INHIBITOR OF APOPTOSIS (IAP) PROTEINS IN EUKARYOTIC EVOLUTION AND DEVELOPMENT: A MODEL OF THEMATIC CONSERVATION (original) (raw)
. Author manuscript; available in PMC: 2009 May 1.
SUMMARY
The past decade and a half has witnessed the discovery of a large, evolutionarily conserved family of cellular genes bearing homology to the prototype baculovirus Inhibitor of Apoptosis (IAP). The logical decision in the field to also refer to these cellular proteins as IAPs fails to do justice to this versatile group of factors that play a wide range of roles in eukaryotic development and homeostasis which include, but are not limited to, the regulation of programmed cell death. Here we describe the shared functional characteristics of several well-characterized IAPs whose defining motifs place them more in the category of multifunctional modular protein interaction domains.
IAPs: a Versatile, Evolutionarily Conserved Family of Intracellular Proteins
The prototype IAP protein was reported in 1994 through a genetic screen designed to identify cytoprotective proteins encoded genes from baculovirus, which primarily infect members of the Lepidoptera order (Birnbaum et al., 1994). Through an elegant complementation approach, an open reading frame was identified from Cydia pomonella granulosis virus, a baculovirus used agriculturally as a commercial pesticide to control infestations of fruit trees with the Codling moth, C. pomonella and which, when expressed in trans, was able to protect insect cells from virus-induced cell death, thus enhancing viral replication and titer (Crook et al., 1993). It is probably fair to state that Lois Miller and colleagues could not have imagined the sweeping implications their original study identifying IAPs would have for a huge scope of disciplines ranging from metazoan development, mitotic regulation, pathogenesis of neoplastic and immunoproliferative diseases, intracellular metal ion trafficking and receptor-initiated cell signaling. At the same time, despite efforts to rename and reclassify this multifaceted family of factors, the term ‘IAP’ has stuck, a fact that frequently causes confusion to those new to the field, primarily because not all IAPs inhibit apoptosis or cell death.
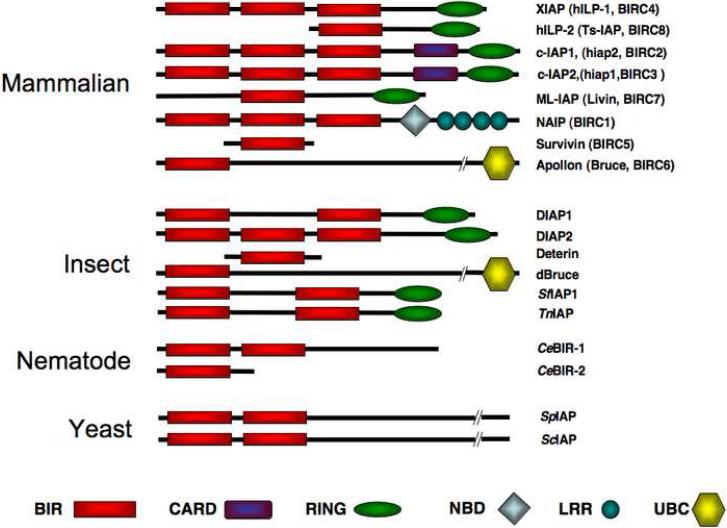
Two prominent structural features of the baculovirus IAPs were originally described: the baculovirus IAP repeat (BIR) and the RING finger domain. The BIR motif is a domain of approximately 65 residues, characterized by an invariant arrangement of conserved cysteines and histidines that adopt a classical zinc coordination configuration (Hinds et al., 1999; Miller, 1999). The BIR is the defining motif of the IAP family, and IAPs contain between one and three BIRs (Figure 1). Early findings suggested that the function of the BIRs was to prevent cell death through the direct binding and inhibition of caspases (Deveraux et al., 1998), the principal effector proteases of the apoptotic program. However, as will be described below, caspase inhibition is just one property of a small subset of BIR domains contained in only a minority of IAP proteins.
Figure 1. Domain structure of the IAP protein family.
The characteristic BIR domains are indicated by red rectangles, CARD domains by purple rectangles, RING domains by green ovals, NBD domains by diamonds, LRR domains by teal circles, and UBC domains by yellow hexagons. DIAP1, DIAP2, Deterin, and dBruce are Drosophila IAPs, while _Sf_IAP1 and _Tn_IAP are lepidopteran IAPs. Abbreviations: IAP, inhibitor of apoptosis; XIAP, X-linked IAP; BIRC, baculoviral IAP repeat containing; hILP, human IAP-like protein; Ts-IAP, testis-specific IAP; c-IAP, cellular IAP; ML-IAP, melanoma-IAP; NAIP, neuronal apoptosis inhibitory protein; DIAP, Drosophila IAP; _Sf_IAP1, Spodoptera frugiperda IAP; _Tn_IAP, Trichoplusia ni IAP; _Ce_BIR-1,-2, Caenorhabditis elegans BIRC _Sp_IAP, Schizosaccharomyces pombe IAP; _Sc_IAP, Saccharomyces cerevisiae IAP; BIR, baculoviral IAP repeat; CARD, caspase recruitment domain; NBD, nucleotide binding oligomerization domain; LRR, leucine rich repeat.
The second structural motif described in the prototype IAPs is the RING domain. RINGs are a specialized subset of zinc finger-like domains, which are found in a variety of proteins in addition to IAPs, including the c-CBL and PML proto-oncoproteins, and the RAG-2 protein involved in immunoglobulin gene rearrangement (Joazeiro and Weissman, 2000). Not all IAPs contain RINGs, but in those that do, the RING is characteristically located at the extreme carboxyl terminus of the protein. Many recent studies on RING-containing proteins, including IAPs, has revealed their involvement in ubiquitination of substrate proteins by functioning as E3 ubiquitin ligases. In many situations ubiquitination catalyzes the proteasome-mediated degradation of target proteins.
Reports of the existence of cellular IAP-like proteins appeared in the literature only two or three years after the discovery of the baculovirus IAPs. The first of these was neuronal apoptosis inhibitory protein (NAIP), which was first identified as a candidate gene potentially disrupted in a class of neurodegenerative diseases known as spinal muscular atrophy (Roy et al., 1995). Shortly after the description of NAIP, numerous cellular IAPs were also identified in an evolutionarily diverse range of organisms, discovered in some cases through sequence homology with the baculovirus IAPs, and in others through biochemical and genetic screens (Hay et al., 1995; Duckett et al., 1996; Rothe et al., 1995; Uren et al., 1996; Ambrosini et al., 1997; Liston et al., 1996). While eight distinct human IAPs have been characterized (Figure 1), in this review we will focus primarily on the five most studied: XIAP, c-IAP1, c-IAP2, NAIP and Survivin.
Caspase Inhibitory Properties of the IAPs
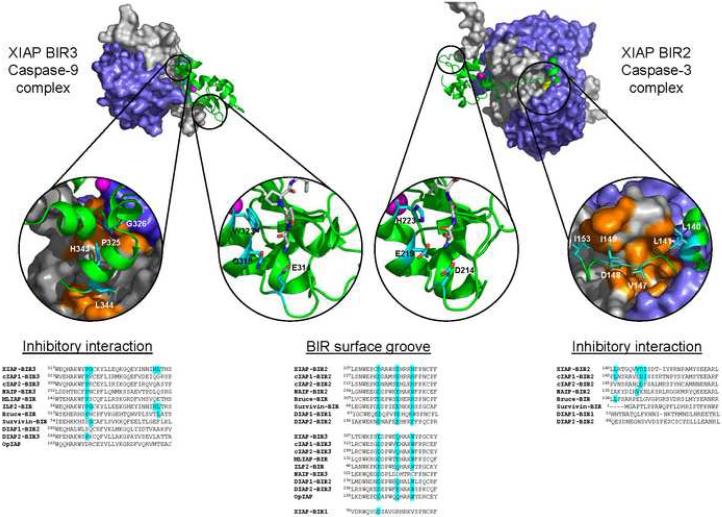
The widely expressed mammalian X-linked IAP (XIAP) was identified by virtue of its sequence homology to the prototype baculoviral IAPs (Duckett et al., 1996; Liston et al., 1996; Uren et al., 1996). However, unlike the baculovirus IAPs, which are composed of two BIRs, XIAP is comprised of three BIRs and a carboxy-terminal RING (Figure 2). XIAP exhibits antiapoptotic properties, and is the only mammalian IAP that directly inhibits the enzymatic activity of caspases-3, 7 and 9 – three proteases that are central to the apoptotic program (Deveraux et al., 1997; Takahashi et al., 1998; Riedl et al., 2001; Shiozaki et al., 2003). A domain comprised of the second BIR (BIR2) and a linker region immediately amino terminal to BIR2 is necessary and sufficient for inhibition of caspases-3 and -7, while the most carboxy-terminal BIR (BIR3), inhibits caspase-9 (Deveraux et al., 1999). Both BIR domains use a two-site binding mechanism for potent caspase inhibition. One of these sites is a conserved surface groove found in most IAP BIR domains. To achieve potent caspase inhibition, the XIAP BIR domain binds and anchors the caspase IAP binding motif (IBM) that is generated following caspase activation (Figure 2). This binding functions as an exosite, thus strengthening inhibitor binding. Although this anchoring interaction is conserved, the mechanism of enzyme inhibition is not. The peptide strand flanking the amino-terminus of XIAP BIR2 binds directly to the active site of caspase-3 and -7 (Figure 2, right) (Chai et al., 2001; Huang et al., 2001; Riedl et al., 2001) . Thus, tight inhibition of the executioner caspases requires two surfaces – a surface groove anchoring motif and an active site-directed inhibitory interaction. As opposed to targeting the enzyme active site directly, the functional inhibitory surface of XIAP BIR3 is a helix immediately following the BIR3 domain (Figure 2, left), which packs against the dimer interface of caspase-9, forcing the protease into an inactive conformation (Shiozaki et al., 2003).
