Friedreich Ataxia: Molecular Mechanisms, Redox Considerations, and Therapeutic Opportunities (original) (raw)
Abstract
Mitochondrial dysfunction and oxidative damage are at the origin of numerous neurodegenerative diseases like Friedreich ataxia and Alzheimer and Parkinson diseases. Friedreich ataxia (FRDA) is the most common hereditary ataxia, with one individual affected in 50,000. This disease is characterized by progressive degeneration of the central and peripheral nervous systems, cardiomyopathy, and increased incidence of diabetes mellitus. FRDA is caused by a dynamic mutation, a GAA trinucleotide repeat expansion, in the first intron of the FXN gene. Fewer than 5% of the patients are heterozygous and carry point mutations in the other allele. The molecular consequences of the GAA triplet expansion is transcription silencing and reduced expression of the encoded mitochondrial protein, frataxin. The precise cellular role of frataxin is not known; however, it is clear now that several mitochondrial functions are not performed correctly in patient cells. The affected functions include respiration, iron–sulfur cluster assembly, iron homeostasis, and maintenance of the redox status. This review highlights the molecular mechanisms that underlie the disease phenotypes and the different hypothesis about the function of frataxin. In addition, we present an overview of the most recent therapeutic approaches for this severe disease that actually has no efficient treatment. Antioxid. Redox Signal. 13, 0000–0000.
- Introduction and History
- Clinical Features and Pathogenesis of Friedreich Ataxia
- Mutations in the FXN Gene Cause FRDA
- Frataxin Is a Unique Protein
- Frataxin Function in Cell Iron Use and Oxidative-Stress Defense
- Frataxin Is Involved in Development, Cell Death, and Cancer
- Therapeutic Approaches for Treatment of Friedreich Ataxia
- Conclusion
I. Introduction and History
Friedreich ataxia (FRDA) was first described in 1863 by Nikolaus Friedreich (107–109) and generally was accepted as a new disease after his publications in 1877 (110, 111). Friedreich described the fundamental clinical and pathologic features of the most common hereditary ataxia, which are age at onset around puberty; degenerative atrophy of the posterior columns of the spinal cord, leading to progressive ataxia, sensory loss, and muscle weakness; and also often observed, scoliosis, foot deformity, and cardiac symptoms. The eponym was proposed for this disease by Brousse in 1882 (233). Eight years later, Ladame (175) published the description of 165 cases. However, the degree of variability in the clinical features of FRDA made controversial the diagnosis and the description of new cases for the next 100 years. Since the description of the disease by Friedreich, a disagreement existed about the distinct nature of FRDA and Charcot-Marie-Tooth disease, and misdiagnosis were frequent (141, 229, 271). Therefore, a clear definition of FRDA was needed, and the Québec Collaborative Group in 1976, based on a clinical study of 50 cases, proposed several criteria essential for diagnosis (123). In a study of 150 cases, Harding (140) agreed with the Québec classification stating that ataxia of all limbs and absence of tendon reflexes were obligatory features, but found these criteria too rigid and difficult to apply in early childhood. She thus proposed a list of criteria for the diagnosis of FRDA shown in Fig. 1. A later clinical review of 12 children confirmed that the Québec criteria were not appropriate in such cases and supported the use of Harding's criteria for early diagnosis (1). Recessive inheritance of the disease was widely accepted at this time, even though occasional reports of pseudodominant cases continue to appear in the literature (153, 230, 318). Nevertheless, the failure to diagnose atypical cases with an overall FRDA-like phenotype, but missing some of the essential diagnostic features, persisted. Whether these cases represented extreme patterns of the disease or different diseases was determined only once the genetic mutation underlying FRDA had been identified.
FIG. 1.
Diagnostic criteria for typical FRDA according to Harding (140).
Given that such clinical variability is unusual for a recessive disorder, several authors suggested that FRDA was caused by mutations in several different genes, with one mutation playing a predominant role (19, 140, 324). In 1988, the gene mutated in FRDA patients was mapped to chromosome 9 by linkage analysis with restriction length polymorphism (RFLP) markers (57). The meticulous work of several research groups allowed the chromosome region to be narrowed to 150 kb at 9q13 (113, 212, 260, 291, 295). Cloning the gene proved problematic because of the reduced level of recombination events occurring close to the FRDA locus and the proximity of this locus to the variable heterochromatic region near the centromere. In 1996, an international collaborative effort successfully identified the X25 gene (now named FXN gene according to the HUGO Gene Nomenclature Committee) and the mutations responsible for FRDA (51). The majority of FRDA patients (96%) were homozygous for an unstable GAA trinucleotide repeat expansion in the first intron of the FXN gene, but a few patients were heterozygous, with point mutations found on the other allele (51, 70). The identification of the FXN gene and of the most frequent mutation provided a valuable tool for the diagnosis of FRDA. Additionally, it demonstrated that the typical and atypical phenotypes of the disease were caused by mutations in the same gene (58, 78, 93, 164).
Before the identification of the gene involved, FRDA was suggested to be an inherited metabolic disease; however, despite the large number of investigations carried out, the findings were contradictory, with no precise biochemical deficiency detected (250). Nevertheless, the mitochondrial nature of the pathology was anticipated before the discovery of the gene (17). The identification of the gene allowed the biochemical defect underlying the disease to be clarified. The expanded GAA repeats cause an abnormal conformation of DNA and a decrease in transcription. The consequence is a reduction in the expression of the FXN gene and a decrease in the encoded protein, frataxin (51). Frataxin localizes to the mitochondria, but its function remains unclear. It is involved in iron homeostasis (iron–sulfur cluster and heme synthesis), respiratory control, and resistance to oxidative stress (15, 40, 183, 215, 262).
A link to oxidative stress was anticipated because of the similarity of phenotypes between FRDA and ataxia with vitamin E deficiency (AVED) (126). The importance of oxidative damage in the pathogenesis of FRDA was further confirmed by the treatment of three patients with idebenone, a free-radical scavenger, and the resulting reduction in myocardial hypertrophy (266). Idebenone remains the best drug for the treatment of FRDA.
Here, we provide a comprehensive review of the clinical features and pathogenesis of Friedreich ataxia, the mutations in the FXN gene causing the disease, the various findings and hypotheses concerning the function of frataxin and an update on therapeutic solutions.
II. Clinical Features and Pathogenesis of Friedreich Ataxia
A. Epidemiology and clinical features
Friedreich ataxia (MIM 229300) is the most common inherited recessive ataxia. It is an autosomal recessive disease with an estimated prevalence in the order of 1:50,000 in white populations (51, 140). The carrier rate has been estimated at 1:120–1:60 (51, 85, 141). The incidence of the disease may be higher in certain populations because of founder effects (35, 80) or consanguineous marriages (261). This disease is found only in individuals of European, North African, Middle Eastern, or Indian origin and is very rare, or inexistent, in sub-Saharan Africans, Amerindians, and Asians (173).
The first symptoms usually appear around puberty, but the age at onset can vary from infancy (2–3 years) to adulthood (after 25 years old). Gait instability and generalized clumsiness are typical presenting symptoms. As described by Friedreich, the main feature of this disease is the progressive and unremitting ataxia. On average, patients lose the ability to walk 10 to 15 years after onset of the disease and need a wheelchair to accomplish daily activities (93, 140, 211). Other main neurologic features of FRDA include dysarthria, tendon areflexia, sensory loss, and pyramidal signs (Fig. 1, Table 1). Two thirds of patients have cardiac symptoms (left ventricular hypertrophy), which contribute to disability and cause premature death (93). Cardiomyopathy is the most frequent cause of death among FRDA patients. Only in some patients do skeletal deformation (such as scoliosis and pes cavus), ocular abnormalities (such as nystagmus, optic atrophy, or fixation instability) and hearing loss develop. Diabetes mellitus is found in 14–19% of patients, and glucose intolerance, in 24–40% (99, 100, 288). Diabetes usually develops at a late stage of the disease, after a mean disease duration of 15 years (97, 140).
Table 1.
Frequency of Clinical Signs (Percentage of Patients) Observed in Harding's Study and in Genetically Confirmed, Homozygous, and Heterozygous, FRDA Patients
| Clinical sign | Harding (140) 1981a | Dürr et al. (93) 1996b | Schöls et al. (282) 1997c | Lamont et al. (179) 1997d | Delatycki et al. (84) 1999e | Cossée et al. (70) 1999f | Cossée et al. (70) 1999g |
|---|---|---|---|---|---|---|---|
| Gait ataxia | – | 100 | 100 | – | 100 | 100 | 96h |
| Limb ataxia | 99 | 99 | 100 | 100 | 100 | – | – |
| Lower-limb areflexia | 99 | 87 | 84 | 87 | 98 | 88 | 74 |
| Decreased vibration sense | 73 | 78 | 83 | 87 | 88 | 88 | 84 |
| Extensor plantar response | 89 | 79 | 95 | 96 | 74 | 91 | 86 |
| Axonal neuropathy | 96 | 98 | 100 | – | – | – | – |
| Dysarthria | 97 | 91 | 100 | 91 | 95 | 90 | 58 |
| Scoliosis | 79 | 60 | 84 | – | 78 | 61 | 75 |
| Cardiomyopathy | 66 | 63 | 89 | 77 | 65 | 70 | 68 |
| Nystagmus | 20 | – | 39 | – | – | 38 | 35 |
| Decreased visual acuity/Optic atrophy | 18 | 13 | 9 | – | – | 5i | 33i |
| Hearing loss | 8 | 13 | 39 | – | – | – | – |
| Amyotrophy | 40 | – | 50 | – | – | – | – |
| Pes cavus | 55 | 55 | 82 | – | 74 | 54 | 73 |
| Diabetes or glucose intolerance | 10 | 32 | 6 | – | 8 | 20 | 9 |
| Percentage atypical | 0 | 24 | 25 | 14 | 8 | – | 36 |
The variability in clinical signs observed in FRDA cases is very extensive and includes age at onset, rate of progression, severity, and duration of the disease. Several atypical FRDA variants [as opposed to “typical” or “classic” FRDA cases that have all the clinical features described by Harding (140)], with an overall FRDA-like phenotype but missing at least one essential diagnostic criterion, are well characterized. Atypical variants thus include the Acadian type, late-onset FRDA (LOFA), and FRDA with retained reflexes (FARR). The Acadian type is observed in a population of French origin living in North America and in their descendants, living in Louisiana, and now called Cajuns. The age at onset is slightly later, and the disease has a milder course of degeneration than does classic FRDA (19, 209). In addition, several clinical signs, including scoliosis, pes cavus, and cardiomyopathy, are frequently less severe in the Acadian type (19, 209).
LOFA cases have all the features of typical FRDA, but disease onset is after 25 years of age (77, 211). Disease progression, as indicated by years from onset to becoming confined to a wheelchair, is slower in LOFA. Comparative studies of patients with LOFA and typical FRDA show an increased occurrence of lower-limb spasticity and retained reflexes and decreased skeletal abnormalities in LOFA patients (27, 67). In some reported cases, age at onset occurs after 40 or even after 60 years old (very late-onset FRDA) (122, 299).
FARR is a variant in which tendon reflexes in the lower limbs are preserved, and the clinical features are generally present but less pronounced (139, 164, 228). FARR cases frequently arise in siblings of patients with typical FRDA cases (98).
B. Pathophysiology
The neuropathology of FRDA shows marked differences in comparison with other hereditary ataxias, and major changes occur in the spinal cord, peripheral nerves, and cerebellum [for a recent review, see (232)]. Neurodegeneration occurs first in the dorsal root ganglia (DRG), with loss of large sensory neurons, followed by degeneration of posterior columns, corticospinal tracts and spinocerebellar tracts of the spinal cord, and the dentate nucleus in the cerebellum.
Sural nerve biopsies of FRDA patients show axonal neuropathy with a profound reduction in the density of large myelinated fibers (152, 259, 267). The density of small myelinated fibers may be normal (267) or moderately reduced (334). The fine unmyelinated fibers are generally well preserved. Onion-bulb complexes may also be present (20, 259). Electrophysiologic abnormalities in peripheral nerves include severe reduction or complete loss of sensory nerve action potentials and slightly decreased nerve-conduction velocities (334). The cellular events leading to the loss of the large myelinated fibers remain unclear, but could include axonal degeneration and demyelination, involving both neurons and Schwann cells (194).
Magnetic resonance imaging of the cervical spinal cord of FRDA patients showed thinning consistent with degeneration of posterior and lateral columns (197). Loss of myelinated fibers and gliosis are characteristic of these regions of the spinal cord (152, 167, 176). Severe neuronal loss also is observed in the Clarke column, with atrophy in the spinocerebellar tracts. The Clarke column is a major relay center for unconscious proprioception, while sensory information is passed to the cerebellar cortex by the spinocerebellar tracts. Neuronal degeneration in these regions leads to the loss of position and vibration senses and abolishes reflexes in FRDA patients. Atrophy also is observed in the corticospinal motor tracts. The pattern of atrophy of the long-tract fibers, severely affected in the distal portions, suggests a “dying back” process (267). The degeneration of corticospinal and pyramidal tracts leads to muscle weakness and extensor plantar responses. In the cerebellum, the dentate nucleus is severely affected, but the cortex is spared during the beginning stages of the disease, until Purkinje cell loss can be observed (166). Progressive atrophy of sensory and cerebellar pathways causes ataxia, dysarthria, gait instability, and profound sensory loss.
Other organs affected in FRDA patients include heart, pancreas, and skeleton. The heart is affected in the majority of patients, the most common cardiac lesion being hypertrophic cardiomyopathy, in which ventricular and interventricular septum walls are thickened. Iron deposition in the myocardium also has been reported (177, 272). In FRDA patients, loss of islet cells causes diabetes, but the signs of autoimmune attack associated with type I diabetes are not observed in these patients (281).
III. Mutations in the FXN Gene Cause FRDA
A. Mapping and cloning of the FXN gene
The gene responsible for FRDA was identified by positional cloning. To map the locus, Chamberlain and colleagues (57) examined 22 European and American families with at least three affected siblings, diagnosed by using the criteria of the Québec Collaborative Group to minimize clinical heterogeneity (123). They mapped the locus to chromosome 9 by genetic linkage to an anonymous marker D9S15 (MCT112 probe) and an interferon-β gene probe (IFNB) and proposed a regional localization in the proximal short arm (9p22-CEN). Genetic linkage to D9S15 was confirmed by the study of another 33 French families, and an additional unmapped RFLP marker was found closely linked to the FRDA locus, D9S5 (112). However, close linkage to the IFNB probe localized to 9p22 was not confirmed (112, 138). The analysis of allelic association to D9S5 and D9S15 revealed linkage disequilibrium between the FRDA locus and the D9S15 RFLP probe (138). In situ hybridization by using D9S5 and D9S15 probes physically assigned these markers to the 9q13-9q21.1 region. The FRDA locus was thus mapped to the proximal long arm of chromosome 9, close to the heterochromatic region (138, 291). These results and others (58) were in favor of locus homogeneity, at least in typical cases of FRDA. The distance between the FRDA locus and D9S5/D9S15 markers was estimated to be < 1 cM, but the absence of recombination between the three loci prevented further genetic ordering. Therefore, a 1-Mb physical map was constructed by PFGE by using YAC and cosmid cloning in the region encompassing D9S5 and D9S15 (114, 115, 320). This map showed that the distance between D9S5 and D9S15 markers was < 260 kb (114). Finally, a meiotic recombination event was found in three large inbred Tunisian families, and by using additional polymorphic markers, D9S15 was excluded (22). The position of the locus was further refined, and the gene order determined, by the discovery of other recombination events. The gene order was thus found to be cen-FRDA-D9S5-D9S15-tel (56). The genomic region containing the FRDA locus was progressively narrowed, by using new polymorphic microsatellite markers, to a 150-kb interval between FR2 and F8101 (212, 260). The only gene within the minimum candidate region was X25 (subsequently named FRDA and now FXN), and the FRDA locus was at last identified (51).
Two reports have shown evidence of genetic heterogeneity in FRDA (62, 168). The patients included in these studies had typical FRDA but did not have mutations in the FXN gene. A second locus, FRDA2, mapping to chromosome 9p23-p11, was thus proposed (62). No other studies have confirmed these observations, but occasional reports appear in the literature of patients who clinically have FRDA symptoms but in whom a mutation in the FXN gene cannot be detected (70, 75, 199, 336).
B. Structure and regulation of the FXN gene
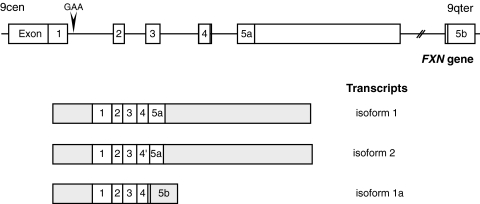
The sequence of the FXN gene was reported by Campuzano and co-workers in 1996 (51). They described a gene composed of seven exons (exons 1–4, 5a, 5b, and 6) encompassing 85 kb of genomic DNA (Fig. 2). The 5′ end of the gene, including the first exon, enclosed an unmethylated CpG island containing several rare restriction-enzyme sites. Northern blot analysis, RNase protection assays, and cDNA cloning showed a major 1.3-kb transcript composed of five exons, 1 to 5a (51). This transcript encoded a protein of 210 amino acids, frataxin (isoform 1, Figs. 2 and 3). A minor alternative transcript, containing exon 5b instead of 5a, followed or not by noncoding exon 6, was also described (51). Exon 5b was located 40 kb downstream of exon 5a. Exon 5b carried an in-frame stop codon, resulting in a transcript that encoded a shorter protein of 171 amino acids and differed in the 11 amino acids of the C-terminus [isoform 1a, Figs. 2 and 3; referred as isoform 2 by Campuzano and associates (51)]. This transcript is very rare, and the expression pattern of frataxin isoform 1a was not studied further. The functional significance of this shorter isoform is questionable, the most conserved domains of frataxin being encoded by exons 4 and 5a.
FIG. 2.
Structure of the FXN gene and transcription maps. The structures of the FXN gene, except for the nontranslated exon 6, and of the three transcripts, are depicted. Transcript encoding isoform 2 results from an alternative splicing at exon 4′). Transcript encoding isoform 1a uses the exon 5b that is located 40 kb downstream of exon 5a. The GAA-repeat expansion in the first intron is indicated. The gray regions are not translated. Exons and introns are represented in different scales.
FIG. 3.
Amino acid sequence comparison of the three human frataxin isoforms. The mature forms are represented in bold, and the C-terminal variant region is boxed (isoform 1, 210 amino acids; isoform 1a, 171 amino acids; and isoform 2, 196 amino acids). Clustal W software was used for sequence alignment.
In an attempt to clone the full-length frataxin cDNA by PCR by using primers targeting the extremities of FXN isoform 1 coding sequence, Pianese and co-workers (242) isolated a third transcript. This transcript is generated by alternative splicing at a second donor splice site in intron 4, resulting in an 8-bp insertion between exons 4 and 5a. This splicing introduces a frameshift with the appearance of a new stop codon in exon 5a. The transcript thus encodes a putative 196-amino acid protein that differs from isoform 1 after residue 160 (isoform 2, Figs. 2 and 3). This transcript is produced at lower levels than isoform 1 and was found in brain, cerebellum, spinal cord, heart, and skeletal muscle (242). No functional data have been reported regarding the potential function of this transcript and of frataxin isoform 2.
The 1,255-bp region extending 5′ from the human FXN open reading frame contains the promoter region (136). This promoter does not contain a TATA sequence but is rich in repetitive elements of different origins; retroelements (_Alu_Jb, _Alu_Y, and L2), and mammalian-wide interspersed repeats (MIR). The Alu and MIR elements significantly enhance the activity of the promoter, and the 221-bp sequence containing the L2-like element is required to drive the luciferase activity of reporter constructs (136). Additionally, an E-box element in the first intron of the gene modulates the activity of the promoter because its deletion causes a significant reduction in reporter detection (137). Comparison of promoter sequences between unaffected individuals, three patients with classic FRDA, and three Acadian-descendent patients did not show any significant differences that could explain the variation in clinical presentation (136).
Little is known about the regulation of the human FXN gene. The E-box sequence binds transcriptional factors belonging to the basic helix-loop-helix family. One protein that may bind to this sequence is the muscle-specific factor Mt, although its biologic importance is not known (137). Recent findings have shown that frataxin expression is iron regulated. The addition of the iron chelator deferoxamine to various human cell lines and to FRDA patient fibroblasts and lymphoblasts leads to a reduced steady-state level of frataxin mRNA and protein (187). Conversely, addition of ferric ammonium citrate or hemin increases frataxin expression (187, 276). Studies based on the use of promoter-luciferase constructs have shown that iron acts on transcriptional regulation, but the key regulators remain to be determined (187).
The mouse FXN gene was reported to be directly regulated by the transcription factor hypoxia-inducible factor 2α (HIF-2α encoded by the Epas1 gene) (227). The frataxin protein and mRNA levels are reduced to < 50% in the liver of knockout _Epas_-/- mice compared with control mice. However, it is not known whether the human FXN gene is also regulated by HIF-2α.
C. Developmental expression of the FXN gene
FXN gene expression and production of the protein, frataxin, are ubiquitous. However, the levels of mRNA and protein show tissue specificity that partially correlates with the main sites of disease. In humans, adults show the highest levels of expression in the heart and spinal cord, with intermediate levels observed in the cerebellum, liver, skeletal muscle, and pancreas and very low levels in the cerebral cortex (51).
Northern blot analysis and RNA in situ hybridization of mouse adult tissues showed that the FXN gene is expressed in the heart, liver, skeletal muscle, kidney, spleen, and thymus, and, to a lesser extent, in the brain and lungs (169). In addition, transcript-distribution studies in mouse embryos have demonstrated that the frataxin gene is developmentally regulated. Expression is not detectable at E8.5, is weak at E12.5, and increases until E16.5, when no further change is observed until the neonatal period (158, 169). At E14.5, expression is high in the ventricular zone of the brain, the anterior horns of the spinal cord, the large neuronal cells in the DRG, and in the granular layer of the cerebellum. In nonneural tissues, FXN mRNA is found in the heart, kidney, and brown fat (158, 169). The major sites of frataxin gene expression in the developing mouse embryo correlate with the major sites of disease, with a few exceptions. In FRDA patients, for example, the posterior columns of the spinal cord are affected, whereas, in the mouse embryo, the major site of expression is observed in the anterior columns (158). Another example is the substantially higher level of frataxin gene expression found in the mouse cortex (158) than in the human brain (51). Additionally, whereas expression levels are high in the adult and fetal mouse kidney, they are very low in the human adult kidney, which is not an organ affected in FRDA (51, 169). In summary, the distribution of frataxin mRNA in mouse and human tissues shows a broader distribution of sites with frataxin gene expression than of sites affected in FRDA disease. The reason for only certain tissues being affected may be that neurons, cardiomyocytes and pancreatic β-cells are particularly highly dependent on mitochondrial metabolism and, being nondividing cells, are not replaced when they die (233). Alternatively, this tissue specificity could be due to somatic instability and accumulation of larger trinucleotide repeat expansions in these cell types, at least for DRG neurons (73).
D. Molecular mechanism of GAA triplet-repeat expansion
FRDA is caused by a distinctive mutation not found in any other disease. In 98% of patient chromosomes, a GAA trinucleotide repeat expansion has been detected in the first intron of the gene (51). GAA repeats are normally found in the human FXN gene and are derived from a poly(A) expansion of the canonic A5TACA6 sequence at the center of an _Alu_Sx sequence flanked by a 13-bp direct repeat (AAAATGGATTTCC) (Fig. 4) (51, 72, 209). Because Alu retrotransposons are primate-genome specific, GAA triplet expansions are thought to have appeared in the primate lineage (159).
FIG. 4.
Comparison between the nucleotide sequences of the FXN Alu element and the _Alu_Sx consensus. The human FXN Alu element contains an expanded A5TACA5 sequence (A6TACA16, bold) followed by the GAA repeats (underlined). The most frequent number of GAA triplets (nine repeats) is represented. The flanking direct repeat is boxed.
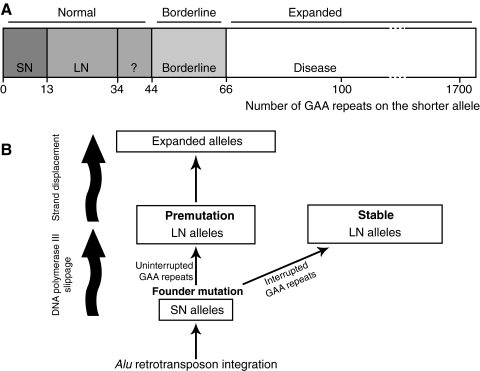
Studies of populations from different parts of the world have shown that half of normal alleles carry nine GAA repeats (72, 159, 209). Two studies (72, 209), composed mainly of European individuals, showed that the GAA motif in normal alleles is polymorphic with a bimodal distribution, 83% having six to 12 repeats (small normal, SN) and 17% having 13 to 34 repeats (large normal, LN). This was confirmed for African American, African, and Syrian populations (159). Interestingly, no alleles with > 10 GAA repeats were found in Papua New Guineans, and only one was found in the Asian population (159), correlating with the absence of disease in these regions (173). A small proportion (<1%) of LN alleles have > 34 GAA repeats, which can be interrupted by a hexanucleotide repeat (GAGGAA) or another sequence in some cases or are continuous in other cases (72, 209). Occasionally, LN alleles containing ≥34 uninterrupted GAA repeats undergo hyperexpansion to produce hundreds of triplets in one generation (72, 209, 282), indicating meiotic instability. However, alleles with interrupted GAA expansions are stable, even those containing >100 GAA repeats (72). LN alleles carrying between 44 and 66 uninterrupted repeats are borderline, having been associated both with healthy carriers and with FRDA symptoms (Fig. 5A) (290). Analysis of intergenerational variability of GAA-repeat number in parent–child transmission to affected and carrier offspring showed that paternal transmission is most often accompanied by a contraction of the repeats, and maternal transmission may result in further expansion or contraction (76, 82, 208, 241, 282). Reversion of the expansion to a normal number of repeats is very rare but has been reported for three different cases (30, 76, 289). Expansion of the triplet repeat in FRDA patient chromosomes ranges from 44 to 1,700 repeats, this number being between 600 and 900 for the majority of chromosomes (Fig. 5A) (51, 93, 96, 97, 211, 290).
FIG. 5.
Sequence and origin of the GAA triplet-repeat expansion in the FXN gene. (A) Number of GAA-triplet repeats in normal, borderline, and expanded alleles. The phenotypic consequence of alleles containing 35 to 43 repeats is not known. Borderline alleles can cause disease with mild phenotypes or not. (B) Model for the origin and evolution of GAA triplet-repeat expansion [see text for explanation; adapted from (209)].
The analysis of normal alleles is fundamental to our understanding of the major mechanisms involved, first, in driving GAA triplet-repeat expansion and, second, in determining the pathogenicity of such expansion. Linkage disequilibrium analysis and haplotype data suggest that SN alleles evolved into expanded alleles through a two-step process (Fig. 5B) (72, 209). LN and expanded alleles are associated frequently with the same haplotypes and rarely with the major haplotypes found in SN alleles, suggesting that LN alleles are derived from one or a small number of ancestral founder mutations (72). The mutation was probably caused by the slipping of DNA polymerase III. Conversely, de novo hyperexpansion from the pool of LN alleles (34 to 60 GAA) carrying uninterrupted rows of repeats (premutation) may represent a reservoir for pathogenic expansion (72, 209, 289). Thus, the overall tendency for contraction of the expanded alleles, which could lead to their disappearance in the population, is compensated for by expansion of the premutation.
Expanded GAA triplet repeats show extensive instability in cultured cells, in the blood, in the central nervous system, in the DRG, the spinal cord, and the heart (30, 73, 74, 210, 211). This leads to somatic mosaicism for expansion sizes when single-cell analysis is performed. Small-pool PCR experiments have shown that expanded GAA triplet repeats are very unstable in the peripheral leukocytes of patients, giving ∼65% variability in size compared with the results obtained with classic PCR (289). The threshold expansion length for the initiation of somatic variability is between 26 and 44 uninterrupted GAA repeats (289), or even just between 40 and 44 repeats (244). DRG degeneration is the primary cause of neurologic problems in FRDA patients. These neurons are highly sensitive to frataxin deficiency, as observed in neuron-specific conditional frataxin-knockout mice (294). In addition, De Biase et al. (73, 74) demonstrated that, specifically in DRG, somatic instability starts after early embryonic development and continues after birth throughout life, resulting in progressive, age-dependent accumulation of larger GAA triplet-repeat expansions. In this study, the DRG was the only tissue analyzed in which long expansions arose more frequently than did contractions of a similar length (73). It is thus possible that DRG somatic instability contributes to disease progression. This was not seen in other regions of the central nervous system, in which disease progression seems to be related to frataxin gene expression rather than to somatic mosaicism (210).
Expanded alleles lead to the inhibition of FXN expression, resulting in decreased levels of frataxin (51, 121). By using RNase protection assays, Bidichandani et al. (29) were able to show a reduced steady-state level of FXN mRNA. They also reported that GAA expanded repeats adopt unusual structures (they predicted triplex) and hindrance of in vitro transcription for sequences containing 79 and 100 GAA repeats, but not for those containing 45 repeats (29). In a study of long tracts of GAA•TTC (150 and 270 repeats), Sakamoto et al. (269) discovered a novel DNA structure, sticky DNA, which results from intramolecular association of triplexes. The other types of non-B DNA structures characterized for the GAA triplet repeats include hairpins and parallel DNA (146, 182).
