Coiled-coil interactions are required for post-Golgi R-SNARE trafficking (original) (raw)
Abstract
The sorting of post-Golgi R-SNAREs (vesicle-associated membrane protein (VAMP)1, 2, 3, 4, 7 and 8) is still poorly understood. To address this, we developed a system to investigate their localization, trafficking and cell-surface levels. Here, we show that the distribution and internalization of VAMPs 3 and 8 are determined solely through a new conserved mechanism that uses coiled-coil interactions, and that VAMP4 does not require these interactions for its trafficking. We propose that VAMPs 3 and 8 are trafficked while in a complex with Q-SNAREs. We also show that the dileucine motif of VAMP4 is required for both its internalization and retrieval to the trans-Golgi network. However, when the dileucine motif is mutated, the construct can still be internalized potentially through coiled-coil interactions with Q-SNAREs.
Keywords: SNARE, VAMP3, VAMP4, VAMP7, VAMP8
Introduction
The human genome encodes at least 39 SNAREs, with each one localized to a defined set of membranes and involved in a specific set of fusion reactions (Jahn & Scheller, 2006). SNAREs, as with all cargo molecules, must be packaged efficiently into transport vesicles and tubules. Failure to sort SNAREs will cause marked defects in membrane transport. Surprisingly, little is known about this process for post-Golgi SNAREs in mammalian cells.
The post-Golgi Q-SNAREs syntaxins, 6, 7, 8, 10, 13, 16, and vti1a and b are packed into clathrin-coated vesicles (CCVs; Peden et al, 2001; Borner et al, 2006). Syntaxin 6, 7, 8 and 13 contain conventional sorting signals (E/DxxxLL or YxxΦ, where Φ is a bulky hydrophobic residue; Watson & Pessin, 2000; Kasai & Akagawa, 2001; however, these signals have not been shown experimentally to engage the machinery of CCVs. It has been shown that vti1b has its own specific clathrin adaptor, EpsinR (Hirst et al, 2004), and that the interaction occurs through the folded Habc domain of vti1b (Miller et al, 2007). This has led to the speculation that many post-Golgi Q-SNAREs might have their own dedicated clathrin adaptors. This would allow SNAREs to be packaged into CCVs without having to compete with cargo containing conventional linear sorting signals.
The post-Golgi R-SNAREs, vesicle-associated membrane proteins (VAMPs) 1, 2, 3, 4, 7 and 8, are also enriched in CCVs (Blondeau et al, 2004; Borner et al, 2006; however, most do not have conventional sorting signals. VAMP7 is packaged into CCVs formed at the cell surface through an interaction with the HIV-1 Rev binding protein (HRB). HRB has been shown to bind to both the adaptor protein 2 (AP-2) appendage domain and the folded amino-terminal longin domain of VAMP7, thus sorting VAMP7 into newly forming CCVs (Chaineau et al, 2008; Pryor et al, 2008). Once VAMP7 has been internalized, it is probably sorted from early endosomes into late endosomes or lysosomes through its interaction with AP-3. Again, this interaction is thought to occur through the longin domain of VAMP7 (Martinez-Arca et al, 2003). VAMP4 is most likely packaged into CCVs through its N-terminal dileucine motif. This motif is essential for AP-1 binding in vitro and is required for the steady-state localization of VAMP4 in vivo (Peden et al, 2001; Hinners et al, 2003). However, it is unclear at what step in the trafficking of VAMP4 that this motif is used. VAMPs 1, 2, 3 and 8 do not contain conventional sorting signals or folded domains as they are too small; so how are they packaged into CCVs?
Results
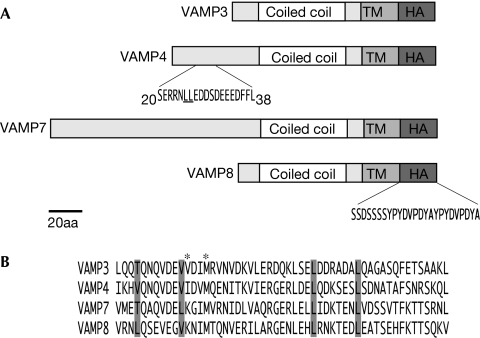
Studying the trafficking of SNAREs is not trivial as expressing and tagging them can cause marked changes in their biology. To overcome this, we used a retroviral system that gives weak-to-moderate expression and screened through various epitope tags, linker domains and antibodies. The R-SNARE constructs used in this study were generated by a two-step PCR that introduced a flexible linker derived from the luminal domain of human VAMP5 and a double haemagglutinin (HA) tag at their carboxy-terminus (Fig 1A). We chose this system because the tagged constructs were efficiently exported from the endoplasmic reticulum, have a localization identical to that of the endogenous SNAREs and were easily detected when expressed at low levels.
