Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145 (original) (raw)
. Author manuscript; available in PMC: 2011 Jul 14.
Published in final edited form as: Proteins. 2010 Jun;78(8):1992–1998. doi: 10.1002/prot.22707
INTRODUCTION
Nuclear pore complexes (NPCs) are large, octagonally symmetric dynamic macromolecular assemblies responsible for exchange of proteins and RNAs between the nucleus and cytoplasm. NPCs are made up of at least 456 polypetides from ~30 distinct nucleoporins.1,2 Several of these components, sharing similar structural motifs, form stable subcomplexes that form a co-axial structure containing two outer rings (the nuclear and cytoplasmic rings), two inner rings, and a membrane ring.3,4 The yeast (Saccharomyces cerevisiae) Nup145 and its human counterpart are unique among the nucleoporins, in that they undergo autoproteolysis to generate functionally distinct proteins.5–9 The human counterpart of Nup145 is expressed as two alternatively spliced mRNA transcripts. The larger 190kDa precursor undergoes post-translational auto-proteolysis at the Phe863-Ser864 peptide bond yielding the 92kDa Nup98 and the 96kDa Nup96.8,9 The smaller 98kDa precursor is also autoproteolysed at an analogous site giving 92kDa Nup98-N and a 6kDa C-terminal fragment, which may form a non-covalent complex.10,11
The yeast Nup145 precursor [Fig. 1(A)] contains twelve repeats of a “GLFG” peptide motif (FG repeats) at its N-terminus, an internal autoproteolytic domain (a region of high conservation with the homologous yeast nucleoporins Nup110 and Nup106, neither of which undergo autoproteolysis), followed by the C-terminal domain.5 Various forms of the FG repeats are present in nearly half of all nucleoporins; they form intrinsically disordered regions implicated in gating mechanisms that control passage of macromolecules through NPCs.12 Nup145 undergoes autoproteolysis at the Phe605-Ser606 peptide bond to generate two functionally distinct proteins, Nup145N and Nup145C.6,7 Subsequently, Nup145C associates with six other proteins to form the heptameric Y-complex,13 a component of the outer rings of the NPC.3,4 Nup145N, on the other hand, can shuttle between the NPC and the nuclear interior. It has been suggested that Nup145N, by analogy with Nup98, carries RNA between the nucleus and the NPC.14,15 Nup145 belongs to a highly conserved family of homologs found throughout eukarya. Curiously, the Phe-Ser autoproteolytic site is not always conserved, resulting in the absence of autoproteolysis for some of these closely related proteins.16,17
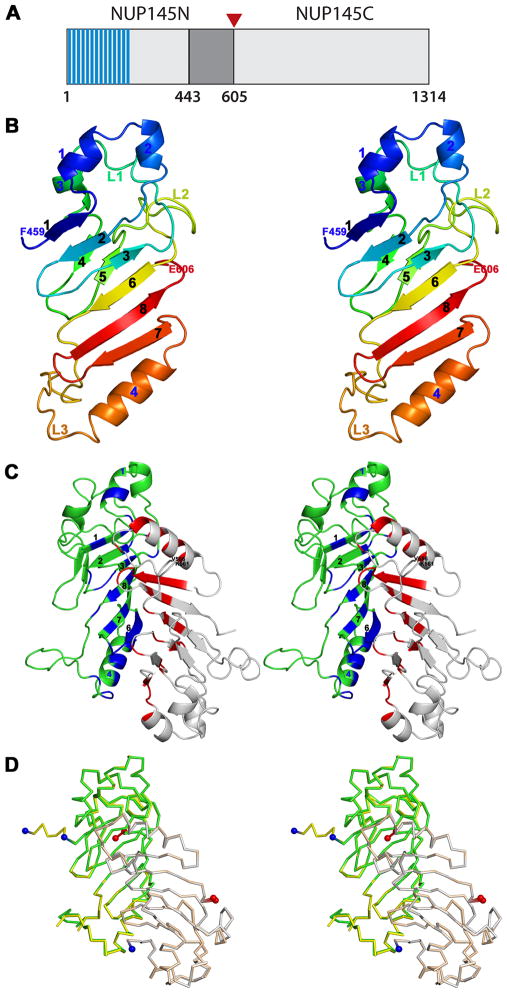
Figure 1.
