Regulation of protein stability by GSK3 mediated phosphorylation (original) (raw)
. Author manuscript; available in PMC: 2010 Feb 18.
Published in final edited form as: Cell Cycle. 2009 Dec 17;8(24):4032–4039. doi: 10.4161/cc.8.24.10111
Abstract
Glycogen synthase kinase-3 (GSK3) plays important roles in numerous signaling pathways that regulate a variety of cellular processes including cell proliferation, differentiation, apoptosis and embryonic development. In the canonical Wnt signaling pathway, GSK3 phosphorylation mediates proteasomal targeting and degradation of β-catenin via the destruction complex. We recently reported a biochemical screen that discovered multiple additional protein substrates whose stability is regulated by Wnt signaling and/or GSK3 and these have important implications for Wnt/GSK3 regulation of different cellular processes.1 In this article, we also present a bio-informatics based screen for proteins whose stability may be controlled by GSK3 and β-Trcp, the SCF E3 ubiquitin ligase that is responsible for β-catenin degradation in the Wnt signaling pathway. Furthermore, we review various GSK3 regulated proteolysis substrates described in the literature. We propose that GSK3 phosphorylation dependent proteolysis is a widespread mechanism that the cell employs to regulate a variety of cell processes in response to signals.
Keywords: GSK3, wnt, degradation, stability, ubiquitination, proteolysis
Introduction
Glycogen synthase kinase-3 (GSK3) was originally identified in the 1980's as a key regulator of glycogen metabolism.2 When blood glucose level is low, GSK3 constitutively phosphorylates glycogen synthase and inhibits its activity. This inhibition is released by increased insulin signaling resulting from elevated blood glucose levels in which case PI3K-Akt signaling cascade inhibits GSK3 activity and glycogen synthase is de-phosphorylated and activated, resulting in the synthesis of glycogen from glucose (reviewed in ref. 3).
Since its discovery, GSK3 has been found to phosphorylate many proteins and play important roles in a variety of cellular processes such as cell proliferation, differentiation, microtubule dynamics, cell cycle and apoptosis.4 In fact, a consensus motif and context based computational analysis of identified in vivo protein phosphorylation sites indicates that GSK3 is one of the kinases with most substrates in the cell.5 In turn, multiple cell signaling pathways are known to inhibit GSK3 activity, such as the PI3K-Akt pathway, the Wnt signaling pathway and the MAPK pathway. The general function and regulation of GSK3 has been the subject of numerous excellent reviews (refs. 4 and 6) and therefore will not be the focus of this article.
An interesting characteristic of GSK3 is that it is usually constitutively active in the cell and inhibited by upstream signals. Phosphorylation by GSK3 usually negatively regulates its downstream substrates, as exemplified by the classic GSK3 regulation of glycogen synthase that was described earlier. In many cases GSK3 phosphorylation earmarks target proteins for ubiquitination and proteolysis, one famous example being the regulation of β-catenin stability in the canonical Wnt signaling pathway. In this article, we will discuss potential Wnt and/or GSK3 regulated protein degradation substrates revealed by a biochemical screen1 and a bioinformatics based search. We will also review other GSK3 regulated proteolytic substrates from literature and propose protein stability control as a major mechanism that GSK3 employs to regulate cellular processes.
GSK3 Regulates β-Catenin Turnover in the Wnt Signaling Pathway
Originally identified using genetic studies in Drosophila and developmental studies in Xenopus,7 the canonical Wnt signaling pathway plays a very important role in numerous processes such as embryonic development, tissue regeneration and carcinogenesis. In the absence of Wnt signaling, casein kinase I (CKI) phosphorylation of β-catenin primes it for subsequent GSK3β phosphorylation at multiple sites at the N-terminus. Phosphorylated β-catenin is then recognized by β-Trcp and rapidly degraded by the 26S proteasome. Other components of the multi-protein “destruction complex” that targets β-catenin for degradation, the tumor suppressor genes APC (adenomatous polyposis coli) and Axin, serve as scaffolding proteins for this reaction. The level of the cytoplasmic and nuclear β-catenin that mediates Wnt signaling is thus kept low in resting cells, resulting in the silence of downstream target genes. In the presence of Wnt ligand the destruction complex is inactivated through a mechanism that is not fully understood (likely dissociation of the complex), allowing β-catenin levels to increase in the cytoplasm. The accumulated β-catenin then translocates into the nucleus and binds to LEF/TCF transcription factors to activate downstream target genes. GSK3 dependent control of β-catenin stability is thus the central regulatory mechanism of the canonical Wnt signaling pathway (reviewed in refs. 8–10).
Wnt Regulated Proteolytic Substrates in Addition to β-Catenin
In addition to β-catenin, levels of Snail, a repressor of E-cadherin gene transcription and Smad1, the BMP signaling mediator, were recently reported to be regulated by the canonical Wnt signaling pathway and GSK3.11–15 Very similar to β-catenin, stability of the Snail protein is controlled by GSK3 dependent phosphorylation followed by β-Trcp mediated ubiquitination and proteosomal degradation in the absence of Wnt signaling. Wnt signal inhibits the GSK3 mediated phosphorylation and consequently increases Snail protein levels, which in turn triggers the epithelial-mesechymal transition (EMT) in certain cell types.12–14 The BMP signaling mediator Smad1 is regulated by Wnt signaling in a similar fashion. However, instead of total Smad1 protein levels, only the BMP/MAPK activated form of Smad1 (C-terminal phosphorylated Smad1) is recognized and regulated by Wnt/GSK3, resulting in a fine control of BMP signaling strength and longevity by Wnt signaling.15 Interestingly, a recent study revealed that the protein stability of another Smad, Smad3, is regulated by Axin and GSK3β but possibly not Wnt signaling.16 Further studies may help to confirm whether Wnt signaling can regulate Smad3 stability in certain contexts.