Figure 1.
Schematic of VAMP-HA constructs. (A) VAMP-HA constructs were generated by two-step PCR (TM, transmembrane domain; HA-serine linker and HA tag). Numbers indicate amino acids positions. The VAMP4 dileucine motif (underlined) was mutated to AA (V4AA). (B) Alignment of the coiled-coil domains of VAMPs 3–8. Conserved hydrophobic residues are highlighted in grey; these residues were mutated to proline to generate the coiled-coil mutants (P). Residues marked with an asterisk were mutated to AA in VAMP3 (ADIA). HA, haemagglutinin; VAMP, vesicle-associated membrane protein.
VAMP4
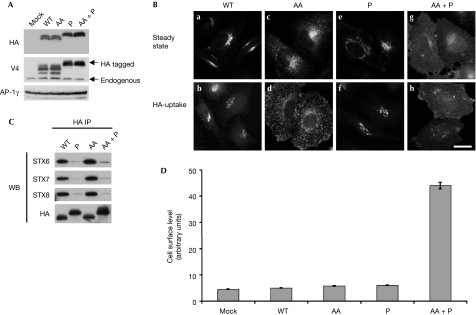
To validate our system, we first characterized the trafficking of VAMP4. VAMP4 has previously been shown to be localized predominantly to the trans-Golgi network (TGN; Steegmaier et al, 1999), is enriched in CCVs and is trafficked through a conserved, conventional dileucine motif in its N-terminal extension (Peden et al, 2001; Zeng et al, 2003). Mutation of the dileucine motif results in a marked alteration of the steady-state distribution of VAMP4 from the TGN to endosomes (Peden et al, 2001). However, it is unclear how this mutation alters the trafficking of VAMP4. To investigate this, we generated stable cell populations expressing either wild-type or dileucine mutant (AA) VAMP4 constructs. The wild-type and dileucine mutant (AA) were both expressed at comparable levels (Fig 2A). As published previously, wild-type VAMP4 is localized predominantly to the TGN and the dileucine mutant (AA) accumulates in endosomal structures, as determined by immunofluorescence microscopy (Fig 2B, compare panel a with c). Interestingly, the dileucine mutant (AA) does not accumulate on the cell surface as measured by flow cytometry (Fig 2D) but is efficiently internalized, as determined through HA antibody uptake (Fig 2B, compare panel b with d). However, the internalized construct fails to be delivered to the TGN (Fig 2B, compare panel b with d). These data also suggest that the VAMP4 dileucine mutant (AA) is being internalized in a new manner.
Figure 2.
VAMP4 can be internalized through its dileucine motif or its coiled-coil domain. (A) Expression levels of VAMP4-HA constructs were determined by Western blotting. Extracts from HeLa cells stably transduced with empty vector (mock), wild-type (WT) and mutant constructs (AA, P and AA+P) were probed with HA, VAMP4 (V4) and γ antibodies (AP-1, γ was used as a loading control). (B) To determine the steady state localization of the VAMP4-HA constructs, immunofluorescence microscopy was performed on fixed cells stained with HA antibodies. For HA-uptake experiments, cells were incubated in the presence of HA antibodies for 4 h at 37°C before being fixed. Scale bar, 10 μm. (C) To measure SNARE complex formation immunoprecipitations (anti-HA) were performed on detergent extracts generated from wild-type and mutant VAMP4-HA cells. Blots were probed with HA, syntaxin (STX) 6, 7 and 8 antibodies. (D) Cell surface levels of VAMP4-HA constructs were measured by flow cytometry. The mean fluorescence for each cell population was calculated. The data shown are the average of three experiments, ±s.e. AA, dileucine mutant; HA, haemagglutinin; IP, immunoprecipitation; P, proline mutant coiled-coil; VAMP, vesicle-associated membrane protein; WB, Western blotting.