A: Schematic representation of the S. cerevisiae Nup145 protein. The “FG” repeats are shaded in blue and the expression construct boundaries of Nup145N(443–605) delimited in gray. The site of autoproteolysis is marked with a red triangle.
B: Stereoview of the Nup145N(443–605) monomer. Cartoon of Chain B is shown as a rainbow from blue to red from N- to C-terminus.
C: Stereoview of the Nup145N(443–605) head-to-tail dimer seen in the tetragonal crystals. Cartoons of Chains A and B are shown in grey and green, respectively. Residues involved in dimeric interactions from Chain B are colored in blue and those from Chain A are colored in red.
D: Stereoview of the superposition of Nup145N(443–605) dimers from two different crystal forms. Nup145N(443–605) forms an identical dimer in both tetragonal and hexagonal crystal forms. All structural superpositions were carried out using SSM.44 The A and B chains of tetragonal form are shown as gray and green ribbons, respectively. Similarly, A and B chains of the hexagonal forms are shown in yellow and wheat, respectively.
Our structural understanding of NPC function has benefited recently from X-ray crystal structures of segments of individual proteins.18 As a part of ongoing efforts at the New York SGX Research Center for Structural Genomics (www.nysgxrc.org) to determine crystal structures of distinct components of the yeast NPC, we report herein structures of Nup145N residues 443–605 [Nup145N(443–605)] from the larger autoproteolytic segment. The monomeric architecture of Nup145N(443–605) is similar to that of the autoproteolytic domain of human Nup98-N and the homologous domain of Nup116. In contrast, Nup145N(443–605) and Nup98-N show different modes of association in the crystalline state. Results of Small Angle X-ray Scattering (SAXS) in solution are consistent with the head-to-tail dimer of Nup145N(443–605), observed in two different crystal forms.
EXPERIMENTAL PROCEDURES
Cloning, expression, and purification of Nup145N(443–605)
The gene encoding Nup145N from Saccharomyces cerevisiae was cloned from the genomic DNA of the yeast strain Sc2601D-5 (American Type Culture Collection, USA). The desired truncation (encoding residues 443–605) was PCR amplified using AATAAACAAGACGGCGAAAATAC and CAAAATGATTGACTTTGAAAGTCC as forward and reverse primers, respectively. The purified PCR product was subsequently TOPO®(Invitrogen, USA) cloned into pSGX3, a derivative of pET26b(+), giving rise to a protein with a non-cleavable C-terminal hexa-histidine tag. The resulting plasmid was transformed into BL21(DE3)-Condon+RIL (Invitrogen, USA) cells for overexpression. Production of Se-Met protein was carried out in 1L of HY media at 22 °C containing 50 μg/mL of kanamycin and 35 μg/mL of chloramphenicol. Protein expression was induced by addition of 0.4 mM IPTG. Cells were harvested after 21 hours by centrifugation at 4 °C.
For purification, the E. coli cell pellet was resuspended in 30mL of cold buffer containing 20 mM Tris HCl pH 8.0, 500 mM NaCl, 25 mM imidazole, and 0.1% (v/v) Tween20 and cells were lysed via sonication. Debris was removed by centrifugation at 4°C. The decanted supernatant was applied to a 5 mL HisTrapHP column (GE Health Care, USA) charged with nickel and pre-equilibrated with 20 mM Tris HCl pH 8.0, 500 mM NaCl, 10% (v/v) glycerol, and 25 mM imidazole. The sample was washed with 5 column volumes (CV) of 20 mM Tris HCl pH 8.0, 500 mM NaCl, 10% (v/v) glycerol, and 40 mM imidazole, and subsequently eluted with 2 CV of same buffer with an imidazole concentration of 250 mM. Eluted protein was passed over a 120 mL Superdex 200 size exclusion column equilibrated with 10 mM HEPES pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, and 5 mM DTT (protein storage buffer). SDS-PAGE analysis showed greater than 95% purity and protein fractions corresponding to the symmetric portion of the size exclusion chromatography profile were pooled for concentration with AMICON spin filters. Concentrated protein aliquots were frozen in liquid nitrogen and stored at −80 °C.
