An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network (original) (raw)
Abstract
Distinctive SWI/SNF-like ATP-dependent chromatin remodeling esBAF complexes are indispensable for the maintenance and pluripotency of mouse embryonic stem (ES) cells [Ho L, et al. (2009) Proc Natl Acad Sci USA 10.1073/pnas.0812889106]. To understand the mechanism underlying the roles of these complexes in ES cells, we performed high-resolution genome-wide mapping of the core ATPase subunit, Brg, using ChIP-Seq technology. We find that esBAF, as represented by Brg, binds to genes encoding components of the core ES transcriptional circuitry, including Polycomb group proteins. esBAF colocalizes extensively with transcription factors Oct4, Sox2 and Nanog genome-wide, and shows distinct functional interactions with Oct4 and Sox2 at its target genes. Surprisingly, no significant colocalization of esBAF with PRC2 complexes, represented by Suz12, is observed. Lastly, esBAF colocalizes with Stat3 and Smad1 genome-wide, consistent with a direct and critical role in LIF and BMP signaling for maintaining self-renewal. Taken together, our studies indicate that esBAF is an essential component of the core pluripotency transcriptional network, and might also be a critical component of the LIF and BMP signaling pathways essential for maintenance of self-renewal and pluripotency.
Keywords: BAF complexes, Brg, SWI/SNF ATP-dependent chromatin remodeling
Embryonic stem cells (ES) maintain an epigenetic state that enables both self-renewal and differentiation into all embryonic lineages (1). Recent studies reveal that in ES cells Oct4, Sox2, Nanog, and Klf4 elaborate a core transcriptional circuitry (2–4), working in coordination with Polycomb complexes (5, 6), microRNAs (7), and histone modification enzymes (8) to stably maintain the expression of pluripotency genes, and to repress lineage determinant genes. This transcriptional circuitry is kept in exquisite balance, because it can be perturbed both by reducing or increasing the levels of core regulators such as Oct4, Sox2, and Nanog, causing ES cells to lose self-renewal ability and/or pluripotency (9–14). At the same time, ATP-dependent chromatin remodeling enzymes in the Tip60/p400 and SWI/SNF families have been recently shown to be crucial for the maintenance and function of ES cells (15). Recent findings by us and others have shown that components of mammalian SWI/SNF (or BAF, Brg/Brahma Associated Factors) complex, Brg, BAF155, and BAF250A are crucial for the proliferation, self-renewal and pluripotency of ES cells (16, 17, 18). ES cells deficient in Brg maintain the expression of Oct4, Sox2, and Nanog for several cell divisions but rapidly lose colony morphology and proliferative capacity characteristic of ES cells (16). Upon prolonged absence of Brg, remaining ES cells down-regulate pluripotency markers such as Oct4 and Sox2 (16, 17), reflecting the complete loss of ES cell identity secondary to the effects of Brg depletion or suggesting that Brg is required to maintain stable expression of these markers over many cell divisions. In addition, the composition of BAF complexes in ES cells (esBAF) is biochemically and functionally specialized. esBAF complexes are defined by the incorporation of Brg but not Brm, BAF155 but not BAF170, and BAF60A but not BAF60C (16). When esBAF complexes are altered by enforced incorporation of BAF170, their ability to maintain ES cell self-renewal is compromised. Hence, specialized esBAF complexes are clearly crucial for stem cell maintenance. However, the mechanism by which BAF complexes establish and maintain self-renewal and pluripotency is not understood.
To understand the mechanism underlying the role of esBAF complexes in pluripotency, we performed expression and genome-wide occupancy studies of esBAF complexes in ES cells. Our studies indicate that a specialized esBAF complex is essential for self-renewal and pluripotency and is a critical component and regulator of the core transcriptional circuitry of ES cells.
Results
ChIP-Seq Analysis of Brg and BAF155 Reveals Enrichment of esBAF Complexes at Transcription Start Sites of Genes.
