Elucidation of Seventeen Human Peripheral Blood B cell Subsets and Quantification of the Tetanus Response Using a Density-Based Method for the Automated Identification of Cell Populations in Multidimensional Flow Cytometry Data (original) (raw)
. Author manuscript; available in PMC: 2011 Apr 29.
Published in final edited form as: Cytometry B Clin Cytom. 2010;78(Suppl 1):S69–S82. doi: 10.1002/cyto.b.20554
Abstract
Background
Advances in multi-parameter flow cytometry (FCM) now allow for the independent detection of larger numbers of fluorochromes on individual cells, generating data with increasingly higher dimensionality. The increased complexity of these data has made it difficult to identify cell populations from high-dimensional FCM data using traditional manual gating strategies based on single-color or two-color displays.
Methods
To address this challenge, we developed a novel program, FLOCK (FLOw Clustering without K), that uses a density-based clustering approach to algorithmically identify biologically relevant cell populations from multiple samples in an unbiased fashion, thereby eliminating operator-dependent variability.
Results
FLOCK was used to objectively identify seventeen distinct B cell subsets in a human peripheral blood sample and to identify and quantify novel plasmablast subsets responding transiently to tetanus and other vaccinations in peripheral blood. FLOCK has been implemented in the publically available Immunology Database and Analysis Portal – ImmPort (http://www.immport.org) for open use by the immunology research community.
Conclusions
FLOCK is able to identify cell subsets in experiments that use multi-parameter flow cytometry through an objective, automated computational approach. The use of algorithms like FLOCK for FCM data analysis obviates the need for subjective and labor intensive manual gating to identify and quantify cell subsets. Novel populations identified by these computational approaches can serve as hypotheses for further experimental study.
Key terms: flow cytometry, density-based analysis, data clustering, tetanus vaccination, B lymphocyte subsets
1. INTRODUCTION
Flow cytometry (FCM) is a method used to detect the presence and amount of cellular constituents in large numbers of single cells allowing for the enumeration and functional characterization of different cell types in complex cellular mixtures (9, 22, 23, 30, 46, 52). FCM can be used to measure the expression of membrane proteins, intracellular and secreted cell constituents, nucleic acids, phospholipids and post-translationally modified forms of proteins allowing FCM to be used to monitor cellular processes like DNA replication, programmed cell death and signal transduction activation (14, 18, 21, 24, 25, 27, 35). Thus, FCM has become an invaluable tool for investigation of basic physiological and biological processes in the experimental lab.
FCM has also become important as a diagnostic tool in the clinical lab (3, 5, 19, 37, 38, 50). For example, Chronic Lymphocytic Leukemia (CLL) and Monoclonal B cell Lymphocytosis (MBL) are both diagnosed based on abnormal patterns of CD5, CD20 and immunoglobulin light chains expression as assessed by FCM, even though the latter may show normal absolute leukocyte counts (32, 36, 44).
In the last decade, FCM has experienced dramatic technical advances in both instrumentation and reagent development such that the simultaneous assessment of 19 fluorescent and light scatter parameters on each individual cell is possible (8, 41). However, the standard methods for analyzing FCM data have not kept pace with these advances in technology. The majority of investigators still rely on iterative manual gating strategies in two-dimensional dot or contour plots to identify cell population subsets, which is somewhat subjective, time-consuming and difficult to reproduce even by the same investigator. In addition, if each of the 19 parameters is simply used to divide cell mixtures into expressing (+) and non-expressing (−) subsets, the number of potential cell populations could be as large as 524,288 (219). Thus, novel techniques are required to eliminate the dependence on manual gating in order to comprehensively analyze these rich datasets and extract biologically relevant findings.
Several groups have proposed new strategies for FCM data analysis, including a series of recent reports based on the modeling of cell population characteristics as mixtures of mathematical distributions (13, 17, 29, 42). Here we describe a model-independent approach – FLOw Clustering without K (FLOCK) – that utilizes grid-based partitioning and density distribution analysis to identify cell populations in high dimensional FCM data. The utility of FLOCK is demonstrated by the identification of seventeen B cell subsets in human peripheral blood and the monitoring of B cell population dynamics induced in response to vaccination.
2. METHODS
2.1 Subjects
Eight healthy adult subjects ages 22 to 65 years (mean 42 ± 15 year) were enrolled in this study at the University of Rochester Medical Center in 2008–2010; three were men and five were women. Peripheral blood mononuclear cells (PBMC) were isolated from heparinized blood by Ficoll-Hypaque density gradient centrifugation (Pharmacia Biotech). Seven subjects received the following vaccinations separately or in combination as a part of routine medical care: tetanus and diptheria toxoids (Td, Sanofi Pasteur, Swiftwater, PA), trivalent influenza vaccine 2009 (Fluzone, Sanofi,Pasteur. Swiftwater, PA), H1N1 monovalent influenza vaccine 2009 (Sanofi Pasteur, Swiftwater, PA), and Hepatitis Aand Hepatitis B (Twinrix, GlaxoSmithKline, Research Triangle Park, NC). PBMC were isolated on days 7 for all vaccination subjects; one subject had blood analyzed pre and post tetanus vaccination on days 5, 6, 7, 8, and 15. All studies were approved by the Research Subjects Review Boards at the University of Rochester Medical Center.
2.2 FCM Analysis
FCM data from three sample sets were analyzed using FLOCK in this manuscript. PBMC from the one untreated subject were stained with the following anti-human antibody staining reagents: IgD-FITC (BD Biosciences, San Diego, CA), CD1c-PE (Miltenyi, Auburn, CA), CD24-PE Alexa610 (Invitrogen), IgG-PE Cy5 (BD), CD3-PerCP Cy5.5 (BD), B220-PE Cy7 (BD), CD38-PacificBlue (BD custom conjugate), live/dead cell-staining PacificOrange Aqua (Invitrogen, Carlsbad, CA), CD27-APC (eBioscience, San Diego, CA), and CD19-APC-Cy7 (BD Biosciences, San Diego, CA). Note that the B220-PE-Cy7 reagent contains the RA3-6B2 monoclonal antibody, which recognizes the B220 glycoform of the CD45R protein. Stained cells were analyzed on an LSRII flow cytometer (Becton Dickinson, Franklin Lakes, NJ). Single conjugated mouse anti-human antibodies were added to Simply Cellular Anti-mouse compensation standard bead (Bangs Laboratories, Fishers, IN) and compensated automatically using Flow jo.8.8.6 (Treestar, Ashland, OR). The dataset used was CD3-CD19+ live lymphocyte gated (~67,000 events after gating) to focus the analysis on B cell populations. Data for CD3, CD19, CD1c and Aqua fluorescence were excluded from the FLOCK analysis.
The second dataset (tetanus study) examined B cell populations in serial PBMC samples from another subject after tetanus vaccination. Samples were taken immediately before vaccination (Day 0) and on Day 5, 6, 7, 8, and 15 after vaccination. PBMC were stained with the following anti-human antibodies to surface markers at 4°C for 20 minutes: IgD-PE (BD Biosciences, San Diego, CA), CXCR4-PE-Cy5 (eBioscience, San Diego, CA), CD3-PE-Cy5.5 (Caltag Laboratories, Carlsbad, CA), CD14-PE-Cy5.5 (Invitrogen, Carlsbad, CA), CD19-PE-Cy7 (BD Biosciences, San Diego, CA), CD38-Pacific Blue (BD Biosciences, San Diego, CA), CD138-APC (Miltenyi Biotec, Auburn, CA), HLA-DR-AlexaF700 (eBioscience, San Diego, CA), and CD27-APC-Alexa750 (eBioscience, San Diego, CA). Cells were then washed and resuspended in Cytoperm/Cytofix (BD Biosciences, San Jose, CA) containing anti-human Ki67-FITC antibodies (Invitrogen, Carlsbad, CA), and incubated for 20 minutes at 4°C. The dataset analyzed using FLOCK included onlyCD3-CD14-CD19+ gated events – Day 0 (13721 events), Day 5 (27832 events), Day 6 (27627 events), Day 7 (32204 events), Day 8 (31640 events), and Day 15 (39208 events).
The third data set (cross vaccine study) included PBMC samples from different subjects collected seven days after vaccination with various combinations of tetanus and diptheria toxoid, trivalent influenza 2009, monovalent H1N1 influenza 2009, Hepatitis Aand Hepatitis B vaccines. Samples were stained as above except excluding HLA-DR-AlexaF700. The dataset analyzed using FLOCK included only CD3-CD14-CD19+ gated events – 21702-8 (61691 events), 21702-19 (40333 events), 21702-6 (48124 events), 25251-134 (89261 events), 3334-139 (23951 events), and 21702-12 (68882 events).