Figure 2. The caspase-binding elements of IAPs.
Structure of the complex between XIAP BIR3 and caspase-9 (left, PDB 1NW9) and XIAP BIR2 with caspase-3 (right, PDB 1I3O). The caspase is in surface representation (large subunit in blue, small subunit in grey). The XIAP BIR domain and flanking region are in green cartoon, with the co-ordinated zinc in pink. Caspase inhibition is achieved via two-binding sites: an anchoring interaction with the BIR surface groove, and an “Inhibitory interaction”. Insets: critical XIAP residues that interact with the caspase are in cyan stick representation. Critical caspase residues that interact with XIAP are in orange surface representation. Primary sequence alignment reveals that the BIR surface groove is common to most IAP BIR domains, and there is overlapping binding specificity between IAP BIR domains. The BIR surface groove binds IAP-binding motif (IBM) containing proteins including caspase-9 and -3 (shown here in inset, N-terminus of caspase small subunit in grey stick), caspase-7, Smac/DIABLO, HtrA2/Omi, Grim, Reaper, Hid and others. Alignment of primary sequence across the “Inhibitory interaction” site demonstrates that XIAP is the only IAP that contains all the critical residues to confer direct inhibition of caspase catalytic activity. Although ILP2 also contains all the caspase-9 inhibitory elements, it is an unstable protein whose endogenous expression is yet to be demonstrated.
Although initial reports suggested other mammalian IAPs could also directly inhibit the proteolytic activity of caspases, this has turned out not to be the case (reviewed in (Eckelman et al., 2006). Comparing the primary sequence of other IAP BIR domains to the XIAP BIRs, it is clear why they do not directly inhibit caspase activity (Figure 2, alignments). Although they share the BIR surface groove that mediates caspase binding, no other IAP (besides the unstable ILP2) contains all the critical residues required for the functional inhibitory interaction. Therefore it is likely that XIAP evolved these specialized caspase-binding flanking regions to specifically inhibit caspase activity, and that the more evolutionarily conserved surface groove confers the BIR domain as a protein-protein binding module. This is analogous to the SH2 and LIM domains of cell signaling proteins. Indeed c-IAP1 and c-IAP2 can bind to mammalian caspases, yet the physiological consequence remains unclear (Eckelman and Salvesen, 2006; Tenev et al., 2004). It is possible that c-IAP1 and c-IAP2 may regulate caspases and other proteins by targeting them for ubiquitination, much like the mechanism utilized by DIAP1 to regulate Drosophila caspases. This redundancy may explain why the _Xiap_-deficient mice, while exhibiting differences from their wild-type counterparts, do not display a more catastrophic phenotype even if XIAP is the only IAP that directly inhibits caspases (Harlin et al., 2001). It also suggests that XIAP is likely to have other functions, as discussed further on. From an evolutionary perspective, the ability of XIAP to regulate apoptosis by direct caspase inhibition may represent a relatively recent acquisition as the family has diversified structurally and functionally to play a wide variety of physiological roles. For this reason, IAPs are also referred to as BIR-domain-containing proteins (BIRPs), to denote the fact that they can play multiple roles within the cell.
Non-caspase inhibitory mammalian IAPs
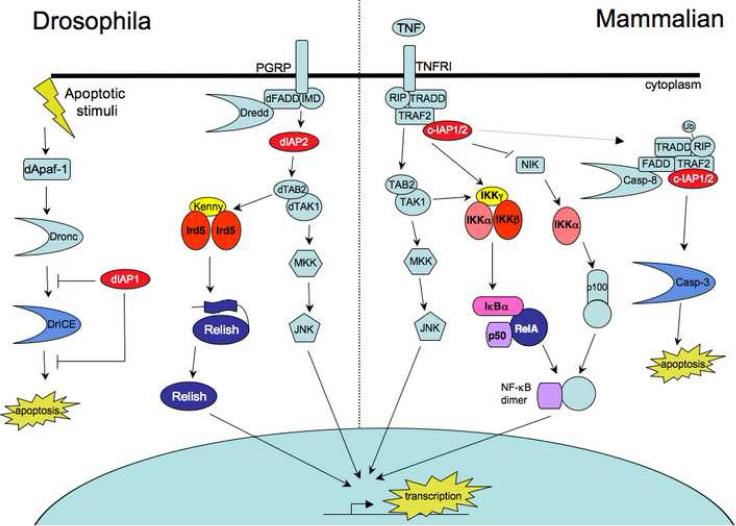
Like XIAP, two other mammalian IAP proteins, c-IAP1 and c-IAP2, have three BIR domains and a carboxy-terminal RING (Figure 1). In terms of homology and function, these two proteins are much more similar to each other than they are to XIAP. These cellular proteins were identified through interactions with the type-2 tumor necrosis factor receptor (TNFR2), perhaps foreshadowing a role for the IAP family as signaling modulators (Rothe et al., 1995). C-IAP1 and c-IAP2 were shown to associate with TNFR2 through interactions with the TNF receptor associated factors, TRAF1 and TRAF2 (Figure 3) . Despite their overall structural similarity, c-IAP1/2 exhibit very different functional properties to XIAP; in contrast to XIAP, the c-IAPs do not inhibit caspases (Eckelman and Salvesen, 2006), and conversely, XIAP does not to bind TRAFs (Roy et al., 1997; Duckett et al., 1998). TNF signaling through TRAFs leads to the activation of nuclear factor-κB (NF-κB), which then mediates inflammatory responses, cell proliferation, and cell survival by inducing transcription of pro-inflammatory and pro-survival genes (Pomerantz and Baltimore, 2002). While the role of c-IAP1 and c-IAP2 in TNF-mediated signaling was unclear, recent findings describe critical roles for the c-IAP proteins in TNF-mediated induction of NF-κB (Petersen et al., 2007; Varfolomeev et al., 2007; Vince et al., 2007; Gaither et al., 2007; Wang et al., 2008). c-IAP1 negatively regulates NF-κB-inducing kinase (NIK), preventing ubiquitination and subsequent processing of the p100 precursor form of NF-κB to the active p52 form (Varfolomeev et al., 2007; Vince et al., 2007). Thus, the physiological role of c-IAP proteins appears to involve inhibition of the non-canonical NF-κB pathway by TNF receptors.
Figure 3. IAP-dependent regulation of conserved Drosophila and mammalian signaling pathways.
Inhibition of apoptosis in Drosophila cells by DIAP1 occurs through binding to the initiator and effector caspases, Dronc and DrICE. Similarly, direct binding and inhibition of caspase-3 in mammalian cells is mediated by XIAP. Mammalian c-IAP1 and c-IAP2 can directly bind caspases but are poor caspase inhibitors, instead acting to regulate apoptosis by indirectly modulating caspase-8 activity. Binding of TNF to its receptor results in recruitment of TRADD, RIP, and TRAF2. The cIAPs also participate in pro-survival signaling through TNFR by associating with TRAF2. C-IAP1 and -2 ubiquitinate RIP1, minimizing association with caspase-8 and preventing apoptosis. Additionally the association of RIP, TRAF2, and cIAP1/2 leads to the activation of TAK and subsequent NF-κB and JNK activation, resulting in enhanced transcription of pro-survival genes. C-IAP1 and -2 can also inhibit NIK kinase and downstream processing of p100, thereby negatively regulating NF-κB activation. Thus, the effects of c-IAP1 and -2-dependent signaling on NF-κB are likely context dependent. A TNFR-like pathway regulates immune responses to microbial infection in Drosophila. Peptidoglycan from Gram-negative bacteria is recognized by peptidoglycan recognition proteins (PGRP), which can activate the IMD signaling pathway. IMD is an insect homolog of mammalian RIP1. Genetic studies place IMD, dFADD, Dredd and DIAP2 upstream of or parallel to dTAK activation. DTAK activates both the JNK and Relish pathways analogously to TAK1 in mammalian cells, promoting induction of anti-microbial peptide genes.
Survivin is the smallest mammalian IAP, comprised of a single BIR (Ambrosini et al., 1997). While its role in modulating caspase activity is complex and likely indirect, it plays a crucial role in embryonic development and in mitotic spindle formation, as discussed in more detail below (Li et al., 1998).
Clues from invertebrate IAPs – a division of labor
IAPs were first identified in baculoviruses, and subsequent phylogenetic studies suggested that the baculoviral IAP genes arose by capture of a host gene early in the evolution of Lepidoptera (Hughes, 2002). Given the vast phylogenetic diversity of the Lepidoptera order, which includes moths, and its dissimilarity to the Dipteran order to which the Drosophila genus belongs, it was not at all obvious that orthologs of the baculovirus IAPs would be found in the fruit fly. Remarkably, however, the Drosophila melanogaster genome was found to encode four IAPs with diverse functions, although mechanisms of cell death regulation have been the most extensively studied (Figure 1). In this section, we discuss the insights gleaned into the IAPs from the well-characterized Drosophila model, and subsequently will expand these observations into a broader discussion of IAP function in other organisms.