The transcription silencing caused by pathologic expansions is due to the formation of non-B DNA structures (primarily triplexes and sticky DNA), persistent RNA•DNA hybrids, or heterochromatin formation [for recent review, see (319)]. The molecular mechanism underlying the inhibition of transcription by sticky DNA involves the sequestration of RNA polymerase by its direct binding to the complex DNA structure (270). Transcription of a GAA•TTC template (88 repeats) by using T7 DNA polymerase showed that the polymerase paused at the distal end of the repeat (134), and that this was tightly linked to RNA•DNA hybrid formation (132). These in vitro studies demonstrated that RNA polymerase is arrested by triplex structures, preventing transcription elongation. Further studies, searching for epigenetic changes in the promoter and intron regions flanking the GAA-repeat expansion, showed increased methylation of specific CpG sites in lymphoblasts (137), peripheral blood (53), and brain and heart tissues (7) from FRDA patients. Additionally, hallmark features of heterochromatin formation, an overall reduction of histone H3 and H4 acetylation levels and increased H3K9 methylation, have all been observed in cell lines and brain tissues from patients (7, 137, 147). Interestingly, histone hypoacetylation was observed only in the vicinity of the GAA expanded repeat but not in the promoter region (147). By using competitive nucleosome-reconstitution assays, Ruan and Wang (265) showed that GAA•TTC duplex and triplex structures reduced the efficiency of nucleosome assembly. Thus, the non-B structures adopted by long tracts of GAA repeats may cause heterochromatin-dependent and -independent gene silencing. Given that all patients carry this mutation, any drug that destabilizes these DNA structures or prevents heterochromatin formation could be a good therapeutic candidate.
The triplex and sticky DNA structures of GAA triplet repeats also affect DNA replication, recombination, and repair (319). It is possible that the genetic instability associated with these trinucleotide expansions occurs during DNA replication. Studies carried out in Escherichia coli and Saccharomyces cerevisiae demonstrated that the presence of a GAA-repeat sequence in the lagging strand of the replication fork led to attenuation of replication, to the occurrence of small slippage events, and to large contractions (146, 171, 244). The minimum number of GAA repeats required for replication stalling in yeast (40 repeats) and the appearance of contractions/deletions in plasmid replication in E. coli (48 repeats) were compatible with the threshold of somatic instability observed in human FRDA patient cells (40–44 repeats) (244, 289). However, these findings did not account for the fact that expansions in borderline and expanded alleles have been observed in carrier and affected individuals. Replication assays of plasmids in COS-1 cells confirmed the increased instability of GAA repeats in the lagging strand previously observed in E. coli and S. cerevisiae (257). Furthermore, depending on the orientation of replication and distance between the origin of replication and the GAA-repeat sequence, no instability, predominant expansion, or both expansion and contraction were observed in these mammalian cells. The somatic instability observed in postmitotic neurons suggests that other mechanisms than replication, such as transcription and postreplicative DNA repair, could be responsible for the triplet-repeat expansions observed in FRDA patients. Recently, Ditch et al. (89) developed a new model for the study of GAA-repeat expansion in human cells and found that transcription through the repeat tracts is a major contributor for expansions. Other uninterrupted sequences of (GAA)44 were found in the human and mouse genomes, but only the FXN alleles carrying GAA expansions of the same size showed a high mutation load, suggesting that somatic instability is locus specific (257).
E. Genotype–phenotype correlations
Several studies described a relation between the size of the GAA-repeat expansion and the presence and timing of several features of the disease. An inverse correlation was found between the size of the smaller expansion and both the age at onset and rate of disease progression, measured as the time until wheelchair confinement (84, 93, 97, 211, 282). The two major complications of the disease are cardiomyopathy and diabetes. Cardiomyopathy frequently arises in patients with large expansions in the smaller allele and is independent of the duration of the disease (84, 93, 97, 211). Diabetes does not appear to be associated with either the number of GAA repeats or the duration of disease (84, 93), but develops during the late stages of disease (97). Other clinical manifestations, such as dysarthria, skeletal deformities, optic atrophy, and hearing loss, show direct correlation to GAA-expansion size (93, 211). Additionally, expansion size has been shown to be associated with the severity of sensory neuropathy (273). Loss of large myelinated fibers (>7 μm) is directly correlated to the duration of disease and is inversely correlated to the GAA-repeat expansion size in the smaller allele. The methylation of two CpG sites in the genome has been directly correlated to the size of the smaller allele and indirectly correlated to the age at onset (53). Residual levels of frataxin vary according to the expansion and cell type. In peripheral blood leukocytes, frataxin levels in patients range from 5 to 30% of normal (121). The size of the smaller allele is inversely correlated to the amount of residual frataxin, providing a potential biochemical basis for the genotype–phenotype correlation with this allele.
Only 37 to 50% of the variation in age at onset is accounted for by the size of the smaller allele (84, 93, 97, 282). Variability among individuals is very high, and it is not possible to predict the clinical severity based only on the GAA mutation. For example, LOFA is the only atypical form of FRDA to be associated with a reduced number of GAA repeats in both alleles; all the other atypical FRDA presentations including, Acadian and FARR, did not show any statistical difference in GAA-expansion size compared with typical cases (211). Several factors may explain the clinical variability observed among individuals with almost identical numbers of repeats. One such factor could be mitotic instability, causing somatic mosaicism of expansion size (210, 211, 289). Mitochondrial haplotype may also affect FRDA phenotype, as described for a population from southern Italy (125).
Mitochondrial oxidative stress is involved in the pathogenesis of several neurodegenerative diseases, including FRDA (198). It is generally accepted that mitochondria-driven reactive oxygen species (ROS) induce mutations in mtDNA (116). The contribution of mtDNA mutations to the FRDA phenotype is poorly documented (145, 149). However, it is possible that these mutations could account for some of the variability observed among individuals carrying similar GAA expansions. A study screening for mutations in the mtDNA noncoding displacement loop (D-loop) in 25 Iranian patients from 12 unrelated families revealed a significantly higher mutation rate (single nucleotide substitutions, mostly transitions) in patients than in controls (149). Additionally, whereas 76% of the patients had deletions of 8.6–10 kb, no deletions were observed in the mtDNA of healthy controls (149). Another study showed that mutations in the genes encoding NADH dehydrogenase subunits were more frequent in patients than in controls and that the number of mutations present in these genes was inversely correlated with the age at onset of the disease (145). Even though many of these mutations are not harmful, the resultant instability of mtDNA demonstrated in these studies may be a predisposing factor and may, in addition to other genetic or environmental risk factors, affect the age at onset and progression of FRDA disease.
In a few cases, a second mutation could not be detected in patients who are heterozygous for GAA expansions and presenting a typical FRDA phenotype (70, 75, 199, 282, 336). Such cases are indicative of locus heterogeneity. However, alternative explanations may be linked to the technical limitations of single-strand conformation polymorphism analysis used to detect point mutations or to the fact that only the coding regions, and not the FXN regulatory region, were searched for mutations. AVED disease caused by mutations in the α-tocopherol transfer protein result in a FRDA-like phenotype in patients, and misdiagnosis can occur if serum levels of vitamin E are not determined (26). It also is possible, in populations in which carrier frequency is high, that the patient has another disease and coincidently is the carrier of a mutation in the FXN gene (283). Another explanation, although unlikely, could be the complete reversion of GAA expansion in one allele to normal size in blood leukocytes, which would have led to a heterozygous diagnosis (30).
The gene causing FRDA was identified in 1996, and data gathered since then have demonstrated that the clinical spectrum is even larger than expected. Among the essential criteria defined by Harding (140), only the progressive limb and gait ataxia has proved to be a consistent feature for all patients, without exception. The recessive autosomal nature of transmission can be difficult to prove, because most cases are sporadic and occur in non-consanguineous families. Of the patients who carry a GAA mutation, ≤25% do not fulfill all of Harding's mandatory criteria, having atypical presentations (93, 199, 282). Patients carrying point mutations also frequently have atypical presentation. Nevertheless, Harding's criteria for diagnosis (140) are the most useful diagnostic tool for clinicians, and >80% of patients presenting all essential FRDA features have homozygous GAA expansion (93, 199). FRDA is a progressive disease, and thus, full clinical presentation is observed only several years after onset, making it difficult to diagnose in the early stages. Delayed or erroneous diagnosis hampers genetic counseling and therapeutic solutions. FRDA should be considered in the differential diagnosis of all types of nondominant ataxias. For these reasons, molecular diagnosis of FRDA should be performed for all patients with not only typical FRDA, but in all cases of idiopathic recessive ataxia.
F. Point mutations
Approximately 2% of the mutations described in FRDA patients are point mutations (51, 70). To date, 43 mutations (missense, nonsense, frameshift, splice-site, and one 2.8-kb deletion) have been detected in the FXN gene of FRDA patients (Table 2, Fig. 6). All patients carrying point mutations are compound heterozygotes with an expanded GAA repeat on the other allele. The lack of patients homozygous for point mutations may be due to the rare occurrence of mutations [estimated as 1:100 million individuals (85) or 1:2,500 FRDA patients (70)]. Conversely, inactivation of the frataxin-encoding gene in mice causes embryonic lethality (71), suggesting that null mutations in humans may also result in a very severe or lethal phenotype.
Table 2.
Mutations Observed in FRDA Compound Heterozygote Patients
| Exon, Intron | DNA mutationa | Type of mutation | Protein changea–c | Range of GAA repeats | FRDA pathologyd | Ethnic origin (number of families) | References |
|---|---|---|---|---|---|---|---|
| Exon 1 | c.1 A>T | Missense | p.M1L (incorrect initiation of translation) | 950 | Typical | USA (1) | (70) |
| c.2 T>C | Missense | p.M1T (incorrect initiation of translation) | 466 | Typical | Sweden (1) | (70) | |
| c.2 delT | Frameshift | p.M1S (incorrect initiation of translation) | ∼800-900 | Atypical (early onset, chorea) | Australia (1) | (333) | |
| c.3 G>T | Missense | p.M1I (incorrect initiation of translation) | 1,066 | Typical | Germany (3) | (69,70,335) | |
| c.3 G>A | Missense | p.M1I (incorrect initiation of translation) | ∼1,000 | Typical(early onset) | USA (1) | (247) | |
| c.11-12 delTC | Frameshift | p.L4RfsX88 | ∼250 | Atypical (chorea, sudden progression) | Malaysia (1) | (296) | |
| c.100 delG | Frameshift | p.A34PfsX42 | 770 | Typical | Italy (1) | (121) | |
| c.104 delC | Frameshift | p.P35HfsX41 | 640 | Typical | Italy (1) | (121) | |
| c.118 C>T | Missense | p.R40C (processing default) | NDe | Typical | USA (1) | (307) | |
| c.118 delC | Frameshift | p.R40VfsX36 | 810 | Typical (early onset) | Spain (1) | (75) | |
| c.157 delCf | Frameshift | p.R53AfsX23 | 733; 170–900 | Typical | France (1), Italy (4) | (70,121) | |
| c.157 insCf | Frameshift | p.R53PfsX40 | 700 | Atypical (late onset) | Poland, Canada (1) | (70) | |
| Intron 1 | c.165 + 1 G>A | Splice donor | Aberrant splicing | ∼1,000 | Atypical onset | Italy (1) | (178) |
| c.165 + 5 G>C | Splice donor | Aberrant splicing (no frataxin detected by WB) | 670 | Atypical (motor neuropathy, optic atrophy) | USA (1) | (200) | |
| Exon 2 | c.202_205 delGTCAinsTTG | Frameshift | p.V68LfsX8 | ∼1,000 | Typical (early onset) | UK (1) | (245) |
| Exon 3 | c.296_297 insT | Frameshift | p.E100RfsX12 | 350 | Atypical(late onset, milder) | Spain (1) | (75) |
| c.317 T>G | Nonsense | p.L106X | NDe | Typical | France (1) | (51) | |
| c.317 T>C | Missense | p.L106S | 840 | Atypical (milder) | USA (1) | (21) | |
| c.317 delT | Nonsense | p.L106X | ∼500 | Typical (early onset) | UK (1) | (245) | |
| c.340_352 del13 | Frameshift | p.A114TfsX15 | ∼1050 | Typical (early onset) | UK (1) | (245) | |
| c.354 C>G | Nonsense | p.Y118X | 640 | Typical | Italy (1) | (121) | |
| c.364 G>T | Missense | p.D122Y | 750 | Atypical (milder) | Germany (1) | (70) | |
| Exon 3/ Intron 3 | c.381_384 delTGGG + c.384 + 1_ + 9 delGTACCTCTT | Frameshift and splice donor | Aberrant splicing | 470 | Typical | Italy (1) | (121) |
| Intron 3 | c.384 + 1 G>A | Splice donor | Aberrant splicing | NDe | Typical | Caucasian (1) | (91) |
| c.384-2 A>G | Splice acceptor | Aberrant splicing | 800 | Typical | Spain (1) | (51,75) | |
| Exon 4 | c.389 G>T | Missense | p.G130V | 800-1330 | Atypical (milder) | Caucasian: USA, Australia, France (4) | (28,70,101) |
| c.438 C>G | Missense | p.N146K | 820 | Atypical (milder) | Germany (1) | (336) | |
| c.443 A>G | Missense | p.Q148R | NDe | Typical | Caucasian (1) | (201) | |
| c.460 A>T | Missense | p.I154F | 625-1010 | Typical | Italy (4) | (51,70,97,121) | |
| c.463 T>C | Missense | p.W155R | 750 | Typical (early onset) | USA (1) | (174) | |
| c.465 G>A | Nonsense | p.W155X | 850 | Typical | Cuba (1) | (75) | |
| c.467 T>C | Missense | p.L156P | 366 | Typical | Sweden (1) | (70) | |
| Intron 4 | c.482 + 2 T>G | Splice donor | Aberrant splicing | ∼600 | Typical | Fiji Islands (1) | (101) |
| c.482 + 3 delA | Splice donor | Aberrant splicing | 731 | Typical | Italy (1) | (70) | |
| Exon 5ag | c.493 C>T | Missense | p.R165C | 380; ∼1000 | Atypical (milder) | Australia, USA (2) | (101,200) |
| c.494 G>C | Missense | p.R165P (isof. 1) p.V168L (isof. 2) | 940; 1100 | Typical(early onset) | Italy (1) | (79) | |
| c.517 T>G | Missense | p.W173G (processing default) | 720; 530-820 | Typical | USA, Italy (2); Italy (3) | (54,70,121) | |
| c.544 C>T | Missense | p.L182F | 730 | Atypical (milder) | Australia (1) | (101) | |
| c.545 T>A | Missense | p.L182H (isof. 1) p.S185T (isof. 2) | 800 | Typical | France (1) | (70) | |
| c.548 A>G | Missense | p.H183R (isof. 1) p.M186V (isof. 2) | 1,000 | Typical | France (1) | (70) | |
| c.557 T>G | Missense | p.L186R (isof. 1) p.W189G (isof. 2) | 920 | Typical | Germany (1) | (336) | |
| c.593 T>G | Missense | p.L198R | NDe | Typical | Ireland (1) | (9) | |
| g.120032_122808 del | Deletion | Deletion of exon 5a | 820 | Typical | Germany (1) | (336) |
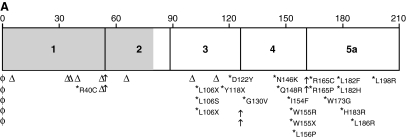
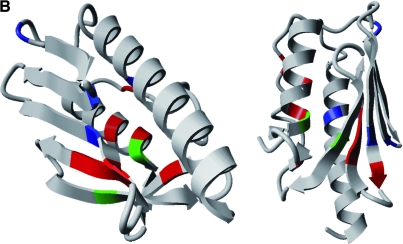
FIG. 6.
Distribution of frataxin point mutations. (A) Mutations in the ATG codon are represented by the symbol φ; frameshifts, by the symbol Δ; and splice-site mutations by arrows. *Missense mutations and the amino acid changes. Regions encoding the mitochondrial addressing sequence are in gray, and those encoding the mature frataxin protein are in white. Most of missense mutations are distributed in the conserved exons 3-5a. (B) Distribution of missense mutations in the frataxin structure. Changes resulting in typical disease presentation are in red, changes resulting in atypical disease presentation are in blue, and those that can result in both typical and atypical disease presentation are in green. The YASARA View software was used to visualize the structure of the human frataxin deposited in Protein Data Bank (PDB ID: 1ekg). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
The effects of genomic mutations on transcript abundance or on the specific protein defect have been addressed in only a few studies. The consequences of mutations on protein sequence can therefore only be speculated on. Half of the mutations identified so far are predicted to lead to the absence or a truncated form of frataxin (Table 2). Five of these mutations affect the translation-initiation codon. It is possible that a second ATG codon located in exon 2 (226 nt downstream) is used in the translation of these mutants. This would result in a translated protein lacking the mitochondrial targeting sequence and the first 20 amino acids encoded by exon 2. Six mutations affect splice-site donors, and one mutation, a splice acceptor site, leading to aberrant splicing and predicted exon skipping. The product resulting from one of these mutations (c.381_384 delTGGG + c.384 + 1_ + 9 delGTACCTCTT) was analyzed with Western blotting. Only a full-length transcript was detected, suggesting that aberrantly spliced mRNA may be unstable and rapidly degraded (121). Four nonsense and nine frameshift mutations introduce premature stop codons, and nearly all of the predicted proteins lack exons 3–5. Only one mutation causing a deletion of 2,776 bp (g.120032_122808 del) was reported. This deletion spans 2,776 bp and encompasses the last 1,315 bp of intron 4, the complete exon 5a sequence, and 971 bp downstream of the FXN gene (336). This mutation is also expected to lead to truncated frataxin. All these mutations are associated with typical FRDA, in some cases with particularly severe phenotypes (Table 2).
A single amino-acid change is predicted for 17 missense mutations (Table 2). These mutations span across exons 1, 3, 4, and 5, with a cluster of mutations found within exons 4 and 5, which correspond to the C-terminal and the most conserved portion of frataxin (Fig. 7A). Most of the patients with these mutations have typical presentations. However, six of the mutations (L106S, D122Y, G130V, N146K, R165C, and L182F) were associated with a milder course of the disease (Table 2, Fig. 6).
FIG. 7.
Frataxin structure and homology. (A) Schematic representation of the structural elements found in human frataxin, as described at www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/ (PDB: 1ekg). A color code identifies the primary structure conservation with cold colors, indicating low conservation, whereas hot colors indicate the most-conserved residues. (B) Surface charges distribution in human frataxin structure (PDB: 1ekg). Negative potential is represented in red, and positive potential is represented in blue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
The most common mutations are those that affect the ATG codon, G130V, and I154F. Haplotype analysis provided evidence for founder events in the cases of M1I (335) and G130V (81) mutations. Patients heterozygous for the G130V mutation have a milder disease presentation. Although onset can be in the early teens, progression is very slow and associated with brisk knee reflexes, moderate ataxia, absence of dysarthria, and absence of diabetes (28, 70). In G130V heterozygotes, frataxin mRNA levels are similar to those in clinically healthy carriers, suggesting that this mutation causes the atypical phenotypes in these patients (28). The I154F mutation was first described in five patients belonging to three families from southern Italy with typical FRDA without any signs of diabetes (51, 97).
The phenotypic features of patients harboring point mutations are frequently typical FRDA, although slightly different phenotypes are sometimes observed, which may cause confusion in the clinical diagnosis. In general, disease is severe in these patients, often being associated with early onset and infrequent signs such as chorea (Table 2). Interestingly dysarthria and diabetes are less frequent. A study comparing the clinical parameters of homozygous and heterozygous patients showed only significant earlier age at onset, less dysarthria, and more-frequent optic disc pallor in those with point mutations (70) (Table 1). This study included a small set of 19 families carrying a total of 14 mutations. The GAA-repeat expansion was significantly larger in heterozygotes than in homozygous patients, making it difficult to evaluate the effect of the point mutations on phenotype. No other recent comparative study exists. A point mutation that results in loss of function of frataxin is generally associated with a severe phenotype. In the case of missense mutations, even in regions that have been conserved through evolution, it cannot be predicted whether the disease will have a mild or severe clinical course. In all cases, the size of the GAA triplet-repeat expansion in the other allele may modulate the effect of the point mutation.
IV. Frataxin Is a Unique Protein
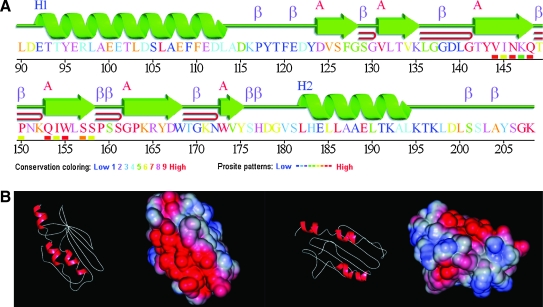
A. Phylogeny and structure of frataxin
Frataxins are small proteins (between 100 and 220 amino acids) that are conserved from gram-negative bacteria to humans (126). All eukaryotic frataxins identified so far, except in the human pathogens Trichomonas vaginalis and Trachipleistophora hominis, are localized to the mitochondrial matrix (50, 126, 169, 189, 309). Trichomonas vaginalis is a protist that inhabits oxygen-poor environments and lacks mitochondria. Their energy metabolism depends on cytosolic glycolysis and pyruvate breakdown in a specialized organelle enclosed by a double membrane, the hydrogenosome. The frataxin protein is targeted to the hydrogenosome, where iron–sulfur (Fe-S) clusters assembly also takes place (90). The microsporidia T. hominis is an obligate intracellular parasite that has mitochondrial remnants called mitosomes. In other microsporidia, frataxin is addressed to the mitosomes, but in T. hominis, it is located in the cytoplasm along with the Fe-S cluster scaffold protein Isu1 (128). No frataxin homologue has been identified in gram-positive bacteria or Archae, suggesting that all eukaryotic frataxins originated at the moment of proteobacteria endosymbiosis and that the gene migrated later from the mitochondrial to the nuclear genome (126).
Owing to its small size, frataxin proved to be readily amenable to structure resolution in solution. Furthermore, the analysis of 1H-15N chemical shifts of amide protons in NMR experiments allowed frataxin-interaction sites to be mapped. Structures were determined for the human frataxin isoform 1 (only residues 91 to 210 because of the autodegradation and proteolysis of the protein's N-terminus; PDB ID: 1dlx and PDB ID: 1ly7) (216), the E. coli frataxin homologue CyaY (PDB ID: 1soy) (218), and the mature yeast frataxin homologue Yfh1 (PDB ID: 1xaq/2ga5) (143) in solution. The crystal structures of the three proteins also are available for the human frataxin (PDB ID: 1ekg) (86), CyaY (PDB ID: 1ew4) (61), and Yfh1 (PDB ID: 2fql) (161). Solution and crystal structures are in remarkable overall agreement and are highly conserved between prokaryotes and eukaryotes. The frataxin fold is unique. It consists of a large, twisted, six-stranded β-antiparallel sheet, flanked by N- and C-terminal α helices (α1 and α2), with no main surface cavity (Fig. 7A), and is described in the CATH database (www.cathdb.info; ID 3.30.920.10) as an “alpha beta 2-layer sandwich”. The only discrepancy between the solution and crystal structures is the presence of a seventh short β strand before the second helix α2 in the crystal structure of Yfh1 but not in the solution structure. Conversely, this strand was observed in solution but not in the crystal structure for the human protein. One major feature of the frataxin structure is the presence of a large patch of negatively charged residues on the helical plane (Fig. 7B). This anionic surface may be involved in iron binding (6). By contrast, the β-sheet surface is almost uncharged and may be involved in protein–protein interactions. Most of the residues conserved during evolution or affected by mutations in FRDA patients are located on this surface.
Although the frataxin fold is considered to be unique, it also has been found in the Nqo15 subunit of the hydrophilic domain of the respiratory complex I from Thermus thermophilus, an extremophile bacterium (277). Similarly, Nqo15 interacts with the other subunits of complex I through the exposed β-sheet surface, and it is possible that it also binds iron. It was thus suggested that Nqo15 may be involved in iron delivery for regeneration of nearby Fe-S clusters (277). The structural analogy between Nqo15 and frataxin may provide new perspectives for the study of frataxin function.
B. Frataxin maturation
Frataxin is translated in cytoplasmic ribosomes (268) and imported into the mitochondria (169), where the targeting sequence is proteolytically removed in a two-step process to produce the mature protein (38, 54). Maturation of yeast and human frataxin depends on the mitochondrial processing peptidase (MPP). MPP first cleaves the precursor to give an intermediate form, followed by conversion of this product to the mature form (Fig. 8) (38, 54, 130). In a yeast two-hybrid assay, the mouse frataxin (N-terminal 4–87 amino acids) was shown to interact with the β-subunit of MPP (170).
FIG. 8.
Maturation of human and yeast frataxin by mitochondrial processing protease. Maturation of precursor (p) frataxin by MPP is a sequential two-step cleavage originating the intermediate (i) and the mature (m) forms. The human frataxin is synthesized as a 210-amino acid precursor, and processing in vitro may originate m56-FXN and m78-FXN, but only the m81-FXN mature form has a functional significance in vivo. For the yeast frataxin (174-amino acid precursor), only one mature form has been detected. It is interesting to note that frataxin proteins show a higher apparent molecular weight on SDS-PAGE gels than predicted because of the acidic nature of the N-terminal α-helix (279). The apparent sizes for FXN are described in (63, 279), and for Yfh1, are described in (38, 131).
The precursor of the human frataxin is initially cleaved between G41 and L42 (RRG↓LRT), as demonstrated by in vitro processing assays with recombinant MPP and N-terminal radiosequencing of the intermediate form (54). Identification of the site involved in the second step of processing to generate the mature form has been less clear. In the study mentioned earlier, Cavadini et al. (54) sequenced the mature form and identified a cleavage site between A55 and S56 (m56-FXN; 17.2 kDa) (54). The recombinant human frataxin purified from E. coli undergoes iron-mediated autoproteolysis, producing another mature form (m78-FXN; 14.7 kDa) (331). However, analysis of the in vivo processing of frataxin in human cells showed a major mature form that was smaller (63). Sequencing by Edman degradation of the immunopurified mature form of precursor frataxin overexpressed in HEK293 cells gave the sequence SGTLGH, suggesting that cleavage occurred between K80 and S81 (m81-FXN; 14.3 kDa) (63). These results were confirmed by MALDI-PMF analysis of the immunopurified mature form from COS-1 cells (279). This mature form co-migrated with endogenous frataxin in fibroblasts, lymphoblasts, and heart tissue in Western blots (63). Moreover, the rescue of aconitase activity deficiency in FRDA patient cells (63) and of the lethal phenotype in frataxin-deficient murine fibroblasts (279) demonstrated that m81-FXN was functional. In summary, in vivo experiments have established that m81-FXN is the normal mitochondrial mature form in living cells. However, m56-FXN and m78-FXN can be produced in vivo when normal processing is impaired or when processing is carried out in vitro (63, 279).
In yeast, the first MPP cleavage site lies between residues Y20 and M21 (RRY↓MIA), removing ∼2 kDa, and the second cleavage is between residues F51 and V52 (KRF↓VES), resulting in the removal of an additional ∼4 kDa (Fig. 8) (130). As for the human frataxin, the cleavage sequences match MPP consensus sequence. Detailed analysis of the two sequences, residues 1–20 (Domain I) and residues 21–51 (Domain II), established that Domain I is the matrix-targeting signal. This domain can be replaced by other mitochondria-addressing peptides without affecting import efficiency or Yfh1 function. In the absence of Domain II, mitochondria-targeting signals cannot mediate import of yeast frataxin. Domain II acts as a spacer, separating the basic Domain I from the mature acidic Yfh1 and thus precluding futile interactions (130).
In addition to MPP, efficient maturation of Yfh1 pre-protein requires the sequential action of the Hsp70-family mitochondrial chaperones, Ssc1 and Ssq1 (165, 315). In the ssc1-3 mutant mitochondria, Yfh1 precursor was not processed to the intermediate or mature forms, indicating that Ssc1 is crucial for translocation of Yfh1 across the inner membrane (315). However, Ssq1 is necessary for efficient processing of the intermediate form by MPP, but the steady-state level of Yfh1 in Δ_ssq1_ mitochondria is only 25% lower than that in the wild-type (165, 315). Another partner from the inner-membrane protein-import machinery, Tim44, is required for binding of Ssc1 to Yfh1 (120).
The pathologic mutations G130V and I154F (corresponding to the G127V and I151F changes on the mouse sequence) were shown, in a yeast two-hybrid system or when expressed in COS cells, to decrease the efficiency of processing of the mouse frataxin without any change in site cleavage (170). However, no differences were observed between these mutants and the wild type when they were processed by recombinant MPP or in isolated mitochondria (54, 131). Two other FXN point mutations, R40C and W173G, affect frataxin maturation. R40C disrupts the consensus sequence recognized by MPP (307), and W173G inhibits the second cleavage step, causing accumulation of the intermediate form (54). A recent study, in which HEK293T cells were transfected with plasmids encoding wild-type or mutated forms (G130V, I154F, W155R, L156P, W173G), showed by Western blotting that the mature form was absent from G130V-, L156P-, and W173G-producing cells and reduced in I154F cells (287).
Two-step cleavage by MPP is very rare and has potential regulatory functions. The human frataxin precursor is cleaved rapidly and quantitatively to the intermediate form, whereas the second cleavage is slower and limits the overall rate of mature frataxin production in rat liver mitochondria (54). It is possible that in human cells, as in yeast, mitochondrial proteins modulate processing of the intermediate form, accounting for some of the variability observed in clinical phenotype between individual patients.