2.3 Tetanus-specific and Total IgG Antibody Secreting Cell (ASC) ELISpot Assay
PBMC were added to 96-well ELISpot plates (MAIPS4510 96 well) coated with tetanus toxoid (1 Lf/mL Cylex Incorporated, Columbia, MD) or anti-human IgG (5μg/mL, Jackson Immunoresearch, West Grove, PA) and incubated overnight. Wells were washed and bound antibodies were detected with alkaline phosphatase-conjugated anti-human IgG antibody (1μg/mL, Jackson Immunoreseach), and reactions developed with VECTOR Blue Alkaline Phosphatase Substrate Kit III (Vector Laboratories, Burlingame, CA). Spots in each well were counted using the CTL immunospot reader (Cellular Technologies Ltd, Shaker Heights, OH).
2.4 Quantitative PCR for Expression of B Cell Transcription Factors
PBMC were isolated from a subject on day 7 after trivalent influenza vaccination and stained with the vaccination panel as described above. Three plasmablast/plasma cell populations identified by FLOCK were sorted and total cellular RNA was isolated using the RNeasy Mini Kit (Qiagen Inc., Valencia, CA) according to the manufacturer's protocol. After DNase I treatment (Invitrogen, Carlsbad, CA), 1 μg of RNA was subjected to reverse transcription using the iScript RT Kit (BioRad Inc., Hercules, CA). Aliquots of the resulting single-stranded cDNA products were included with BLIMP-1, Pax5, or GAPDH primers (6) in a 20 μl PCR reaction using HotStarTaq PCR Kit (Qiagen Inc., Valencia, CA), and amplified using a Bio-Rad MyCycler (Bio-Rad, Hercules, CA) with the following conditions: after an initial step of 95°C for 5 min, 40 cycles of 95°C for 30 sec, 55°C for 30 sec, and 68°C for 1 min, ending with a final extension step of 68°C for 5 min.
2.5 FLOCK
There are five major components in a FLOCK analysis pipeline: data preprocessing, grid-based density clustering, cross-sample comparison, result visualization, and population statistics calculations.
2.5.1 Data Preprocessing
Data preprocessing consists of FCS file conversion and data normalization. FCS files used in this paper have been compensated before preprocessing. Binary .fcs files were converted to tab-delimited ASCII text format using the channel number export function in the FlowJo software package (Tree Star, Inc.). The exported data constitute a data matrix table with each row corresponding to an event (cell) and each column to either a light scatter parameter such as FSC or a fluorescence emission channel such as FL1.
As the exported matrix columns have different value ranges, they could potentially contribute differently to the similarity of events being measured and used for clustering. To balance their contributions to the similarity measure, each column in the data matrix was normalized. Two normalization methods have been used successfully in our studies. For the peripheral B cell subset analysis, the data matrix was column-wise z-score normalized. Z-score normalization has been shown to be effective in classifying hematopoietic bone marrow cell populations (57). For the tetanus study, the data matrix was column-wise, 0–1, min-max normalized so that the Stone’s binning method (51) could be used. The formulas for the z-score and the min-max normalization are:
where S is the original value, S’ the normalized value, μand σ the average and the standard deviation of the data column of S, respectively, Min and Max are the smallest and the largest value of the data column of S. Although normalized data are used for event clustering, original values are used for visualization and population statistics calculations.
2.5.2 Grid-based Density Clustering
The core of the FLOCK algorithm begins by first partitioning each dimension into equally sized bins, which results in partition of the _n_-dimensional space into “hyper-regions”. Figure 1A shows a 2D mock example of how the data space is partitioned. However, in the FLOCK algorithm, partitioning is applied to all dimensions available in the dataset simultaneously. Each dimension of the data space is partitioned into the same number of equal-sized bins. Therefore a _d_-dimensional data space is partitioned into t*d hyper-regions with t equal-sized bins in each dimension.
Figure 1 . Algorithmic components of FLOCK.
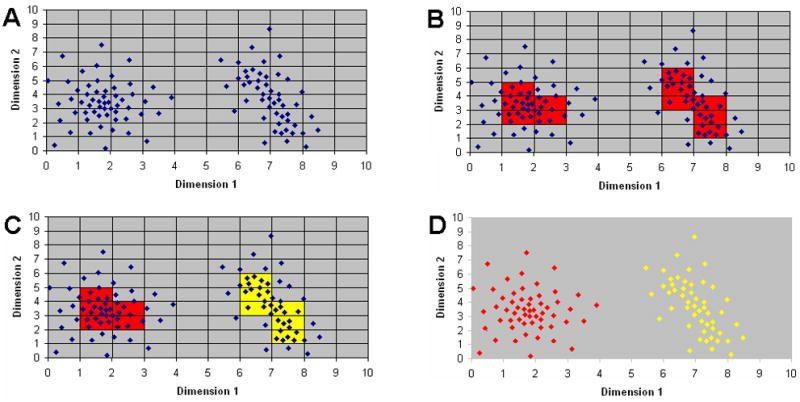
The FLOCKalgorithm consists of four core steps: hyper-grid creation, identifying dense hyper-regions, merging neighboring dense hyper-regions, clustering based on centroids derived from the merged dense hyper-regions. A) A gridding example. Each dimension corresponds to one measured characteristics (e.g. fluorescence at a given wavelength). Each dot is one event (e.g. data about a cell) with characteristic values (e.g. fluorescence intensity) as its coordinates. A number of equal-sized partitions (in this example the number is ten) are created on each dimension to generate a hyper-grid. B) Dense data hyper-regions. Density of a data hyper-region is calculated based on the number of event inside the region. A density threshold is determined by a data-driven approach described in the Methods section. Hyper-regions having larger number of events than the density threshold are considered dense. In this example, the density threshold is set at two and the resulting dense hyper-regions are marked in red. C) Merging dense hyper-regions. Dense hyper-regions are merged if they neighbor each other. The number of merged hyper-region groups (in this example the number is two, highlighted in red and yellow) is used as the number of final clusters, and the centroid of the events in a given hyper-region group calculated. D) Final clustering. Dots (events) are assigned to the closest centroid of the merged dense region group based on Euclidean distance to form the two populations.
In the second step, each hyper-region is assessed to determine the number of events present, and any hyper-region in which the number of events exceeds a certain threshold is labeled as being “dense” (Figure 1B). Equal-sized binning generates hyper-regions of equal volume. Therefore we can define the density of a hyper-region as the number of events in the region. A density threshold is used to distinguish a dense hyper-region from sparse and empty hyper-regions. As the density threshold increases, the number of dense hyper-regions decreases.
In the third step, dense hyper-regions adjacent to each other in _n_-dimensional space (two dense hyper-regions are adjacent if they are next to and contiguous with each other in one dimension and have the same position on all other dimensions) are grouped together into dense hyper-region groups, and the centroid (a point whose coordinates are the averages of the corresponding coordinates of a given set of point) representing all events in each dense hyper-region group determined (Figure 1C).
Finally, the dense hyper-region centroids are used to assign each event to its closest centroid based on Euclidean distance (Figure 1D). Other distance metric (e.g. Mannahalobis distance) could be used, but these have not yet been tested to determine their affect on the FLOCK results. Then the centroids are updated based on the member events of the dense regions and the cluster membership is computed again with the new centroids. This procedure can be regarded as a modified _K_-means procedure with only a few iterations. As initial centroids are not randomly generated, but rather based on data density, FLOCK quickly converges to a stable result during the centroid recalculation. This approach provides for two important advantages to traditional K-means– it determines – the number of populations in the high dimensional space from the number of dense hyper-region groups and it arrives at an estimated location of the center of the density distribution for each.
Two parameters need to be provided to execute the core of the FLOCK algorithm the number of partitions used to divide each dimension (# of bins) and the value of the density threshold cut-off. Both parameters appear to be dependent on a number of characteristics being measured, i.e. the number of events captured and the distribution of the data in the _n_-dimensional space. In order to automatically select the number of partitions, two different methods were employed. The first method is based on our observation of the growth of non-empty hyper-regions generated, which initially increases as the number of bins increases, but then starts to decrease when the number of bins used per dimension is larger than necessary. For the peripheral B cell subset analysis, we chose a bin number to maximize the ratio of the number of non-empty hyper-regions versus the number of bins to ensure that distinct populations are separated into different hyper- regions. For the tetanus data set, Stone’s method (51, 54) was used to optimize the bias-variance trade-off for different number of bins, which allows FLOCK to partition the space sufficiently in order to ensure that distinct cell populations are separated, while avoiding excessive partitioning that would eliminate the density characteristics of the data space.