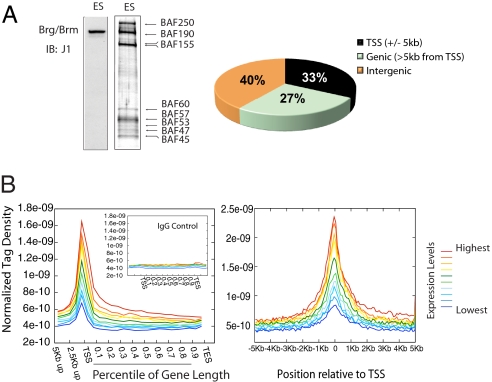
To elucidate the essential role of esBAF complexes in pluripotency, we performed genome-wide ChIP-Seq analysis with the anti-Brg/Brm J1 antibody (Fig. 1A Left) using Solexa sequencing technology and obtained a total of ≈12.2 million 25-base pair tags that mapped to unique genomic locations. Because Brg is the exclusive ATPase subunit of esBAF (16) and is not found outside of BAF complexes (19), we consider Brg to be representative of esBAF complexes in our following analyses. Using the TIROE algorithm developed by Jothi and colleagues [see supporting information (SI) Methods], we identified 10,559 regions of Brg enrichment under stringent conditions (p <E-10, see SI Methods). In contrast to most transcription factors, which have small footprints (20, 21), the average footprint of esBAF, determined by the median length of Brg-bound regions, is 6.2 Kb (Fig. S1_A_), suggesting that the complex is present in multiple copies at each location with the overall effect of coating its target chromatin. By calculating the cumulative footprint of Brg across the genome, we estimate that Brg is bound to 4% of the entire genome. Sixty percent of Brg-bound regions are located in genic or promoter regions of 5,630 distinct genes, and 40% were detected in intergenic regions (Fig. 1A Right). Analysis of average Brg tag density distribution across gene units [defined as the gene body plus 5 Kb upstream of transcription start site (TSS)] revealed enrichment of Brg occupancy near the TSS (Fig. 1B Left), with peak enrichment directly over the TSS when analyzed at higher resolution (Fig. 1B Right). However, for any individual gene, peak binding was often also found upstream of the TSS or within the gene body (Fig. 2B). In addition, the absolute expression levels of Brg-bound genes [by analysis of E14 ES cell expression data from Loh et al. (26)] correlated positively with the degree of Brg occupancy, i.e., most highly expressed genes are most likely to be bound by Brg (Fig. 1B and Fig. S1_B_).
Fig. 1.
High-resolution genome-wide analysis of esBAF occupancy. (A) (Left) Specificity of J1 antibody used for ChIP-Seq both by immunoblotting of ES nuclear extracts and immunoprecipitation of BAF complexes from ES extracts and visualization by silver stain. (Right) Distribution of Brg-bound regions throughout the genome. TSS, transcription start site; genic regions are defined as 5 Kb downstream of TSS to the end of an annotated gene. (TES; transcription end site.) (B) Average distribution of Brg-normalized tag density across a gene unit (Left). Higher-resolution analysis of average tag density surrounding the TSS (Right). In each plot, genes were classified into 10 groups based on expression levels in ES cells (highest to lowest represented by colored lines), and average tag density across the gene unit was plotted for each group.
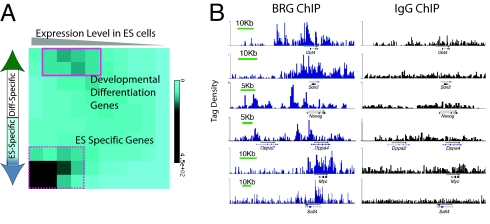
Fig. 2.
esBAF complexes occupy genes of the core transcription network. (A) All Brg-bound genes were placed in a matrix to determine their expression in ES cells (x axis), and whether they are ES-specific or differentiation-specific according to their direction of regulation as ES cells differentiate to day 14 embryoid bodies (embryoid/ES fold change). Intensity of the heat map represents the absolute numbers of genes falling in each square of the matrix, as indicated by color bar on the right. In this matrix, y-axis represents n = 17,030 genes that had unique transcripts represented by probes on Affymetrix's MOE430_2 expression arrays minus ∼15% genes with low expression levels in both ES and differentiated cells, and x-axis represents 5,630 genes that are bound by Brg (see SI Methods). (B) Brg occupancy on genes of the core ES cell circuitry from the University of California, Santa Cruz (UCSC) genome browser. Brg ChIP (Left) and IgG ChIP (Right) plotted on comparable tag density axes.