C. Cellular function of frataxin
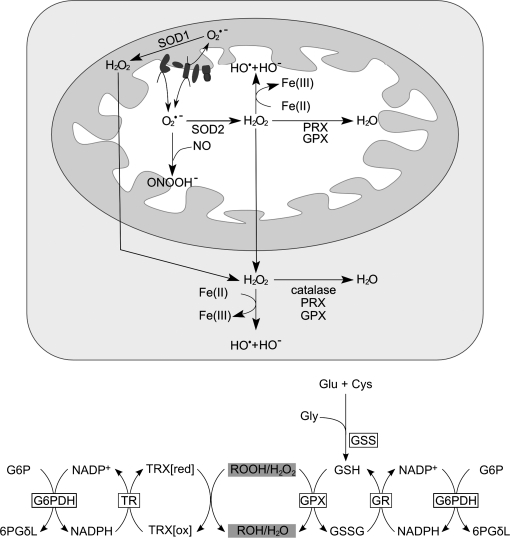
The first clues about the role of frataxin came from the analysis of a S. cerevisiae frataxin-deficient mutant (Δ_yfh1_) (15, 104, 169, 323). The phenotypes observed in this mutant were severe growth deficit on fermentable substrates; reduced rate of respiration and impaired growth on glycerol and ethanol; accumulation of petite cells (complete or partial loss of mtDNA); high sensitivity to hydrogen peroxide and copper; low cytosolic iron level and constitutive activation of the high-affinity iron-transport system at the plasma membrane; and mitochondrial iron content > 10 times that of the wild type (15, 104, 323). These observations led to the first hypothesis that frataxin regulates mitochondrial iron efflux (15). Accordingly, when Yfh1 was reintroduced in the Δ_yfh1_ mutant, the accumulated iron was exported back into the cytoplasm (252). This hypothesis was never confirmed, but these observations and the finding of iron accumulation in the heart tissue of FRDA patients (177, 272) have established a role for frataxin in iron homeostasis. At the same time, Rötig et al. (262) found selective deficiencies of the respiratory chain complexes I, II, and III and of both mitochondrial and cytosolic aconitase activities in heart biopsies from patients with hyperthrophic cardiomyopathy (262). These enzymes all have Fe-S clusters in their active sites, which are exquisitely sensitive to ROS. These data thus suggested that the alteration in iron homeostasis caused by frataxin deficiency resulted from increased mitochondrial iron and ROS production by the Fenton reaction, leading to inactivation of Fe-S clusters, mtDNA damage, and hypersensitivity to oxidative stress (15, 262). Several subsequent studies were consistent with the idea that iron accumulation in the mitochondria was responsible for the abnormalities observed in frataxin-deficient cells (60, 103, 266). However, addition of an iron chelator to the culture media restored normal intramitochondrial iron content in Δ_yfh1_ cells without increasing aconitase activity, suggesting a more direct role for Yfh1 in Fe-S cluster biogenesis (103). Data obtained from knockout and conditional mouse models also demonstrated that the deficiency in Fe-S cluster enzyme activity occurs before iron accumulation in the mitochondria (71, 249). In a more recent study, Mühlenhoff et al. (215) suggested that the primary function of yeast frataxin is the maturation of Fe-S cluster proteins and that the other phenotypes are derived from this main function (215).
Another interesting hypothesis first proposed by Isaya and colleagues (4), is that frataxin may be an iron-binding protein that stores iron in a bioavailable, nontoxic form for heme and Fe-S cluster synthesis.
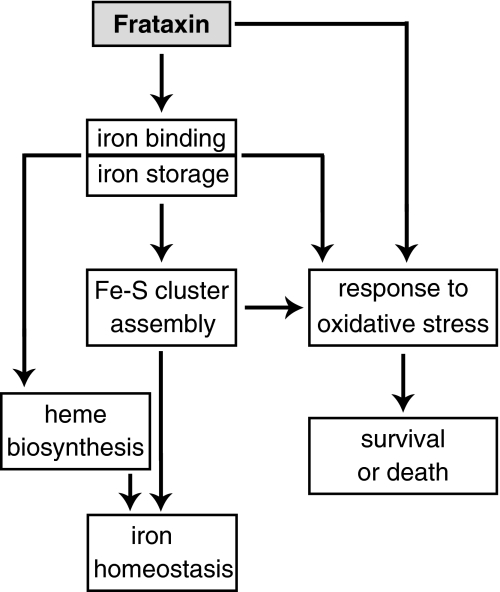
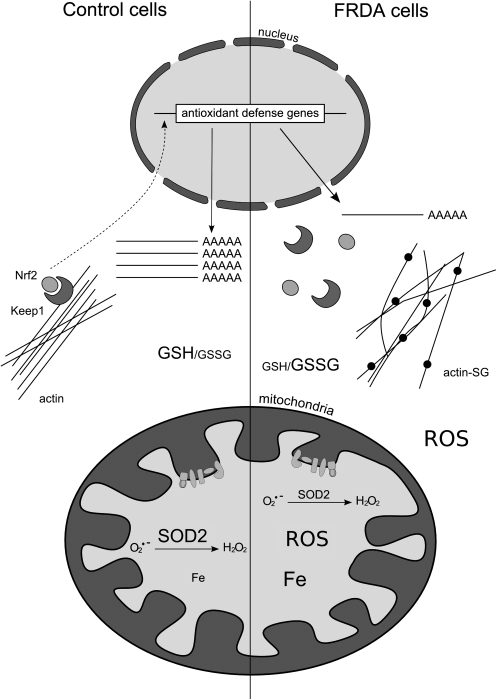
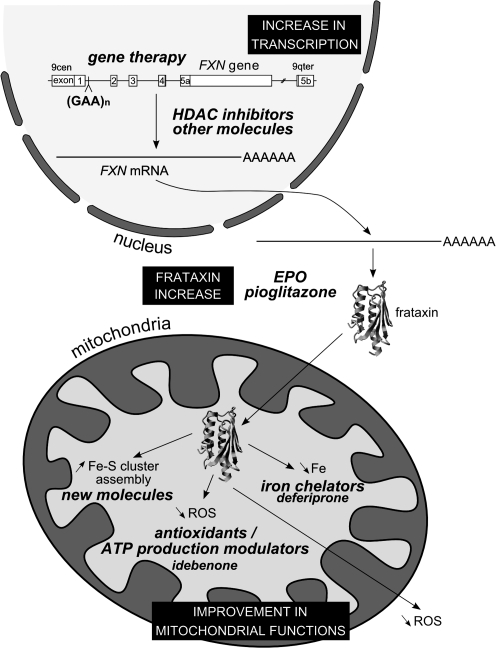
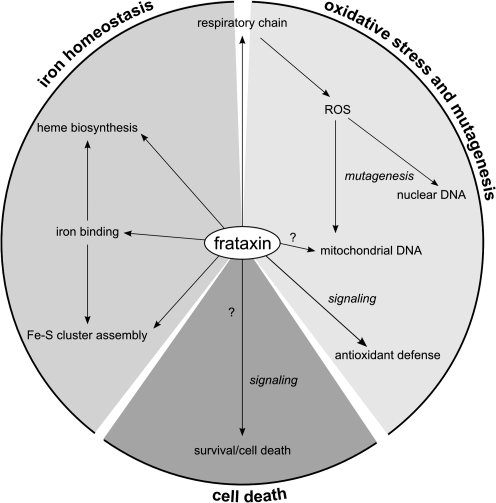
The precise function of frataxin remains unclear, but recent efforts have led to significant advances in this area. It is clear that frataxin is involved in mitochondrial iron use and that this function is important for maintenance of overall cellular iron homeostasis and redox status. In the following sections, we discuss the findings and hypotheses concerning the role of frataxin in greater detail: Fe-S cluster and heme biosynthesis; iron binding and storage; and response to oxidative stress and survival (Fig. 9).
FIG. 9.
Hypothesis on the cellular functions of frataxin. Frataxin is an iron-binding protein involved in mitochondrial iron storage or iron use or both. These functions are important for maintenance of the overall cellular iron homeostasis and redox status. Frataxin deficiency causes impairment of Fe-S cluster and heme biosynthesis, oxidative stress, and cell death.
V. Frataxin Function in Cell Iron Use and Oxidative-Stress Defense
A. Frataxin is critical for Fe-S cluster assembly
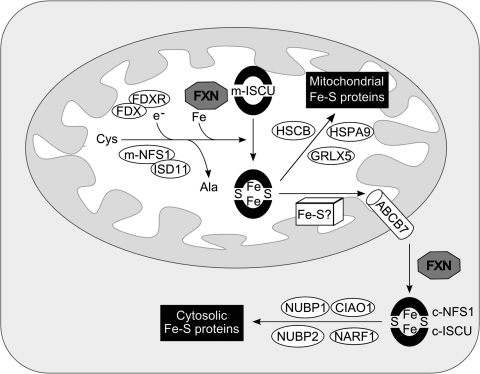
A deficit in Fe-S cluster proteins is commonly observed in organisms lacking frataxin, with the exception of bacteria (184, 314), including yeasts, protists, plants, flies, and mammals (11, 44, 71, 192, 262, 323). Fe-S clusters appeared early in evolution as prosthetic groups essential for many fundamental cellular processes, including respiration, replication, and DNA repair and translation [for recent reviews, see (188, 264)]. The most common Fe-S clusters in eukaryotic cells are [2Fe-2S] and [4Fe-4S], which are involved mainly in electron-transfer reactions. Biogenesis of the Fe-S clusters in eukaryotes requires two sets of molecular assembly machineries: the mitochondrial Fe-S cluster–assembly machinery (ISC) and the cytosolic Fe-S cluster–assembly machinery (CIA). Assembly of mitochondrial Fe-S cluster holoproteins requires only the ISC machinery, but nuclear and cytosolic Fe-S cluster proteins require both ISC and CIA.
The overall ISC assembly process can be divided in two steps. The first step is de novo synthesis, starting by the release of one sulfur atom from cysteine by cysteine desulfurase (yeast and human Nfs1/Isd11 complex in vivo), and the transfer to scaffold proteins (Isu1/2 in yeast and ISCU in humans) through a direct protein/protein interaction and iron coordination. Fe-S cluster assembly on the scaffold protein Isu1 also requires electron transfer in the presence of ferredoxin and ferredoxin reductase (Yah1 and Arh1 in yeast and FDX1 and FDXR in humans).
The second step of biogenesis is the transfer of the newly formed cluster from scaffold proteins and its assembly into apoproteins. This requires the Hsp70 ATPase Ssq1 and the DnaJ-like Jac1 (HSPA9 and HSCB in humans) chaperones. It is thought that energy from ATP hydrolysis drives conformational changes in scaffold proteins, facilitating cluster dissociation and transfer to recipient proteins. Other components have also been implicated in Fe-S cluster assembly and are shown in Fig. 10.
FIG. 10.
A model for Fe-S cluster protein assembly in human cells. In mitochondria and cytosol of mammalian cells, cysteine desulfurases (m-NFS1 and c-NFS1) remove sulfur from free cysteine and transfer it to the scaffold ISCU proteins (m-ISCU and c-ISCU). The proposed function of frataxin is to deliver iron to the desulfurase/scaffold complex for de novo biogenesis of [2Fe-2S] and [4Fe-4S] clusters in the mitochondria. This synthesis also requires the redox proteins ferrredoxin (FDX) and ferredoxin reductase (FDXR). The clusters are transiently bound to the scaffolds before being released and incorporated into recipient apoproteins. These steps are facilitated by the HSPA9 and HSCB chaperones in the mitochondria, and possibly by the NUBP1, NUBP2, NARF1, and CIAO1 chaperones in the cytosol. The cytosolic assembly of Fe-S clusters requires an unknown precursor exported from the mitochondria by the ABCB7 transporter.
Several lines of evidence strongly suggest that frataxin is directly involved in iron delivery for de novo Fe-S cluster biosynthesis in yeast and human cells. A synthetic lethal screen, identifying a functional interaction between YFH1 and ISU1, suggested that these genes are functionally related (254). Another study showed the yeast frataxin to bind specifically the core of the ISC-assembly complex, Nfs1/Isu1, an interaction that was enhanced by the addition of iron (124). In vitro studies using the mature form of human frataxin demonstrated that holofrataxin (six to seven iron ions per frataxin molecule) interacts with apoISCU, and that two iron atoms are transferred for the assembly of a [2Fe-2S] cluster (329). Additionally, by immunoprecipitation of tagged proteins in HEK293T cells, Shan et al. (287) showed that human frataxin interacts with ISD11, a component of the NFS1/ISCU scaffold complex, and with the chaperone HSPA9. The interaction between frataxin and ISD11 was abolished in I154F mutants and diminished in W155R mutants (287). These observations have been confirmed in yeast; the interaction of Yfh1 with the ISC assembly machinery is mediated by direct binding to Isd11 or Isu1 (185, 317). Several regions at the surface of frataxin have been implicated in this interaction; the acidic residues of helix α1 and the β1 strand are required, and also the β3 strand for Yfh1 (105, 150, 317).
The E. coli frataxin CyaY interacts only with the cysteine desulfurase IscS, as shown in affinity chromatography experiments; however, CyaY-Fe(III) was found to serve as an iron donor for Fe-S cluster formation on the IscU scaffold in vitro (180). Another protein, IscA, is able to recruit and deliver iron for Fe-S assembly. In the presence of the thioredoxin reductase system required to mimic intracellular redox potential, CyaY, unlike IscA, failed to bind iron, suggesting that IscA was the iron donor for cluster synthesis (88). However, when hydrogen peroxide was added, the iron-binding thiol groups of IscA became oxidized, preventing iron binding, whereas CyaY was able to bind iron at low affinity (88). Based on these findings, Ding et al. (88) suggested that, under normal physiologic conditions, IscA is the iron donor for Fe-S biogenesis and that CyaY serves as an iron chaperone to sequester redox-active free iron and alleviate oxidative damage under conditions of oxidative stress (88). The iscA gene forms part of the operon iscRSUA; therefore, the role of IscA as the iron donor would imply that this operon encodes the complete set of ISC components in E. coli. Recently, the monomeric form of CyaY was shown to function as an iron-dependent inhibitor of Fe-S cluster formation through the binding to IscS (5). Therefore, E. coli frataxin could be an iron sensor and negatively regulate Fe-S cluster assembly in conditions of iron deficiency (5). Although this seems to provide a very attractive hypothesis, the interaction with the cysteine desulfurase complex in eukaryotes is mediated by physical interaction with ISD11 (185, 287). Nevertheless, the accumulated data may suggest that frataxin plays a regulatory role instead of having a direct function in Fe-S cluster biogenesis.
Frataxin can have an extramitochondrial localization in human cells (3, 64). The human colon adenocarcinoma cell line Caco-2 is widely used for intestinal epithelial differentiation studies. In these cells, frataxin is found outside the mitochondria, and protein levels increase during differentiation (3). Immunoprecipitation experiments have shown an interaction between frataxin and the cytosolic ISCU1 scaffold protein in differentiated cells only. These findings suggest that frataxin may also be involved in the assembly of cytosolic and nuclear Fe-S clusters (3).
Frataxin also can act as an iron chaperone in converting the oxidative damaged [3Fe-4S] cluster into the active [4Fe-4S] cluster of aconitase (40). Furthermore, interaction of frataxin with aconitase, in the presence of citrate, protects the cluster from oxidation, reducing the risk of enzyme inactivation (40).
Microarray studies revealed a number of differences in the gene-expression profiles obtained from yeast, mouse, and three human cell types. In yeast, expression of the genes encoding Isu1/2 was found to be upregulated in the Δ_yfh1_ mutant, suggesting a genetic link between frataxin and ISC biogenesis (106). However, a recent study showed that ISCU and NFS1 gene expression is repressed in the cardiac tissue of the muscle creatine kinase knockout mouse (MCK) (151). Another study, using fibroblasts and lymphoblasts from FRDA patients and a neural NT2 cell line with frataxin RNAi knockdown, showed significantly reduced levels of transcription for seven genes involved in sulfur amino acid and Fe-S cluster biosynthesis in these cells, none of which encoded components of the essential ISC assembly machinery (303). Four of these downregulated genes are involved in serine synthesis. Serine is condensed with homocysteine to produce cystathionine in a reaction catalyzed by cystathionine β-synthase. Cystathionine is then cleaved by cystathionase to form cysteine. Expression of the gene encoding cystathionase also is repressed. Thus, consistent with the downregulation of these genes, biochemical data showed that steady-state levels of cysteine, serine, homocysteine, and cystathionine were reduced in the frataxin-deficient cell types (303).
Recent data show that the yeast Pichia guilliermondii Δ_yfh1_ mutant displays organic sulfur auxotrophy (251). Of note, cysteine desulfurase and cystathionine β-synthase have the same cofactor, pyridoxal phosphate (PLP). Our unpublished results show lower levels of intracellular PLP in yeast Δ_yfh1_ cells and FRDA patient fibroblasts than in controls. Taken together, these findings suggest that cysteine and PLP deficiency may contribute to decreased de novo Fe-S cluster synthesis. This is consistent with the Duby et al. (92) proposal that Yfh1 plays an important but not essential role in this process, because Fe-S cluster biogenesis can occur in the absence of frataxin, even though at a reduced level (92).
B. Frataxin is involved in heme biosynthesis
Heme is an iron-containing tetrapyrrole ring that is used as a cofactor by many cytochromes, enzymes, and other hemoproteins, involved mainly in respiration and oxygen transport and sensing. The heme biosynthetic pathway occurs in eight sequential steps; the first step occurs in the mitochondria, followed by four steps in the cytoplasm, with the last three steps taking place back in the mitochondria. In a study of 13 FRDA patients tested for free erythrocyte protoporphyrin, all patients had levels above the normal range, indicating heme deficiency in these patients caused by inhibition of ferrochelatase activity and leading to ineffective and persistent erythropoiesis (213). A cellular deficit in heme metabolism was first reported in the S. cerevisiae Δ_yfh1_ strain (104, 183). The mutant had 20-fold less total heme content compared with the wild type, and low-temperature spectra of whole cells revealed a near absence of cytochrome signals (a + a3, b, and c) (183). Zinc protoporphyrin was visible in these cells, suggesting that ferrochelatase, which catalyzes the end step of heme biosynthesis, incorporated zinc instead of iron into protoporphyrin IX. A detailed analysis of ferrochelatase levels demonstrated repression of the encoding gene HEM15 and reduction of total protein levels to < 25% of wild type in frataxin-deficient cells (183). In vivo studies showed that the unavailability of iron, rather than the decrease in ferrochelatase activity, accounted for the lack of heme and the presence of Zn-protoporphyrin in Δ_yfh1_ cells (183). These results were later confirmed in FRDA patient lymphoblasts (280).
A specific interaction (K_D ∼40 n_M) between recombinant Yfh1 and Hem15 proteins was seen in the absence of iron by using plasmon surface resonance (183). These observations were completed by in vitro experiments using the human frataxin and ferrochelatase proteins (330). In these studies, high-affinity binding of frataxin to ferrochelatase (frataxin monomer/ferrochelatase dimer stoichiometry) was iron dependent and stimulated ferrochelatase activity, suggesting a physiologic role of frataxin in iron delivery to ferrochelatase (330). The contact surface of yeast and human frataxin was mapped by NMR and was shown to involve predominantly the helical plane and β1 strand, hence overlapping with the putative iron-binding domain (25, 143). For Yfh1, but not for human frataxin, the β6-loop-α2 region and the surface of the β-sheet were also found to be involved in the interaction (143). Neutral residues in the β6-loop region might provide a hydrophobic patch on the protein, facilitating binding to the hydrophobic external part of ferrochelatase (143). The specific Yfh1 and human frataxin residues that showed a chemical-shift perturbation as a result of complex formation with ferrochelatase are shown in Fig. 11. These data suggest a model in which holofrataxin docks to the ferrochelatase dimer mostly through helical surface residues, with complex formation allowing ferrous iron delivery for heme synthesis (25, 143).
FIG. 11.
Mapping of frataxin residues that interact with ferrochelatase. (A) FXN and Yfh1 amino acid sequence alignment showing the frataxin residues, identified by NMR spectroscopy, that show a chemical-shift perturbation as a result of complex formation with ferrochelatase (25, 143). (B) Residue visualization on the human frataxin structure by using the YASARA View software (PDB ID: 1ekg). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
In mammalian cells, but not in yeast cells, ferrochelatase has an Fe-S cluster. Absence of this cofactor would therefore be expected to result in inactivation of the enzyme in mammals. However, normal levels of ferrochelatase activity were observed in FRDA patient lymphoblasts (280) and in HeLa cells with frataxin depletion by RNAi (298). Despite normal ferrochelatase activity, HPLC analysis and heme staining revealed that heme a and c levels were reduced and protoporphyrin IX levels were increased in FRDA lymphoblasts and in a frataxin-deficient human oligodendroglial cell line (220, 280). Furthermore, gene-transcription analyses using various human and mouse cell types demonstrated that several steps of heme biosynthesis were affected by frataxin deficiency (151, 220, 280). The genes encoding the 5-aminolevulinate dehydratase, porphobilinogen deaminase, uroporphyrinogen III synthase, coproporphyrinogen oxidase, and ferrochelatase were found to be downregulated.
C. Iron homeostasis
Disruption of cellular iron homeostasis, resulting in mitochondrial accumulation and cytosolic depletion, is characteristic of frataxin deficiency. Massive iron accumulation (reaching levels > 10 times), as observed in mitochondria of S. cerevisiae Δ_yfh1_ cells (15, 104, 323), is exceptional even among yeasts. In human and mouse tissues, the increased iron level resulting from frataxin insufficiency does not reach much more than twice the normal level. Several mouse models of FRDA have been constructed, and iron accumulation was not frequently observed. No significant iron deposits were seen in the complete knockout (71), the neuron knockout (neuron-specific enolase promoter, NSE model) (249), the knockin (homozygous insertion of 230 GAA-repeat expansions in the first intron of the mouse FXN gene) (207), or the Cb/Br models (cerebellum/brain; specific neuronal expression of tamoxifen-dependent recombinase Cre-ERT under the mouse Prion protein promoter, with ablation of frataxin expression in adult mice) (294). However, iron accumulates in the heart of 10-week-old MCK mice (249) and iron deposition and lipofuscin accumulation were detected in the DRG and heart of the humanized mouse model after 1 year of age (YAC transgenic mice containing the human FXN gene with 190 GAA repeats) (8). Studies of tissues and cells obtained from FRDA patients have occasionally given inconsistent results. Iron accumulation has been found in the heart (36, 177, 206, 272), liver (36), and spleen (36), but not in the DRG, spinal cord, skeletal muscle, cerebellum, peripheral nerves, or pancreas (36). By using magnetic resonance imaging, Waldvogel et al. (316) found increased iron in the dentate nucleus of the cerebellum in 12 FRDA patients. However, these findings were not confirmed by biochemical analyses of autopsy tissue from nine patients; no significant differences in total iron and ferritin content were observed between patients and normal controls (166). In a later study, total iron levels in the DRG from three patients also were found to be in the normal range (167). Additionally, serum iron and ferritin concentrations, measured in 10 FRDA patients, were found similar to normal controls (322). Studies of cultured fibroblast and lymphoblast cells from patients or mouse models, however, showed a modest increase of mitochondrial iron content (46, 302, 326). Overall, these results do not suggest a global iron overload but rather accumulation in specific tissues, such as the heart. The potential relation between the increased iron observed in the heart and development of cardiomyopathy in FRDA patients remains unclear (36). Recently, Huang et al. (151) proposed a model explaining iron overload in the mitochondria and iron scarcity in the cytosol by using microarray analysis of RNA extracted from the heart tissue of MCK mice. They found induction of the genes encoding the transferrin receptor 1 (increasing cellular iron uptake from circulating transferrin) and the mitochondrial iron importer mitoferrin-2, and repression of the gene encoding the cell-membrane iron exporter ferroportin 1. Additionally, the expression of genes encoding proteins involved in iron use, like in Fe-S clusters and heme biosynthesis, was decreased (151).
In S. cerevisiae, a strong relation is found between Fe-S cluster status and the regulation of iron uptake and homeostasis (239). Iron uptake is controlled by the transcriptional activator Aft1, which shuttles from the cytosol to the nucleus on iron starvation. This signaling pathway is dependent on an iron-sensor complex formed by Grx3/4, Fra2, and a [2Fe-2S] cluster (186). Frataxin-deficient cells, as observed in other mutants with impaired Fe-S cluster biogenesis and in iron-deprived cells, upregulate Aft1-dependent genes (15, 239). Interestingly, the frataxin-deficient mutant of the respiratory yeast Candida albicans, which uses transcriptional repression, rather than activation, for the regulation of iron uptake, accumulates iron in the mitochondria at much lower levels than does S. cerevisiae (275).
In addition to the transcriptional deregulation of the iron-regulon genes, a defect in Fe-S cluster assembly in yeast leads to mislocalization of the iron taken up by the cells, which accumulates in the mitochondria instead of in its normal storage compartment, the vacuole. Consequently, all yeast mutants with impaired Fe-S cluster biogenesis display a high level of iron uptake and accumulate iron specifically in their mitochondria (264). The molecular mechanisms leading to the mislocalization of iron in yeast cells with defective Fe-S assembly are unknown. In mammalian cells, Fe-S clusters also play a major role, through the IRP proteins, in transcriptional and posttranscriptional regulation of iron homeostasis (214). For example, inactivation of human ISD11, a protein involved in mitochondrial Fe-S cluster biogenesis, results in disruption of iron homeostasis, through increased binding activity of IRP1 and increased protein levels of IRP2 (292). However, the strong link between Fe-S cluster status and the intracellular distribution of iron observed in yeast has not been observed for mammalian cells. These observations may thus explain the greater mitochondrial iron accumulation in Δ_yfh1_ yeast cells than in other frataxin-deficient organisms.
Mitochondrial iron accumulation is common to all yeast mutants with defective Fe-S cluster assembly, irrespective of frataxin abundance. It is therefore important to distinguish between yeast phenotypes that are specifically related to the lack of frataxin and those that are more generally related to defects in Fe-S cluster assembly. Only a few of the phenotypes observed in yeast are specifically related to the lack of frataxin. For example, anaerobiosis has a beneficial effect on cell growth in Δ_yfh1_ cells, but not in other mutants with defective Fe-S cluster biogenesis, suggesting a specific role of frataxin in cellular oxygen handling (39).
The surplus iron in frataxin-deficient cells may exacerbate oxidative stress through the Fenton reaction (15, 45, 234, 262). Whether this is the case is unclear. In human cells, the expression of mitochondrial ferritin or the addition of deferiprone leads to a reduction in ROS accumulation and in cell death and to an increase in the activity of Fe-S cluster enzymes (49, 160). In yeast, preventing mitochondrial iron accumulation in Δ_yfh1_ cells by overexpressing CCC1 (which encodes a vacuolar iron importer), by adding chelators to the growth medium or by disrupting the transcriptional activator gene AFT1, improves cellular respiration and decreases the amount of oxidatively modified proteins in the mitochondria (60, 103). Moreover, the heterologous expression of human mitochondrial ferritin in Δ_yfh1_ yeast cells attenuates the deleterious phenotypes of this mutant (48). Some suppressor strains of Δ_yfh1_, however, become resistant to oxidative stress while still accumulating iron in the mitochondria (183). Additionally, Δ_yfh1_ cells grow better when excess iron is added to the medium than when grown in iron-restricted conditions (275). These findings question the toxicity of the accumulated iron in mitochondria.
The chemical form of mitochondrial iron in primary fibroblasts from FRDA patients was analyzed by x-ray absorption spectroscopy (246). Most of the iron in patient fibroblasts exists as ferrihydrite associated with mitochondrial ferritin (at levels 3 times higher than control cells). Ferrous iron is present at a low level in these cells (246). Although we cannot exclude the fact that this small pool of iron may contribute to ROS production, most of the mitochondrial iron is thus in a non–redox-active form. Accordingly, Sturm et al. (300) did not find any difference in the mitochondrial labile (chelatable and reactive) iron pool in fibroblasts and lymphoblasts between patients and controls. Mössbauer spectroscopy studies have shown that the mitochondrial iron in yeast Δ_yfh1_ cells forms amorphous nanoparticles of ferric phosphate (183). Similar findings have been reported for other Fe-S cluster mutants that accumulate iron in the mitochondria (204, 205). In vivo experiments demonstrated that this iron is not available for heme synthesis (183) or as a cofactor for heterologous expression of E. coli FeSOD (R. Santos, unpublished data). Additionally, unlike iron in the mitochondria of other iron-accumulating mutants, this iron is not “Sod2-reactive,” meaning that it cannot compete with manganese for binding to Sod2 (328). Thus, most of the iron accumulated in Δ_yfh1_ mitochondria is probably not redox active.
As well as mitochondrial iron accumulation, the effects of concurrent cytosolic depletion should be considered. In FRDA lymphoblasts, the posttranscriptional regulators IRP1 and IRP2 (displaying aconitase activity in iron-replete conditions) show increased IRE-binding activities, indicative of low levels of iron in the cytosol and oxidative stress (187). Li et al. (187) also demonstrated that frataxin expression is increased by iron and substantially reduced by addition of the iron chelator deferoxamine. Iron-dependent expression of YFH1 also was observed in yeast cells (275, 286). These results suggest that cytosolic iron depletion exacerbates the repression of frataxin expression, initially due to FXN gene silencing, thereby promoting the pathogenesis of the disease.
D. Iron-binding properties and oligomerization of frataxin
Given that the lack of frataxin results in mitochondrial iron accumulation, it was quickly predicted that frataxin may directly interact with iron. The iron-binding capacity of frataxin was first determined for yeast frataxin (4) and then for the human and bacterial homologues (6, 25, 34, 55, 150). Furthermore, the ability of the bacterial and yeast frataxin to oligomerize is tightly related to their iron-binding capacity. These proteins generally form highly soluble monomers, but in the presence of a large excess of iron, they can oligomerize or aggregate (6). The human frataxin protein seems to behave differently, because the monomer assembles in an iron-independent manner through stable protein–protein interactions mediated by the nonconserved N-terminal region of the protein (225, 226). Frataxin can bind iron in vitro in the monomeric (65), trimeric (161), or oligomeric/multimeric form (278). Based on these observations, and consistent with determinations of the iron/protein ratio, frataxin was suggested to play a role as an iron donor [chaperoning Fe(II)] for biologic processes (317, 329, 330), as an iron-storage protein (4, 117), or both (118, 161).
The biochemical and structural aspects of iron binding by frataxin have been extensively studied [reviewed in (24)] Structural attributes may account for the iron-binding properties; the plane formed by the two terminal α-helices includes highly conserved exposed acidic residues (glutamic and aspartic acids), forming a patch on the protein that could be involved in cation binding (Fig. 7B) (24). Studies with bacterial, yeast, and human frataxin show that the monomeric CyaY and Yfh1 proteins can bind two Fe(II) atoms (no metal–metal interaction) with dissociation constants of 3.8 and 3.0 μ_M_, respectively, and that human frataxin binds seven Fe(II) atoms with a dissociation constant of 55 μ_M_ (34, 65, 329). Human frataxin was found to have the lowest affinity for iron (329). In some cases, no evidence of interaction between human frataxin and iron could be detected, leading to the suggestion that the putative iron-storage property of the frataxin family is a side function that has been lost during evolution (6).