DIAP1 was originally identified as an allele of thread through a genetic screen designed to identify modulators of cell death (Hay et al., 1995). DIAP1 contains two tandemly repeated BIRs and a RING domain at the extreme carboxyl terminus. The use of genetic and biochemical approaches to dissect DIAP1 function has generated strong evidence for this IAP in regulating cell death. The RING domain of DIAP1 promotes ubiquitylation of DIAP1 itself and DRONC, the ortholog of mammalian caspase-9, resulting in inactivation, although whether caspase degradation occurs in vivo is unclear (Wilson et al., 2002). During larval development, intrinsic death signaling initiated by DRONC appears to be constitutively activated at a low level, but cell death is minimized by physiological expression of DIAP1 (Muro et al., 2002; Rodriguez et al., 2002). RNAi depletion of DIAP1 in the Drosophila S2 cell line or a diap1 null mutation in flies resulted in widespread caspase-dependent cell death in the absence of exogenous signals (Goyal et al., 2000; Meier et al., 2000; Wang et al., 1999). In contrast, Drosophila zygotes with a Dronc null mutation exhibit widespread defects in programmed cell death, resulting in markedly abnormal development (Chew et al., 2004; Xu et al., 2005). A tractable in vivo model of programmed cell death is the developing eye, which undergoes temporally and spatially restricted apoptosis during differentiation, regulated by the initiator caspase, DRONC, and the IAP antagonists Reaper (Rpr) and Head-involution defective (Hid), to produce the precise patterning of the ommatidia (Hay et al., 1995). Rpr and Hid were the first IAP antagonists identified, forming a novel protein family characterized by an N-terminal conserved region termed the IAP-binding motif (IBM), that now includes Grim and Sickle (Kornbluth and White, 2005). The Drosophila IBM containing proteins promote cell death through multiple mechanisms, including competing with caspases for binding to DIAP1 and suppressing DIAP1 translation (Holley et al., 2002; Yoo et al., 2002). Overexpression of DRONC, Rpr or Hid in the eye altered retinal structures causing aberrant eye phenotypes that could be rescued by co-expression of DIAP1 (Hay et al., 1995; Meier et al., 2000). Notably, although overexpression of DIAP2 in the eye suppressed programmed cell death, DIAP2 mutant flies did not display the early lethality phenotype observed in DIAP1 null animals (Huh et al., 2007; Leulier et al., 2006). These results suggest that conclusions about the physiological function of the IAPs may be more clearly drawn using loss-of-function approaches.
DIAP1 binds Dronc through the BIR2 domain, which was required to inhibit Dronc-induced apoptosis in the developing eye (Chai et al., 2003; Wilson et al., 2002). In contrast, a DIAP1 RING domain mutant protein still bound to DRONC, Rpr and Hid, but failed to ubiquitinate DRONC or to ameliorate cell death caused by DRONC overexpression (Wilson et al., 2002). Levels of DIAP1 itself are modulated by regulatory proteins, such as Rpr, which directs DIAP1 auto-ubiquitination and degradation via recruitment of the ubiquitin conjugating enzyme, UbcD1 (Ryoo et al., 2002). In addition, DIAP1 degradation is promoted by the pro-apoptotic E2 ubiquitin conjugase-like protein, Morgue; mutations in morgue suppress cell death in the Drosophila eye (Hays et al., 2002; Wing et al., 2002). Thus, the ratio of DIAP1 to pro-apoptotic proteins in individual components of the eye such as Morgue and DRONC may serve as a biological rheostat for determining which cells undergo cell death. In this case, the rheostat could be sensitized to the levels of different pro- vs. anti-apoptotic proteins, where the levels are controlled through a ubiquitination cascade mediated by DIAP1 and other ubiquitin modifying machinery. Consistent with the rheostat model, heterozygosity of diap1 exacerbates the eye ablation phenotype caused by ectopic DRONC expression, while heterozygosity of dronc ameliorates cell death caused by overexpression of Rpr or Hid (Meier et al., 2000). Although the Drosophila genome encodes other IAP proteins, it is evident that DIAP1 interacts uniquely with modulators of apoptosis to perform a non-redundant cytoprotective function during development. Notably, a recent report from Montell and colleagues has revealed a novel function for DIAP1 in border cell migration in the fly ovary (Geisbrecht and Montell, 2004). A subset of follicle cells migrate to the border between the oocyte and surrounding nurse cells in a Rac-dependent manner, and are thus termed border cells. These cells require DIAP1 for proper migration, but not for protection from cell death. Overall, the wealth of data from these Drosophila studies unequivocally implicate DIAP1 in protecting cells from programmed cell death, but also suggest additional roles for DIAP1 in developmental contexts independent of cell survival.
DIAP2 and Deterin were both discovered by DNA sequence homology searches; DIAP2 contains three BIRs and a carboxy-terminal RING and is important for innate immune function. In contrast to the clear role of DIAP1 in regulating cell death during development, DIAP2-null flies do not exhibit an embryonic phenotype, although they may exhibit increased sensitivity to some apoptotic stimuli (Huh et al., 2007; Leulier et al., 2006; Ribeiro et al., 2007). However, diap2 was identified in several genetic screens as a candidate regulator of the invertebrate immune response, specifically the immune deficiency (imd) pathway (Gesellchen et al., 2005; Kleino et al., 2005; Leulier et al., 2006). Imd, an invertebrate homolog of the mammalian signaling adaptor TNF receptor interacting protein (RIP), controls a pathway required for induction of antimicrobial peptides in response to infection; imd mutant flies are peculiarly susceptible to infection by Gram-negative bacteria (Lemaitre et al., 1995) (Figure 3). Triggered by bacterial peptidoglycan, the Imd pathway activates the Dredd caspase, feeding into evolutionarily conserved signaling modules such as the MAP3K, dTAK, and the Drosophila IKK complex (Cherry and Silverman, 2006). Imd-dependent signaling required the DIAP2 RING domain, as well as dTAB2, a homolog of mammalian TAB1 that binds the BIR1 domain of XIAP (Huh et al., 2007; Lu et al., 2007). Epistasis studies placed dIAP2 parallel to dTAK1, which is upstream of both JNK and NF-κB pathways (Gesellchen et al., 2005; Kleino et al., 2005). Although overexpression of DIAP2 resulted in association with Reaper and Hid and inhibition of apoptosis, DIAP2 null flies exhibited no apparent cell death related phenotypes in addition to the immune deficiency (Huh et al., 2007). Instead, DIAP2 itself is regulated by increased expression of Rpr or Hid, indicating that DIAP2 might be a node for integrating external signals through the Imd pathway, with the internal state of cellular stress (Huh et al., 2007). In the immune signaling paradigm, DIAP2 primarily serves as a signal transducer rather than a modulator of cell death.
Deterin and dBruce in Drosophila, and in other organisms as discussed in further mechanistic detail below, appear to be regulators of cytokinesis that are essential in specific cellular contexts. Deterin is a small, Survivin-like IAP that plays a key role in mitotic spindle formation and cell cycle progression (Jones et al., 2000). In contrast, dBruce is a giant (>500 kDa) protein, displaying a single BIR and a UBC-like E2 ubiquitin ligase domain, that is critical for sperm differentiation (Arama et al., 2003; Vernooy et al., 2002). Overall, the Drosophila model has provided important insights into the pro-survival function of IAPs such as DIAP1, but has also pointed to an expanded model of IAP function, where apoptosis is but one of many molecular programs controlled by these versatile signaling regulators.
Cell cycle
Mitotic regulation by IAPs was first discovered in transformed cells, where Altieri and colleagues reported that expression of Survivin increased from G1 to S phase, and was highly expressed in G2/M (Li et al., 1998). Their data demonstrated that Survivin associated with microtubules, and disruption of the association led to caspase-3 processing and mitotic catastrophe, providing a link between the integrity of the mitotic apparatus and cell death. Mammalian Survivin consists of a single BIR domain, and similar IAP-related sequences have also been characterized in lower invertebrates, specifically in the nematode C. elegans, whose genome encodes two BIR-containing proteins, and in both budding and fission yeast, each of which encode a single IAP-like protein (Fraser et al., 1999; Uren et al., 1999; Li et al., 2000). Comparisons with mammalian IAPs, coupled with the fact that yeast do not contain classic caspases with clear apoptotic substrates, suggest that BIR-containing proteins may have originally evolved to control non-apoptotic functions, such as cytokinesis and mitotic spindle formation.
Bir1p, the Saccharomyces cerevisiae protein homologous to Survivin, was found in a two-hybrid screen for proteins interacting with components of the yeast kinetochore, a structure that directs spindle fiber attachment to chromosomes to facilitate separation of sister chromatids during mitosis (Yoon and Carbon, 1999). Interestingly, a bir1 null mutant strain of S. cerevisiae in the haploid state did not have noticeable growth or spindle morphology defects, however, over time Bir1p-deficient yeast cultures showed instability of a yeast minichromosome, indicating chromosome missegregation. The specific role of the BIR motifs in Bir1p-mediated chromosome segregation remains somewhat unclear, since overexpression of a truncated Bir1p lacking the BIR motifs in the bir1Δ mutant rescued stability of a reporter minichromosome. Combining the bir1Δ allele with mutations in the genes encoding kinetochore proteins, Cbf1p or Ctf19p, led to synthetic lethality, emphasizing the contribution of Bir1p to kinetochore function.
