Hydrogen Peroxide–Mediated Activation of MAP Kinase 6 Modulates Nitric Oxide Biosynthesis and Signal Transduction in Arabidopsis (original) (raw)
Nitric oxide has been proposed to act as a signal for numerous physiological and developmental processes in plants. This work describes the involvement of the mitogen-activated protein kinase (MPK) cascade in nitric oxide synthesis via nitrate reductase. It demonstrates H2O2-mediated MPK6 activation of nitric oxide production and signal transduction during root development in Arabidopsis.
Abstract
Nitric oxide (NO) is a bioactive molecule that functions in numerous physiological and developmental processes in plants, including lateral root development. In this study, we used biochemical and genetic approaches to analyze the function of Arabidopsis thaliana mitogen-activated protein kinase 6 (MPK6) in the regulation of NO synthesis in response to hydrogen peroxide (H2O2) during lateral root development. In both mpk6 mutants studied, H2O2-induced NO synthesis and nitrate reductase (NR) activity were decreased dramatically. Furthermore, one NR isoform, NIA2, was required for the MPK6-mediated production of NO induced by H2O2. Notably, NIA2 interacted physically with MPK6 in vitro and in vivo and also served as a substrate of MPK6. Phosphorylation of NIA2 by MPK6 led to an increase in NR activity, and Ser-627 was identified as the putative phosphorylation site on NIA2. Phenotypical analysis revealed that mpk6-2 and mpk6-3 seedlings produce more and longer lateral roots than wild-type plants did after application of the NO donor sodium nitroprusside or H2O2. These data support strongly a function of MPK6 in modulating NO production and signal transduction in response to H2O2 during Arabidopsis root development.
INTRODUCTION
Nitric oxide (NO) has been characterized recently as an important signal molecule that mediates many developmental and physiological processes in plants, which include seed germination, lateral root initiation, flowering, stomatal closure, and responses to abiotic stresses (He et al., 2005; Simpson, 2005; Libourel et al., 2006; Lombardo et al., 2006; Neill et al., 2008). In plant cells, NO is produced mainly via two distinct enzymatic pathways: the Arg-dependent nitric oxide synthase (NOS) pathway and the nitrite-dependent nitrate reductase (NR) pathway. In addition, nonenzymatic processes contribute to the synthesis of NO in plants (Neill et al., 2003, 2008; Wilson et al., 2007). Although previous findings have indicated the existence of NOS in plants, no gene or protein with sequence homology to known mammalian-type NOS has been found (Guo et al., 2003; Crawford, 2006). In Arabidopsis thaliana, NOS1/NOA1, which was identified originally as a potential NOS (Guo et al., 2003), was found to be unable to bind and oxidize Arg to NO and was later shown to be a circularly permuted GTPase (Moreau et al., 2008).
NR is the only enzyme that has been shown to catalyze the production of NO in plant defense responses. Although the primary function of NR in plants is to convert nitrate to nitrite, it can also convert nitrite to NO in vitro and in vivo (Dean and Harper, 1988; Desikan et al., 2002). Bursts of NO that are induced by auxins, elicitors, abscisic acid (ABA), or hydrogen peroxide (H2O2) are dependent on NR activity (Bright et al., 2006; Yamamoto-Katou et al., 2006; Kolbert et al., 2008). In Arabidopsis, NR is encoded by two genes, NIA1 and NIA2 (Campbell, 1999). Deficiency of NIA1 and NIA2 results in a significant reduction in NO synthesis (Bright et al., 2006; Modolo et al., 2006). Further investigations have revealed that NIA1 and NIA2 contribute differently to the synthesis of NO in different tissues. During stomatal closure induced by ABA, NIA1 plays the major role in NO production (Bright et al., 2006), although NIA1 is known to be less abundant and less active than NIA2 in seedlings (Wilkinson and Crawford, 1991).
Mitogen-activated protein kinase (MAPK) cascades are conserved pathways that transduce environmental stimuli into intracellular responses in many organisms, including humans, Drosophila melanogaster, yeast, and plants. Each MAPK cascade is composed of three kinases. MAPKs are activated through phosphorylation by upstream MAPK kinases (MAPKKs), which are in turn activated by MAPKK kinases (MAPKKKs) (Ichimura et al., 2000). In Arabidopsis, there are 20 MAPKs, 10 MAPKKs, and ~60 MAPKKKs (Ichimura et al., 2000). It is well documented that MAPK cascades play key roles in innate immunity and environmental stress responses and are also involved in the regulation of plant growth and development (Tena et al., 2001; Zhang and Klessig, 2001; Jonak et al., 2002; Mishra et al., 2006; Colcombet and Hirt, 2008). Mitogen-activated protein kinase 6 (MPK6) is a well-characterized MAPK in Arabidopsis. It can be activated by various abiotic and biotic stresses, which include low temperature, low humidity, hyperosmolarity, touch, wounding, microbial elicitors, and oxidative stress (Ichimura et al., 2000; Nuhse et al., 2000; Desikan et al., 2001; Yuasa et al., 2001). MPK6 is also involved in the signaling and responses of the plant hormones ethylene and jasmonic acid (Liu and Zhang, 2004; Seo et al., 2007; Takahashi et al., 2007; Joo et al., 2008; Yoo et al., 2008). Emerging evidence has also indicated a novel function of MPK6 in plant growth and development. For example, MPK6 and MPK3 are required for specification of cell fate during stomatal development, the formation of anther lobes and differentiation of anther cells, and development of ovules (Wang et al., 2007; Hord et al., 2008; Wang et al., 2008). Several substrates of MPK6 have been identified by biochemical and genetic analyses in Arabidopsis; these include the rate-limiting enzyme of ethylene biosynthesis, 1-aminocyclopropane-1-carboxylic acid synthase (ACS) (Liu and Zhang, 2004), the transcriptional regulators ethylene-insensitive 3 (Yoo et al., 2008) and ethylene response factor 104 (Bethke et al., 2009), and PHOS32 (a 32-kD phosphorylated protein) (Merkouropoulos et al., 2008). Furthermore, 39 proteins were isolated as potential substrates of MPK6 from protein microarrays that included 1690 Arabidopsis proteins, which indicates that MPK6 acts as a universal regulator in plant stress responses, as well as during growth and development (Feilner et al., 2005).
Previous findings have indicated that NO can activate MAPK cascades; for example, it activates salicylic acid–induced protein kinase (SIPK) in tobacco (Nicotiana tabacum) and a 46-kD MAPK in maize (Zea mays) and Arabidopsis (Clarke et al., 2000; Kumar and Klessig, 2000; Zhang et al., 2007a). Surprisingly, recent evidence also indicates that MAPKs play a critical role in regulating the production of NO (Asai et al., 2008). In tobacco, the elicitor INF-1, produced by Phytophthora infestans, can induce the activation of MAP or ERK kinases (MEK) MEK1 and MEK2 and also enhances the production of NO (Yamamoto et al., 2004). Constitutive activation of MEK2 enhances NO synthesis dramatically. Furthermore, silencing of SIPK and the SIPK homolog, NTF4, a MAPK that is activated by MEK2 in tobacco, markedly reduces the production of NO. Synthesis of NO that is induced by INF1 and Solanum tuberosum (St) MEK2DD is decreased significantly in plants in which NOA1 is silenced, which suggests that NOA1 is involved in the process (Asai et al., 2008). However, tungstate, an inhibitor of NR, can also suppress the production of NO that is induced by INF1 and St MEK2DD, which suggests that NR also participates in the NO burst. Although the regulation of NO synthesis might involve the posttranscriptional modification of NR, details of the mechanism remain unclear.
In the study reported here, we investigated the roles of the MAPK cascade in the production of NO induced by H2O2 in Arabidopsis. We found that MPK6 was involved in regulating H2O2-induced synthesis of NO. We identified a direct interaction between MPK6 and NIA2 in vitro and in vivo and demonstrated that MPK6 could phosphorylate NIA2, with Ser-627 being the putative phosphorylation site of NIA2. Phosphorylation of NIA2 by MPK6 not only increased the activity of NIA2 and the production of NO dramatically, but also led to morphological changes in the root system of Arabidopsis. Our findings suggest a novel model for the involvement of MPK6 in NO synthesis and signal transduction during root development.
RESULTS
mpk6 Mutants Are Defective in H2O2-Induced NO Generation
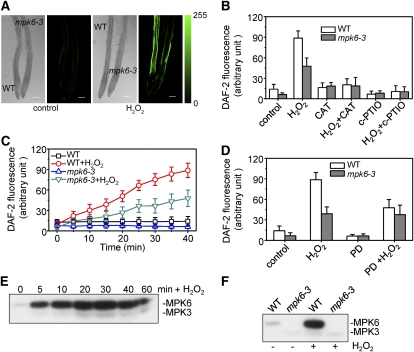
A recent study has indicated that MAPK signaling regulates the production of NO and NADPH oxidase–dependent oxidative bursts in tobacco (Asai et al., 2008). In Arabidopsis, MPK6 is the functional ortholog of the tobacco protein SIPK (Tena et al., 2001; Ichimura et al., 2002), and MPK6 can be activated by oxidative stress (Desikan et al., 2001; Yuasa et al., 2001). To investigate the role of MPK6 in NO synthesis, we measured the H2O2-induced production of NO in the roots of _MPK6-_deficient mutants using the NO-specific fluorescent indicator diaminofluorescein diacetate (DAF-2 DA). DAF-2 DA is membrane permeable and is deacetylated by intracellular esterases to nonfluorescent 4,5-diaminofluorescein (DAF-2). However, in the presence of NO and O2, DAF-2 is converted to the fluorescent triazole derivative DAF-2 T. This increases the quantum yield of fluorescence >180-fold, which allows the measurement of NO levels in the cell cytoplasm (Kojima et al., 1998). The roots of seedlings were loaded with DAF-2 DA, and changes in fluorescence induced by NO were examined by confocal laser scanning microscopy. Exogenous application of H2O2 enhanced the relative fluorescent intensity of DAF-2 in root cells of both mpk6-3 and wild-type seedlings; the relative fluorescent intensity corresponds to the intensity for each pixel averaged over a whole root (Figure 1A). When catalase (CAT), a widely used H2O2 scavenger, was applied alone or together with 10 μM H2O2, H2O2-induced fluorescence was almost completely abolished (Figure 1B). Furthermore, application of 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (c-PTIO), which is a specific scavenger of NO, eliminated the DAF-2 fluorescence induced by H2O2 in the wild type and mpk6-3 (Figure 1B). These results suggest that DAF-2 fluorescence intensity reflects the concentration of NO in the roots.
Figure 1.
mpk6 Mutants Produce Less Nitric Oxide than Wild-Type Plants in Response to H2O2.
(A) Images of H2O2-induced NO production in wild-type and mpk6-3 plants. Roots from wild-type and mpk6-3 mutants were loaded with DAF-2 DA, and NO synthesis was monitored after the addition of 10 μM H2O2 or a water control. The micrographs show pairs of representative bright-field and fluorescence images of the roots of wild-type and mpk6-3 plants in six independent experiments. Bar = 100 μm.
(B) Effects of CAT and c-PTIO on DAF-2 fluorescence in wild-type and mpk6-3 plants in the presence and absence of H2O2. Control represents wild-type and mutants treated with water. Average pixel intensities of DAF-2 DA fluorescence were calculated after addition of CAT or c-PTIO for 30 min. Values are means (±sd) from whole root regions (from maturation zone to meristematic zone) from six independent experiments.
(C) Time course of NO production expressed as the pixel intensities of DAF-2 DA fluorescence in the roots of wild-type and mpk6-3 plants before and after addition of H2O2. Wild-type and mpk6-3 plants were treated with 10 μM H2O2 for the times indicated. Signal intensities were quantified over whole roots from micrographs taken under confocal laser scanning microscopy. Values are means (±sd) from six independent experiments.
(D) Effects of PD 098059 on NO production in wild-type and mpk6-3 plants in the presence and absence of H2O2. Average pixel intensities of DAF-2 DA fluorescence were calculated after addition of PD 098059, H2O2, or water control for 30 min. Values are means (±sd) from whole root regions from six independent experiments.
(E) H2O2-induced activation of MPK6 and MPK3 in vivo. Proteins were extracted from wild-type seedlings treated with 2 mM H2O2 for the time indicated. In-gel kinase assays were performed as described in Methods, and MBP was used as the substrate.
(F) Mutation of mpk6 abolishes H2O2-activated MPK6. The wild-type and mpk6-3 plants were treated with 2 mM H2O2 for 20 min, and the activity of MPK6 was detected by in-gel kinase assay using MBP as the substrate.