The other parameter to select is the density threshold cut-off. Different cut-off values lead to different clustering results. The one that gives rise to the best Silhouette value (47) in the clustering is used. To avoid exhaustively testing each possible cut-off value we employed several different threshold selection methods, and the method that generates the best Silhouette value is chosen. For the peripheral B cell subset analysis, an ad hoc method designed to distinguish the dense hyper-regions from background based on the average density of the hyper-regions was used. For the tetanus data set, the density threshold cut-off was selected based on the minimum description length (MDL) principle (2, 45) that is commonly used to identify the best cut-off value within a data sequence. We have also developed another ad hoc method to identify the inflexion point of the decrease of the number of dense hyper-regions as the cut-off increases, which usually generates a lower density threshold value than the MDL principle and is more effective at identifying sparse cell populations. The use of these different approaches for density threshold estimation allows FLOCK to be tailored for each data set and to identify both relatively rare and relatively abundant cell populations.
The FLOCK algorithm has been implemented in the Immunology Database and Analysis Portal (ImmPort; http://www.immport.org). The runtime of a single FLOCK analysis is largely dependent on the number of events in a single data file. A relatively large file of ~2 million events returned results in less than 20 minutes; more typical files in the range of 10 thousand to 100 thousand events return results in less than 2 minutes.
2.5.3 Visualization and Statistics
The visualization module of FLOCK as implemented in ImmPort supports _2_-dimensional dot plots with each population colored uniquely. An individual population can be viewed in its own color with other populations in white as background. Users can navigate the multidimensional space by changing the dimensions or populations that are displayed.
Expression profiles of a population are presented that indicate the approximate expression level of each marker for the population. To generate the profile, FLOCK partitions each dimension into four bins with equal width. The four bins, from lowest values to highest values, are denoted with integers 1, 2, 3, and 4, respectively. The bin a population belongs to is decided by the majority vote of population events, and therefore each population’s profile is represented with a set of the integers –one integer (from 1–4) per marker.
Statistics calculated in FLOCK include population proportions, mean fluorescence intensity (MFI), cell count, standard deviation of the population on each parameter, and coefficient of variation.
2.6 Cross-sample Comparison
For the analysis of the tetanus and cross vaccine studies, the group of population centroids generated by FLOCK analysis of a single sample was applied to multiple samples to facilitate population comparison. Each event is assigned to its closest centroid in each sample. Populations generated in such a way have the same ID and color across all samples. For these data, the FCM results from the tetanus study Day 6 sample was used to identify the initial centroid locations.
3. RESULTS
3.1 Seventeen B Cell Subsets
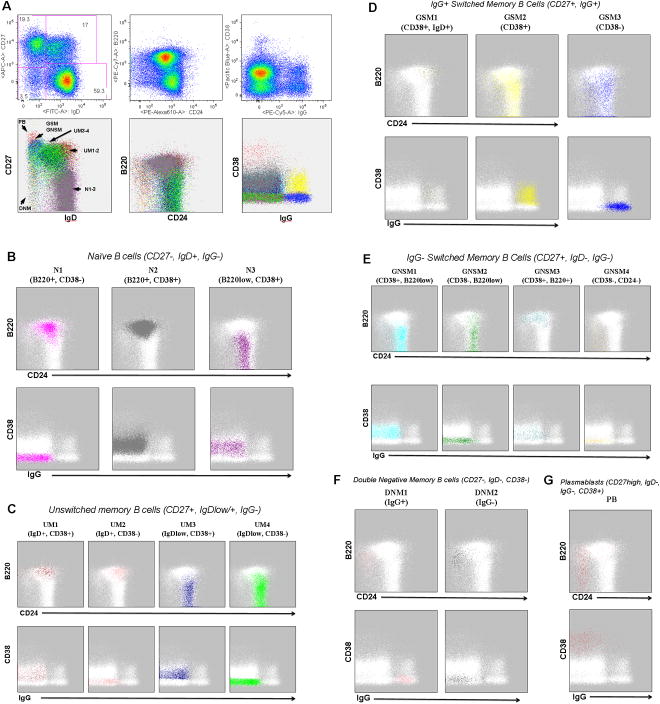
FLOCK was used as an objective method for the identification of B cell subsets in a dataset derived from the staining of a peripheral blood mononuclear cell (PBMC) sample derived from a normal human volunteer with reagents selected to identify various B lymphocyte (CD19+, CD3-) cell types based on six cell characteristics - expression of IgD, IgG, CD27, CD38, CD24, and B220. Seventeen distinct cell populations were identified by FLOCK using these characteristics. Figure 2A shows three two-dimensional dot plot combinations (IgD vs CD27; CD24 vs B220; IgG vs CD38) with cell events in a given B cell population subset highlighted in the same color. Six distinct regions can be observed from IgD versus CD27 dot plots, which appear to match previously defined B cell subsets (1, 20, 53, 55):
Figure 2 . Analysis of B cell populations in normal blood using FLOCK.
Following file conversion, data for IgD, IgG, CD24, B220, CD38 and CD27 fluorescence intensity was extracted and used for FLOCK analysis with automated parameter selection. Data has been CD3-CD19+ pre-gated. A) Two-dimensional dot plot displays for IgD vs. CD27, CD24 vs B220, and IgG vs CD38. The first row shows FlowJo displays of selected two-dimensional dot plots with manual gating applied to the IgD vs CD27 display; gated population proportions are provided. FLOCK-identified populations are displayed in the second row with each of the FLOCK-identified cell subsets highlighted in a different color. The six regions of the IgD vs CD27 dot plot corresponding to previously defined naïve and memory B cell subsets are highlighted - N (naïve), UM (unswitched memory), GSM (IgG positive switched memory), GNSM (IgG negative switched memory), (DNM (double negative memory), and PB (plasmablasts)). CD24 vs B220 and IgG vs CD38 dot plots showing individual populations for each of the naïve (B), unswitched memory (C), IgG positive switched memory (D), IgG negative switched memory (E), double negative memory (F), and plasmablasts (G). The IgD vs CD27 plots for the individual populations are included in Supplemental Figure 1.
- CD27−, IgD+ (naïve B cells – N);
- CD27+, IgD+ (unswitched memory B cells –UM);
- CD27+, IgDlow (unswitched memory B cells –UM);
- CD27+, IgD- (switched memory B cells – GSM & GNSM);
- CD27−, IgD- (double negative memory B cells – DNSM);
- CD27high, IgD- (plasmablasts – PB).
FLOCK was able to further identify additional subsets within each of these regions from the differential expression of the remaining markers.
While an IgD+CD27- phenotype is commonly used to globally define naïve B cells, there is mounting evidence demonstrating heterogeneity within this population, including the presence of early (T1 and T2) and late (T3, also called pre-naïve) transitional cells (28, 39). As shown in Figure 2B, FLOCK is indeed able to further subdivide this larger population into several discreet subsets based on the expression pattern of the remaining markers: N1 (B220+, CD38−), N2 (B220+, CD38+), and N3 (B220low, CD38+). The N2 subset was found to be the most abundant B cell subset in this peripheral blood sample at 48.9% of the CD19+ cells (see Table 1 for population proportions for all B cell subsets). Figure 2B also documents how FLOCK can take advantage of the differential expression of additional markers (in this case the relative expression of CD24) to provide a clean differentiation of naïve subsets that are identified in the literature as Bm1 (N1) and Bm2 (N2) based on the expression of IgD and CD38 (10, 40, 49, 55). These findings contrast with the recent report by Caraux in which circulating naïve B cells were reported as being CD38− (12). However, we found that FLOCK was able to clearly delineate low-level expression of CD38 in N2 and N3 populations as distinct from the negative expression in N1. Similar distinctions in CD38 expression were found in several memory B cell subsets as well (see below). This feature powerfully illustrates the ability of FLOCK to identify within multicolor data small populations previously recognized using conventional analysis with a limited number of markers. Additional scrutiny of these subpopulations using the relative expression of CD24 and CD38 suggest that CD24++, CD38++ T1/T2 transitional cells are likely included within the N2 population. (This important fraction, which has been proposed to include IL-10-producing regulatory B cells and to correlate with good clinical responses to B cell depletion in SLE, has been identified and quantified by FLOCK within the larger IgD+, CD27- N2 population on the basis of the expression of other markers in other samples (data not shown)).
Table 1.