Screen for Targets of the Wnt Signaling Pathway Destruction Complex in Addition to β-Catenin
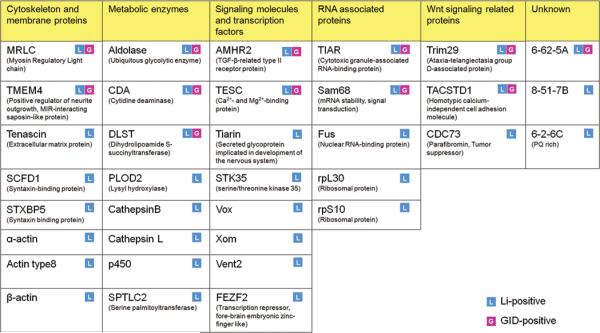
We recently reported a screen for potential Wnt and/or GSK3 regulated protein degradation substrates using in vitro expression cloning technique and biochemical reconstitution in a Xenopus egg cytoplasmic extract.1 Using LiCl as a general GSK3 inhibitor or GID (the GSK3 interacting domain of Axin) protein as specific inhibitor of GSK3 activity in the Wnt signaling pathway, we screened pools of cDNAs encoding ~10,000 polypeptides and identified 35 potential GSK3 regulated proteolytic substrates that responded to LiCl in the screen. 12 of the 35 candidates responded to both LiCl and GID, and to other destruction complex inhibitors such as Dsh (dishevelled) and AxinΔRGS (axin that misses the APC-interacting RGS domain), and are thus strong candidates for Wnt regulation. Wnt regulation of selected candidates was verified by expression in vivo in Xenopus embryos. The other 23 candidates responded to LiCl but not GID in the screen, and are thus potential GSK3 protein substrates but not Wnt regulated (Fig. 1 and ref. 1).
Figure 1.
Potential GSK3 phosphorylation dependent protein degradation substrates identified in biochemical screen (modified from Fig. 3 in ref. 1). 35 novel proteolytic targets of Wnt and/or GSK3 identified in the screen are grouped according to their cellular functions. Proteins that are known to play roles in the canonical wnt signaling pathway are separated into one group.
This study provided evidence that Wnt signaling regulates the stability of multiple proteins, and therefore various cellular processes, in additional to β-catenin mediated gene expression. Many of the identified substrates interact with each other or have similar cellular functions, indicating Wnt and/or GSK3 regulation of those cellular functions. One example includes MRLC (myosin regulatory light chain) and TMEM4 (or MASP for MIR-interacting saposin-like protein). MRLC is a main regulator of myosin contractility and cell motility and its activation has been known to be regulated by multiple signaling pathways.17–19 Wnt signaling has also been known to regulate MRLC activity and cell motility, although through the noncanonical Wnt pathways.20 Little is known about TMEM4, but it has been reported to interact with MRLC and positively regulate its activity and stability.21,22 Finding MRLC and TMEM4 in the screen indicates that the canonical Wnt signaling pathway may also control cell motility in some cell contexts through the destruction complex mediated regulation of MRLC and TMEM4 stability. Interestingly, components of the destruction complex, GSK3 and APC, have been implicated in regulation of cell polarity and cell migration through their regulation of microtubule polarity and dynamics.23,24
Another group of interesting targets identified in the screen include the RNA binding proteins TIAR (T-cell restricted intracellular antigen-related protein) and Sam68, a STAR (signal transduction and activation of RNA) family protein. TIAR has been shown to bind to U-rich sequences near 5' splice sites of pre-mRNAs and modulates alternative splicing.25,26 Indeed, one bioinformatics study estimates that ~15% of alternative cassette exons in the genome are regulated by TIA1/TIAR, suggesting a widespread role of TIAR in the regulation of alternative splicing.27 Sam68 has been reported to regulate the splicing of genes such as CD44 and Bcl-x, and its activity is regulated by cellular signaling and phosphorylation.28,29 Identifying TIAR and Sam68 as potential Wnt signaling regulated proteolysis substrates therefore raise the exciting possibility that Wnt signaling may regulate mRNA splicing and processing through regulation of the stability of these proteins.
Some proteins identified as Wnt regulated destruction complex targets in the screen also play roles in the Wnt signaling pathway. Two of the LiCl and GID positive targets, Trim29 and TACSTD1, have recently been reported to be positive regulators of β-catenin signaling. TACSTD1, also called EpCAM, is a membrane protein that is overexpressed in cancer cells and was one of the first identified cancer antigens.30–32 It undergoes proteolytic cleavage upon extracellular domain homophilic binding and releases the intracellular domain (EpICD). EpICD then enters the nucleus and binds to transcription factors including β-catenin and Lef-1, and activates downstream gene expression.33 Trim29, also known as ATDC (ataxiatelangiectasia group D complementing gene), was found to positively regulate Dishevelled levels and inhibit the destruction complex, thus stabilizing β-catenin.34 Potential Wnt regulation of the stability of EpCAM and ATDC therefore suggests that there exists feedback control of the canonical Wnt/β-catenin signaling pathway, probably resulting in fine control of the signaling strength and length in various cellular contexts.
Bioinformatics Based Search for Potential GSK3/β-Trcp Regulated Proteins
Both β-catenin and Snail contain a “DSGxxS/TxxxS/T” motif that is necessary for their regulation by Wnt signaling.12 DSG(X)2+nS is the recognition site for the E3 ubiquitin ligase β-Trcp: when the serine in the DSG motif is phosphorylated, β-Trcp binds to this motif and adds ubiquitin onto a lysine usually located upstream (reviewed in ref. 35). “S/TxxxS/Tp” is the GSK3 consensus phosphorylation site, in which the second (c-terminal) serine or threonine is usually primed by phosphorylation by another kinase before GSK3 phosphorylates the N-terminal serine/threonine. In many cases there are tandem repeats of this consensus motif, resulting in processive phosphorylation as in the case of β-catenin (reviewed in ref. 6).
We hypothesized that proteins containing the DSGxxS β-Trcp binding motif followed by tandem GSK3 consensus sites are likely regulated by GSK3 and β-Trcp in a similar fashion as β-catenin and Snail. With this hypothesis, we searched the UniProtKB/Swiss-Prot protein database for a “D/ESGxxS/TxxxS/TxxxS/T” motif. We identified 605 hits in 579 sequences in the database that contains ~500,000 sequence entries (release 57.4, eukaryota). As important regulatory amino acid motifs are usually evolutionarily conserved, we blasted each hit sequence against its homolog and identified ones that contained motifs that were evolutionarily conserved among numerous species. This narrowed the list down to 38 potential protein substrates regulated by GSK3 and β-Trcp. They can be included in functional groups as shown in Table 1, indicating potential GSK3/β-Trcp regulation of the respective cellular processes.