As shown in Figure 1, NO was synthesized more rapidly after H2O2 treatment in wild-type than in mpk6-3 roots (Figures 1A and 1C, significantly different according to the Steel-Dwass test, P ≤ 0.05). The effect of H2O2 on promotion of NO production in a time-dependent manner (Figure 1C) was significant (P ≤ 0.05) at a concentration of H2O2 > 10 μM in roots of the wild type. The pixel intensity of root cells from mpk6-3 was 40% lower than that from the wild type after 20 min of treatment with H2O2 (Figures 1A and 1C, significantly different according to the Steel-Dwass test, P ≤ 0.05). In the presence of 0.5 to 2 mM H2O2, NO generation in the wild type and mpk6-3 mutants was further enhanced (see Supplemental Figure 1A online).
In mammalian systems, PD 098059 has been used widely as an inhibitor of specific MAPKKs (and thus activation of MAPKs) to establish the physiological role of MAPKs in various types of cell (Alessi et al., 1995). When roots were preincubated with 50 μM PD 098059 for 30 min, the increase in NO production induced by H2O2 was suppressed by 46% in the wild type (P ≤ 0.05), whereas the fluorescence intensity in mpk6-3 was almost unaffected by treatment with PD 098059 (P ≤ 0.05) (Figure 1D). NO production of the roots in the wild type and mpk6-3 treated with PD 098059 alone was almost equal to that of untreated controls. The data suggest that the H2O2-mediated NO generation is dependent on the activation of MPK6.
Encouraged by the above results, we assayed the MPK6 kinase activity induced by H2O2 by measuring the levels of phosphorylation in wild-type and mpk6-3 mutant plants that were exposed to different concentrations of exogenous H2O2. Exogenous application of H2O2 promoted the kinase activity of MPK6 in a dose-dependent manner (see Supplemental Figure 1B online). The obvious promotion of MPK kinase activity was observed at 30 min after treatment with 10 μM H2O2 (see Supplemental Figure 1B online). Under the same conditions, the NO production was significantly increased (Figure 1C). Application of 2 mM H2O2 resulted in the strongest activation of MPK6 (Figure 1E; see Supplemental Figure 1B online). Consistent with the above results, application of H2O2 to wild-type plants stimulated MPK6 activity significantly in a time-dependent manner (Figure 1E). The activation of MPK6 by H2O2 was rapid, with the maximum level of phosphorylation occurring 30 min after the addition of H2O2 (Figure 1E). Activation of MPK6 was still detectable at 60 min. In contrast with MPK6 activity, MPK3 activity was weak and relatively unstimulated by H2O2 under the same experimental conditions (Figure 1E).
Rapid activation of MPK6 by H2O2 was abolished completely in the mpk6-3 mutant (Figure 1F; see Supplemental Figure 1B online). These data show that regulation of H2O2-induced NO production is disrupted in mpk6-3, indicating that active MPK6 is required for H2O2-induced NO synthesis in Arabidopsis.
NIA2 Is Required for the MPK6-Mediated Increase in NO Production Induced by H2O2
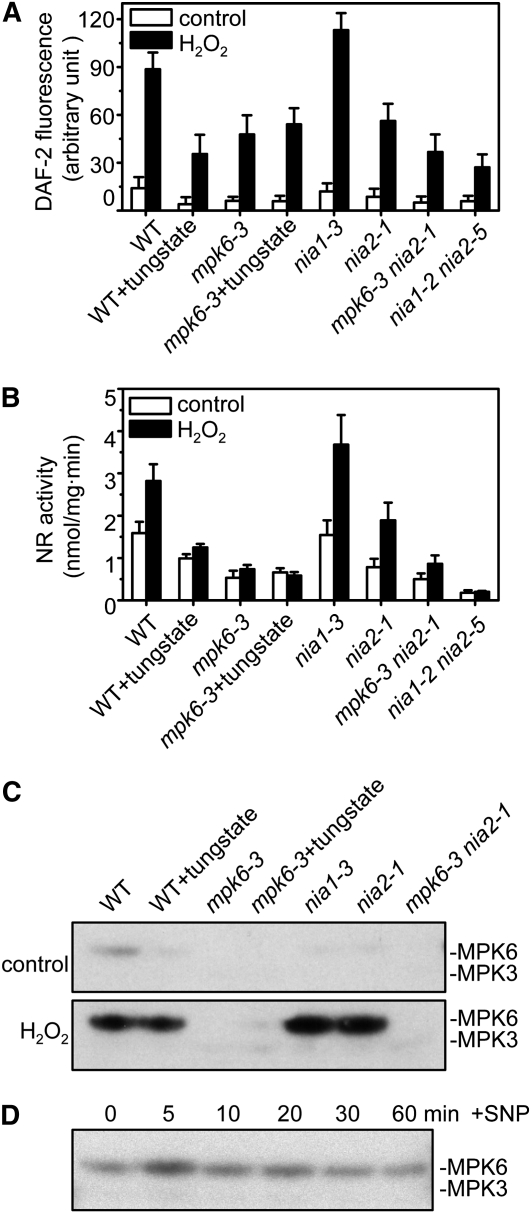
As mentioned above, NOS and NR are the two potential enzymatic sources of NO in planta (Desikan et al., 2002; Guo et al., 2003). However, the identity of Arabidopsis NOA1 as an authentic NOS has been questioned recently (Moreau et al., 2008). To determine whether the decrease in H2O2-induced NO production in mpk6-3 was due to a change in NR activity, NO production was measured after treatment with tungstate (an inhibitor of NR). Tungstate treatment resulted in a 61% reduction in H2O2-induced NO production in wild-type plants (Figure 2A, black column 1 versus black column 2, significantly different according to the Steel-Dwass test, P ≤ 0.05). The amount of NO produced in response to H2O2 in the kinase-deficient mutant mpk6-3 was similar to that produced in wild-type plants pretreated with tungstate (black column 2 versus black column 3). These data suggest that MPK6 is involved in NO synthesis induced by H2O2 through the action of NR.
Figure 2.
Involvement of NIA2 in MPK6-Modulated NO Biosynthesis in Response to H2O2.
(A) NO production in the wild type, mpk6-3, and nia single or mpk6-3 nia2-1 and nia1-2 nia2-5 double mutants. Average pixel intensities of DAF-2 DA fluorescence were measured at 30 min after exposure to exogenous 10 μM H2O2 or a water control. For tungstate treatment, seedlings were loaded with 200 μM tungstate for 30 min before addition of H2O2. Values are means (±sd) from whole root regions from six independent experiments.
(B) H2O2-induced increase in NR activity in the wild type, mpk6-3, and nia single or mpk6-3 nia2-1 and nia1-2 nia2-5 double mutants. Arabidopsis plants were treated with 10 μM H2O2 or a water control for 30 min. Each value represents the mean (±sd) of three independent experiments.
(C) Effects of H2O2 on MPK6 phosphorylation in wild-type, mpk6-3, nia, and mpk6-3 nia2-1 mutants. The wild-type and mutant plants were treated with 2 mM H2O2 or a water control for 20 min. For tungstate treatment, seedlings were loaded with 200 μM tungstate for 30 min before addition of H2O2. The activity of MPK6 was detected by in-gel kinase assay using MBP as the substrate.
(D) Effects of the NO donor SNP on MPK6 phosphorylation. Proteins were extracted from wild-type seedlings treated with 20 μM SNP at the indicated times. In-gel kinase assays were performed with MBP as the substrate.
Given that NR is encoded by two genes, NIA1 and NIA2, in Arabidopsis (Campbell, 1999), deficiency of both NIA1 and NIA2 results in significant reduction in NO generation (Bright et al., 2006; Modolo et al., 2006). To test which gene functions in MPK6-mediated NO synthesis, the T-DNA insertion mutants nia1-3, nia1-4, and nia2-1, nia2-2 and the double mutant mpk6-3 nia2-1, mpk6-3 nia2-2 were used for measurements of NR activity and NO production. Since similar results were obtained with these double mutants, we presented the data only from the analysis of nia1-2, nia2-1, and nia2-1 mpk6-3. Real-time RT-PCR analysis indicated that the genes NIA1, NIA2, and MPK6 were knocked out in these single and double mutants (see Supplemental Figure 2 and Supplemental Methods online). In the nia1 mutants, H2O2-induced production of NO was almost unaffected, perhaps even higher than that in the wild type (Figure 2A, black column 5 versus black column 1), whereas NO production was inhibited dramatically in the nia2 mutants (37.5%, P ≤ 0.05; black column 6 versus black column 1). In the nia1-2 nia2-5 double mutant, the level of NO production was even lower than in the single mutant and was almost equal to that in the wild-type plants treated with tungstate (Figure 2A, the last black column versus black column 2). These results suggest that NIA2 plays an important role in nitrite-dependent NO synthesis in response to exogenous H2O2 in roots.
Next, we measured the NR activity in wild-type, mpk6-3, and _NIA_-deficient plants. As mentioned above, tungstate treatment has been used to produce nonfunctional NR. In our working conditions, NR activity in wild-type and mpk6-3 mutants was almost inhibited for 30 min after 200 μM tungstate treatment (see Supplemental Figure 4 online; Figure 2B, columns 2 and 4). Furthermore, the activity of NR was rapidly enhanced in wild-type, nia1-3, and nia2-1 plants when exogenous H2O2 was applied (significantly different according to the Steel-Dwass test, P ≤ 0.05) (Figure 2B, the black column versus the white column). An increase in total NR activity of 177, 238, and 161% was observed in wild-type, nia1-3, and nia2-1 seedlings, respectively, in response to H2O2 treatment. By contrast, there was only a slight increase in total NR activity in mpk6-3 and mpk6-3 nia2-1 in response to treatment with H2O2. Regardless of the total NR activity or level of NO production, these increases were significantly lower than those observed in wild-type and nia1-3 plants. As mentioned above, in the mpk6-3 nia2-1 double mutant, the level of NO synthesis after exposure to H2O2 was very similar to that measured in the mpk6-3 and nia2-1 single mutants, which indicated that it is mainly the NIA2 isoform that is responsible for the production of NO in response to H2O2 in roots. The nia1-2 nia2-5 double mutant, which lacked both NIA genes, showed diminished NR activity (Figure 2B, last column), but NO production was still detectable, which indicated that NO might also be generated nonenzymatically or by an as yet unknown source in root cells (Crawford, 2006).
To confirm that NR activity and NO synthesis induced by H2O2 correlate with the activation of MPK6, we also determined the level of H2O2-induced MPK6 activity in wild-type and mpk6 and nia mutant plants. As shown in Figure 2C, H2O2 increased the level of MPK6 phosphorylation in wild-type plants, nia mutants, and wild-type plants treated with tungstate. The activation of MPK6 in response to H2O2 was abolished completely in the mpk6-3 and mpk6-3 nia2-1 mutants. Furthermore, after treatment with the NO donor sodium nitroprusside (SNP) for the times indicated (5 to 60 min), slight activation of MPK6 was observed by in-gel kinase assay (Figure 2D), indicating that there might be a feedback loop regulating MPK6 activity. The H2O2-mediated MPK6 activation correlated well with NO synthesis and NR activity, but the expression of NIA2 was not affected (see Supplemental Figure 3 online), which suggests that MPK6 is involved in the NR-mediated production of NO via posttranscriptional modulation.
MPK6 Interacts with NIA2
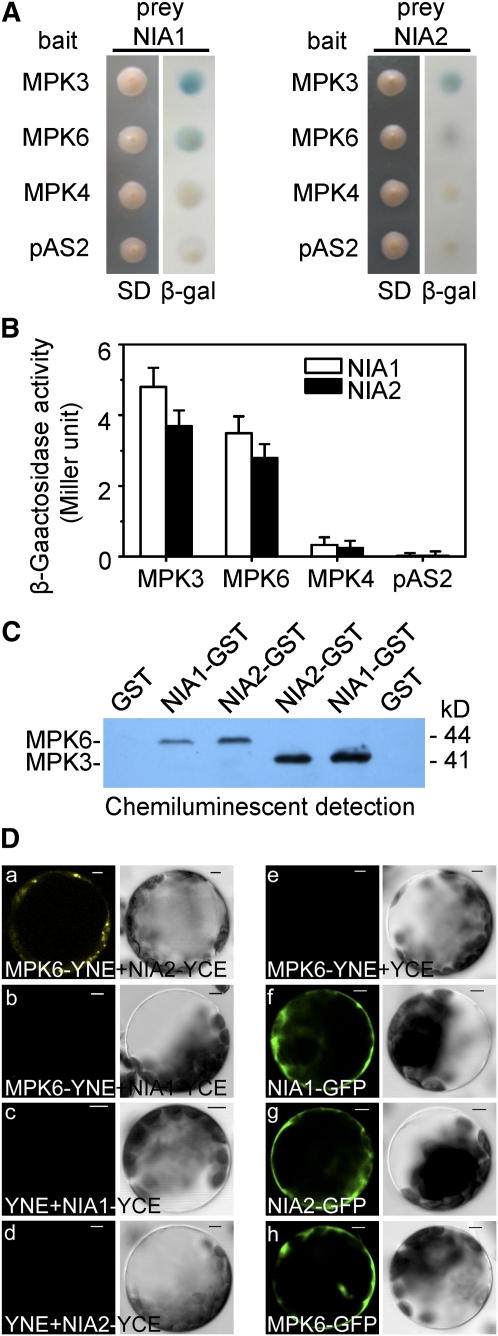
After we established that MPK6 was involved in the regulation of H2O2-induced NO synthesis catalyzed by NIA2, we investigated whether MPK6 interacted with NIA in vitro or in vivo. Cotransformation of yeast Y190 with these constructs (pAS-MPK6 or pAS-MPK3 with pACT-NIA1 or -NIA2) yielded His/Trp/Leu auxotrophs that were also positive for β-galactosidase expression (Figure 3A), which demonstrated that MPK6 and MPK3 interacted physically with NIA1 and NIA2. In addition, cotransformation of a construct that expressed the control bait protein MPK4 with pACT-NIA1 or NIA2 did not result in detectable β-galactosidase activity. These results show that MPK6 and MPK3 interact with both NIA1 and NIA2 in the yeast two-hybrid system (Figures 3A and 3B).