Analysis of the protein sequence of VAMP4 did not reveal any other known sorting signals. A possible explanation for this observation is that the VAMP4 dileucine mutant (AA) is being trafficked while it is in a complex with Q-SNAREs. VAMP4 has been shown to form SNARE complexes with syntaxins 6, 13 and vti1a (Brandhorst et al, 2006). Interestingly, these SNAREs are also enriched in CCVs. To investigate this possibility, we introduced proline residues at conserved positions in the coiled-coil domain of VAMP4 (Fig 1B). Analysis of this sequence by secondary structure prediction algorithms indicated that these mutations should disrupt the ability of VAMP4 to form SNARE–SNARE interactions (data not shown). Next, we generated stable cell populations expressing the VAMP4 proline (P) mutant, and a construct that contains both the dileucine and the proline mutations (AA+P). To confirm whether these mutations disrupt SNARE complex formation, immunoprecipitations were performed from these cells and probed for several Q-SNAREs (Fig 2C). The wild-type and dileucine mutant (AA) constructs are able to interact with the Q-SNARE syntaxin 6 as shown previously (Steegmaier et al, 1999). In addition, they are also able to interact with syntaxins 7 and 8. As predicted, the constructs containing the proline mutations (P and AA+P) fail to interact with the Q-SNAREs. We then investigated what effect these mutations had on the trafficking of VAMP4. The VAMP4 proline mutant (P), similar to wild-type VAMP4, is localized to the TGN (Fig 2B, panels e and f) and does not accumulate on the cell surface (Fig 2D). However, the construct containing both the dileucine and proline mutations (AA+P) accumulates on the cell surface (Fig 2D) and fails to be internalized (Fig 2B, panel h). To estimate the cell surface pool of these constructs Proteinase K digestion experiments were performed (supplementary Fig 1A online). VAMP4 wild-type, VAMP4 dileucine (AA) and VAMP4 proline mutant (P) constructs do not have a significant cell surface pool. However, at steady state approximately 48% of the double mutant (AA+P) is redistributed to the cell surface.
VAMPs 3, 7 and 8
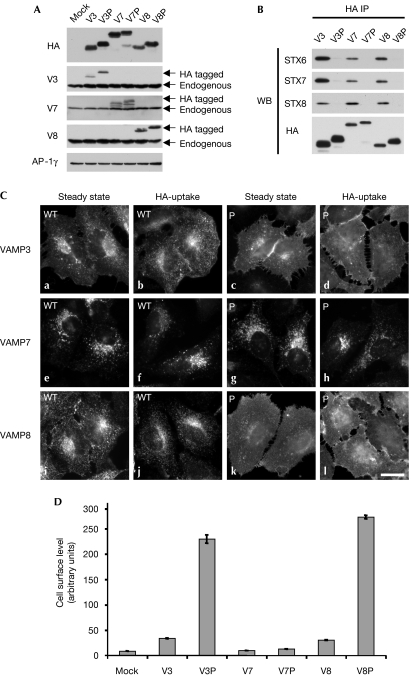
Although the trafficking itinerary of VAMPs 3 and 8 remains unclear, they are probably internalized from the cell surface through CCVs. In agreement with this, depletion of the clathrin heavy chain causes a significant accumulation of VAMPs 3 and 8 on the cell surface (supplementary Fig 2 online). However, it is not known how they are recognized by the machinery of the CCV. We wondered whether they might be sorted through coiled-coil interactions, such as the VAMP4 dileucine mutant (AA). This would allow VAMPs 3 and 8 to be trafficked efficiently and internalized even though they do not have conventional sorting signals. VAMP3 has been shown to form SNARE complexes with syntaxins 2, 6, 7, 13 and Synaptosomal-associated protein 23 (SNAP23; Stow et al, 2006); VAMP8 has been shown to form SNARE complexes with syntaxins 7, 8 and vti1b (Pryor et al, 2004). Interestingly, a significant proportion of these SNAREs—syntaxins 6, 7, 8, 13 and vti1b—has been shown to be enriched in CCVs (Borner et al, 2006). To test whether VAMPs 3 and 8 are being trafficked through their interactions with Q-SNAREs, we inserted proline residues into the conserved positions in their coiled-coil domains (Fig 1B). Analysis of these sequences by secondary structure prediction indicated that these mutations should disrupt the ability of VAMPs 3 and 8 to form SNARE complexes (data not shown). In agreement with these predictions, in vitro SNARE complex formation experiments show that these proline mutations greatly reduce the ability of the mutant R-SNAREs to form SNARE complexes (supplementary Fig 3A,B online). Next, we generated stable cell populations expressing wild-type and proline mutant (P) versions of VAMPs 3 and 8. In addition, we also generated stable cell populations expressing wild-type and proline mutant (P) forms of VAMP7. We predicted that the proline form of VAMP7 should traffic normally if it could engage directly the machinery of the CCV. Constructs of VAMPs 3, 7 and 8 are expressed at levels lower than the endogenus R-SNAREs (Fig 3A). As with the VAMP4 proline mutants, VAMPs 3, 7 and 8 proline mutants (P) fail to co-immunoprecipitate with the Q-SNAREs, syntaxin 6, 7 and 8 (Fig 3B).