esBAF complexes are characterized by the inclusion of BAF155 and exclusion of BAF170 (16). In addition, BAF155 is absolutely required for the maintenance of ES cells (16). Hence, we expected Brg and BAF155 to occupy their target sites in a similar manner. Indeed, similar to Brg, BAF155 exhibits strong enrichment of binding near the TSS (Fig. S2_A_). In addition, as expected, a comparison of BAF155 and Brg binding reveals that the 2 proteins have a very similar binding profile on their target genes (Fig. S2_B_), confirming that Brg and BAF155 act in coordination in the context of esBAF at their target genes.
esBAF Complexes Occupy Genes of the Core Pluripotency Transcriptional Network.
By comparing the expression levels of genes in ES cells with that in fully differentiated ES cells (day 14 embryoid bodies) (22), we found that Brg is enriched at highly expressed, ES cell- specific genes that are down-regulated during differentiation (Fig. 2A, dashed square and Fig. S1_C_) and at low-to-moderately expressed genes that undergo up-regulation during differentiation, i.e., developmental or lineage-determinant genes (Fig. 2A, solid square). Within the former class of ES cell-specific genes, we examined binding to all genes known to have a role in pluripotency. Remarkably, each of the ES cell master regulator genes, including Oct4, Sox2, Nanog, Dppa2, Dppa4, Sall4, and Myc show extensive Brg (Fig. 2B) and BAF155 (Fig. S2_B_) binding. Furthermore, genes known to be critical for ES cells (Klf4, Rif1) also show statistically significant Brg binding (Fig. S3_A_). Within the class of developmental or lineage-determinant genes, we found that esBAF complexes bound many genes related to neuronal development (Fig. S3_B_). This observation suggests that Brg might have a role in preventing premature expression of these developmental genes in ES cells. Together, these observations implicate esBAF complexes as a central component of the core transcriptional circuitry of ES cells.
esBAFs Cooccupy Target Genes of Master Regulators Oct4, Sox2, and Nanog.
If esBAF were a component of the core circuitry, one might expect it to occupy genes that are targets of ES cell master regulators Oct4, Sox2, and Nanog. To test this hypothesis, we used published ChIP-Seq datasets for Oct4, Sox2, and Nanog (21) and calculated the fraction of annotated genes (n = 21,352) that bound Brg, and also Sox2, Oct4, or Nanog within the gene body plus 5 Kb upstream of the TSS. Remarkably, 67% of Oct4 target genes, 74% of Sox2 target genes, and 65% of Nanog target genes are also bound by Brg (Table 1). The observed number of genes bound by Brg and each of these factors is significantly higher than expected by random coincidence (Table 1), suggesting functional interactions between Brg and components of the core circuitry. Similar trends of enrichment were observed when we defined cooccupancy as physical colocalization i.e., overlap of the binding regions of Brg and specific factors (Table 1). In addition, similar analyses using data from Chen et al. (21) revealed high degrees of colocalization of Brg with the bone morphogenetic protein (BMP) and leukemia inhibitory factor (LIF) signaling pathway transcription factors Smad1 (89%) and Stat3 (70%), respectively (Table 1). BMP and LIF signaling are crucial to ES cell maintenance (23) and have been proposed to oppose mitogen-activated protein kinase ERK1/2-induced differentiation (24). These findings implicate esBAF as a core component of the ES transcriptional circuitry, not only by functional interactions with master regulators Oct4, Sox2, and Nanog but also with effectors of the LIF and BMP signaling pathway.
Table 1.
Overall enrichment of ES regulator (X)-binding sites within or in proximity to Brg-bound regions
| Genes bound by X | Common target genes | Physical overlap | ||||
|---|---|---|---|---|---|---|
| Genes with both Brg and X within the gene unit | Genes where X binds within Brg enrichment regions | |||||
| Percentage | Obs/exp fold | P | Percentage | Obs/exp fold | P | |
| Brg | 100 | 100 | ||||
| Oct4 | 67 | 2.15 | 1.91 E−286 | 56 | 1.82 | 1.25E−146 |
| Sox2 | 74 | 2.39 | 0.00E+00 | 61 | 1.92 | 3.78E−189 |
| Nanog | 65 | 2.09 | 0.00E+00 | 51 | 1.64 | 1.12E−173 |
| Suz12 | 30 | 0.98 | 0.239 | 18 | 0.58 | 1.63E−56 |
| STAT3 | 70 | 2.25 | 2.20E−224 | 54 | 1.75 | 2.66E−82 |
| Smad1 | 89 | 2.88 | 6.45E−176 | 82 | 2.64 | 7.17E−131 |
esBAF Represses Developmental Genes and Refines the Core Transcriptional Circuitry by Programmatic Interactions with Oct4 and Sox2.
