-suppressed genes. Hence, H2Bub may coordinately regulate small clusters of adjacent genes, although this does not appear to encompass larger chromatin regions.
Remarkably, genes constitutively suppressed by RNF20 and presumably by H2Bub were significantly enriched in compact chromatin fibers and were associated with higher Pol II occupancy and higher levels of H2Bub, H3K4me3, and H3K9/14ac, suggesting that many of them are constitutively “on” but are transcribed at rather low rates, perhaps owing to unfavorable chromatin context. The enhanced transcription of those genes upon ablation of RNF20 suggests that H2Bub may play a direct role in maintaining this “low gear” state. However, we cannot rule out an indirect effect of ubH2B; e.g., through regulation of genes whose products contribute to the attenuated transcription rates or to the unfavorable chromatin context. The unique features of the RNF20-suppressed genes may poise them to become rapidly activated in response to appropriate signals, owing to the fact that the transcriptional machinery and the associated histone modifications such as H3K4 methylation and H3 acetylation are already well in place. Indeed, this subgroup comprises several early response genes, activated within minutes after stimulation with serum or growth factors (Amit et al. 2007). It is tempting to speculate that H2B ubiquitylation helps maintain highly inducible genes in a transcriptionally restrained state. H2Bub may thus play a buffering role, ensuring that such pivotal genes are not expressed accidentally in the absence of a legitimate activating signal, yet can be rapidly turned on when such a signal is delivered. The dynamic nature of H2B ubiquitylation makes it a particularly attractive candidate for controlling such rapid switches. In support of this conjecture, induction of numerous EGF-responsive genes became more rapid, robust, and prolonged upon H2Bub ablation. Loss or attenuation of this restraint might unleash the expression of these otherwise tightly regulated genes. It is therefore perhaps not surprising that depletion of RNF20 and H2Bub elicits traits suggestive of a more transformed phenotype both in vitro and in vivo, and that RNF20 promoter hypermethylation is observed in human tumors.
The identities of the genes whose expression is affected by RNF20 depletion offer numerous potential mechanistic explanations for the proposed tumor suppressor activity of RNF20. Notably, RNF20 is required in order to maintain optimal expression of p53, whose loss of function is a most frequent event in cancer. RNF20 also sustains the expression of the TP53BP1 gene encoding 53BP1 (Supplemental Table 1), a pivotal component of the cellular response to double-strand DNA breaks; loss of 53BP1 function promotes genomic instability and facilitates cancer progression (Morales et al. 2006; Clarke et al. 2007). Moreover, reduced transcription of histone H2A and H2B, while maintaining high expression of the other histones, may also contribute to genome instability, perhaps by perturbing orderly nucleosome assembly (Prado et al. 2004; Prado and Aguilera 2005).
On the other hand, RNF20 restrains the transcription of genes whose excessive expression is conducive to malignancy. In addition to c-myc, several other proto-oncogenes, including c-Fos and FOSL2 (Fra-2), are also negatively regulated by RNF20. FOSL2/Fra-2 has been specifically implicated in the enhancement of cancer cell invasion and migration (Milde-Langosch et al. 2008). Likewise IL-8, whose induction by EGF is accelerated by RNF20 knockdown (Fig. 3B) promotes proliferation and invasiveness of cancer cells (Yao et al. 2007a, b). The CTEN gene is also implicated in cell migration (Katz et al. 2007); although only mildly affected in HeLa cells (data not shown), CTEN induction by EGF was significantly augmented by RNF20 depletion in MCF10A (Supplemental Fig. 8). Another interesting RNF20 target is ITGA6 (Supplemental Table 1), encoding α 6 integrin, which contributes to cancer progression (Chung and Mercurio 2004) and promotes motility and invasion (Yang et al. 2008). Moreover, α 6 integrin interacts with the EGF receptor and augments EGF-induced signaling (Bill et al. 2004; Edick et al. 2007), possibly explaining some of the biological effects documented in Figure 5. Finally, RNF20 depletion up-regulates SIRT1 expression (Supplemental Table 1), which is expected to further attenuate p53 function (Brooks and Gu 2008). Thus, RNF20 activity augments expression of growth-restricting genes while restraining expression of growth-promoting genes.