Figure 3.
VAMPs 3 and 8 are trafficked through their coiled-coil domains. (A) Expression levels of VAMP-HA constructs were determined by Western blotting. Extracts generated from HeLa cells stably transduced with empty vector (mock), wild-type and proline mutant coiled-coil (P) constructs, were probed with HA, VAMPs 3, 7, 8 and γ antibodies (AP-1, γ was used as a loading control). (B) To measure SNARE complex formation immunoprecipitations (IPs; anti-HA) were performed on detergent extracts generated from wild-type or mutant VAMP-HA cells. Blots were probed with HA, syntaxin (STX) 6, 7 and 8 antibodies. (C) To determine the steady state localization of the VAMP-HA constructs, immunofluorescence microscopy was performed on fixed cells stained with HA antibodies. For HA-uptake experiments, cells were incubated in the presence of HA antibodies for 4 h at 37 °C before being fixed. Scale bar, 10 μm. (D) Cells surface levels of VAMP-HA constructs were measured by flow cytometry. The mean fluorescence for each cell population was calculated. The data shown are the average of three experiments, ±s.e. HA, haemagglutinin; P, proline mutant coiled-coil; VAMP, vesicle-associated membrane protein; WB, Western blotting.
Then we determined where these constructs were localized within the cell. If VAMPs 3 and 8 are normally trafficked through coiled-coil interactions, we would predict that the proline mutant versions of these proteins should be grossly mislocalized. The wild-type VAMP3 construct is localized to the cell surface and endosomes, and is efficiently internalized (Fig 3C, panels a and b) as reported previously (Teter et al, 1998). However, the VAMP3 proline mutant (P) is markedly redistributed to the cell surface (Fig 3C, panel c; 3D) and fails to be internalized (Fig 3C, panel d). As with the VAMP3 proline mutant (P), the VAMP8 proline mutant (P) is markedly redistributed to the plasma membrane (Fig 3C, compare panels i–k; 3D) and fails to be internalized (Fig 3C, compare panels j–l). As predicted, the trafficking of the VAMP7 proline mutant (P) does not seem to be affected as, similar to the wild-type construct, it is localized to late endosomes and does not accumulate on the cell surface (Fig 3C, compare panels e and f with g and h; 3D). To estimate the cell surface pool of these constructs, Proteinase K digestion experiments were performed (supplementary Fig 1B online). VAMP3, VAMP7, VAMP7 proline (P) and VAMP8 constructs only have a modest cell surface pool. However, VAMP3 proline (56%) and VAMP8 proline (58%) were significantly redistributed in the cell surface.