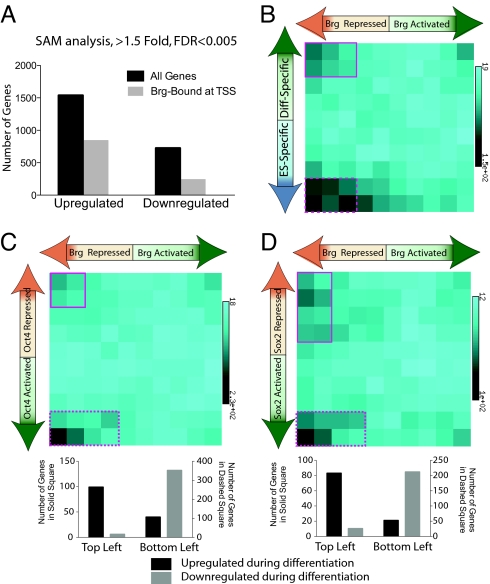
To determine how esBAF complexes regulate the core circuitry, as suggested by occupancy studies, we performed microarray analysis of BrgshRNA (16) ES cells 96 h after transfection of knockdown constructs. To minimize off-target hits, we defined a stringent set of Brg target genes as genes that underwent >1.5-fold expression change in Brg shRNA knockdown compared with control (FDR <0.005) and have Brg-bound regions within the gene body and/or 5 Kb upstream of TSS (Fig. 3A). Analysis of this group of Brg-bound, Brg-regulated targets demonstrates that Brg acts most commonly as a repressor, and in a programmatic manner that would not be expected if it were a general transcription regulator. First, we found that Brg repressed a significant number of developmental genes (Fig. 3B, solid box), such as EphrinB4, Fgf5, Fgfr1, VEGF, NFATc4, and others. Unexpectedly, Brg also repressed many genes expressed only in ES cells (Fig. 3B, dashed box). This implies that ES cell-specific genes are tonically repressed by Brg, possibly to refine and maintain their expression within the correct threshold. The elevated expression of many of these ES cell markers after Brg knockdown was confirmed by Q-PCR analysis (Fig. S4). We propose that refining the expression of this class of genes is essential because increasing their levels can lead to disruption of the core circuitry (11). In contrast to the refinement function of Brg, master regulators such as Oct4 and Sox2 regulate the core circuitry by activating ES-specific genes and repressing differentiation-associated genes (Fig. S1_D_). When we examined the functional interaction between Brg and the ES cell master regulators Oct4 and Sox2 at their target genes by comparing Brg target genes with published expression datasets of Oct4 knockdown and Sox2 knockout ES cells (12, 25), we observed a predominantly antagonistic interaction between Brg and Oct4/Sox2 (Fig. 3 C and D, dashed square), i.e., genes that are activated by Oct4/Sox2 are repressed by Brg (mostly ES cell-specific genes, Fig. 3 C and D Lower). Brg and Oct4/Sox2 also work in coordination to repress certain genes that are mostly differentiation-associated genes (Fig. 3 C and D, solid square and Lower). This indicated that esBAF complexes function in some contexts to enhance the transcriptional repression of Oct4/Sox2 target genes, whereas in other contexts to oppose transcriptional activation by Oct4/Sox2. Hence, esBAF appears to have a dual role in the ES cell circuitry: keeping developmental genes repressed to prevent premature differentiation and optimizing the levels of ES cell-specific genes within limits to prevent perturbation of the core circuitry.
Fig. 3.