Further studies have revealed a wealth of mechanistic information about how Bir1p and its homologs direct chromosome segregation during cell division. Bir1p assembles in a chromosomal passenger protein complex (CPC) that includes INCENP, Aurora-B kinase and Borealin (Ruchaud et al., 2007; Yoon and Carbon, 1999). The Aurora-B kinase performs the enzymatic activity of the CPC, phosphorylating Survivin and other targets such as histone H3; however, all four proteins of the CPC are essential for mitosis (Jeyaprakash et al., 2007). During prophase, the CPC accumulates on condensing chromosomes, and is then targeted to the inner centromeres in a Survivin-dependent manner. The association of Survivin with centromeres requires K63 ubiquitylation, and is dynamically regulated by opposing activities of UFD1 and the de-ubiquitinating enzyme, hFAM, on Survivin (Vong et al., 2005). Upon transition to metaphase, the complex migrates to the spindle midzone, mediating centromeric cohesion and attachment to spindle microtubules. The CPC appears to act as a sensor of mechanical tension between centromeres and microtubules of the spindle, leading to proper chromosome segregation (Fraser et al., 1999; Sandall et al., 2006; Speliotes et al., 2000). High fidelity in chromosome segregation is a critical element of basic cellular function, and thus it could be predicted that Survivin would be involved in many aspects of development.
Regulation of cell division by Survivin homologs has turned out to be highly relevant in multicellular organisms, which have facilitated analysis of Survivin-like proteins in complex developmental processes. The nematode, Caenorhabditis elegans, is a genetically tractable model system with which to dissect the precisely regulated processes of cell division, lineage determination and cell death during embryonic development. In fact, genetic screens in C. elegans identified key determinants of apoptosis that have formed the basis of the current paradigm of cell death regulation (Lettre and Hengartner, 2006). Genomic analysis of C. elegans identified only two BIR proteins, BIR-1 and BIR-2, leading to the hypothesis that these proteins might regulate cell death or division in the nematode (1998). Inducible overexpression of either BIR-1 or BIR-2 did not suppress caspase-mediated cell death in the anterior pharynx of L3-L4 larvae (Fraser et al., 1999). Inhibition of BIR-1 expression in C. elegans by RNAi did not affect apoptosis in adult germ line, but instead resulted in early lethality of the embryos and a failure to complete cytokinesis (Fraser et al., 1999). Expression of mammalian Survivin in the bir1 RNAi treated embryos partially suppressed the cytokinesis defect, increasing cellularity. These genetic experiments revealed an important role for BIR-domain proteins in directing the cell division machinery in more complex organisms, and demonstrated that mammalian Survivin participated in mitotic regulation. Since then, the physiological role of Survivin in mammalian development has been amply documented. Mice lacking Survivin exhibit embryonic lethality around embryonic day 4.5, characterized by grossly abnormal nuclear morphology and defects in cytokinesis (Uren et al., 2000). The multinucleate phenotype of Survivin-deficient animals suggests that regulation of chromosome separation and cytokinesis is the predominant mechanism by which Survivin promotes early embryonic development.
The early lethality phenotype of animals lacking Survivin initially prevented analysis of the role of Survivin in other developmental processes. To determine if Survivin might contribute to differentiation and organ development, several groups employed a conditional knockout strategy, crossing animals with an allele of Survivin flanked by loxP sites with different cre recombinase transgenes. Survivin depletion in different contexts yielded phenotypes that implicated aberrant cell cycle regulation, or decreased cell survival to be discussed later in this review. Cell cycle defects were primarily associated with loss of Survivin in hematopoietic lineages. Survivin is expressed in hematopoietic stem cells and lymphocytes, as well as other adult cells types, including vascular endothelial cells (Leung et al., 2007). Cre-mediated deletion of Survivin in adult animals resulted in lethality within 2 weeks, marked by anemia, reduction in the hematopoietic stem cell and progenitor compartments and substantial loss of cellularity in the bone marrow and spleen (Leung et al., 2007). Immunophenotyping analysis of erythroid differentiation established that mature erythrocytes, which had exited the cell cycle, were unaffected in Survivin-depleted mice compared to control animals, but the highly proliferative erythroid precursor populations were notably decreased. Survivin-deficient erythroblasts exhibited an altered cell cycle profile and polyploidy, with more cells in G1 phase and fewer cells in S phase than control erythroblasts. This study provided evidence that in vivo, Survivin is involved in hematopoietic proliferation and differentiation (Leung et al., 2007). Further evidence of the importance of cell cycle regulation by Survivin in hematopoietic cells came from studies in which Survivin was depleted specifically in the T-cell lineage. Deletion of a floxed allele of Survivin in thymocytes, driven by an lck-cre transgene, blocked early differentiation of T cell precursors at the CD4−CD8− stage (Okada et al., 2004). Cell cycle arrest and defects in spindle formation were observed in Survivin-deficient thymocytes, concomitant with increased cell death, which could not be rescued by the introduction of a bcl-2 transgene. Survivin depletion at later stages of thymocyte development, mediated by a CD4-cre transgene, allowed thymocytes to differentiate past the CD4−CD8− stage, but caused a substantial decrease in peripheral T cell populations (Xing et al., 2004). In these Survivin-depleted mice, no increase in T lymphocyte apoptosis was observed, but the Survivin-deficient peripheral T cells were unable to proliferate in response to mitogenic signals, arresting in G2/M phase. Therefore, among the IAP proteins, Survivin uniquely plays a role in cell cycle regulation that is conserved from yeast to mammals and may represent the earliest function of the BIR domain.
More recently, studies have revealed that BRUCE (also termed Apollon or BIRC6), a large multidomain IAP present in most higher eukaryotes but not in yeast, also has an essential function in the cell cycle, directing final resolution of the midbody channel that connects two dividing cells (Pohl and Jentsch, 2008). BRUCE is a noncanonical IAP, as the protein exhibits both E2 ubiquitin conjugating and E3 ubiquitin ligase activity towards targets like SMAC (Bartke et al., 2004). However, targets of BRUCE appear primarily monoubiquitinated, suggesting that the physiological function of BRUCE may not be to direct target proteins for proteasomal degradation, but rather to modify their function. Interestingly, BRUCE is required for recruitment of membrane vesicles to the midbody ring, a circular phase-dense structure between the two daughter cells, that acts as a platform for BRUCE-dependent ubiquitination (Pohl and Jentsch, 2008). BRUCE is essential for mammalian development, as BRUCE-deficient embryos exhibit growth retardation at embryonic day 14, apparently due to defects in maturation of the placenta (Hitz et al., 2005; Lotz et al., 2004; Ren et al., 2005). The three different laboratories constructed distinct BRUCE targeting alleles, and although all reported a similar embryonic lethality phenotype, the groups differed in their interpretation of whether the embryonic lethality in BRUCE-deficient mice was due to altered cell survival or defective cytokinesis or both. Nevertheless, the available evidence strongly suggests that like Survivin, BRUCE can play a role in regulation of the cell cycle.
Signaling and cell survival
Proteins of the mammalian IAP family exhibit one of two predominant patterns of expression that may reflect differing roles in development. Survivin is expressed during embryogenesis and in many tumors, but to a lesser degree in normal adult animals, whereas XIAP, c-IAP1, c-IAP2, and NAIP are broadly expressed in adult tissues (Verhagen et al., 2001). Of the IAP proteins, data from diverse model organisms suggests that DIAP1, c-IAP1 and Survivin are required early in development, participating in both apoptosis-independent and – dependent processes. In vertebrates, IAPs also promote cell survival in organ and tissue development. Experimental vertebrate models with fewer IAP genes and excellent genetic tractability, such as Danio rerio, have provided insights into IAP function that may have been obscured by gene duplication and redundancy in mammals. A zebrafish strain containing a null mutation in the ciap1 (birc2) gene was identified in a forward genetic screen for mutants with abnormal vascular phenotypes (Santoro et al., 2007). The mutation was originally christened tomato (tom) due to the observed vascular hemorrhage, blood pooling and vascular regression that occurred between 54 and 60 hours post fertilization (hpf). Ciap1 was expressed in the vasculature as early as 48 hpf, supporting a role for c-IAP1 in preventing apoptosis and regression of the vascular endothelium. CIAP-1 mutants in either the BIR1 domain, which interacts with the TNFR adapter TRAF2, or the RING domain failed to rescue the tom mutant phenotype. Activation of NF-κB, which can occur through TNFR, was also required for vascular integrity in the zebrafish, as pharmacological inhibition of NF-κB or the upstream IKK complex resulted in hemorrhage and vascular instability, mimicking some aspects of the tom phenotype. Overexpression of the IKK kinase, NEMO, rescued apoptosis of the vascular endothelium in the tom mutants, emphasizing a signaling function of c-IAP1 in promoting vascular homeostasis through the NF-κB pathway. TNFR signaling regulates many aspects of cell physiology, including differentiation, apoptosis or survival, and in mammalian cells involves both c-IAP1 and c-IAP2 (Samuel et al., 2006; Wang et al., 2008) (Figure 3). No evidence has been reported for a defect in vascular integrity or any other developmental process in mice deficient in c-IAP1 or c-IAP2, although the high degree of amino acid conservation between the two proteins suggests the possibility of at least partial redundancy. The close proximity of the ciap1/birc2 and ciap2/birc3 genes in mammalian genomes, likely arising from a gene duplication (Rajcan-Separovic et al., 1996), has thus far prevented the generation of a double mutant to determine if these IAPs have an important and overlapping function in mammalian embryogenesis. In further support of a role for IAPs as key regulators of vasculogenesis, morpholinos targeting Survivin-1 injected into 1−4 cell zebrafish embryos increased apoptosis in the brain and neural tube, as well as in axial vasculature, resulting in perturbations in angiogenesis (Ma et al., 2007). Moreover, _tie2_-cre mediated deletion of a conditional Survivin allele in the endothelial lineage caused lethality in mice at embryonic day 9.5, characterized by peripheral hemorrhaging, abnormal heart development, and endothelial cell defects (Zwerts et al., 2007). Since c-IAP1 and Survivin differ substantially in their domain structure and contain distinct BIR subtypes, these IAPs may act through multiple mechanisms to modulate survival of the vascular endothelium and vessel homeostasis. Overall, regulation of endothelial cell survival, particularly during vascular development, appears to be a critical function of IAPs in embryogenesis.
