Structure of an Arrestin2-Clathrin Complex Reveals a Novel Clathrin Binding Domain That Modulates Receptor Trafficking (original) (raw)
Abstract
Non-visual arrestins play a pivotal role as adaptor proteins in regulating the signaling and trafficking of multiple classes of receptors. Although arrestin interaction with clathrin, AP-2, and phosphoinositides contributes to receptor trafficking, little is known about the configuration and dynamics of these interactions. Here, we identify a novel interface between arrestin2 and clathrin through x-ray diffraction analysis. The intrinsically disordered clathrin binding box of arrestin2 interacts with a groove between blades 1 and 2 in the clathrin β-propeller domain, whereas an 8-amino acid splice loop found solely in the long isoform of arrestin2 (arrestin2L) interacts with a binding pocket formed by blades 4 and 5 in clathrin. The apposition of the two binding sites in arrestin2L suggests that they are exclusive and may function in higher order macromolecular structures. Biochemical analysis demonstrates direct binding of clathrin to the splice loop in arrestin2L, whereas functional analysis reveals that both binding domains contribute to the receptor-dependent redistribution of arrestin2L to clathrin-coated pits. Mutagenesis studies reveal that the clathrin binding motif in the splice loop is (L/I)2G_X_L. Taken together, these data provide a framework for understanding the dynamic interactions between arrestin2 and clathrin and reveal an essential role for this interaction in arrestin-mediated endocytosis.
Many transmembrane signaling systems consist of specific G protein-coupled receptors (GPCRs)3 that transduce a diverse array of extracellular stimuli into intracellular signaling events (1). GPCRs modulate the activity of numerous effector molecules and regulate multiple biological functions including neurotransmission, sensory perception, cardiovascular function, development, and cell growth and differentiation (2). To ensure that extracellular stimuli are translated into intracellular signals of appropriate magnitude and duration, these signaling cascades are tightly regulated. GPCRs are subject to three principle modes of regulation; 1) desensitization, in which a receptor becomes refractory to continued stimuli; 2) endocytosis, where receptors are removed from the cell surface; 3) down-regulation, where total receptor levels are decreased (3, 4). Agonist-dependent regulation is primarily mediated by GPCR kinases that specifically phosphorylate activated GPCRs and initiate the recruitment of arrestins. Arrestins are divided into two major classes, visual and non-visual, based on their localization and function. The non-visual arrestins, arrestin2 and 3 (also termed β-arrestin1 and -2, respectively), are broadly distributed and function in multiple processes including GPCR desensitization, trafficking, and signaling (4–6).
Initial structural insight on arrestins was provided by the x-ray crystal structure of bovine arrestin1 (7, 8), whereas the crystal structures of C-terminal-truncated (9) and wild type (10) bovine arrestin2 and salamander arrestin4 (11) have also been solved. In general, arrestins are composed of two major domains made up of β strands and connecting loops that are held together by a polar core region consisting of buried salt bridges. It has been proposed that arrestins adopt an active conformation upon binding to phosphorylated receptors, which disrupts the polar core resulting in the release of the C-terminal tail (12). Disruption of the polar core by point mutation of Arg-169 generates a constitutively active arrestin2, which mimics the active state. This mutated arrestin binds to the β2-adrenergic receptor (β2AR) in a phosphorylation-independent manner, induces internalization of a δ-opioid receptor lacking phosphorylation sites (13), and has increased binding to clathrin and AP-2 (14).
A role for non-visual arrestins in GPCR endocytosis was first described for the β2AR (15, 16), although it is now evident that arrestins regulate the trafficking of multiple GPCRs as well as additional classes of receptors (4). An early step in this process involves arrestin binding to an activated phosphorylated receptor that enhances arrestin interaction with the endocytic proteins, clathrin, and AP-2 (16, 17). An additional important step in this process involves arrestin interaction with phosphoinositides such as phosphatidylinositol diphosphate and trisphosphate (18). Although the dynamics of these interactions have not been studied, arrestin2 and -3 have been shown to interact specifically and stoichiometrically with clathrin (16). Furthermore, fluorescence microscopy reveals that activated β2AR, arrestin2, clathrin, and AP-2 all colocalize upon receptor stimulation (16). The primary clathrin binding determinant in arrestin2, LIELD, spans residues 376–380 and is located in an extended disordered loop that immediately precedes the final C-terminal β-strand (10, 19). This region, the clathrin binding box, is consistent with a consensus motif, LϕXϕ(D/E) (where ϕ is a bulky hydrophobic residue, and X represents any polar amino acid), established in other clathrin-binding proteins including AP-2 (20), AP180 (21), amphiphysin (22), and epsin (23). Importantly, the mutation of this motif in arrestin3 and its deletion in arrestin2 significantly disrupts clathrin binding and receptor endocytosis (14, 19). A mutagenesis study of clathrin localized an arrestin binding site to the N-terminal domain of the clathrin heavy chain, specifically residues Glu-89, Lys-96, and Lys-98 (24). Moreover, a crystal structure of clathrin-(1–363) in complex with an arrestin3 peptide (residues 369–381) supports the mutagenesis data and the predicted location of the arrestin-clathrin interaction site (25).
To further elucidate the mechanisms involved in mediating arrestin/clathrin interaction, we have determined the crystal structure of clathrin with the short (arrestin2S) and long (arrestin2L) isoforms of arrestin2, which differ by an 8-amino acid insert between β strands 18 and 19 (26). Our results identify an additional and unique interaction encoded in the arrestin2L isoform that is distinct from the previously well characterized interaction involving the LϕXϕ(D/E) motif. Specifically, we observe that the 8 amino acid splice loop in arrestin2L interacts with a pocket formed by blades 4 and 5 in clathrin. Biochemical and cell biological analysis confirm a role for both binding sites in arrestin2L/clathrin interaction and demonstrate an essential role of these interactions in arrestin-mediated GPCR endocytosis.
EXPERIMENTAL PROCEDURES
Purification of Arrestin and Clathrin Terminal Domains
The cDNA of bovine wild type arrestin2L, arrestin2S, arrestin2L-ΔLIELD, arrestin2S-ΔLIELD, arrestin2L-(1–393), and arrestin2S-(1–385) was cloned into pTrcHisB using NcoI and HindIII, which results in removal of the His tag from the vector, and transformed into BL21(DE3)pLysS cells. Each construct was purified as previously described with minor modifications (10, 24). Selenomethionyl-labeled clathrin-(1–363) was prepared and purified similar to that previously described (25, 27) except the cells were grown in a selenomethionine minimum media (28).
Crystallization of Arrestin2-Clathrin Complexes
Arrestin2S-(1–385) and clathrin-(1–363) were mixed to a final concentration of ∼400 μm and centrifuged at 16,000 × g for 30 min at 4 °C to remove any aggregates. Initial screening for crystallization conditions was performed in a 96-well format using screening reagents from Hampton Research (San Diego, CA) and Qiagen (Valencia, CA). Crystals were grown by hanging drop in a 24-well plate with drops consisting of 2.5 μl of the protein mixture, 2 μl of reservoir (0.1 m Bis-Tris propane, pH 7.0, 4–4.5 m ammonium acetate), and 0.5 μl of 45% (v/v) ethylene glycol and equilibration against 0.5 ml of the reservoir for 3 weeks. Crystals were transferred to a new plate and further grown in fresh mother liquor using 60% of the original protein concentration. Crystals of dimensions 200 × 100 × 100 μm were transferred into a solution containing 25% (v/v) ethylene glycol and flash-cooled in liquid nitrogen.
Arrestin2L-(1–393) and selenomethionine-labeled clathrin-(1–363) were mixed at a final concentration of ∼400 μm. Hanging drops were generated by mixing 2.5 μl of the protein complex, 2 μl of the reservoir solution (0.1 m Bicine, pH 9.0, 7% (v/v) _O_-(2-aminopropyl)-O′-(2-methoxyethyl) polypropylene glycol 500, 6% (w/v) polyethylene glycol 8000, 4% (v/v) acetone, and 1% (v/v) ethylene glycol solution), and 0.5 μl of 0.1 m strontium chloride and were equilibrated against 0.5 ml of the reservoir. Crystals with dimensions 400 × 100 × 100 μm grew over a period of 1 week. The crystals were then transferred into a solution containing 25% (v/v) ethylene glycol and flash-cooled in liquid nitrogen.