Figure 3.
MPK6 Interacts with NIA2 in Vitro and in Vivo.
(A) MPK6 and MPK3 interacted with NIA1 and NIA2 in the yeast two-hybrid system. Yeast strains that contained pAS-MPK6 or pAS-MPK3 as bait and pACT-NIA1/2 as prey were grown for 48 h on synthetic defined (SD) medium that lacked Trp and Leu (left panel) and were assayed for LacZ expression by a filter-lift assay for β-galactosidase activity (β-gal; right panel). pAS-MPK4 and the empty bait vector were used as negative controls. A blue color indicates interaction. β-gal, β-galactosidase activity.
(B) Quantitative analysis of β-galactosidase activity of the yeast strains in liquid culture showing the interaction between MPK3, MPK6, and MPK4 and NIA1 or NIA2. Values are means of data from three independent experiments. Error bars indicate sd.
(C) MPK3 and MPK6 proteins labeled with biotinylated-Lys were pulled down by GST-NIA1 and GST-NIA2 but not by GST.
(D) NIA2, but not NIA1, interacted with MPK6 in vivo as determined by BiFC. (a) The YFP signal in the cytoplasm indicates a positive interaction between MPK6 and NIA2; (b) no YFP signal was detected in protoplasts cotransformed with MPK6-YNE and NIA1-YCE; (c) and (d) no YFP signal was detected in protoplasts cotransformed with SPYNE and NIA2-YCE or NIA1-YCE; (e) no YFP signal was detected in protoplasts cotransformed with MPK6-YNE and SPYCE; (f) to (h) localization of GFP signals from NIA1, NIA2, and MPK6 fused with GFP. Left panels, fluorescence images under confocal microscopy; right panels, bright-field images of the cells. Bars = 5 μm.
To confirm the interaction between MPK6 and NIA1/NIA2, we also performed a glutathione _S_-transferase (GST) pull-down assay. MPK6 protein labeled with biotinylated Lys was incubated together with GST-NIA1 or GST-NIA2 fusion protein, using GST alone as the negative control. As shown in Figure 3C, NIA1 and NIA2 could bind to MPK6 and MPK3, but GST could not. The binding of NIA1 to MPK6 and NIA2 to MPK3 was weaker than that of NIA2 and NIA1, respectively (Figure 3C).
To determine whether MPK6 and NIA2 interacted in vivo, we monitored the association of transiently expressed MPK6 and NIA2 in protoplasts of Arabidopsis leaves using bimolecular fluorescence complementation (BiFC) (Walter et al., 2004). To carry out BiFC, the coding region of yellow fluorescent protein (YFP) was split between two vectors: the N-terminal 155 amino acids were encoded by pSPYNE, and the C-terminal 84 amino acids were encoded by pSPYCE. Fusion of these two segments to two separate proteins that interact resulted in the reconstitution of YFP fluorescence when the proteins were coexpressed. MPK6 and NIA1 or NIA2 were cloned into pSPYNE and pSPYCE to give constructs that encoded the proteins MPK6-YNE and NIA1-YCE or NIA2-YCE, respectively. MPK6-YNE and NIA2-YCE were coexpressed in protoplasts of Arabidopsis leaves. As expected, we observed reconstituted fluorescence in the cytoplasm (Figure 3D, a). However, coexpression of MPK6-YNE and NIA1-YCE did not reconstitute fluorescence (Figure 3D, b). In addition, no YFP signal was detected with the combinations of MPK6-YNE and pSPYCE or NIA1-YCE/NIA2-YCE and pSPYNE (Figure 3D, c to e). Consistent with the localization of the interaction signal, confocal microscopy of protoplasts transformed with MPK6, NIA2, or NIA1 fused with green fluorescent protein (GFP) showed that the MPK6, NIA1, and NIA2 fusion proteins were also localized in the cytoplasm. These results confirmed that MPK6 and NIA2 can interact in planta.
Interaction of MPK6 and NIA2 Requires Hinge 2 and FAD Domains
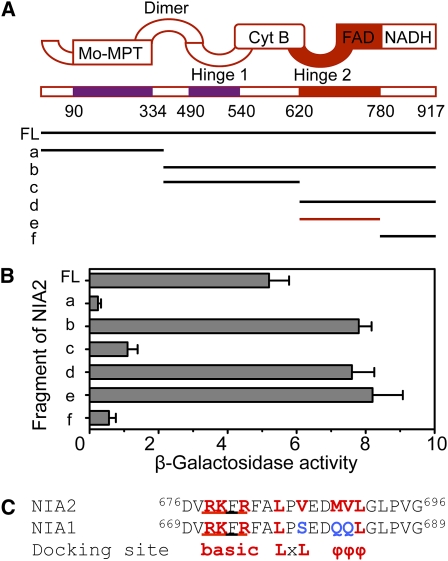
To identify the domains of NIA2 that interacted with MPK6, seven fragments that covered various regions of NIA2 were tested for their ability to interact with MPK6 (Figure 4A). The C-terminal region (amino acids 621 to 917), which contained hinge 2, FAD, and NADH binding domains, could interact with MPK6, but the N-terminal region (amino acids 1 to 334), which contained an Mo-molybdopterin domain, and the middle region (amino acids 335 to 620), which contained dimer, hinge 1, and cytochrome b domains, could not (Figure 4B). Furthermore, deletion of the NADH binding domain (amino acids 781 to 917) did not affect the interaction between C-terminal regions with MPK6 protein, which suggested that the hinge 2 and FAD domains (amino acids 621 to 780) were required for recognition of and interaction with MPK6.
Figure 4.
MPK6 Interacts with the Hinge 2 and FAD Domains of NIA2.
(A) Schematic representation of seven fragments that covered various regions of the NIA2 protein and were used for yeast two-hybrid assays. FL, full-length NIA2 protein (amino acids 1 to 917). (a) N-terminal region and Mo-molybdopterin domain (amino acids 1 to 334); (b) all regions except those contained in fragment a (amino acids 335 to 917); (c) dimer, hinge 1, and cytochrome b domains (amino acids 335 to 620); (d) hinge 2, FAD, and NADH binding domains (amino acids 621 to 917); (e) hinge 2 and FAD domains (amino acids 621 to 780); (f) NADH binding domain (amino acids 781 to 917).
(B) Quantitative analysis of β-galactosidase activity of the yeast strains in liquid culture showing the interaction between MPK6 and the seven fragments of NIA2. Values are means (±sd) of data from three independent experiments. The results show that fragment e is required for interaction between MPK6 and NIA2.
(C) Sequence of the putative docking domain in NIA1 and NIA2. The first amino acid in each sequence used for alignment is indicated, and conserved residues are highlighted (red). A triplet of conserved hydrophobic residues (MVL) in NIA2 corresponded to QQL in NIA1 (blue). f represents a hydrophobic amino acid.
It is well known that two regions within the substrate are involved in recognition and binding by MAPK: docking sequences and the phosphorylation site (Sharrocks et al., 2000). Generally, sequence analysis indicated that the docking sequence contains a region rich in basic amino acids, an LxL motif, and sometimes a triplet of hydrophobic residues (Jacobs et al., 1999; Sharrocks et al., 2000). The hinge 2 and FAD binding domain of NIA2 contain a typical RKFRFALPVEDMVLGL sequence (Figure 4C), which is similar to the MAPK docking sequence in mammals and plants (Kiegerl et al., 2000; Sharrocks et al., 2000). Four potential sites of phosphorylation by MAPK (Ser-627, Thr-649, Thr-715, and Thr-799), where Ser or Thr residues are followed by Pro, are located in this region, which indicates that NIA2 could be a substrate of MPK6 in Arabidopsis. However, the triplet of conserved hydrophobic residues (MVL) of NIA2 corresponded to QQL in NIA1 at the C-terminal end of docking motif (Figure 4C), which is not a hydrophobic triplet. In addition, NIA1 lacks the obvious LxL motif. Perhaps these differences may determine that NIA2, not NIA1, interacts with MPK6 in vivo.
Phosphorylation of NIA2 by MPK6 Enhances NR Activity and NO Production
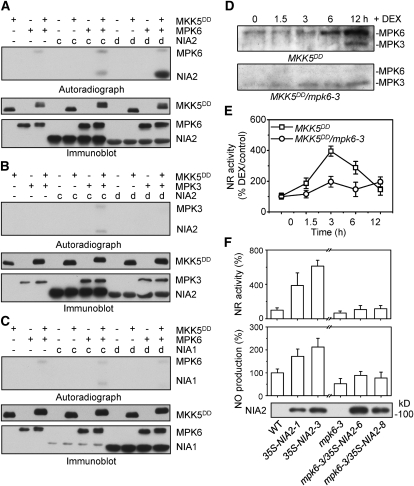
Previous studies have indicated that the activity of NR can be regulated by phosphorylation of a Ser residue in the hinge 1 region (Su et al., 1996; Kanamaru et al., 1999). On the basis of previous results and those presented above, we hypothesized that NIA2 might be a substrate of MPK6 and a target of the MAPK cascade. To test this hypothesis, we performed an in vitro phosphorylation assay. The recombinant His-tagged C-terminal fragments NIA2c (amino acids 335 to 620) and NIA2d (amino acids 621 to 917), as well as NIA1c (amino acids 338 to 623) and NIA1d (amino acids 624 to 917), were used as substrates. As shown in Figure 5A, recombinant MPK6 that was activated by MKK5DD, which is a constitutively active MAPKK, could phosphorylate NIA2d but not NIA2c. However, NIA1d and NIA1c could not be phosphorylated by activated MPK6 (Figure 5C). In addition, MPK3 could phosphorylate neither NIA2c nor NIA2d (Figure 5B). These data demonstrate that MPK6 is the kinase responsible for the phosphorylation of NIA2.
Figure 5.
Phosphorylation of NIA1 and NIA2 by MPK3 and MPK6.
(A) and (B) Activated MPK6 phosphorylated NIA2 in the C-terminal region, but activated MPK3 could not phosphorylate NIA2. Fragments c and d of NIA2 are described in Figure 4 and correspond to amino acids 335 to 620 and 621 to 917, respectively. Recombinant His-tagged MPK6 was activated and then used to phosphorylate His-tagged NIA2c and NIA2d in the presence of [γ-32P]ATP. After electrophoresis, the phosphorylated proteins were visualized by autoradiography. Reactions with various components omitted (−) were used as controls. Immunoblotting with anti-Flag and anti-His antibodies was performed to show the loading of Flag-tagged MKK5DD and His-tagged MPK6, NIA2c, and NIA2d, respectively.
(C) Activated MPK6 could not phosphorylate NIA1 in the C-terminal region. The assay was performed as described in (A).
(D) Treatment with DEX enhanced the phosphorylation of MPK6 in MKK5DD mutants. Protein extracts from MKK5DD and MKK5DD/mpk6-3 seedlings were treated with 2 μM DEX for the indicated times, and 10 μg protein extract was used for SDS-PAGE. Activated MPK6 and MPK3 were detected by immunoblotting using the phospho-p44/42 MAPK antibody.
(E) Phosphorylation of NIA2 by MPK6 increases NR activity. MKK5DD and MKK5DD/mpk6-3 seedlings were treated with DEX (2 mM). The NR activity in each mutant was determined at the indicated time point. NR activity was measured as described in Figure 2B. Each value represents the mean (±sd) of three independent experiments.
(F) MPK6 is required for full activation of NR and NO generation. The NR activity (top panel) and NO production (middle panel) in 35S-NIA2 and mpk6-3/35S-NIA2 transgenic plants were measured as described in Figure 2. Each value represents the mean (±sd) of three independent experiments. The NR activity and NO production in the wild type were set as 100%. Protein extract (10 μg) from 35S-NIA2 and mpk6-3/35S-NIA2 transgenic plants was separated by electrophoresis, and immunoblotting with anti-Flag antibody was performed to determine the amount of Flag-tagged NIA2 proteins (bottom panel).