Table 1.
Potential GSK3/β-Trcp regulated proteolysis substrates
| Cell motility and cytoskeleton | Transcription factors | Cell signaling | Protein modification | Adaptor/scaffold | GTPase/GEF | Unknown |
|---|---|---|---|---|---|---|
| ABLIM2 | NACA | BNIP3 | WNK2 | Gephyrin | FGD3 | BAZ2B |
| Cappuccino | DMRT | EPOR | ARD1 | DOK7 | RAP GEF1 | Clorf172 |
| KIF1B | β-catenin | I10R1 | BRD3 | JIP3 | TBC15 | FAM135A |
| KIF1C | Snail | LZTS2 | KDIS | AKAP 220 | PRIC3 | |
| Ninein | WWTR1 | NP1L3 | PLCH1 | TRI67 | ||
| Plakoglobin | ZBT20 | PRP16 | YQ013 | |||
| Unc-84 | ZN395 |
Both β-catenin and Snail were, by design, identified in the search. Interestingly, the search identified FGD3 (faciogenital dysplasia 3), a putative Cdc42 guanine nucleotide exchange factor (GEF) whose stability was reported to be regulated by GSK3 and β-Trcp36 and plakoglobin, which was known to be regulated by Wnt signaling in a similar manner as β-catenin.37–39 These two positive hits serve as validation for the screen.
The search also identified many interesting potential GSK3/β-Trcp targets. NACα has been reported to be regulated by GSK3 phosphorylation dependent ubiquitination but not the DSG motif and β-Trcp (Table 2). Further studies may help clarify this apparent discrepancy. Additionally, AKAP220 (A-kinase anchor protein 220) and Ninein have been found to bind GSK3β, and EPO-R (Erythropoietin receptor precursor) is known to be β-Trcp substrate, which makes them strong candidates for GSK3 and β-Trcp regulated proteolysis.40–42 There are also two hits, ARD1 (N-terminal acetyltransferase complex subunit) and LZTS2 (Leucine zipper putative tumor suppressor 2), which interact with β-catenin and play roles in the canonical Wnt signaling pathway.43,44
Table 2.
Known GSK3 regulated proteolysis substrates
| Protein | GSK phosphorylation site | Upstream signal | E3 Ubiquitin ligase | References | |
|---|---|---|---|---|---|
| Site | Sequence | ||||
| β-catenin | S33, S37, S41 | LDSGIHSGATSTAPSLS | Wnt | β-Trcp | 37–39, 57, 58 |
| Snail | S96, S100, S104 | DSGKSSQPPSPP | Wnt | β-Trcp | 11–14 |
| Smad1 | S191, S198, T202, S210 | NSSYPNSPGSSSSTYPHSPTSSDPGSPF | Wnt | Smurf1 | 15 |
| Hath1 | S54 | ELSLLDSTD | Wnt | - | 59 |
| Smad3 | T66 | CITIPRSLD | Axin involved | - | 16 |
| SRC-3 | S505 | VHSPMASSG | Akt | Fbw7α | 54 |
| BCL-3 | S394, S398 | SSSPSQSPP | Akt | 52, 53 | |
| p21 | T57 | DLSLSCTLV | ATR, Akt | Skp2 | 60–62 |
| S114 | TETPLEGDF | ||||
| HIF-1α | S551, T555 | PFSTQDTDL | hypoxia, PI3-Akt | - | 63, 64 |
| S589 | SASPESASP | ||||
| Mcl-1 | S155, 159 | NTSTDGSLPSTPP | PI3K-Akt | β-Trcp | 65–68 |
| c-Jun | T239 | GETPPLSPI | PI3K-Akt | Fbw7 | 69 |
| c-Myc | T58 | LPTPPLSPS | Ras-PI3K-Akt | Fbw7 | 70–73 |
| Cyclin D1 | T286 | ACTPTDVRD | Ras-PI3K-Akt | 74 | |
| Cyclin E | T380 | LLTPPQSGK | - | Fbw7 | 75 |
| SREBP | T426, S430 | TLTPPPSDA | insulin signaling | Fbw7 | 76 |
| Cdc25a | S76 | MGSSESTDS | - | β-Trcp | 77 |
| FGD1 | S283, S287 | RDSGIDSIS | - | β-Trcp | 78 |
| FGD3 | S72, S76 | RDSGIDSPS | - | β-Trcp | 36 |
| c-Myb | T572 | LMTPVSED | - | Fbw7 | 71 |
| mCRY2 | S553 | LSSGPASPK | - | Fbxl3 | 79 |
| NaCα | T159 | TQTPTVQEE | - | - | 80 |
| MafA | S61, T57, T53, S49 | PGSLSSTPLSTPCSSVPSSPS | - | - | 55, 56 |
| IPF1/PDX1 | S61, S66? | QGSPPDISPY | - | - | 81 |
| PS1 CTF | S397 | KASATASGD | - | - | 82 |
| NFκB1 (cleavage) | S903, 907 | AHSLPLSPA | TNFα | - | 49 |
| Ci (cleavage) | S852, S888 | MQSRRSSQS, GCSRRSSQM | Hedgehog | Slimb/β-Trcp (cleavage) | 45–48 |
GSK3 Regulated Proteolysis Substrates Described in the Literature
GSK3 has been found to regulate the proteolysis of an increasing number of proteins (Table 2). In the absence of upstream inhibitory signals, GSK3 phosphorylation of serine or threonine creates binding sites for phosphorylation-dependent E3 ubiquitin ligases such as Fbw7 and β-Trcp, allowing subsequent ubiquitination and proteolysis of the substrates. In many cases GSK3 needs a priming phosphorylation by another kinase, thus allowing additional regulatory input from other signaling pathways. The regulated proteins include transcription factors and signaling proteins with various cellular functions including cell proliferation, differentiation, apoptosis and response to stress. Growth signals and mitogens typically inhibit GSK3 activity, resulting in the increase of substrate protein levels and activation of respective downstream cellular events.