Crystallization, data collection, and structure determination
Nup145N(443–605) was subjected to crystallization screening with the Classics, Classics II, and PEGs kits (Qiagen, USA) using a Phoenix Liquid Handling System (Art Robins, USA) via sitting drop vapor diffusion at 21 oC (11.8 mg/ml; 0.3 μL protein + 0.3 μL reservoir solution). Tetragonal crystals grew in 100 mM Hepes pH 7.5, 25% PEG 3350, and 200 mM sodium chloride (Classics II suite; condition number 72) and were cryo-protected by addition of ethylene glycol [final concentration~20% (v/v)] and flash frozen by immersion in liquid nitrogen. Hexagonal crystals were obtained in 1600 mM tri-Sodium Citrate dehydrate (Classics suite; condition number 45) and cryo-protected by addition of ethylene glycol [final concentration ~ 20% (v/v)]. Diffraction data were recorded using the LRL-CAT 31-ID beamline at the Advanced Photon Source (APS) and processed with MOSFLM19 and SCALA (CCP4)20. Structures were determined independently following similar procedures. All eight expected Se atoms were located with SHELXD21 and phases were calculated using SHELXE as implemented in HKL2MAP.22 Initial model building was carried out with ARP/wARP23, followed by manual rebuilding with COOT.24 Atomic models of Nup145N(443–605) refined to convergence using REFMAC525 show excellent stereochemistry (Table 1). Structural analyses were carried out using COOT and CCP4 and illustrations were prepared using PyMol.26
Table I.
Crystallographics Statistics
| Data collection | Nup145N (443–605) | |
|---|---|---|
| PDB code | 3KEP | 3KES |
| Space group | P43212 | P61 |
| Unit-cell dimensions a, c (Å) | 91.0, 108.4 | 135.1, 56.2 |
| Wavelength (Å) | 0.97929 | 0.97929 |
| Matthew’s coefficient (Å3/Da) | 2.78 | 3.66 |
| Solvent content (%) | 56 | 66 |
| Resolution (Å) | 28.78–1.82 (1.91–1.82)a | 25.92–2.10 (2.21–2.10)a |
| Number of unique reflections | 41497 (5943) | 34126 (4940) |
| Completeness (%) | 99.9 (100.0) | 99.2 (99.0) |
| Rsymm (%) | 9.5 (54.6) | 8.9 (50.2) |
| Multiplicity | 29.1 (29.4) | 21.7 (19.8) |
| 〈I/σ(I)〈 | 22.9 (8.8) | 21.2 (8.2) |
| Refinement | ||
| R-factor (%) | 21.3 | 19 |
| Rmath image%) | 24.4 | 23 |
| Root mean square deviations from ideal values | ||
| Bond length (Å) | 0.02 | 0.019 |
| Bond angles (°) | 1.66 | 1.49 |
| Ramachandran Plot28 | ||
| MolProbity29 residues in | ||
| Favored region (%) | 96.2 | 96.2 |
| Allowed region (%) | 100 | 100 |
Size-exclusion chromatography with Multi-angle laser light scattering (SEC-MALLS)
SEC-MALLS was used to determine molecular weight in solution with an analytical gel filtration column (Wyatt Technologies-05S5; 7.8 mm × 300 mm; pore size = 150A) connected to a binary LC pumping system (Agilent 1100), temperature controlled auto-sampler, a UV-visible detector, DAWN®- TREOS™ three-angle static light scattering detector (Wyatt Technology, USA), and an Optilab® rEX differential refractometer equipped with a Peltier temperature-regulated flow cell, maintained at 25 °C (Wyatt Technology, USA). The LC system and light scattering detector (LSD) was controlled by ChemStation (Agilent Technologies, USA) and Astra V software (Wyatt Technology, USA), respectively. The column and LSD was pre-equilibrated with the protein storage buffer. The experiment was performed at room temperature at a flow rate of 0.5 mL/min for 40min. Approximately 100 μg of Nup145N(443–605), at concentrations of 11.8, 22.7 and 46.0 mg/mL, were injected and monitored at a wavelength of 280nm. The molecular mass from light scattering data was calculated using a specific refractive index increment (dn/dc) value of 0.183 mL/g that is universal for proteins.