Despite the strong developmental phenotypes associated with defects in vertebrate IAPs, relatively little evidence connects these phenotypes to direct or indirect inhibition of caspase-dependent cell death, as has been described in Drosophila. One study reported that specific deletion of Survivin in murine neural progenitor cells at embryonic day 10.5 resulted in increased multi-focal apoptosis of neuronal precursors and dramatically altered brain size and architecture (Jiang et al., 2005). Neuronal apoptosis in the brains of these Survivin-depleted mice was associated with an increase in caspase-3 and -9 activity, which did not appear to be accompanied by cell cycle arrest. However, the activation of caspases might be an indirect consequence of Survivin loss, as a more direct role for Survivin in inhibition of apoptosis has not been clearly demonstrated.
Of all the IAP proteins, the strongest case can be made for XIAP to be a bona fide caspase inhibitor. Since XIAP has also been shown to modulate signaling through the TGF-β receptor and the NF-κB pathway, both of which are implicated in embryonic development, it is therefore somewhat surprising that no striking developmental phenotype was found in XIAP-deficient mice until recently, although XIAP protein is detected in many adult tissues, including spleen, liver, kidney and mammary gland (Harlin et al., 2001; Olayioye et al., 2005). During gestation, XIAP expression in the mammary glands increased, peaking at embryonic day 18, and at late stages of pregnancy, lobuloalveolar development was delayed in xiap−/− animals (Olayioye et al., 2005). No evidence of apoptosis in the mammary gland of XIAP-deficient pregnant mice was found, and lactation occurred normally. However, XIAP could be demonstrated to play a physiological role in adult mammals, directing copper homeostasis by regulating COMMD proteins and contributing to anti-microbial immunity (discussed below) by mechanisms that are not yet well defined, but appear to involve signal transduction rather than protection from apoptosis (Bauler et al., 2008; Mufti et al., 2006; Rigaud et al., 2006). Much of our mechanistic understanding of IAP-dependent regulation of apoptosis has been inferred from studies in cell culture, and interrogation of IAP function using animal models supports a cytoprotective role for IAP proteins in embryogenesis. However, it is more difficult to interpret the precise mechanisms by which individual IAPs promote cell survival in development. Although increased cell death was observed in many cases where IAP expression or function was perturbed, cell death may have been an indirect result of dysfunctional signaling or mitotic regulation, as opposed to lack of direct inhibition of the cell death machinery. Overall, data generated using multiple approaches and animal models point to the IAP family as powerfully diverse modulators of cellular function, employing cell division, signaling and cytoprotective regulatory mechanisms to direct development.
Innate Immunity
An emerging body of work in Drosophila has suggested that IAP proteins may serve an underappreciated role in modulating innate immunity to infection. As described above, DIAP2 potentiates the Imd signaling cascade responsible for upregulating anti-microbial peptides upon infection (Gesellchen et al., 2005; Huh et al., 2007; Kleino et al., 2005; Leulier et al., 2006). Innate immune signaling pathways are well conserved from humans to Drosophila, leading to the hypothesis that mammalian IAP proteins also regulate innate immunity (Hoffmann and Reichhart, 2002).
Mammalian orthologs of proteins of the Imd pathway, such as FADD and RIP, are found as part of the TNFR signaling module and have been implicated in immune defense against intracellular pathogens (Balachandran et al., 2004; Chin et al., 2002; Kobayashi et al., 2002) (Figure 3). However, the role of IAPs in mammalian immunity is still incompletely understood. The neuronal apoptosis inhibitor protein NAIP5, which contains a leucine rich repeat (LRR) domain not characteristic of the IAP family, nucleates the assembly and activation of a caspase-1 activating, IL-1β processing protein complex termed the inflammasome during infection of murine macrophages by the intracellular bacterial pathogen Legionella pneumophila, (Coers et al., 2007; Lamkanfi et al., 2007; Molofsky et al., 2006; Ren et al., 2006; Wright et al., 2003; Zamboni et al., 2006) (Figure 1). While it is yet unclear whether NAIP5 is itself an innate immune sensor, coordinated signaling by NAIP5 and another LRR-containing protein, IPAF, is required for detection of bacterial flagellin within the host cytosol (Coers et al., 2007). Mice with mutations in the Naip5/Birc1e locus fail to restrict L. pneumophila infection in mice (Diez et al., 2003; Wright et al., 2003). Caspase-1 activation and IL-1β production are characteristic of an inflammatory cell death termed pyroptosis, that shares some aspects of apoptosis, but is more commonly observed in the context of microbial infection (Fink and Cookson, 2005). In the context of L. pneumophila infection, NAIP5 appears to promote cell death by activating caspase-1 rather than acting to promote survival as we have described for other IAP family members. However, depending on the strength of the inflammatory stimulus, infected macrophages can either upregulate autophagy, emerging as an important innate immune defense, or are driven down an inflammatory cell death pathway (Swanson and Molofsky, 2005). These data strongly support a role for NAIP5 in innate immune signaling, and have led to the hypothesis that NAIP5 may act as a rheostat that determines cellular responses based on the signal strength and context. Based on its domain structure, NAIP5 may also be considered part of the Nod-like receptors (NLR), a family of cytosolic proteins that play an important role in sensing both self and non-self (i.e., microbial) danger signals (Kanneganti et al., 2007). Thus, the hypothesis that regulation of innate immune signaling is a common attribute of many of the mammalian IAP proteins remains to be fully investigated.
Consistent with the idea that IAPs play an important role in immunomodulation, mutations in XIAP were found in a cohort of patients with X-linked proliferative syndrome (XLP), a primary immunodeficiency characterized by lymphoproliferation in response to infection by Epstein-Barr virus (Rigaud et al., 2006). XIAP-deficient XLP patients have fewer natural killer T cells, and T lymphocytes derived from these patients were more susceptible to apoptosis upon T cell receptor signaling. However, mice lacking XIAP have similar numbers of NKT cells compared to wildtype mice (Bauler et al., 2008; Rigaud et al., 2006). Thus, XIAP may have a role in promoting NKT cell development or survival in humans that is not reflected in the murine model. However, we have recently found that XIAP is required in mice for innate immunity to infection by the intracellular bacterial pathogen, Listeria monocytogenes (Bauler et al., 2008). XIAP potentiated JNK activation in response to the presence of bacteria in the cytosol, leading to amplification of pro-inflammatory cytokine production. Lastly, XIAP was necessary for integration of external Toll-like receptor and cytosolic NLR signaling to produce synergistic cytokine output. Notably, there are clear parallels in the molecular players involved in the mammalian NLR signaling pathway and the Drosophila Imd pathway, both of which sense bacterial peptidoglycan, emphasizing a critical and conserved contribution of IAPs to immune signaling (Cherry and Silverman, 2006; Girardin et al., 2002).
Although there is yet relatively little published evidence for innate immune regulation by the cIAPs, c-IAP2-deficient mice were more resistant to lipopolysaccharide (LPS)-induced sepsis, a syndrome mediated by pro-inflammatory cytokines (Conte et al., 2006). Wildtype macrophages upregulate expression of c-IAP2 upon treatment with LPS, and were protected from apoptosis in this model of septic shock while c-IAP2-deficient macrophage populations were reported to exhibit increased cell death. Thus, c-IAP2 appears to contribute to macrophage survival in a pro-inflammatory model of innate immune signaling, but the mechanism by which c-IAP2 prevents macrophage cell death is still unclear, and may be an indirect effect of cytokine regulation. A recent study demonstrated that c-IAP1 and c-IAP2 influence cancer cell survival through K48 and K63 ubiquitin modification of RIP1, reducing formation of a pro-apoptotic RIP1/caspase-8 complex (Bertrand et al., 2008). Kinases of the RIP family modulate innate immune signaling in Drosophila (IMD), and in mammalian cells (RIP2/RICK), inducing antimicrobial peptides or proinflammatory cytokines upon microbial infection (Figure 3) (Girardin et al., 2002). It is intriguing to speculate that the cIAPs may also ubiquitinate RIP2 in mammalian cells, enhancing NF-κB-dependent proinflammatory cytokine responses. Taken together, these data provide some evidence to suggest that immunomodulation is an important function of the IAP family. Further studies will be required to define both anti-apoptotic and apoptosis-independent signaling mechanisms by which individual IAPs modulate immunity.