To characterize further the MAPK pathway involved in the regulation of NR activity and NO synthesis, we measured NR activity in the conditional gain-of-function (dexamethasone [DEX]-inducible) MKK5DD transgenic plants and in mpk6-3/MKK5DD transgenic plants (Ren et al., 2008). Following DEX treatment, the constitutively active MKK5DD induced the activation of MPK6 in a time-dependent manner (Figure 5D, top panel), which resulted in a corresponding rapid increase in NR activity when compared with the same line treated with water as a control (Figure 5E). However, DEX could not stimulate MPK6 activation (Figure 5D, bottom panel) or NR activity (Figure 5E) in MKK5DD/mpk6-3 plants. In addition, NR activity was increased significantly (612%) in 35S-NIA2 transgenic plants, which overexpressed NIA2 (Figure 5F, column 3 versus column 1, P ≤ 0.05). Similar results were obtained with a second line that overexpressed NIA2 (Figure 5F, column 2). By contrast, even when NIA2 was overexpressed in an _MPK6_-deficient background, as shown by immunoblotting (Figure 5F, bottom panel), the NR activity only elevated slightly when compared with wild-type or mpk6 mutants (Figure 5F, top panel). Similar results were obtained for the NO production in these transgenic plants (Figure 5F, middle panel). Overexpression of _NIA2_-enhanced NO production and mutations in MPK6 prevented the increase of NO production, even in the background of NIA2 overexpression. These data suggest that the MKK5/MPK6 pathway mediates the activation of NR through phosphorylation of NIA2.
Phenotypic Characterization of the mpk6 Mutants Responsive to NO and H2O2
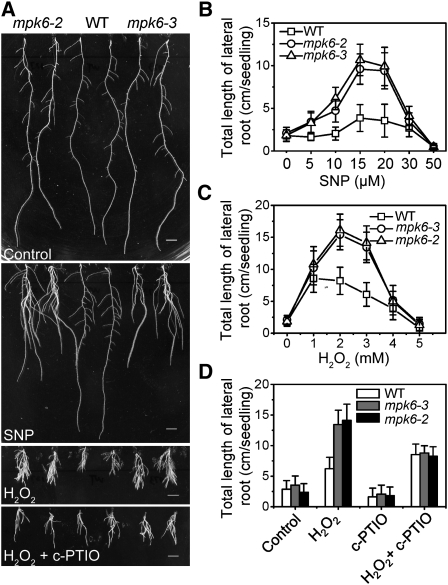
The results of several previous studies have suggested that NO affects lateral root development in a dose-dependent manner (Correa-Aragunde et al., 2004). In wild-type Arabidopsis plants, treatment with SNP enhanced the initiation and growth of lateral roots in a concentration-dependent manner (Figures 6A and 6B). At concentrations ≤20 μM SNP, mpk6-2 and mpk6-3 were more sensitive to SNP than the wild-type plants regarding promotion of lateral root growth. On average, the total length of lateral roots in the mutant plants was ~2.6-fold longer than that in the wild type (Figure 6B, significantly different according to the Steel-Dwass test, P ≤ 0.05). By contrast, in treatments with ≥20 μM SNP, the growth-promoting effects of SNP in lateral roots were gradually decreased. Compared with untreated seedling roots, treatment with 50 μM SNP significantly inhibited growth of lateral roots (P ≤ 0.05) (Figure 6B).
Figure 6.
Changes in Morphology of the Root System in Wild-Type and mpk6-3 Plants in Response to NO and H2O2.
(A) The number and length of lateral roots was greater in mpk6 mutant plants than in wild-type plants after treatment with SNP or H2O2. Five-day-old seedlings of the wild type, mpk6-2, and mpk6-3 were transferred to vertical MS plates containing (top to bottom): no additions (control); 15 μM SNP, 2.5 mM H2O2, or 2.5 mM H2O2 plus 0.5 mM c-PTIO. The photographs were taken at 10 d (top two panels) or 15 d (bottom two panels) after transfer. Bar = 0.5 cm.
(B) Average length of the lateral roots of all plants on MS medium supplemented with SNP at the indicated concentrations. Data represent means (±sd) of three independent experiments (~20 plants per point).
(C) Average length of the lateral roots of each type of plant on MS medium supplemented with H2O2 at the indicated concentrations. Data represent means (±sd) of three independent experiments (at least 30 plants).
(D) c-PTIO treatment abolished the H2O2-induced differences in lateral roots between the wild type and mpk6 mutant. Five-day-old seedlings of the wild type, mpk6-2, and mpk6-3 were transferred to MS plates (control) or vertical MS plates that contained 2.5 mM H2O2 and 0.5 mM c-PTIO. Data represent means (±sd) of three independent experiments (at least 30 plants).
Next, we examined carefully the phenotypes of the mpk6 mutant after treatment with exogenous H2O2. Under control conditions, mpk6 and wild-type plants showed similar rates of germination and growth. However, after 2 weeks of H2O2 treatment, mpk6-2 and mpk6-3 seedlings had produced more and longer lateral roots than wild-type seedlings (Figures 6A and 6C, significantly different according to the Steel-Dwass test, P ≤ 0.05). For example, with 2 mM H2O2, the mean length of lateral roots in the wild type was 8.21 ± 2.14 cm, whereas in mpk6-2 and mpk6-3, the mean lengths were 15.4 ± 2.35 and 16.1 ± 2.64 cm, respectively. The effects of H2O2 on lateral root elongation were dose dependent, and H2O2 concentrations of 4 mM or more inhibited lateral root initiation entirely in both wild-type and mpk6 mutant plants. These results reveal that lateral root development in mpk6-2 and mpk6-3 seedlings is more sensitive to H2O2 than that in wild-type seedlings.
To test whether the root phenotype of the mpk6 mutants resulted from a deficiency of NR activity, tungstate was used to inhibit the total NR activity in both wild-type and mpk6 plants. Application of tungstate totally abolished the difference in the lateral root phenotype between wild-type and mpk6 plants in response to H2O2 (see Supplemental Figure 5 online). Furthermore, application of the NO scavenger c-PTIO eliminated the inhibition by H2O2 partially and also abolished the lateral root phenotype of mpk6-2 and mpk6-3 (Figures 6A and 6E), which suggests that the lateral root phenotypes of mpk6 mutants are critically dependent on the concentration of NO.
Ser-627 of NIA2 Is a Putative Phosphorylation Site and Its Modifications Affect NR Activity and Lateral Root Development
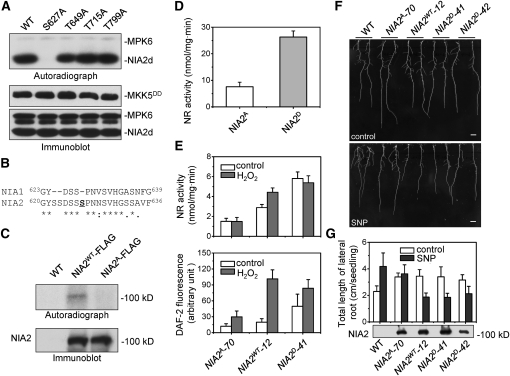
To identify which amino acid residue is required for MPK6 phosphorylation, recombinant NIA2d proteins were generated by mutating four putative Ser (S) or Thr (T) residues to Ala (A) by site-directed mutagenesis and were used as substrates for MPK6 phosphorylation assay. As shown in Figure 7A, MPK6 could not phosphorylate the S627A mutant, whereas T649A, T715A, and T799A mutants could still be phosphorylated. This result indicated that the Ser-627 residue in hinge 2 domain of NIA2 is the unique MPK6 phosphorylation site. Interestingly, NIA1 lacks several adjacent Ser residues in this region, which may be required for MPK6 recognization and phosphorylation (Figure 7B).
Figure 7.
Ser-627 of NIA2 Was a Putative Site for MPK6 Phosphorylation and Essential for the Efficient Activation of NR and Development of Lateral Roots.
(A) Ser-627 in the hinge 2 domain of NIA2 is the MPK6 phosphorylation site. Recombinant His-tagged MPK6 was activated and then used to phosphorylate His-tagged NIA2d protein with either the S627A, T649A, T715A, or the T799A mutation in the presence of [γ-32P]ATP. After electrophoresis, the phosphorylated proteins were visualized by autoradiography. Immunoblotting with anti-Flag and anti-His antibodies was performed to show the loading of Flag-tagged MKK5DD and His-tagged MPK6 and NIA2d, respectively.
(B) Sequence alignment shows three Ser residues adjacent to the phosphorylation site are missing in NIA1 protein. The first amino acid in each sequence used for alignment is indicated, and the phosphorylation site is underlined.
(C) Phosphorylation of native NIA2 protein by MPK6. Flag-tagged full-length NIA2WT and NIA2S627A protein were immunoprecipitated from DEX-treated NIA2WT and NIA2A transgenic plants and were used for the phosphorylation assay as described in (A). Only wild-type NIA2 protein could be phosphorylated by MPK6. Immunoblotting with anti-Flag antibodies was performed to show the loading of NIA2 protein.
(D) NR activity was stimulated by the phosphorylation of NIA2-Ser627. Five micrograms of purified NIA2A and NIA2D protein from P. pastoris was used for NR activity assay as described in Figure 2B. Each value represents the mean (±sd) of three independent experiments.
(E) Phosphorylation and substitution of Ser-627 of NIA2 affected NR activity and NO production. The NR activity (top panel) and NO production (bottom panel) in NIA2A, NIA2WT, and NIA2D transgenic plants treated with 10 μM H2O2 or a water control were measured as described in Figure 2B. Each value represents the mean (±sd) of three independent experiments.
(F) Photography of transgenic plants containing the modifications of Ser-627 of NIA2 on an MS agar plate (control, top) and an MS agar plate supplemented with 15 μM SNP (bottom). Five-day-old seedlings of the wild-type, NIA2A, NIA2WT, and NIA2D transgenic plants were transferred to the plate. The photographs were taken 10 d after the transfer. Bar = 0.5 cm.
(G) Average of the total length of lateral roots of wild-type and transgenic plants on MS medium (control) or MS supplemented with 15 μM SNP. Data represent means (±sd) of three independent experiments (~20 plants per point).
To confirm Ser-627 as the specific phosphorylation site in NIA2, Arabidopsis transgenic plants were generated by introducing recombinant FLAG-tagged full length NIA2WT and NIA2A, which has Ser-627 replaced by Ala. NIA2WT and NIA2A proteins were immunoprecipitated from these transgenic seedlings and served as MPK6 substrates in vitro phosphorylation assays. Although both mutants produced a complete NR protein, activated MPK6 could phosphorylate only NIA2WT, not NIA2A mutant protein (Figure 7C, top panel). For the nontransgenic plants, no FLAG-tagged protein was immunoprecipitated and no phosphorylated band was observed. Alternatively, the same results were obtained when MYC-tagged NIA2WT and NIA2A expressed in Pichia pastoris (see Supplemental Figure 6 online).
To determine whether phosphorylation of Ser-627 affects NR activity, Ser-627 was also replaced by Asp (D), creating NIA2D, which mimics NIA2 phosphorylated at Ser-627. Consistent with above results, the NR activity in the yeast cells containing NIA2D was increased by 3.5-fold compared with that of NIA2A (Figure 7D). Similarly, the phosphomimic NIA2D showed substantially greater NR activity in transgenic plants (Figure 7E, top panel). A 5.15-fold increase in NO production was observed in NIA2WT plants in response to H2O2 treatment. By contrast, there was only a slight increase in NO content in NIA2A plants (Figure 7E, bottom panel). This increase was significantly lower than that observed in NIA2D. By contrast, the much higher NR activity and NO production in NIA2D transgenic plants were observed even without H2O2 treatment.
To investigate whether modification of Ser-627 in NIA2 also has a function in planta, 5-d-old seedlings of overexpression lines harboring replacements of Ser-627 were transferred to Murashige and Skoog (MS) medium without or with 15 μM SNP. It was previously reported that lateral root growth in Arabidopsis was induced by SNP treatment (Correa-Aragunde et al., 2004). Under our experimental conditions, lateral roots of wild-type seedlings treated with 15 μM SNP for 7 d were markedly longer than those without SNP; their length increased from 2.29 ± 0.42 to 4.18 ± 1.01 cm (Figures 7F and 7G). Similarly, lateral root length in all overexpression lines of NIA2 (NIA2A, NIA2WT, and NIA2D) was increased to ~37.5 to 50.5% compared with the wild type. The increase in lateral root length in NIA2 overexpression lines is associated with the increased NO production and higher NR activity (Figures 5F and 7E). Surprisingly, following SNP treatment, both the number and length of lateral roots in NIA2D, the Ser-627 phosphomimic, were decreased compared with those of NIA2A plants (3.63 ± 0.68 cm in NIA2A versus 1.87 ± 0.31 cm in NIA2D) (Figures 7F and 7G, significantly different according to the Steel-Dwass test, P ≤ 0.05). This stands in contrast with wild-type plants in which significantly longer lateral roots were observed. This inhibited lateral root growth suggests an increased NO concentration in NIA2D seedlings, presumably due to their increased sensitivity to exogenous NO derived from SNP. These results suggest that Ser-627 of NIA2 is essential site for MPK6 phosphorylation.