Small Angle X-ray Scattering (SAXS)
SAXS measurements of Nup145N(443–605) were carried out with Beamline 4–2 at the Stanford Synchrotron Radiation Lightsource (SSRL) and the SIBYLS Beamline 12.3.1 at the Advanced Light Source (ALS) yielding essentially identical results. At SSRL, we used an automatic sample delivery system equipped with a 1.5 mm-diameter thin-wall quartz capillary within which a sample aliquot oscillated in the X-ray beam to minimize radiation damage. It was placed at 1.7 m from a Rayonix225 (MAR-USA, USA) CCD detector with a binned pixel size of 293 μm × 293 μm. Ten 3 sec exposures were made for each protein sample maintained at 10 °C. Each of the 10 diffraction images were scaled using the transmitted beam intensity, azimuthally integrated, and averaged to obtain fully processed data in the form of intensity versus q [q = 4πsin(θ)/λ, θ = one-half of the scattering angle; λ = X-ray wavelength]. The buffer profile was obtained in the same manner and subtracted from a protein profile. SAXS profiles of Nup145N(443–605) were recorded at protein concentrations of 0.5, 1.0, 2.0, 5.0, and 11.8 mg/mL in protein storage buffer. Moreover, mild concentration dependence was removed by extrapolating to the zero concentration. We merged the average of the lower scattering angle parts (q < 0.15A−1) of the lower concentration profiles (0.5–2.0 mg/mL) and the average of the higher scattering angle parts (**_q_** > 0.12A−1) of the higher concentration (5.0–11.8 mg/mL) profiles, to obtain the final, merged experimental SAXS profile. The shape of Nup145N(443–605) was calculated from this merged experimental SAXS profile by running DAMMIF27 10 times individually, followed by superposition and averaging with DAMAVER.28 The oligomeric state of Nup145N(443–605) was calculated from the merged experimental SAXS profile using OLIGOMER.29 The merged experimental SAXS profile was also compared with profiles calculated for the monomer (Chain B) and the crystallographic dimer of Nup145N(443–605) with IMP30 and CRYSOL.31
RESULTS AND DISCUSSION
Structure of Nup145N(443–605)
The crystal structure of Nup145N(443–605) has been determined at 1.82Å resolution [Fig. 1(B), Table 1]. The tetragonal crystal form (_P_43212) has two molecules in the asymmetric unit. The B chain could be traced continuously from Phe459 to Phe605. In the A chain, residues 557–560 appear disordered. Otherwise, the A and B chains are very similar with a root mean square deviation (r.m.s.d.) of 0.4Å for 144 Cα atomic pairs. The structure of Nup145N(443–605) contains two anti-parallel β-sheets capped by α-helices [Fig. 1(B)], as predicted based on its sequence similarity with Nup98.32 A six stranded β-sheet is formed by β1-β2-β3-β6-β8-β7 and a two stranded β-sheet is formed by β4–β5. Helices α1, α2, and α3 form a cap near the N-terminus and α4 caps the six stranded β-sheet near the C-terminus. Nup145N(443–605) possesses three long loops: L1 (between β3-β4), L2 (between β5-β6), and L3 (between β6-α4). Autoproteolysis of Nup145 occurs at the Phe605-Ser606 peptide bond.6,7 Autoproteolytic events result from N-O or N-S acyl shift catalyzed by Ser/Thr or Cys side chains.33 In our structure, the Phe605-Ser606 bond is replaced by Phe605-Glu606 (Glu606 is a cloning artifact derived from the expression plasmid) and, thus, the electron density between Phe605 and Glu606 is continuous in both A and B chains.