IAP Antagonists
As discussed above, the protoype IAP antagonist proteins were identified in Drosphila through an analysis of the H99 locus, which encoded Reaper, Hid and Grim (Chen et al., 1996; Grether et al., 1995; White et al., 1994). Additionally, two elegant biochemical studies led to the independent identification of a mammalian IAP binding protein designated Smac (second mitochondrial activator of caspases; DIABLO in the mouse) (Du et al., 2000; Verhagen et al., 2000). Smac/DIABLO is a nuclear-encoded, mitochondrially localized protein, that is released into the cytosol following an apoptotic trigger. Structural studies have shown that the amino-terminal four residues of the mature Smac/DIABLO protein, the so-called IAP binding motif (IBM) are necessary and sufficient for binding to XIAP, an event which can lead to the competitive displacement of XIAP from bound caspases and so augment intracellular caspase activation (Liu et al., 2000; Wu et al., 2000). Thus, Smac/DIABLO is a pro-apoptotic molecule that can function to neutralize the cytoprotective effects of XIAP (Vaux and Silke, 2003). Interestingly, Smac/DIABLO also exhibits a high affinity for other IAP family members that do not inhibit caspases (Yang and Du, 2004). The consequences of these interactions are less clear, but a dynamic association appears to exist between Smac/DIABLO and the IAPs that can trigger the autoubiquitination of the IAP in question, or conversely the ubiquitination of Smac/DIABLO itself; however, the kinetic details of these events are currently not well understood. Recent work has focused on the exploitation of this interaction by developing small-molecule synthetic compounds that mimic the IBM, and these may have great promise and therapeutic potential for the treatment of neoplastic and proliferative disease (Petersen et al., 2007; Varfolomeev et al., 2007; Vince et al., 2007).
Concluding Remarks
The exponential increase in our understanding of the biology of the IAPs has illuminated their physiological roles in a wide variety of cellular processes including development, intracellular signaling, mitosis and immunological responses. Recent studies are removing the misconception that primary function of the IAPs is to suppress apoptosis; indeed the ability of XIAP to directly inhibit caspases appears to be a unique function restricted to that IAP, and others appear to participate largely in cellular processes that involve ubiquitination. Since the identification of IAP antagonists including Reaper in Drosophila, followed by the discovery of Smac/DIABLO in mammals, numerous IAP-interacting proteins have been identified (Verhagen et al., 2007), many of which interact by virtue of their IBM, strongly suggesting that the interactions between IBM-containing proteins and IAPs evolved prior to the acquisition of the ability of XIAP to inhibit caspases (Wing et al., 2001; Wright and Clem, 2001; Wu et al., 2001). Thus the IAP-neutralizing properties of IBM-containing proteins are likely to have a wide range of functions encompassing, but certainly not limited to, regulating cell death. The fact that IBM-bearing proteins are so prolific in number suggests a fine specificity for signaling under physiological conditions that has yet to be realized. Overall, the IAPs are emerging as a family of signal modulators that may act as hubs to integrate and translate molecular information into the appropriate biological currency of death, inflammation or differentiation.
ACKNOWLEDGEMENTS
We apologize to our many colleagues whose work could not be cited and discussed due to space limitations. We thank Rebecca Csomos, Stefanie Galban, Casey Wright, and members of the Duckett and O'Riordan labs for their insightful suggestions and critical reading of this review. This work was supported in part by Crohn's and Colitis Foundation of America and Ellison Medical Foundation awards to M.X.D.O., and NIH R01 GM067827 and a Sandler Program for Asthma Research Senior Investigator Award to C.S.D. L.D.B is a predoctoral fellow of the American Heart Association. C.S.D. and M.X.D.O. are consultants for Aegera Therapeutics Inc.; F.L.S. is an employee of Apoptos, Inc.
REFERENCES
- Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell. 2003;4:687–697. doi: 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- Balachandran S, Thomas E, Barber GN. A FADD-dependent innate immune mechanism in mammalian cells. Nature. 2004;432:401–405. doi: 10.1038/nature03124. [DOI] [PubMed] [Google Scholar]
- Bartke T, Pohl C, Pyrowolakis G, Jentsch S. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol Cell. 2004;14:801–811. doi: 10.1016/j.molcel.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Bauler LD, Duckett CS, O'Riordan MX. XIAP regulates cytosol-specific innate immunity to Listeria infection. PLoS Pathog. 2008;4:e1000142. doi: 10.1371/journal.ppat.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ, Barker PA. cIAP1 and cIAP2 Facilitate Cancer Cell Survival by Functioning as E3 Ligases that Promote RIP1 Ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Birnbaum MJ, Clem RJ, Miller LK. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J Virol. 1994;68:2521–2528. doi: 10.1128/jvi.68.4.2521-2528.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J, Yan N, Huh JR, Wu JW, Li W, Hay BA, Shi Y. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat Struct Biol. 2003;10:892–898. doi: 10.1038/nsb989. [DOI] [PubMed] [Google Scholar]
- Chai J, Shiozaki E, Srinivasula SM, Wu Q, Dataa P, Alnemri ES, Shi Y. Structural basis of caspase-7 inhibition by XIAP. Cell. 2001;104:769–780. doi: 10.1016/s0092-8674(01)00272-0. [DOI] [PubMed] [Google Scholar]
- Chen P, Nordstrom W, Gish B, Abrams JM. grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- Cherry S, Silverman N. Host-pathogen interactions in drosophila: new tricks from an old friend. Nat Immunol. 2006;7:911–917. doi: 10.1038/ni1388. [DOI] [PubMed] [Google Scholar]
- Chew SK, Akdemir F, Chen P, Lu WJ, Mills K, Daish T, Kumar S, Rodriguez A, Abrams JM. The apical caspase dronc governs programmed and unprogrammed cell death in Drosophila. Dev Cell. 2004;7:897–907. doi: 10.1016/j.devcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Chin AI, Dempsey PW, Bruhn K, Miller JF, Xu Y, Cheng G. Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature. 2002;416:190–194. doi: 10.1038/416190a. [DOI] [PubMed] [Google Scholar]
- Coers J, Vance RE, Fontana MF, Dietrich WF. Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signalling pathways. Cell Microbiol. 2007;9:2344–2357. doi: 10.1111/j.1462-5822.2007.00963.x. [DOI] [PubMed] [Google Scholar]
- Conte D, Holcik M, Lefebvre CA, Lacasse E, Picketts DJ, Wright KE, Korneluk RG. Inhibitor of apoptosis protein cIAP2 is essential for lipopolysaccharide-induced macrophage survival. Mol Cell Biol. 2006;26:699–708. doi: 10.1128/MCB.26.2.699-708.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook NE, Clem RJ, Miller LK. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux QL, Roy N, Stennicke HR, Van Arsdale T, Zhou Q, Srinivasula SM, Alnemri ES, Salvesen GS, Reed JC. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 1998;17:2215–2223. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- Diez E, Lee SH, Gauthier S, Yaraghi Z, Tremblay M, Vidal S, Gros P. Birc1e is the gene within the Lgn1 locus associated with resistance to Legionella pneumophila. Nat Genet. 2003;33:55–60. doi: 10.1038/ng1065. [DOI] [PubMed] [Google Scholar]
- Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- Duckett CS, Li F, Wang Y, Tomaselli KJ, Thompson CB, Armstrong RC. Human IAP-like protein regulates programmed cell death downstream of Bcl-xL and cytochrome c. Mol. Cell. Biol. 1998;18:608–615. doi: 10.1128/mcb.18.1.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett CS, Nava VE, Gedrich RW, Clem RJ, Van Dongen JL, Gilfillan MC, Shiels H, Hardwick JM, Thompson CB. A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J. 1996;15:2685–2694. [PMC free article] [PubMed] [Google Scholar]
- Eckelman BP, Salvesen GS. The human anti-apoptotic proteins cIAP1 and cIAP2 bind but do not inhibit caspases. J Biol Chem. 2006;281:3254–3260. doi: 10.1074/jbc.M510863200. [DOI] [PubMed] [Google Scholar]
- Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, James C, Evan GI, Hengartner MO. Caenorhabditis elegans inhibitor of apoptosis protein (IAP) homologue BIR-1 plays a conserved role in cytokinesis. Curr Biol. 1999;9:292–301. doi: 10.1016/s0960-9822(99)80137-7. [DOI] [PubMed] [Google Scholar]
- Gaither A, Porter D, Yao Y, Borawski J, Yang G, Donovan J, Sage D, Slisz J, Tran M, Straub C, Ramsey T, Iourgenko V, Huang A, Chen Y, Schlegel R, Labow M, Fawell S, Sellers WR, Zawel L. A Smac mimetic rescue screen reveals roles for inhibitor of apoptosis proteins in tumor necrosis factor-α signaling. Cancer Res. 2007;67:11493–11498. doi: 10.1158/0008-5472.CAN-07-5173. [DOI] [PubMed] [Google Scholar]