DISCUSSION
In Arabidopsis, MPK6 is involved in a number of signaling pathways; it activates downstream partner proteins in response to various stresses and during various developmental processes (Ichimura et al., 2000; Nuhse et al., 2000; Yuasa et al., 2001). The biochemical and genetic data obtained in this study reveal a key function of MPK6 in NO biosynthesis and signal transduction during root development. The involvement of NO in lateral root development, described herein, is likely to be a result of the modulation of NIA2 activity by MPK6.
MPK6 Activity Appears to Be Redox Regulated in Root Cells
It has been reported that reactive oxygen species (ROS) act upstream of the MAPK cascade in plant development and stress responses (Lu et al., 2002; Yoshioka et al., 2003; Desikan et al., 2004; Zhang et al., 2006). We found that H2O2 significantly stimulated MPK6 activity in an in-gel kinase assay (Figures 1E and 1F). More and longer lateral roots were observed in mpk6 plants than in wild-type plants after treatment with H2O2 (Figure 6). These data indicate that the MPK6 cascade might be part of a redox-sensitive signaling pathway.
In HeLa cells treated with lysophosphatidic acid, a potential signaling molecule that stimulates cell proliferation, H2O2 mediates the activation of the epidermal growth factor receptor and MAPK (Cunnick et al., 1998). The kinase-activating role of H2O2 in these cells was demonstrated by blocking the production of H2O2 with diphenylene iodonium, an inhibitor of NADPH oxidase, by preventing the accumulation of H2O2 with _N-_acetylcysteine or CAT, or by introducing exogenous H2O2 (Cunnick et al., 1998; Greene et al., 2000; Rhee et al., 2000). In Arabidopsis, MPK3 and MPK6 have been shown to be activated by H2O2 via the activation of NPK1-related protein kinase 1, an MAPKKK (Kovtun et al., 2000). The observations that H2O2 strongly induces Arabidopsis NDP kinase 2 (NDPK2) and that NDPK2 interacts specifically with MPK3 as well as MPK6 (Moon et al., 2003) suggest that oxidative stress activates NDPK2, possibly by activating the MAPK cascade. Alternatively, changes in redox state can be detected by protein Tyr phosphatase 1, which can inactivate MPK6 (Gupta and Luan, 2003). Here, it is important to consider an important link between the phosphorylation- and redox-based signaling pathways. Many kinases have a conserved Cys pair in their active domain, and oxidation of this pair potentiates their activity (Sitia and Molteni, 2004). Thus, it is easy to envision how redox changes could affect many aspects of cell physiology. We expected that the MPK6 cascade might also be redox regulated. Indeed, we found that MPK6 was activated by H2O2 and that more NO accumulated in response to H2O2 (Figure 1). A feedback loop situation appears to exist whereby NO can activate the MAPKs, as has been reported previously (Clarke et al., 2000; Kumar and Klessig, 2000; Zhang et al., 2007a). In fact, NO transiently activated MPK6 in our working conditions (Figure 2D). The findings support the idea that changes in the redox state might be the molecular switch for activation of MPK6. A possible explanation for this is that a feedback loop regulates the redox state of cells to activate MAPK cascade.
NIA2 Is Responsible for the Production of NO in Root Cells in Response to H2O2
The genetic data have demonstrated that NR, which catalyzes the conversion of nitrite to NO in a reaction that depends on NADP(H), was the major source of NO in plants (Yamasaki and Sakihama, 2000; Rockel et al., 2002; Bright et al., 2006). In plant cells, the amount of NR is regulated at the transcriptional and posttranscriptional level; for example, the expression of Arabidopsis NIA1 and NIA2 is induced by light, nitrate, and sucrose (Cheng et al., 1992; Campbell, 1999). The activity of NR protein is also regulated at a posttranscriptional level; for example, Ser-534 in hinge 1 region of Arabidopsis NIA2 can be phosphorylated by a calcium-dependent protein kinase (Su et al., 1996). Deficiency of NIA1 had almost no affect on NR activity and NO synthesis in the roots (Figure 2B), whereas, in mpk6-3 and nia2-1 mutants, total NR activity and NO synthesis increased only slightly in response to H2O2 treatment. In the mpk6-3 nia2-1 double mutant, the level of NO synthesis was very similar to that in the mpk6-3 and nia2-1 single mutants following treatment with H2O2. These data imply strongly that NIA2 is responsible for the production of NO in root cells in response to exogenous H2O2.
Although our data showed that H2O2-induced NO production in roots resulted mainly from MPK6 activating NR by phosphorylation of Ser-627 in the hinge 2 region of NIA2, this does not rule out the alternative pathway to stimulate NO generation. The fact that NO production was not completely abolished by mutation of mpk6 or by addition of the MAPKK inhibitor to wild-type plants (Figures 1A and 1D) indeed implies that there is an alternative route to activate NO production. It is well documented that the regulatory Ser-534 phosphorylation site, occurring in a region of hinge 1 in the NR molecule (Campbell, 2001), is important for binding of a 14-3-3 protein, leading to a catalytically inactive complex (NR+pSer534+14-3-3) that modulates NR activity in response to light and other environmental signals (Su et al., 1996; Moorhead et al., 1996). A recent report indicated that oxidation of surface-exposed Met-538 in NR can inhibit the phosphorylation of nearby sites (Ser-534) (Hardin et al., 2009), suggesting that Met-538 oxidation may play a role as a redox switch in regulation of NR phosphorylation.
Guard cells of the nia1 nia2 double mutant do not generate NO in response to H2O2, and stomatal apertures in nia1 nia2 leaves treated with H2O2 do not differ from those in untreated controls (Bright et al., 2006), suggesting that NO production via NR is required for H2O2-mediated stomatal closure. However, previous data have indicated that the nia1 dissociation insertion mutant (nia1::Ds nia1::DS) (Parinov et al., 1999; Wang et al., 2004) appears to be deficient in ABA and H2O2-induced stomatal closure and NO generation, whereas the nia2 deletion mutant responds normally to ABA and H2O2 (Bright et al., 2006). The simplest explanation is that NIA1 might mediate ABA- or H2O2-activated NO synthesis in guard cells.
Given that NIA1 and NIA2 are both expressed in guard cells, it is likely that protein–protein interactions and differential activities of these proteins are important for their different roles. Indeed, we found a strong interaction between MPK6 and NIA2, but not MPK6 and NIA1, in vivo (Figure 3D). In Arabidopsis, NIA2 represents ~90% of the total NR activity (Wilkinson and Crawford, 1991), and NIA2 is expressed mainly in roots rather than leaves (Cheng et al., 1991). Thus, although NIA1 is required for effective ABA signal transduction in guard cells during stomatal closure (Bright et al., 2006), our data suggest that NIA2 is the main target of MPK6 with respect to NO biosynthesis in root cells. The fact that significant NR activity was observed in nia2 mutants (Figure 2B) might be because whole seedlings were used in the measurement. NIA1 and NIA2 have similar amino acid sequences, but there are distinct regions that differ between the two proteins and that influence the protein partners with which they interact with and their activation characteristics. In addition, NIA1 and NIA2 are expressed in different tissues. These differences might explain why NIA1 and NIA2 have distinct signal transduction pathways and roles in N assimilation.
NIA2 Is the Target for the MKK5-MPK6 Module in Root Cells
Complete MAPK cascades that function in innate immunity, ethylene signaling, and stomatal development in plants have been elucidated (Asai et al., 2002; Ouaked et al., 2003; Bergmann et al., 2004; Wang et al., 2007). All these MAPK cascades involve MPK6 and/or MPK3, which indicates that the MPK3/MPK6 module has important multifunctional abilities. It has been well documented that ROS act upstream of MAPK cascades and that H2O2 induces the synthesis of NO (Lu et al., 2002; Yoshioka et al., 2003; Desikan et al., 2004; Bright et al., 2006; Zhang et al., 2006). We extend these observations to establish that H2O2 and NO signaling are linked through the activation of MPK6.
We found that MPK6 interacted strongly with NIA2, and NR activity was increased by H2O2 treatment in a manner dependent on MPK6 (Figure 2C). We demonstrated that NIA2, not NIA1, was the substrate of MPK6 (Figure 5) and that the S627A mutant of NIA2 failed to be phosphorylated by MPK6 (Figure 7C; see Supplemental Figure 6 online). Intriguingly, phosphorylation of NIA2 also dramatically increased NR activity and NO production in root cells (Figure 7E). These results clearly demonstrate that NIA2 is the substrate of MPK6, and phosphorylation of NIA2 is required for H2O2-induced NO production. As mentioned above, NR activity is known to be regulated by reversible phosphorylation (Su et al., 1996; Campbell, 1999). This reversible mechanism allows NR activity to be modulated more rapidly than can be achieved by protein degradation or synthesis (Campbell, 2001; Kaiser and Huber, 2001). Thus, regulation of NR by the MPK6-mediated cascade may play a key role in the posttranslational regulation of nitrate reduction and NO production, but more detailed studies are needed to establish the mechanisms involved.
Our analysis of the role of MPK6 activation in response to H2O2 helps us to position H2O2 and NO in the signaling scheme for specific responses. Although it is well established that H2O2 induces the synthesis and accumulation of NO, it also has been suggested that NO modulates H2O2 levels in the guard cells of Vicia faba (She et al., 2004; He et al., 2005). However, NO does not appear to induce the production of H2O2 in Arabidopsis guard cells (Bright et al., 2006) or in maize mesophyll cells (Zhang et al., 2007a). In addition, we did not observe this NO-induced H2O2 production in V. faba guard cells (Lü et al., 2005). Thus, the function and relationship between NO and H2O2 remain topics of debate. Here, we found that H2O2 induced the rapid activation of MPK6 (Figures 1C and 1D), which resulted in increase in NR activity and NO production.
NO has long been defined as a positive regulator in lateral root development when applied exogenously (Pagnussat et al., 2002; Correa-Aragunde et al., 2004). Surprisingly, unlike the NO promotion effects seen in wild-type plants, NIA2D transgenic plants showed the growth retardation phenotypes of lateral root in response to exogenously applied NO (Figures 7F and 7G). One possibility is that there is a bell-shaped curve of dose response for NO, similar to that for cytokinin in root growth (Werner et al., 2001). Indeed, bell-shaped NO response curve for lateral root growth in mpk6 mutants was observed (Figure 6B). Because the level of endogenous NO might be nearly optimal for growth in NIA2 overexpression lines, supplying the NIA2D plants with exogenous NO (15 μM SNP) may cause overly high concentrations of NO that may be inhibitory. Similarly, without SNP treatment, lateral root growth in all overexpression lines of NIA2 (NIA2A, NIA2WT, and NIA2D) was strongly stimulated compared with the wild type (Figures 7F and 7G). These data are consistent with the previous observations (Pagnussat et al., 2002; Correa-Aragunde et al., 2004). By contrast, we did not find the differences in lateral root growth between wild-type and transgenic plants after application of H2O2 (see Supplemental Figure 7 online), probably due to the consistent activation of NR in the overexpression lines. Together, these data strongly support this notion that lateral root growth may require a minimum concentration of NO to promote growth but that root growth is inhibited in NIA2D by SNP concentrations that promote elongation in the wild type.
Similarly, the observation that mpk6 mutants have longer lateral roots in response to H2O2 compared with the wild type seems contradictory to the conclusion that mpk6 mutants have reduced NR and NO synthesis (Figure 6C), which is not the case. As mentioned above, a threshold concentration of NO must be reached to initiate the growth response. Beyond the optimum concentration, NO becomes inhibitory. By this logic, the NO concentration in mpk6 mutants should be below optimal for lateral root growth, since low concentrations of SNP caused a stimulation of lateral root growth (Figure 6B). Consistent with this, external nitrate stimulates the elongation of lateral roots (Zhang et al., 1999, 2007b; Linkohr et al., 2002). In the case of mpk6 mutants, lateral root growth appears to be mediated in part by nitrate. On the other hand, NO levels in the wild type after application H2O2 is ~2-fold higher than that in mpk6 mutants (Figures 1B and 2A). Thus, accumulation of supraoptimal NO levels was caused not only through activation of MPK6 by H2O2 but also through the positive feedback loop of NO (Figure 2D). However, the precise threshold that NO stimulates or inhibits lateral root growth and its exact mechanisms require further investigation.