Nup145N(443–605) forms a tight dimer in the crystal [Fig. 1(C)]. An identical dimer of Nup145N(443–605) was observed in our hexagonal (_P_61) crystal form, the structure of which was independently determined with Se-Met experimental phases (Table 1 and [Fig. 1(D)]). Monomers of the tetragonal and hexagonal crystal forms superimposed well with r.m.s.d. = 0.37Å for 145 Cα atom pairs. The dimers are also very similar (r.m.s.d. = 0.45 Å for 288 Cα atom pairs, [Fig. 1(D)]). The total buried surface area on complex formation estimated with PISA34 is ~1900Å2. This head-to-tail dimer is stabilized in part by extensive interactions between the β6-strand of one protomer and the α4 helix of its dimer counterpart. In addition, all strands of the six stranded β-sheet contribute to the dimer interface [Fig. 1(C)]. The geometry and amino acid composition of protein-protein interfaces can be helpful in distinguishing specific from non-specific complexes.35 NOXclass36 analysis predicted Nup145N(443–605) to be a “biological” dimer with 99% confidence and further classified it as an obligate dimer with 71% confidence. The gap volume index (an indicator of shape complementarity within a protein-protein interface) for the Nup145N(443–605) dimer in the asymmetric unit is 3.26Å, which is less than the average value of 4.0A observed for known obligate dimers.36
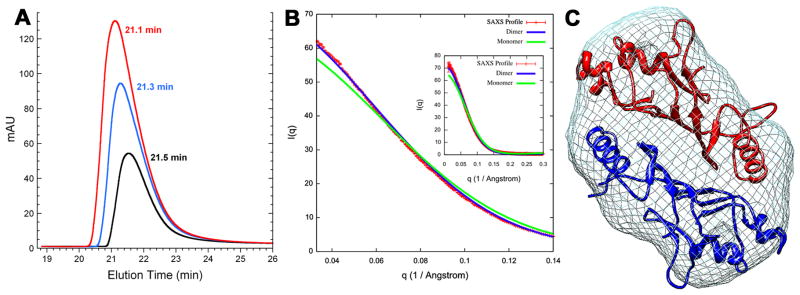
These findings prompted us to investigate the oligomerization state of Nup145N(443–605) in solution using SEC-MALLS and SAXS. Although Nup145N(443–605) eluted as an apparent monomer on the gel filtration column [Fig. 2(A)], light scattering measurements revealed a mixture of monomer and dimer species (data not shown). SAXS analyses showed: (i) the merged experimental SAXS profile matches the SAXS profile calculated for the crystallographic dimer of Nup145N(443–605) [Fig. 2(B)]; (ii) a measured radius of gyration (Rg) of 22.1 ± 0.01A, obtained with AutoRg,37 is close to the value of 21.5A calculated from the structure of crystallographic dimer (for reference, the calculated Rg for the Chain B monomer is 18.0A); (iii) the “ab initio shape” reconstructed from the merged experimental SAXS profile with DAMMIF27 and GASBOR38 matches the overall shape of the Nup145N(443–605) head-to-tail dimer observed in the crystal [Fig. 2(C)]; and, (iv) based on the merged experimental SAXS profile, OLIGOMER29 estimates a dimer fraction of ~98%. Thus, our SAXS analyses of the solution behavior of Nup145N(443–605) are consistent with the head-to-tail dimer observed in both crystal forms.
Figure 2.
A: Analytical gel filtration profile of Nup145N(443–605) at concentrations of 11.8 (black), 22.7 (blue), and 46.0mg/ml (red) with elution times 21.5, 21.3, and 21.1min, respectively.
B: Comparison of the merged experimental SAXS profile (red) of Nup145N(443–605) with SAXS profiles computed by IMP27 for the crystallographic dimer (blue) and monomer (green) structures (inset shows the SAXS profiles across the entire measured resolution range).
C: The shape of Nup145N(443–605) represented as mesh derived from the experimental SAXS profile.