Structural studies on the oligomerization of frataxin have focused mainly on the yeast protein, because of the tendency of human frataxin to polymerize and form higher-order structures spontaneously and independent of iron (6, 55). Yeast frataxin monomers can self-assemble in vitro in an iron-dependent manner to yield a macromolecular complex with physical features consistent with a role in iron storage (117). The assembly of yeast frataxin in vitro is a stepwise process that requires the presence of Fe(II). It proceeds through the formation of several stable intermediates: α1 → α3 → α6 → α12 → α24 → α48 (4, 117). In this model, the trimer represents the basic structural and functional unit of the 24- or 48-subunit frataxin oligomer (161, 235). Fe(II) is converted into Fe(III) during oligomerization, through two sequential oxygen-dependent iron-oxidation reactions: a ferroxidase reaction catalyzed by frataxin induces the first assembly step (α1 → α3), followed by a slower autoxidation reaction that promotes the assembly of higher-order oligomers yielding ferritin-like particles (α24 or α48) (234, 235). Frataxin monomers are arranged into trimers within these particles, which contain up to 2,400 iron atoms that exist predominantly as ferrihydrite (4, 117, 161, 224, 278). Stabilization of yeast frataxin oligomers seems to be mediated by interactions between the ferrihydrite crystallites, formed at separate mineralization sites, with disassembly of the frataxin oligomers occurring on reduction of their ferric iron core (235). Consistent with this, the crystal structure of the trimer showed the contact region between monomers to form a channel that may accommodate the metal ions (161). The iron-binding properties of human frataxin have been studied less extensively. Although its oligomerization is iron independent (6, 55), human frataxin overproduced in E. coli can also assemble into a stable homooligomer with ferroxidase activity (226), which can bind ∼10 iron atoms per frataxin molecule (55).
Frataxin has been compared with ferritin, despite major structural differences, because both proteins can convert redox-active iron into an inert mineral through ferroxidation activity (278). These findings led several authors to suggest that frataxin not only promotes the biogenesis of iron-containing proteins (through monomeric and/or trimeric forms of the protein), but also detoxifies surplus iron in the mitochondria (through the formation of ferritin-like frataxin particles), thus serving as a major antioxidant (118, 119, 185).
The iron-storage properties associated with frataxin oligomerization in vitro are well established; however, it remains unclear whether these properties are relevant to in vivo function. In vivo oligomerization of Yfh1 is induced by iron, heat stress, and overproduction of the monomer (119, 286), but iron storage in frataxin multimers has never been shown to accompany the increase in oligomerization in these conditions. Moreover, Aloria et al. (10) observed that a Yfh1 mutant, defective in iron-induced oligomerization in vitro, was still able functionally to replace the wild-type protein in vivo, even when produced at very low levels. Nevertheless, it remains possible that frataxin oligomerization may be required in vivo under stringent conditions, such as exposure to stress (118, 119). The observation that mitochondrial iron accumulates in frataxin-deficient cells seems to support the hypothesis of frataxin being involved in mitochondrial iron storage (117). Moreover, the expression of human mitochondrial ferritin in frataxin-deficient yeast cells attenuates the deleterious phenotypes associated with the lack of frataxin (48, 49). However, many different yeast mutants, most of which with impaired [Fe-S] cluster assembly, accumulate large amounts of iron in their mitochondria, although they possess a functional frataxin protein (263). It is possible that expression of human ferritin in these mutants would also improve cell viability. Finally, frataxin is not an abundant protein, with a concentration in yeast of ∼1,000–1,500 molecules per cell under standard conditions (286). Thus, the iron concentration in the mitochondrial matrix greatly exceeds the iron-binding capacity of frataxin (286).
E. Regulation of cellular antioxidant defenses
Frataxin deficiency causes pathologic oxidative stress in cells from FRDA patients and in all eukaryotic organisms studied (8, 11, 44, 59, 275, 309). Oxidative stress is therefore a central feature of the disease and a potential target for therapy. Several antioxidant drugs are currently being tested in clinical trials (see VII.B). Biomarkers of oxidative stress have been discovered in urine and blood samples from FRDA patients. Levels of urinary 8-hydroxy-2′-deoxyguanosine, a marker of DNA oxidative damage (284), and plasma malondialdehyde, a product of lipid peroxidation (37, 95), are higher in patients than in controls. Nonetheless, conflicting results have been obtained, as a difference could not be found in urinary 8-hydroxy-2′-deoxyguanosine (87) and F2-isoprostanes (217) levels between patients and controls in these studies. Oxidative stress due to frataxin deficiency is frequently associated with iron accumulation in the mitochondria, as discussed earlier (see V.D). However, the misregulation of antioxidant enzymatic defense mechanisms has been observed in frataxin-deficient cells, which may result in ROS accumulation and oxidative stress (59, 157, 237, 305).
ROS are produced in several cellular compartments but the large majority, estimated at 90%, comes from mitochondrial respiration [for reviews, see (16, 102, 148, 325)]. A small proportion of the electrons passing through the electron-transport chain, mostly at complexes I and III, react with molecular oxygen to produce superoxide anion (O2•−), which can be converted into other ROS species either enzymatically or nonenzymatically (Fig. 12). Superoxide dismutases (SODs) are the first line of defense against ROS, converting O2•− into hydrogen peroxide (H2O2). Eukaryotic cells have two SODs, one in the mitochondrial matrix that uses manganese as a cofactor (Sod2 or MnSOD), and a copper–zinc SOD located in the cytoplasm and the mitochondrial intermembrane space (Sod1 or CuZnSOD). Mammalian cells have a third SOD isoform, an extracellular CuZnSOD. Superoxide can inactivate [4Fe-4S] cluster-containing enzymes such as aconitase, releasing iron and thereby increasing the intracellular free-iron pool. This favors the Fenton reaction [Fe(II) + H2O2 → Fe(III) + HO− + HO•], in which H2O2 reacts with ferrous iron to produce the hydroxyl radical (HO•), which can damage any biologic macromolecule. Excess O2•− also can react with nitric oxide (NO) to generate the deleterious peroxynitrite (ONOO−). Detoxification of H2O2 involves the action of diverse scavenging enzymes such as catalases, glutathione peroxidases (GPXs), or peroxiredoxins. The glutathione tripeptide (GSH/GSSG; l-γ-glutamyl-cysteinyl-glycine) is a major antioxidant molecule in eukaryotic cells. Peroxides are reduced by GPXs through a reaction in which GSH is simultaneously oxidized to GSSG. GSH is regenerated from GSSG by glutathione reductase by using NADPH as the electron donor (Fig. 12) (45). The cellular response to oxidative stress involves the induction of detoxifying enzymes such as SODs and GPXs, an increase in GSH and NADPH synthesis, a decrease in the GSH/GSSG ratio, and glutathionylation of target proteins (45). The cellular thiol redox status is maintained by the glutathione/glutaredoxin and thioredoxin/thioredoxin reductase systems, which reduce the oxidized sulfydryl groups of proteins (Fig. 12). Even though ROS and reactive nitrogen species (RNS) can cause cellular damage, some have other important roles. H2O2 can be produced by several enzymes in different compartments and functions as a signaling molecule in growth, apoptosis, and aging (127). NO is an important messenger in neuronal communication in the central nervous system, despite also being a mediator of neurotoxicity in several disorders (45). Under normal physiologic conditions, the rate of ROS and RNS production is compensated by the activity of scavenging enzymes and small antioxidant molecules (α-tocopherol, ascorbic acid, and glutathione). An imbalance between ROS and RNS formation and antioxidant defense mechanisms results in increased oxidative stress. Fibroblasts from FRDA patients, unlike those from healthy controls, are sensitive to low doses of H2O2, oligomycin, and iron and do not upregulate SODs in response to oxidative-stress inducers (59, 157, 237, 302, 326).
FIG. 12.
ROS production and cellular antioxidant defense enzymes. Superoxide anion (O2•−) is produced by complexes I and III of the electron-transport chain and converted into hydrogen peroxide (H2O2) by superoxide dismutases (SOD) or into peroxynitrite (ONOO−) by reacting with nitric oxide (NO). H2O2 can react with ferrous iron to produce the hydroxyl radical (HO•). Glutathione (GSH/GSSG) is a tripeptide synthesized in two steps from glutamic acid, cysteine, and glycine. H2O2 and other peroxides are detoxified by glutathione peroxidases (GPXs), which oxidizes glutathione. GSSG is reduced to GSH by glutathione reductase (GR) by using electrons from NADPH. NADPH is regenerated by the pentose phosphate pathway enzymes, glucose 6-phosphate dehydrogenase (G6PDH), and 6-phosphogluconate dehydrogenase. Other enzymes that scavenge H2O2 and peroxides are catalases and peroxiredoxins (PRXs). Peroxiredoxins also can scavenge ONOO−. The cellular thiol redox status is maintained by the thioredoxin (TRX)/thioredoxin reductase (TR) and glutathione/glutaredoxin systems by reducing the oxidized sulfhydryl groups of proteins.
Several studies using blood or cells from FRDA patients, or yeast, suggest that frataxin deficiency leads to the impairment of glutathione homeostasis (13, 221, 236, 243). A considerable reduction in free glutathione levels (15-fold) and a significant increase in the glutathione bound to hemoglobin (twofold) in erythrocytes were observed in the blood of FRDA patients (243). The glutathione-dependent redox status of frataxin-deficient cells was studied in detail in the yeast model (13). In Δ_yfh1_ cells, the total glutathione concentration and the GSH/GSSG ratio are significantly lower, and GPX activity is higher than in wild-type cells. Although the pentose phosphate pathway (NADPH-producing pathway in the cell) is stimulated (glucose-6-phosphate dehydrogenase activity being 3 times higher than in the wild type), the NADPH/NADP+ pool remains low in mutant cells. In contrast to these marked metabolic changes, no significant difference was observed in the expression of genes involved in glutathione-dependent systems between Δ_yfh1_ and control cells. These findings suggest that frataxin deficiency in yeast results in the remodeling of glutathione-dependent defense systems and in cell adaptation to chronic oxidative stress (13). Interestingly, the addition of iron, but not of an iron chelator, to the culture medium increases total GSH levels and decreases the activity levels of GPX and glucose-6-phosphate dehydrogenase (13). Additionally, GPX activity has been shown to be induced and GSSG concentration increased in cultured fibroblasts and lymphoblasts from FRDA patients, suggesting that these cells are in a state of oxidative stress (221, 236, 237, 303). Consistent with a twofold increase in glutathionylated hemoglobin (243) and an 83% increase in gluthatione _S_-transferase activity (305) in patients' blood, Pastore et al. (236) found that the glutathione pool is shifted toward its protein-bound form in patient fibroblasts. Immunoprecipitation of fibroblast lysates with anti-actin antibody and probing with anti-GSH antibody confirmed that actin glutathionylation levels were 4.7 times higher in patient fibroblasts than in normal control cells (236). The glutathionylation of actin results in the disassembly of filaments in vivo, which is reversible by glutathione treatment. In contrast to observations in the yeast model, addition of iron to FRDA fibroblasts leads to increased protein-bound glutathione (236). These results were confirmed in autopsy samples from the spinal cords of four patients (297). Immunostaining with an antibody that specifically recognizes protein-bound glutathione showed stronger staining in patient samples (in gray matter neurons and in white matter cells and axons) than in controls, suggesting a higher level of glutathionylated proteins in FRDA patients. This study also showed abnormal microtubule dynamics in patient autopsy samples (297).
It was established several years ago that FRDA fibroblasts are sensitive to oxidants (59, 157), but the signaling defect in the pathway leading to SOD induction was discovered only recently (237). Impairment of mitochondrial SOD2 induction with iron treatment does not appear to involve the transcription factor NF-κB (157). Paupe et al. (237) demonstrated that the Nrf2-dependent Phase II antioxidant pathway is defective in frataxin-deficient fibroblasts. Under normal conditions, the activity of the transcription factor Nrf2 is regulated by the actin-associated Keap1 protein, which sequesters Nrf2 in the cytoplasm and promotes its degradation through ubiquitination (223). Under conditions of stress, this interaction is disrupted, and Nrf2 is translocated to the nucleus, where it binds to DNA sequences of the _cis_-acting ARE (antioxidant-responsive element), activating the expression of Phase II antioxidant genes (223). These encode proteins including SODs, catalase, glutathione _S_-transferase, and NADH quinone oxidoreductase (223, 237). In FRDA fibroblasts treated with oligomycin or _t_BHQ, Nrf2 fails to translocate to the nucleus, and none of these genes is induced (237). These results are consistent with the observation of the disorganization of actin fibers in patient fibroblasts and the consequent dissociation of Keap1 and Nrf2 from actin. Similar results were obtained in an shRNA frataxin-depleted neuronal model (neuroblastoma-derived cell line) (237). Taken together, these findings suggest that constitutive oxidative stress due to frataxin deficiency causes changes in the glutathione pools, resulting in increased actin glutathionylation and altered cytoskeletal dynamics and thus impairing the induction of Phase II antioxidant defense pathways (Fig. 13).
FIG. 13.
Frataxin deficiency leads to oxidative stress. In frataxin-deficient cells, the Nrf2-dependent Phase II antioxidant defense pathway is impaired. Actin is glutathionylated, and actin fibers are disorganized and not associated with Keap1 and Nrf2. Consequently, expression of the genes controlled by Nrf2, such as the mitochondrial SOD2, is not induced on treatment of frataxin-deficient cells with oxidants. The total glutathione concentration may be decreased, in addition to increase in the levels of the oxidized form (GSSG) and to more glutathione bound to proteins.
In parallel to reduced cell antioxidant defense mechanisms, frataxin deficiency also seems to lead to an increase in ROS accumulation (154, 221, 274, 275, 286). It remains unclear, however, which molecular species among ROS and RNS are accumulated in frataxin-deficient cells and are responsible for the observed damage. Additionally, little is known about the regulation of the enzymes that produce ROS and RNS in FRDA patient cells or in other models. In patient lymphoblasts, the reduced activity of respiratory chain, specifically of complex III and cytochrome c, leads to accumulation of O2•− (221).
Other studies suggest that H2O2 is the most likely cause of oxidative stress. The treatment of patient fibroblasts with a catalase mimetic, Euk134, rescues the Nrf2 signaling pathway (237); and the expression of H2O2-detoxifying enzymes, but not SOD1 or SOD2, in Drosophila rescues the deleterious phenotypes of frataxin deficiency (12). Several different enzymes in the cell can produce H2O2 (127). One of these enzymes, the mitochondrial outer membrane monoamine oxidase (MAO) A, was found induced in frataxin-deficient HEK293 T-Rex cells (193). MAOs catalyze the oxidative deamination of biogenic amines to generate the reaction products H2O2 and aldehyde. Oxidative stress and apoptosis are thereby induced, resulting in neuronal degeneration (219).
The role of NO production and the contribution of RNS to oxidative damage in frataxin-deficient cells has yet to be determined. Given that NO is a major signaling molecule in the central nervous system, it may be of particular interest to evaluate its role in the pathophysiology of the disease.
F. Mitochondrial and nuclear genome integrity
Loss of mitochondrial DNA was one of the first phenotypes observed in yeast Δ_yfh1_ cells, the so-called petite phenotype (rho- and rho° cells) (15, 104, 323). On downregulation of YFH1 expression, by using a tightly regulated GAL1 promoter, it was shown that iron accumulation and protein oxidative damage precede the appearance of petite cells (163). In this study, cells with acute frataxin deficiency displayed the petite phenotype once they had accumulated at least five lesions per mitochondrial genome, corresponding to 10 generations under glucose repression. Complete loss of mtDNA (rho° cells) was observed four to five generations later (163). In cells in which frataxin was still present, but at reduced levels, lesions were detected after 16 generations with only a limited loss of mtDNA after 22 generations, indicative of a slow accumulation of damage (163). Low levels of frataxin correspond to the situation observed in FRDA patients, and mtDNA lesions have been observed (36, 145, 149). Analysis of tissues from biopsies or autopsies of patients with FRDA have shown a reduced ratio of total mtDNA to 18S rDNA in the skeletal muscle (60%), the heart (33%), cerebellum (27%), and DRG (18%) (36).
Mutations in mtDNA can lead to impairment of oxidative phosphorylation, causing several disorders, such as mitochondrial myopathies and cardiomyopathy. In vivo analysis of cardiac and skeletal muscle bioenergetics of FRDA patients by using phosphorus magnetic resonance spectroscopy showed interesting findings. Lodi et al. (190) demonstrated that in skeletal muscle of FRDA patients, the maximal rate of mitochondrial ATP production was significantly lower than that in healthy controls or in patients with unrelated muscular diseases. In addition, the deficit in ATP production correlated with the number of GAA triplet repeats in the smaller allele, strongly suggesting that oxidative phosphorylation is affected by a decrease in frataxin level. The cardiac bioenergetics, measured as the phosphocreatine-to-ATP and inorganic phosphate–to-phosphocreatine ratios, are abnormal in FRDA patients, even in the absence of left ventricular hypertrophy (41, 191). These results imply that energy-metabolism deficit in cardiac tissue could be the cause of cardiomyopathy in FRDA patients. No relation was found between the GAA-repeat length and the myocardial energy (191).
A growing body of evidence highlights the importance of normal mitochondrial metabolism in nuclear genome integrity. Like the other petite mutants, Δ_yfh1_ cells are unable to grow on respiratory substrates. However, they grow more slowly on fermentable carbon sources. This observation encouraged Karthikeyan et al. (162) to investigate whether frataxin deficiency leads to nuclear DNA damage. They found evidence of chromosomal instability, with the Δ_yfh1_ mutant showing higher levels of illegitimate mating, a sixfold higher rate of spontaneous recombination, and a twofold higher mutation rate than controls. Frataxin deficiency also led to increased sensitivity to the DNA-alkylating methyl methanesulfonate and to the replication inhibitor hydroxyurea than to rho° control cells (162). Furthermore, the DNA damage-inducible promoter from the DIN1 gene was upregulated in mutant cells, and addition of the antioxidant N_-acetylcysteine suppressed the induction of DIN1 expression, suggesting that the nuclear damage was due to increased ROS. Consistent with this, deletion of the glutathione peroxidase gene GPX1 in Δ_yfh1 cells led to a marked increase in the nuclear mutation rate, as determined by canavanine resistance (162). These results led the authors to suggest that the substantial spontaneous nuclear damage in yeast frataxin-deficient cells was caused by H2O2 generated in the mitochondria (162).
A recent study challenges this hypothesis and suggests that mitochondrial dysfunction promotes nuclear genome instability by inhibiting the assembly of Fe-S cluster proteins that are required for maintenance of nuclear genome integrity (311). Veatch et al. (311) described the crisis events that follow loss of mtDNA in yeast; they observed cell-cycle arrest, progressive loss of viability, and selection for nuclear mutations that improve growth in the absence of mtDNA. Loss of mtDNA led to a reduction in the inner mitochondrial membrane electrochemical potential (ΔΨ), a transcriptional profile characteristic of iron starvation and intracellular Aft1-dependent iron accumulation, an increase in oxidative protein damage, and impaired mitochondrial and nuclear/cytosolic Fe-S cluster assembly. By using Δ_aft1_ mutant cells, which do not accumulate iron, they concluded that increased iron and oxidative damage may play a role in the crisis after mtDNA loss but are not required for the increase in nuclear genomic instability.
VI. Frataxin Is Involved in Development, Cell Death, and Cancer
A. Development in model organisms
As discussed earlier (see III.F), no FRDA patient has yet been found to be homozygous for point mutations, suggesting an essential role for frataxin in development. Inactivation of the frataxin gene in mice causes embryonic lethality at day E6.5, a few days after implantation (for details on frataxin expression in mouse, see III.C) (71). Frataxin also was reported to be necessary for differentiation in cell cultures. The mouse embryonic carcinoma P19 cells can differentiate into a variety of cell types, and frataxin was found to be required for neuronal, but not for cardiomyocyte, differentiation (274).
The Drosophila melanogaster dfh gene, encoding the frataxin homologue, is expressed at a low level throughout development, from early embryonic stages to adults (11, 52). Frataxin protein levels are highest in late embryos, diminishing in larvae and pupae, and increasing to modest levels in young adults (11). A peak in dfh mRNA expression is thus observed in 6- to 12-h embryos, corresponding to the time at which most tissues differentiate (52). RNA in situ hybridization has shown a ubiquitous distribution of dfh transcripts in embryos (52). The effects of downregulating dfh expression by using a GAL4-UAS transgene (RNAi-based technology) differ between larvae and adults (11). Larvae exhibit retarded development, leading to a prolonged larval phase, and impaired metamorphosis to become adults at 25°C. At 18°C, 1–2% of larvae develop into adults, but most die within 3–4 days; the remaining flies survive ≤ 40 days. The silencing of dfh in the peripheral nervous system, but not in motor neurons, imposes a reduction of 40% in adult life span without any defect in pre-adult development (11). A ubiquitous overexpression of dfh also leads to death of all individuals before pupae eclosion from puparium because of alterations in the development of embryonic muscles, peripheral nervous system, and the heart (189). These results show that changing the level of cellular frataxin in Drosophila affects the same tissues as those observed in FRDA patients. At the biochemical level, the effects of dfh silencing include decreased activity of Fe-S cluster-containing enzymes, disruption of intracellular iron homeostasis, and sensitivity to oxidative stress (11, 189). Overexpression of dfh resulted in inhibition of mitochondrial aconitase under hyperoxia, but not in normal oxygen conditions (189).
Caenorhabditis elegans transgenic strains carrying fusions of the frataxin-encoding gene (frh-1) and the green fluorescent protein showed localization of frataxin to the muscles, gut, pharynx, spermatheca, and head neurons (308). As observed for other organisms, frh-1 knockout in C. elegans results in developmental arrest at the L2/L3 larval stage (312). Data obtained on the effect of frataxin knockdown on nematode life span have been inconsistent (309, 312, 332). Ventura et al. (312) reported that the life span was ∼25% greater than normal, despite their small size, reduced fertility, and sensitivity to the superoxide generator juglone. By contrast, other groups have described a shorter life span, associated with impaired respiration and increased sensitivity to paraquat (another superoxide generator) (309, 332). These results were reconciled by using an RNAi dilution strategy to obtain different degrees of frataxin gene expression and of four genes involved in the electron-transport chain (255). The observed phenotypes were dose dependent, with the inhibition of mitochondrial function increasing the life span until a certain threshold, beyond which cell viability is severely compromised (255). Recently, p53/CEP-1 was found to mediate these opposing effects in response to the level of mitochondrial bioenergetic stress (313).
In the plant Arabidopsis thaliana, frataxin knockout results in early embryonic lethality (44, 310). Mutant atfh-1 plants with < 50% of normal frataxin levels show retarded growth, without any morphologic abnormalities in roots, leaves, and flowers, and impaired fructification (44). The reduced fruit fresh weight and number of seeds per fruit observed in these plants correlated well with the pattern of frataxin gene expression, which was higher in flowers than in roots or leaves (44, 310). These higher levels of expression in certain organs correlate with their energy requirement. Flowers have high energy demands for anther development and pollen maturation and also have an increased number of mitochondria and a higher respiration rate (44). Mutant plants show decreased activity of the Fe-S cluster enzymes aconitase and succinate dehydrogenase, improved CO2 assimilation rates, iron accumulation, increased ROS and nitric oxide production, and induction of an oxidative stress–defense response (44, 196).
In lower eukaryotes, frataxin deficiency also can be lethal. In our hands, deletion of the S. cerevisiae YFH1 gene in BY4741 genetic background is viable in anaerobiosis but lethal under aerobic conditions (R. Santos, unpublished data). Additionally, deletion of the frataxin-encoding gene in the wild-type C. albicans SC5314 strain leads to death within a month after generation of the mutant (S. Lefevre, unpublished data). These findings thus show that the metabolic functions regulated by frataxin are essential for the normal development and life span of several different organisms.
B. Susceptibility to cell death and neuron degeneration
The major sites of neurodegeneration in FRDA patients are the dorsal root ganglia, the dorsal roots of the spinal cord, and the dentate nucleus in the cerebellum (232). It is not known why these regions of the nervous system are particularly sensitive to frataxin deficiency. Available data have been obtained through the analysis of autopsy or biopsy tissues from patients with variable disease duration. It is, therefore, difficult to elucidate the events that initiate neuron degeneration and the pathway leading to cell death (152, 167). Unfortunately, murine models fully reproducing the pathophysiology of FRDA have not yet been successfully developed [for recent review, see (248)]. The Cb inducible knockout model, however, presents several FRDA symptoms, with repression of frataxin expression in adult mice leading to progressive sensory and cerebellar ataxia (294). Loss of large myelinated fibers and an increased number of small unmyelinated fibers in the DRG, dorsal roots, and peripheral nerves is characteristic of FRDA disease (152, 167). In the Cb mouse mutant, the earliest pathologic feature detected was DRG neuronal degeneration, specifically of the large sensory neurons, but the myelin sheet was preserved (294). A large number of vacuoles and accumulation of lipofuscin can be observed in degenerating neurons. These observations led Simon et al. (248, 294) to suggest that degeneration was mediated by autophagy; however, no specific markers were used to determine whether the vacuoles were indeed autophagosomes. Consistent with this hypothesis, they found no evidence of apoptosis by TUNEL staining in the DRG neurons (294). Degeneration of large sensory neurons harboring giant vacuoles also was observed in the humanized mouse model, but with secondary demyelination (7).
Autophagy is a survival response of cells subjected to nutrient deprivation or other stresses; in certain conditions, induction of autophagy is not sufficient, and cells die (94, 172). It is not clear, however, if cell death occurs by autophagy or by another pathway with morphologic features characteristic of autophagy (172). Neurons are postmitotic cells that depend on autophagy for survival (181). Taken together, it is possible that autophagy is induced to protect frataxin-deficient DRG neurons against stress, but degeneration is due to additional pathways, like apoptosis. The apoptosis is frequently observed in cultured cells and has been detected in mouse knockout embryos at E6.75 (71). Several studies show that frataxin deficiency, in fibroblasts and lymphoblasts from FRDA patients or in different cell lines, renders cells more susceptible to apoptosis (64, 156, 160, 240, 258, 274, 285, 327). A significant increase in the transcription of genes involved in apoptosis was detected by microarray analysis of FRDA fibroblasts and lymphoblasts, a neural NT2 cell line knocked down for frataxin expression (303), and in the heart tissue of the MCK mouse (151). Additionally, caspases 3 and 9 were more strongly activated in frataxin-deficient cells than in controls (258, 326). Further studies are needed to understand the potential role of autophagy and apoptosis in neuron degeneration in FRDA patients. Such studies require biopsy samples from patients before or soon after manifestation of the first symptoms.
Oligodendrocytes and Schwann cells are glial cells specialized in the myelination of axons in the central and peripheral nervous systems, respectively. These cells may also contribute to the preservation of axon integrity [for recent review, see (222)]. Two recent reports suggest that frataxin deficiency affects Schwann cells and that neuronal loss occurs as a secondary event (167, 194). Analysis of FRDA patient autopsy samples shows Schwann cells in the dorsal roots of the spinal cord to be abnormally small and few in number, with inappropriate myelination of the thin fibers (167). The comparison of DRG, oligodendroglial, and Schwann cell lines with frataxin knockdown by siRNA demonstrated that frataxin depletion has no effect on DRG, but inhibits the proliferation of oligodendrocytes and has significant effects on the Schwann cells (194). Frataxin deficiency in oligodendrocytes and Schwann cells blocks cell-cycle progression at G2M. In Schwann cells, but not in oligodendrocytes, this is followed by an inflammatory response and an increase in cell death (apoptotic and necrotic death). Treatment of these cells with antiinflammatory and antiapoptotic drugs rescues the death phenotype (194). Altogether, these results suggest that cell-cycle arrest, inflammation, and death of oligodendrocytes and particularly Schwann cells could result in demyelination and axon degeneration.
C. Cancer
Cancer is not a clinical feature of FA disease, and a clear causal connection between frataxin expression and cancer has not been established in vivo. However, several lines of evidence suggest that frataxin acts as a tumor suppressor by increasing cellular oxidative metabolism (285) and antioxidant defense mechanisms (293). Frataxin gene knockout in murine hepatocytes leads to the development of multiple hepatic tumors in mice (304). Liver cells in these mice show the classic phenotypes associated with frataxin deficiency, including reduced respiration rate, decreased activity of Fe-S enzymes, and increased sensitivity to oxidative stress (304). These cells also show induction of Bax-dependent and p53-independent apoptotic pathways. Concurrently, overexpression of frataxin in murine 3T3L1 cells was shown to increase thiol-dependent oxidative stress defense mechanisms and to reduce tumor formation in nude mice (293). Consistent with these findings, colon cancer cells stably overexpressing frataxin have an increased respiration rate, increased mitochondrial membrane potential, and increased ATP levels and aconitase activity, but do not accumulate ROS (285). When these cells are injected into nude mice, tumors are significantly smaller than those formed after injection of control cancer cells, suggesting that the increased oxidative metabolism induced by frataxin overexpression inhibits expansion of cancer cells in vivo (285). The phosphorylation of the tumor suppressor p38 MAP kinase seems to be dependent on the presence of frataxin and could mediate the antiproliferative effect of frataxin in tumor cells (285, 304).
Treatment of patient cancers with chemotherapy drugs like paclitaxel and cisplatin causes peripheral neuropathy. Interestingly, Melli et al. (203) showed that the antioxidant α-lipoic acid protects DRG and Schwann cells from mitochondrial damage induced by these drugs, and that its effect is dependent on the increased expression of frataxin.