Brg represses developmental genes and refines ES-specific genes by cooperating with Oct4 and Sox2. (A) SAM analysis of microarray data from Brg knockdown (KD) ES cells compared with control knockdown, 96 h after transfection of constructs. Genes that were significantly changed were further classified as Brg-bound within the gene unit (gray bars) or not (black bars). (B) Brg-bound genes were placed in a matrix with fold change after Brg KD on the x axis against EB/ES fold (i.e., differentiation- or ES-specific) on the y axis to ascertain whether Brg-dependent target genes are ES-specific, or differentiation associated, and in which direction they are changed upon Brg KD. (C) Brg-bound genes were placed in a matrix with fold change after Brg KD on the x axis against observed fold change of the same genes afternOct4 KD (25) or Sox2 KO (D) (12) in ES cells on the y axis to determine the functional interaction between Oct4/Sox2 and Brg. Bar graphs indicate the natural developmental fate of genes within the top left (solid) squares and bottom left (dashed) squares by comparing their fold change between ES cells and day 14 embryoid bodies.
esBAF Opposes Polycomb Complexes by Direct Repression of Subunits of the PRC1 Complex.
Several studies have implicated Polycomb repressive complexes in the maintenance of ES cells (5). Because Brg is closely related to Brm, whose homologue in Drosophila (Brahma) is a classical trithorax group protein, we hypothesized that esBAFs might functionally interact with PcG on target promoters as suggested from biochemical studies (35). Surprisingly, no significant cooccupancy with Brg was detected for Polycomb [represented by Suz12, from (7)] on a genome-wide level (Table 1). In fact, the observed physical colocalization of Suz12 and Brg (Table 1) suggests that the 2 complexes cooccupy genomic locations less than that which would be expected by chance (P value < 1.63E−56). This is reminiscent of the mutually exclusive pattern of PcG and Brm on Drosophila polytene chromosomes (26). These observations suggest that esBAF might not generally oppose PcG proteins by widespread and direct interactions on target promoters in the ES-cell state but does not rule out other mechanisms of functional antagonism. Indeed, when we examined the level of expression of polycomb subunits after knocking down Brg, several subunits including Bmi1, Cbx7, Ring1, and Phc1 and Phc2 showed increased expression after 96 h of Brg knockdown (Fig. S5_A_). In addition, each of these genes contains a statistically significant Brg-bound region (Fig. S5_B_) suggesting that esBAF opposes PcG action in ES cells by directly binding to PcG genes and repressing their transcription.
Discussion
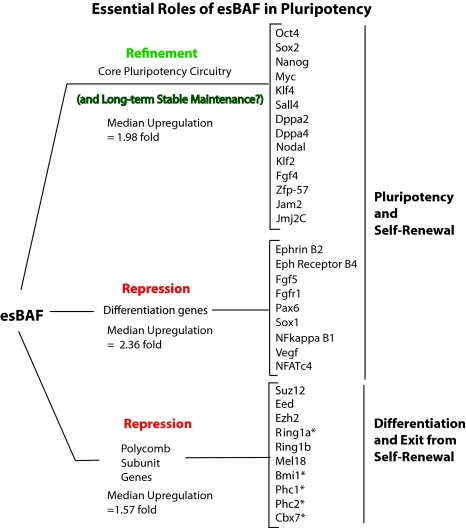
Our studies indicate that BAF complexes form an integral part of the core pluripotency transcriptional network of ES cells (Fig. 4). In a companion article, we show that such complexes in ES cells are functionally and compositionally specialized (16). Such esBAF complexes are critical for the self-renewal of ES cells and maintenance of the ES cell state by means of regulation of the core transcriptional circuitry.
Fig. 4.
Proposed roles of esBAFs in the regulation of the core circuitry. In one strategy esBAF functionally interacts with Sox2 and Oct4 to refine the levels of ES-specific genes, or to repress the expression of some differentiation genes. In a second strategy that probably facilitates exit from the pluripotent state, esBAF directly regulates the expression of some PcG proteins. The median fold change of genes in each group is noted. Each of the genes on the right are direct targets of esBAF complexes by ChIP-Seq. *, PcG genes that were ditectably upregulated by microarray analysis.
esBAF Is an Essential Regulatory Component of the Transcriptional Circuitry of Pluripotency.