Another possibility is that the retardation of lateral root growth in the wild type is attributed mainly to the cell death and cell damage under H2O2 treatment. Previous reports indicated that ethylene biosynthesis was positively regulated by MPK6 through rate-limiting ACC synthase isoforms, ACS2 and ACS6 (Liu and Zhang, 2004; Joo et al., 2008). Exogenous ethylene increased superoxide anion (O2–)-dependent cell death, whereas impairment of ethylene perception and plant development, including smaller, more erect rosettes, altered leaf shape, and short roots in ozone oversensitive radical-induced cell death 1 (rcd1) (Overmyer et al., 2000; Ahlfors et al., 2004). Early accumulation of ethylene stimulates spreading of cell death (Tuominen et al., 2004). On the other hand, it has been reported in tobacco plants that NO and peroxynitrite inhibit the activities of CAT and ascorbate peroxidase, the two major H2O2-scavenging enzymes of plants (Clark et al., 2000). This breakdown of the antioxidant system would lead to an overproduction of H2O2 and toxicity in the plant cells. Therefore, both ethylene and ROS seem to exert their effects by mediating cell death and toxicity to cells, resulting in a consequently stronger growth retardation of lateral roots in wild-type plants.
Consistently, the evidence that c-PTIO obliterated the H2O2-induced difference in the morphology and number of lateral roots of root system between wild-type and mpk6 mutants suggests that the phenotype differences between wild-type and mpk6 mutants resulted from the NO generation. However, under these conditions, both wild-type and mpk6 mutants still display severe growth retardation phenotypes (Figures 6B and 6C). A major reason for the pleiotropic phenotypes of plants in response to H2O2 is the result of ABA, auxin, and ethylene signaling and/or an indirect effect of cell damage or photoinhibition (Rao et al., 1997; Meinhard et al., 2002; De Cnodder et al., 2005; Tarantino et al., 2005; Nakagami et al., 2006).
Previous evidence has demonstrated that mutation of MEKK1, the kinase upstream of MPK6 (Asai et al., 2002), results in fewer lateral roots and shorter root hairs in Arabidopsis (Nakagami et al., 2006) and that NR serves as a source of NO for the induction of lateral root development by indole-3-butyric acid (Lombardo et al., 2006; Kolbert et al., 2008). Therefore, it is likely that NO derived from MPK6-mediated NR activity is the main original source of NO that affects lateral root development. Together, these data give solid support to the hypothesis that MPK6 stimulates the NR-associated production of NO that regulates lateral root development. This also shows that an H2O2 signaling event lies upstream of NO biosynthesis. However, our results do not rule out the possibility that MPK6 upregulates NR activity indirectly through another mechanism that is as yet unidentified.
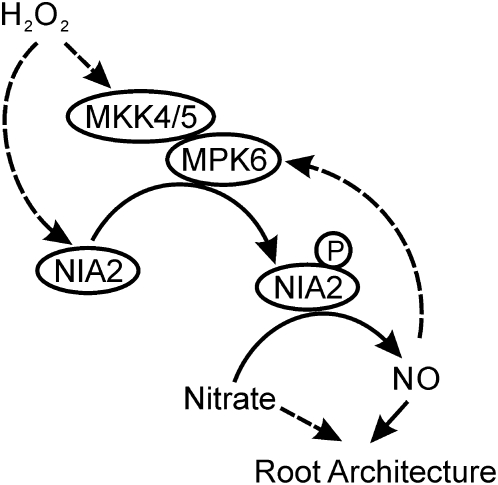
Therefore, we propose a pathway for the regulation of NO biosynthesis that involves the modulation of NIA2 by MPK6 (Figure 8). When the intracellular levels of H2O2 increase, MPK6 is activated, which in turn leads to the phosphorylation of NIA2 at Ser-627. Phosphorylation of NIA2 by MPK6 dramatically increases the activity of NIA2 and the production of NO and also results in morphological changes in the root system. Both from data reported previously (Clarke et al., 2000; Kumar and Klessig, 2000; Zhang et al., 2007a) and transiently activation of MPK6 by SNP, we infer that a feedback loop exists in which NO could activate the MAPKs. The alternative way for the regulation of NIA2 activity is that H2O2 could directly modify the surface-exposed Met-538 in NR and inhibit the phosphorylation of nearby sites (Ser-534) (Hardin et al., 2009). Our data suggest that the increase in H2O2-induced NO production that is dependent on the MAPK cascade could represent an amplification loop in NO signaling during root development.
Figure 8.
Model for the Putative Pathway of NO Biosynthesis and Signal Transduction Mediated by H2O2-Activated MPK6 in Arabidopsis.
The solid and dotted arrows indicate the positive regulation based on our data and the previous results, respectively. Please see the text for a detailed description of this model.
METHODS
Selection of T-DNA Insertion Mutants and Generation of Double Mutants
The T-DNA insertion lines for MAPK used in this study were mpk6-2 (Salk_073907) and mpk6-3 (Salk_127507), which were the same as those used previously (Liu and Zhang, 2004). The T-DNA insertion lines for NIA used in this study were nia1-3 (Salk_004164), nia1-4 (Salk_071547), nia2-1 (Salk_138297), and nia2-2 (Salk_088070). The double mutant of NIA used in this study was nia1-2 nia2-5 (Wang et al., 2004). The primers to check the T-DNA insertions were designed by the SIGnAL iSect tool (http://singal.salk.edu/tdnaprimers.2.html), and PCR was performed using genomic DNA from seedlings (see Supplemental Table 1 online). To generate double mutants, the alleles mpk6-2 and mpk6-3 were both crossed with nia2-1, creating nia2-1 mpk6-2 and nia2-1 mpk6-3. Because similar results were obtained with these double mutants, we present data only from the analysis of nia2-1 mpk6-3.
PCR
PCR amplifications were performed according to standard protocols in thermocyclers using Taq DNA polymerase or Pyrobest DNA polymerase (TaKaRa). For cDNA synthesis, total RNA from Arabidopsis thaliana seedlings was extracted by the TRIzol method, and reverse transcription was performed using 5 μg of total RNA and SuperScript II reverse transcriptase (Invitrogen). Full-length or fragment coding regions of NIA1, NIA2, MPK3, or MPK6 were amplified and inserted into the corresponding vector. The primers and vectors used for constructions are listed in Supplemental Table 1 online.
Transgenic Plants
The coding region of NIA2 was amplified from Arabidopsis ecotype Columbia-0 using primers that contained _Kpn_I and _Sal_I sites. PCR fragments were cloned together with an N-terminal Flag epitope into the vector pCAMBIA1205 under the control of the cauliflower mosaic virus 35S promoter. The constructs were introduced into Agrobacterium tumefaciens strain LBA4404 and transformed by floral infiltration into wild-type Arabidopsis and the mpk6-3 mutant.
To create transgenic plants mutated at Ser627, the wild-type and mutant coding regions of NIA2 were cloned into an N-terminal Flag-epitope vector pTA7002, which contains a DEX-inducible promoter (Ren et al., 2008).
Detection of Endogenous NO Production
Seedlings were incubated in the cell-permeable fluorescent probe DAF-2 DA (Calbiochem) at a concentration of 5 μM in incubation buffer (50 mM KCl and 10 mM MES-KOH, pH 7.2) for 30 min. For inhibitor assays, seedlings were preincubated with the NO scavenger c-PTIO or the hydrogen peroxide scavenger CAT. Roots were then washed three times with fresh buffer and examined by epifluorescence (DAF-2 DA excitation 488 nm and emission 500 to 560 nm) and bright-field microscopy using a Fluoview FV1000 microscope (Olympus). To extract quantitative data, pixel values were measured over root regions, which were located manually on confocal images and calculated using ImageJ 1.41 software (http://rsbweb.nih.gov/ij).
GST Pull-Down Assays
For GST pull-down assays, the coding regions of NIA1 and NIA2 were amplified by PCR with primers that contained appropriate restriction sites, and the amplified fragments were inserted into the plasmid pGEX-6P1 (Amersham Biosciences). The recombinant NIA1-GST and NIA2-GST fusion proteins were expressed using the pGEX-6P1-NIA1 and -NIA2 vectors. The coding regions of MPK6 and MPK3 were amplified by PCR with primers that contained appropriate restriction sites, and the amplified fragments were inserted into the plasmids pCITE-4a-c(+) (Novagen). MPK3 and MPK6 proteins that were biotinylated on Lys residues were produced from pCITE-MPK3 and pCITE-MPK6 using an in vitro transcription and translation assay kit (TNT Quick Coupled Transcription/Translation system; Promega) with incorporation and detection of Transcend Biotinylated Lys tRNA (Promega) according to the manufacturer’s instructions.
Yeast Two-Hybrid System
For yeast two-hybrid assays, the coding regions of MPK3, MPK6, and MPK4 were amplified by PCR with primers that contained appropriate restriction sites. The amplified fragments were inserted into the plasmid pAS2 (Clontech), which contains the GAL4 DNA binding domain, producing the constructs pAS-MPK3, pAS-MPK6, and pAS-MPK4, which encoded the bait constructs. The full-length coding regions and different fragments of NIA1 and NIA2 (Figure 4) were cloned in frame between corresponding restriction sites into the pACT2 vector, which contains the GAL4 activation domain, creating the prey plasmids pACT2-NIA1, pACT2-NIA2, pACT2-NIA2-a (amino acids 1 to 334), pACT2-NIA2-b (amino acids 335 to 917), pACT2-NIA2-c (amino acids 335 to 620), pACT2-NIA2-d (amino acids 621 to 917), pACT2-NIA2-e (amino acids 621 to 780), and pACT2-NIA2-f (amino acids 781 to 917). Yeast two-hybrid interaction assays were performed as described (Song et al., 2005; Miao et al., 2006). Competent cells of Saccharomyces cerevisiae strain Y190 (Clontech) were transformed simultaneously with pAS-MPK6/3/4 and pACT2-NIA1/2 or pACT2-NIA2-a to pACT2-NIA2-f. Yeast cells that had been cotransformed with pAS2 and pACT2 with no inserts were used as negative controls.
BiFC Assay
To measure in vivo interactions, the coding regions of MPK6, NIA1, and NIA2 were amplified by PCR with primers that contained appropriate restriction sites, and the amplified fragments were inserted into the plasmids pSPYNE and pSPYCE, which contain DNA encoding the N-terminal and C-terminal region of YFP, respectively (Walter et al., 2004), to form pSPYNE-MPK6, pSPYCE-NIA1, and pSPYCE-NIA2, respectively. Protoplasts isolated from Arabidopsis leaves were transformed with the following combinations of plasmids: pSPYNE-MPK6 and pSPYCE, pSPYNE and pSPYCE-NIA1/2, or pSPYNE-MPK6 and pSPYCE-NIA1/2, according to the previous protocols (Walter et al., 2004). For the GFP constructs, the coding regions of MPK6, NIA1, and NIA2 were amplified by PCR and inserted into the modified plasmid pHBT-GFP-NOS (Sheen, 2001) to form pMPK6-GFP, pNIA1-GFP, and pNIA2-GFP, respectively. The protoplast transient expression assay was performed as described (Sheen, 2001). After incubation for 16 to 20 h, the fluorescence of the protoplasts was measured with a confocal laser scanning microscope FV1000 (Olympus). All figures show representative images from three independent experiments.
Recombinant Protein Expression in Escherichia coli
For in vitro protein expression, the DNA regions containing the coding region of MPK6 and NIA1/2 were amplified and inserted in frame into the plasmid pET-28a (Novagen). Mutations of NIA2d were introduced by QuickChange site-directed mutagenesis (Stratagene). pET-MPK6 and a construct that expressed Flag-tagged MKK5DD (Ren et al., 2002) were introduced into E. coli BL21(DE3) cells. The recombinant His-tagged proteins were purified using Ni-NTA agarose (Qiagen) according to the manufacturer’s protocol.
Immunoprecipitation for Full-Length NIA2
To obtain the native NIA2 proteins, 2-week-old seedlings of NIA2WT and NIA2A were incubated in 15 μM DEX for 24 h, and total protein was extracted from seedlings by grinding in IP buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM Na3VO4, 1 mM NaF, 10 mM β-glycerophosphate, 1 μg/mL leupeptin, 1 μg/mL aprotinin, and 0.1% Triton X-100). Two hundred micrograms of total protein was add to 20 μL anti-FLAG M2 affinity gel (Sigma-Aldrich) and incubated at 4°C for 4 h on a rocker. After washing with TBS (20 mM Tris-HCl, pH 7.5, and 150 mM NaCl) three times, agarose bead-protein complexes were used for kinase assays.