VII. Therapeutic Approaches for Treatment of Friedreich Ataxia
Recent advances in understanding how GAA trinucleotide repeats repress FXN gene expression and frataxin function has led to several new therapeutic approaches (Fig. 14). Idebenone was the first drug used to treat FRDA patients. It exerts its beneficial effects by ameliorating hypertrophic cardiomyopathy (202). This drug is now in Phase III clinical trials in the United States and Europe and is being tested for its potential effects on patient neurologic status. Phase II trials are presently being undertaken for other drugs that improve mitochondrial function or increase antioxidant defense levels (e.g., pioglitazone). Other approaches include reducing iron-mediated toxicity (e.g., deferiprone, Phase II trial), increasing frataxin expression (e.g., polyamides or erythropoietin), and gene therapy. However, no known pharmacologic treatment cures, or even slows, FRDA disease. Given that frataxin function remains to be elucidated, high-throughput screening with different cellular models is being used to search for new drugs (47). These screens are hampered by the instability of frataxin-deficient cells, resulting in nonreproducible results for the various steps involved in hit confirmation (47). Long-term adaptation of mammalian cell lines and yeast cells to frataxin deficiency may arise through epigenetic changes and metabolism remodeling or an increased rate of chromosomal mutation (162). Calmels et al. (46) thus constructed a new cellular model based on murine fibroblasts producing the human frataxin mutants, G130V and I154F, which stably reproduce the phenotypes associated with frataxin deficiency. These models seem very promising, although not knowing the function of frataxin can cause difficulties in the interpretation of high-throughput screens. The development of induced pluripotent stem cells from patient cells could be an alternative approach.
FIG. 14.
Therapeutic strategies for the treatment of FRDA. No efficient therapy is now available to treat patients. Given that the frataxin function is not known, targets for therapy are based on main phenotypes of frataxin deficiency, including antioxidant defense (idebenone, CoQ10 plus vitamin E), improvement of respiration (idebenone, CoQ10 plus vitamin E), reduction of the mitochondrial iron pools (deferiprone), and increase in frataxin protein (erythropoietin EPO, pioglitazone). A major goal is the finding of chemical drugs that alleviate the Fe-S cluster deficiency. Recent strategies target the GAA-expansion triplet repeat (sticky DNA structure and heterochromatin) and intend to increase the FXN gene expression (HDAC inhibitors, polyamides). Other therapeutic strategies focus on replacement of the mutated gene by gene therapy.
As a final consideration, it is possible that the only drugs that will treat FRDA patients effectively are those that restore Fe-S cluster assembly. For recent reviews on disease progression and treatment, see (83, 144, 202, 283, 306).
A. Evaluation of disease progression
Clinical trials of drugs to treat FRDA patients are complicated by intrinsic features of this disease. Disease progression is slow and clinical variability among patients is extensive. Tools that accurately evaluate the progression of this disease are therefore needed to be able to detect small beneficial effects of potential treatments in trials with small numbers of patients [for recent review, see (83)]. Two scales are commonly used to score the neurologic status of FRDA patients, the International Cooperative Ataxia Rating Scale (ICARS), and the Friedreich Ataxia Rating Scale (FARS). The ICARS has been used in clinical trials for idebenone and deferiprone, but is not appropriate for the evaluation of disease progression in patients with a long duration of disease (83, 283). The FARS scale seems to be more sensitive to changes in disease progression but is more labor intensive (83, 283). A new scale (Scale for Assessment of Rating of Ataxia, SARA), validated on dominant ataxias, is quick to administer, gives very good results, and is applicable to FRDA (42, 83, 283). This scale was used for the assessment of neurologic status in an open-label study of erythropoietin effect in FRDA patients (32). The SARA scale may be the preferred choice for future clinical trials because of its high interrater and test–retest reliability and the ease of administration by clinicians (42).
B. Antioxidants and oxidative phosphorylation
Treatment of patients with idebenone or a combination of coenzyme Q10 (CoQ10) with vitamin E aims to improve mitochondrial function and to reduce oxidative stress. Vitamin E is a natural cellular lipid-soluble antioxidant that is highly abundant in nuclear and mitochondrial membranes. CoQ10 is an electron carrier in the respiratory chain and is involved in the reduction of oxidized vitamin E. Treatment of patients with vitamin E showed a therapeutic effect only when used in combination with CoQ10 (66, 231). A 4-year open-label trial of 10 patients treated with 2,100 IU/day vitamin E and 400 mg/day CoQ10 showed improvement in cardiac and skeletal muscle bioenergetics, even though no apparent benefit was observed on cardiomyopathy (142). Additionally, neurologic ICARS scores were stable during the trial, suggesting that no progression of the disease occurred over this period (142). However, a limitation of this study was that ICARS scores were obtained by comparison with cross-sectional data to predict FRDA disease progression.
Idebenone is a short-chain CoQ10 analogue that has a dual functions in the cell; it acts as an antioxidant, by protecting membrane lipids from peroxidation, and it stimulates oxidative phosphorylation and ATP production by carrying electrons from complexes I and II to complex III in the electron-transport chain (202). Idebenone was first identified as a candidate for treatment of FRDA in a work by Rustin and colleagues (266). They showed that idebenone protected the respiratory complex II from iron inactivation and decreased lipoperoxidation in heart homogenates from patients with valvular stenosis. Furthermore, treatment of three patients with 5 mg/kg/day idebenone for 4 to 9 months resulted in reduction of myocardial hypertrophy (266). This work was followed by several clinical studies, most of which used the same idebenone concentration, confirming a positive effect on cardiomyopathy [for recent review, see (202, 231)]. An open-label trial showed a decrease in the levels of the urinary marker of oxidative stress 8-hydroxy-2′-deoxyguanosine in eight patients treated for 8 weeks with 5 mg/kg/day idebenone (284). Unexpectedly, no difference was found in the baseline urinary levels of 8-hydroxy-2′-deoxyguanosine between controls and young patients enrolled in a randomized, double-blind, and placebo-controlled trial using three doses of idebenone for 6 months (87). However, this study suggested that treatment with high-dose idebenone improves neurologic functions (87). Idebenone, even at high doses, appears to be a safe and well-tolerated drug. Currently, two Phase III trials are being carried out in Europe and the United States.
Other CoQ10 analogues have been developed. One such product, MitoQ, specifically targets mitochondria and protects patient fibroblasts from endogenous oxidative stress with a high efficiency (155). Another drug, EPI-A0001 or α-tocopherol quinone, also improves the mitochondrial functions in vitro, and a Phase II clinical trial is ongoing.
C. Iron chelators
Iron chelation is one of the major potential strategies for treatment of this disease and has been the focus of intensive research. The pathogenesis of FRDA seems to involve an imbalance in the intracellular accumulation of iron, with mitochondrial accumulation and relative cytosolic depletion, rather than an overall accumulation of iron in the cell (see V.D). Consistent with this, iron and ferritin levels were found to be normal, and transferrin receptor concentration increased, in the serum of FRDA patients (321, 322). Rustin et al. (266) showed that Fe(II), but not Fe(III), inactivates complex II in the heart tissue of patients and that this effect can be rescued by the addition of deferoxamine. However, cytosolic and mitochondrial aconitase activity levels are decreased by addition of this iron chelator (187, 266). Aconitase is an iron homeostasis regulator in the cell. The inhibition of aconitase activity is counteracted by the induction of IRE-binding activity, which can lead to further depletion of cytosolic iron in frataxin-deficient cells (187). Additionally, deferoxamine decreases frataxin mRNA and protein levels (187). In conclusion, although deferoxamine is commonly used in clinical settings, findings thus far obtained do not support its use in treatment of FRDA patients. The only potential useful chelators are those that can act on pools of labile iron within intracellular compartments, without depleting aconitase activity and other cellular iron-enzymes or transferrin-bound iron from the plasma. This has led to the development of mitochondria- targeted iron chelators (256).
The mitochondrial iron chelator deferiprone is currently being tested in a Phase II trial, in Europe, Australia, and Canada, directed toward improvement of neurologic abilities. This trial is based on previous results of a pilot study carried out in France, in which nine adolescent patients were given low-dose deferiprone (20 to 30 mg/kg/day) for 6 months (31). Iron accumulation in the brain was assessed by magnetic resonance imaging in these patients. Chelation treatment was found to reduce iron accumulation in specific areas of the brain (dentate nucleus) and to improve neurologic status (ICARS score); however, three of 13 patients were withdrawn from the study because of side effects (31). Despite these promising findings, studies of various human cells treated with deferiprone have given conflicting results. Kakhlon et al. (160) showed that treatment of frataxin-depleted HEK293 T-Rex cells with 50 μ_M_ deferiprone restored mitochondrial redox potential, reduced ROS accumulation and apoptosis, and increased aconitase activity. In contrast, Gonçalves et al. (129) found that treatment of patient fibroblasts with 150 μ_M_ deferiprone abolished aconitase activity and inhibited cellular growth.
D. Molecules that increase frataxin levels
The expansion of GAA repeats in the first intron of the FXN gene forms non-B DNA structures, such as triplex and sticky DNA, and induces histone deacetylation, causing a decrease in gene transcription (319). Several drugs are being developed that target non-B DNA formation or inhibit histone deacetylase activity. Polyamides specifically bind to GAA tracts and increase transcription and frataxin levels in FRDA patient lymphoblasts (43). Oligodeoxyribonucleotides designed to block triplex formation result in increased full-length transcript in vitro (133). Thus, the discovery of cell-permeable small molecules that target non-B type DNA conformations may have potential therapeutic value and as such have been of particular interest in recent studies (23, 135).
The gene silencing of expanded FXN alleles is characterized by hypoacetylation of histones H3 and H4 and trimethylation of H3 at K9 (7, 137, 147). Histone deacetylase (HDAC) inhibitors, by increasing histone acetylation, may change silent heterochromatin to an active chromatin conformation and restore the normal function of silenced genes. Several HDAC inhibitors were tested in patient lymphoblasts in one study. Of the inhibitors tested, only the benzamide derivative BML-210 and its analogues were found to be nontoxic and to increase the frataxin levels to those observed for FRDA carriers (147). Another derivative of this chemical family, compound 106, corrected for frataxin deficiency in knockin mice (KIKI), in which the mouse FXN gene contains 230 repeats in the first intron (253). In addition, the global gene-expression profile of KIKI mice treated with compound 106 resembled the expression pattern of control animals (253). These findings suggest that these compounds may be promising potential candidates for the treatment of FRDA disease, although no data on toxicity in humans are available.
Other molecules have been reported to increase frataxin levels in cells. Although several of these molecules have no therapeutic interest (e.g., cisplatin or 3-nitroproprionic acid), some display potential beneficial effects. Recombinant human erythropoietin significantly increases frataxin levels in primary lymphocytes and fibroblasts from patients (2, 301). Interestingly, no increase in FXN mRNA was observed, suggesting that recombinant human erythropoietin acts at the posttranscriptional level (2). This drug has been tested in small open-label clinical pilot trials in Austria; the results show an improvement in FARS and SARA scores and a decrease in oxidative-stress biomarkers in the urine and blood of patients (8-hydroxy-2′-deoxyguanosine and peroxides) (32, 33). Side effects include enhancement of the hematopoietic response in patients and a frequent need for phlebotomy (33).
Treatment of the neuroblastoma cell line SKNBE and FRDA patient fibroblasts with Azelaoyl-PAF increases intracellular frataxin levels by a factor of two (195). Azelaoyl-PAF is an agonist of the peroxisome proliferator–activated receptor gamma (PPARγ), which belongs to the nuclear receptor family of ligand-activated transcription factors and regulates adipocyte differentiation and lipid storage. Recently, the PPARγ coactivator PGC-1α was found to be downregulated in cells from FRDA patients and in tissues from KIKO mice (68). These results suggest that modulation of PPARγ could be another potential therapeutic strategy. Earlier in 2006, P. Rustin (14) had proposed that pioglitazone, a commercially available PPARγ agonist that can cross the blood–brain barrier, could be useful for treatment of FRDA because of its ability to increase fatty acid oxidation and mitochondrial function and decrease ROS accumulation and inflammation (14). A Phase III clinical trial has already been started in France, with an expected outcome of improved neurologic function.
VIII. Conclusion
Friedreich ataxia is a neurologic disease caused by a dynamic mutation, a somatic and germline unstable trinucleotide repeat expansion, in the first intron of the FXN gene. Several other neuromuscular disorders are caused by unstable repeats, including fragile X syndrome (FRAXA), myotonic dystrophy, Huntington disease, and spinocerebellar ataxias. However, FRDA is the only disease known to be caused by a GAA trinucleotide repeat expansion. FRDA shares a high degree of similarity with other diseases caused by dynamic mutations, particularly with FRAXA, in terms of the molecular mechanism of repeat expansion, intergenerational transmission, and cellular effects of the mutation (238). FRAXA is caused by a CGG repeat expansion in the 5′ untranslated region of the FMR1 gene. This causes aberrant methylation of the CpG island in the regulatory region and decreased histone acetylation, resulting in loss of expression of FMRP protein. As observed for FRDA, a pool of genes carrying premutations (55–200 repeats) is responsible for the emergence of expanded alleles (>200 repeats). Understanding the biology of unstable trinucleotide repeats is essential to developing new therapeutic strategies to treat these diseases. In the case of FRDA, the tendency of GAA repeats to contract offers a potential target, because finding drugs that stimulate the contraction of these repeats could efficiently treat the patients.
The pathophysiology of FRDA is associated with mitochondrial dysfunction. However, the pathway leading to damage and cell death is not understood, and this is because the frataxin function remains unclear. Several mitochondrial abnormalities have been proposed as the primary defect in FRDA patients, and these include deficiencies in the activities of several enzymes (lipoamide dehydrogenase, pyruvate carboxylase, malic enzyme, α-ketoglutarate dehydrogenase, pyruvate dehydrogenase complex, and the respiratory chain) (17, 18). It is now clear that impairment of Fe-S cluster protein biogenesis, disruption of iron homeostasis, and oxidative stress are the central cellular features of this disease (Fig. 15). Improved knowledge of the physiology of frataxin-deficient cells is needed to understand why and how the DRG and dentate nucleus neurons degenerate. This is likely to depend on the elucidation of frataxin function.
FIG. 15.
Comprehensive model of frataxin function. Frataxin is an iron-binding protein implicated in the delivery of iron to Fe-S cluster assembly and heme synthesis and, by doing so, regulates mitochondrial respiration and cellular iron homeostasis. Impaired regulation of antioxidant defenses, decreased respiration, iron accumulation in the mitochondria, and possibly induction of ROS-producing enzymes causes ROS accumulation in the cell, which results in mutagenesis and oxidative stress. Frataxin is implicated, most likely, indirectly in the signaling of antioxidant defense systems and in the pathways that lead to survival or to death.
FRDA is a recessive disease that occurs in families with no known history of the disease. Initiation of neuronal death precedes the appearance of the first symptoms; thus, damage will already have occurred before diagnosis is confirmed. A major aim in treating FRDA disease is to develop new therapies that not only block disease progression but also induce neuron regeneration.
Abbreviations Used
AVED
ataxia with vitamin E deficiency
cAMP
cyclic adenosine monophosphate
CIA
cytosolic iron–sulfur cluster assembly machinery
CoQ10
coenzyme Q10
D-loop
mtDNA displacement loop
DNA
deoxyribonucleic acid
DRG
dorsal root ganglia
FARR
FRDA with retained reflexes
FARS
Friedreich Ataxia Rating Scale
Fe-S clusters
iron–sulfur clusters
FRAXA
fragile X syndrome
FRDA
Friedreich ataxia
HDAC
histone deacetylase
HO•
hydroxyl anion
H2O2
hydrogen peroxide
ICARS
International Cooperative Ataxia Rating Scale
IFNB
interferon-β gene RFLP probe
ISC
iron–sulfur cluster
KIKI
knockin mice with homozygous insertion of 230 GAA-repeat expansion in the first intron of the mouse FXN gene
LN
large normal FXN allele
LOFA
late-onset FRDA
MCK
knockout mouse model using the muscle creatine kinase promoter
MIRs
mammalian-wide interspersed repeats
MPP
mitochondrial processing peptidase
mtDNA
mitochondrial DNA
NMR
nuclear magnetic resonance
NO
nitric oxide
NSE
knockout mouse model using the neuron-specific enolase promoter
O2•-
superoxide anion
ONOO-
peroxynitrite
PCR
polymerase chain reaction
PDB
Protein Data Bank (www.pdb.org)
PFGE
pulsed-field gel electrophoresis
PLP
pyridoxal phosphate
PPARγ
peroxisome proliferator–activated receptor gamma
RFLP
restriction length polymorphism
RNA
ribonucleic acid
RNAi
RNA interference
RNS
reactive nitrogen species
ROS
reactive oxygen species
SARA
Scale for Assessment of Rating of Ataxia
SDS-PAGE
sodium dodecylsulfate polyacrylamide gel electrophoresis
SN
small normal FXN allele
_t_BHQ
_tert_-butylhydroquinone
TUNEL staining
terminal deoxynucleotidyl transferase–mediated dUTP-biotin nick end labeling method
YAC
yeast artificial chromosome
Footnotes
Reviewing Editors: Raffaele Lodi, Daniele Marmolino, Thomas Meier, Fiorella Piemonte, Timothy Stemmler, and Natascia Ventura
Acknowledgments
We thank A. Dancis and P. Rustin for interesting discussions over the years. The laboratory was funded by the French government, Centre National de la Recherche Scientifique (CNRS), and Agence Nationale de la Recherche-Maladies Rares (ANR-06-MRAR-025-01), and also by FARA (Friedreich's Ataxia Research Alliance, Exton, PA). D.S. and A.S. were supported by the French Friedreich's ataxia patient organization (AFAF, Association Française de l'Ataxie de Friedreich).
References
- 1.Ackroyd RS. Finnegan JA. Green SH. Friedreich's ataxia: a clinical review with neurophysiological and echocardiographic findings. Arch Dis Child. 1984;59:217–221. doi: 10.1136/adc.59.3.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acquaviva F. Castaldo I. Filla A. Giacchetti M. Marmolino D. Monticelli A. Pinelli M. Saccà F. Cocozza S. Recombinant human erythropoietin increases frataxin protein expression without increasing mRNA expression. Cerebellum. 2008;7:360–365. doi: 10.1007/s12311-008-0036-x. [DOI] [PubMed] [Google Scholar]
- 3.Acquaviva F. De Biase I. Nezi L. Ruggiero G. Tatangelo F. Pisano C. Monticelli A. Garbi C. Acquaviva AM. Cocozza S. Extra-mitochondrial localisation of frataxin and its association with IscU1 during enterocyte-like differentiation of the human colon adenocarcinoma cell line Caco-2. J Cell Sci. 2005;118:3917–3924. doi: 10.1242/jcs.02516. [DOI] [PubMed] [Google Scholar]
- 4.Adamec J. Rusnak F. Owen WG. Naylor S. Benson LM. Gacy AM. Isaya G. Iron-dependent self-assembly of recombinant yeast frataxin: implications for Friedreich ataxia. Am J Hum Genet. 2000;67:549–562. doi: 10.1086/303056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adinolfi S. Iannuzzi C. Prischi F. Pastore C. Iametti S. Martin SR. Bonomi F. Pastore A. Bacterial frataxin CyaY is the gatekeeper of iron-sulfur cluster formation catalyzed by IscS. Nat Struct Mol Biol. 2009;16:390–396. doi: 10.1038/nsmb.1579. [DOI] [PubMed] [Google Scholar]
- 6.Adinolfi S. Trifuoggi M. Politou AS. Martin S. Pastore A. A structural approach to understanding the iron-binding properties of phylogenetically different frataxins. Hum Mol Genet. 2002;11:1865–1877. doi: 10.1093/hmg/11.16.1865. [DOI] [PubMed] [Google Scholar]
- 7.Al-Mahdawi S. Pinto R. Ismail O. Varshney D. Lymperi S. Sandi C. Trabzuni D. Pook M. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum Mol Genet. 2008;17:735–746. doi: 10.1093/hmg/ddm346. [DOI] [PubMed] [Google Scholar]
- 8.Al-Mahdawi S. Pinto RM. Varshney D. Lawrence L. Lowrie MB. Hughes S. Webster Z. Blake J. Cooper JM. King R. Pook MA. GAA repeat expansion mutation mouse models of Friedreich ataxia exhibit oxidative stress leading to progressive neuronal and cardiac pathology. Genomics. 2006;88:580–590. doi: 10.1016/j.ygeno.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Mahdawi S. Pook M. Chamberlain S. A novel missense mutation (L198R) in the Friedreich's ataxia gene. Hum Mutat. 2000;16:95. doi: 10.1002/1098-1004(200007)16:1<95::AID-HUMU29>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 10.Aloria K. Schilke B. Andrew A. Craig EA. Iron-induced oligomerization of yeast frataxin homologue Yfh1 is dispensable in vivo. EMBO Rep. 2004;5:1096–1101. doi: 10.1038/sj.embor.7400272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson PR. Kirby K. Hilliker AJ. Phillips JP. RNAi-mediated suppression of the mitochondrial iron chaperone, frataxin, in Drosophila. Hum Mol Genet. 2005;14:3397–3405. doi: 10.1093/hmg/ddi367. [DOI] [PubMed] [Google Scholar]
- 12.Anderson PR. Kirby K. Orr WC. Hilliker AJ. Phillips JP. Hydrogen peroxide scavenging rescues frataxin deficiency in a Drosophila model of Friedreich's ataxia. Proc Natl Acad Sci U S A. 2008;105:611–616. doi: 10.1073/pnas.0709691105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auchère F. Santos R. Planamente S. Lesuisse E. Camadro J. Glutathione-dependent redox status of frataxin-deficient cells in a yeast model of Friedreich's ataxia. Hum Mol Genet. 2008;17:2790–2802. doi: 10.1093/hmg/ddn178. [DOI] [PubMed] [Google Scholar]
- 14.Babady NE. Carelle N. Wells RD. Rouault TA. Hirano M. Lynch DR. Delatycki MB. Wilson RB. Isaya G. Puccio H. Advancements in the pathophysiology of Friedreich's ataxia and new prospects for treatments. Mol Genet Metab. 2007;92:23–35. doi: 10.1016/j.ymgme.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babcock M. de Silva D. Oaks R. Davis-Kaplan S. Jiralerspong S. Montermini L. Pandolfo M. Kaplan J. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 16.Balaban RS. Nemoto S. Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Barbeau A. Friedreich's ataxia 1980: an overview of the physiopathology. Can J Neurol Sci. 1980;7:455–468. doi: 10.1017/s0317167100023064. [DOI] [PubMed] [Google Scholar]
- 18.Barbeau A. Friedreich's disease 1982: etiologic hypotheses a personal analysis. Can J Neurol Sci. 1982;9:243–263. doi: 10.1017/s0317167100044048. [DOI] [PubMed] [Google Scholar]
- 19.Barbeau A. Roy M. Sadibelouiz M. Wilensky MA. Recessive ataxia in Acadians and “Cajuns.”. Can J Neurol Sci. 1984;11:526–533. doi: 10.1017/s0317167100034995. [DOI] [PubMed] [Google Scholar]
- 20.Barreira A. Marques Júnior W. Sweeney M. Davis M. Chimelli L. Paçó-Larson M. Wood N. A family with Friedreich ataxia and onion-bulb formations at sural nerve biopsy. Ann N Y Acad Sci. 1999;883:466–468. [PubMed] [Google Scholar]
- 21.Bartolo C. Mendell JR. Prior TW. Identification of a missense mutation in a Friedreich's ataxia patient: implications for diagnosis and carrier studies. Am J Med Genet. 1998;79:396–399. [PubMed] [Google Scholar]
- 22.Belal S. Panayides K. Sirugo G. Ben Hamida C. Ioannou P. Hentati F. Beckmann J. Koenig M. Mandel J. Ben Hamida M. Middleton L. Study of large inbred Friedreich ataxia families reveals a recombination between D9S15 and the disease locus. Am J Hum Genet. 1992;51:1372–1376. [PMC free article] [PubMed] [Google Scholar]
- 23.Belotserkovskii BP. Liu R. Hanawalt PC. Peptide nucleic acid (PNA) binding and its effect on in vitro transcription in Friedreich's ataxia triplet repeats. Mol Carcinog. 2009;48:299–308. doi: 10.1002/mc.20486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bencze KZ. Kondapalli KC. Cook JD. McMahon S. Millán-Pacheco C. Pastor N. Stemmler TL. The structure and function of frataxin. Crit Rev Biochem Mol Biol. 2006;41:269–291. doi: 10.1080/10409230600846058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bencze KZ. Yoon T. Millán-Pacheco C. Bradley PB. Pastor N. Cowan JA. Stemmler TL. Human frataxin: iron and ferrochelatase binding surface. Chem Commun. 2007;18:1798–1800. doi: 10.1039/b703195e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benomar A. Yahyaoui M. Meggouh F. Bouhouche A. Boutchich M. Bouslam N. Zaim A. Schmitt M. Belaidi H. Ouazzani R. Chkili T. Koenig M. Clinical comparison between AVED patients with 744 del A mutation and Friedreich ataxia with GAA expansion in 15 Moroccan families. J Neurol Sci. 2002;198:25–29. doi: 10.1016/s0022-510x(02)00057-6. [DOI] [PubMed] [Google Scholar]
- 27.Bhidayasiri R. Perlman SL. Pulst SM. Geschwind DH. Late-onset Friedreich ataxia: phenotypic analysis, magnetic resonance imaging findings, and review of the literature. Arch Neurol. 2005;62:1865–1869. doi: 10.1001/archneur.62.12.1865. [DOI] [PubMed] [Google Scholar]