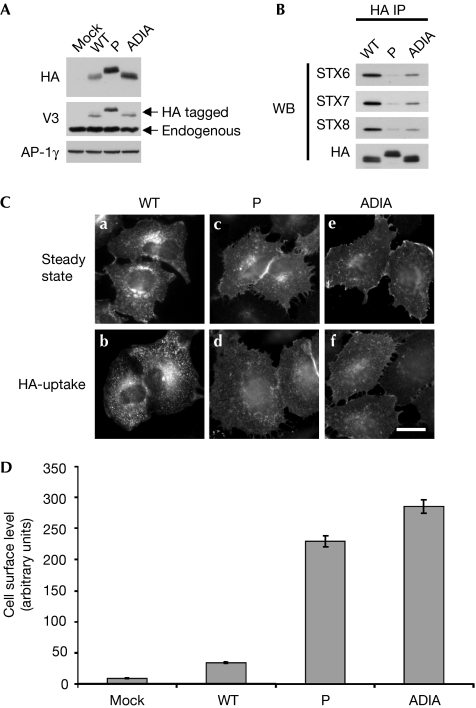
It has been proposed that VAMP2 and the yeast homologues, Snc1/2, contain a putative sorting motif (residues VDIM) in their coiled-coil domains (Grote et al, 1995; Lewis et al, 2000). Mutation of these residues results in the failure of VAMP2 to be packaged into synaptic vesicles and causes the accumulation of Snc1/2 on the cell surface. An alternative explanation of these observations is that the mutation of conserved residues in their coiled-coil domains is likely to disrupt SNARE–SNARE interactions. VAMPs 3, 4, 7 and 8 are highly conserved at these residues (Fig 1B). To determine whether these residues are a sorting signal or are required for SNARE complex formation, we generated a VAMP3 construct (ADIA) that contained the same mutation (26VDIM29 to 26ADIA29) as described previously for VAMP2. Secondary structure prediction for this construct suggested that the ADIA mutation would still be able to form a SNARE complex (data not shown). However, this was not the case as, when in vitro SNARE complex formation experiments were performed, this mutation greatly reduced SNARE complex formation (supplementary Fig 3C,D online). We then generated stable cell populations expressing either wild-type, proline and ADIA constructs. The proline and ADIA mutant constructs are expressed at comparable levels (Fig 4A). To validate the in vitro SNARE complex formation experiments, immunoprecipitations were performed from wild-type, proline and ADIA cell populations and probed for syntaxin 6, 7 and 8. The immunoprecipitations show that the ADIA mutation markedly reduces the ability of VAMP3 to interact with Q-SNAREs in vivo (Fig 4B). Similar to the VAMP3 proline mutant, the ADIA mutant is mislocalized to the cell surface (Fig 4C, panel e; 4D) where it is then unable to be internalized (Fig 4C, panel f).
Figure 4.
Mutation of VDIM to ADIA in the coiled-coil domain of VAMP3 causes defects in SNARE complex formation and trafficking. (A) Expression levels of VAMP3-HA constructs were determined by Western blotting. Extracts generated from HeLa cells stably transduced with empty vector (mock), wild-type (WT), coiled-coil mutant (P) and 26VDIM29 to 26ADIA29 mutant (ADIA) constructs were probed with HA, VAMP3 and γ antibodies (AP-1, γ was used as a loading control). (B) To measure SNARE complex formation immunoprecipitations (IP; anti-HA) were performed on detergent extracts generated from HeLa cells stably expressing wild-type and mutant VAMP3-HA constructs. Blots were probed with HA, syntaxin (STX) 6, 7 and 8 antibodies. (C) To determine the steady state localization of the VAMP3-HA constructs, immunofluorescence microscopy was performed on fixed cells stained with HA antibodies. For HA-uptake experiments, cells were incubated in the presence of HA antibodies for 4 h at 37 °C before being fixed. Scale bar, 10 μm. (D) Cells surface levels of VAMP3-HA constructs were measured by flow cytometry. The mean fluorescence for each cell population was calculated. The data shown are the average of three experiments, ±s.e. HA, haemagglutinin; P, proline mutant coiled-coil; VAMP, vesicle-associated membrane protein; WB, Western blotting.
Discussion
In summary, we have identified a new conserved mechanism for defining SNARE localization. We have shown that coiled-coil interactions are required for the correct localization and internalization of VAMPs 3 and 8, suggesting that they might be trafficked while in a complex with Q-SNAREs. We feel that this hypothesis seems plausible as Q-SNAREs can bind directly to sorting machinery (Hirst et al, 2004) and they are also enriched in CCVs (Peden et al, 2001; Borner et al, 2006). In addition, it has been shown in vitro that the clathrin adaptor, EpsinR, can bind to the Q-SNARE, vti1b, while it is in a SNARE complex (Miller et al, 2007). Packaging of SNARE complexes into CCVs would potentially simplify their trafficking because not all of the SNAREs would require a sorting signal.