In some cases GSK3 phosphorylation dependent ubiquitination does not lead to full degradation but rather proteolytic cleavage processing of the substrate, as in the case of transcription activator Ci (Cubitus interruptus) in the Hedgehog (Hh) signaling pathway (described in more detail in the next section).45–48 Although less well-studied, GSK3 phosphorylation and subsequent ubiquitination may also regulate p105 to p50 protein cleavage processing in the NFκB pathway.49
In some signaling events, phosphorylation by GSK3 plays both positive and negative roles on the same substrates, as in the case of MafA and SRC-3. MafA is a basic leucine zipper (bZip) family transcription factor that promotes oncogenic transformation in embryonic fibroblasts and insulin expression in β cells.50 SRC-3 is a steroid receptor co-activator that regulates cell growth.51 Phosphorylation by GSK3 induces both activation and degradation of MafA and SRC-3. Another GSK3 regulated proteolysis substrate, BCL-3, may also be regulated through a similar mechanism.52,53 This tightly coupled activation and degradation creates a `rapid spike' of downstream substrate activity by limiting the temporal length of activation that may be necessary for strict regulation.54–56
GSK3 Regulated Proteolysis in Major Signaling Pathways during Embryonic Patterning
GSK3 regulation of β-catenin in the canonical Wnt signaling pathway has long been known to play important roles in embryonic development and tumorigenesis.83,84 GSK3 has also been found to function similarly in the Hedgehog (Hh) signaling pathway, which plays important roles in many developmental processes and diseases (reviewed in ref. 85). In the absence of the Hh signal, GSK3 phosphorylation results in the Slimb/β-Trcp dependent ubiquitination of transcription activator Ci. As we mentioned earlier, ubiquitination does not lead to full degradation but rather cleavage of the full-length Ci protein (Ci155) into a truncated form (Ci75), which acts as a repressor of Hh downstream genes. Hh signaling blocks this process through an unknown mechanism, resulting in the activation of downstream gene expression. The similar roles of GSK3 in Wnt signaling, TGFβ/BMP signaling and Hh signaling pathways suggest that GSK3 regulated protein proteolysis plays important roles in the regulation of cell proliferation and differentiation in embryonic development.
Interestingly, we identified Vent/Vox family protein members as potential GSK3 but not Wnt signaling regulated protein degradation substrates in the biochemical screen.1 Vent/Vox family homeobox transcription repressors are known antagonists of dorsalizing signals in early embryonic patterning in Xenopus and zebrafish, and their transcription has been reported to be regulated by BMP and Wnt signaling.86–88 Another study also reported that the stability of one Vent/Vox family protein, Xom, is regulated during Xenopus embryonic development, although the evidence suggested it was through a kinase different from GSK3.89 If GSK3 is confirmed to regulate the stability of Vent/Vox family proteins, it will be very interesting to identify the upstream signaling pathways that regulate GSK3 activity in this process.
GSK3 Regulated Proteolysis in Cell Cycle and Proliferation
GSK3 has been known to play an inhibitory role in cell cycle progression and cell proliferation, at least partly through its regulation of the stability of cyclin E, cyclin D1, cdc25A and c-Myc.74,75,77 Cyclin D1 and cyclin E protein levels dictate the G1 to S transition and has been known to be tightly regulated by mitogens and cell cycle signals (reviewed in ref. 90). GSK3 phosphorylation mediates rapid degradation of both cyclin D1 and cyclin E. Ras signal inactivates GSK3 through the PI3K-Akt pathway and results in accumulation of stabilized cyclins, triggering cell cycle progression.74,75 It may also lead to the accumulation of Cdc25A phosphatase, another GSK3 regulated protein degradation substrate, and activate cyclin-dependent protein kinases (Cdks).77 At the same time, mitogen signaling also inhibits the GSK3 mediated degradation of c-Myc, resulting in the activation of its target genes, including cyclin D1, cyclin E and other cell cycle mediators.70,72 GSK3 thus has both direct and indirect roles in regulation of cell cycle progression. Interestingly, GSK3 activity is high in quiescent cells and cells in G1 phase, but lowers as cells progress into S phase, reflecting upstream regulation of its activity in cell cycle.77
GSK3 Regulated Proteolysis in Stress Response and Apoptosis
In line with its role in cell cycle regulation, GSK3 regulated proteolysis also plays important roles in the cellular response to stress stimuli such as nutrient deprivation or DNA damage. One example is the regulation of p21cip1 (p21) turnover in response to UV irradiation. Following low dose UV irradiation p21 is rapidly degraded through ubiquitination, which releases its inhibition of PCNA (proliferating cell nuclear antigen, a cofactor of DNA polymerase δ) and allows DNA repair.61 Increase of GSK3 activity after UV irradiation was found to be responsible for triggering p21 degradation in this process.60 GSK3 mediated degradation of cdc25A may also play a role in inducing cell cycle arrest after UV irradiation, thus permitting more time for DNA repair.77
GSK3 plays pro-apoptotic roles in the cell, as its overexpression induces apoptosis and inhibition protects cells against apoptotic stimuli.91,92 It has recently been found to negatively regulate the protein stability of Mcl-1, a Bcl-2 like anti-apoptotic protein that antagonize the effects of pro-apoptotic Bcl-2 family proteins on mitochondrial outer membrane permeabilization (MOMP) and cytochrome c release.93 Cell stimuli such as growth factors or increased glucose level inhibits GSK3 activity through PI3K-Akt and promotes cell survival, while cellular stress such as UV irradiation increases GSK3 activity and triggers apoptosis.65,66,68 It is conceivable that GSK3 serves as a central kinase that integrates multiple positive and negative stimuli for cell survival, and triggers different cellular response through regulation of the turnover of various signaling protein substrates.