- Geisbrecht ER, Montell DJ. A role for Drosophila IAP1-mediated caspase inhibition in Rac-dependent cell migration. Cell. 2004;118:111–125. doi: 10.1016/j.cell.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Gesellchen V, Kuttenkeuler D, Steckel M, Pelte N, Boutros M. An RNA interference screen identifies Inhibitor of Apoptosis Protein 2 as a regulator of innate immune signalling in Drosophila. EMBO Rep. 2005;6:979–984. doi: 10.1038/sj.embor.7400530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin SE, Sansonetti PJ, Philpott DJ. Intracellular vs extracellular recognition of pathogens--common concepts in mammals and flies. Trends Microbiol. 2002;10:193–199. doi: 10.1016/s0966-842x(02)02334-x. [DOI] [PubMed] [Google Scholar]
- Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 2000;19:589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Harlin H, Reffey SB, Duckett CS, Lindsten T, Thompson CB. Characterization of XIAP-deficient mice. Mol Cell Biol. 2001;21:3604–3608. doi: 10.1128/MCB.21.10.3604-3608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Hays R, Wickline L, Cagan R. Morgue mediates apoptosis in the Drosophila melanogaster retina by promoting degradation of DIAP1. Nat Cell Biol. 2002;4:425–431. doi: 10.1038/ncb794. [DOI] [PubMed] [Google Scholar]
- Hinds MG, Norton RS, Vaux DL, Day CL. Solution structure of a baculoviral inhibitor of apoptosis (IAP) repeat. Nat Struct Biol. 1999;6:648–651. doi: 10.1038/10701. [DOI] [PubMed] [Google Scholar]
- Hitz C, Vogt-Weisenhorn D, Ruiz P, Wurst W, Floss T. Progressive loss of the spongiotrophoblast layer of Birc6/Bruce mutants results in embryonic lethality. Genesis. 2005;42:91–103. doi: 10.1002/gene.20128. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA, Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nat Immunol. 2002;3:121–126. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- Holley CL, Olson MR, Colon-Ramos DA, Kornbluth S. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat Cell Biol. 2002;4:439–444. doi: 10.1038/ncb798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Park YC, Rich RL, Segal D, Myszka DG, Wu H. Structural Basis of Caspase Inhibition by XIAP. Differential Roles of the Linker versus the BIR Domain. Cell. 2001;104:781–790. [PubMed] [Google Scholar]
- Hughes AL. Evolution of inhibitors of apoptosis in baculoviruses and their insect hosts. Infect Genet Evol. 2002;2:3–10. doi: 10.1016/s1567-1348(02)00040-0. [DOI] [PubMed] [Google Scholar]
- Huh JR, Foe I, Muro I, Chen CH, Seol JH, Yoo SJ, Guo M, Park JM, Hay BA. The Drosophila inhibitor of apoptosis (IAP) DIAP2 is dispensable for cell survival, required for the innate immune response to gram-negative bacterial infection, and can be negatively regulated by the reaper/hid/grim family of IAP-binding apoptosis inducers. J Biol Chem. 2007;282:2056–2068. doi: 10.1074/jbc.M608051200. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash AA, Klein UR, Lindner D, Ebert J, Nigg EA, Conti E. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell. 2007;131:271–285. doi: 10.1016/j.cell.2007.07.045. [DOI] [PubMed] [Google Scholar]
- Jiang Y, de Bruin A, Caldas H, Fangusaro J, Hayes J, Conway EM, Robinson ML, Altura RA. Essential role for survivin in early brain development. J Neurosci. 2005;25:6962–6970. doi: 10.1523/JNEUROSCI.1446-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- Jones G, Jones D, Zhou L, Steller H, Chu Y. Deterin, A New Inhibitor of Apoptosis From Drosophila Melanogaster. J Biol Chem. 2000;275:22157–22165. doi: 10.1074/jbc.M000369200. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27:549–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Kleino A, Valanne S, Ulvila J, Kallio J, Myllymaki H, Enwald H, Stoven S, Poidevin M, Ueda R, Hultmark D, Lemaitre B, Ramet M. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. EMBO J. 2005;24:3423–3434. doi: 10.1038/sj.emboj.7600807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, Janeway CA, Medzhitov R, Flavell RA. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- Kornbluth S, White K. Apoptosis in Drosophila: neither fish nor fowl (nor man, nor worm). J Cell Sci. 2005;118:1779–1787. doi: 10.1242/jcs.02377. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Amer A, Kanneganti TD, Munoz-Planillo R, Chen G, Vandenabeele P, Fortier A, Gros P, Nunez G. The Nod-like receptor family member Naip5/Birc1e restricts Legionella pneumophila growth independently of caspase-1 activation. J Immunol. 2007;178:8022–8027. doi: 10.4049/jimmunol.178.12.8022. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart JM, Hoffmann JA. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci U S A. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G, Hengartner MO. Developmental apoptosis in C. elegans: a complex CEDnario. Nat Rev Mol Cell Biol. 2006;7:97–108. doi: 10.1038/nrm1836. [DOI] [PubMed] [Google Scholar]
- Leulier F, Lhocine N, Lemaitre B, Meier P. The Drosophila inhibitor of apoptosis protein DIAP2 functions in innate immunity and is essential to resist gram-negative bacterial infection. Mol Cell Biol. 2006;26:7821–7831. doi: 10.1128/MCB.00548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CG, Xu Y, Mularski B, Liu H, Gurbuxani S, Crispino JD. Requirements for survivin in terminal differentiation of erythroid cells and maintenance of hematopoietic stem and progenitor cells. J Exp Med. 2007;204:1603–1611. doi: 10.1084/jem.20062395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- Li F, Flanary PL, Altieri DC, Dohlman HG. Cell division regulation by BIR1, a member of the inhibitor of apoptosis family in yeast. J Biol Chem. 2000;275:6707–6711. doi: 10.1074/jbc.275.10.6707. [DOI] [PubMed] [Google Scholar]
- Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton-Horvat G, Farahani R, McLean M, Ikeda J-E, MacKenzie A, Korneluk RG. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature. 1996;379:349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- Liu Z, Sun C, Olejniczak ET, Meadows RP, Betz SF, Oost T, Herrmann J, Wu JC, Fesik SW. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature. 2000;408:1004–1008. doi: 10.1038/35050006. [DOI] [PubMed] [Google Scholar]
- Lotz K, Pyrowolakis G, Jentsch S. BRUCE, a giant E2/E3 ubiquitin ligase and inhibitor of apoptosis protein of the trans-Golgi network, is required for normal placenta development and mouse survival. Mol Cell Biol. 2004;24:9339–9350. doi: 10.1128/MCB.24.21.9339-9350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Lin SC, Huang Y, Kang YJ, Rich R, Lo YC, Myszka D, Han J, Wu H. XIAP induces NF-κB activation via the BIR1/TAB1 interaction and BIR1 dimerization. Mol Cell. 2007;26:689–702. doi: 10.1016/j.molcel.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AC, Lin R, Chan PK, Leung JC, Chan LY, Meng A, Verfaillie CM, Liang R, Leung AY. The role of survivin in angiogenesis during zebrafish embryonic development. BMC Dev Biol. 2007;7:50. doi: 10.1186/1471-213X-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier P, Silke J, Leevers SJ, Evan GI. The Drosophila caspase DRONC is regulated by DIAP1. EMBO J. 2000;19:598–611. doi: 10.1093/emboj/19.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LK. An exegesis of IAPs: salvation and surprises from BIR motifs. Trends Cell Biol. 1999;9:323–328. doi: 10.1016/s0962-8924(99)01609-8. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Byrne BG, Whitfield NN, Madigan CA, Fuse ET, Tateda K, Swanson MS. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med. 2006;203:1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mufti AR, Burstein E, Csomos RA, Graf PC, Wilkinson JC, Dick RD, Challa M, Son JK, Bratton SB, Su GL, Brewer GJ, Jakob U, Duckett CS. XIAP Is a copper binding protein deregulated in Wilson's disease and other copper toxicosis disorders. Mol Cell. 2006;21:775–785. doi: 10.1016/j.molcel.2006.01.033. [DOI] [PubMed] [Google Scholar]
- Muro I, Hay BA, Clem RJ. The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J Biol Chem. 2002;277:49644–49650. doi: 10.1074/jbc.M203464200. [DOI] [PubMed] [Google Scholar]
- Okada H, Bakal C, Shahinian A, Elia A, Wakeham A, Suh WK, Duncan GS, Ciofani M, Rottapel R, Zuniga-Pflucker JC, Mak TW. Survivin loss in thymocytes triggers p53-mediated growth arrest and p53-independent cell death. J Exp Med. 2004;199:399–410. doi: 10.1084/jem.20032092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olayioye MA, Kaufmann H, Pakusch M, Vaux DL, Lindeman GJ, Visvader JE. XIAP-deficiency leads to delayed lobuloalveolar development in the mammary gland. Cell Death Differ. 2005;12:87–90. doi: 10.1038/sj.cdd.4401524. [DOI] [PubMed] [Google Scholar]
- Petersen SL, Wang L, Yalcin-Chin A, Li L, Peyton M, Minna J, Harran P, Wang X. Autocrine TNFαsignaling renders human cancer cells susceptible to Smac-mimetic-induced apoptosis. Cancer Cell. 2007;12:445–456. doi: 10.1016/j.ccr.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl C, Jentsch S. Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell. 2008;132:832–845. doi: 10.1016/j.cell.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Pomerantz JL, Baltimore D. Two pathways to NF-kappaB. Mol Cell. 2002;10:693–695. doi: 10.1016/s1097-2765(02)00697-4. [DOI] [PubMed] [Google Scholar]
- Rajcan-Separovic E, Liston P, Lefebvre C, Korneluk RG. Assignment of human inhibitor of apoptosis protein (IAP) genes xiap, hiap-1, and hiap-2 to chromosomes Xq25 and 11q22-q23 by fluorescence in situ hybridization. Genomics. 1996;37:404–406. doi: 10.1006/geno.1996.0579. [DOI] [PubMed] [Google Scholar]