Our genome-wide studies point to the specificity in the action of esBAF complexes. Although binding of esBAF complexes was widespread and occurred on nearly 1/4 of all annotated genes in ES cells, all known genes that contribute to pluripotency, and other ES cell-specific genes were selective targets. Colocalization analyses examining the colocalization of Brg with Oct4, Sox2, or Nanog, reveal high degrees of target gene overlap. This suggests that in addition to direct transcriptional regulation of these factors, esBAF complexes might functionally interact with Oct4, Sox2, and Nanog at their common target genes. In support of these findings, we found that endogenous Brg interacts with endogenous Sox2 and Oct4 in ES cells (16). Because Oct4 binds to DNA with Sox2 (27, 28), esBAF might be recruited by Oct4/Sox2 heterodimers. Alternatively, esBAF might engage chromatin through its multiple histone recognition and DNA-binding domains and remodel its target sites to enable binding of Oct4/Sox2. In addition, the striking colocalization of Brg with Smad1 and Stat3 is noteworthy because BMP and LIF signaling pathways oppose Erk1/2-mediated differentiation of ES cells (24) and might partly account for the requirement of Brg in ES cell maintenance. Last, we also found that all subunits of the Polycomb Repressive Complex 2 and almost all of the subunits of Polycomb Repressive Complex 1 showed clear and statistically significant Brg binding. esBAF complexes appear to oppose PcG proteins, as might be expected from a TrxG complex, by means of repression, and thus provide a potential means of exit from the pluripotent state. The lack of significant colocalization between PcG and esBAF complexes genome-wide appears (Table 1) at first to be contradictory to previous reports that PcG colocalizes extensively with Oct4, Sox2, and Nanog in human ES cells (6). However, such colocalization occurs only for a subset of PcG target genes that encode for developmental regulators that are coordinately repressed by these proteins (6). On a genome-wide scale in mouse ES cells, we do not observe significant colocalization between PcG and Brg even though Brg colocalizes extensively with Oct4, Sox2, and Nanog, indicating that, whereas the functional interaction between esBAFs with Oct4, Sox and Nanog is general, this is not the case with PcG complexes.
esBAF Refines the Elements of the Core Pluripotency Circuitry.
Analysis of Brg-dependent target genes indicate that esBAF acts highly programmatically both in coordination with the core ES cell circuitry and, unexpectedly, also in opposition to it. Brg represses the expression of certain genes involved in development and differentiation, presumably to prevent premature differentiation. We initially expected esBAF complexes to be required for the continuous expression of pluripotency markers, which would explain its critical role in ES cells. However, depletion of Brg by shRNA and by a conditional allele (16) revealed that the expression of pluripotency markers Oct4, Sox2, and Nanog is maintained for many cell divisions in the absence of Brg and that their down-regulation occurs only after prolonged depletion of Brg, long after Brg-deficient ES cells lose colony morphology and self-renewal (16). In contrast, upon acute depletion of Brg, many ES-specific genes, including master regulators such as Nanog, are up-regulated in expression. This indicates that esBAF functions normally to tonically repress the expression of ES cell-specific genes, such that in the absence of Brg, the expression of these genes is further elevated. The repressive function of esBAF on pluripotency genes has also been reported by Knott and colleagues (17), who observed derepression of Oct4 in the trophectoderm of Brg-depleted blastocysts and down-regulation of pluripotency genes in ES cells only after prolonged depletion of Brg. Hence, esBAFs regulate the circuitry by refining the levels of ES cell-specific genes, particularly transcription factors, because elevated levels of transcription factors such as Oct4 and Nanog can lead to perturbation of the circuitry and result in differentiation of ES cells or inhibit differentiation and development (10, 11). Acute reduction of esBAF complexes might cause an initial perturbation of the circuitry because of increased expression of ES cell-specific and developmental genes, which eventually leads to the dismantling of the core circuitry and the loss of ES cell determinants such as Oct4, Sox2 and Nanog. The refining function of esBAF might also enable the destabilization of the circuit to facilitate exit from the self-renewal state during differentiation and to prevent the formation of tumors.