Various Upstream Signals Regulate GSK3 Activity in its Regulation of Protein Stability
The upstream signals that control GSK3 activity in regulation of many of these established or potential GSK3 regulated proteolysis protein substrates are still unknown. The protein targets that responded to both LiCl and GID protein in the biochemical screen are likely to be regulated by Wnt signaling (Fig. 1). On the contrary, the targets that responded to LiCl but not GID in the screen are regulated by GSK3 but not Wnt signaling, as LiCl acts as general GSK3 inhibitor while GID is specific to the destruction complex in our assay.1
Different upstream signals may regulate GSK3 activity through different mechanisms. For example, the PI3K-Akt pathway induces the inhibitory Serine 9 phosphorylation of GSK3β (Serine 21 in GSK3α) while Wnt signaling does not act through this phosphorylation. Instead, it may inhibit the GSK3 activity by dissociating the destruction complex, thus prohibiting GSK3 from reaching its targets.4,6,94 It will be very interesting to see how specific upstream signals and cell stimuli regulate GSK3 activity and affect the stability of specific substrates, and how different upstream signals may converge on GSK3 and mediate crosstalk between each other to orchestrate cellular response to external and internal signals.
Conclusion
GSK3 phosphorylates a great number of proteins and resides in the middle of many important signaling pathways that respond to cellular stimuli such as growth factors and stress. GSK3 has been found to regulate the ubiquitination and proteolysis of a number of important signaling proteins or transcription factors. The results from our recent screens indicate that there are many more proteins whose stability may be regulated by GSK3 phosphorylation. Protein stability control is thus emerging as a major mechanism that GSK3 employs to modulate cellular processes. Moreover, many different signaling pathways utilize this GSK3 dependent mechanism to target proteins for turnover. We speculate that GSK3 is a conserved kinase that regulates protein turnover in response to cellular signals, and in turn regulates a variety of cell processes such as cell proliferation, differentiation, apoptosis, embryonic patterning and tumorigenesis.
Glossary
Abbreviations
GSK3
glycogen synthase kinase-3
SCF
Skp1-Cul1-F-box-protein
PI3K
phosphoinositide-3 kinase
MAPK
mitogen-activated protein kinase
CKI
casein kinase I
APC
adenomatous polyposis coli
LEF/TCF
lymphoid enhancer factor/T-cell factor
EMT
epithelial-mesechymal transition
BMP
bone morphogenetic protein
Dsh
dishevelled
MRLC
myosin regulatory light chain
TMEM4
transmembrane protein 4
TIAR
T-cell restricted intracellular antigen-related protein
TACSTD1
tumor-associated calcium signal transducer 1
ATDC
ataxia-telangiectasia group D complementing gene
FGD3
faciogenital dysplasia 3
GEF
guanine nucleotide exchange factor
AKAP220
A-kinase anchor protein 220
EPO-R
erythropoietin receptor precursor
ARD1
ADP-ribosylation factor domain protein 1
LZTS2
leucine zipper putative tumor suppressor 2
Ci
cubitus interruptus
Hh
Hedgehog
NFκB
nuclear factor kappa-light-chain-enhancer of activated B cells
MafA
musculoaponeurotic fibrosarcoma oncogene homolog A
SRC-3
steroid receptor coactivator protein 3
BCL-3
B-cell CLL/lymphoma 3
CDK
cyclin-dependent protein kinase
MOMP
mitochondrial outer membrane permeabilization
PCNA
proliferating cell nuclear antigen
References
- 1.Kim NG, Xu C, Gumbiner BM. Identification of targets of the Wnt pathway destruction complex in addition to beta-catenin. Proc Natl Acad Sci USA. 2009;106:5165–70. doi: 10.1073/pnas.0810185106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Embi N, Rylatt DB, Cohen P. Glycogen synthase kinase-3 from rabbit skeletal muscle. Separation from cyclic-AMP-dependent protein kinase and phosphorylase kinase. Eur J Biochem. 1980;107:519–27. [PubMed] [Google Scholar]
- 3.Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2:769–76. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 4.Frame S, Cohen P. GSK3 takes centre stage more than 20 years after its discovery. Biochem J. 2001;359:1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–26. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116:1175–86. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegfried E, Chou TB, Perrimon N. wingless signaling acts through zeste-white 3, the Drosophila homolog of glycogen synthase kinase-3, to regulate engrailed and establish cell fate. Cell. 1992;71:1167–79. doi: 10.1016/s0092-8674(05)80065-0. [DOI] [PubMed] [Google Scholar]
- 8.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 9.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 10.Huang H, He X. Wnt/beta-catenin signaling: new (and old) players and new insights. Curr Opin Cell Biol. 2008;20:119–25. doi: 10.1016/j.ceb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doble BW, Woodgett JR. Role of glycogen synthase kinase-3 in cell fate and epithelial-mesenchymal transitions. Cells Tissues Organs. 2007;185:73–84. doi: 10.1159/000101306. [DOI] [PubMed] [Google Scholar]
- 12.Yook JI, Li XY, Ota I, Fearon ER, Weiss SJ. Wnt-dependent regulation of the E-cadherin repressor snail. J Biol Chem. 2005;280:11740–8. doi: 10.1074/jbc.M413878200. [DOI] [PubMed] [Google Scholar]
- 13.Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol. 2006;8:1398–406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 14.Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6:931–40. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 15.Fuentealba LC, Eivers E, Ikeda A, Hurtado C, Kuroda H, Pera EM, et al. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell. 2007;131:980–93. doi: 10.1016/j.cell.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo X, Ramirez A, Waddell DS, Li Z, Liu X, Wang XF. Axin and GSK3-control Smad3 protein stability and modulate TGF-signaling. Genes Dev. 2008;22:106–20. doi: 10.1101/gad.1590908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamm KE, Stull JT. Dedicated myosin light chain kinases with diverse cellular functions. J Biol Chem. 2001;276:4527–30. doi: 10.1074/jbc.R000028200. [DOI] [PubMed] [Google Scholar]