Comparison of Nup145N(443–605) with yeast Nup116 and human Nup98
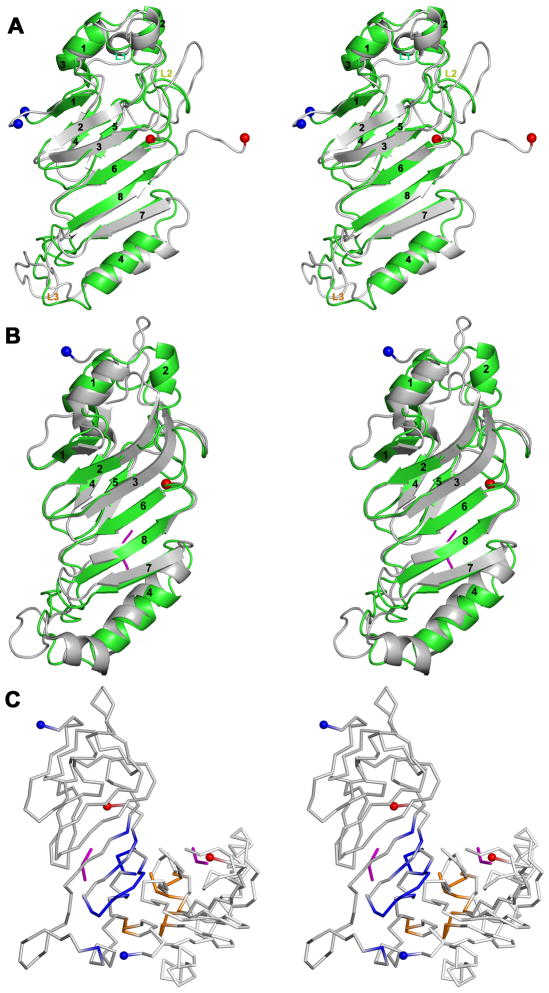
The yeast nucleoporins Nup100 and Nup116 arose by duplication and divergence events from a gene ancestral to Nup145, with both gene products lacking the regions homologous to Nup145C.5,39 The solution NMR structure of yeast Nup116 (PDB Code 2AIV)40 revealed a similar overall structure to that of Nup145N(443–605) [Fig. 3(A); r.m.s.d. = 2.5Å for 117 Cα atom pairs with 32% sequence identity], although it does not appear to be a dimer in solution. The Nup116 structure revealed a remarkable conformational flexibility of the loop regions and the α4 helix (along the helical axis).37 Differences between the two structures include the conformation of loop L3, which projects toward the six stranded β-sheet in Nup145N(443–605). Loop L3 of Nup116 adopts a conformation that is intermediate between those seen in Nup145N and human Nup98 (see below). Similarly, loop L2 of Nup145N projects toward the core of the protein as compared to L2 of Nup116. These conformational differences in the loops of Nup145N and Nup116 may explain, at least in part, their differing positions within the NPC. Nup116 is found mainly on the cytoplasmic face of the NPC, whereas Nup145N is found mainly on the nuclear face.1,3
Figure 3.
A: Stereoview of Nup145N(443–605, green) superposed on a representative NMR structure of Nup116 (PDB Code 2AIV; grey).
B: Stereoview of the Nup145N(443–605, chain B; green) superposed on Nup98-N (grey) from the Nup98-N:C-terminal fragment complex (PDB Code 2Q5Y).
C: Stereoview of the dimeric association of Nup98-N in the Nup98-N:C-terminal fragment complex. The ordered C-terminal fragment tri-peptides are shown in magenta. Residues contributing to the dimer interface from the A and C chains are colored blue and orange, respectively. The N- and C-terminal residues are shown on the cartoons as blue and red spheres, respectively.
Structures for the autoproteolytic domain of human Nup98 have been determined both as a Nup98-N:C-terminal fragment complex (PDB Code 2Q5Y) and as a non-cleavable Ser864Ala mutant (PDB Code 2Q5X).7,8 The structures of the Nup98-N and Nup145N(443–605) monomers are similar [Fig. 3(B); r.m.s.d. = 2.1Å for 128 Cα atom pairs with 39% sequence identity]. Noticeable differences between the two structures include some longer β-strands in the case of Nup98-N and conformations of the loops connecting the secondary structural elements. In particular, the L3 loop between β6-strand and α4-helix [residues 554–566 of Nup145N(443–605)] projects toward the protein core. Conversely, structurally equivalent residues of this loop in Nup98 project away from the protein core. The structure of the Nup98-N:C-terminal fragment complex contains two copies of the complex in the asymmetric unit. The surface area buried by association of the A and C chains of Nup98-N:C-terminal fragment complex [Fig. 2(C)] is 1147Å2 [compared to ~1900Å2 buried within the Nup145N(443–605) dimer]. The two molecules of Nup98-N in the Nup98-N:C-terminal fragment complex associate quite differently from that seen for the Nup145N(443–605) head-to-tail dimer [Fig. 1(C)]. The α4-helix is involved in mediating extensive interactions within the dimeric interface of Nup145N(443–605). However, the α4-helix of the Nup98-N:C-terminal fragment complex, which is involved in binding to the tail peptide, contributes much less to the protein-protein interaction [Fig. 2(C)]. NOXclass34 analysis suggests that the Nup98-N dimer observed in the Nup98-N:C-terminal fragment complex crystal does not occur in nature (biological dimer < 2% confidence, non-obligate dimer 68% confidence). Thus, the autoproteolytic domains of Nup98 and Nup145N associate differently in their respective crystallographic preparations.