Data Collection, Structure Determination, and Refinement
The diffraction quality of the crystals was analyzed using an Xcalibur PX Ultra diffractometer (Oxford Instruments, Concord, MA). Both data sets were collected at the National Synchrotron Light Source, beamline X4A, using an ADSC Quantum 4 CCD detector. The crystals were cooled to 100 K during collection. Anomalous data for the arrestin2L and clathrin crystals were collected using inverse beam geometry (29). Data were indexed and scaled using HKL2000 package (30). The B factor from the Wilson plot for the Arr2S/clathrin data was 24.9 Å2. The resolution maximum for the Arr2L/clathrin data was too low to derive a meaningful B-factor from the Wilson Plot.
For the arrestin 2L-clathrin complex, the multiwavelength anomalous diffraction phases were obtained using SOLVE/RESOLVE (31). 10 of 14 sites within clathrin were identified with the initial figure of merit of 0.47, and each site had a height-to-σ ratio better than 9. After density modification using Resolve, the figure of merit was 0.75 with a correlation coefficient of 0.84. At this point, clathrin and arrestin (stripped of water and cofactors) were placed in the unit cell using 1C9I and 1JSY as models. After an initial refinement using Refmac (F(s)/σ > 0), the splice loop was visible and subsequently built into the model. Iterative cycles of model building and refinement were carried out using CNS (32), CCP4 (33), Refmac (34), O (35), and COOT (36).
Molecular replacement for the Arr2S/clathrin structure was performed using Phaser (37) and Amore (38) with search models PDB codes 1C9I for clathrin and 1ZSH for arrestin. Solvent and heteroatoms were removed. The clathrin molecule was placed first. The top rotation Z score using data from 15 to 3 Å was 15.8 with a translation Z score of 31.0. The initial R-factor using data from 30 to 2.2 Å was 48.3%. Using Amore, a similar solution was found. The rotational correlation coefficient was 18.6%, and the R-factor was 53.6%. The next best solution was 15.9 and 54.4%, respectively. After translation and fitting, the correlation coefficient and R-factor values were 41.6 and 47.2%, respectively. The arrestin model was placed next. The rotation Z score was 12.3 for the top solution with a translation Z score of 23.3. Using Amore, the top solution produced a correlation coefficient of 18.1% and an R-factor of 53.4%. The next best solution had a correlation coefficient of 13.1% and an R-factor of 54.4%. After rigid body fitting the values were 34.6 and 48.0%. The combination of the two molecules produced few side chain clashes and had an initial R-factor of 40.0%. Iterative cycles of model building and refinement were carried out using CNS (32), CCP4 (33), Refmac (34), O (35), and COOT (36).
To differentiate the interaction of the clathrin binding box of arrestin with clathrin, the splice loop in Arr2L with clathrin, and adventitious crystal lattice contacts, we used the PISA server to estimate the free energy of each potential interface (EMBL-EBI). First, we noted that the calculated free energy of the clathrin binding box and clathrin from either Arr2S (Δ_G_ = −6.9 kcal/mol) or Arr2L (Δ_G_ = −6.5 kcal/mol) compared well to calculated values from the published structure, 1C9I (Δ_G_ = −6.7 and −6.2 kcal/mol, two molecules in the asymmetric unit). Next, we observed that the interaction between the splice loop of Arr2L and clathrin had a free energy of Δ_G_ = −4.1 kcal/mol. In this analysis, we also found an interface between the two arrestin molecules, Δ_G_ = −6.5 kcal/mol. This involved a symmetric interaction where the loops located at the C-terminal domain of each arrestin interacted with loops at the C- and N-terminal bridge. This interface is also observed in the bovine arrestin crystal structure, 1G4M but not in the 1ZSH or 1JSY structures. Although equivalent in terms of calculated free energy, the interaction between the clathrin binding box or the Arr2L splice loop and clathrin was free of salt bridges. The interfaces observed in the arrestin-only structures differed in that they contained a substantial number of salt bridges. Although we have performed site-directed mutagenesis vis à vis the splice loop and clathrin (and these support that the interaction is biologically relevant), we have not made point mutations within the other interfaces to substantiate whether the increase in the number of salt bridges is consistent with lattice contacts or is biologically significant. It is important to note, however, that sedimentation equilibrium experiments of the individual components suggest that each is predominantly monomeric.
Biochemical Glutathione S-Transferase (GST) Pulldown Assays
Purification and assay procedures were used as described previously (14). Wild type and mutant GST-clathrin-(1–363) were expressed and purified on glutathione-agarose beads. Wild type and mutant arrestin2 were purified as described previously (10) and diluted to a concentration of 10 μm. Briefly, 75 pmol of GST and GST-clathrin (1–363) and 10 pmol of purified arrestin were mixed in a total volume of 200 μl of binding buffer (final concentrations of 375 and 50 nm, respectively), incubated, washed, and eluted by adding SDS sample buffer. The samples were resolved by SDS-PAGE and then transferred to nitrocellulose. The amount of arrestin bound to GST-clathrin was quantitated using an Odyssey infrared scanner (Li-Cor Biotechnology, Lincoln, NE) with an arrestin polyclonal antibody and rhodamine-conjugated anti-rabbit IgG (Rockland Immunochemicals, Gilbertsville, PA). All GST pulldown experiments were repeated >10 times and analyzed using Prism (GraphPad, San Diego, CA). p values of unpaired t tests are shown in bar graphs.
The C-terminal tail (residues 319–418) of arrestin2L-ΔLIELD with BamHI and SmaI sites at the 5′ and 3′ ends, respectively, was generated by PCR and cloned into pGEX-4T-2 (GE Healthcare). A series of single mutations was made using QuikChange (Stratagene, La Jolla, CA). For constructs containing multiple mutations, a silent mutation was generated in pGEX-4T-2-arrestin2L-(319–418)-ΔLIELD to create an XbaI site, and Xbal-SmaI-treated PCR products were subcloned into pGEX-4T-2. All constructs were verified by nucleotide sequencing. Wild type and mutant GST-arrestin2L-(319–418)-ΔLIELD were expressed and purified on glutathione agarose beads. Clathrin-(1–363) was purified as described above and diluted to a concentration of 10 μm. Briefly, 150 pmol of GST or GST-arrestin2L-(319–418)-ΔLIELD and 20 pmol of purified clathrin-(1–363) were mixed in a total volume of 200 μl of binding buffer (final concentrations of 750 and 100 nm, respectively), incubated, washed, and eluted by adding SDS sample buffer. The samples were resolved by SDS-PAGE and transferred to nitrocellulose. The amount of clathrin bound to GST-arrestin2L-(319–418)-ΔLIELD was detected using anti-clathrin monoclonal antibodies (BD Biosciences) and quantified using an Odyssey scanner.
Binding Analysis by Surface Plasmon Resonance
Purified arrestin2L, arrestin2S, and arrestin2S-ΔLIELD were coupled to separate cells of a CM5 sensor chip using an amine coupling kit and BIAcore 3000 instrument (GE Healthcare). Maximum immobilization of arrestins (about 10,000 response units) was obtained with a programmed procedure from the instrument. Clathrin heavy chain was purified from bovine calf brain (Pel-Freeze, Rogers, AK) as previously described (16). Sedimentation velocity experiments using a ProteomeLab XL analytical ultracentrifuge (Beckman Coulter, Fullerton, CA) demonstrated that purified clathrin heavy chain had a sedimentation coefficient of 6.6 S (193-kDa molecular mass) with a minor contamination of clathrin light chain, and adaptin subunits were determined using Sedfit (NIH, Bethesda, MD). Binding experiments were performed using purified clathrin (125 nm to 2 μm) in a running buffer (0.5 m Tris, 0.5 m MES, pH 7.0) with 30 μl/min flow rate at 25 °C. The chip was regenerated with 45 μl of 10 mm glycine, pH 5.6, with a flow rate of 90 μl/min after each injection. A cell labeled with arrestin2S-ΔLIELD was used as a negative control.