Expression of NIA2 Protein in Pichia pastoris
Expression of NIA2 protein in P. pastoris was performed as described previously (Su et al., 1996). Wild-type and mutant forms of NIA2 cDNA with a C-terminal MYC-epitope were directly inserted into the _Bam_HI/_Eco_RI site of the P. pastoris expression vector pPIC9K (Invitrogen). The expression clones were transformed into P. pastoris strain SMD1168 by electroporation. Growth media and conditions for selection of transformants were as described by the manufacturer. HIS+ and G418 transformants were selected. NIA2 proteins were induced in MM medium (0.35% yeast nitrogen base with 1% ammonium sulfate, 0.5% methanol, and 4× 10 to 5% biotin). Two grams of yeast cells were collected in 2 mL IP buffer, and an equal volume of glass beads was added. The mixture was vortexed for 10 min and centrifuged, and the supernatants were used for immunoprecipitation with anti-c-MYC agarose affinity gel (Sigma-Aldrich). After washing with TBS three times, and then washed with kinase buffer or NR extraction buffer one more time, NIA2 proteins were eluted with c-Myc peptide and used for kinase assays or NR activity assays.
Phosphorylation Assay
The in vitro kinase assay was performed as described previously (Liu and Zhang, 2004). Recombinant His-tagged MPK6 (10 μg) was activated by incubation with recombinant MKK5DD (1 μg) in the presence of 50 μM ATP in 50 μL of reaction buffer (20 mM HEPES, pH 7.5, 10 mM MgCl2, and 1 mM DTT) at 25°C for 30 min. Activated MPK6 was used to phosphorylate recombinant protein purified either from E. coli or P. pastoris (1:10 enzyme:substrate ratio) or 10 μL anti-Flag beads-protein complex in the same reaction buffer, with 50 μM ATP and [γ-32P]ATP (0.1 μCi per reaction). The reactions were stopped by the addition of SDS loading buffer after 60 min. The phosphorylated NIA2c and NIA2d were visualized by autoradiography after separation on a 10% SDS polyacrylamide gel. After electrophoresis, the gel was exposed to a Kodak X-Omat film for 60 min. For enzymes used in the immunoblot assay, phosphorylation was performed without the addition of [γ-32P]ATP.
In-Gel Kinase Assay and Immunobloting
Total protein was extracted from seedlings by grinding in extraction buffer (100 mM HEPES, pH 7.5, 5 mM EDTA, 5 mM EGTA, 10 mM Na3VO4, 10 mM NaF, 50 mM β-glycerophosphate, 10 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 5 μg/mL leupeptin, 5 μg/mL aprotinin, and 5% glycerol). After centrifugation at 18,000_g_ for 40 min, supernatants were transferred into clean tubes, quickly frozen in liquid nitrogen, and stored at −80°C until analyzed. The concentration of protein extracts was determined according to the method of Bradford with BSA as the standard (Bradford, 1976).
The in-gel kinase assay was performed as described previously (Zhang and Klessig, 1997). Aliquots of extract that contained 10 μg protein were electrophoresed on 10% SDS-polyacrylamide gels that contained 2 mg/mL myelin basic protein (MBP) in the separating gel as a substrate for the kinases. After electrophoresis, the gel was washed three times for 20 min at room temperature with washing buffer (25 mM Tris, pH 7.5, 0.5 mM DTT, 0.1 mM Na3VO4, 5 mM NaF, 0.5 mg/mL BSA, and 0.1% Triton X-100) and incubated at 4°C overnight, with three changes of renaturing buffer (25 mM Tris, pH 7.5, 1 mM DTT, 0.1 mM Na3VO4, and 5 mM NaF). The gel was incubated at room temperature in 30 mL reaction buffer (25 mM Tris, pH 7.5, 2 mM EGTA, 12 mM MgCl2, 1 mM DTT, and 0.1 mM Na3VO4) with 200 nM ATP plus 50 μCi of [γ-32P]ATP for 60 min, after which the reaction was stopped by transferring the gel into 5% (w/v) trichloroacetic acid and 1% (w/v) sodium pyrophosphate. The gel was washed in the same solution for at least 6 h with five changes of buffer. The gel was dried and exposed to Kodak X-Omat film for 10 h.
For immunoblotting, 10 μg total proteins was separated by electrophoresis on 10% SDS-polyacrylamide gels, and the proteins were transferred to polyvinylidene difluoride membranes (Roche) by electrophoretic transfer. After blocking with blocking buffer (20 mM Tris, pH 7.5, 30 mM NaCl, and 0.05% Tween 20) at room temperature, the membranes were incubated with monoclonal anti-Flag M2 antibody, monoclonal antipolyhistidine antibody (Sigma-Aldrich), or phospho-P44/42 MAPK antibody (Cell Signaling Technology). After washing three times, the membranes were incubated with horseradish peroxidase–conjugated secondary antibody and visualized using lumi-light protein gel blotting substrate (Roche) following the manufacturer’s instructions.
Analysis of Root Architecture
Seeds of wild-type Arabidopsis and the mpk6 mutant were germinated and grown on MS plates (Murashige and Skoog, 1962) that contained 2% sucrose and 0.8% phytogel (Sigma-Aldrich). Five-day-old seedlings were transferred to MS plates that contained different concentrations of H2O2, SNP, or tungstate. For root morphology assays of NIA2A, NIA2D, and NIA2WT transgenic plants, 5-d-old seedlings were transferred to MS plates that contained 0.02 or 0.1 μM DEX and 15 μM SNP. Photographs were taken and the total length of lateral roots of each seedling were determined at the indicated times after transfer.
Measurement of NR Activity
Wild-type and mutant plants were grown on MS medium at 22°C with a 16-h-light photoperiod for 2 weeks. Seedlings were harvested 30 min after being sprayed with 10 μM H2O2 for 30 min. Samples of 0.2 g of control and H2O2-treated seedlings were ground to a fine powder in liquid nitrogen and homogenized in 1 mL NR extraction buffer (50 mM MOPS-NaOH, pH 7.5, 10 mM MgCl2, 1 mM EDTA, 5 mM DTT, 0.1% Triton X-100, 0.4 μg/mL leupeptin, and 1 μg/mL pepstatin). The mixture was vortexed and then centrifuged at 14,000 rpm for 10 min at 4°C. The supernatant was used directly for NR assays as described previously (Huber et al., 1992; Su et al., 1996). The concentration of protein in the extracts was determined by Bradford assay with BSA as the standard, and 20 μg protein extract was used for the assay of NR activity.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative databases under the following accession numbers: MKK5, At3g21220; MPK3, At3g45640; MPK4, At4g01370; MPK6, At2g43790; NIA1, At1g77760; and NIA2, At1g37130. T-DNA insertion lines used here are mpk6-2 (Salk_073907, germplasm 4676495), mpk6-3 (Salk_127507, germplasm 4845066), nia1-3 (Salk_004164, germplasm 4514312), nia1-4 (Salk_071547, germplasm 4674135), nia2-1 (Salk_138297, germplasm 4855856), nia2-2 (Salk_088070, germplasm 4690658), and nia1-2 nia2-5 (germplasm CS6512).
Supplemental Data
The following materials are available in the online version of this article.
- Supplemental Figure 1.. NO Biosynthesis and MPK6 Activity after Application of H2O2.
- Supplemental Figure 2.. Expression of NIA1, NIA2, and MPK6 in T-DNA Insertion Mutants and Double Mutants.
- Supplemental Figure 3.. Expression of NIA2 in Wild-Type and mpk6-3 in Response to Exogenous H2O2.
- Supplemental Figure 4.. Effects of Tungstate and H2O2 on NR Activity in Wild-Type Plants.
- Supplemental Figure 5.. Effects of Tungstate on H2O2-Induced Lateral Root Growth.
- Supplemental Figure 6.. Phosphorylation of Full-Length NIA2 Protein by MPK6.
- Supplemental Figure 7.. The Length of Lateral roots in Wild-Type and NIA2 Transgenic Plants in Response to H2O2.
- Supplemental Table 1.. Primers Used in This Study.
- Supplemental Methods. Quantitative RT-PCR .
Acknowledgments
We thank Y. Guo for technical help and discussion and Sophie Song for reading of the manuscript. This work was supported by the National Natural Science Foundation of China (30625005, 90817106, and 30600344).
References
- Ahlfors R., Macioszek V., Rudd J., Brosché M., Schlichting R., Scheel D., Kangasjarvi J. (2004). Stress hormone-independent activation and nuclear translocation of mitogen-activated protein kinases in Arabidopsis thaliana during ozone exposure. Plant J. 40: 512–522 [DOI] [PubMed] [Google Scholar]
- Alessi D.R., Cuenda A., Cohen P., Dudley D.T., Saltiel A.R. (1995). PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 270: 27489–27494 [DOI] [PubMed] [Google Scholar]
- Asai S., Ohta K., Yoshioka H. (2008). MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20: 1390–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.-L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Bergmann D.C., Lukowitz W., Somerville C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bright J., Desikan R., Hancock J.T., Weir I.S., Neill S.J. (2006). ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 45: 113–122 [DOI] [PubMed] [Google Scholar]
- Campbell W.H. (1999). Nitrate reductase structure, function and regulation: Bridging the gap between biochemistry and physiology. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 277–303 [DOI] [PubMed] [Google Scholar]
- Campbell W.H. (2001). Structure and function of eukaryotic NAD(P)H:nitrate reductase. Cell. Mol. Life Sci. 58: 194–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.L., Acedo G.N., Cristinsin M., Conkling M.A. (1992). Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc. Natl. Acad. Sci. USA 89: 1861–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.-L., Acedo G.N., Dewdney J., Goodman H.M., Conkling M.A. (1991). Differential expression of the two Arabidopsis nitrate reductase genes. Plant Physiol. 96: 275–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D., Durner J., Navarre D.A., Klessig D.F. (2000). Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Mol. Plant Microbe Interact. 13: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Clarke A., Desikan R., Hurst R.D., Hancock J.T., Neill S.J. (2000). NO way back: Nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J. 24: 667–677 [DOI] [PubMed] [Google Scholar]
- Colcombet J., Hirt H. (2008). Arabidopsis MAPKs: A complex signalling network involved in multiple biological processes. Biochem. J. 413: 217–226 [DOI] [PubMed] [Google Scholar]
- Correa-Aragunde N., Graziano M., Lamattina L. (2004). Nitric oxide plays a central role in determining lateral root development in tomato. Planta 218: 900–905 [DOI] [PubMed] [Google Scholar]
- Crawford N.M. (2006). Mechanisms for nitric oxide synthesis in plants. J. Exp. Bot. 57: 471–478 [DOI] [PubMed] [Google Scholar]
- Cunnick J.M., Dorsey J.F., Standley T., Turkson J., Kraker A.J., Fry D.W., Jove R., Wu J. (1998). Role of tyrosine kinase activity of epidermal growth factor receptor in the lysophosphatidic acid-stimulated mitogen-activated protein kinase pathway. J. Biol. Chem. 273: 14468–14475 [DOI] [PubMed] [Google Scholar]
- De Cnodder T., Vissenberg K., Van Der Straeten D., Verbelen J.P. (2005). Regulation of cell length in the Arabidopsis thaliana root by the ethylene precursor 1-aminocyclopropane- 1-carboxylic acid: A matter of apoplastic reactions. New Phytol. 168: 541–550 [DOI] [PubMed] [Google Scholar]
- Dean J.V., Harper J.E. (1988). The conversion of nitrite to nitrogen oxide(s) by the constitutive NAD(P)H-nitrate reductase enzyme from soybean. Plant Physiol. 88: 389–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R., Cheung M.-K., Bright J., Henson D., Hancock J.T., Neill S.J. (2004). ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J. Exp. Bot. 55: 205–212 [DOI] [PubMed] [Google Scholar]
- Desikan R., Griffiths R., Hancock J., Neill S. (2002). A new role for an old enzyme: Nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 99: 16314–16318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R., Hancock J.T., Ichimura K., Shinozaki K., Neill S.J. (2001). Harpin induces activation of the Arabidopsis mitogen-activated protein kinases AtMPK4 and AtMPK6. Plant Physiol. 126: 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilner T., et al. (2005). High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol. Cell. Proteomics 4: 1558–1568 [DOI] [PubMed] [Google Scholar]
- Greene E.L., Velarde V., Jaffa A.A. (2000). Role of reactive oxygen species in bradykinin-induced mitogen-activated protein kinase and c-fos induction in vascular cells. Hypertension 35: 942–947 [DOI] [PubMed] [Google Scholar]
- Guo F.-Q., Okamoto M., Crawford N.M. (2003). Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302: 100–103 [DOI] [PubMed] [Google Scholar]
- Gupta R., Luan S. (2003). Redox control of protein tyrosine phosphatases and mitogen-activated protein kinases in plants. Plant Physiol. 132: 1149–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin S.C., Larue C.T., Oh M.H., Jain V., Huber S.C. (2009). Coupling oxidative signals to protein phosphorylation via methionine oxidation in Arabidopsis. Biochem. J. 422: 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Xu H., She X., Song X., Zhao W. (2005). The role and the interrelationship of hydrogen peroxide and nitric oxide in the UV-B-induced stomatal closure in broad bean. Funct. Plant Biol. 32: 237–247 [DOI] [PubMed] [Google Scholar]