At present, we do not know what Q-SNAREs are required for the trafficking of VAMPs 3 and 8. VAMP8 has been shown to form complexes with the endosomal SNARES, syntaxin 7, 8 and vti1b (Pryor et al, 2004). We have also shown that VAMP8 is able to interact with syntaxin 6. Syntaxin 6, 7, 8 and vti1b are all enriched in CCVs (Borner et al, 2006) and syntaxin 6, 7 and 8 are thought to contain conventional sorting signals (Watson & Pessin, 2000; Kasai & Akagawa, 2001). In addition, VAMPs 3 and 8 can also interact with syntaxin 4, which is localized at the plasma membrane (Polgar et al, 2002). However, it is unclear whether this interaction could facilitate their internalization, as it is not known if syntaxin 4 is packaged into CCVs. Owing to the promiscuous nature of SNARE interactions it is likely that VAMPs 3 and 8 are internalized, while they are complexed with several different Q-SNAREs. Therefore it is unlikely that the inhibition of any one of these interactions will cause a block in the internalization. In agreement with this, downregulation of Q-SNAREs does not markedly alter the cell surface levels of the VAMP4 dileucine mutant (supplementary Fig 4 online).
It is also worth noting that our data do not eliminate the possibility that the trafficking defects observed in the proline mutants of VAMPs 3 and 8 might be caused by the disruption of an interaction with an unknown sorting molecule rather than reflecting the loss of an interaction with a Q-SNARE required for their trafficking. This issue is not straightforward to address as mutations that block SNARE–SNARE interactions might also block the binding of an unknown sorting protein. At present, one of the most likely candidates for this unknown sorting molecule is Clathrin assembly lymphoid myeloid leukaemia protein (CALM). CALM is a ubiquitously expressed CCV accessory protein that has been shown to be involved in the trafficking of VAMP2 (Harel et al, 2008). However, depletion of CALM only has a modest effect on the internalization of VAMP2, and it is unclear whether CALM has any role in the trafficking of VAMPs 3 and 8.
Finally, we have shown that the VAMP4 dileucine mutant does not accumulate on the cell surface as reported previously (Tran et al, 2007), but is internalized potentially through Q-SNARE interactions. It is unclear why there is a discrepancy between our data and the work by Tran et al (2007); we are now investigating this in more detail. Interestingly, the internalized dileucine mutant construct fails to be delivered to the TGN, suggesting that the dileucine motif is also required for this step. These data raise the possibility that the AP-1 adaptor complex might have a role in the retrieval of VAMP4 to the TGN as suggested previously (Peden et al, 2001).
Methods
Antibody reagents. Antibodies were either generated in-house or purchased from commercial suppliers. See supplementary information online for additional details.
Constructs and retroviral infections. VAMP-HA constructs were cloned into a modified version of the retroviral expression vector pLXIN (Clontech, Mountain View, CA, USA). Retroviral infections were performed as described previously (Peden et al, 2004). See supplementary information online for additional details.
Western blotting and immunoprecipitations. Cells for Western blotting were resuspended in the extraction buffer, centrifuged at 16,000_g_ for 15 min at 4°C. The supernantants were then normalized for protein concentration (Bradford Assay, BIO-RAD, Hercules, CA, USA), boiled in reducing SDS sample buffer, separated by polyacrylamide gel electrophoresis and analysed by Western blotting. Immunoprecipitations were performed on detergent extracts generated from cell populations pre-treated with _N_-ethylmaleimide (100 μM) for 30 min at 37°C. For additional details see supplementary information online.
Flow cytometry. To measure the cell surface levels of HA-tagged constructs, transduced cell populations were trypsinized, washed in DME and incubated on ice for 1 h with 0.5 μg of anti-HA-Alexa Fluor 647 per 500,000 cells. The samples were then washed two times with DME and resuspended in DME containing the vital stain, 7AAD. The samples were analysed on a BD FACSCalibur (approximately 10,000 cells were gated for forward/side scatter and 7AAD exclusion). The mean fluorescence for each sample was then calculated using the software FlowJo (Tree Star, Inc., Ashland, OR, USA). Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
I thank David Owen, Paul Luzio, Jennifer Hirst and Scottie Robinson for comments on this paper and helpful discussions. Dr Peden was funded by a Career Development Award from the Medical Research Council.
Footnotes
The authors declare that they have no conflict of interest.