Total Internal Reflection Fluorescence (TIRF) Microscopy and Image Analysis
Arrestin constructs were C-terminal-tagged with enhanced GFP as described previously (39) and expressed by transient transfection using Effectene (Qiagen) of a previously characterized clone of stably transfected HEK293 cells expressing FLAG-tagged β2AR (40). Cells were imaged 72–96 h after transfection, and GFP fluorescence was used to select cells expressing the indicated arrestin-GFP constructs at similar levels for subsequent analysis. M1 anti-FLAG monoclonal antibody, conjugated to Alexa 555 (Invitrogen), was used to specifically label FLAG-β2AR present in the plasma membrane as described previously (40). Live imaging was carried out using a Nikon TE-2000 microscope with a 100 × 1.49 NA TIRF objective, equipped for dual color through-the-objective TIRF illumination. 488- and 568-nm laser lines were used as light sources for imaging arrestin-GFP and Alexa555-FLAG-β2AR, respectively. Sequential images were collected every 3 s using a Cascade II EM-CCD camera operated in the electron multiplying mode and in the linear range of detection (Photometrics, Tucson, AZ). The collected images were exported as stacks of 16-bit TIFF images and analyzed using ImageJ. To measure the magnitude of enrichment of arrestin in individual clusters, an average projection was made of 30 sequential frames from each series, starting from when clustering was first apparent. A circular mask three pixels in diameter, which corresponded to the average apparent size of a cluster projected on the CCD, was used to manually select each cluster. In cases where arrestin clusters were not apparent (because of low levels of net concentration), clusters were selected based on receptor fluorescence. The average arrestin fluorescence in each cluster was measured and normalized to the average fluorescence observed in the cell before agonist addition. To measure the rate of enrichment of arrestin in clusters, the whole cell was selected, and the maximum arrestin fluorescence was calculated in all frames of the stack. In the same cells, to estimate the loss of receptors from the plasma membrane outside of clusters, the minimum receptor fluorescence was similarly calculated. Data analysis and graphing were done using Excel (Microsoft, Redmond, WA) and Prism (GraphPad, La Jolla, CA).
RESULTS
Cellular Function of Arrestin2S and Arrestin2L, the Two Splice Forms of Arrestin2
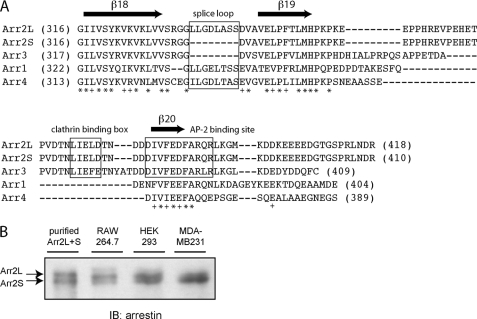
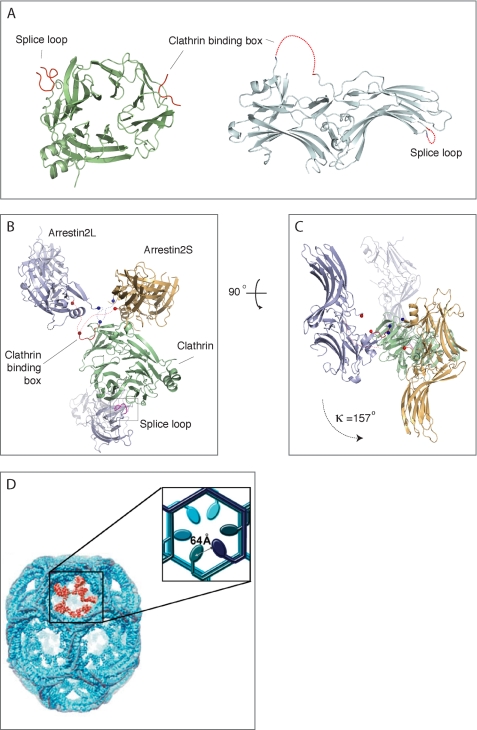
Previous studies identified two splice forms of arrestin2 (termed long (L) and short (S)) that differ by an 8-amino acid insert between β strands 18 and 19 in arrestin2 (26) (Fig. 1A). Although there was no significant difference in the ability of these isoforms to bind to the β2AR and M2 muscarinic acetylcholine receptors (41), there appeared to be differential expression of these proteins. Arrestin2S was found primarily in the lung, liver, spleen, kidney, and pituitary, whereas arrestin2L was expressed in the retina, pineal, and various regions of the brain (26). To extend these studies we characterized arrestin2S and 2L expression in a few commonly used cell lines. HEK293 cells appear to have comparable levels of arrestin2S and 2L, whereas Raw264.7 macrophages primarily express arrestin2L, and MDA-MB 231 breast cancer cells primarily express arrestin2S (Fig. 1B).
FIGURE 1.
Sequence alignment of arrestins and analysis of arrestin2 expression. A, sequence alignment of bovine visual arrestins (arrestin1 and -4) and non-visual arrestins (arrestin2L, -2S, and -3) was performed by ClustalW (EMBL-EBI). The LϕXϕ(D/E) motif, splice loop, and AP-2 binding site are boxed, and at the bottom of the alignment, identical residues are noted with an asterisk, and conserved residues are noted with a +. B, purified arrestin2L and arrestin2S were electrophoresed along with lysates from RAW 264.7 (0.5 μg), HEK293 (30 μg), and MDA-MB 231 (30 μg) cells on an 8% polyacrylamide gel using a large format vertical gel (Bio-Rad). Proteins were transferred to nitrocellulose paper and detected using an arrestin peptide polyclonal antibody (58). IB, immunoblot.
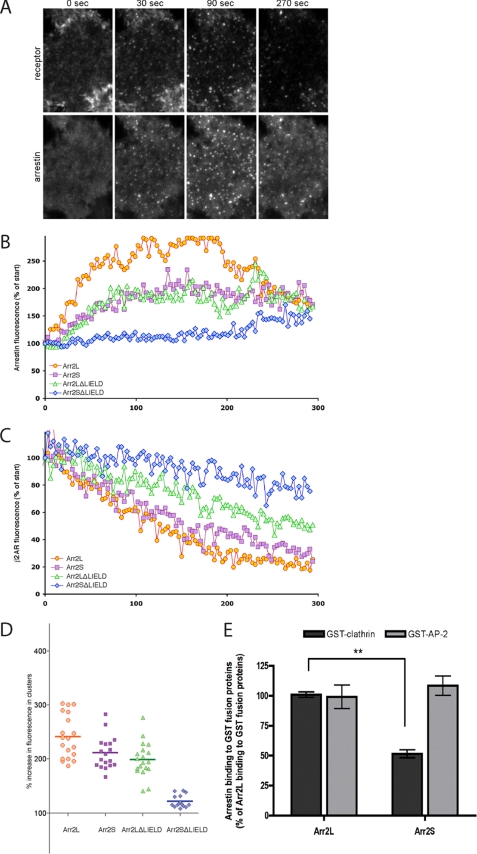
To evaluate potential biological differences between arrestin2S and -2L, we examined arrestin and GPCR trafficking in living cells using TIRF microscopy to selectively visualize endocytic events occurring in the plasma membrane (40). HEK293 cells stably expressing a FLAG-tagged β2AR were transiently transfected with GFP-tagged arrestin constructs, and GFP fluorescence intensity was used to select cells expressing similar amounts of mutant arrestin for analysis. Receptors present in the plasma membrane were labeled with Alexa555-conjugated anti-FLAG monoclonal antibody, and the β-adrenergic agonist isoproterenol was added to the imaging medium to promote receptor activation. Agonist-induced redistribution of arrestin-GFP and internalization of surface-labeled FLAG-β2AR were visualized in parallel by dual channel TIRF imaging (Fig. 2A). Arrestin2L-GFP exhibited rapid and robust redistribution to clathrin-coated pits upon receptor activation, whereas arrestin2S-GFP redistributed less extensively (Fig. 2B). We also examined arrestins lacking the clathrin binding box (LIELD) to evaluate the effect of disrupting clathrin binding on arrestin redistribution. Interestingly, arrestin2L-ΔLIELD-GFP effectively redistributed to clathrin-coated pits, whereas arrestin2S-ΔLIELD-GFP was greatly impaired in agonist-induced redistribution (Fig. 2B). These changes correlated well with differences in β2AR internalization, indicated by the disappearance of Alexa555 fluorescence, which was most extensive in cells expressing arrestin2L, reduced in arrestin-2S-expressing cells, further reduced in arrestin2L-ΔLIELD cells, and largely absent in cells expressing arrestin2S-ΔLIELD (Fig. 2C). Further analysis of arrestin-GFP dynamics was performed by quantifying arrestin-GFP concentration in discrete fluorescent clusters representing individual clathrin-coated pits and then compiling averaged results from individual coated pit measurements across multiple cells (Fig. 2D). Arrestin2L showed an ∼2.4-fold enrichment in coated pits upon receptor activation, arrestin2S was ∼2.1-fold, arrestin2L-ΔLIELD was ∼2.0-fold, and arrestin2S-ΔLIELD produced a nearly complete loss of arrestin enrichment after agonist-induced activation of receptors (Fig. 2D). These results reveal that arrestin2L effectively localizes in clathrin-coated pits even in the absence of the clathrin binding box, thus demonstrating that the eight-amino acid splice loop can mediate arrestin redistribution to coated pits.