- Ren J, Shi M, Liu R, Yang QH, Johnson T, Skarnes WC, Du C. The Birc6 (Bruce) gene regulates p53 and the mitochondrial pathway of apoptosis and is essential for mouse embryonic development. Proc Natl Acad Sci U S A. 2005;102:565–570. doi: 10.1073/pnas.0408744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2006;2:e18. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro PS, Kuranaga E, Tenev T, Leulier F, Miura M, Meier P. DIAP2 functions as a mechanism-based regulator of drICE that contributes to the caspase activity threshold in living cells. J Cell Biol. 2007;179:1467–1480. doi: 10.1083/jcb.200706027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Renatus M, Schwarzenbacher R, Zhou Q, Sun C, Fesik SW, Liddington RC, Salvesen GS. Structural basis for the inhibition of caspase-3 by XIAP. Cell. 2001;104:791–800. doi: 10.1016/s0092-8674(01)00274-4. [DOI] [PubMed] [Google Scholar]
- Rigaud S, Fondaneche MC, Lambert N, Pasquier B, Mateo V, Soulas P, Galicier L, Le Deist F, Rieux-Laucat F, Revy P, Fischer A, de Saint Basile G, Latour S. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 2006;444:110–114. doi: 10.1038/nature05257. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Chen P, Oliver H, Abrams JM. Unrestrained caspase-dependent cell death caused by loss of Diap1 function requires the Drosophila Apaf-1 homolog, Dark. EMBO J. 2002;21:2189–2197. doi: 10.1093/emboj/21.9.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe M, Pan M-G, Henzel WJ, Ayres TM, Goeddel DV. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- Roy N, Mahadevan MS, McLean M, Shutler G, Yaraghi Z, Farahani R, Baird S, Besner-Johnston A, Lefebvre C, Kang X. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell. 1995;80:167–178. doi: 10.1016/0092-8674(95)90461-1. et, a. [DOI] [PubMed] [Google Scholar]
- Roy N, Deveraux QL, Takahashi R, Salvesen GS, Reed JC. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol. 2002;4:432–438. doi: 10.1038/ncb795. [DOI] [PubMed] [Google Scholar]
- Samuel T, Welsh K, Lober T, Togo SH, Zapata JM, Reed JC. Distinct BIR domains of cIAP1 mediate binding to and ubiquitination of tumor necrosis factor receptor-associated factor 2 and second mitochondrial activator of caspases. J Biol Chem. 2006;281:1080–1090. doi: 10.1074/jbc.M509381200. [DOI] [PubMed] [Google Scholar]
- Sandall S, Severin F, McLeod IX, Yates J. R. r., Oegema K, Hyman A, Desai A. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 2006;127:1179–1191. doi: 10.1016/j.cell.2006.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro MM, Samuel T, Mitchell T, Reed JC, Stainier DY. Birc2 (cIap1) regulates endothelial cell integrity and blood vessel homeostasis. Nat Genet. 2007;39:1397–1402. doi: 10.1038/ng.2007.8. [DOI] [PubMed] [Google Scholar]
- Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11:519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- Speliotes EK, Uren A, Vaux D, Horvitz HR. The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol Cell. 2000;6:211–223. doi: 10.1016/s1097-2765(00)00023-x. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Molofsky AB. Autophagy and inflammatory cell death, partners of innate immunity. Autophagy. 2005;1:174–176. doi: 10.4161/auto.1.3.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R, Deveraux Q, Tamm I, Welsh K, Assa-Munt N, Salvesen GS, Reed JC. A single BIR domain of XIAP sufficient for inhibiting caspases. J. Biol. Chem. 1998;273:7787–7790. doi: 10.1074/jbc.273.14.7787. [DOI] [PubMed] [Google Scholar]
- Tenev T, Zachariou A, Wilson R, Ditzel M, Meier P. IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat Cell Biol. 2004 doi: 10.1038/ncb1204. [DOI] [PubMed] [Google Scholar]
- Uren A, Pakusch M, Hawkins C, Puls KL, Vaux DL. Cloning and expression of apoptosis inhibitory proteins homologs that function to inhibit apoptosis and/or bind tumor necrosis factor receptor-associated factors. Proc. Natl. Acad. Sci. USA. 1996;93:4974–4978. doi: 10.1073/pnas.93.10.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, Beilharz T, O'Connell MJ, Bugg SJ, van Driel R, Vaux DL, Lithgow T. Role for yeast inhibitor of apoptosis (IAP)-like proteins in cell division. Proc Natl Acad Sci U S A. 1999;96:10170–10175. doi: 10.1073/pnas.96.18.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL, Choo KH. Survivin and the inner centromere protein INCENP show similar cell- cycle localization and gene knockout phenotype. Curr Biol. 2000;10:1319–1328. doi: 10.1016/s0960-9822(00)00769-7. [DOI] [PubMed] [Google Scholar]
- Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJ, Flygare JA, Fairbrother WJ, Deshayes K, Dixit VM, Vucic D. IAP antagonists induce autoubiquitination of c-IAPs, NF-| B activation, and TNF〈-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Silke J. Mammalian mitochondrial IAP binding proteins. Biochem Biophys Res Commun. 2003;304:499–504. doi: 10.1016/s0006-291x(03)00622-3. [DOI] [PubMed] [Google Scholar]
- Verhagen AM, Coulson EJ, Vaux DL. Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biol. 2001;2:3009–3019. doi: 10.1186/gb-2001-2-7-reviews3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- Verhagen AM, Kratina TK, Hawkins CJ, Silke J, Ekert PG, Vaux DL. Identification of mammalian mitochondrial proteins that interact with IAPs via N-terminal IAP binding motifs. Cell Death Differ. 2007;14:348–357. doi: 10.1038/sj.cdd.4402001. [DOI] [PubMed] [Google Scholar]
- Vernooy SY, Chow V, Su J, Verbrugghe K, Yang J, Cole S, Olson MR, Hay BA. Drosophila bruce can potently suppress rpr- and grim-dependent but not hid-dependent cell death. Curr Biol. 2002;12:1164–1168. doi: 10.1016/s0960-9822(02)00935-1. [DOI] [PubMed] [Google Scholar]
- Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, Benetatos CA, Chunduru SK, Condon SM, McKinlay M, Brink R, Leverkus M, Tergaonkar V, Schneider P, Callus BA, Koentgen F, Vaux DL, Silke J. IAP antagonists target cIAP1 to induce TNF〈-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- Vong QP, Cao K, Li HY, Iglesias PA, Zheng Y. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science. 2005;310:1499–1504. doi: 10.1126/science.1120160. [DOI] [PubMed] [Google Scholar]
- Wang L, Du F, Wang X. TNF-〈 induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, Steller H, Meier P. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- Wing JP, Schreader BA, Takakazu Y, Wang Y, Andrews PS, Huseinovic N, Dong CK, Ogdahl JL, Schwartz LM, White K, Nambu JR. Drosophila Morgue is an F box/ubiquitin conjugase domain protein important for grim-reaper mediated apoptosis. Nat Cell Biol. 2002;4:451–456. doi: 10.1038/ncb800. [DOI] [PubMed] [Google Scholar]
- Wing JP, Schwartz LM, Nambu JR. The RHG motifs of Drosophila Reaper and Grim are important for their distinct cell death-inducing abilities. Mech Dev. 2001;102:193–203. doi: 10.1016/s0925-4773(01)00316-1. [DOI] [PubMed] [Google Scholar]
- Wright CW, Clem RJ. Sequence requirements for hid binding and apoptosis regulation in the anti-apoptotic baculovirus protein Op-IAP: hid binds Op-IAP in a manner similar to Smac binding of XIAP. J Biol Chem. 2001;277:2454–2462. doi: 10.1074/jbc.M110500200. [DOI] [PubMed] [Google Scholar]
- Wright EK, Goodart SA, Growney JD, Hadinoto V, Endrizzi MG, Long EM, Sadigh K, Abney AL, Bernstein-Hanley I, Dietrich WF. Naip5 affects host susceptibility to the intracellular pathogen Legionella pneumophila. Curr Biol. 2003;13:27–36. doi: 10.1016/s0960-9822(02)01359-3. [DOI] [PubMed] [Google Scholar]
- Wu G, Chai J, Suber TL, Wu JW, Du C, Wang X, Shi Y. Structural basis of IAP recognition by Smac/DIABLO. Nature. 2000;408:1008–1012. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
- Wu JW, Cocina AE, Chai J, Hay BA, Shi Y. Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides. Mol Cell. 2001;8:95–104. doi: 10.1016/s1097-2765(01)00282-9. [DOI] [PubMed] [Google Scholar]
- Xing Z, Conway EM, Kang C, Winoto A. Essential role of survivin, an inhibitor of apoptosis protein, in T cell development, maturation, and homeostasis. J Exp Med. 2004;199:69–80. doi: 10.1084/jem.20031588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Li Y, Arcaro M, Lackey M, Bergmann A. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development. 2005;132:2125–2134. doi: 10.1242/dev.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang QH, Du C. Smac/DIABLO selectively reduces the levels of c-IAP1 and c-IAP2 but not that of XIAP and livin in HeLa cells. J Biol Chem. 2004;279:16963–16970. doi: 10.1074/jbc.M401253200. [DOI] [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, Feldman RM, Clem RJ, Muller HA, Hay BA. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- Yoon HJ, Carbon J. Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc Natl Acad Sci U S A. 1999;96:13208–13213. doi: 10.1073/pnas.96.23.13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni DS, Kobayashi KS, Kohlsdorf T, Ogura Y, Long EM, Vance RE, Kuida K, Mariathasan S, Dixit VM, Flavell RA, Dietrich WF, Roy CR. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- Zwerts F, Lupu F, De Vriese A, Pollefeyt S, Moons L, Altura RA, Jiang Y, Maxwell PH, Hill P, Oh H, Rieker C, Collen D, Conway SJ, Conway EM. Lack of endothelial cell survivin causes embryonic defects in angiogenesis, cardiogenesis, and neural tube closure. Blood. 2007;109:4742–4752. doi: 10.1182/blood-2006-06-028068. [DOI] [PMC free article] [PubMed] [Google Scholar]