- 18.Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005;15:371–7. doi: 10.1016/j.tcb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe T, Hosoya H, Yonemura S. Regulation of myosin II dynamics by phosphorylation and dephosphorylation of its light chain in epithelial cells. Mol Biol Cell. 2007;18:605–16. doi: 10.1091/mbc.E06-07-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlessinger K, Hall A, Tolwinski N. Wnt signaling pathways meet Rho GTPases. Genes Dev. 2009;23:265–77. doi: 10.1101/gad.1760809. [DOI] [PubMed] [Google Scholar]
- 21.Bornhauser BC, Lindholm D. MSAP enhances migration of C6 glioma cells through phosphorylation of the myosin regulatory light chain. Cell Mol Life Sci. 2005;62:1260–6. doi: 10.1007/s00018-005-5055-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bornhauser BC, Olsson PA, Lindholm D. MSAP is a novel MIR-interacting protein that enhances neurite outgrowth and increases myosin regulatory light chain. J Biol Chem. 2003;278:35412–20. doi: 10.1074/jbc.M306271200. [DOI] [PubMed] [Google Scholar]
- 23.Etienne-Manneville S, Manneville JB, Nicholls S, Ferenczi MA, Hall A. Cdc42 and Par6-PKCzeta regulate the spatially localized association of Dlg1 and APC to control cell polarization. J Cell Biol. 2005;170:895–901. doi: 10.1083/jcb.200412172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nathke IS, Adams CL, Polakis P, Sellin JH, Nelson WJ. The adenomatous polyposis coli tumor suppressor protein localizes to plasma membrane sites involved in active cell migration. J Cell Biol. 1996;134:165–79. doi: 10.1083/jcb.134.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu H, Hasman RA, Young KM, Kedersha NL, Lou H. U1 snRNP-dependent function of TIAR in the regulation of alternative RNA processing of the human calcitonin/CGRP pre-mRNA. Mol Cell Biol. 2003;23:5959–71. doi: 10.1128/MCB.23.17.5959-5971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu H, Hinman MN, Hasman RA, Mehta P, Lou H. Regulation of neuron-specific alternative splicing of neurofibromatosis type 1 pre-mRNA. Mol Cell Biol. 2008;28:1240–51. doi: 10.1128/MCB.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aznarez I, Barash Y, Shai O, He D, Zielenski J, Tsui LC, et al. A systematic analysis of intronic sequences downstream of 5' splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation. Genome Res. 2008;18:1247–58. doi: 10.1101/gr.073155.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matter N, Herrlich P, Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–5. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 29.Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol. 2007;176:929–39. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herlyn M, Steplewski Z, Herlyn D, Koprowski H. Colorectal carcinoma-specific antigen: detection by means of monoclonal antibodies. Proc Natl Acad Sci USA. 1979;76:1438–42. doi: 10.1073/pnas.76.3.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edwards DP, Grzyb KT, Dressler LG, Mansel RE, Zava DT, Sledge GW, Jr, et al. Monoclonal antibody identification and characterization of a Mr 43,000 membrane glycoprotein associated with human breast cancer. Cancer Res. 1986;46:1306–17. [PubMed] [Google Scholar]
- 32.Ross AH, Herlyn D, Iliopoulos D, Koprowski H. Isolation and characterization of a carcinoma-associated antigen. Biochem Biophys Res Commun. 1986;135:297–303. doi: 10.1016/0006-291x(86)90976-9. [DOI] [PubMed] [Google Scholar]
- 33.Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11:162–71. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Heidt DG, Lee CJ, Yang H, Logsdon CD, Zhang L, et al. Oncogenic function of ATDC in pancreatic cancer through Wnt pathway activation and beta-catenin stabilization. Cancer Cell. 2009;15:207–19. doi: 10.1016/j.ccr.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuchs SY, Spiegelman VS, Kumar KG. The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene. 2004;23:2028–36. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- 36.Hayakawa M, Matsushima M, Hagiwara H, Oshima T, Fujino T, Ando K, et al. Novel insights into FGD3, a putative GEF for Cdc42, that undergoes SCF(FWD1/beta-TrCP)-mediated proteasomal degradation analogous to that of its homologue FGD1 but regulates cell morphology and motility differently from FGD1. Genes Cells. 2008;13:329–42. doi: 10.1111/j.1365-2443.2008.01168.x. [DOI] [PubMed] [Google Scholar]
- 37.Hinck L, Nelson WJ, Papkoff J. Wnt-1 modulates cell-cell adhesion in mammalian cells by stabilizing beta-catenin binding to the cell adhesion protein cadherin. J Cell Biol. 1994;124:729–41. doi: 10.1083/jcb.124.5.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papkoff J, Rubinfeld B, Schryver B, Polakis P. Wnt-1 regulates free pools of catenins and stabilizes APC-catenin complexes. Mol Cell Biol. 1996;16:2128–34. doi: 10.1128/mcb.16.5.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kodama S, Ikeda S, Asahara T, Kishida M, Kikuchi A. Axin directly interacts with plakoglobin and regulates its stability. J Biol Chem. 1999;274:27682–8. doi: 10.1074/jbc.274.39.27682. [DOI] [PubMed] [Google Scholar]
- 40.Hong YR, Chen CH, Chang JH, Wang S, Sy WD, Chou CK, et al. Cloning and characterization of a novel human ninein protein that interacts with the glycogen synthase kinase 3beta. Biochim Biophys Acta. 2000;1492:513–6. doi: 10.1016/s0167-4781(00)00127-5. [DOI] [PubMed] [Google Scholar]