FIGURE 2.
Recruitment of various arrestins to clathrin-coated pits upon GPCR activation. A, montage of FLAG-β2AR and arrestin2S-GFP localization by TIRF microscopy from 0 to 270 s after agonist treatment. B, fluorescence traces from example cells showing the rate of enrichment of the indicated versions of arrestin2-GFP in clusters. The maximum arrestin fluorescence, normalized to the fluorescence before agonist addition, is plotted over time. C, the rate of depletion of surface receptor fluorescence in the same cells, normalized to the surface fluorescence before agonist addition, is shown. β_2-AR_, β2AR. D, the average increase of arrestin2-GFP fluorescence in clusters was calculated per cell (n > 16 cells in each case) and is shown with the mean value. E, binding of purified wild type arrestin2L (Arr2L) and arrestin2S (Arr2S) to GST-clathrin-(1–363) and GST-β2-adaptin-(700–937) was detected and quantified using the Odyssey system. The bars represent the mean ± S.E. from n independent experiments: GST-clathrin (50) and GST-β2-adaptin (13) with Arr2L and Arr2S.
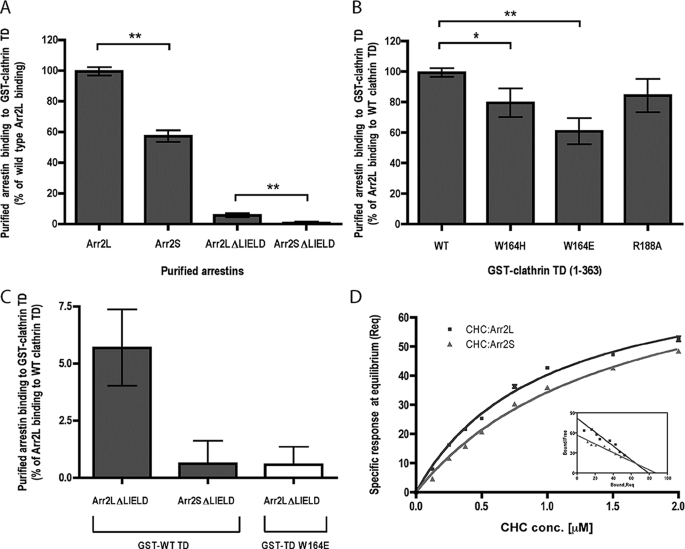
The non-visual arrestins have been shown to bind to clathrin and the β-adaptin subunit of AP-2, and both of these interactions are critical for arrestin-promoted endocytosis of GPCRs (14). To test whether there are differences in arrestin2L and 2S in clathrin or AP-2 binding, we evaluated the ability of the arrestins to bind to GST-clathrin-(1–363) and GST-β2-adaptin-(700-937). These studies reveal a significant difference in binding to clathrin with arrestin2S binding only ∼50% as well as arrestin2L (Fig. 2E). In contrast, there was no difference in the ability of these proteins to bind to β2-adaptin (Fig. 2E).
Molecular Structure of Arrestin Isoforms and Clathrin Complexes
We next focused on evaluating the interaction of various arrestin2 and clathrin variants. C-terminal-truncated arrestin2S (residues 1–385) and arrestin2L (residues 1–393) were found to bind better to GST-clathrin-(1–363) compared with full-length arrestin2 (supplemental Fig. 1_A_ and data not shown), whereas arrestin2L-(1–393) bound better to GST-clathrin-(1–363) than to either GST-clathrin-(1–494) or GST-clathrin-(1–579) (supplemental Fig. 1_B_). The binding of C-terminal-truncated arrestin2S and arrestin2L to clathrin-(1–363) also resulted in a mobility shift on native polyacrylamide gels, suggesting formation of a stable arrestin-clathrin complex (supplemental Figs. 1, C and D).
To provide molecular insight into arrestin2-clathrin interaction, we obtained crystals of both isoforms of arrestin2 bound to clathrin. In both crystal forms we observe that the clathrin binding box of arrestin2 interacts with clathrin between blades 1 and 2. Interestingly, in the long isoform of arrestin2, we observed a second novel interaction through the eight-amino acid splice loop that is uniquely present in arrestin2L and a shallow hydrophobic pocket in clathrin formed by blades 4 and 5. Below, we first describe the interaction between the short form of arrestin2 and clathrin followed by the long form of arrestin2 and clathrin.
Arrestin2S-Clathrin Complex
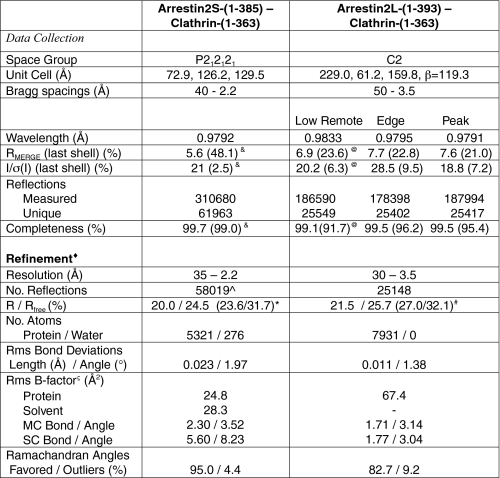
Initially, a small crystal of arrestin2S-(1–385)-clathrin-(1–363), obtained by mixing the proteins at a 1:1 molar ratio, was used for macroseeding and yielded an orthorhombic crystal of P212121 space group (a = 72.9 Å, b = 126.2 Å, c = 129.5 Å) that diffracted to Bragg spacings of 2.2 Å (Table 1). The structure of the complex was determined by molecular replacement using the Phaser and Amore programs with search models of 1ZSH for arrestin2 (10) and 1C9I for clathrin (25). The molecular structure of the arrestin2S-clathrin complex shows a 1:1 interaction with a topology that positions clathrin on arrestin2 opposite to the saddle regions involved in receptor binding (Fig. 3A, supplemental Fig. 2). The solvent content is 65%, and the average B-factor for the arrestin2S-clathrin complex is 24.8 Å2. The average B-factor for the solvent is 28.3 Å2. There are no significant conformational changes in the clathrin or arrestin compared with structures of the apo forms (1C9I and 1JSY, respectively). The mean deviation is 0.49 Å for 354 Cα atoms in clathrin and 1.96 Å in arrestin for 315 Cα atoms. The mean deviation of the N-terminal domain of arrestin2 (residues 5–173) to 1JSY and other published arrestins (1G4M and 1ZSH) is less than 1.0 Å, whereas the mean deviation for the C-terminal domain (residue 174–345) ranges from 1.8 to 2.5 Å. The loops spanning residues 65–72, 90–95, and 130–138 is disordered and not clearly visible in the maps.
TABLE 1.
Crystallographic data and refinement statistics
& indicates 2.20–2.25 Å; @ indicates 3.49–3.61 Å; * indicates 2.20–2.24 Å; # indicates 3.50–3.55 Å. ∧, does not include reflections held separate for Rfree calculations; ♦, data cutoff used in refinement: |Fobs|/σ(F) > 0; ς B factor from Wilson Plot of Arrestin2S-Clathrin is 24.9 (Å2).
FIGURE 3.
Structure of an arrestin2S-(1–385) complex with clathrin-(1–363). A, ribbon model of the complex shows a molecule of arrestin2S-(1–385) (wheat) bound to a molecule of clathrin-(1–363) (green) using the LϕXϕ(D/E) motif (red). Unstructured areas of the C-terminal loop are depicted as dotted lines, and each end-point of the area is marked with colored spheres. The red spheres connect the C terminus of residue 349 to residue 366 (∼27 Å), and the blue spheres connect residues 372 and 378 (∼14 Å). Although it is possible that the clathrin binding box originates from one arrestin and extends to an adjacent crystallographic arrestin (as there is no clear electron density for residues 350–365), the distance between the termini of the arrestin core and the clathrin binding box suggests that the loop originates and returns from the same arrestin core. B, stereoview of the interface between arrestin2S-(1–385) and clathrin. Residues of the LϕXϕ(D/E) motif and some of the key residues in the hydrophobic pocket of clathrin are numbered as black and green, respectively. C, B is depicted as a one-dimensional map with possible hydrogen bonding (dotted lines) and hydrophobic interactions (bold dotted lines).