The lack of an acute need for Brg for transcription at most ES cell-specific genes, in contrast with the continuous transcriptional need for Swi2 in yeast (29) appears contradictory. This disparity might reflect the stability of the core circuitry, which once established, can be maintained by intrinsic positive feedback independent of esBAF for several days. However, it is possible that esBAF is required for the initial establishment of the pluripotent circuitry, given that Brg is a maternal effect gene found to be essential for zygotic gene activation (30) and somatic nuclear reprogramming (31). Once established, the core circuitry might become less dependent on the activity of esBAF.
In conclusion, our proteomic (16) and genomic studies indicate that a specialized complex, esBAF, is tailored to interact with ES cell-specific transcription factors and to participate in the core pluripotency transcriptional circuitry and provide the essential groundwork for further investigation into the involvement of esBAF Complexes in regulating the pluripotent chromatin landscape.
Materials and Methods
Culture of ES Cells.
All experiments were performed with E14Tg2a murine embryonic stem cells cultured under standard feeder-free conditions. For details, please refer to SI Methods.
Microarray Profiling of Gene Expression.
E14 ES cells were separately transfected with BrgshRNA#2, BAF155shRNA, and shGFP Control with Lipofectamine 2000 (Invitrogen). Transfected ES cells were exposed to 1.5 μg/mL puromycin and harvested at 96 h for RNA isolation. Samples were prepared and hybridized onto Affymetrix Mouse 430_2 gene arrays according to the manufacturer's instructions. Microarray data were normalized and summarized with RMA algorithm (32), and subsequently analyzed for statistical significance with SAM package (33).
ChIP-Sequencing and Data Analysis.
ChIP was performed with affinity purified J1 antisera [Khavari et al. (34)]. Because 76% of the spectra obtained from mass spectrometry of purified complexes (Fig. 2A) were from known components of the complex, we estimate that the maximum false-positive rate of our ChIP-Seq analysis is 24%. ChIP samples were sequenced and the 25-bp reads were mapped to the mouse genome (mm8 assembly) by using the Solexa Analysis Pipeline. Approximately 12.2 million reads from J1 ChIP and 14.5 million reads from IgG ChIP that mapped to unique genomic locations were processed further by using algorithms developed by Jothi et al. (20) to identify 10,559 Brg-binding regions. More details on the data analysis and statistical significance can be found in SI Methods.
Supplementary Material
Supporting Information
Acknowledgments.
We are grateful to Jiang Wu, Andrew Yoo, and Elena Gallo for critical comments on the manuscript. This work was supported by the Howard Hughes Medical Institute, NIH grants NS046789, Hd55391 and AI060037 and the JDRF (G.R.C.) and the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health (R.J., K.C., and K.Z). L.H. was funded by the Agency of Science, Technology and Research of Singapore.
Footnotes
The authors declare no conflict of interest.
Data deposition: Primary sequencing and microarray data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE14344).
References
- 1.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang J, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 5.Boyer L, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 6.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 9.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 10.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 11.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 12.Masui S, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 13.Yasuda SY, et al. NANOG maintains self-renewal of primate ES cells in the absence of a feeder layer. Genes Cells. 2006;11:1115–1123. doi: 10.1111/j.1365-2443.2006.01000.x. [DOI] [PubMed] [Google Scholar]
- 14.Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 15.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60–p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho L, et al. An embryonic stem cell chromatin remodeling complex, esBAF is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2008 doi: 10.1634/stemcells.2008-0710. in press. [DOI] [PubMed] [Google Scholar]
- 18.Yan Z, et al. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–1165. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lessard J, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 22.Sene K, et al. Gene function in early mouse embryonic stem cell differentiation. BMC Genomics. 2007;8:85. doi: 10.1186/1471-2164-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 24.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loh Y, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 26.Mohrmann L, et al. Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol Cell Biol. 2004;24:3077–3088. doi: 10.1128/MCB.24.8.3077-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chew JL, et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodda DJ, et al. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 2005;28:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 29.Biggar SR, Crabtree GR. Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J. 1999;18:2254–2264. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bultman SJ, et al. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006;20:1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansis C, Barreto G, Maltry N, Niehrs C. Nuclear reprogramming of human somatic cells by Xenopus egg extract requires BRG1. Curr Biol. 2004;14:1475–1480. doi: 10.1016/j.cub.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 32.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 33.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 35.Shao Z, et al. Stablization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information