- 41.Tanji C, Yamamoto H, Yorioka N, Kohno N, Kikuchi K, Kikuchi A. A-kinase anchoring protein AKAP220 binds to glycogen synthase kinase-3beta (GSK-3beta) and mediates protein kinase A-dependent inhibition of GSK-3beta. J Biol Chem. 2002;277:36955–61. doi: 10.1074/jbc.M206210200. [DOI] [PubMed] [Google Scholar]
- 42.Meyer L, Deau B, Forejtnikova H, Dumenil D, Margottin-Goguet F, Lacombe C, et al. beta-Trcp mediates ubiquitination and degradation of the erythropoietin receptor and controls cell proliferation. Blood. 2007;109:5215–22. doi: 10.1182/blood-2006-10-055350. [DOI] [PubMed] [Google Scholar]
- 43.Hyun Hwa C, Hye Joon J, Ji Sun S, Yong Chan B, Jin Sup J. Crossregulation of beta-catenin/Tcf pathway by NFkappaB is mediated by lzts2 in human adipose tissue-derived mesenchymal stem cells. Biochim Biophys Acta. 2008;1783:419–28. doi: 10.1016/j.bbamcr.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Thyssen G, Li TH, Lehmann L, Zhuo M, Sharma M, Sun Z. LZTS2 is a novel beta-catenin-interacting protein and regulates the nuclear export of beta-catenin. Mol Cell Biol. 2006;26:8857–67. doi: 10.1128/MCB.01031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jia J, Amanai K, Wang G, Tang J, Wang B, Jiang J. Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature. 2002;416:548–52. doi: 10.1038/nature733. [DOI] [PubMed] [Google Scholar]
- 46.Jia J, Zhang L, Zhang Q, Tong C, Wang B, Hou F, et al. Phosphorylation by double-time/CKIepsilon and CKIalpha targets cubitus interruptus for Slimb/beta-TRCP-mediated proteolytic processing. Dev Cell. 2005;9:819–30. doi: 10.1016/j.devcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Price MA, Kalderon D. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell. 2002;108:823–35. doi: 10.1016/s0092-8674(02)00664-5. [DOI] [PubMed] [Google Scholar]
- 48.Smelkinson MG, Zhou Q, Kalderon D. Regulation of Ci-SCFSlimb binding, Ci proteolysis, and hedgehog pathway activity by Ci phosphorylation. Dev Cell. 2007;13:481–95. doi: 10.1016/j.devcel.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demarchi F, Bertoli C, Sandy P, Schneider C. Glycogen synthase kinase-3 beta regulates NFkappaB1/p105 stability. J Biol Chem. 2003;278:39583–90. doi: 10.1074/jbc.M305676200. [DOI] [PubMed] [Google Scholar]
- 50.Aramata S, Han SI, Kataoka K. Roles and regulation of transcription factor MafA in islet beta-cells. Endocr J. 2007;54:659–66. doi: 10.1507/endocrj.kr-101. [DOI] [PubMed] [Google Scholar]
- 51.Suen CS, Berrodin TJ, Mastroeni R, Cheskis BJ, Lyttle CR, Frail DE. A transcriptional coactivator, steroid receptor coactivator-3, selectively augments steroid receptor transcriptional activity. J Biol Chem. 1998;273:27645–53. doi: 10.1074/jbc.273.42.27645. [DOI] [PubMed] [Google Scholar]
- 52.Viatour P, Merville MP, Bours V, Chariot A. Protein phosphorylation as a key mechanism for the regulation of BCL-3 activity. Cell Cycle. 2004;3:1498–501. doi: 10.4161/cc.3.12.1328. [DOI] [PubMed] [Google Scholar]
- 53.Viatour P, Dejardin E, Warnier M, Lair F, Claudio E, Bureau F, et al. GSK3-mediated BCL-3 phosphorylation modulates its degradation and its oncogenicity. Mol Cell. 2004;16:35–45. doi: 10.1016/j.molcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 54.Wu RC, Feng Q, Lonard DM, O'Malley BW. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 2007;129:1125–40. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 55.Han SI, Aramata S, Yasuda K, Kataoka K. MafA stability in pancreatic beta cells is regulated by glucose and is dependent on its constitutive phosphorylation at multiple sites by glycogen synthase kinase 3. Mol Cell Biol. 2007;27:6593–605. doi: 10.1128/MCB.01573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rocques N, Abou Zeid N, Sii-Felice K, Lecoin L, Felder-Schmittbuhl MP, Eychene A, et al. GSK-3-mediated phosphorylation enhances Maf-transforming activity. Mol Cell. 2007;28:584–97. doi: 10.1016/j.molcel.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 57.Latres E, Chiaur DS, Pagano M. The human F box protein beta-Trcp associates with the Cul1/Skp1 complex and regulates the stability of beta-catenin. Oncogene. 1999;18:849–54. doi: 10.1038/sj.onc.1202653. [DOI] [PubMed] [Google Scholar]
- 58.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsuchiya K, Nakamura T, Okamoto R, Kanai T, Watanabe M. Reciprocal targeting of Hath1 and beta-catenin by Wnt glycogen synthase kinase 3beta in human colon cancer. Gastroenterology. 2007;132:208–20. doi: 10.1053/j.gastro.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 60.Lee JY, Yu SJ, Park YG, Kim J, Sohn J. Glycogen synthase kinase 3beta phosphorylates p21WAF1/CIP1 for proteasomal degradation after UV irradiation. Mol Cell Biol. 2007;27:3187–98. doi: 10.1128/MCB.01461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bendjennat M, Boulaire J, Jascur T, Brickner H, Barbier V, Sarasin A, et al. UV irradiation triggers ubiquitin-dependent degradation of p21(WAF1) to promote DNA repair. Cell. 2003;114:599–610. doi: 10.1016/j.cell.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 62.Rossig L, Badorff C, Holzmann Y, Zeiher AM, Dimmeler S. Glycogen synthase kinase-3 couples AKT-dependent signaling to the regulation of p21Cip1 degradation. J Biol Chem. 2002;277:9684–9. doi: 10.1074/jbc.M106157200. [DOI] [PubMed] [Google Scholar]
- 63.Flugel D, Gorlach A, Michiels C, Kietzmann T. Glycogen synthase kinase 3 phosphorylates hypoxia-inducible factor 1alpha and mediates its destabilization in a VHL-independent manner. Mol Cell Biol. 2007;27:3253–65. doi: 10.1128/MCB.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mottet D, Dumont V, Deccache Y, Demazy C, Ninane N, Raes M, et al. Regulation of hypoxiainducible factor-1alpha protein level during hypoxic conditions by the phosphatidylinositol 3-kinase/Akt/glycogen synthase kinase 3beta pathway in HepG2 cells. J Biol Chem. 2003;278:31277–85. doi: 10.1074/jbc.M300763200. [DOI] [PubMed] [Google Scholar]