The extended clathrin binding box, TNLIELD, which lies in a large loop between β strands 19 and 20 of arrestin, is clearly visible in the difference maps and was built into the density using the arrestin3 peptide sequence (25) as a guide. There was, however, no clear connection between the clathrin binding box and the core of arrestin (at contour levels of σ > 0.5) (Fig. 3A). The detailed structure of the binding interface shows Ile-369 and Leu-371 in the LϕXϕ(D/E) motif of arrestin2 interacting with a hydrophobic pocket formed by blades 1 and 2 of the β-propeller structure of the clathrin terminal domain (Figs. 3, B and C). The root mean square difference of all atoms between the clathrin binding boxes of arrestin2 and arrestin3 bound to clathrin is 0.53 Å. There was a slight shift in the backbone position of Leu-371 of arrestin2 in our structure compared with the arrestin3 peptide (25). This difference likely stems from differences in the Leu and Phe side chains and potentially alters the interaction of Asp-372 with clathrin. In addition, Asn-367, the residue N-terminal to the LϕXϕ(D/E) motif, forms a hydrogen bond with Gln-89 in clathrin and together with hydrogen bonds between Asp-372 in arrestin2 and Lys-96 in clathrin helps to position the hydrophobic interface between the proteins.
Arrestin2L-Clathrin Complex
The long isoform of arrestin bound to clathrin produced a monoclinic crystal of C2 space group (a = 229.0 Å, b = 61.2 Å, c = 159.8 Å, β = 119.3°) that diffracted well to 3.5 Å (Table 1). To obtain experimental phasing, we labeled the clathrin with selenomethionine because it contains 14 methionines (compared with 4 for arrestin2L). The initial electron density maps were readily interpretable and indicated an unusual 2:1 arrestin to clathrin stoichiometry. The solvent content was 68%, and the average B-factor for the arrestin2L/clathrin complex was 67.4 Å2 (Table 1). To facilitate the model building, we placed the arrestin2 model (1JSY) and clathrin model (1C9I) in the experimental electron density maps. We observed minor differences between the core regions of each component (e.g. no large conformational changes between the arrestin lobes). The mean deviation between Cα carbons of clathrin from the complex and 1C9I was 0.50 Å for 357 residues, for one arrestin2L in the asymmetric unit and 1JSY, it was 1.03 Å for 339 atoms, and for the second arrestin2L in the asymmetric unit and 1JSY it was 1.01 Å for 300 residues. Similar to the arrestin2S-clathrin structure, a number of loops in the arrestin models are mostly disordered, and despite extensive attempts to model these loops at lower contour levels, their inclusion invariably led to worse refinement statistics and afforded stereochemical violations. Thus, we limited our model to residues that are clearly defined in the electron density maps at 1σ or greater.
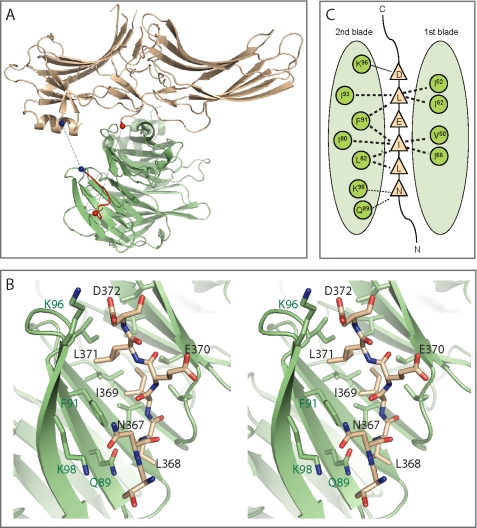
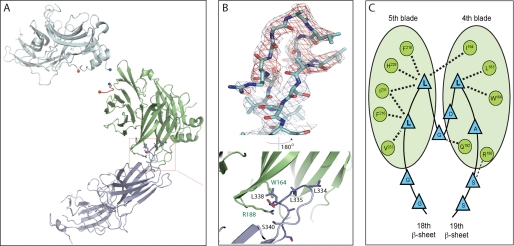
The refined structure shows that one arrestin molecule interacts with clathrin via the LϕXϕ(D/E) motif and blades 1 and 2 and that the other arrestin interacts with clathrin through the 8-amino acid splice loop uniquely present in arrestin2L and a shallow hydrophobic pocket on clathrin formed by blades 4 and 5 (Figs. 4, A and B). The interface between the additional loop in arrestin2L and blades 4 and 5 of clathrin is primarily because of hydrophobic interactions. Residues Gly-333, Leu-335, Gly-336, Asp-337, Leu-338, and Ser-340 in arrestin2L and Trp-164, Leu-183, Arg-188, Val-190, Gln-192, Ile-194, Ile-231, Glu-232, Thr-235, and Lys-245 in clathrin define the solvent excluded interface (using a cut-off of 1.4 Å radius for water) (Fig. 4C). As depicted in Fig. 4A, the two arrestin binding sites are on opposing sides of the clathrin β-propeller domain. Likewise, the clathrin binding loops of arrestin are also on the opposing domains. This geometry suggests that each site is independent and that these sites are functionally relevant in higher order structures (e.g. clathrin-coated pits). It is likely that the observed 2:1 arrestin2L to clathrin stoichiometry was because of crystal packing and that this would be different in cells.
FIGURE 4.
Structure of an arrestin2L-(1–393) complex with clathrin-(1–363). A, ribbon model of the complex shows a molecule of clathrin-(1–363) (green) with two molecules of arrestin2L-(1–393) (cyan) using two independent binding interfaces. The boxed area (red) is an additional interface using the splice loop of arrestin2L. B, multiwavelength anomalous diffraction electron density maps (σ = 1.5) showing the splice loop of arrestin2L (top) and detailed structure of the boxed area from A with labeled key residues of arrestin2L and clathrin (bottom; black and green, respectively). C, B is depicted as a one-dimensional map noting possible hydrogen bonding (dotted lines) and hydrophobic interactions (bold dotted lines).
Interaction of Arrestin2 and the Clathrin Terminal Domain
The interaction between clathrin and the eight-amino acid splice loop in arrestin2L was further tested using biochemical approaches. This was done by analyzing the ability of various purified arrestins to bind to GST-clathrin-(1–363) (Fig. 5A). The absence of the splice loop in arrestin2S reduced clathrin binding by ∼45%, whereas deletion of the LϕXϕ(D/E) motif from arrestin-2L (Arr2L-ΔLIELD) reduced clathrin binding by ∼94%. Deletion of the splice loop and LϕXϕ(D/E) motif (Arr2S-ΔLIELD) caused a >99% loss of clathrin binding (Figs. 5, A and C). These data demonstrate that the splice loop in arrestin2L is a functional clathrin binding site, although the LϕXϕ(D/E) motif appears to be the major site for clathrin binding in arrestin2L.
FIGURE 5.
Binding of arrestin2 to wild type and mutated GST-clathrin-(1–363). A, binding of purified wild type arrestin2L (Arr2L), arrestin2S (Arr2S), arrestin2L-ΔLIELD (Arr2L_Δ_LIELD), and arrestin2S-ΔLIELD (Arr2S_Δ_LIELD) to GST-clathrin-(1–363) was detected and quantified using the Odyssey system. The bars represent the mean ± S.E. from n independent experiments: arrestin2L (53), arrestin2S (71), arrestin2L-ΔLIELD (67), and arrestin2S-ΔLIELD (40). Statistical analysis was performed using an unpaired t test (**, p < 0.001) using Prism. TD, terminal domain. B, binding of purified wild type arrestin2L to wild type or mutated (W164H, W164E, and R188A) GST-clathrin-(1–363) was performed as described under “Experimental Procedures.” The bars represent the mean ± S.E. from 14 independent experiments. Statistical analysis was performed using an unpaired t test (*, p < 0.005; **, p < 0.001) using Prism. C, binding of purified arrestin2L-ΔLIELD and arrestin2S-ΔLIELD to wild type (gray bar, data are from Fig. 5A) or W164E mutant GST-clathrin-(1–363) (white bar). Note the difference in the y axis scale in A and C. D, specific binding of clathrin heavy chain (CHC) to arrestin2L (Arr2L) and 2S (Arr2S) at steady-state level (Req) was measured by BIAevaluation 3.0 and analyzed as a Scatchard plot (inset). Kd values for arrestin2L (0.98 ± 0.01 μm) and arrestin 2S (2.1 ± 0.4 μm) were determined using a steady-state affinity model from 3 independent experiments.