Seventeen peripheral blood B cell subsets
| Populationa | Colorb | CD27c | IgDc | IgGc | CD38c | CD24c | B220c | Proportiond | Putative cell typea |
|---|---|---|---|---|---|---|---|---|---|
| N1 | Magenta | − | + | − | − | − | + | 4.69% | naïve (CD38−) |
| N2 | Gray | − | + | − | + | int | + | 48.94% | naïve (CD38+) |
| N3 | Purple | − | + | − | + | + | low | 4.41% | naïve (CD38+B220low) |
| UM1 | Darkred | + | + | − | + | + | + | 1.55% | unswitched memory (CD38+) |
| UM2 | Salmon | + | + | − | − | + | + | 0.94% | unswitched memory (CD38−) |
| UM3 | Darkblue | + | int | − | + | + | low | 6.16% | IgDlow unswitched memory (CD38+) |
| UM4 | Green | + | int | − | − | + | low | 11.50% | IgDlow unswitched memory (CD38−) |
| GSM1 | Grayishgreen | + | + | + | + | + | + | 0.36% | switching memory (IgD+IgG+CD38+) |
| GSM2 | Yellow | + | − | + | + | + | low | 4.05% | switched memory (CD38+) |
| GSM3 | Blue | + | − | + | − | + | low | 4.40% | switched memory (CD38−) |
| GNSM1 | Cyan | + | − | − | + | + | low | 4.84% | IgD-IgG- memory |
| GNSM2 | Darkgreen | + | − | − | − | + | low | 3.84% | IgD-IgG- memory |
| GNSM3 | Teal | + | − | − | + | + | + | 1.30% | IgD-IgG- memory |
| GNSM4 | Orange | + | − | − | − | − | low | 0.51% | IgD-IgG- memory |
| DNSM1 | Pink | − | − | + | − | − | + | 0.85% | double negative memory (IgG+) |
| DNSM2 | Darkgray | − | − | − | − | − | + | 0.91% | double negative memory (IgG−) |
| PB | Red | high | − | − | high | − | low | 0.75% | plasmablasts |
Unswitched memory B cells, also designated by different investigators as natural memory or marginal zone-like B cells, are characterized by expression of both CD27 and IgD. These unswitched memory B cells were further subdivided into four distinct subsets by FLOCK based on the relative amount of IgD (UM1 - 4, Figure 2A), B220 and CD38 (Figure 2C). Interestingly, the majority of UM cells (UM3 & UM4) belong to the IgDint fraction and are B220low, a feature that has been proposed to coincide with memory cell differentiation of B220+ naïve cells, and which these cells share with isotype switched CD27+ memory cells. In contrast, the remainder of CD27+ UM cells (UM1 & UM2) express levels of IgD and B220 comparable to naïve cells, a feature suggesting that they might represent an earlier differentiation stage of UM cells. Of note, as before for naïve B cells, distinct CD38+ and CD38- fractions are readily discriminated by FLOCK within both IgD+ and IgDint fractions (UM1/UM2 and UM3/UM4, respectively).
Memory B cells that have lost surface IgD expression through AID-mediated switch recombination to downstream isotypes like IgG and IgA (CD27+, IgD-) are considered isotype switched memory cells that by and large express surface IgA or IgG. These cells could be first subdivided based on expression of IgG into IgG+ switched memory B cells (GSM, Figure 2D) and IgG- switched (largely IgA+ according to our own published and unpublished data) memory B cells (GNSM, Figure 2E). These two groups could be further subdivided based on the relative expression of CD38, B220 and CD24. Consistent with previous studies, the majority of GSM cells downregulate B220 expression. One CD27+, IgG+ population was found to express IgD as well (GSM1). This population likely represents B cells that have recently undergone class switch recombination to the gamma heavy chain locus, but still retain residual IgD protein. Of note, these unique cells also retain expression of B220, a feature possibly suggesting an early/incomplete differentiation stage from naïve B cells. As for previous populations, CD38 presence or absence could also be resolved with FLOCK analysis for both GSM and GNSM populations. Based on CD38 expression, it appears that the combination of GSM2, GNSM1 and GNSM3 would correspond to the traditional early Bm5 subset (IgD-, CD38+) and the combination of GSM3, GNSM2 and GNSM4 would correspond to the late Bm5 subset (IgD-, CD38-) (49, 55). Some interesting differences however were detected between these two subsets of switched memory cells. Thus, no IgD co-expression was detected in any GNSM cells. Moreover, while the majority of GNSM cells behave like the majority of GSM cells, the GNSM3 subset retained expression of B220. Further analysis will be needed to explore the interesting possibility that these cells could represent the relatively rare IgM-only memory subset identified by others. Such possibility would be consistent with the B220 expression and in keeping with the early differentiation stage proposed for these cells (26). Similarly, since no reagents for the detection of Ig mu heavy chain or kappa and lambda light chains were included, the presence of an IgD-only, lambda positive B cell subset (5) cannot be addressed from these data. Finally, GNSM cells also contained one of only two peripheral blood subsets with negative expression of both CD24 and CD38 (GNSM4).
Cells lacking expression of both CD27 and IgD (predominantly expressing either IgG or IgA and containing a significant degree of somatic hypermutation) have been reported by different groups. We have reported that patients with SLE are characterized by large expansion of these cells; significant expansions have also been reported in patients with HIV infection and malaria (34, 55, 56). As for other populations, these so-called double negative memory B cells (DNM1 - 2) that express low levels of CD27 and lack expression of CD38 and IgD were also found in both IgG+ and IgG- varieties (Figure 2F). In contrast to conventional memory cells and in keeping with our previous observations, these cells express higher levels of B220 and are devoid of CD24 expression.
Finally, the plasmablast subset (PB) can be easily delineated based on the relatively high level of both CD27 and CD38 expression (Figure 2A & G). PB were found to be predominantly B220-, CD24- and surface Ig−.
In the end, each of the seventeen B cell subsets identified by FLOCK can be distinguished from every other population, based on some combination of staining parameters, and thus represents a distinct B cell subset in peripheral blood. In addition, although not necessarily symmetric, the distributions of events for each population in each dimension appear unimodal indicating that these subsets cannot be further subdivided based on the data available.
3.2 B Cell Responses to Vaccination
As a test of the utility of this strategy for automated cell population identification in FCM data, FLOCK was applied to a dataset from multiple peripheral blood samples before and after tetanus vaccination. In order to support direct cross-sample comparison, FLOCK was first applied to the Day 6 sample, and the centroid locations used to cluster each of the remaining data files.
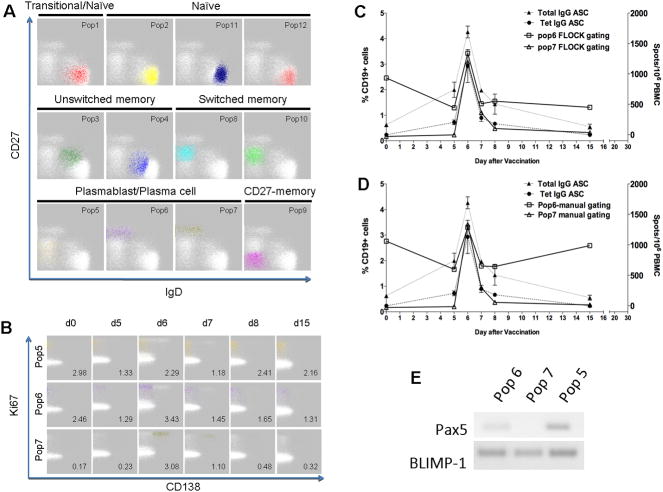
Twelve distinct CD19+ cell populations were identified in this dataset using reagents designed to identify particular plasmablast and plasma cell populations (Figure 3A). (Plots of all CD19+ cell populations are provided in Supplemental Figure 2. Supplemental Table 1 lists the expression profiles and population proportions generated by FLOCK for the identified populations). Again, FLOCK was able to identify conventional B cell subsets as observed in IgD vs CD27 plots (49). Populations #1, #2, #11 and #12 correspond to mature naïve (B cells that have not yet encountered their cognate antigen) and, perhaps, transitional B cells in that they express IgD but lack CD27 (Figure 3A). These four naïve B cell populations are distinguishable by differential expression of other cell surface proteins. For example, Population #1 expresses CD38, suggesting that it may represent a transitional B cell subset. Population #2 expresses low levels of CD138; Population #11 lacks HLA-DR expression; while Population #12 expresses HLA-DR (Supplemental Figure 2). The functional significance of these expression differences is not known at this time and requires further investigation.
Figure 3 . Analysis of B cell responses to tetanus vaccination.
B cell subsets identified using CD19, CD27, IgD, CD38, CD138, CXCR4, and HLA-DR expression from peripheral blood of a subject pre (day 0) and post tetanus vaccination (Day 5, 6, 7, 8 and 15). (A) Twelve populations identified by FLOCK analysis as shown by CD27 and IgD plots of the Day 6 sample, and further characterized in Supplemental Figure 1. (B) Three CD19+ plasmablast-like subsets were identified - Pop5: CD138−, CD38+, Ki67+, Pop6: CD138−, CD38hi, Ki67+, and Pop7: CD138+, CD38hi, Ki67+. (C, D) Total IgG and tetanus-specific IgG antibody-secreting cells (ASC) were identified by Elispot assay, and Pop 6 and 7 plasmablast-like subsets were identified by FLOCK and manual gating, respectively. (E) Pax5 and BLIMP-1 mRNA expression in Populations #5, #6, and #7.