The behavior of Nup145N and Nup98 highlight the dynamic nature of NPC constituents, as they both associate with NPCs and reside within the nucleus.41 The relevance of the observed Nup145N dimer to NPC biogenesis and function is not yet clear, although SAXS analyses of Nup145N(443–605), presented herein, provide indirect support for dimer formation within the NPC and/or in the nucleus of yeast. Further biophysical and cell biological characterization of Nup145N is required to determine whether or not Nup145N and/or Nup98-N function as a dimer in vivo.
Acknowledgments
We thank members of the Rout and Sali laboratories for their help and advice. We are also grateful to Drs. John Tainer and Michal Hammel for help in using the SIBYLS beamline 12.3.1 at ALS. Funding for the NYSGXRC was provided by NIH Grant U54 GM074945 (PI: S.K. Burley). Additional funding for this work was provided by NIH R01 GM062427 (MPR) and NIH U54 RR022220 (JP, JFM, JDF, AS, and MPR). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Basic Energy Sciences. Access to the LRL-CAT beam line facilities at Sector 31 of the APS was provided by Eli Lilly, which operates the facility. SSRL operation is funded by the U.S. Department of Energy, Office of Basic Energy Sciences and the SSRL Structural Molecular Biology program by the U.S. Department of Energy, Office of Biological and Environmental Research and the NIH National Center for Research Resources, Biotechnology program. Use of the SIBYLS beamline 12.3.1 at ALS was supported in part by the Office of Science, Office of Biological and Environmental Research, U.S. Department of Energy, under Contract DE-AC02-05CH11231.
Footnotes
Protein Data Bank Codes
Atomic coordinates and structure factors of the tetragonal and hexagonal crystal forms of Nup145N(443–605) were deposited to the PDB on 26 October 2009 with accession codes 1KEP and 1KES, respectively. The NYSGXRC target identifier for yeast Nup145 in TargetDB (http://targetdb.pdb.org) is “NYSGXRC-15145a”. Expression clone sequences and selected interim experimental results are available in PepcDB (http://pepcdb.pdb.org/).
References
- 1.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao YM, Chait BT. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D’Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alber F, Dokudovskaya S, Veenhoff LM, Zhang WH, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 4.Alber F, Dokudovskaya S, Veenhoff LM, Zhang WZ, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Rout MP, Sali A. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 5.Wente SR, Bobel G. Nup145 encodes a yeast glycine-luecine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J Cell Biol. 1994;125:955–969. doi: 10.1083/jcb.125.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teixeira MT, Siniossoglou S, Podtelejnikov S, Benichou JC, Mann M, Dujon B, Hurt Ed, Fabre E. Two functionally distinct domain generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J. 1997;16:5086–5097. doi: 10.1093/emboj/16.16.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teixeira MT, Fabre E, Dujon B. Self-catalyzed cleavage of the yeast nucleoporin Nup145p precursor. J Biol Chem. 1999;274:32439–32444. doi: 10.1074/jbc.274.45.32439. [DOI] [PubMed] [Google Scholar]
- 8.Rosenblum JS, Blobel G. Autoproteolysis in nucleoporin biogenesis. Proc Natl Acad Sci USA. 1999;96:11370–11375. doi: 10.1073/pnas.96.20.11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fontoura BMA, Blobel G, Matunis MJ. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J Cell Biol. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodel EA, Hodel MR, Griffis ER, Hennig KA, Ratner GA, Xu S, Powers MA. The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98. Mol Cell. 2002;10:347–358. doi: 10.1016/s1097-2765(02)00589-0. [DOI] [PubMed] [Google Scholar]