To confirm a role for the arrestin2L splice loop in clathrin binding, point mutations were generated in the clathrin terminal domain at the residues that mediate interaction with the loop based on the structural model (Fig. 4B). Mutation of Trp-164 in clathrin to a histidine (W164H) or glutamate (W164E) resulted in a 20–40% decrease, respectively, in arrestin2L binding to GST-clathrin-(1–363), whereas mutation of Arg-188 in clathrin to an alanine (R188A) caused an ∼15% decrease in arrestin2L binding (Fig. 5B). Because our structural model and biochemical studies suggest two independent interactions between arrestin2L and clathrin, we also tested whether the ability of arrestin2L-ΔLIELD to bind to clathrin can be further reduced by mutation of Trp-164 in clathrin. This complementary disruption between arrestin2L and clathrin completely abolished arrestin2L-ΔLIELD binding to clathrin and provided the same loss as arrestin2S-ΔLIELD binding to wild type clathrin (Fig. 5C). We also compared the ability of arrestin2S and arrestin2L to bind to purified clathrin using surface plasmon resonance. These studies revealed an ∼2-fold difference in affinity with arrestin2L binding with a Kd = 0.98 ± 0.01 μm, whereas arrestin2S bound with a Kd = 2.1 ± 0.4 μm (Fig. 5D). Although these studies suggest that the eight-amino acid insert in arrestin2L contributes to clathrin binding, we believe that this is a low affinity site and is unlikely to be saturated in any of our assays except for the crystallography. The lack of two-site binding in our surface plasmon resonance results with arrestin2L may reflect the lack of saturation of the low affinity site or the possibility that a 1:1 complex is achieved when both sites are occupied.
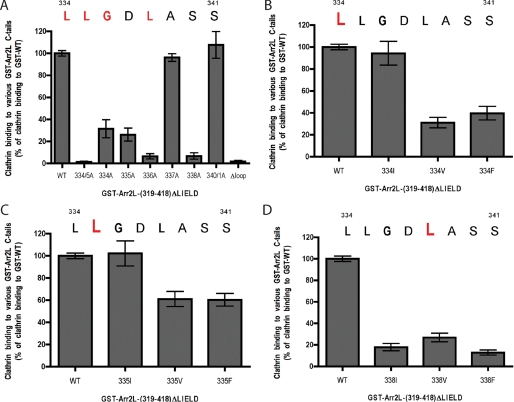
To define the specific residues in the eight-amino acid insert in arrestin2L that mediate clathrin binding, we studied clathrin binding to a GST fusion protein containing residues 319–418 of arrestin2L with the clathrin binding box deleted. Clathrin binds effectively to this construct, and this binding was completely dependent on the eight-amino acid insert (Fig. 6A, compare WT versus Δloop). Mutation of Leu-334 and Leu-335 together completely disrupted clathrin binding, whereas mutation of the individual leucines reduced binding by ∼70% (Fig. 6A). Similarly, G336A and L338A mutations also completely disrupted binding, whereas mutation of Asp-337 or Ser-340 and -341 had no effect on clathrin binding. Additional mutagenesis of the leucine residues revealed that Ile could effectively substitute for Leu-334 (Fig. 6B) and Leu-335 (Fig. 6C), whereas Ile, Val, or Phe could not substitute for Leu-338 (Fig. 6D). Taken together, these studies demonstrate that (L/I)2G_X_L (where X is any amino acid) functions as a clathrin binding motif in arrestin2L.
FIGURE 6.
Binding of clathrin terminal domain to wild type and mutated GST-arrestin2L-(319–418)-ΔLIELD. A, binding of purified clathrin-(1–363) to wild type or mutated GST-arrestin2L-(319–418)-ΔLIELD was detected and quantified using the Odyssey system. The bars represent the mean ± S.E. from n independent experiments: WT (52), L334/5A (22), L334A (32), L335A (40), G336A (16), D337A (19), L338A (24), S340/1A (15), and Δloop (19). B, binding of purified clathrin-(1–363) to wild type or mutated GST-arrestin2L-(319–418)-ΔLIELD was detected and quantified as in panel A. The bars represent the mean ± S.E. from n independent experiments: WT (20), L334I (9), L334V (6), and L334F (8). C, binding of purified clathrin-(1–363) to wild type or mutated GST-arrestin2L-(319–418)-ΔLIELD was detected and quantified as in panel A. The bars represent the mean ± S.E. from n independent experiments: WT (20), L335I (12), L335V (8), and L335F (8). D, binding of purified clathrin-(1–363) to wild type or mutated GST-arrestin2L-(319–418)-ΔLIELD was detected and quantified as in panel A. The bars represent the mean ± S.E. from n independent experiments: WT (37), L338I (24), L338V (9), and L338F (26).
We next searched the protein data base to see if there were additional proteins that contain the (L/I)2G_X_L motif. Surprisingly, there are a large number of proteins that contain such a motif, some of which are shown in supplemental Fig. 3. These include the visual arrestins, the β-adaptin subunits of AP-1 and AP-2, synaptotagmin, synaptojanin, and many other proteins that have been shown to have a role in trafficking. Although it is unclear at this point whether any of these proteins utilize the (L/I)2G_X_L motif to interact with clathrin, this at least raises the possibility that this is a widely utilized clathrin binding domain.
To identify additional residues in arrestin2 that might contact and orient clathrin binding, we superimposed the clathrin β-propeller domain from the two different crystal forms (Fig. 7). First, crystallographic symmetry mates of arrestin2 near the clathrin binding box (<10 Å) from the high resolution structure were generated. Then clathrin from the high resolution structure (arrestin2S) was superimposed on the clathrin in the low resolution structure (arrestin2L) using least square fitting (42). The same transformation matrix was then applied to the arrestin symmetry mates. We found that the relative position of the “equivalent” arrestins between the two crystals differed significantly, with the polar angle between these arrestins being κ = 157° after the clathrin superposition (Figs. 7, B and C). This indicates that the interaction between arrestin2 and clathrin that occurs via the LϕXϕ(D/E) motif does not confer a specific geometry or orientation. This may play an important role in facilitating arrestin interaction with clathrin in the highly organized clathrin lattice found in coated pits.
FIGURE 7.
Comparison of arrestin position in clathrin superposition. A, top view of the clathrin terminal domain shows two distinct interactions with arrestin2L, which are depicted as red ribbons (left). The side view of arrestin2L shows two separated clathrin binding sites, the LϕXϕ(D/E) motif and splice loop (right, red dotted lines). B, side view; arrestins (cyan, low resolution; wheat, high resolution) near the clathrin binding box (red ribbon) do not superimpose upon the superposition of clathrin (green) from the different crystal forms. Also shown is the second arrestin with a splice loop. Each end point of flanking regions in unstructured C-terminal loop is marked and connected as blue balls and a dotted line for the N-terminal area and red balls and a dotted line for the C-terminal area. C, top view; same as B but rotated 90° in a plane and looking down at approximate rotation axis relating the two arrestins. D, electron microscopy image of a clathrin barrel with a set of clathrin terminal domains shown in red (from Edeling et al. (59)). The enlarged inset is a schematic representation of the six terminal domains within the cluster, depicting a distance of ∼64 Å between terminal domains. The distance between terminal domains was measured using the measurement function in PyMol using PDB code 1XI4 for the clathrin D6 coat (48).
DISCUSSION
We present here biochemical, cellular, and diffraction data that define a functional role for the splice loop in the long isoform of arrestin2 in clathrin binding and GPCR trafficking. Although we find that arrestin2S and arrestin2L bind to clathrin via a conserved LϕXϕ(D/E) motif as expected, we also observe a second independent binding site on clathrin through an eight-amino acid loop that is uniquely present in arrestin2L. The ability of this loop to interact with clathrin was confirmed through biochemical and cell-based analysis. This is the first demonstration of a functional role for this region of arrestin2, as previous studies had not detected any differences between arrestin2L and arrestin2S in binding to GPCRs (41). In addition, no previous studies tested whether this loop plays a role in GPCR trafficking. It is interesting to note, however, that previous work that measured arrestin binding to clathrin mutants that were disrupted in the LϕXϕ(D/E) binding pocket (e.g. F91A, K96E, and K98E) reported a complete disruption of arrestin3 binding but only a 65–80% loss of arrestin2L binding (24). Thus, although not noted in the paper, these previous results also suggest that a second clathrin binding domain is present in arrestin2L. It is also worth noting that the eight-amino acid splice loop in arrestin2L is also present in the two visual arrestins, arrestin1 and -4, but is absent in arrestin3. This raises the intriguing possibility that mammalian visual arrestins might bind to clathrin, although this eight-amino acid loop might also provide a hydrophobic surface for interaction with other proteins. Although there is no evidence that mammalian opsins undergo endocytosis, arrestin-promoted endocytosis of rhodopsin has been observed in Drosophila, although this appears to be primarily mediated by arrestin interaction with AP-2 (43, 44).