References
- Blondeau F et al. (2004) Tandem MS analysis of brain clathrin-coated vesicles reveals their critical involvement in synaptic vesicle recycling. Proc Natl Acad Sci USA 101: 3833–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GH, Harbour M, Hester S, Lilley KS, Robinson MS (2006) Comparative proteomics of clathrin-coated vesicles. J Cell Biol 175: 571–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandhorst D, Zwilling D, Rizzoli SO, Lippert U, Lang T, Jahn R (2006) Homotypic fusion of early endosomes: SNAREs do not determine fusion specificity. Proc Natl Acad Sci USA 103: 2701–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaineau M, Danglot L, Proux-Gillardeaux V, Galli T (2008) Role of HRB in clathrin-dependent endocytosis. J Biol Chem 283: 34365–34373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote E, Hao JC, Bennett MK, Kelly RB (1995) A targeting signal in VAMP regulating transport to synaptic vesicles. Cell 81: 581–589 [DOI] [PubMed] [Google Scholar]
- Harel A, Wu F, Mattson MP, Morris CM, Yao PJ (2008) Evidence for CALM in directing VAMP2 trafficking. Traffic 9: 417–429 [DOI] [PubMed] [Google Scholar]
- Hinners I, Wendler F, Fei H, Thomas L, Thomas G, Tooze SA (2003) AP-1 recruitment to VAMP4 is modulated by phosphorylation-dependent binding of PACS-1. EMBO Rep 4: 1182–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Miller SE, Taylor MJ, von Mollard GF, Robinson MS (2004) EpsinR is an adaptor for the SNARE protein Vti1b. Mol Biol Cell 15: 5593–5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Scheller RH (2006) SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol 7: 631–643 [DOI] [PubMed] [Google Scholar]
- Kasai K, Akagawa K (2001) Roles of the cytoplasmic and transmembrane domains of syntaxins in intracellular localization and trafficking. J Cell Sci 114: 3115–3124 [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HR (2000) Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell 11: 23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Arca S et al. (2003) A dual mechanism controlling the localization and function of exocytic v-SNAREs. Proc Natl Acad Sci USA 100: 9011–9016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SE, Collins BM, McCoy AJ, Robinson MS, Owen DJ (2007) A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles. Nature 450: 570–574 [DOI] [PubMed] [Google Scholar]
- Peden AA, Park GY, Scheller RH (2001) The Di-leucine motif of vesicle-associated membrane protein 4 is required for its localization and AP-1 binding. J Biol Chem 276: 49183–49187 [DOI] [PubMed] [Google Scholar]
- Peden AA, Oorschot V, Hesser BA, Austin CD, Scheller RH, Klumperman J (2004) Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J Cell Biol 164: 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgar J, Chung SH, Reed GL (2002) Vesicle-associated membrane protein 3 (VAMP-3) and VAMP-8 are present in human platelets and are required for granule secretion. Blood 100: 1081–1083 [DOI] [PubMed] [Google Scholar]
- Pryor PR, Mullock BM, Bright NA, Lindsay MR, Gray SR, Richardson SC, Stewart A, James DE, Piper RC, Luzio JP (2004) Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep 5: 590–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor PR, Jackson L, Gray SR, Edeling MA, Thompson A, Sanderson CM, Evans PR, Owen DJ, Luzio JP (2008) Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell 134: 817–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegmaier M, Klumperman J, Foletti DL, Yoo JS, Scheller RH (1999) Vesicle-associated membrane protein 4 is implicated in trans-Golgi network vesicle trafficking. Mol Biol Cell 10: 1957–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow JL, Manderson AP, Murray RZ (2006) SNAREing immunity: the role of SNAREs in the immune system. Nat Rev Immunol 6: 919–929 [DOI] [PubMed] [Google Scholar]
- Teter K, Chandy G, Quinones B, Pereyra K, Machen T, Moore HP (1998) Cellubrevin-targeted fluorescence uncovers heterogeneity in the recycling endosomes. J Biol Chem 273: 19625–19633 [DOI] [PubMed] [Google Scholar]
- Tran TH, Zeng Q, Hong W (2007) VAMP4 cycles from the cell surface to the trans-Golgi network via sorting and recycling endosomes. J Cell Sci 120: 1028–1041 [DOI] [PubMed] [Google Scholar]
- Watson RT, Pessin JE (2000) Functional cooperation of two independent targeting domains in syntaxin 6 is required for its efficient localization in the trans-golgi network of 3T3L1 adipocytes. J Biol Chem 275: 1261–1268 [DOI] [PubMed] [Google Scholar]
- Zeng Q, Tran TT, Tan HX, Hong W (2003) The cytoplasmic domain of Vamp4 and Vamp5 is responsible for their correct subcellular targeting: the N-terminal extension of VAMP4 contains a dominant autonomous targeting signal for the trans-Golgi network. J Biol Chem 278: 23046–23054 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information