Population #3 corresponds to conventional unswitched memory B cells, which express both CD27 and IgD; Population #4 may constitute a subset in transition between naïve and unswitched memory B cells. Populations #8 and #10 correspond to class switched memory B cells, which express CD27 but lack IgD; again these two sub-populations can be distinguished based on HLA-DR expression. Population # 9 lacks IgD and CD27 expression suggestive of the double-negative switched memory phenotype (55).
The traditional plasmablast subset has been described as being CD38high and CD27high. Interestingly, FLOCK identifies 3 different plasmablast/plasma cell populations using this 10-parameter panel. All three of these plasmablast/plasma cell subsets are positive for intracellular Ki67, a marker of recent proliferation. Population #6 (CD19+, IgD-, CD27high, CD38high, CD138-) expresses high levels of CD27 and CD38 but lacks CD138, consistent with the traditional plasmablast phenotype. In contrast, Population #7 (CD19+, IgD-, CD27high, CD38high, CD138+) not only expresses high levels of CD27 and CD38 but also CD138, a marker of long-lived plasma cells, suggesting the mobilization of a novel plasma cell subset in blood after vaccination. In addition, a third putative plasmablast subset, Population #5 (CD19+, IgD−, CD27+, CD38+, CD138−), was identified by FLOCK analysis.
Plasmablast/plasma cell populations #5, #6 and #7 show interesting dynamics in response to tetanus vaccination (Figure 3B). Population #6 has the characteristic phenotype of the traditional plasmablast subset and shows an oscillatory response in which population proportions dip by Day 5 only to rise again on Day 6 before falling on Day 7. In contrast, Population #7 (plasma cells) starts out low and remains low at Day 5, and then shows a dramatic but transient spike on Day 6. By clearly segregating these population subsets, FLOCK was able to demonstrate that these cells responding to vaccination express higher levels of Ki67, suggesting that the responding cells are proliferating, and also exhibit cytokine-secreting effector functions (data not shown). In contrast to the prevailing notion of old CD138+ plasma cells displaced from the bone marrow after vaccination, these data suggest that circulating CD138+ plasma cells are newly generated, as shown by Ki67 positive staining, and are active participants in the newly induced immune response (43). Population #5 does not express the characteristic high levels of CD27 (Figure 3A) and CD38 (Supplemental Figure 2) of the traditional plasmablasts and plasma cells but shows the same kind of oscillatory response as Population #6 (plasmablasts). Based on the gene expression studies described below, this subset may represent an early plasmablast precursor based on its phenotype.
The results of FLOCK were similar to the results of populations identified using manual gating (Supplemental Figure 3; Figure 3C, D). In addition, tetanus-specific IgG antibody secreting cells (ASCs) and total IgG ASCs were enumerated and compared with plasmablast populations identified by flow cytometry using FLOCK or manual analysis. The proportions of Population #7 identified by FLOCK closely match the results obtained from manual gating for CD138-expressing, CD38+ plasma cells (Figure 3C, D). The dynamic pattern of Population #7 proportions also closely paralleled the levels of total IgG-secreting and tetanus-specific plasma cells as quantified by ELISPOT.
Plasma cell differentiation is initiated by down-regulation of Pax5 and up-regulation of BLIMP-1. To validate that these 3 novel plasmablast-like populations are indeed of plasma cell lineage, Populations #5, 6, and 7 were sorted from the PBMC of a patient on day 7 after trivalent inactivated influenza vaccine (2009) vaccination and Pax5 and BLIMP-1 mRNA expression measured (Figure 3E). Population #7 demonstrates a lack of Pax5 and high levels of BLIMP-1 expression, evidence of the fully-differentiated plasma cell phenotype; whereas Population #6 has low Pax5 and high BLIMP-1 levels suggesting an intermediary phenotype. Lastly, Population #5 has high levels of both Pax5 and BLIMP-1 suggesting that this novel population might correspond to an early plasmablast population that has acquired BLIMP-1 expression but has not yet downregulated Pax5. Thus, FLOCK was able to discover previously characterized and novel plasmablast/plasma cell populations.
Finally, to demonstrate the reproducibility of cell population identification using FLOCK and to further validate the presence of these 3 distinct plasmablast/plasma cell populations in PBMC, an additional 6 subjects who received routine vaccinations with tetanus and diptheria toxoid, trivalent influenza vaccine 2009, H1N1 monovalent influenza vaccine 2009, Hepatitis A, and Hepatitis B were analyzed 7 days post-vaccination. These 3 plasmablast/plasma cell populations were identified in all 6 subjects with varying percentages (Figure 4A) – Population #5 (1.14–2.79%), Population #6 (1.86–9.68%), and Population #7 (0.62–9.1%). The population proportions derived from FLOCK analysis were compared with the results from manual gating (Figure 4B). For Populations #6 and #7, the results are highly correlated, with an r > 0.99. For Population #5, the correlation is less strong; this likely relates to the overlapping nature of the staining distributions in the CD38 and CD138 dimensions, which is difficult to address using manual gating in 2D plots. However, taken together, these data suggest that automated population identification using FLOCK is highly reproducible and demonstrate that the FLOCK program can accurately and objectively identify relevant human B cell populations in clinical samples without the need for labor-intensive manual gating.
Figure 4 . Reproducible detection of plasmablast/plasma cell populations in vaccination subjects using FLOCK.
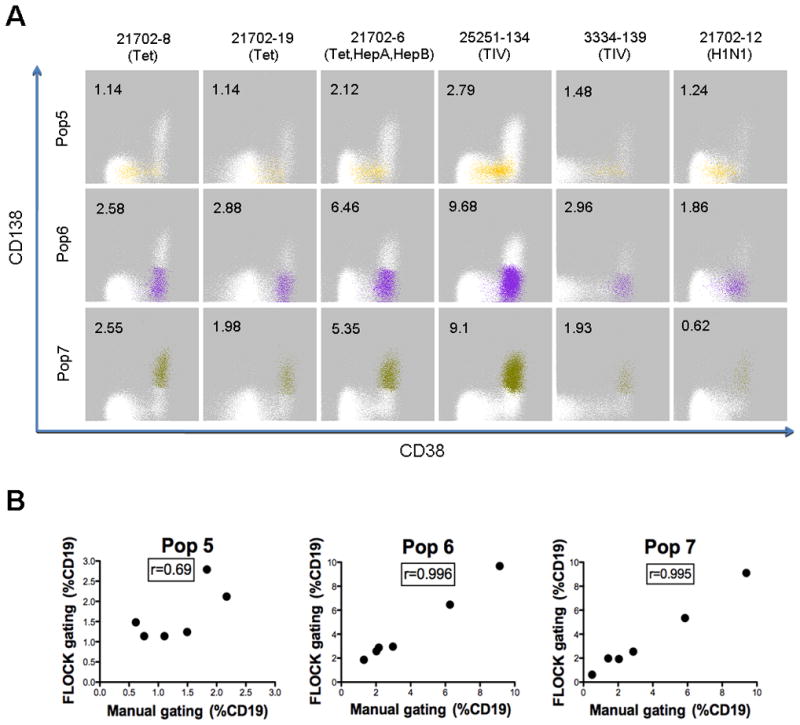
The centroid locations for the B cell subsets identified in the tetanus study (Figure 3) were used to identify the plasmablast (Pop5 & Pop6) and plasma cell (Pop7) subsets in six independent subjects vaccinated with the indicated vaccines – tetanus and diptheria toxoids (Tet), trivalent influenza vaccine 2009 (TIV), H1N1 monovalent influenza vaccine 2009 (H1N1), Hepatitis A(HepA), and Hepatitis B (HepB). (A) CD38 versus CD138 dot plots with individual cell events for the specific cell populations color-coded. Percentages of population events in the CD19+CD3- gated parent population are indicated. (B) Population percentages determined using FLOCK and manual gating are compared. Pearson two-tailed correlation coefficient (r) was used to assess the degree of correlation.
4. DISCUSSION
4.1 Automated Population Identification using FLOCK
Recent progress in FCM data analysis has mainly focused on model-based approaches, including the applications of Bayes mixture Gaussian models (13], t-mixture models as implemented in flowClust (29) and skew-t models as implemented in FLAME (Flow Analysis with Automated Multivariate Estimation) (42). User-driven clustering procedures for FCM population identification have also been described (57). The main advantage of model-based clustering is the robustness in identifying populations that follow the predefined models, if the model parameters can be estimated in an optimal way without becoming trapped in a local minimum solution. However, it remains an open question if model-based approaches can solve the high-dimensional clustering problem. Estimating and refining model parameters is a slow procedure for high-dimensional data set. In addition, the populations may not follow the distributional models when they become sparse in the high-dimensional space due to the curse of dimensionality.