Down-regulation of USP3, capable of deubiquitylating histones H2A and H2B, results in DNA strand breaks and cell cycle perturbations (Nicassio et al. 2007). This suggests that disruption of the normal control of histone ubiquitylation can give rise to genome instability. In the case of USP3, the long-term cancer-promoting effects of such disruption are presumably blunted by induction of a potent DNA damage response (DDR) (Nicassio et al. 2007). However, if such imbalance is brought about by reduced RNF20 activity, the ensuing genome damage may not be properly managed because of the concurrent reduced expression of p53 and 53BP1. The notion that histone H2B ubiquitylation may underpin a tumor suppressor mechanism is further strengthened by the recent finding that the histone H2B deubiquitylating enzyme USP22 is required for cell cycle progression and, when overexpressed, may exert oncogenic effects (Nicassio et al. 2007; Zhang et al. 2008).
Together, our findings strongly suggest that RNF20 is a candidate tumor suppressor, exerting its effects via selective modulation of defined subsets of genes possessing distinct chromatin features. Elaboration of the underlying molecular mechanisms remains an appealing challenge, as does the further elucidation of the links between histone H2B ubiquitylation and cancer.
Materials and methods
Cell culture and siRNA transfections
Human cervical carcinoma HeLa cells and mouse NIH3T3 fibroblasts were grown in DMEM with 10% bovine serum supplemented with antibiotics, and MCF10A normal human mammary epithelium-derived cells were grown in DMEMF12 medium supplemented with antibiotics as well as 10 μg/mL insulin, 0.1 μg/mL cholera toxin, 0.5 μg/mL hydrocortisone, 5% heat-inactivated horse serum, and 10 ng/mL EGF. Oligonucleotides for the siRNA experiments were carried out with Dharmafect 1 reagent (HeLa) or Dharmafect 4 (MCF10A) according to the manufacturer’s protocol.
Recombinant lentiviruses
Lentivirus-based shRNA vectors for luciferase and RNF20 were purchased from Sigma. Lentiviruses were produced as described previously (Huang et al. 2006). Sequences of the short hairpins are listed in Supplemental Table 4.
Tissue specimens
Breast carcinomas of patients treated at the Masaryk Memorial Cancer Institute were collected from surgically treated women without neoadjuvant treatment and clinically documented distant metastases (Nenutil et al. 2005) upon informed consent confirming the availability of redundant tissue samples for research. Lumpectomy or mastectomy resection specimens were obtained within 20 min of surgical removal and were immediately evaluated by a pathologist. Tissue pieces of ∼3 mm ×3 mm × 8 mm were cut from redundant tumor tissue after standard surgicopathological processing, snap-frozen in liquid nitrogen, and stored at −80°C. All samples were stored for no longer than 2 years, and were thawed once immediately before DNA/RNA preparation. A paired sample of macroscopically uninvolved breast tissue, maximally removed from the tumor region, was processed in the same manner. Tissue samples were evaluated by a pathologist using sections from formalin-fixed paraffin embedded blocks, obtained in parallel to the frozen samples. For tumor samples, the percentage of tumor cells had to be >50%. For nontumorous tissue the samples were free of in-situ carcinoma and contained at least 5% epithelial cells together with a variable proportion of connective tissue and fat.
DNA isolation was performed using Qiagen DNeasy Blood and Tissue Kit (Qiagen) according to Animal Tissue Spin Column protocol.
Antibodies
The following commercial antibodies were used: anti-H2B (07-371, Millipore); anti-H3 (ab1791, Abcam); anti-Acetyl-H3 (06-599, Millipore), anti-H3K4me3 (ab8580, Abcam), anti-PolII (ab5408, Abcam). Anti-RNF20 (Novus Biologicals, NB100-2242) and a noncommercial anti-RNF20 antibody (Y. Shiloh, unpubl.) were used for ChIP and Western blots, respectively. Anti-H2Bub is described in Minsky et al. (2008).