- 28.Bidichandani SI. Ashizawa T. Patel PI. Atypical Friedreich ataxia caused by compound heterozygosity for a novel missense mutation and the GAA triplet-repeat expansion. Am J Hum Genet. 1997;60:1251–1256. [PMC free article] [PubMed] [Google Scholar]
- 29.Bidichandani SI. Ashizawa T. Patel PI. The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am J Hum Genet. 1998;62:111–121. doi: 10.1086/301680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bidichandani SI. Purandare SM. Taylor EE. Gumin G. Machkhas H. Harati Y. Gibbs RA. Ashizawa T. Patel PI. Somatic sequence variation at the Friedreich ataxia locus includes complete contraction of the expanded GAA triplet repeat, significant length variation in serially passaged lymphoblasts and enhanced mutagenesis in the flanking sequence. Hum Mol Genet. 1999;8:2425–2436. doi: 10.1093/hmg/8.13.2425. [DOI] [PubMed] [Google Scholar]
- 31.Boddaert N. Le Quan Sang KH. Rötig A. Leroy-Willig A. Gallet S. Brunelle F. Sidi D. Thalabard JC. Munnich A. Cabantchik ZI. Selective iron chelation in Friedreich ataxia: biologic and clinical implications. Blood. 2007;110:401–408. doi: 10.1182/blood-2006-12-065433. [DOI] [PubMed] [Google Scholar]
- 32.Boesch S. Sturm B. Hering S. Goldenberg H. Poewe W. Scheiber-Mojdehkar B. Friedreich's ataxia: clinical pilot trial with recombinant human erythropoietin. Ann Neurol. 2007;62:521–524. doi: 10.1002/ana.21177. [DOI] [PubMed] [Google Scholar]
- 33.Boesch S. Sturm B. Hering S. Scheiber-Mojdehkar B. Steinkellner H. Goldenberg H. Poewe W. Neurological effects of recombinant human erythropoietin in Friedreich's ataxia: a clinical pilot trial. Mov Disord. 2008;23:1940–1944. doi: 10.1002/mds.22294. [DOI] [PubMed] [Google Scholar]
- 34.Bou-Abdallah F. Adinolfi S. Pastore A. Laue TM. Dennis Chasteen N. Iron binding and oxidation kinetics in frataxin CyaY of Escherichia coli. J Mol Biol. 2004;341:605–615. doi: 10.1016/j.jmb.2004.05.072. [DOI] [PubMed] [Google Scholar]
- 35.Bouchard JP. Barbeau A. Bouchard R. Paquet M. Bouchard RW. A cluster of Friedreich's ataxia in Rimouski, Québec. Can J Neurol Sci. 1976;6:205–208. doi: 10.1017/s0317167100119651. [DOI] [PubMed] [Google Scholar]
- 36.Bradley JL. Blake JC. Chamberlain S. Thomas PK. Cooper JM. Schapira AH. Clinical, biochemical and molecular genetic correlations in Friedreich's ataxia. Hum Mol Genet. 2000;9:275–282. doi: 10.1093/hmg/9.2.275. [DOI] [PubMed] [Google Scholar]
- 37.Bradley JL. Homayoun S. Hart PE. Schapira AH. Cooper JM. Role of oxidative damage in Friedreich's ataxia. Neurochem Res. 2004;29:561–567. doi: 10.1023/b:nere.0000014826.00881.c3. [DOI] [PubMed] [Google Scholar]
- 38.Branda SS. Cavadini P. Adamec J. Kalousek F. Taroni F. Isaya G. Yeast and human frataxin are processed to mature form in two sequential steps by the mitochondrial processing peptidase. J Biol Chem. 1999;274:22763–22769. doi: 10.1074/jbc.274.32.22763. [DOI] [PubMed] [Google Scholar]
- 39.Bulteau A. Dancis A. Gareil M. Montagne J. Camadro J. Lesuisse E. Oxidative stress and protease dysfunction in the yeast model of Friedreich ataxia. Free Radic Biol Med. 2007;42:1561–1570. doi: 10.1016/j.freeradbiomed.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 40.Bulteau AL. O'Neill HA. Kennedy MC. Ikeda-Saito M. Isaya G. Szweda LI. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science. 2004;305:242–245. doi: 10.1126/science.1098991. [DOI] [PubMed] [Google Scholar]
- 41.Bunse M. Bit-Avragim N. Riefflin A. Perrot A. Schmidt O. Kreuz FR. Dietz R. Jung WI. Osterziel KJ. Cardiac energetics correlates to myocardial hypertrophy in Friedreich's ataxia. Ann Neurol. 2003;53:121–123. doi: 10.1002/ana.10419. [DOI] [PubMed] [Google Scholar]
- 42.Bürk K. Mälzig U. Wolf S. Heck S. Dimitriadis K. Schmitz-Hübsch T. Hering S. Lindig TM. Haug V. Timmann D. Degen I. Kruse B. Dörr JM. Ratzka S. Ivo A. Schöls L. Boesch S. Klockgether T. Klopstock T. Schulz JB. Comparison of three clinical rating scales in Friedreich ataxia (FRDA) Mov Disord. 2009;24:1779–1784. doi: 10.1002/mds.22660. [DOI] [PubMed] [Google Scholar]
- 43.Burnett R. Melander C. Puckett JW. Son LS. Wells RD. Dervan PB. Gottesfeld JM. DNA sequence-specific polyamides alleviate transcription inhibition associated with long GAA.TTC repeats in Friedreich's ataxia. Proc Natl Acad Sci U S A. 2006;103:11497–11502. doi: 10.1073/pnas.0604939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Busi MV. Maliandi MV. Valdez H. Clemente M. Zabaleta EJ. Araya A. Gomez-Casati DF. Deficiency of Arabidopsis thaliana frataxin alters activity of mitochondrial Fe-S proteins and induces oxidative stress. Plant J. 2006;48:873–882. doi: 10.1111/j.1365-313X.2006.02923.x. [DOI] [PubMed] [Google Scholar]
- 45.Calabrese V. Lodi R. Tonon C. D'Agata V. Sapienza M. Scapagnini G. Mangiameli A. Pennisi G. Stella AM. Butterfield DA. Oxidative stress, mitochondrial dysfunction and cellular stress response in Friedreich's ataxia. J Neurol Sci. 2005;233:145–162. doi: 10.1016/j.jns.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 46.Calmels N. Schmucker S. Wattenhofer-Donzé M. Martelli A. Vaucamps N. Reutenauer L. Messaddeq N. Bouton C. Koenig M. Puccio H. The first cellular models based on frataxin missense mutations that reproduce spontaneously the defects associated with Friedreich ataxia. PLoS One. 2009;4:e6379. doi: 10.1371/journal.pone.0006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calmels N. Seznec H. Villa P. Reutenauer L. Hibert M. Haiech J. Rustin P. Koenig M. Puccio H. Limitations in a frataxin knockdown cell model for Friedreich ataxia in a high-throughput drug screen. BMC Neurol. 2009;9:46. doi: 10.1186/1471-2377-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campanella A. Isaya G. O'Neill HA. Santambrogio P. Cozzi A. Arosio P. Levi S. The expression of human mitochondrial ferritin rescues respiratory function in frataxin-deficient yeast. Hum Mol Genet. 2004;13:2279–2288. doi: 10.1093/hmg/ddh232. [DOI] [PubMed] [Google Scholar]
- 49.Campanella A. Rovelli E. Santambrogio P. Cozzi A. Taroni F. Levi S. Mitochondrial ferritin limits oxidative damage regulating mitochondrial iron availability: hypothesis for a protective role in Friedreich ataxia. Hum Mol Genet. 2009;18:1–11. doi: 10.1093/hmg/ddn308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campuzano V. Montermini L. Lutz Y. Cova L. Hindelang C. Jiralerspong S. Trottier Y. Kish SJ. Faucheux B. Trouillas P. Authier FJ. Durr A. Mandel JL. Vescovi A. Pandolfo M. Koenig M. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum Mol Genet. 1997;6:1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- 51.Campuzano V. Montermini L. Moltò MD. Pianese L. Cossee M. Cavalcanti F. Monros E. Rodius F. Duclos F. Monticelli A. Zara F. Cañizares J. Koutnikova H. Bidichandani SI. Gellera C. Brice A. Trouillas P. De Michele G. Filla A. De Frutos R. Palau F. Patel PI. Di Donato S. Mandel JL. Cocozza S. Koenig M. Pandolfo M. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 52.Cañizares J. Blanca JM. Navarro JA. Monrós E. Palau F. Moltó MD. dfh is a Drosophila homolog of the Friedreich's ataxia disease gene. Gene. 2000;256:35–42. doi: 10.1016/s0378-1119(00)00343-7. [DOI] [PubMed] [Google Scholar]
- 53.Castaldo I. Pinelli M. Monticelli A. Acquaviva F. Giacchetti M. Filla A. Sacchetti S. Keller S. Avvedimento VE. Chiariotti L. Cocozza S. DNA methylation in intron 1 of the frataxin gene is related to GAA repeat length and age of onset in Friedreich ataxia patients. J Med Genet. 2008;45:808–812. doi: 10.1136/jmg.2008.058594. [DOI] [PubMed] [Google Scholar]
- 54.Cavadini P. Adamec J. Taroni F. Gakh O. Isaya G. Two-step processing of human frataxin by mitochondrial processing peptidase: precursor and intermediate forms are cleaved at different rates. J Biol Chem. 2000;275:41469–41475. doi: 10.1074/jbc.M006539200. [DOI] [PubMed] [Google Scholar]
- 55.Cavadini P. O'Neill HA. Benada O. Isaya G. Assembly and iron-binding properties of human frataxin, the protein deficient in Friedreich ataxia. Hum Mol Genet. 2002;11:217–227. doi: 10.1093/hmg/11.3.217. [DOI] [PubMed] [Google Scholar]
- 56.Chamberlain S. Farrall M. Shaw J. Wilkes D. Carvajal J. Hillerman R. Doudney K. Harding A. Williamson R. Sirugo G. Fujita R. Koenig M. Mandel J. Palau F. Monros E. Vilchez J. Prieto F. Richter A. Vanasse M. Melancon S. Cocozza S. Redolfi E. Cavalcanti F. Pianese L. Filla A. DiDonato S. Pandolfo M. Genetic recombination events which position the Friedreich ataxia locus proximal to the D9S15/D9S5 linkage group on chromosome 9q. Am J Hum Genet. 1993;52:99–109. [PMC free article] [PubMed] [Google Scholar]
- 57.Chamberlain S. Shaw J. Rowland A. Wallis J. South S. Nakamura Y. von Gabain A. Farrall M. Williamson R. Mapping of mutation causing Friedreich's ataxia to human chromosome 9. Nature. 1988;334:248–250. doi: 10.1038/334248a0. [DOI] [PubMed] [Google Scholar]
- 58.Chamberlain S. Shaw J. Wallis J. Rowland A. Chow L. Farrall M. Keats B. Richter A. Roy M. Melancon S. Deufel T. Berciano J. Williamson R. Genetic homogeneity at the Friedreich ataxia locus on chromosome 9. Am J Hum Genet. 1989;44:518–521. [PMC free article] [PubMed] [Google Scholar]
- 59.Chantrel-Groussard K. Geromel V. Puccio H. Koenig M. Munnich A. Rotig A. Rustin P. Disabled early recruitment of antioxidant defenses in Friedreich's ataxia. Hum Mol Genet. 2001;10:2061–2067. doi: 10.1093/hmg/10.19.2061. [DOI] [PubMed] [Google Scholar]
- 60.Chen OS. Kaplan J. CCC1 suppresses mitochondrial damage in the yeast model of Friedreich's ataxia by limiting mitochondrial iron accumulation. J Biol Chem. 2000;275:7626–7632. doi: 10.1074/jbc.275.11.7626. [DOI] [PubMed] [Google Scholar]
- 61.Cho SJ. Lee MG. Yang JK. Lee JY. Song HK. Suh SW. Crystal structure of Escherichia coli CyaY protein reveals a previously unidentified fold for the evolutionarily conserved frataxin family. Proc Natl Acad Sci U S A. 2000;97:8932–8937. doi: 10.1073/pnas.160270897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Christodoulou K. Deymeer F. Serdaroglu P. Ozdemir C. Poda M. Georgiou DM. Ioannou P. Tsingis M. Zamba E. Middleton LT. Mapping of the second Friedreich's ataxia (FRDA2) locus to chromosome 9p23-p11: evidence for further locus heterogeneity. Neurogenetics. 2001;3:127–132. doi: 10.1007/s100480100112. [DOI] [PubMed] [Google Scholar]
- 63.Condò I. Ventura N. Malisan F. Rufini A. Tomassini B. Testi R. In vivo maturation of human frataxin. Hum Mol Genet. 2007;16:1534–1540. doi: 10.1093/hmg/ddm102. [DOI] [PubMed] [Google Scholar]
- 64.Condò I. Ventura N. Malisan F. Tomassini B. Testi R. A pool of extramitochondrial frataxin that promotes cell survival. J Biol Chem. 2006;281:16750–16756. doi: 10.1074/jbc.M511960200. [DOI] [PubMed] [Google Scholar]
- 65.Cook JD. Bencze KZ. Jankovic AD. Crater AK. Busch CN. Bradley PB. Stemmler AJ. Spaller MR. Stemmler TL. Monomeric yeast frataxin is an iron-binding protein. Biochemistry. 2006;45:7767–7767. doi: 10.1021/bi060424r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cooper JM. Schapira AH. Friedreich's ataxia: coenzyme Q10 and vitamin E therapy. Mitochondrion. 2007;7S:S127–S135. doi: 10.1016/j.mito.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 67.Coppola G. De Michele G. Cavalcanti F. Pianese L. Perretti A. Santoro L. Vita G. Toscano A. Amboni M. Grimaldi G. Salvatore E. Caruso G. Filla A. Why do some Friedreich's ataxia patients retain tendon reflexes? A clinical, neurophysiological and molecular study. J Neurol. 1999;246:353–357. doi: 10.1007/s004150050362. [DOI] [PubMed] [Google Scholar]
- 68.Coppola G. Marmolino D. Lu D. Wang Q. Cnop M. Rai M. Acquaviva F. Cocozza S. Pandolfo M. Geschwind DH. Functional genomic analysis of frataxin deficiency reveals tissue-specific alterations and identifies the PPARγ pathway as a therapeutic target in Friedreich's ataxia. Hum Mol Genet. 2009;18:2452–2461. doi: 10.1093/hmg/ddp183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cossée M. Campuzano V. Koutnikova H. Fischbeck K. Mandel JL. Koenig M. Bidichandani SI. Patel PI. Moltò MD. Cañizares J. De Frutos R. Pianese L. Cavalcanti F. Monticelli A. Cocozza S. Montermini L. Pandolfo M. Frataxin fracas. Nat Genet. 1997;15:337–338. doi: 10.1038/ng0497-337. [DOI] [PubMed] [Google Scholar]
- 70.Cossée M. Dürr A. Schmitt M. Dahl N. Trouillas P. Allinson P. Kostrzewa M. Nivelon-Chevallier A. Gustavson KH. Kohlschütter A. Müller U. Mandel JL. Brice A. Koenig M. Cavalcanti F. Tammaro A. De Michele G. Filla A. Cocozza S. Labuda M. Montermini L. Poirier J. Pandolfo M. Friedreich's ataxia: point mutations and clinical presentation of compound heterozygotes. Ann Neurol. 1999;45:200–206. doi: 10.1002/1531-8249(199902)45:2<200::aid-ana10>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 71.Cossée M. Puccio H. Gansmuller A. Koutnikova H. Dierich A. LeMeur M. Fischbeck K. Dollé P. Koenig M. Inactivation of the Friedreich ataxia mouse gene leads to early embryonic lethality without iron accumulation. Hum Mol Genet. 2000;9:1219–1226. doi: 10.1093/hmg/9.8.1219. [DOI] [PubMed] [Google Scholar]
- 72.Cossée M. Schmitt M. Campuzano V. Reutenauer L. Moutou C. Mandel JL. Koenig M. Evolution of the Friedreich's ataxia trinucleotide repeat expansion: founder effect and premutations. Proc Natl Acad Sci U S A. 1997;94:7452–7457. doi: 10.1073/pnas.94.14.7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De Biase I. Rasmussen A. Endres D. Al-Mahdawi S. Monticelli A. Cocozza S. Pook M. Bidichandani SI. Progressive GAA expansions in dorsal root ganglia of Friedreich's ataxia patients. Ann Neurol. 2007;61:55–60. doi: 10.1002/ana.21052. [DOI] [PubMed] [Google Scholar]
- 74.De Biase I. Rasmussen A. Monticelli A. Al-Mahdawi S. Pook M. Cocozza S. Bidichandani SI. Somatic instability of the expanded GAA triplet-repeat sequence in Friedreich ataxia progresses throughout life. Genomics. 2007;90:1–5. doi: 10.1016/j.ygeno.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 75.De Castro M. García-Planells J. Monrós E. Cañizares J. Vázquez-Manrique R. Vílchez JJ. Urtasun M. Lucas M. Navarro G. Izquierdo G. Moltó MD. Palau F. Genotype and phenotype analysis of Friedreich's ataxia compound heterozygous patients. Hum Genet. 2000;106:86–92. doi: 10.1007/s004399900201. [DOI] [PubMed] [Google Scholar]
- 76.De Michele G. Cavalcanti F. Criscuolo C. Pianese L. Monticelli A. Filla A. Cocozza S. Parental gender, age at birth and expansion length influence GAA repeat intergenerational instability in the X25 gene: pedigree studies and analysis of sperm from patients with Friedreich's ataxia. Hum Mol Genet. 1998;7:1901–1906. doi: 10.1093/hmg/7.12.1901. [DOI] [PubMed] [Google Scholar]
- 77.De Michele G. Filla A. Barbieri F. Perretti A. Santoro L. Trombetta L. Santorelli F. Campanella G. Late onset recessive ataxia with Friedreich's disease phenotype. J Neurol Neurosurg Psychiatry. 1989;52:1398–1401. doi: 10.1136/jnnp.52.12.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Michele G. Filla A. Cavalcanti F. Di Maio L. Pianese L. Castaldo I. Calabrese O. Monticelli A. Varrone S. Campanella G. Leone M. Pandolfo M. Cocozza S. Late onset Friedreich's disease: clinical features and mapping of mutation to the FRDA locus. J Neurol Neurosurg Psychiatry. 1994;57:977–979. doi: 10.1136/jnnp.57.8.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.De Michele G. Filla A. Cavalcanti F. Tammaro A. Monticelli A. Pianese L. Di Salle F. Perreti A. Santoro L. Caruso G. Cocozza S. Atypical Friedreich ataxia phenotype associated with a novel missense mutation in the X25 gene. Neurology. 2000;54:496–499. doi: 10.1212/wnl.54.2.496. [DOI] [PubMed] [Google Scholar]
- 80.Dean G. Chamberlain S. Middleton L. Friedreich's ataxia in Kathikas-Arodhes, Cyprus. Lancet. 1988;1:587. doi: 10.1016/s0140-6736(88)91378-5. [DOI] [PubMed] [Google Scholar]
- 81.Delatycki M. Knight M. Koenig M. Cossée M. Williamson R. Forrest S. G130V, a common FRDA point mutation, appears to have arisen from a common founder. Hum Genet. 1999;105:343–346. doi: 10.1007/s004399900142. [DOI] [PubMed] [Google Scholar]
- 82.Delatycki M. Paris D. Gardner R. Forshaw K. Nicholson G. Nassif N. Williamson R. Forrest S. Sperm DNA analysis in a Friedreich ataxia premutation carrier suggests both meiotic and mitotic expansion in the FRDA gene. J Med Genet. 1998;35:713–716. doi: 10.1136/jmg.35.9.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Delatycki MB. Evaluating the progression of Friedreich ataxia and its treatment. J Neurol. 2009;256(suppl 1):36–41. doi: 10.1007/s00415-009-1007-y. [DOI] [PubMed] [Google Scholar]
- 84.Delatycki MB. Paris DB. Gardner RJ. Nicholson GA. Nassif N. Storey E. MacMillan JC. Collins V. Williamson R. Forrest SM. Clinical and genetic study of Friedreich ataxia in an Australian population. Am J Med Genet. 1999;87:168–174. doi: 10.1002/(sici)1096-8628(19991119)87:2<168::aid-ajmg8>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 85.Delatycki MB. Williamson R. Forrest SM. Friedreich ataxia: an overview. J Med Genet. 2000;37:1–8. doi: 10.1136/jmg.37.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dhe-Paganon S. Shigeta R. Chi YI. Ristow M. Shoelson SE. Crystal structure of human frataxin. J Biol Chem. 2000;275:30753–30756. doi: 10.1074/jbc.C000407200. [DOI] [PubMed] [Google Scholar]
- 87.Di Prospero NA. Baker A. Jeffries N. Fischbeck KH. Neurological effects of high-dose idebenone in patients with Friedreich's ataxia: a randomised, placebo-controlled trial. Lancet Neurol. 2007;6:878–886. doi: 10.1016/S1474-4422(07)70220-X. [DOI] [PubMed] [Google Scholar]
- 88.Ding H. Yang J. Coleman LC. Yeung S. Distinct iron binding property of two putative iron donors for the iron-sulfur cluster assembly: IscA and the bacterial frataxin ortholog CyaY under physiological and oxidative stress conditions. J Biol Chem. 2007;282:7997–8004. doi: 10.1074/jbc.M609665200. [DOI] [PubMed] [Google Scholar]
- 89.Ditch S. Sammarco MC. Banerjee A. Grabczyk E. Progressive GAA.TTC repeat expansion in human cell lines. PLoS Genet. 2009;5:e1000704. doi: 10.1371/journal.pgen.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dolezal P. Dancis A. Lesuisse E. Sutak R. Hrdý I. Embley TM. Tachezy J. Frataxin, a conserved mitochondrial protein, in the hydrogenosome of Trichomonas vaginalis. Eukaryot Cell. 2007;6:1431–1438. doi: 10.1128/EC.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Doudney K. Pook M. Al-Mahdawi S. Carvajal J. Hillerman R. Chamberlain S. A novel splice site mutation (384 + 1G-A) in the Friedreich's ataxia gene. Hum Mutat. 1997;11:415. [Google Scholar]
- 92.Duby G. Foury F. Ramazzotti A. Herrmann J. Lutz T. A non-essential function for yeast frataxin in iron-sulfur cluster assembly. Hum Mol Genet. 2002;11:2635–2643. doi: 10.1093/hmg/11.21.2635. [DOI] [PubMed] [Google Scholar]
- 93.Dürr A. Cossee M. Agid Y. Campuzano V. Mignard C. Penet C. Mandel JL. Brice A. Koenig M. Clinical and genetic abnormalities in patients with Friedreich's ataxia. N Engl J Med. 1996;335:1169–1175. doi: 10.1056/NEJM199610173351601. [DOI] [PubMed] [Google Scholar]
- 94.Eisenberg-Lerner A. Bialik S. Simon HU. Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16:966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- 95.Emond M. Lepage G. Vanasse M. Pandolfo M. Increased levels of plasma malondialdehyde in Friedreich ataxia. Neurology. 2000;55:1752–1753. doi: 10.1212/wnl.55.11.1752. [DOI] [PubMed] [Google Scholar]
- 96.Epplen C. Epplen JT. Frank G. Miterski B. Santos EJ. Schols L. Differential stability of the (GAA)n tract in the Friedreich ataxia (STM7) gene. Hum Genet. 1997;99:834–836. doi: 10.1007/s004390050458. [DOI] [PubMed] [Google Scholar]
- 97.Filla A. De Michele G. Cavalcanti F. Pianese L. Monticelli A. Campanella G. Cocozza S. The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am J Hum Genet. 1996;59:554–560. [PMC free article] [PubMed] [Google Scholar]
- 98.Filla A. De Michele G. Cavalcanti F. Santorelli F. Santoro L. Campanella G. Intrafamilial phenotype variation in Friedreich's disease: possible exceptions to diagnostic criteria. J Neurol. 1991;238:147–150. doi: 10.1007/BF00319681. [DOI] [PubMed] [Google Scholar]
- 99.Filla A. DeMichele G. Caruso G. Marconi R. Campanella G. Genetic data and natural history of Friedreich's disease: a study of 80 Italian patients. J Neurol. 1990;237:345–351. doi: 10.1007/BF00315657. [DOI] [PubMed] [Google Scholar]
- 100.Finocchiaro G. Baio G. Micossi P. Pozza G. di Donato S. Glucose metabolism alterations in Friedreich's ataxia. Neurology. 1988;38:1292–1296. doi: 10.1212/wnl.38.8.1292. [DOI] [PubMed] [Google Scholar]
- 101.Forrest SM. Knight M. Delatycki MB. Paris D. Williamson R. King J. Yeung L. Nassif N. Nicholson GA. The correlation of clinical phenotype in Friedreich ataxia with the site of point mutations in the FRDA gene. Neurogenetics. 1998;1:253–257. doi: 10.1007/s100480050037. [DOI] [PubMed] [Google Scholar]
- 102.Fourquet S. Huang ME. D'Autreaux B. Toledano MB. The dual functions of thiol-based peroxidases in H2O2 scavenging and signaling. Antioxid Redox Signal. 2008;10:1565–1576. doi: 10.1089/ars.2008.2049. [DOI] [PubMed] [Google Scholar]
- 103.Foury F. Low iron concentration and aconitase deficiency in a yeast frataxin homologue deficient strain. FEBS Lett. 1999;456:281–284. doi: 10.1016/s0014-5793(99)00961-8. [DOI] [PubMed] [Google Scholar]
- 104.Foury F. Cazzalini O. Deletion of the yeast homologue of the human gene associated with Friedreich's ataxia elicits iron accumulation in mitochondria. FEBS Lett. 1997;411:373–377. doi: 10.1016/s0014-5793(97)00734-5. [DOI] [PubMed] [Google Scholar]
- 105.Foury F. Pastore A. Trincal M. Acidic residues of yeast frataxin have an essential role in Fe-S cluster assembly. EMBO Rep. 2007;8:194–199. doi: 10.1038/sj.embor.7400881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Foury F. Talibi D. Mitochondrial control of iron homeostasis: a genome wide analysis of gene expression in a yeast frataxin-deficient strain. J Biol Chem. 2001;276:7762–7768. doi: 10.1074/jbc.M005804200. [DOI] [PubMed] [Google Scholar]
- 107.Friedreich N. Über degenerative Atrophie der spinalen Hinterstränge. Virchow's Arch Pathol Anat. 1863;26:433–459. [Google Scholar]
- 108.Friedreich N. Über degenerative Atrophie der spinalen Hinterstränge. Virchow's Arch Pathol Anat. 1863;26:391–419. [Google Scholar]
- 109.Friedreich N. Über degenerative Atrophie der spinalen Hinterstränge. Virchow's Arch Pathol Anat. 1863;27:1–26. [Google Scholar]
- 110.Friedreich N. Über Ataxie mit besonderer Berücksichtigung der hereditären Formen. Virchow's Arch Pathol Anat. 1876;68:145–245. [Google Scholar]
- 111.Friedreich N. Über Ataxie mit besonderer Berücksichtigung der hereditären Formen. Virchow's Arch Pathol Anat. 1877;70:140–142. [Google Scholar]
- 112.Fujita R. Agid Y. Trouillas P. Seck A. Tommasi-Davenas C. Driesel AJ. Olek K. Grzeschik KH. Nakamura Y. Mandel JL. Hanauer A. Confirmation of linkage of Friedreich ataxia to chromosome 9 and identification of a new closely linked marker. Genomics. 1989;4:110–111. doi: 10.1016/0888-7543(89)90323-6. [DOI] [PubMed] [Google Scholar]
- 113.Fujita R. Hanauer A. Sirugo G. Heilig R. Mandel JL. Additional polymorphisms at marker loci D9S5 and D9S15 generate extended haplotypes in linkage disequilibrium with Friedreich ataxia. Proc Natl Acad Sci U S A. 1990;87:1796–1800. doi: 10.1073/pnas.87.5.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fujita R. Hanauer A. Vincent A. Mandel J. Koenig M. Physical mapping of two loci (D9S5 and D9S15) tightly linked to Friedreich ataxia locus (FRDA) and identification of nearby CpG islands by pulse-field gel electrophoresis. Genomics. 1991;10:915–920. doi: 10.1016/0888-7543(91)90179-i. [DOI] [PubMed] [Google Scholar]
- 115.Fujita R. Sirugo G. Duclos F. Abderrahim H. Le Paslier D. Cohen D. Brownstein B. Schlessinger D. Mandel J. Koenig M. A 530kb YAC contig tightly linked to the Friedreich ataxia locus contains five CpG clusters and a new highly polymorphic microsatellite. Hum Genet. 1992;89:531–538. doi: 10.1007/BF00219179. [DOI] [PubMed] [Google Scholar]
- 116.Fukui H. Moraes CT. The mitochondrial impairment, oxidative stress and neurodegeneration connection: reality or just an attractive hypothesis? Trends Neurosci. 2008;31:251–256. doi: 10.1016/j.tins.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gakh O. Adamec J. Gacy AM. Twesten RD. Owen WG. Isaya G. Physical evidence that yeast frataxin is an iron storage protein. Biochemistry. 2002;41:6798–6804. doi: 10.1021/bi025566+. [DOI] [PubMed] [Google Scholar]
- 118.Gakh O. Park S. Liu G. Macomber L. Imlay JA. Ferreira GC. Isaya G. Mitochondrial iron detoxification is a primary function of frataxin that limits oxidative damage and preserves cell longevity. Hum Mol Genet. 2006;15:467–479. doi: 10.1093/hmg/ddi461. [DOI] [PubMed] [Google Scholar]
- 119.Gakh O. Smith IV DY. Isaya G. Assembly of the iron-binding protein frataxin in Saccharomyces cerevisiae responds to dynamic changes in mitochondrial iron influx and stress level. J Biol Chem. 2008;283:31500–31510. doi: 10.1074/jbc.M805415200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Geissler A. Krimmer T. Schönfisch B. Meijer M. Rassow J. Biogenesis of the yeast frataxin homolog Yfh1p: Tim44-dependent transfer to mtHsp70 facilitates folding of newly imported proteins in mitochondria. Eur J Biochem. 2000;267:3167–3180. doi: 10.1046/j.1432-1327.2000.01334.x. [DOI] [PubMed] [Google Scholar]
- 121.Gellera C. Castellotti B. Mariotti C. Mineri R. Seveso V. Didonato S. Taroni F. Frataxin gene point mutations in Italian Friedreich ataxia patients. Neurogenetics. 2007;8:289–299. doi: 10.1007/s10048-007-0101-5. [DOI] [PubMed] [Google Scholar]
- 122.Gellera C. Pareyson D. Castellotti B. Mazzucchelli F. Zappacosta B. Pandolfo M. Di Donato S. Very late onset Friedreich's ataxia without cardiomyopathy is associated with limited GAA expansion in the X25 gene. Neurology. 1997;49:1153–1155. doi: 10.1212/wnl.49.4.1153. [DOI] [PubMed] [Google Scholar]
- 123.Geoffroy G. Barbeau A. Breton G. Lemieux B. Aube M. Leger C. Bouchard JP. Clinical description and roentgenologic evaluation of patients with Friedreich's ataxia. Can J Neurol Sci. 1976;3:279–286. doi: 10.1017/s0317167100025464. [DOI] [PubMed] [Google Scholar]
- 124.Gerber J. Mühlenhoff U. Lill R. An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 2003;4:906–911. doi: 10.1038/sj.embor.embor918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Giacchetti M. Monticelli A. De Biase I. Pianese L. Turano M. Filla A. De Michele G. Cocozza S. Mitochondrial DNA haplogroups influence the Friedreich's ataxia phenotype. J Med Genet. 2004;41:293–295. doi: 10.1136/jmg.2003.015289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gibson TJ. Koonin EV. Musco G. Pastore A. Bork P. Friedreich's ataxia protein: phylogenetic evidence for mitochondrial dysfunction. Trends Neurosci. 1996;19:465–468. doi: 10.1016/S0166-2236(96)20054-2. [DOI] [PubMed] [Google Scholar]