Unlike model-based clustering, grid-based density clustering used in FLOCK partitions the data based on the closeness among events and the density variance. The analysis is conducted on data regions generated from the grid, which makes the whole procedure highly efficient as the number of data regions is much smaller than the number of events. Also the grid structure helps FLOCK rapidly locate the central areas of cell population distributions, which, as Bradley (11) and Ding (16) have pointed out, is critical for obtaining high-dimensional data clustering because optimization-based procedures are much more likely to become trapped in local minima with high-dimensional distributions than with low-dimensional distributions. In this respect, FLOCK could be complementary to recent model-based methods as the results of FLOCK could be used to provide the model-based approaches with initial parameters. An additional advantage to the FLOCK approach to cross-sample comparison is that the approach appears to be relatively robust to slight difference in fluorescence intensity of cell population between samples. This feature is especially useful for studies in which differences in sample preparation and staining procedures might generate technical variability in fluorescence that are not biologically relevant.
In the studies reported here, FLOCK was able to identify cell subsets that represented as few as 0.17% (23 events) of CD19+ cells in the complex PBMC mixture (Population #7 in the tetanus Day 0 sample). Visual inspection of the cell events in Figure 3B suggest that these are real. Thus, if a lower limit of 20 events is used as a confidence cutoff, a minimum of 20,000 (gated) events would need to be collected to identify cell populations present at a proportion of 0.1% or greater.
As the number of staining dimensions increases, the high dimensional space becomes increasing sparse, making it difficult to find population clusters based on density. We have recently developed a modification to the core FLOCK algorithm that incorporates a dimension selection step that focuses the initial clustering on the most informative 4–6 dimensions, followed by a refinement of the results based on data from other dimensions. Using this modification, FLOCK is able to handle datasets up to the 19-dimension limit of the current cytometer instrumentation (data not shown)
One of the main obstacles for the assessment of FLOCK is that there are no gold standards of cell population identity. In this paper we attempted to use population histograms to evaluate the results of FLOCK-based population identification. Populations identified by FLOCK showed unimodal distributions for all parameters measured in the experiment; 2-dimensional dot plots demonstrated the uniqueness of each population subset. Since these are objective characteristics that are independent from the criteria used in the FLOCK clustering algorithm (full-dimensional closeness and density), the results of FLOCK are validated using criteria that are not incorporated in the algorithm itself. Ultimately, it will be up to the biologists to determine their functional relevance. At least FLOCK will provide a more objective perspective for the understanding of FCM data instead of relying solely on human interpretation to supervise the data classification. To this end, a density-based approach seems the best candidate as it assumes very little about the data distributions but only uses the inherent density variation to partition the data.
Collaborative efforts have been made to provide an objective way to compare and evaluate the FCM data clustering methods, and also to establish guidance about appropriate use and application of these methods. These efforts have recently resulted in the open project FlowCAP (Flow Cytometry: Critical Assessment of Population Identification Methods, http://flowcap.flowsite.org/). FlowCAP is seeking and collecting FCM data to test different computational methods. Additional evaluation of FLOCK will be performed within the FlowCAP project once sufficient data sets have been collected.
4.2 B Cell Subsets and the B Cell Recall Response
The value of FLOCK is illustrated by two main features reflected in this manuscript, namely the automated recognition of multiple populations from data sets generated by multicolor flow analysis and the ability to identify unheralded populations that were not be readily suspected with iterative bi-dimensional gating. Indeed, by and large, the human B cell literature is confined to the classification of B cell subsets based on the relative expression of IgD/CD27/CD38/CD24, typically in 2 to 4-color combinations. In contrast, the multicolor approach used in our studies allowed FLOCK to consistently identify 17 well-defined subsets in human peripheral blood (the results shown in Figure 2 and Table 1 were corroborated in 4 additional unimmunized healthy subjects; results not shown). Thus, FLOCK identified major parental populations conventionally defined by the relative expression of IgD and CD27 (naïve, unswitched memory, switched memory, CD27- switched memory and plasmablasts). But, in addition to these classical B cell subsets, FLOCK was able to further subdivide these populations based on the expression of additional cell surface markers –CD38, CD24 and B220 - that contribute important information regarding the developmental and activation state of B cells. In contrast to previous studies, the ability to accurately segregate these subsets using FLOCK will allow their consistent and more objective quantification in longitudinal studies and across samples obtained in multicenter studies, thereby providing reliable information to understand the homeostasis of the different B cells subsets and the significance of their perturbation in different clinical settings, including both physiological and pathological immune responses (31, 48). Furthermore, this approach will greatly facilitate the isolation of individual subsets for the ex vivo and in vitro study of their biology and function.
In the tetanus study, FLOCK was able to identify several B cell subsets that responded to vaccination in a recall response following well-established kinetics patterns. While validating the reproducibility of FLOCK measurements, these studies also powerfully illustrate the ability of FLOCK to provide detailed phenotypic characterization of predetermined populations and to demonstrate new functional properties. The latter capability is perhaps best encapsulated in the recognition by FLOCK of the high levels of Ki67 universally present in the CD38high plasmablast (Population #6) and plasma cell (Population #7) populations also expressing high levels of CD27. The expression of Ki67 in both the CD138- plasmablast and CD138+ plasma cell populations in normal peripheral blood is consistent with a recent report by Caraux et al. (12). Moreover the longitudinal studies demonstrated the ability of FLOCK to uncover a previously unrecognized candidate population (Population #5 –CD38+, CD138-) likely to represent an early stage of plasma cell differentiation in the context of an antigen-specific acute recall response. This finding will now permit detailed studies of clonal relatedness between the different populations as well as analysis of antigen-affinity and affinity maturation across the different antibody-secreting cell subsets in order to understand the rules that govern the maturation of plasma cells and their selection into long-lived compartments (indicated at least in part by the differential expression of CXCR4 (4, 43)).
In the cross vaccine study, the proportions of the three plasmablast/plasma cell subsets varied dramatically between subjects, as much as 15 fold for the plasma cell (Population #7) population. Given the relatively small sample size in this pilot study it is difficult to determine if this variability in vaccine response is due to genetic differences in the human subjects enrolled in the study, differences in the prior vaccination or pathogen exposure history of the subjects, or differences in the vaccine formulations. The fact that the strongest response and one of the weakest responses were observed in the two subjects receiving the trivalent influenza vaccine 2009 argues against the latter explanation. It will be interesting to determine if the absolute levels of one or more of these plasmablast/plasma cell populations, or their dynamics, will serve as an effective surrogate marker of protective immunity following vaccination.
4.3 Monoclonal B Cell Lymphocytosis (MBL) and the Mapping of B Cell Subsets
One challenges that arises from these studies is how to compare the 17 B cell populations identified in normal peripheral blood by FLOCK using this combination of staining reagents compare with other studies investigating B cell subsets in other normal and pathologic conditions, including MBL. Unfortunately, this is a difficult question to address because reagent panels used to distinguish between normal B cell subsets are not exactly the same as those used for disease diagnosis. Markers that are useful for disease diagnosis are often selected based on their abnormality of expression. For example, MBL was recognized partly through the investigation of chronic lymphocytic leukemia, with both characterized by abnormalities in the expression pattern of CD5, CD20 and immunoglobulin light chains. More recently, Nieto et al (36) used an 8-marker panel, including CD20, CD5, CD23, and kappa and lambda immunoglobulin light chains to study CLL-like MBL and found that these cells could be routinely detected in peripheral blood of healthy subjects. Unfortunately, the markers used in the analysis reported here were selected to highlight differences in normal memory B cell populations, and did not include CD5 and CD20. This raises the general challenge of comparing results from separate studies in which distinct, but overlapping sets of cellular markers are used. Indeed, this challenge even arose when we tried to compare the populations identified in the three data sets analyzed in this study, which is why different naming conventions were used. Collaborative efforts may be necessary in order to develop strategies to determine the relationships between cell populations identified in different studies, perhaps replacing less informative markers in our study (e.g. CD24, CD1c) with markers better suited to identify CLL-like MBL cells (e.g. CD5, CD20)(36) and immature/transitional early B cells (e.g. CD10) (12).
In order to address this challenge, one of our near-term goals is to link FLOCK results to the Cell Ontology (7, 15, 33). The Cell Ontology is being developed to provide a standard description of normal cell types based on experimentally-defined properties. In the case of hematopoietic cells, the expression of cell surface and intracellular proteins is being used as the major property defining distinct cell types in the ontology. The fluorescence profile of a population identified by FLOCK indicates the relative expression of the marker protein in the population, and thus could be used to link FLOCK results with Cell Ontology definitions. This would facilitate the management and exchange of cell population data among different labs. And since cell types are represented as a hierarchy of type-subtype relations in the ontology, it will be possible to understand the relationship of cell types identified at different levels of granularity.