RNA purification, real-rime qPCR, and microarray hybridization
RNA purification and qRT–PCR quantification were as described (Minsky et al. 2008). Primers for qRT–PCR analysis are listed in Supplemental Table 5. For oligonucleotide microarray hybridization, RNA was extracted using Qiagen RNeasy Mini Kit, according to the manufacturer’s protocol. Ten micrograms of RNA were labeled and hybridized to an Affymetrix HG-U133A oligonucleotide array. Expression value for each gene was calculated using Affymetrix Microarray software 5.0. Average intensity difference values were normalized across the sample set.
Microarray analysis and definition of RNF20-regulated genes
Log2 ratio (RNF20/LacZ) values were averaged over the two experiments and used to define genes as unaffected, up-regulated, or down-regulated in response to RNF20 depletion. Since log2 ratios are inflated for genes with lower LacZ expression levels, we normalized the log2 ratio of each gene based on the log2 ratios of the 1000 genes with the most similar LacZ levels: Xi′ = (Xi − mi)/si, where Xi is the log2 ratio of the i′th gene, mi, and si are the mean and standard deviation of the log2 ratios of 1000 genes with LacZ expression levels most similar to that of the i′th gene, and Xi′ is the normalized log2 ratio of the i′th gene. Up-regulated (or down-regulated) genes were defined with normalized log2 ratio thresholds of 2.5 (or −2.5) corresponding to _P_-value of 0.0062; for genes with two different probe sets, we lowered the threshold to 2 for both probes, while genes with three or more probes had to pass a threshold of 1.5.
ChIP, DNA array hybridization, and high-throughput sequencing
ChIP was performed as described (Minsky and Oren 2004). For oligonucleotide microarray hybridization, 1 μg of DNA was labeled and hybridized to a custom tiling array. High-throughput sequencing by the Solexa technology was performed by Illumina (http://www.illumina.com) on 20 ng of DNA from H2Bub ChIP. Sequences were aligned to the genome using Illumina’s alignment program.
Custom ChIP array and data analysis
We designed a custom Nimblegen array to interrogate histone modifications at ∼3000 selected genes, including those up-regulated, down-regulated and unaffected by RNF20 (∼1000 genes from each class). For each gene, 17 probes of 60 bp were designed: 15 probes for the region of −2 kb to +3 kb with respect to the TSS, at spacing of 300 bp, and one probe each for the middle and end of the coding region, respectively. Probes were selected by scanning a 200-bp region around each position of interest and choosing the best 60-bp probe according to a scoring system including %GC, lack of low-complexity sequences (e.g., polyA), lack of self complementarity, and low similarity to the rest of the genome. Each modification (or Pol II) was compared with a control sample (H3/H2B for histone modifications or input for Pol II). The resulting log2 ratio of each probe was normalized for %GC by subtracting the mean and dividing by the standard deviation of all probes with similar %GC.
Fluorescence-activated cell sorting (FACS) analysis
FACS-assisted cell cycle analysis for DNA content was performed as described (Aylon et al. 2006). For BrdU incorporation assay, Bromodeoxyuridine (BrdU; 10 μM final concentration) was added for 45 min. Cells were detached with trypsin, collected, washed once with ice-cold PBS, fixed in 70% Ethanol/HBSS overnight at 4°C, washed, rehydrated, and then stained with anti-BrdU antibodies followed by propidium iodide. Samples were analyzed by flow cytometry using a FACS sorter (Becton Dickinson).
Cell migration and transformation assays
Cell migration analysis was as described (Katz et al. 2007). Soft agar assay was as described (Milyavsky et al. 2003), and pictures were taken at 14 d. Images were converted to black and white and inverted. WaterShed segmentation defined individual colonies. Areas and intensities were quantified with a self-written quantification program (Z. Kam, unpubl.). Two fields from each condition were quantified.
Tumorigenicity in nude mice
Experiments were performed with 6- to 8-wk-old athymic nude mice using a minimum of five animals per treatment group. Mice were maintained in laminar flow rooms with constant temperature and humidity. Experimental protocols were approved by The Institutional Animal Care and Use Committee (Weizmann Institute). NIH-3T3 cells stably expressing lentivirus-encoded RNF20-specific shRNA (shRNF20, clone #1) or shLuciferase (2 × 106 cells per mouse), were inoculated subcutaneously into the left flank. Tumor growth was monitored by caliper measurement.