- 127.Giorgio M. Trinei M. Migliaccio E. Pelicci PG. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? Nat Rev Mol Cell Biol. 2007;8:722–728. doi: 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
- 128.Goldberg AV. Molik S. Tsaousis AD. Neumann K. Kuhnke G. Delbac F. Vivares CP. Hirt RP. Lill R. Embley TM. Localization and functionality of microsporidian iron-sulphur cluster assembly proteins. Nature. 2008;452:624–629. doi: 10.1038/nature06606. [DOI] [PubMed] [Google Scholar]
- 129.Goncalves S. Paupe V. Dassa EP. Rustin P. Deferiprone targets aconitase: implication for Friedreich's ataxia treatment. BMC Neurol. 2008;8:20. doi: 10.1186/1471-2377-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gordon DM. Kogan M. Knight SA. Dancis A. Pain D. Distinct roles for two N-terminal cleaved domains in mitochondrial import of the yeast frataxin homolog, Yfh1p. Hum Mol Genet. 2001;10:259–269. doi: 10.1093/hmg/10.3.259. [DOI] [PubMed] [Google Scholar]
- 131.Gordon DM. Shi Q. Dancis A. Pain D. Maturation of frataxin within mammalian and yeast mitochondria: one-step processing by matrix processing peptidase. Hum Mol Genet. 1999;8:2255–2262. doi: 10.1093/hmg/8.12.2255. [DOI] [PubMed] [Google Scholar]
- 132.Grabczyk E. Mancuso M. Sammarco MC. A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 2007;35:5351–5359. doi: 10.1093/nar/gkm589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Grabczyk E. Usdin K. Alleviating transcript insufficiency caused by Friedreich's ataxia triplet repeats. Nucleic Acids Res. 2000;28:4930–4937. doi: 10.1093/nar/28.24.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Grabczyk E. Usdin K. The GAA.TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Grant L. Sun J. Xu H. Subramony SH. Chaires JB. Hebert MD. Rational selection of small molecules that increase transcription through the GAA repeats found in Friedreich's ataxia. FEBS Lett. 2006;580:5399–5405. doi: 10.1016/j.febslet.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Greene E. Entezam A. Kumari D. Usdin K. Ancient repeated DNA elements and the regulation of the human frataxin promoter. Genomics. 2005;85:221–230. doi: 10.1016/j.ygeno.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 137.Greene E. Mahishi L. Entezam A. Kumari D. Usdin K. Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res. 2007;35:3383–3390. doi: 10.1093/nar/gkm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hanauer A. Chery M. Fujita R. Driesel AJ. Gilgenkrantz S. Mandel JL. The Friedreich ataxia gene is assigned to chromosome 9q13-q21 by mapping of tightly linked markers and shows linkage disequilibrium with D9S15. Am J Hum Genet. 1990;46:133–137. [PMC free article] [PubMed] [Google Scholar]
- 139.Harding AE. Early onset cerebellar ataxia with retained tendon reflexes: a clinical and genetic study of a disorder distinct from Friedreich's ataxia. J Neurol Neurosurg Psychiatry. 1981;44:503–508. doi: 10.1136/jnnp.44.6.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Harding AE. Friedreich's ataxia: a clinical and genetic study of 90 families with an analysis of early diagnostic criteria and interfamilial clustering of clinical features. Brain. 1981;104:589–620. doi: 10.1093/brain/104.3.589. [DOI] [PubMed] [Google Scholar]
- 141.Harding AE. Zilkha KJ. “Pseudo-dominant” inheritance in Friedreich's ataxia. J Med Genet. 1981;18:285–287. doi: 10.1136/jmg.18.4.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hart PE. Lodi R. Rajagopalan B. Bradley JL. Crilley JG. Turner C. Blamire AM. Manners D. Styles P. Schapira AH. Cooper JM. Antioxidant treatment of patients with Friedreich ataxia: four-year follow-up. Arch Neurol. 2005;62:621–626. doi: 10.1001/archneur.62.4.621. [DOI] [PubMed] [Google Scholar]
- 143.He Y. Alam SL. Proteasa SV. Zhang Y. Lesuisse E. Dancis A. Stemmler TL. Yeast frataxin solution structure, iron binding, and ferrochelatase interaction. Biochemistry. 2004;43:16254–16262. doi: 10.1021/bi0488193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hebert MD. Targeting the gene in Friedreich ataxia. Biochimie. 2008;90:1131–1139. doi: 10.1016/j.biochi.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 145.Heidari M. Houshmand M. Hosseinkhani S. Nafissi S. Scheiber-Mojdehkar B. Khatami M. A novel mitochondrial heteroplasmic C13806A point mutation associated with Iranian Friedreich's ataxia. Cell Mol Neurobiol. 2009;29:225–233. doi: 10.1007/s10571-008-9315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Heidenfelder BL. Makhov AM. Topal MD. Hairpin formation in Friedreich's ataxia triplet repeat expansion. J Biol Chem. 2003;278:2425–2431. doi: 10.1074/jbc.M210643200. [DOI] [PubMed] [Google Scholar]
- 147.Herman D. Jenssen K. Burnett R. Soragni E. Perlman SL. Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat Chem Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 148.Herrero E. Ros J. Bellí G. Cabiscol E. Redox control and oxidative stress in yeast cells. Biochim Biophys Acta. 2008;1780:1217–1235. doi: 10.1016/j.bbagen.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 149.Houshmand M. Panahi MSS. Nafisi S. Soltanzadeh A. Alkandari FM. Identification and sizing of GAA trinucleotide repeat expansion, investigation for D-loop variations and mitochondrial deletions in Iranian patients with Friedreich's ataxia. Mitochondrion. 2006;6:82–88. doi: 10.1016/j.mito.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 150.Huang J. Dizin E. Cowan JA. Mapping iron binding sites on human frataxin: implications for cluster assembly on the ISU Fe-S cluster scaffold protein. J Biol Inorg Chem. 2008;13:825–836. doi: 10.1007/s00775-008-0369-4. [DOI] [PubMed] [Google Scholar]
- 151.Huang ML. Becker EM. Whitnall M. Rahmanto YS. Ponka P. Richardson DR. Elucidation of the mechanism of mitochondrial iron loading in Friedreich's ataxia by analysis of a mouse mutant. Proc Natl Acad Sci U S A. 2009;106:16381–16386. doi: 10.1073/pnas.0906784106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hughes JT. Brownell B. Hewer RL. The peripheral sensory pathway in Friedreich's ataxia: an examination by light and electron microscopy of the posterior nerve roots, posterior root ganglia, and peripheral sensory nerves in cases of Friedreich's ataxia. Brain. 1968;91:803–818. doi: 10.1093/brain/91.4.803. [DOI] [PubMed] [Google Scholar]
- 153.Illarioshkin SN. Bagieva GK. Klyushnikov SA. Ovchinnikov IV. Markova ED. Ivanova-Smolenskaya IA. Different phenotypes of Friedreich's ataxia within one “pseudo-dominant” genealogy: relationships between trinucleotide (GAA) repeat lengths and clinical features. Eur J Neurol. 2000;7:535–540. doi: 10.1046/j.1468-1331.2000.t01-1-00113.x. [DOI] [PubMed] [Google Scholar]
- 154.Irazusta V. Cabiscol E. Reverter-Branchat G. Ros J. Tamarit J. Manganese is the link between frataxin and iron-sulfur deficiency in the yeast model of Friedreich ataxia. J Biol Chem. 2006;281:12227–12232. doi: 10.1074/jbc.M511649200. [DOI] [PubMed] [Google Scholar]
- 155.Jauslin ML. Meier T. Smith RA. Murphy MP. Mitochondria-targeted antioxidants protect Friedreich ataxia fibroblasts from endogenous oxidative stress more effectively than untargeted antioxidants. FASEB J. 2003;17:1972–1974. doi: 10.1096/fj.03-0240fje. [DOI] [PubMed] [Google Scholar]
- 156.Jauslin ML. Wirth T. Meier T. Schoumacher F. A cellular model for Friedreich ataxia reveals small-molecule glutathione peroxidase mimetics as novel treatment strategy. Hum Mol Genet. 2002;11:3055–3063. doi: 10.1093/hmg/11.24.3055. [DOI] [PubMed] [Google Scholar]
- 157.Jiralerspong S. Ge B. Hudson TJ. Pandolfo M. Manganese superoxide dismutase induction by iron is impaired in Friedreich ataxia cells. FEBS Lett. 2001;509:101–105. doi: 10.1016/s0014-5793(01)03140-4. [DOI] [PubMed] [Google Scholar]
- 158.Jiralerspong S. Liu Y. Montermini L. Stifani S. Pandolfo M. Frataxin shows developmentally regulated tissue-specific expression in the mouse embryo. Neurobiol Dis. 1997;4:103–113. doi: 10.1006/nbdi.1997.0139. [DOI] [PubMed] [Google Scholar]
- 159.Justice CM. Den Z. Nguyen SV. Stoneking M. Deininger PL. Batzer MA. Keats BJ. Phylogenetic analysis of the Friedreich ataxia GAA trinucleotide repeat. J Mol Evol. 2001;52:232–238. doi: 10.1007/s002390010151. [DOI] [PubMed] [Google Scholar]
- 160.Kakhlon O. Manning H. Breuer W. Melamed-Book N. Lu C. Cortopassi G. Munnich A. Cabantchik ZI. Cell functions impaired by frataxin deficiency are restored by drug-mediated iron relocation. Blood. 2008;112:5219–5227. doi: 10.1182/blood-2008-06-161919. [DOI] [PubMed] [Google Scholar]
- 161.Karlberg T. Schagerlöf U. Gakh O. Park S. Ryde U. Lindahl M. Leath K. Garman E. Isaya G. Al-Karadaghi S. The structures of frataxin oligomers reveal the mechanism for the delivery and detoxification of iron. Structure. 2006;14:1535–1546. doi: 10.1016/j.str.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 162.Karthikeyan G. Lewis LK. Resnick MA. The mitochondrial protein frataxin prevents nuclear damage. Hum Mol Genet. 2002;11:1351–1362. doi: 10.1093/hmg/11.11.1351. [DOI] [PubMed] [Google Scholar]
- 163.Karthikeyan G. Santos JH. Graziewicz MA. Copeland WC. Isaya G. Van Houten B. Resnick MA. Reduction in frataxin causes progressive accumulation of mitochondrial damage. Hum Mol Genet. 2003;12:3331–3342. doi: 10.1093/hmg/ddg349. [DOI] [PubMed] [Google Scholar]
- 164.Klockgether T. Zühlke C. Schulz JB. Bürk K. Fetter M. Dittmann H. Skalej M. Dichgans J. Friedreich's ataxia with retained tendon reflexes: molecular genetics, clinical neurophysiology, and magnetic resonance imaging. Neurology. 1996;46:118–121. doi: 10.1212/wnl.46.1.118. [DOI] [PubMed] [Google Scholar]
- 165.Knight SAB. Sepuri NBV. Pain D. Dancis A. Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J Biol Chem. 1998;273:18389–18393. doi: 10.1074/jbc.273.29.18389. [DOI] [PubMed] [Google Scholar]
- 166.Koeppen AH. Michael SC. Knutson MD. Haile DJ. Qian J. Levi S. Santambrogio P. Garrick MD. Lamarche JB. The dentate nucleus in Friedreich's ataxia: the role of iron-responsive proteins. Acta Neuropathol. 2007;114:163–173. doi: 10.1007/s00401-007-0220-y. [DOI] [PubMed] [Google Scholar]
- 167.Koeppen AH. Morral JA. Davis AN. Qian J. Petrocine SV. Knutson MD. Gibson WM. Cusack MJ. Li D. The dorsal root ganglion in Friedreich's ataxia. Acta Neuropathol. 2009;118:763–776. doi: 10.1007/s00401-009-0589-x. [DOI] [PubMed] [Google Scholar]
- 168.Kostrzewa M. Klockgether T. Damian MS. Muller U. Locus heterogeneity in Friedreich ataxia. Neurogenetics. 1997;1:43–47. doi: 10.1007/s100480050007. [DOI] [PubMed] [Google Scholar]
- 169.Koutnikova H. Campuzano V. Foury F. Dolle P. Cazzalini O. Koenig M. Studies of human, mouse and yeast homologues indicate a mitochondrial function for frataxin. Nat Genet. 1997;16:345–351. doi: 10.1038/ng0897-345. [DOI] [PubMed] [Google Scholar]
- 170.Koutnikova H. Campuzano V. Koenig M. Maturation of wild-type and mutated frataxin by the mitochondrial processing peptidase. Hum Mol Genet. 1998;7:1485–1489. doi: 10.1093/hmg/7.9.1485. [DOI] [PubMed] [Google Scholar]
- 171.Krasilnikova MM. Mirkin SM. Replication stalling at Friedreich's ataxia (GAA)n repeats in vivo. Mol Cell Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kroemer G. Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Labuda M. Labuda D. Miranda C. Poirier J. Soong BW. Barucha NE. Pandolfo M. Unique origin and specific ethnic distribution of the Friedreich ataxia GAA expansion. Neurology. 2000;54:2322–2324. doi: 10.1212/wnl.54.12.2322. [DOI] [PubMed] [Google Scholar]
- 174.Labuda M. Poirier J. Pandolfo M. A missense mutation (W155R) in an American patient with Friedreich ataxia. Hum Mutat. 1999;13:506. [Google Scholar]
- 175.Ladame P. Friedreich's disease. Brain. 1890;13:467–537. [Google Scholar]
- 176.Lamarche J. Luneau C. Lemieux B. Ultrastructural observations on spinal ganglion biopsy in Friedreich's ataxia: a preliminary report. Can J Neurol Sci. 1982;9:137–139. doi: 10.1017/s0317167100043833. [DOI] [PubMed] [Google Scholar]
- 177.Lamarche JB. Shapcott D. Côté M. Lemieux B. Cardiac iron deposits in Friedreich's ataxia. In: Lechtenberg R, editor. Handbook of Cerebellar Diseases. New York: Marcel Dekker; 1993. pp. 453–458. [Google Scholar]
- 178.Lamba L. Ciotti P. Giribaldi G. Di Maria E. Varese A. Di Stadio M. Schenone A. Mandich P. Bellone E. Friedreich's ataxia: a new mutation in two compound heterozygous siblings with unusual clinical onset. Eur Neurol. 2009;61:240–243. doi: 10.1159/000198417. [DOI] [PubMed] [Google Scholar]
- 179.Lamont PJ. Davis MB. Wood NW. Identification and sizing of the GAA trinucleotide repeat expansion of Friedreich's ataxia in 56 patients: clinical and genetic correlates. Brain. 1997;120:673–680. doi: 10.1093/brain/120.4.673. [DOI] [PubMed] [Google Scholar]
- 180.Layer G. Ollagnier-de Choudens S. Sanakis Y. Fontecave M. Iron-sulfur cluster biosynthesis: characterization of Escherichia coli CyaY as an iron donor for the assembly of [2Fe-2S] clusters in the scaffold IscU. J Biol Chem. 2006;281:16256–16263. doi: 10.1074/jbc.M513569200. [DOI] [PubMed] [Google Scholar]
- 181.Lee JA. Autophagy in neurodegeneration: two sides of the same coin. BMB Rep. 2009;42:324–330. doi: 10.5483/bmbrep.2009.42.6.324. [DOI] [PubMed] [Google Scholar]
- 182.LeProust EM. Pearson CE. Sinden RR. Gao X. Unexpected formation of parallel duplex in GAA and TTC trinucleotide repeats of Friedreich's ataxia. J Mol Biol. 2000;302:1063–1080. doi: 10.1006/jmbi.2000.4073. [DOI] [PubMed] [Google Scholar]
- 183.Lesuisse E. Santos R. Matzanke BF. Knight SA. Camadro JM. Dancis A. Iron use for haeme synthesis is under control of the yeast frataxin homologue (Yfh1) Hum Mol Genet. 2003;12:879–889. doi: 10.1093/hmg/ddg096. [DOI] [PubMed] [Google Scholar]
- 184.Li DS. Ohshima K. Jiralerspong S. Bojanowski MW. Pandolfo M. Knock-out of the cyaY gene in Escherichia coli does not affect cellular iron content and sensitivity to oxidants. FEBS Lett. 1999;456:13–16. doi: 10.1016/s0014-5793(99)00896-0. [DOI] [PubMed] [Google Scholar]
- 185.Li H. Gakh O. Smith IV DY. Isaya G. Oligomeric yeast frataxin drives assembly of core machinery for mitochondrial iron-sulfur cluster synthesis. J Biol Chem. 2009;284:21971–21980. doi: 10.1074/jbc.M109.011197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Li H. Mapolelo DT. Dingra NN. Naik SG. Lees NS. Hoffman BM. Riggs-Gelasco PJ. Huynh BH. Johnson MK. Outten CE. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry. 2009;48:9569–9581. doi: 10.1021/bi901182w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Li K. Besse EK. Ha D. Kovtunovych G. Rouault TA. Iron-dependent regulation of frataxin expression: implications for treatment of Friedreich ataxia. Hum Mol Genet. 2008;17:2265–2273. doi: 10.1093/hmg/ddn127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 189.Llorens JV. Navarro JA. Martínez-Sebastián MJ. Baylies MK. Schneuwly S. Botella JA. Moltó MD. Causative role of oxidative stress in a Drosophila model of Friedreich ataxia. FASEB J. 2007;21:333–344. doi: 10.1096/fj.05-5709com. [DOI] [PubMed] [Google Scholar]
- 190.Lodi R. Cooper JM. Bradley JL. Manners D. Styles P. Taylor DJ. Schapira AH. Deficit of in vivo mitochondrial ATP production in patients with Friedreich ataxia. Proc Natl Acad Sci U S A. 1999;96:11492–11495. doi: 10.1073/pnas.96.20.11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lodi R. Hart PE. Rajagopalan B. Taylor DJ. Crilley JG. Bradley JL. Blamire AM. Manners D. Styles P. Schapira AH. Cooper JM. Antioxidant treatment improves in vivo cardiac and skeletal muscle bioenergetics in patients with Friedreich's ataxia. Ann Neurol. 2001;49:590–596. [PubMed] [Google Scholar]
- 192.Long S. Jirku M. Mach J. Ginger ML. Sutak R. Richardson D. Tachezy J. Lukeš J. Ancestral roles of eukaryotic frataxin: mitochondrial frataxin function and heterologous expression of hydrogenosomal Trichomonas homologues in trypanosomes. Mol Microbiol. 2008;69:94–109. doi: 10.1111/j.1365-2958.2008.06260.x. [DOI] [PubMed] [Google Scholar]
- 193.Lu C. Cortopassi G. Frataxin knockdown causes loss of cytoplasmic iron-sulfur cluster functions, redox alterations and induction of heme transcripts. Arch Biochem Biophys. 2007;457:111–122. doi: 10.1016/j.abb.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lu C. Schoenfeld R. Shan Y. Tsai HJ. Hammock B. Cortopassi G. Frataxin deficiency induces Schwann cell inflammation and death. Biochim Biophys Acta. 2009;1792:1052–1061. doi: 10.1016/j.bbadis.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Marmolino D. Acquaviva F. Pinelli M. Monticelli A. Castaldo I. Filla A. Cocozza S. PPAR-γ agonist Azelaoyl PAF increases frataxin protein and mRNA expression: new implications for the Friedreich's ataxia therapy. Cerebellum. 2009;8:98–103. doi: 10.1007/s12311-008-0087-z. [DOI] [PubMed] [Google Scholar]
- 196.Martin M. Colman MJ. Gómez-Casati DF. Lamattina L. Zabaleta EJ. Nitric oxide accumulation is required to protect against iron-mediated oxidative stress in frataxin-deficient Arabidopsis plants. FEBS Lett. 2009;583:542–548. doi: 10.1016/j.febslet.2008.12.039. [DOI] [PubMed] [Google Scholar]
- 197.Mascalchi M. Salvi F. Piacentini S. Bartolozzi C. Friedreich's ataxia: MR findings involving the cervical portion of the spinal cord. AJR Am J Roentgenol. 1994;163:187–191. doi: 10.2214/ajr.163.1.8010211. [DOI] [PubMed] [Google Scholar]
- 198.Mattson MP. Gleichmann M. Cheng A. Mitochondria in neuroplasticity and neurological disorders. Neuron. 2008;60:748–766. doi: 10.1016/j.neuron.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.McCabe D. Ryan F. Moore D. McQuaid S. King M. Kelly A. Daly K. Barton D. Murphy R. Typical Friedreich's ataxia without GAA expansions and GAA expansion without typical Friedreich's ataxia. J Neurol. 2000;247:346–355. doi: 10.1007/s004150050601. [DOI] [PubMed] [Google Scholar]
- 200.McCormack ML. Guttmann RP. Schumann M. Farmer JM. Stolle CA. Campuzano V. Koenig M. Lynch DR. Frataxin point mutations in two patients with Friedreich's ataxia and unusual clinical features. J Neurol Neurosurg Psychiatry. 2000;68:661–664. doi: 10.1136/jnnp.68.5.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.McDaniel D. Woodley C. Langford L. Subromany S. A novel frataxin mutation and unusual heterozygote expansion associated with a very late onset case of FA. Am J Hum Genet. 2003;73:258. [Google Scholar]
- 202.Meier T. Buyse G. Idebenone: an emerging therapy for Friedreich ataxia. J Neurol. 2009;256(suppl 1):25–30. doi: 10.1007/s00415-009-1005-0. [DOI] [PubMed] [Google Scholar]
- 203.Melli G. Taiana M. Camozzi F. Triolo D. Podini P. Quattrini A. Taroni F. Lauria G. Alpha-lipoic acid prevents mitochondrial damage and neurotoxicity in experimental chemotherapy neuropathy. Exp Neurol. 2008;214:276–284. doi: 10.1016/j.expneurol.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 204.Miao R. Kim H. Koppolu UM. Ellis EA. Scott RA. Lindahl PA. Biophysical characterization of the iron in mitochondria from Atm1p-depleted Saccharomyces cerevisiae. Biochemistry. 2009;48:9556–9568. doi: 10.1021/bi901110n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Miao R. Martinho M. Morales JG. Kim H. Ellis EA. Lill R. Hendrich MP. Münck E. Lindahl PA. EPR and Mössbauer spectroscopy of intact mitochondria isolated from Yah1p-depleted Saccharomyces cerevisiae. Biochemistry. 2008;47:9888–9899. doi: 10.1021/bi801047q. [DOI] [PubMed] [Google Scholar]
- 206.Michael S. Petrocine SV. Qian J. Lamarche JB. Knutson MD. Garrick MD. Koeppen AH. Iron and iron-responsive proteins in the cardiomyopathy of Friedreich's ataxia. Cerebellum. 2006;5:257–267. doi: 10.1080/14734220600913246. [DOI] [PubMed] [Google Scholar]
- 207.Miranda CJ. Santos MM. Ohshima K. Smith J. Li L. Bunting M. Cossee M. Koenig M. Sequeiros J. Kaplan J. Pandolfo M. Frataxin knockin mouse. FEBS Lett. 2002;512:291–297. doi: 10.1016/s0014-5793(02)02251-2. [DOI] [PubMed] [Google Scholar]
- 208.Monrós E. Moltó M. Martínez F. Cañizares J. Blanca J. Vílchez J. Prieto F. de Frutos R. Palau F. Phenotype correlation and intergenerational dynamics of the Friedreich ataxia GAA trinucleotide repeat. Am J Hum Genet. 1997;61:101–110. doi: 10.1086/513887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Montermini L. Andermann E. Labuda M. Richter A. Pandolfo M. Cavalcanti F. Pianese L. Iodice L. Farina G. Monticelli A. Turano M. Filla A. De Michele G. Cocozza S. The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum Mol Genet. 1997;6:1261–1266. doi: 10.1093/hmg/6.8.1261. [DOI] [PubMed] [Google Scholar]
- 210.Montermini L. Kish SJ. Jiralerspong S. Lamarche JB. Pandolfo M. Somatic mosaicism for Friedreich's ataxia GAA triplet repeat expansions in the central nervous system. Neurology. 1997;49:606–610. doi: 10.1212/wnl.49.2.606. [DOI] [PubMed] [Google Scholar]
- 211.Montermini L. Richter A. Morgan K. Justice CM. Julien D. Castellotti B. Mercier J. Poirier J. Capozzoli F. Bouchard JP. Lemieux B. Mathieu J. Vanasse M. Seni MH. Graham G. Andermann F. Andermann E. Melancon SB. Keats BJ. Di Donato S. Pandolfo M. Phenotypic variability in Friedreich ataxia: role of the associated GAA triplet repeat expansion. Ann Neurol. 1997;41:675–682. doi: 10.1002/ana.410410518. [DOI] [PubMed] [Google Scholar]
- 212.Montermini L. Rodius F. Pianese L. Moltò MD. Cossée M. Campuzano V. Cavalcanti F. Monticelli A. Palau F. Gyapay G. Wenhert M. Zara F. Patel PI. Cocozza S. Koenig M. Pandolfo M. The Friedreich ataxia critical region spans a 150-kb interval on chromosome 9q13. Am J Hum Genet. 1995;57:1061–1067. [PMC free article] [PubMed] [Google Scholar]
- 213.Morgan RO. Naglie G. Horrobin DF. Barbeau A. Erythrocyte protoporphyrin levels in patients with Friedreich's and other ataxias. Can J Neurol Sci. 1979;6:227–232. doi: 10.1017/s0317167100119699. [DOI] [PubMed] [Google Scholar]
- 214.Muckenthaler MU. Galy B. Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu Rev Nutr. 2008;28:197–213. doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- 215.Mühlenhoff U. Richhardt N. Ristow M. Kispal G. Lill R. The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum Mol Genet. 2002;11:2025–2036. doi: 10.1093/hmg/11.17.2025. [DOI] [PubMed] [Google Scholar]
- 216.Musco G. Stier G. Kolmerer B. Adinolfi S. Martin S. Frenkiel T. Gibson T. Pastore A. Towards a structural understanding of Friedreich's ataxia: the solution structure of frataxin. Structure. 2000;8:695–707. doi: 10.1016/s0969-2126(00)00158-1. [DOI] [PubMed] [Google Scholar]
- 217.Myers LM. Lynch DR. Farmer JM. Friedman LS. Lawson JA. Wilson RB. Urinary isoprostanes in Friedreich ataxia: lack of correlation with disease features. Mov Disord. 2008;23:1920–1922. doi: 10.1002/mds.22038. [DOI] [PubMed] [Google Scholar]
- 218.Nair M. Adinolfi S. Pastore C. Kelly G. Temussi P. Pastore A. Solution structure of the bacterial frataxin ortholog, CyaY: mapping the iron binding sites. Structure. 2004;12:2037–2048. doi: 10.1016/j.str.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 219.Naoi M. Maruyama W. Akao Y. Yi H. Yamaoka Y. Involvement of type A monoamine oxidase in neurodegeneration: regulation of mitochondrial signaling leading to cell death or neuroprotection. J Neural Transm Suppl. 2006;71:67–77. doi: 10.1007/978-3-211-33328-0_8. [DOI] [PubMed] [Google Scholar]
- 220.Napoli E. Morin D. Bernhardt R. Buckpitt A. Cortopassi G. Hemin rescues adrenodoxin, heme a and cytochrome oxidase activity in frataxin-deficient oligodendroglioma cells. Biochim Biophys Acta. 2007;1772:773–780. doi: 10.1016/j.bbadis.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 221.Napoli E. Taroni F. Cortopassi GA. Frataxin, iron-sulfur clusters, heme, ROS, and aging. Antioxid Redox Signal. 2006;8:506–516. doi: 10.1089/ars.2006.8.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Nave KA. Trapp BD. Axon-glial signaling and the glial support of axon function. Annu Rev Neurosci. 2008;31:535–561. doi: 10.1146/annurev.neuro.30.051606.094309. [DOI] [PubMed] [Google Scholar]
- 223.Nguyen T. Nioi P. Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Nichol H. Gakh O. O'Neill HA. Pickering IJ. Isaya G. George GN. Structure of frataxin iron cores: an X-ray absorption spectroscopic study. Biochemistry. 2003;42:5971–5976. doi: 10.1021/bi027021l. [DOI] [PubMed] [Google Scholar]
- 225.O'Neill HA. Gakh O. Isaya G. Supramolecular assemblies of human frataxin are formed via subunit-subunit interactions mediated by a non-conserved amino-terminal region. J Mol Biol. 2005;345:433–439. doi: 10.1016/j.jmb.2004.10.074. [DOI] [PubMed] [Google Scholar]
- 226.O'Neill HA. Gakh O. Park S. Cui J. Mooney SM. Sampson M. Ferreira GC. Isaya G. Assembly of human frataxin is a mechanism for detoxifying redox-active iron. Biochemistry. 2005;44:537–545. doi: 10.1021/bi048459j. [DOI] [PubMed] [Google Scholar]
- 227.Oktay Y. Dioum E. Matsuzaki S. Ding K. Yan LJ. Haller RG. Szweda LI. Garcia JA. Hypoxia-inducible factor 2a regulates expression of the mitochondrial aconitase chaperone protein frataxin. J Biol Chem. 2007;282:11750–11756. doi: 10.1074/jbc.M611133200. [DOI] [PubMed] [Google Scholar]
- 228.Palau F. De Michele G. Vilchez JJ. Pandolfo M. Monrós E. Cocozza S. Smeyers P. Lopez-Arlandis J. Campanella G. Di Donato S. Filla A. Early-onset ataxia with cardiomyopathy and retained tendon reflexes maps to the Friedreich's ataxia locus on chromosome 9q. Ann Neurol. 1995;37:359–362. doi: 10.1002/ana.410370312. [DOI] [PubMed] [Google Scholar]
- 229.Panas M. Kalfakis N. Karadima G. Davaki P. Vassilopoulos D. Friedreich's ataxia mimicking hereditary motor and sensory neuropathy. J Neurol. 2002;249:1583–1586. doi: 10.1007/s00415-002-0902-2. [DOI] [PubMed] [Google Scholar]
- 230.Panas M. Kalfakis N. Vassilopoulos D. Pseudodominant Friedreich's ataxia with phenotypic heterogeneity. Acta Neurol Scand. 2007;115:364–366. doi: 10.1111/j.1600-0404.2007.00734.x. [DOI] [PubMed] [Google Scholar]
- 231.Pandolfo M. Drug insight: antioxidant therapy in inherited ataxias. Nat Clin Pract Neurol. 2008;4:86–96. doi: 10.1038/ncpneuro0704. [DOI] [PubMed] [Google Scholar]
- 232.Pandolfo M. Friedreich ataxia: the clinical picture. J Neurol. 2009;256(suppl 1):3–8. doi: 10.1007/s00415-009-1002-3. [DOI] [PubMed] [Google Scholar]
- 233.Pandolfo M. Koenig M. Friedreich's ataxia. In: Wells RD, editor. Genetic Instabilities and Hereditary Neurological Diseases. New York: Academic Press; 1998. pp. 373–398. [Google Scholar]