- 65.Morel C, Carlson SM, White FM, Davis RJ. Mcl-1 integrates the opposing actions of signaling pathways that mediate survival and apoptosis. Mol Cell Biol. 2009;29:3845–52. doi: 10.1128/MCB.00279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Y, Altman BJ, Coloff JL, Herman CE, Jacobs SR, Wieman HL, et al. Glycogen synthase kinase 3alpha and 3beta mediate a glucose-sensitive anti-apoptotic signaling pathway to stabilize Mcl-1. Mol Cell Biol. 2007;27:4328–39. doi: 10.1128/MCB.00153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding Q, He X, Hsu JM, Xia W, Chen CT, Li LY, et al. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol. 2007;27:4006–17. doi: 10.1128/MCB.00620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2006;21:749–60. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 69.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 70.Gregory MA, Qi Y, Hann SR. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J Biol Chem. 2003;278:51606–12. doi: 10.1074/jbc.M310722200. [DOI] [PubMed] [Google Scholar]
- 71.Kitagawa K, Hiramatsu Y, Uchida C, Isobe T, Hattori T, Oda T, et al. Fbw7 promotes ubiquitin-dependent degradation of c-Myb: involvement of GSK3-mediated phosphorylation of Thr-572 in mouse c-Myb. Oncogene. 2009;28:2393–405. doi: 10.1038/onc.2009.111. [DOI] [PubMed] [Google Scholar]
- 72.Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101:9085–90. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–14. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Welcker M, Singer J, Loeb KR, Grim J, Bloecher A, Gurien-West M, et al. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol Cell. 2003;12:381–92. doi: 10.1016/s1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
- 76.Sundqvist A, Bengoechea-Alonso MT, Ye X, Lukiyanchuk V, Jin J, Harper JW, et al. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7) Cell Metab. 2005;1:379–91. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 77.Kang T, Wei Y, Honaker Y, Yamaguchi H, Appella E, Hung MC, et al. GSK-3beta targets Cdc25A for ubiquitin-mediated proteolysis, and GSK-3beta inactivation correlates with Cdc25A overproduction in human cancers. Cancer Cell. 2008;13:36–47. doi: 10.1016/j.ccr.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hayakawa M, Kitagawa H, Miyazawa K, Kitagawa M, Kikugawa K. The FWD1/beta-TrCP-mediated degradation pathway establishes a `turning off switch' of a Cdc42 guanine nucleotide exchange factor, FGD1. Genes Cells. 2005;10:241–51. doi: 10.1111/j.1365-2443.2005.00834.x. [DOI] [PubMed] [Google Scholar]
- 79.Harada Y, Sakai M, Kurabayashi N, Hirota T, Fukada Y. Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3beta. J Biol Chem. 2005;280:31714–21. doi: 10.1074/jbc.M506225200. [DOI] [PubMed] [Google Scholar]
- 80.Quelo I, Akhouayri O, Prud'homme J, St-Arnaud R. GSK3beta-dependent phosphorylation of the alpha NAC coactivator regulates its nuclear translocation and proteasome-mediated degradation. Biochemistry. 2004;43:2906–14. doi: 10.1021/bi036256+. [DOI] [PubMed] [Google Scholar]
- 81.Boucher MJ, Selander L, Carlsson L, Edlund H. Phosphorylation marks IPF1/PDX1 protein for degradation by glycogen synthase kinase 3-dependent mechanisms. J Biol Chem. 2006;281:6395–403. doi: 10.1074/jbc.M511597200. [DOI] [PubMed] [Google Scholar]
- 82.Kirschenbaum F, Hsu SC, Cordell B, McCarthy JV. Glycogen synthase kinase-3beta regulates presenilin 1 C-terminal fragment levels. J Biol Chem. 2001;276:30701–7. doi: 10.1074/jbc.M102849200. [DOI] [PubMed] [Google Scholar]
- 83.He X, Saint-Jeannet JP, Woodgett JR, Varmus HE, Dawid IB. Glycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos. Nature. 1995;374:617–22. doi: 10.1038/374617a0. [DOI] [PubMed] [Google Scholar]
- 84.Pierce SB, Kimelman D. Regulation of Spemann organizer formation by the intracellular kinase Xgsk-3. Development. 1995;121:755–65. doi: 10.1242/dev.121.3.755. [DOI] [PubMed] [Google Scholar]
- 85.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–12. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schmidt JE, von Dassow G, Kimelman D. Regulation of dorsal-ventral patterning: the ventralizing effects of the novel Xenopus homeobox gene Vox. Development. 1996;122:1711–21. doi: 10.1242/dev.122.6.1711. [DOI] [PubMed] [Google Scholar]
- 87.Onichtchouk D, Glinka A, Niehrs C. Requirement for Xvent-1 and Xvent-2 gene function in dorsoventral patterning of Xenopus mesoderm. Development. 1998;125:1447–56. doi: 10.1242/dev.125.8.1447. [DOI] [PubMed] [Google Scholar]
- 88.Ramel MC, Lekven AC. Repression of the vertebrate organizer by Wnt8 is mediated by Vent and Vox. Development. 2004;131:3991–4000. doi: 10.1242/dev.01277. [DOI] [PubMed] [Google Scholar]
- 89.Zhu Z, Kirschner M. Regulated proteolysis of Xom mediates dorsoventral pattern formation during early Xenopus development. Dev Cell. 2002;3:557–68. doi: 10.1016/s1534-5807(02)00270-8. [DOI] [PubMed] [Google Scholar]
- 90.Sherr CJ. Mammalian G1 cyclins. Cell. 1993;73:1059–65. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 91.Cross DA, Culbert AA, Chalmers KA, Facci L, Skaper SD, Reith AD. Selective small-molecule inhibitors of glycogen synthase kinase-3 activity protect primary neurones from death. J Neurochem. 2001;77:94–102. doi: 10.1046/j.1471-4159.2001.t01-1-00251.x. [DOI] [PubMed] [Google Scholar]
- 92.Pap M, Cooper GM. Role of glycogen synthase kinase-3 in the phosphatidylinositol 3-Kinase/Akt cell survival pathway. J Biol Chem. 1998;273:19929–32. doi: 10.1074/jbc.273.32.19929. [DOI] [PubMed] [Google Scholar]
- 93.Michels J, Johnson PW, Packham G. Mcl-1. Int J Biochem Cell Biol. 2005;37:267–71. doi: 10.1016/j.biocel.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 94.Dominguez I, Green JB. Missing links in GSK3 regulation. Dev Biol. 2001;235:303–13. doi: 10.1006/dbio.2001.0317. [DOI] [PubMed] [Google Scholar]