Arrestin2L clearly contains two distinct domains that mediate interaction with clathrin. The LϕXϕ(D/E) motif mediates interaction with a shallow hydrophobic groove between blades 1 and 2 in the clathrin β-propeller domain and is utilized by many clathrin-binding proteins including the β1, β2, and β3 subunits of the APs, amphiphysin 1 and 2, AP180, LAP, and epsin1 (25, 45). In fact, it appears that some proteins may actually compete for binding to the LϕXϕ(D/E) binding site in clathrin (25). Because arrestin2L can still bind to clathrin and promote β2AR endocytosis even without the LϕXϕ(D/E) motif, this suggests that arrestin2L might be able to bind to clathrin even in the presence of other clathrin-binding proteins. This has some analogy to amphiphysin, which not only contains a LϕXϕ(D/E) motif but also a W box motif (PW_XX_W) that interacts with the membrane proximal surface of the clathrin terminal domain at a site ∼23 Å from the LϕXϕ(D/E) binding site (46). It has been proposed that these two motifs enable amphiphysin to bind more tightly to clathrin compared with proteins that only contain a LϕXϕ(D/E) motif and that this helps to preferentially localize amphiphysin to the periphery of the clathrin lattice. One clear distinction between amphiphysin and arrestin, however, is that the two clathrin binding domains in arrestin2L are ∼68 Å apart, and our structural analysis suggests that the two domains on an individual arrestin molecule would not simultaneously bind to an individual clathrin molecule. Cryoelectron microscopy of a clathrin lattice reveals that overlapping clathrin triskelion within the lattice results in a distance of ∼64 Å between adjacent clathrin terminal domains (47–49) (Fig. 7D). Considering these distances and the flexible movement of terminal domains, it is possible that the two independent binding motifs on one arrestin2L could bridge two molecules of clathrin in the lattice. This would enable arrestin2L to more effectively localize in clathrin-coated pits and mediate receptor endocytosis compared with arrestin2S. This hypothesis is consistent with the functional analysis of arrestin and GPCR trafficking, which showed significant differences between arrestin2L and arrestin2S in their ability to concentrate in coated pits and to promote endocytosis of the β2AR after agonist-induced activation of receptors (Fig. 2).
Although the (L/I)2G_X_L motif in arrestin2L clearly has the ability to bind to clathrin (Figs. 5 and 6) and mediate arrestin redistribution to clathrin-coated pits (Fig. 2B) and β2AR endocytosis (Fig. 2C), a general role for this motif has not been established. Nevertheless, this motif is found in a variety of trafficking proteins (supplemental Fig. 3) and, thus, might contribute to clathrin binding. However, there are two features that may be important for the ability of this motif to function in arrestin2L. The first is that this motif is contained within an unstructured loop that connects two β-strands that are fixed in position. This structural arrangement may well be critical for its ability to bind to clathrin. The second feature is that this motif cannot function in isolation. Although this motif can bind to clathrin in the absence of the clathrin binding box, arrestin2 also needs to interact with AP-2 to effectively mediate β2AR endocytosis (Refs. 14 and 50 and data not shown). Thus, non-visual arrestins need to bind to clathrin and AP-2 to localize in clathrin-coated pits and regulate endocytosis.
Although the x-ray crystal structures of arrestin2S, arrestin2L, and clathrin provide important new insight into the interfaces and potential dynamics of these protein-protein interactions, the precise mechanisms involved in GPCR endocytosis remain incompletely defined. The first step in this process appears to be the association of cytoplasmic arrestin with an activated phosphorylated receptor at the plasma membrane (4, 12). This interaction is proposed to disrupt the polar core and mediate a significant conformational change in arrestin that results in a rearrangement of the N-terminal domain and release of the C-terminal tail. In vitro studies support this model and reveal that a “constitutively active” arrestin2 (arrestin2-R169E) has enhanced binding to clathrin and AP-2 (14), whereas a phosphopeptide that binds to arrestin3 induces altered accessibility of the N- and C-terminal domains of arrestin to trypsin and enhanced binding to clathrin (51). Similarly, mutation of specific residues within the arrestin2 N- and C-terminal domains also appears to relieve the normal constraints that regulate arrestin binding to clathrin (52). In contrast, only minor differences between arrestin2 and arrestin2-R169E were observed by hydrogen/deuterium exchange analysis (53). Similarly, small-angle x-ray scattering showed no significant changes in conformation between wild type arrestin1, constitutively active arrestin1-R175Q, and arrestin1 with a bound phospho-peptide from rhodopsin (54). The lack of a significant conformational change upon “activation” is further supported by the demonstration that the x-ray crystal structures of wild type arrestin2, arrestin2-(1–393), and constitutively active arrestin2-(1–382) are virtually identical (9, 10). Taken together, these studies suggest that disruption of the polar core likely causes only minor conformational changes in the arrangement of the N and C domains and/or local conformational changes in the C-terminal tail. However, these changes appear to destabilize the basal conformation enough to facilitate binding to clathrin and the β-adaptin subunit of AP-2 and that the binding of arrestin2 to AP-2 is what stabilizes a significant conformational change in arrestin. This hypothesis is supported by the observation that a peptide containing the D_XX_F_XX_(F/L)_XXX_R motif from arrestin2 or ARH (autosomal recessive hypercholesterolemia) undergoes a conformational change to an α-helix when bound to the β2-adaptin appendage domain (55–57). Because this AP-2 binding region in arrestin2 is part of β strand 20, this suggests that disruption of some polar core contacts destabilizes β strand 20, resulting in enhanced interaction with β2-adaptin and the conformational change of this region of arrestin. To test this possibility, the structure of higher order complexes that contain arrestin2, β2-adaptin, clathrin, and possibly a GPCR will need to be solved.
Supplementary Material
Supplemental Data
Acknowledgments
We thank Dr. Vsevolod Gurevich for the bacterial expression construct for arrestin2S, Drs. Yuri Sykulev and Tatiana Mareeva for help with the surface plasmon resonance studies, and the staff of beamline X4 and X6 at the National Synchrotron Light Source.
*
This work was supported, in whole or in part, by National Institutes of Health Grants GM047417 and GM068857 (to J. L. B.) and DA010711 (M. v. Z.). This project was also funded, in part, under a grant with the Pennsylvania Department of Health.
The atomic coordinates and structure factors (codes 3GC3 and 3GD1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
3
The abbreviations used are:
GPCR
G protein-coupled receptor
AP-2
clathrin adaptor protein complex 2
GST
glutathione _S_-transferase
GFP
green fluorescent protein
TIRF
total internal reflection fluorescence
Bis-Tris
2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
Bicine
N,_N_-bis(2-hydroxyethyl)glycine
MES
4-morpholineethanesulfonic acid
Arr2S
arrestin2S
Arr2L
arrestin2L
WT
wild type
β2AR
β2-adrenergic receptor.