- 234.Park S. Gakh O. Mooney SM. Isaya G. The ferroxidase activity of yeast frataxin. J Biol Chem. 2002;277:38589–38595. doi: 10.1074/jbc.M206711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Park S. Gakh O. O'Neill HA. Mangravita A. Nichol H. Ferreira GC. Isaya G. Yeast frataxin sequentially chaperones and stores iron by coupling protein assembly with iron oxidation. J Biol Chem. 2003;278:31340–31351. doi: 10.1074/jbc.M303158200. [DOI] [PubMed] [Google Scholar]
- 236.Pastore A. Tozzi G. Gaeta LM. Bertini E. Serafini V. Di Cesare S. Bonetto V. Casoni F. Carrozzo R. Federici G. Piemonte F. Actin glutathionylation increases in fibroblasts of patients with Friedreich's ataxia: a potential role in the pathogenesis of the disease. J Biol Chem. 2003;278:42588–42595. doi: 10.1074/jbc.M301872200. [DOI] [PubMed] [Google Scholar]
- 237.Paupe V. Dassa E. Goncalves S. Auchère F. Lönn M. Holmgren A. Rustin P. Impaired nuclear Nrf2 translocation undermines the oxidative stress response in Friedreich ataxia. PLoS One. 2009;4:e4253. doi: 10.1371/journal.pone.0004253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Pellegrini L. Scorrano L. A cut short to death: Parl and Opa1 in the regulation of mitochondrial morphology and apoptosis. Cell Death Differ. 2007;14:1275–1284. doi: 10.1038/sj.cdd.4402145. [DOI] [PubMed] [Google Scholar]
- 239.Philpott CC. Protchenko O. Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:20–27. doi: 10.1128/EC.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Pianese L. Busino L. De Biase I. De Cristofaro T. Lo Casale MS. Giuliano P. Monticelli A. Turano M. Criscuolo C. Filla A. Varrone S. Cocozza S. Up-regulation of c-Jun N-terminal kinase pathway in Friedreich's ataxia cells. Hum Mol Genet. 2002;11:2989–2996. doi: 10.1093/hmg/11.23.2989. [DOI] [PubMed] [Google Scholar]
- 241.Pianese L. Cavalcanti F. De Michele G. Filla A. Campanella G. Calabrese O. Castaldo I. Monticelli A. Cocozza S. The effect of parental gender on the GAA dynamic mutation in the FRDA gene. Am J Hum Genet. 1997;60:460–463. [PMC free article] [PubMed] [Google Scholar]
- 242.Pianese L. Tammaro A. Turano M. De Biase I. Monticelli A. Cocozza S. Identification of a novel transcript of X25, the human gene involved in Friedreich ataxia. Neurosci Lett. 2002;320:137–140. doi: 10.1016/s0304-3940(02)00048-4. [DOI] [PubMed] [Google Scholar]
- 243.Piemonte F. Pastore A. Tozzi G. Tagliacozzi D. Santorelli FM. Carrozzo R. Casali C. Damiano M. Federici G. Bertini E. Glutathione in blood of patients with Friedreich's ataxia. Eur J Clin Invest. 2001;31:1007–1011. doi: 10.1046/j.1365-2362.2001.00922.x. [DOI] [PubMed] [Google Scholar]
- 244.Pollard LM. Sharma R. Gomez M. Shah S. Delatycki MB. Pianese L. Monticelli A. Keats BJ. Bidichandani SI. Replication-mediated instability of the GAA triplet repeat mutation in Friedreich ataxia. Nucleic Acids Res. 2004;32:5962–5971. doi: 10.1093/nar/gkh933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Pook MA. Al-Mahdawi SA. Thomas NH. Appleton R. Norman A. Mountford R. Chamberlain S. Identification of three novel frameshift mutations in patients with Friedreich's ataxia. J Med Genet. 2000;37:E38–E42. doi: 10.1136/jmg.37.11.e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Popescu BF. Pickering IJ. George GN. Nichol H. The chemical form of mitochondrial iron in Friedreich's ataxia. J Inorg Biochem. 2007;101:957–966. doi: 10.1016/j.jinorgbio.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 247.Potter NT. Miller CA. Anderson IJ. Mutation detection in an equivocal case of Friedreich's ataxia. Pediatr Neurol. 2000;22:413–415. doi: 10.1016/s0887-8994(00)00136-3. [DOI] [PubMed] [Google Scholar]
- 248.Puccio H. Multicellular models of Friedreich ataxia. J Neurol. 2009;256(suppl 1):18–24. doi: 10.1007/s00415-009-1004-1. [DOI] [PubMed] [Google Scholar]
- 249.Puccio H. Simon D. Cossée M. Criqui-Filipe P. Tiziano F. Melki J. Hindelang C. Matyas R. Rustin P. Koenig M. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat Genet. 2001;27:181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- 250.Purkiss P. Baraitser M. Borud O. Chalmers RA. Biochemical and clinical studies of Friedreich's ataxia. J Neurol Neurosurg Psychiatry. 1981;44:574–582. doi: 10.1136/jnnp.44.7.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Pynyaha YV. Boretsky YR. Fedorovych DV. Fayura LR. Levkiv AI. Ubiyvovk VM. Protchenko OV. Philpott CC. Sibirny AA. Deficiency in frataxin homologue YFH1 in the yeast Pichia guilliermondii leads to missregulation of iron acquisition and riboflavin biosynthesis and affects sulfate assimilation. Biometals. 2009;22:1051–1061. doi: 10.1007/s10534-009-9256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Radisky DC. Babcock MC. Kaplan J. The yeast frataxin homologue mediates mitochondrial iron efflux: evidence for a mitochondrial iron cycle. J Biol Chem. 1999;274:4497–4499. doi: 10.1074/jbc.274.8.4497. [DOI] [PubMed] [Google Scholar]
- 253.Rai M. Soragni E. Jenssen K. Burnett R. Herman D. Coppola G. Geschwind DH. Gottesfeld JM. Pandolfo M. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS One. 2008;3:e1958. doi: 10.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Ramazzotti A. Vanmansart V. Foury F. Mitochondrial functional interactions between frataxin and Isu1p, the iron-sulfur cluster scaffold protein, in Saccharomyces cerevisiae. FEBS Lett. 2004;557:215–220. doi: 10.1016/s0014-5793(03)01498-4. [DOI] [PubMed] [Google Scholar]
- 255.Rea SL. Ventura N. Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Richardson DR. Friedreich's ataxia: iron chelators that target the mitochondrion as a therapeutic strategy? Expert Opin Invest Drugs. 2003;12:235–245. doi: 10.1517/13543784.12.2.235. [DOI] [PubMed] [Google Scholar]
- 257.Rindler PM. Clark RM. Pollard LM. De Biase I. Bidichandani SI. Replication in mammalian cells recapitulates the locus-specific differences in somatic instability of genomic GAA triplet-repeats. Nucleic Acids Res. 2006;34:6352–6361. doi: 10.1093/nar/gkl846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Ristow M. Mulder H. Pomplun D. Schulz TJ. Müller-Schmehl K. Krause A. Fex M. Puccio H. Müller J. Isken F. Spranger J. Müller-Wieland D. Magnuson MA. Möhlig M. Koenig M. Pfeiffer AF. Frataxin deficiency in pancreatic islets causes diabetes due to loss of ß cell mass. J Clin Invest. 2003;112:527–534. doi: 10.1172/JCI18107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Rizzuto N. Monaco S. Moretto G. Galiazzo-Rizzuto S. Fiaschi A. Forti A. De Maria R. Friedreich's ataxia: a light- and electron-microscopic study of peripheral nerve biopsies. Acta Neuropathol Suppl. 1981;7:344–347. doi: 10.1007/978-3-642-81553-9_98. [DOI] [PubMed] [Google Scholar]
- 260.Rodius F. Duclos F. Wrogemann K. Le Paslier D. Ougen P. Billault A. Belal S. Musenger C. Brice A. Dürr A. Mignard C. Sirugo G. Weissenbach J. Cohen D. Hentati F. Hamida M. Mandel JL. Koenig M. Recombinations in individuals homozygous by descent localize the Friedreich ataxia locus in a cloned 450-kb interval. Am J Hum Genet. 1994;54:1050–1059. [PMC free article] [PubMed] [Google Scholar]
- 261.Romeo G. Menozzi P. Ferlini A. Fadda S. Di Donato S. Uziel G. Lucci B. Capodaglio L. Filla A. Campanella G. Incidence of Friedreich ataxia in Italy estimated from consanguineous marriages. Am J Hum Genet. 1983;35:523–529. [PMC free article] [PubMed] [Google Scholar]
- 262.Rötig A. de Lonlay P. Chretien D. Foury F. Koenig M. Sidi D. Munnich A. Rustin P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 263.Rouault TA. Tong WH. Iron-sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat Rev Mol Cell Biol. 2005;6:345–351. doi: 10.1038/nrm1620. [DOI] [PubMed] [Google Scholar]
- 264.Rouault TA. Tong WH. Iron-sulfur cluster biogenesis and human disease. Trends Genet. 2008;24:398–407. doi: 10.1016/j.tig.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Ruan H. Wang YH. Friedreich's ataxia GAA.TTC duplex and GAA.GAA.TTC triplex structures exclude nucleosome assembly. J Mol Biol. 2008;383:292–300. doi: 10.1016/j.jmb.2008.08.053. [DOI] [PubMed] [Google Scholar]
- 266.Rustin P. von Kleist-Retzow JC. Chantrel-Groussard K. Sidi D. Munnich A. Rotig A. Effect of idebenone on cardiomyopathy in Friedreich's ataxia: a preliminary study. Lancet. 1999;354:477–479. doi: 10.1016/S0140-6736(99)01341-0. [DOI] [PubMed] [Google Scholar]
- 267.Said G. Marion M. Selva J. Jamet C. Hypotrophic and dying-back nerve fibers in Friedreich's ataxia. Neurology. 1986;36:1292–1299. doi: 10.1212/wnl.36.10.1292. [DOI] [PubMed] [Google Scholar]
- 268.Saint-Georges Y. Garcia M. Delaveau T. Jourdren L. Le Crom S. Lemoine S. Tanty V. Devaux F. Jacq C. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS One. 2008;3:e2293. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Sakamoto N. Chastain PD. Parniewski P. Ohshima K. Pandolfo M. Griffith JD. Wells RD. Sticky DNA: self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich's ataxia. Mol Cell. 1999;3:465–475. doi: 10.1016/s1097-2765(00)80474-8. [DOI] [PubMed] [Google Scholar]
- 270.Sakamoto N. Ohshima K. Montermini L. Pandolfo M. Wells RD. Sticky DNA, a self-associated complex formed at long GAA.TTC repeats in intron 1 of the frataxin gene, inhibits transcription. J Biol Chem. 2001;276:27171–27177. doi: 10.1074/jbc.M101879200. [DOI] [PubMed] [Google Scholar]
- 271.Salisachs P. Findley LJ. Codina M. La Torre P. Martinez-Lage JM. A case of Charcot-Marie-Tooth disease mimicking Friedreich's ataxia: is there any association between Friedreich's ataxia and Charcot-Marie-Tooth disease? Can J Neurol Sci. 1982;9:99–103. doi: 10.1017/s0317167100043778. [DOI] [PubMed] [Google Scholar]
- 272.Sanchez-Casis G. Cote M. Barbeau A. Pathology of the heart in Friedreich's ataxia: review of the literature and report of one case. Can J Neurol Sci. 1976;3:349–354. doi: 10.1017/s0317167100025580. [DOI] [PubMed] [Google Scholar]
- 273.Santoro L. De Michele G. Perretti A. Crisci C. Cocozza S. Cavalcanti F. Ragno M. Monticelli A. Filla A. Caruso G. Relation between trinucleotide GAA repeat length and sensory neuropathy in Friedreich's ataxia. J Neurol Neurosurg Psychiatry. 1999;66:93–96. doi: 10.1136/jnnp.66.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Santos MM. Ohshima K. Pandolfo M. Frataxin deficiency enhances apoptosis in cells differentiating into neuroectoderm. Hum Mol Genet. 2001;10:1935–1944. doi: 10.1093/hmg/10.18.1935. [DOI] [PubMed] [Google Scholar]
- 275.Santos R. Buisson N. Knight SA. Dancis A. Camadro JM. Lesuisse E. Candida albicans lacking the frataxin homologue: a relevant yeast model for studying the role of frataxin. Mol Microbiol. 2004;54:507–519. doi: 10.1111/j.1365-2958.2004.04281.x. [DOI] [PubMed] [Google Scholar]
- 276.Sarsero JP. Li L. Wardan H. Sitte K. Williamson R. Ioannou PA. Upregulation of expression from the FRDA genomic locus for the therapy of Friedreich ataxia. J Gene Med. 2003;5:72–81. doi: 10.1002/jgm.320. [DOI] [PubMed] [Google Scholar]
- 277.Sazanov LA. Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 278.Schagerlöf U. Elmlund H. Gakh O. Nordlund G. Hebert H. Lindahl M. Isaya G. Al-Karadaghi S. Structural basis of the iron storage function of frataxin from single-particle reconstruction of the iron-loaded oligomer. Biochemistry. 2008;47:4948–4954. doi: 10.1021/bi800052m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Schmucker S. Argentini M. Carelle-Calmels N. Martelli A. Puccio H. The in vivo mitochondrial two-step maturation of human frataxin. Hum Mol Genet. 2008;17:3521–3531. doi: 10.1093/hmg/ddn244. [DOI] [PubMed] [Google Scholar]
- 280.Schoenfeld RA. Napoli E. Wong A. Zhan S. Reutenauer L. Morin D. Buckpitt AR. Taroni F. Lonnerdal B. Ristow M. Puccio H. Cortopassi GA. Frataxin deficiency alters heme pathway transcripts and decreases mitochondrial heme metabolites in mammalian cells. Hum Mol Genet. 2005;14:3787–3799. doi: 10.1093/hmg/ddi393. [DOI] [PubMed] [Google Scholar]
- 281.Schoenle EJ. Boltshauser EJ. Baekkeskov S. Landin Olsson M. Torresani T. von Felten A. Preclinical and manifest diabetes mellitus in young patients with Friedreich's ataxia: no evidence of immune process behind the islet cell destruction. Diabetologia. 1989;32:378–381. doi: 10.1007/BF00277262. [DOI] [PubMed] [Google Scholar]
- 282.Schöls L. Amoiridis G. Przuntek H. Frank G. Epplen JT. Epplen C. Friedreich's ataxia: revision of the phenotype according to molecular genetics. Brain. 1997;120:2131–2140. doi: 10.1093/brain/120.12.2131. [DOI] [PubMed] [Google Scholar]
- 283.Schulz JB. Boesch S. Bürk K. Dürr A. Giunti P. Mariotti C. Pousset F. Schöls L. Vankan P. Pandolfo M. Diagnosis and treatment of Friedreich ataxia: a European perspective. Nat Rev Neurol. 2009;5:222–234. doi: 10.1038/nrneurol.2009.26. [DOI] [PubMed] [Google Scholar]
- 284.Schulz JB. Dehmer T. Schöls L. Mende H. Hardt C. Vorgerd M. Bürk K. Matson W. Dichgans J. Beal MF. Bogdanov MB. Oxidative stress in patients with Friedreich ataxia. Neurology. 2000;55:1719–1721. doi: 10.1212/wnl.55.11.1719. [DOI] [PubMed] [Google Scholar]
- 285.Schulz TJ. Thierbach R. Voigt A. Drewes G. Mietzner BH. Steinberg P. Pfeiffer AF. Ristow M. Induction of oxidative metabolism by mitochondrial frataxin inhibits cancer growth: Otto Warburg revisited. J Biol Chem. 2006;281:977–981. doi: 10.1074/jbc.M511064200. [DOI] [PubMed] [Google Scholar]
- 286.Seguin A. Bayot A. Dancis A. Rogowska-Wrzesinska A. Auchère F. Camadro JM. Bulteau AL. Lesuisse E. Overexpression of the yeast frataxin homolog (Yfh1): contrasting effects on iron-sulfur cluster assembly, heme synthesis and resistance to oxidative stress. Mitochondrion. 2009;9:130–138. doi: 10.1016/j.mito.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 287.Shan Y. Napoli E. Cortopassi G. Mitochondrial frataxin interacts with ISD11 of the NFS1/ISCU complex and multiple mitochondrial chaperones. Hum Mol Genet. 2007;16:929–941. doi: 10.1093/hmg/ddm038. [DOI] [PubMed] [Google Scholar]
- 288.Shapcott D. Melancon S. Butterworth RF. Khoury K. Collu R. Breton G. Geoffroy G. Lemieux B. Barbeau A. Glucose and insulin metabolism in Friedreich's ataxia. Can J Neurol Sci. 1976;3:361–364. doi: 10.1017/s0317167100025609. [DOI] [PubMed] [Google Scholar]
- 289.Sharma R. Bhatti S. Gomez M. Clark RM. Murray C. Ashizawa T. Bidichandani SI. The GAA triplet-repeat sequence in Friedreich ataxia shows a high level of somatic instability in vivo, with a significant predilection for large contractions. Hum Mol Genet. 2002;11:2175–2187. doi: 10.1093/hmg/11.18.2175. [DOI] [PubMed] [Google Scholar]
- 290.Sharma R. De Biase I. Gómez M. Delatycki M. Ashizawa T. Bidichandani S. Friedreich ataxia in carriers of unstable borderline GAA triplet-repeat alleles. Ann Neurol. 2004;56:898–901. doi: 10.1002/ana.20333. [DOI] [PubMed] [Google Scholar]
- 291.Shaw J. Lichter P. Driesel AJ. Williamson R. Chamberlain S. Regional localisation of the Friedreich ataxia locus to human chromosome 9q13-q21.1. Cytogenet Cell Genet. 1990;53:221–224. doi: 10.1159/000132936. [DOI] [PubMed] [Google Scholar]
- 292.Shi Y. Ghosh MC. Tong WH. Rouault TA. Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Hum Mol Genet. 2009;18:3014–3025. doi: 10.1093/hmg/ddp239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Shoichet SA. Bäumer AT. Stamenkovic D. Sauer H. Pfeiffer AF. Kahn CR. Müller-Wieland D. Richter C. Ristow M. Frataxin promotes antioxidant defense in a thiol-dependent manner resulting in diminished malignant transformation in vitro. Hum Mol Genet. 2002;11:815–821. doi: 10.1093/hmg/11.7.815. [DOI] [PubMed] [Google Scholar]
- 294.Simon D. Seznec H. Gansmuller A. Carelle N. Weber P. Metzger D. Rustin P. Koenig M. Puccio H. Friedreich ataxia mouse models with progressive cerebellar and sensory ataxia reveal autophagic neurodegeneration in dorsal root ganglia. J Neurosci. 2004;24:1987–1995. doi: 10.1523/JNEUROSCI.4549-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Sirugo G. Cocozza S. Brice A. Cavalcanti F. De Michele G. Dones I. Filla A. Koenig M. Lorenzetti D. Monticelli A. Pianese L. Redolfi E. Rousseau F. Mandel J-L. Di Donato S. Pandolfo M. Linkage disequilibrium analysis of Friedreich's ataxia in 140 Caucasian families: positioning of the disease locus and evaluation of allelic heterogeneity. Eur J Hum Genet. 1993;1:133–143. doi: 10.1159/000472400. [DOI] [PubMed] [Google Scholar]
- 296.Spacey SD. Szczygielski BI. Young SP. Hukin J. Selby K. Snutch TP. Malaysian siblings with friedreich ataxia and chorea: a novel deletion in the frataxin gene. Can J Neurol Sci. 2004;31:383–386. doi: 10.1017/s0317167100003498. [DOI] [PubMed] [Google Scholar]
- 297.Sparaco M. Gaeta LM. Santorelli FM. Passarelli C. Tozzi G. Bertini E. Simonati A. Scaravilli F. Taroni F. Duyckaerts C. Feleppa M. Piemonte F. Friedreich's ataxia: oxidative stress and cytoskeletal abnormalities. J Neurol Sci. 2009;287:111–118. doi: 10.1016/j.jns.2009.08.052. [DOI] [PubMed] [Google Scholar]
- 298.Stehling O. Elsasser HP. Bruckel B. Muhlenhoff U. Lill R. Iron-sulfur protein maturation in human cells: evidence for a function of frataxin. Hum Mol Genet. 2004;13:3007–3015. doi: 10.1093/hmg/ddh324. [DOI] [PubMed] [Google Scholar]
- 299.Stolle CA. Frackelton EC. McCallum J. Farmer JM. Tsou A. Wilson RB. Lynch DR. Novel, complex interruptions of the GAA repeat in small, expanded alleles of two affected siblings with late-onset Friedreich ataxia. Mov Disord. 2008;23:1303–1306. doi: 10.1002/mds.22012. [DOI] [PubMed] [Google Scholar]
- 300.Sturm B. Bistrich U. Schranzhofer M. Sarsero JP. Rauen U. Scheiber-Mojdehkar B. de Groot H. Ioannou P. Petrat F. Friedreich's ataxia, no changes in mitochondrial labile iron in human lymphoblasts and fibroblasts: a decrease in antioxidative capacity? J Biol Chem. 2005;280:6701–6708. doi: 10.1074/jbc.M408717200. [DOI] [PubMed] [Google Scholar]
- 301.Sturm B. Stupphann D. Kaun C. Boesch S. Schranzhofer M. Wojta J. Goldenberg H. Scheiber-Mojdehkar B. Recombinant human erythropoietin: effects on frataxin expression in vitro. Eur J Clin Invest. 2005;35:711–717. doi: 10.1111/j.1365-2362.2005.01568.x. [DOI] [PubMed] [Google Scholar]
- 302.Tan G. Chen LS. Lonnerdal B. Gellera C. Taroni FA. Cortopassi GA. Frataxin expression rescues mitochondrial dysfunctions in FRDA cells. Hum Mol Genet. 2001;10:2099–2107. doi: 10.1093/hmg/10.19.2099. [DOI] [PubMed] [Google Scholar]
- 303.Tan G. Napoli E. Taroni F. Cortopassi G. Decreased expression of genes involved in sulfur amino acid metabolism in frataxin-deficient cells. Hum Mol Genet. 2003;12:1699–1711. doi: 10.1093/hmg/ddg187. [DOI] [PubMed] [Google Scholar]
- 304.Thierbach R. Schulz TJ. Isken F. Voigt A. Mietzner B. Drewes G. von Kleist-Retzow JC. Wiesner RJ. Magnuson MA. Puccio H. Pfeiffer AF. Steinberg P. Ristow M. Targeted disruption of hepatic frataxin expression causes impaired mitochondrial function, decreased life span and tumor growth in mice. Hum Mol Genet. 2005;14:3857–3864. doi: 10.1093/hmg/ddi410. [DOI] [PubMed] [Google Scholar]
- 305.Tozzi G. Nuccetelli M. Lo Bello M. Bernardini S. Bellincampi L. Ballerini S. Gaeta LM. Casali C. Pastore A. Federici G. Bertini E. Piemonte F. Antioxidant enzymes in blood of patients with Friedreich's ataxia. Arch Dis Child. 2002;86:376–379. doi: 10.1136/adc.86.5.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Tsou AY. Friedman LS. Wilson RB. Lynch DR. Pharmacotherapy for Friedreich ataxia. CNS Drugs. 2009;23:213–223. doi: 10.2165/00023210-200923030-00003. [DOI] [PubMed] [Google Scholar]
- 307.Van Driest SL. Gakh O. Ommen SR. Isaya G. Ackerman MJ. Molecular and functional characterization of a human frataxin mutation found in hypertrophic cardiomyopathy. Mol Genet Metab. 2005;85:280–285. doi: 10.1016/j.ymgme.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 308.Vázquez-Manrique RP. González-Cabo P. Ortiz-Martín I. Ros S. Baylis HA. Palau F. The frataxin-encoding operon of Caenorhabditis elegans shows complex structure and regulation. Genomics. 2007;89:392–401. doi: 10.1016/j.ygeno.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 309.Vázquez-Manrique RP. González-Cabo P. Ros S. Aziz H. Baylis HA. Palau F. Reduction of Caenorhabditis elegans frataxin increases sensitivity to oxidative stress, reduces lifespan, and causes lethality in a mitochondrial complex II mutant. FASEB J. 2006;20:172–174. doi: 10.1096/fj.05-4212fje. [DOI] [PubMed] [Google Scholar]
- 310.Vazzola V. Losa A. Soave C. Murgia I. Knockout of frataxin gene causes embryo lethality in Arabidopsis. FEBS Lett. 2007;581:667–672. doi: 10.1016/j.febslet.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 311.Veatch JR. McMurray MA. Nelson ZW. Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137:1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Ventura N. Rea S. Henderson ST. Condo I. Johnson TE. Testi R. Reduced expression of frataxin extends the lifespan of Caenorhabditis elegans. Aging Cell. 2005;4:109–112. doi: 10.1111/j.1474-9726.2005.00149.x. [DOI] [PubMed] [Google Scholar]
- 313.Ventura N. Rea SL. Schiavi A. Torgovnick A. Testi R. Johnson TE. p53/CEP-1 increases or decreases lifespan, depending on level of mitochondrial bioenergetic stress. Aging Cell. 2009;8:380–393. doi: 10.1111/j.1474-9726.2009.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Vivas E. Skovran E. Downs DM. Salmonella enterica strains lacking the frataxin homolog CyaY show defects in Fe-S cluster metabolism in vivo. J Bacteriol. 2006;188:1175–1179. doi: 10.1128/JB.188.3.1175-1179.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Voisine C. Schilke B. Ohlson M. Beinert H. Marszalek J. Craig EA. Role of the mitochondrial Hsp70s, Ssc1 and Ssq1, in the maturation of Yfh1. Mol Cell Biol. 2000;20:3677–3684. doi: 10.1128/mcb.20.10.3677-3684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Waldvogel D. van Gelderen P. Hallett M. Increased iron in the dentate nucleus of patients with Friedrich's ataxia. Ann Neurol. 1999;46:123–125. doi: 10.1002/1531-8249(199907)46:1<123::aid-ana19>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 317.Wang T. Craig EA. Binding of yeast frataxin to the scaffold for Fe-S cluster biogenesis, Isu. J Biol Chem. 2008;283:12674–12679. doi: 10.1074/jbc.M800399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Webb S. Doudney K. Pook M. Chamberlain S. Hutchinson M. A family with pseudodominant Friedreich's ataxia showing marked variation of phenotype between affected siblings. J Neurol Neurosurg Psychiatry. 1999;67:217–219. doi: 10.1136/jnnp.67.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Wells RD. DNA triplexes and Friedreich ataxia. FASEB J. 2008;22:1625–1634. doi: 10.1096/fj.07-097857. [DOI] [PubMed] [Google Scholar]
- 320.Wilkes D. Shaw J. Anand R. Riley J. Winter P. Wallis J. Driesel A. Williamson R. Chamberlain S. Identification of CpG islands in a physical map encompassing the Friedreich's ataxia locus. Genomics. 1991;9:90–95. doi: 10.1016/0888-7543(91)90224-3. [DOI] [PubMed] [Google Scholar]
- 321.Wilson RB. Lynch DR. Farmer JM. Brooks DG. Fischbeck KH. Increased serum transferrin receptor concentrations in Friedreich ataxia. Ann Neurol. 2000;47:659–661. [PubMed] [Google Scholar]
- 322.Wilson RB. Lynch DR. Fischbeck KH. Normal serum iron and ferritin concentrations in patients with Friedreich's ataxia. Ann Neurol. 1998;44:132–134. doi: 10.1002/ana.410440121. [DOI] [PubMed] [Google Scholar]
- 323.Wilson RB. Roof DM. Respiratory deficiency due to loss of mitochondrial DNA in yeast lacking the frataxin homologue. Nat Genet. 1997;16:352–357. doi: 10.1038/ng0897-352. [DOI] [PubMed] [Google Scholar]
- 324.Winter RM. Harding AE. Baraitser M. Bravery MB. Intrafamilial correlation in Friedreich's ataxia. Clin Genet. 1981;20:419–427. doi: 10.1111/j.1399-0004.1981.tb01052.x. [DOI] [PubMed] [Google Scholar]
- 325.Winterbourn CC. Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;145:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 326.Wong A. Yang J. Cavadini P. Gellera C. Lonnerdal B. Taroni F. Cortopassi G. The Friedreich's ataxia mutation confers cellular sensitivity to oxidant stress which is rescued by chelators of iron and calcium and inhibitors of apoptosis. Hum Mol Genet. 1999;8:425–430. doi: 10.1093/hmg/8.3.425. [DOI] [PubMed] [Google Scholar]
- 327.Wong A. Yang J. Danielson S. Gellera C. Taroni F. Cortopassi G. Sensitivity of FRDA lymphoblasts to salts of transition metal ions. Antioxid Redox Signal. 2000;2:461–465. doi: 10.1089/15230860050192233. [DOI] [PubMed] [Google Scholar]
- 328.Yang M. Cobine PA. Molik S. Naranuntarat A. Lill R. Winge DR. Culotta VC. The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2. EMBO J. 2006;25:1775–1783. doi: 10.1038/sj.emboj.7601064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Yoon T. Cowan JA. Iron-sulfur cluster biosynthesis: characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J Am Chem Soc. 2003;125:6078–6084. doi: 10.1021/ja027967i. [DOI] [PubMed] [Google Scholar]
- 330.Yoon T. Cowan JA. Frataxin-mediated iron delivery to ferrochelatase in the final step of heme biosynthesis. J Biol Chem. 2004;279:25943–25946. doi: 10.1074/jbc.C400107200. [DOI] [PubMed] [Google Scholar]
- 331.Yoon T. Dizin E. Cowan JA. N-terminal iron-mediated self-cleavage of human frataxin: regulation of iron binding and complex formation with target proteins. J Biol Inorg Chem. 2007;12:535–542. doi: 10.1007/s00775-007-0205-2. [DOI] [PubMed] [Google Scholar]
- 332.Zarse K. Schulz TJ. Birringer M. Ristow M. Impaired respiration is positively correlated with decreased life span in Caenorhabditis elegans models of Friedreich ataxia. FASEB J. 2007;21:1271–1275. doi: 10.1096/fj.06-6994com. [DOI] [PubMed] [Google Scholar]
- 333.Zhu D. Burke C. Leslie A. Nicholson G. Friedreich's ataxia with chorea and myoclonus caused by a compound heterozygosity for a novel deletion and the trinucleotide GAA expansion. Mov Disord. 2002;17:585–589. doi: 10.1002/mds.10175. [DOI] [PubMed] [Google Scholar]
- 334.Zouari M. Feki M. Ben Hamida C. Larnaout A. Turki I. Belal S. Mebazaa A. Ben Hamida M. Hentati F. Electrophysiology and nerve biopsy: comparative study in Friedreich's ataxia and Friedreich's ataxia phenotype with vitamin E deficiency. Neuromus Disord. 1998;8:416–425. doi: 10.1016/s0960-8966(98)00051-0. [DOI] [PubMed] [Google Scholar]
- 335.Zühlke C. Laccone F. Cossée M. Kohlschutter A. Koenig M. Schwinger E. Mutation of the start codon in the FRDA1 gene: linkage analysis of three pedigrees with the ATG to ATT transversion points to a unique common ancestor. Hum Genet. 1998;103:102–105. doi: 10.1007/s004390050791. [DOI] [PubMed] [Google Scholar]
- 336.Zühlke CH. Dalski A. Habeck M. Straube K. Hedrich K. Hoeltzenbein M. Konstanzer A. Hellenbroich Y. Schwinger E. Extension of the mutation spectrum in Friedreich's ataxia: detection of an exon deletion and novel missense mutations. Eur J Hum Genet. 2004;12:979–982. doi: 10.1038/sj.ejhg.5201257. [DOI] [PubMed] [Google Scholar]