5. CONCLUSIONS
FLOCK is a computational method that automatically identifies cell populations in multi-dimensional flow cytometry data. Based on a variety of assessment characteristics, FLOCK appears to objectively identify distinct cell types, including both previously characterized B cell subsets and novel subgroups of cells of unknown significance. As the main FCM analytical software developed in the context of the Bioinformatics Integration Support Contract (BISC) project, FLOCK has been implemented within Immunology Database and Analysis Portal -ImmPort (http://www.immport.org) environment, and is freely available for scientific research purposes. While FLOCK was developed for the analysis of FCM data, the clustering strategy developed can also be applied to other data mining problems that have previously relied on K-means to perform cluster analysis. Ultimately, the use of objective algorithmic approaches like FLOCK should play an important role in the improvement of clinical diagnostics based on flow cytometry.
Supplementary Material
Supp Fig S1
Supp Fig S2
Supp Fig S3
Supp Material
Supp Table S1
Acknowledgments
This work was supported in part by the U.S. National Institutes of Health N01-AI40076 (Bioinformatics Integration Support Contract), N01-AI50029 (Rochester Program for Biodefense of Immunocompromised Populations), and U19-AI56390 (Rochester Autoimmunity Center of Excellence).
LITERATURE CITED
- 1.Agematsu K, Nagumo H, Yang F, Nakazawa T, Fukushima K, Ito S, Sugita K, Mori T, Kobata T, Morimoto C, Komiyama A. B cell subpopulations separated by CD27 and crucial collaboration of CD27+ B cells and helper T cells in immunoglobulin production. European Journal of Immunology. 1997;27:2073–2079. doi: 10.1002/eji.1830270835. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal R, Gehrke J, Gunopulos D, Raghavan P. Automated subspace clustering of high-dimensional data for data mining applications. Proceedings of SIGMOD Conference on Management of Data; 1998. pp. 94–105. [Google Scholar]
- 3.Al-Mawali A, Gillis D, Lewis I. The role of multiparameter flow cytometry for detection of minimal residual disease in acute myeloid leukemia. American Journal of Clinical Pathology. 2009;131:16–26. doi: 10.1309/AJCP5TSD3DZXFLCX. [DOI] [PubMed] [Google Scholar]
- 4.Arce S, Luger E, Muehlinghaus G, Cassese G, Hauser A, Horst A, Lehnert K, Odendahl M, Hönemann D, Heller K-D, Kleinschmidt H, Berek C, Dörner T, Krenn V, Hiepe F, Bargou R, Radbruch A, Manz RA. CD38 low IgG-secreting cells are precursors of various CD38 high-expressing plasma cell populations. Journal of Leukocyte Biology. 2004;75:1022–1028. doi: 10.1189/jlb.0603279. [DOI] [PubMed] [Google Scholar]
- 5.Aurran-Schleinitz T, Telford W, Perfetto S, Caporaso N, Wilson W, Stetler-Stevenson MA, Zenger VE, Abbasi F, Marti GE. Identification of a new monoclonal B-cell subset in unaffected first-degree relatives in familial chronic lymphocytic leukemia. Leukemia. 2005;19:2339–2341. doi: 10.1038/sj.leu.2403980. [DOI] [PubMed] [Google Scholar]
- 6.Avery DT, Deenick EK, Ma CS, Suryani S, Simpson N, Chew GY, Chan TD, Palendira U, Bustamante J, Boisson-Dupuis S, Choo S, Bleasel KE, Peake J, King C, French MA, Engelhard D, Al-Hajjar S, Al-Muhsen S, Magdorf K, Roesler J, Arkwright PD, Hissaria P, Riminton DS, Wong M, Brink R, Fulcher DA, Casanova JL, Cook MC, Tangye SG. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. Journal of Experimental Medicine. 2010;207:155–71. doi: 10.1084/jem.20091706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biology. 2005;6(2):R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumgarth N, Roederer M. A practical approach to multicolor flow cytometry for immunophenotyping. Journal of Immunological Methods. 2000;243:77–97. doi: 10.1016/s0022-1759(00)00229-5. [DOI] [PubMed] [Google Scholar]
- 9.Bohn B. Flow Cytometry: A novel approach for the quantitative analysis of receptor-ligand interactions on surfaces of living cells. Molecular and Cellular Endocrinology. 1980;20:1–15. doi: 10.1016/0303-7207(80)90090-8. [DOI] [PubMed] [Google Scholar]
- 10.Bohnhorst J, Bjorgan MB, Thoen JE, Natvig JB, Thompson KM. Bm1-bm5 classification of peripheral blood b cells reveals circulating germinal center founder cells in healthy individuals and disturbance in the b cell subpopulations in patients with primary sjogren's syndrome. Journal of Immunology. 2001;167:3610–3618. doi: 10.4049/jimmunol.167.7.3610. [DOI] [PubMed] [Google Scholar]
- 11.Bradley PS, Fayyad UM. Refining initial points for K-means clustering. Proceedings of the Fifteenth International Conference on Machine Learning; 1998. pp. 91–99. [Google Scholar]
- 12.Caraux A, Klein B, Paiva B, Bret C, Schmitz A, Fuhler GM, Bos NA, Johnsen HE, Orfao A, Perez-Andres M for the Myeloma Stem Cell Network (MSCNET) Circulating human B and plasma cells. Age-associated changes in counts and detailed characterization of circulating normal CD138– and CD138+ plasma cells. Haematologica. 2010;95(6):1004–1015. doi: 10.3324/haematol.2009.018689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan C, Feng F, Ottinger J, Foster D, West M, Kepler TB. Statistical mixture modeling for cell subtype identification in flow cytometry. Cytometry Part A. 2008;73(8):693–701. doi: 10.1002/cyto.a.20583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darzynkiewicz Z, Huang X. Analysis of cellular DNA content by flow cytometry. Current Protocols in Immunology. 2004:5.7.1–5.7.18. doi: 10.1002/0471142735.im0507s60. [DOI] [PubMed] [Google Scholar]
- 15.Diehl AD, Augustine AD, Blake JA, Cowell LG, Gold ES, Gondré-Lewis TA, Masci AM, Meehan TF, Morel PA, Nijnik A, Peters B, Pulendran B, Scheuermann RH, Zand MS, Mungall CM NIAID Cell Ontology Working Group. Hematopoietic cells types: prototype for a revised cell ontology. Journal of Biomedical Informatics. 2010 doi: 10.1016/j.jbi.2010.01.006. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding C, He X, Zha H, Simon HD. Adaptive dimension reduction for clustering high dimensional data. Proceedings of IEEE International Conference on Data Mining; 2002. pp. 147–154. [Google Scholar]
- 17.Finn WG, Carter KM, Raich R, Stoolman LM, Hero AO. Analysis of clinical flow cytometric immunophenotyping data by clustering on statistical manifolds: treating flow cytometry data as high-dimensional objects. Cytometry Part B. 2009;76B:1–7. doi: 10.1002/cyto.b.20435. [DOI] [PubMed] [Google Scholar]
- 18.Fox R, Aubert M. Flow cytometric detection of activated caspase-3. Methods in Molecular Biology. 2008;414:47–56. doi: 10.1007/978-1-59745-339-4_5. [DOI] [PubMed] [Google Scholar]
- 19.Fuhrmann S, Streitz M, Kern F. How flow cytometry is changing the study of TB immunology and clinical diagnosis. Cytometry Part A. 2008;73A:1100–1106. doi: 10.1002/cyto.a.20614. [DOI] [PubMed] [Google Scholar]
- 20.Fulcher DA, Basten A. B cell life span: A review. Immunology and Cell Biology. 1997;75:446–455. doi: 10.1038/icb.1997.69. [DOI] [PubMed] [Google Scholar]
- 21.Hammill A, Uhr JW, Scheuermann RH. Annexin V staining due to loss of membrane asymmetry can be reversible and precede commitment to apoptotic death. Experimental Cell Research. 1999;251:16–21. doi: 10.1006/excr.1999.4581. [DOI] [PubMed] [Google Scholar]
- 22.Herzenberg LA, Tung J, Moore WA, Herzenberg LA, Parks DR. Interpreting flow cytometry data: a guide for the perplexed. Nature Immunology. 2006;7(7):681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- 23.Hulett HR, Bonner WA, Barrett J, Herzenberg LA. Cell sorting: Automated separation of mammalian cells as a function of intracellular fluorescence. Science. 1969;166:747–749. doi: 10.1126/science.166.3906.747. [DOI] [PubMed] [Google Scholar]
- 24.Ilangumaran S, Finan D, Rottapel R. Flow cytometric analysis of cytokine receptor signal transduction. Journal of Immunological Methods. 2003;278:221–234. doi: 10.1016/s0022-1759(03)00177-7. [DOI] [PubMed] [Google Scholar]