Methylation-specific PCR
Methylation-specific PCR was as described (Herman et al. 1996). Values were normalized for the total amount of genomic DNA in the sample, determined with the aid of Alu-specific primers; these primers are refractory to methylation status. Percentage methylation was determined by using the universal methylated human DNA standard (SssI methylated DNA, Ct. No. D5011, Zymo Research; 250 ng per reaction) as a 100% reference standard. The primers used were RNF20 methylated forward, CGTTAGGTATTAGCGAGGTTC; RNF20 methylated reverse, AAAATAACGTACGAAAAAAACACG.
Acknowledgments
We are grateful to Sylvia Wilder and Efrat Lidor-Nili for great help and discussions, Hershel Safer for help in analysis of solexa data, Shirley Horn-Saban for DNA array hybridization, and Zvi Kam for help with image quantification. This work was supported in part by the Dr. Miriam and Sheldon Adelson Medical Research Foundation, grant R37 CA40099 from the National Cancer Institute, a Prostate Cancer Foundation (Israel) Center of Excellence, and the Yad Abraham Center for Cancer Diagnosis and Therapy. B.V. and P.H. were supported in part by grant 301/08/1468 from the Grant Agency of the Czech Republic and grant MZ0MOU2005.
Footnotes
References
- Amit I., Citri A., Shay T., Lu Y., Katz M., Zhang F., Tarcic G., Siwak D., Lahad J., Jacob-Hirsch J., et al. A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- Aylon Y., Michael D., Shmueli A., Yabuta N., Nojima H., Oren M. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes & Dev. 2006;20:2687–2700. doi: 10.1101/gad.1447006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bill H.M., Knudsen B., Moores S.L., Muthuswamy S.K., Rao V.R., Brugge J.S., Miranti C.K. Epidermal growth factor receptor-dependent regulation of integrin-mediated signaling and cell cycle entry in epithelial cells. Mol. Cell. Biol. 2004;24:8586–8599. doi: 10.1128/MCB.24.19.8586-8599.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs S.D., Xiao T., Sun Z.W., Caldwell J.A., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D. Gene silencing: Trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- Brooks C.L., Gu W. p53 activation: A case against Sir. Cancer Cell. 2008;13:377–378. doi: 10.1016/j.ccr.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J., Mercurio A.M. Contributions of the α6 integrins to breast carcinoma survival and progression. Mol. Cells. 2004;17:203–209. [PubMed] [Google Scholar]
- Clarke A.R., Jones N., Pryde F., Adachi Y., Sansom O.J. 53BP1 deficiency in intestinal enterocytes does not alter the immediate response to ionizing radiation, but leads to increased nuclear area consistent with polyploidy. Oncogene. 2007;26:6349–6355. doi: 10.1038/sj.onc.1210457. [DOI] [PubMed] [Google Scholar]
- Dover J., Schneider J., Tawiah-Boateng M.A., Wood A., Dean K., Johnston M., Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Edick M.J., Tesfay L., Lamb L.E., Knudsen B.S., Miranti C.K. Inhibition of integrin-mediated crosstalk with epidermal growth factor receptor/Erk or Src signaling pathways in autophagic prostate epithelial cells induces caspase-independent death. Mol. Biol. Cell. 2007;18:2481–2490. doi: 10.1091/mbc.E06-04-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga M.F., Esteller M. Towards the human cancer epigenome: A first draft of histone modifications. Cell Cycle. 2005;4:1377–1381. doi: 10.4161/cc.4.10.2113. [DOI] [PubMed] [Google Scholar]
- Gilbert N., Boyle S., Fiegler H., Woodfine K., Carter N.P., Bickmore W.A. Chromatin architecture of the human genome: Gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Glinsky G.V., Berezovska O., Glinskii A.B. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Invest. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry K.W., Wyce A., Lo W.S., Duggan L.J., Emre N.C., Kao C.F., Pillus L., Shilatifard A., Osley M.A., Berger S.L. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes & Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman J.G., Graff J.R., Myohanen S., Nelkin B.D., Baylin S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Kahana A., Gottschling D.E., Prakash L., Liebman S.W. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:6693–6699. doi: 10.1128/mcb.17.11.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Perez-Burgos L., Placek B.J., Sengupta R., Richter M., Dorsey J.A., Kubicek S., Opravil S., Jenuwein T., Berger S.L. Repression of p53 activity by Smyd2- mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- Hwang W.W., Venkatasubrahmanyam S., Ianculescu A.G., Tong A., Boone C., Madhani H.D. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kao C.F., Hillyer C., Tsukuda T., Henry K., Berger S., Osley M.A. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes & Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M., Amit I., Citri A., Shay T., Carvalho S., Lavi S., Milanezi F., Lyass L., Amariglio N., Jacob-Hirsch J., et al. A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell migration. Nat. Cell Biol. 2007;9:961–969. doi: 10.1038/ncb1622. [DOI] [PubMed] [Google Scholar]
- Kim J., Hake S.B., Roeder R.G. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol. Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Kuroda M., Oikawa K., Yoshida K., Takeuchi A., Takeuchi M., Usui M., Umezawa A., Mukai K. Effects of 3-methylcholanthrene on the transcriptional activity and mRNA accumulation of the oncogene hWAPL. Cancer Lett. 2005;221:21–28. doi: 10.1016/j.canlet.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Laribee R.N., Fuchs S.M., Strahl B.D. H2B ubiquitylation in transcriptional control: A FACT-finding mission. Genes & Dev. 2007;21:737–743. doi: 10.1101/gad.1541507. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Shukla A., Schneider J., Swanson S.K., Washburn M.P., Florens L., Bhaumik S.R., Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- Milde-Langosch K., Janke S., Wagner I., Schroder C., Streichert T., Bamberger A.M., Janicke F., Loning T. Role of Fra-2 in breast cancer: Influence on tumor cell invasion and motility. Breast Cancer Res. Treat. 2008;107:337–347. doi: 10.1007/s10549-007-9559-y. [DOI] [PubMed] [Google Scholar]
- Milyavsky M., Shats I., Erez N., Tang X., Senderovich S., Meerson A., Tabach Y., Goldfinger N., Ginsberg D., Harris C.C., et al. Prolonged culture of telomerase- immortalized human fibroblasts leads to a premalignant phenotype. Cancer Res. 2003;63:7147–7157. [PubMed] [Google Scholar]
- Minsky N., Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol. Cell. 2004;16:631–639. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Minsky N., Shema E., Field Y., Schuster M., Segal E., Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell. Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- Morales J.C., Franco S., Murphy M.M., Bassing C.H., Mills K.D., Adams M.M., Walsh N.C., Manis J.P., Rassidakis G.Z., Alt F.W., et al. 53BP1 and p53 synergize to suppress genomic instability and lymphomagenesis. Proc. Natl. Acad. Sci. 2006;103:3310–3315. doi: 10.1073/pnas.0511259103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutiu A.I., Hoke S.M., Genereaux J., Liang G., Brandl C.J. The role of histone ubiquitylation and deubiquitylation in gene expression as determined by the analysis of an HTB1(K123R) Saccharomyces cerevisiae strain. Mol. Genet. Genomics. 2007;277:491–506. doi: 10.1007/s00438-007-0212-6. [DOI] [PubMed] [Google Scholar]
- Nenutil R., Smardova J., Pavlova S., Hanzelkova Z., Muller P., Fabian P., Hrstka R., Janotova P., Radina M., Lane D.P., et al. Discriminating functional and non-functional p53 in human tumours by p53 and MDM2 immunohistochemistry. J. Pathol. 2005;207:251–259. doi: 10.1002/path.1838. [DOI] [PubMed] [Google Scholar]
- Nicassio F., Corrado N., Vissers J.H., Areces L.B., Bergink S., Marteijn J.A., Geverts B., Houtsmuller A.B., Vermeulen W., Di Fiore P.P., et al. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Osley M.A. Regulation of histone H2A and H2B ubiquitylation. Brief Funct. Genomic Proteomic. 2006;5:179–189. doi: 10.1093/bfgp/ell022. [DOI] [PubMed] [Google Scholar]
- Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Phelps E.D., Updike D.L., Bullen E.C., Grammas P., Howard E.W. Transcriptional and posttranscriptional regulation of angiopoietin-2 expression mediated by IGF and PDGF in vascular smooth muscle cells. Am. J. Physiol. Cell Physiol. 2006;290:C352–C361. doi: 10.1152/ajpcell.00050.2005. [DOI] [PubMed] [Google Scholar]
- Prado F., Aguilera A. Partial depletion of histone H4 increases homologous recombination-mediated genetic instability. Mol. Cell. Biol. 2005;25:1526–1536. doi: 10.1128/MCB.25.4.1526-1536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F., Cortes-Ledesma F., Aguilera A. The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange. EMBO Rep. 2004;5:497–502. doi: 10.1038/sj.embor.7400128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones D.E., Cui K., Cuddapah S., Roh T.Y., Barski A., Wang Z., Wei G., Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Shukla A., Bhaumik S.R. H2B-K123 ubiquitination stimulates RNAPII elongation independent of H3-K4 methylation. Biochem. Biophys. Res. Commun. 2007;359:214–220. doi: 10.1016/j.bbrc.2007.05.105. [DOI] [PubMed] [Google Scholar]
- Sun Z.W., Allis C.D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Tanny J.C., Erdjument-Bromage H., Tempst P., Allis C.D. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes & Dev. 2007;21:835–847. doi: 10.1101/gad.1516207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F., Gottschling D.E. Genome-wide histone modifications: Gaining specificity by preventing promiscuity. Curr. Opin. Cell Biol. 2002;14:756–762. doi: 10.1016/s0955-0674(02)00393-9. [DOI] [PubMed] [Google Scholar]
- Varambally S., Yu J., Laxman B., Rhodes D.R., Mehra R., Tomlins S.A., Shah R.B., Chandran U., Monzon F.A., Becich M.J., et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Weake V.M., Workman J.L. Histone ubiquitination: Triggering gene activity. Mol. Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Weake V.M., Lee K.K., Guelman S., Lin C.H., Seidel C., Abmayr S.M., Workman J.L. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J. 2008;27:394–405. doi: 10.1038/sj.emboj.7601966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M., Fiegl H., Egle D., Mueller-Holzner E., Spizzo G., Marth C., Weisenberger D.J., Campan M., Young J., Jacobs I., et al. Epigenetic stem cell signature in cancer. Nat. Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Wyce A., Xiao T., Whelan K.A., Kosman C., Walter W., Eick D., Hughes T.R., Krogan N.J., Strahl B.D., Berger S.L. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol. Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Xiao T., Kao C.F., Krogan N.J., Sun Z.W., Greenblatt J.F., Osley M.A., Strahl B.D. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.H., Richardson A.L., Torres-Arzayus M.I., Zhou P., Sharma C., Kazarov A.R., Andzelm M.M., Strominger J.L., Brown M., Hemler M.E. CD151 accelerates breast cancer by regulating α 6 integrin function, signaling, and molecular organization. Cancer Res. 2008;68:3204–3213. doi: 10.1158/0008-5472.CAN-07-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Lin Y., Chua M.S., Ye C.S., Bi J., Li W., Zhu Y.F., Wang S.M. Interleukin-8 modulates growth and invasiveness of estrogen receptor-negative breast cancer cells. Int. J. Cancer. 2007a;121:1949–1957. doi: 10.1002/ijc.22930. [DOI] [PubMed] [Google Scholar]
- Yao C., Lin Y., Ye C.S., Bi J., Zhu Y.F., Wang S.M. Role of interleukin-8 in the progression of estrogen receptor-negative breast cancer. Chin. Med. J. (Engl.) 2007b;120:1766–1772. [PubMed] [Google Scholar]
- Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes & Dev. 2003;17:2733–2740. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- Zhang X.Y., Varthi M., Sykes S.M., Phillips C., Warzecha C., Zhu W., Wyce A., Thorne A.W., Berger S.L., McMahon S.B. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Lang G., Ito S., Bonnet J., Metzger E., Sawatsubashi S., Suzuki E., Le Guezennec X., Stunnenberg H.G., Krasnov A., et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Zhu B., Zheng Y., Pham A.D., Mandal S.S., Erdjument-Bromage H., Tempst P., Reinberg D. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol. Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]