- 11.Sun Y, Guo H-C. Structural constraints on autoprocessing of the human nucleoporin Nup98. Protein Sci. 2008;17:494–505. doi: 10.1110/ps.073311808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 13.Leksa NC, Brohawn SG, Schwartz TU. The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture. Structure. 2009;17:1082–1091. doi: 10.1016/j.str.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabre E, Boelens WC, Wimmer C, Mattaj IW, Hurt EdC. Nup145p is required for nuclear export of mRNA and binds homopolymeric RNA in vitro via a novel conserved motif. Cell. 1994;78:275–289. doi: 10.1016/0092-8674(94)90297-6. [DOI] [PubMed] [Google Scholar]
- 15.Blevins MB, Smith AM, Phillips EM, Powers MA. Complex formation among the RNA export proteins Nup98, Rae1/Gle2 and TAP. J Biol Chem. 2003;278:20979–20988. doi: 10.1074/jbc.M302061200. [DOI] [PubMed] [Google Scholar]
- 16.Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics. 2009;8.9:2119–2130. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brohawn S, Patridge JR, Whittle JRR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leslie AGW, Brick P, Wonacott AJ. An improved program package for the measurement of oscillation photographs. CCP4 News. 1986;18:33–39. [Google Scholar]
- 20.Collaborative Computing Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 21.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 22.Pape T, Schneider TR. HKL2MAP: a graphical user interface for phasing with SHELX program. J Appl Cryst. 2004;37:843–844. [Google Scholar]
- 23.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nature Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. COOT: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the Maximum-Likelihood Method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.DeLano WL. The PyMOL Molecular Graphics System. 2002 http://pymol.sourceforge.net.
- 27.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC- based system for small-angle scattering data analysis. J Appl Cryst. 2003;36:1277–1282. [Google Scholar]
- 30.Forster F, Webb B, Krukenberg KA, Tsuruta H, Agard DA, Sali A. Integration of small-angle X-ray scattering data into structural modeling of proteins and their assemblies. J Mol Biol. 2008;382:1089–1106. doi: 10.1016/j.jmb.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Svergun DI, Barberato C, Koch MHJ. CRYSOL - a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
- 32.Devos DD, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci USA. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perler FB, Xu M, Paulus H. Protein splicing and autoproteolysis mechanisms. Curr Opin Chem Biol. 1997;1:292–299. doi: 10.1016/s1367-5931(97)80065-8. [DOI] [PubMed] [Google Scholar]
- 34.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. ( http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) [DOI] [PubMed]
- 35.Bahadur R, Chakrabarti P, Rodier F, Janin J. A dissection of specific and non-specific protein-protein interfaces. J Mol Biol. 2004;336:943–955. doi: 10.1016/j.jmb.2003.12.073. [DOI] [PubMed] [Google Scholar]
- 36.Zhu H, Domingues FS, Sommer I, Lengauer T. NOXclass: prediction of protein-protein interaction types. BMC Bioinformatics. 2006;7:27. doi: 10.1186/1471-2105-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konarev PV, Kikhney AG, Franke D, Petoukhov MV, Svergun DI. Automated SAXS Data Processing on the X33 Beamline. HASY LAB Annual Report. 2007;Part II:339–340. [Google Scholar]
- 38.Svergun DI, Petoukhov MV, Koch MHJ. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wente SR, Rout MP, Blobel G. A new family of yeast nuclear pore complex proteins. J Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson MA, Park S, Sun ZJ, Silve PA, Wagner G, Hogle JM. Multiple conformations in the ligand-binding site of yeast nuclear pore-targeting domain of Nup116p. J Biol Chem. 2005;280:35723–35732. doi: 10.1074/jbc.M505068200. [DOI] [PubMed] [Google Scholar]
- 41.Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell. 2002;13:1282–1297. doi: 10.1091/mbc.01-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramakrishnan C, Ramachandran GN. Stereochemical criteria for polypeptide and protein chain conformations. II. Allowed conformations for a pair of peptide units. Biophys J. 1965;5:909–933. doi: 10.1016/S0006-3495(65)86759-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucl Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]