- 25.June CH, Moore JS. Measurement of intracellular Ions flow cytometry. Current Protocols in Immunology. 2004:5.5.1–5.5.20. doi: 10.1002/0471142735.im0505s64. [DOI] [PubMed] [Google Scholar]
- 26.Klein U, Kuppers R, Rajewsky K. Human IgM+IgD+ B cells, the major B cell subset in the peripheral blood, express V kappa genes with no or little somatic mutation throughout life. European Journal of Immunology. 1993;23:3272–3277. doi: 10.1002/eji.1830231232. [DOI] [PubMed] [Google Scholar]
- 27.Lee BW, Olin MR, Johnson GL, Griffin RJ. In vitro and in vivo apoptosis detection using membrane permeant fluorescent-labeled inhibitors of caspases. Methods in Molecular Biology. 2008;414:109–135. doi: 10.1007/978-1-59745-339-4_10. [DOI] [PubMed] [Google Scholar]
- 28.Lee J, Kuchen S, Fischer R, Chang S, Lipsky PE. Identification and characterization of a human CD5+ pre-naive B cell population. Journal of Immunology. 2009;182:4116–4126. doi: 10.4049/jimmunol.0803391. [DOI] [PubMed] [Google Scholar]
- 29.Lo K, Brinkman RR, Gottardo R. Automated gating of flow cytometry data via robust model-based clustering. Cytometry A. 2008;73A:321–332. doi: 10.1002/cyto.a.20531. [DOI] [PubMed] [Google Scholar]
- 30.Loken MR, Stall AM. Flow cytometry as an analytical and reparative tool in immunology. Journal of Immunological Methods. 1982;50:R85–R112. doi: 10.1016/0022-1759(82)90161-2. [DOI] [PubMed] [Google Scholar]
- 31.Manjarrez-Orduno N, Quach TD, Sanz I. B cells and immunological tolerance. Journal of Investigative Dermatologyl. 2009;129:278–288. doi: 10.1038/jid.2008.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marti G, Abbasi F, Raveche E, Rawstron AC, Ghia P, Aurran T, Caporaso N, Shim YK, Vogt RF. Overview of monoclonal B-cell lymphocytosis. British Journal of Haematology. 2007;139:701–708. doi: 10.1111/j.1365-2141.2007.06865.x. [DOI] [PubMed] [Google Scholar]
- 33.Masci AM, Arighi CN, Diehl AD, Lieberman AE, Mungall C, Scheuermann RH, Smith B, Cowell LG. An improved ontological representation of dendritic cells as a paradigm for all cell types. BMC Bioinformatics. 2009;10:70. doi: 10.1186/1471-2105-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moir S, Ho J, Malaspina A, Wang W, DiPoto AC, O’Shea MA, Roby G, Kottilil S, Arthos J, Proschan MA, Chun T-W, Fauci AS. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. Journal of Experimental Medicine. 2008;205(8):1797–1805. doi: 10.1084/jem.20072683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muppidi J, Porter M, Siegel RM. Measurement of apoptosis and other forms of cell death. Current Protocols in Immunology. 2004:3.17.1–3.17.36. doi: 10.1002/0471142735.im0317s59. [DOI] [PubMed] [Google Scholar]
- 36.Nieto WG, Almeida J, Romero A, Teodosio C, Lopez A, Henriques AF, Sánchez ML, Jara-Acevedo M, Rasillo A, González M, Fernandez-Navarro P, Vega T, Orfao A the Primary Health Care Group of Salamanca for the Study of MBL. Increased frequency (12%) of circulating chronic lymphocytic leukemia-like B-cell clones in healthy subjects using highly sensitive multicolor flow cytometry approach. Blood. 2009;114:33–37. doi: 10.1182/blood-2009-01-197368. [DOI] [PubMed] [Google Scholar]
- 37.Ogata K. Diagnostic flow cytometry for low-grade myelodysplastic syndromes. Hematological Oncology. 2008;26:193–198. doi: 10.1002/hon.857. [DOI] [PubMed] [Google Scholar]
- 38.Oliveira JB, Notarangelo LD, Fleisher TA. Applications of flow cytometry for the study of primary immune deficiencies. Current Opinion in Allergy and Clinical Immunology. 2008;8:499–1021. doi: 10.1097/ACI.0b013e328312c790. [DOI] [PubMed] [Google Scholar]
- 39.Palanichamy A, Barnard J, Zheng B, Owen T, Quach T, Wei C, Looney RJ, Sanz I, Anolik JH. Novel human transitional B cell populations revealed by B cell depletion therapy. Journal of Immunology. 2009;182:5982–5993. doi: 10.4049/jimmunol.0801859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pascual V, Liu YJ, Magalski A, de Bouteiller O, Banchereau J, Capra JD. Analysis of somatic mutation in five B cell subsets of human tonsil. Journal of Experimental Medicine. 1994;180(1):329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nature Reviews Immunology. 2004;4(8):648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 42.Pyne S, Hu X, Wang K, Rossin E, Lin T, Maier LM, Baecher-Allan C, McLachlan GJ, Tamayo P, Hafler DA, De Jager PL, Mesirov JP. Automated high-dimensional flow cytometric data analysis. PNAS. 2009;106(21):8519–8524. doi: 10.1073/pnas.0903028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Radbruch A, Muehlinghaus G, Luger EO, Inamine A, Smith KGC, Dörner T, Hiepe F. Competence and competition: the challenge of becoming a long-lived plasma cell. Nature Reviews Immunology. 2006;6:741–750. doi: 10.1038/nri1886. [DOI] [PubMed] [Google Scholar]
- 44.Rawstron AC, Bennett FL, O’Connor SJM, Kwok M, Fenton JAL, Plummer M, de Tute R, Owen RG, Richards SJ, Jack AS, Hillmen P. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. New England Journal of Medicine. 2008;359:575–583. doi: 10.1056/NEJMoa075290. [DOI] [PubMed] [Google Scholar]
- 45.Rissanen J. Stochastic Complexity in Statistical Inquiry. World Scientific Publishing, Inc; 1989. p. 250. [Google Scholar]
- 46.Robinson JP. Encyclopedia of Biomaterials and Biomedical Engineering. New York: Marcel Dekker, Inc; 2004. Flow Cytometry; pp. 630–640. [Google Scholar]
- 47.Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. Journal of Computational and Applied Mathematics. 1987;20:53–65. [Google Scholar]
- 48.Sanz I, Lee FE. B cells as therapeutic targets in SLE. Nature Reviews Rheumatology. 2010 doi: 10.1038/nrrheum.2010.68. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanz I, Wei C, Lee FE, Anolik JH. Phenotypic and functional heterogeneity of human memory B cells. Seminars in Immunology. 2008;20(1):67–82. doi: 10.1016/j.smim.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stetler-Stevenson M. Flow cytometric immunophenotyping: Emerging as an important diagnostic tool in the evaluation of cytopenic patients. Leukemia Research. 2009;33:1020–1021. doi: 10.1016/j.leukres.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stone CJ. An asymptotically histogram selection rule. In: Neyman J, editor. Proceedings of Second Berkeley Symposium; 1984. pp. 513–520. [Google Scholar]
- 52.Tung JW, Parks DR, Moore WA, Herzenberg LA, Herzenberg LA. New approaches to fluorescence compensation and visualization of FACS data. Clinical Immunology. 2004;110:277–283. doi: 10.1016/j.clim.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 53.Warnatz K, Denz A, Drager R, Braun M, Groth C, Wolff-Vorbeck G, Eibel H, Schlesier M, Peter HH. Severe deficiency of switched memory B cells (CD27+IgM-IgD-) in subgroups of patients with common variable immunodeficiency: a new approach to classify a heterogeneous disease. Blood. 2002;99(5):1544–1551. doi: 10.1182/blood.v99.5.1544. [DOI] [PubMed] [Google Scholar]
- 54.Wasserman L. All of Statistics: A Concise Course in Statistical Inference. Springer-Verlag, Inc; 2004. p. 442. [Google Scholar]
- 55.Wei C, Anolik J, Cappione A, Zheng B, Pugh-Bernard A, Brooks J, Lee EH, Milner EC, Sanz I. A new population of cells lacking expression of CD27 represents a notable component of the B cell memory compartment in systemic lupus erythematosus. Journal of Immunology. 2007;178(10):6624–6633. doi: 10.4049/jimmunol.178.10.6624. [DOI] [PubMed] [Google Scholar]
- 56.Weiss GE, Crompton PD, Li S, Walsh LA, Moir S, Traore B, Kayentao K, Ongoiba A, Doumbo OK, Pierce SK. Atypical memory B cells are greatly expanded in individuals living in a malaria- endemic area. Journal of Immunology. 2009;183:2176–2182. doi: 10.4049/jimmunol.0901297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zamir E, Geiger B, Cohen N, Kam Z, Katz B-Z. Resolving and classifying haematopoietic bone-marrow cell populations by multi-dimensional analysis of flow-cytometry data. British Journal of Haematology. 2005;129:420–431. doi: 10.1111/j.1365-2141.2005.05471.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp Fig S1
Supp Fig S2
Supp Fig S3
Supp Material
Supp Table S1