REFERENCES
- 1.Dohlman H. G., Thorner J., Caron M. G., Lefkowitz R. J. (1991) Annu. Rev. Biochem. 60, 653–688 [DOI] [PubMed] [Google Scholar]
- 2.Marinissen M. J., Gutkind J. S. (2001) Trends Pharmacol. Sci. 22, 368–376 [DOI] [PubMed] [Google Scholar]
- 3.Ferguson S. S. (2001) Pharmacol. Rev. 53, 1–24 [PubMed] [Google Scholar]
- 4.Moore C. A., Milano S. K., Benovic J. L. (2007) Annu. Rev. Physiol. 69, 451–482 [DOI] [PubMed] [Google Scholar]
- 5.Krupnick J. G., Benovic J. L. (1998) Annu. Rev. Pharmacol. Toxicol. 38, 289–319 [DOI] [PubMed] [Google Scholar]
- 6.DeWire S. M., Ahn S., Lefkowitz R. J., Shenoy S. K. (2007) Annu. Rev. Physiol. 69, 483–510 [DOI] [PubMed] [Google Scholar]
- 7.Granzin J., Wilden U., Choe H. W., Labahn J., Krafft B., Büldt G. (1998) Nature 391, 918–921 [DOI] [PubMed] [Google Scholar]
- 8.Hirsch J. A., Schubert C., Gurevich V. V., Sigler P. B. (1999) Cell 97, 257–269 [DOI] [PubMed] [Google Scholar]
- 9.Han M., Gurevich V. V., Vishnivetskiy S. A., Sigler P. B., Schubert C. (2001) Structure 9, 869–880 [DOI] [PubMed] [Google Scholar]
- 10.Milano S. K., Pace H. C., Kim Y. M., Brenner C., Benovic J. L. (2002) Biochemistry 41, 3321–3328 [DOI] [PubMed] [Google Scholar]
- 11.Sutton R. B., Vishnivetskiy S. A., Robert J., Hanson S. M., Raman D., Knox B. E., Kono M., Navarro J., Gurevich V. V. (2005) J. Mol. Biol. 354, 1069–1080 [DOI] [PubMed] [Google Scholar]
- 12.Gurevich V. V., Gurevich E. V. (2006) Pharmacol. Ther. 110, 465–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovoor A., Celver J., Abdryashitov R. I., Chavkin C., Gurevich V. V. (1999) J. Biol. Chem. 274, 6831–6834 [DOI] [PubMed] [Google Scholar]
- 14.Kim Y. M., Benovic J. L. (2002) J. Biol. Chem. 277, 30760–30768 [DOI] [PubMed] [Google Scholar]
- 15.Ferguson S. S., Downey W. E., 3rd, Colapietro A. M., Barak L. S., Ménard L., Caron M. G. (1996) Science 271, 363–366 [DOI] [PubMed] [Google Scholar]
- 16.Goodman O. B., Jr., Krupnick J. G., Santini F., Gurevich V. V., Penn R. B., Gagnon A. W., Keen J. H., Benovic J. L. (1996) Nature 383, 447–450 [DOI] [PubMed] [Google Scholar]
- 17.Laporte S. A., Oakley R. H., Zhang J., Holt J. A., Ferguson S. S., Caron M. G., Barak L. S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3712–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaidarov I., Krupnick J. G., Falck J. R., Benovic J. L., Keen J. H. (1999) EMBO J. 18, 871–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krupnick J. G., Goodman O. B., Jr., Keen J. H., Benovic J. L. (1997) J. Biol. Chem. 272, 15011–15016 [DOI] [PubMed] [Google Scholar]
- 20.Shih W., Gallusser A., Kirchhausen T. (1995) J. Biol. Chem. 270, 31083–31090 [DOI] [PubMed] [Google Scholar]
- 21.Morris S. A., Schröder S., Plessmann U., Weber K., Ungewickell E. (1993) EMBO J. 12, 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramjaun A. R., Micheva K. D., Bouchelet I., McPherson P. S. (1997) J. Biol. Chem. 272, 16700–16706 [DOI] [PubMed] [Google Scholar]
- 23.Drake M. T., Downs M. A., Traub L. M. (2000) J. Biol. Chem. 275, 6479–6489 [DOI] [PubMed] [Google Scholar]
- 24.Goodman O. B., Jr., Krupnick J. G., Gurevich V. V., Benovic J. L., Keen J. H. (1997) J. Biol. Chem. 272, 15017–15022 [DOI] [PubMed] [Google Scholar]
- 25.ter Haar E., Harrison S. C., Kirchhausen T. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sterne-Marr R., Gurevich V. V., Goldsmith P., Bodine R. C., Sanders C., Donoso L. A., Benovic J. L. (1993) J. Biol. Chem. 268, 15640–15648 [PubMed] [Google Scholar]
- 27.ter Haar E., Musacchio A., Harrison S. C., Kirchhausen T. (1998) Cell 95, 563–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendrickson W. A., Horton J. R., LeMaster D. M. (1990) EMBO J. 9, 1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendrickson W. A. (1991) Science 254, 51–58 [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31.Terwilliger T. C. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2144–2149 [DOI] [PubMed] [Google Scholar]
- 32.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 33.CCP4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 34.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 35.Jones T. A. (1978) J. Appl. Crystallogr. 11, 268–272 [Google Scholar]
- 36.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 37.McCoy A. J. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapani S., Navaza J. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penn R. B., Pascual R. M., Kim Y. M., Mundell S. J., Krymskaya V. P., Panettieri R. A., Jr., Benovic J. L. (2001) J. Biol. Chem. 276, 32648–32656 [DOI] [PubMed] [Google Scholar]
- 40.Puthenveedu M. A., von Zastrow M. (2006) Cell 127, 113–124 [DOI] [PubMed] [Google Scholar]
- 41.Gurevich V. V., Dion S. B., Onorato J. J., Ptasienski J., Kim C. M., Sterne-Marr R., Hosey M. M., Benovic J. L. (1995) J. Biol. Chem. 270, 720–731 [DOI] [PubMed] [Google Scholar]
- 42.Kabsch W. (1976) Acta Crystallogr. A 32, 922–923 [Google Scholar]
- 43.Orem N. R., Xia L., Dolph P. J. (2006) J. Cell Sci. 119, 3141–3148 [DOI] [PubMed] [Google Scholar]
- 44.Wang T., Montell C. (2007) Pflugers Arch. 454, 821–847 [DOI] [PubMed] [Google Scholar]
- 45.Kirchhausen T. (1999) Annu. Rev. Cell Dev. Biol. 15, 705–732 [DOI] [PubMed] [Google Scholar]
- 46.Miele A. E., Watson P. J., Evans P. R., Traub L. M., Owen D. J. (2004) Nat. Struct. Mol. Biol. 11, 242–248 [DOI] [PubMed] [Google Scholar]
- 47.Kirchhausen T. (2000) Annu. Rev. Biochem. 69, 699–727 [DOI] [PubMed] [Google Scholar]
- 48.Fotin A., Cheng Y., Sliz P., Grigorieff N., Harrison S. C., Kirchhausen T., Walz T. (2004) Nature 432, 573–579 [DOI] [PubMed] [Google Scholar]
- 49.Cheng Y., Boll W., Kirchhausen T., Harrison S. C., Walz T. (2007) J. Mol. Biol. 365, 892–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laporte S. A., Oakley R. H., Holt J. A., Barak L. S., Caron M. G. (2000) J. Biol. Chem. 275, 23120–23126 [DOI] [PubMed] [Google Scholar]
- 51.Xiao K., Shenoy S. K., Nobles K., Lefkowitz R. J. (2004) J. Biol. Chem. 279, 55744–55753 [DOI] [PubMed] [Google Scholar]
- 52.Kern R. C., Kang D. S., Benovic J. L. (2009) Biochemistry 48, 7190–7200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carter J. M., Gurevich V. V., Prossnitz E. R., Engen J. R. (2005) J. Mol. Biol. 351, 865–878 [DOI] [PubMed] [Google Scholar]
- 54.Shilton B. H., McDowell J. H., Smith W. C., Hargrave P. A. (2002) Eur. J. Biochem. 269, 3801–3809 [DOI] [PubMed] [Google Scholar]
- 55.Edeling M. A., Mishra S. K., Keyel P. A., Steinhauser A. L., Collins B. M., Roth R., Heuser J. E., Owen D. J., Traub L. M. (2006) Dev. Cell 10, 329–342 [DOI] [PubMed] [Google Scholar]
- 56.Schmid E. M., Ford M. G., Burtey A., Praefcke G. J., Peak-Chew S. Y., Mills I. G., Benmerah A., McMahon H. T. (2006) PLoS Biol. 4, e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burtey A., Schmid E. M., Ford M. G., Rappoport J. Z., Scott M. G., Marullo S., Simon S. M., McMahon H. T., Benmerah A. (2007) Traffic 8, 914–931 [DOI] [PubMed] [Google Scholar]
- 58.Palmitessa A., Hess H. A., Bany I. A., Kim Y. M., Koelle M. R., Benovic J. L. (2005) J. Biol. Chem. 280, 24649–24662 [DOI] [PubMed] [Google Scholar]
- 59.Edeling M. A., Smith C., Owen D. (2006) Nat. Rev. Mol. Cell Biol. 7, 32–44 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data