- Hord C.L.H., Sun Y.-J., Pillitteri L.J., Torii K.U., Wang H., Zhang S., Ma H. (2008). Regulation of Arabidopsis early anther development by the mitogen-activated protein kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol. Plant 1: 645–658 [DOI] [PubMed] [Google Scholar]
- Huber J.L., Huber S.C., Campbell W.H., Redinbaugh M.G. (1992). Reversible light/dark modulation of spinach leaf nitrate reductase activity involves protein phosphorylation. Arch. Biochem. Biophys. 296: 58–65 [DOI] [PubMed] [Google Scholar]
- Ichimura K., Mizoguchi T., Yoshida R., Yuasa T., Shinozaki K. (2000). Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 24: 655–665 [DOI] [PubMed] [Google Scholar]
- Ichimura K., Shinozaki K., Tena G., Sheen J., Henry Y., Champion A., Kreis M., Zhang S., Hirt H.; MAPK Group (2002). Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Jacobs D., Glossip D., Xing H., Muslin A.J., Kornfeld K. (1999). Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 13: 163–175 [PMC free article] [PubMed] [Google Scholar]
- Jonak C., Okrész L., Bögre L., Hirt H. (2002). Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 5: 415–424 [DOI] [PubMed] [Google Scholar]
- Joo S., Liu Y., Lueth A., Zhang S. (2008). MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 54: 129–140 [DOI] [PubMed] [Google Scholar]
- Kaiser W.M., Huber S.C. (2001). Post-translational regulation of nitrate reductase: Mechanism, physiological relevance and environmental triggers. J. Exp. Bot. 52: 1981–1989 [DOI] [PubMed] [Google Scholar]
- Kanamaru K., Wang R., Su W., Crawford N.M. (1999). Ser-534 in the hinge 1 region of Arabidopsis nitrate reductase is conditionally required for binding of 14-3-3 proteins and in vitro inhibition. J. Biol. Chem. 274: 4160–4165 [DOI] [PubMed] [Google Scholar]
- Kiegerl S., Cardinale F., Siligan C., Gross A., Baudouin E., Liwosz A., Eklöf S., Till S., Bögre L., Hirt H., Meskiene I. (2000). SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell 12: 2247–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H., Nakatsubo N., Kikuchi K., Kawahara S., Kirino Y., Nagoshi H., Hirata Y., Nagano T. (1998). Detection and imaging of nitric oxide with novel fluorescent indicators: Diaminofluoresceins. Anal. Chem. 70: 2446–2453 [DOI] [PubMed] [Google Scholar]
- Kolbert Z., Bartha B., Erdei L. (2008). Exogenous auxin-induced NO synthesis is nitrate reductase-associated in Arabidopsis thaliana root primordia. J. Plant Physiol. 165: 967–975 [DOI] [PubMed] [Google Scholar]
- Kovtun Y., Chiu W.L., Tena G., Sheen J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97: 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Klessig D.F. (2000). Differential induction of tobacco MAP kinases by the defense signals nitric oxide, salicylic acid, ethylene, and jasmonic acid. Mol. Plant Microbe Interact. 13: 347–351 [DOI] [PubMed] [Google Scholar]
- Lü D., Zhang X., Jiang J., An G., Zhang L., Song C.-P. (2005). NO may function in the downstream of H2O2 in ABA-induced stomatal closure in Vicia faba L. J. Plant Physiol. Mol. Biol. 31: 62–70 [PubMed] [Google Scholar]
- Libourel I.G., Bethke P.C., De Michele R., Jones R.L. (2006). Nitric oxide gas stimulates germination of dormant Arabidopsis seeds: Use of a flow-through apparatus for delivery of nitric oxide. Planta 223: 813–820 [DOI] [PubMed] [Google Scholar]
- Linkohr B.I., Williamson L.C., Fitter A.H., Leyser H.M.O. (2002). Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 29: 751–760 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo M.C., Graziano M., Polacco J.C., Lamattina L. (2006). Nitric oxide functions as a positive regulator of root hair development. Plant Signal. Behav. 1: 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Han M.H., Guevara-Garcia A., Fedoroff N.V. (2002). Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc. Natl. Acad. Sci. USA 99: 15812–15817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhard M., Rodriguez P.L., Grill E. (2002). The sensitivity of ABI2 to hydrogen peroxide links the abscisic acid-response regulator to redox signalling. Planta 214: 775–782 [DOI] [PubMed] [Google Scholar]
- Merkouropoulos G., Andreasson E., Hess D., Boller T., Peck S.C. (2008). An Arabidopsis protein phosphorylated in response to microbial elicitation, AtPHOS32, is a substrate of MAP kinases 3 and 6. J. Biol. Chem. 283: 10493–10499 [DOI] [PubMed] [Google Scholar]
- Miao Y., Lv D., Wang P., Wang X.-C., Chen J., Miao C., Song C.-P. (2006). An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18: 2749–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N.S., Tuteja R., Tuteja N. (2006). Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 452: 55–68 [DOI] [PubMed] [Google Scholar]
- Modolo L.V., Augusto O., Almeida I.M.G., Pinto-Maglio C.A.F., Oliveira H.C., Seligman K., Salgado I. (2006). Decreased arginine and nitrite levels in nitrate reductase-deficient Arabidopsis thaliana plants impair nitric oxide synthesis and the hypersensitive response to Pseudomonas syringae. Plant Sci. 171: 34–40 [Google Scholar]
- Moon H., et al. (2003). NDP kinase 2 interacts with two oxidative stress-activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proc. Natl. Acad. Sci. USA 100: 358–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead G., Douglas P., Morrice N., Scarabel M., Aitken A., MacKintosh C. (1996). Phosphorylated nitrate reductase from spinach leaves is inhibited by 14-3-3 proteins and activated by fusicoccin. Curr. Biol. 6: 1104–1113 [DOI] [PubMed] [Google Scholar]
- Moreau M., Lee G.I., Wang Y., Crane B.R., Klessig D.F. (2008). AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J. Biol. Chem. 283: 32957–32967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Nakagami H., Soukupová H., Schikora A., Zárský V., Hirt H. (2006). A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 281: 38697–38704 [DOI] [PubMed] [Google Scholar]
- Neill S., Barros R., Bright J., Desikan R., Hancock J., Harrison J., Morris P., Ribeiro D., Wilson I. (2008). Nitric oxide, stomatal closure, and abiotic stress. J. Exp. Bot. 59: 165–176 [DOI] [PubMed] [Google Scholar]
- Neill S.J., Desikan R., Hancock J.T. (2003). Nitric oxide signalling in plants. New Phytol. 159: 11–35 [DOI] [PubMed] [Google Scholar]
- Nühse T.S., Peck S.C., Hirt H., Boller T. (2000). Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J. Biol. Chem. 275: 7521–7526 [DOI] [PubMed] [Google Scholar]
- Ouaked F., Rozhon W., Lecourieux D., Hirt H. (2003). A MAPK pathway mediates ethylene signaling in plants. EMBO J. 22: 1282–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer K., Tuominen H., Kettunen R., Betz C., Langebartels C., Sandermann H., Jr, Kangasjarvi J. (2000). Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12: 1849–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat G.C., Simontacchi M., Puntarulo S., Lamattina L. (2002). Nitric oxide is required for root organogenesis. Plant Physiol. 129: 954–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov S., Sevugan M., Ye D., Yang W.-C., Kumaran M., Sundaresan V. (1999). Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11: 2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M.V., Paliyath G., Ormrod D.P., Murr D.P., Watkins C.B. (1997). Influence of salicylic acid on H2O2 production, oxidative stress, and H2O2-metabolizing enzymes. Salicylic acid-mediated oxidative damage requires H2O2. Plant Physiol. 115: 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Liu Y., Yang K.-Y., Han L., Mao G., Glazebrook J., Zhang S. (2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 5638–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Yang H., Zhang S. (2002). Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 277: 559–565 [DOI] [PubMed] [Google Scholar]
- Rhee S.G., Bae Y.S., Lee S.-R., Kwon J. (2000). Hydrogen peroxide: a key messenger that modulates protein phosphorylation through cysteine oxidation. Sci. STKE 2000: pe1. [DOI] [PubMed] [Google Scholar]
- Rockel P., Strube F., Rockel A., Wildt J., Kaiser W.M. (2002). Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 53: 103–110 [PubMed] [Google Scholar]
- Seo S., Katou S., Seto H., Gomi K., Ohashi Y. (2007). The mitogen-activated protein kinases WIPK and SIPK regulate the levels of jasmonic and salicylic acids in wounded tobacco plants. Plant J. 49: 899–909 [DOI] [PubMed] [Google Scholar]
- Sharrocks A.D., Yang S.-H., Galanis A. (2000). Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci. 25: 448–453 [DOI] [PubMed] [Google Scholar]
- She X.-P., Song X., He J. (2004). Role and relationship of nitric oxide and hydrogen peroxide in light/dark-regulated stomatal movement in Vicia faba. Acta Bot. Sin. 46: 1292–1300 [Google Scholar]
- Sheen J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Simpson G.G. (2005). NO flowering. Bioessays 27: 239–241 [DOI] [PubMed] [Google Scholar]
- Sitia R., Molteni S.N. (2004). Stress, protein (mis)folding, and signaling: the redox connection. Sci. STKE 2004: pe27. [DOI] [PubMed] [Google Scholar]
- Song C.-P., Agarwal M., Ohta M., Guo Y., Halfter U., Wang P., Zhu J.-K. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17: 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Huber S.C., Crawford N.M. (1996). Identification in vitro of a post-translational regulatory site in the hinge 1 region of Arabidopsis nitrate reductase. Plant Cell 8: 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F., Yoshida R., Ichimura K., Mizoguchi T., Seo S., Yonezawa M., Maruyama K., Yamaguchi-Shinozaki K., Shinozaki K. (2007). The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 19: 805–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantino D., Vannini C., Bracale M., Campa M., Soave C., Murgia I. (2005). Antisense reduction of thylakoidal ascorbate peroxidase in Arabidopsis enhances paraquat-induced photooxidative stress and nitric oxide-induced cell death. Planta 221: 757–765 [DOI] [PubMed] [Google Scholar]
- Tena G., Asai T., Chiu W.L., Sheen J. (2001). Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4: 392–400 [DOI] [PubMed] [Google Scholar]
- Tuominen H., Overmyer K., Keinänen M., Kollist H., Kangasjärvi J. (2004). Mutual antagonism of ethylene and jasmonic acid regulates ozone-induced spreading cell death in Arabidopsis. Plant J. 39: 59–69 [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang H., Liu Y., Bruffett K., Lee J., Hause G., Walker J.C., Zhang S. (2008). Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 20: 602–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in _Arabidopsi_s. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Tischner R., Gutiérrez R.A., Hoffman M., Xing X., Chen M., Coruzzi G., Crawford N.M. (2004). Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 136: 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Motyka V., Strnad M., Schmülling T. (2001). Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 98: 10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson J.Q., Crawford N.M. (1991). Identification of the Arabidopsis CHL3 gene as the nitrate reductase structural gene NIA2. Plant Cell 3: 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I.D., Neill S.J., Hancock J.T. (2008). Nitric oxide synthesis and signalling in plants. Plant Cell Environ. 31: 622–631 [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Katou S., Yoshioka H., Doke N., Kawakita K. (2004). Involvement of nitric oxide generation in hypersensitive cell death induced by elicitin in tobacco cell suspension culture. J. Gen. Plant Pathol. 70: 85–92 [Google Scholar]
- Yamamoto-Katou A., Katou S., Yoshioka H., Doke N., Kawakita K. (2006). Nitrate reductase is responsible for elicitin-induced nitric oxide production in Nicotiana benthamiana. Plant Cell Physiol. 47: 726–735 [DOI] [PubMed] [Google Scholar]
- Yamasaki H., Sakihama Y. (2000). Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: In vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett. 468: 89–92 [DOI] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Tena G., Xiong Y., Sheen J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H., Numata N., Nakajima K., Katou S., Kawakita K., Rowland O., Jones J.D.G., Doke N. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15: 706–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa T., Ichimura K., Mizoguchi T., Shinozaki K. (2001). Oxidative stress activates ATMPK6, an Arabidopsis homologue of MAP kinase. Plant Cell Physiol. 42: 1012–1016 [DOI] [PubMed] [Google Scholar]
- Zhang A., Jiang M., Zhang J., Ding H., Xu S., Hu X., Tan M. (2007a). Nitric oxide induced by hydrogen peroxide mediates abscisic acid-induced activation of the mitogen-activated protein kinase cascade involved in antioxidant defense in maize leaves. New Phytol. 175: 36–50 [DOI] [PubMed] [Google Scholar]
- Zhang A., Jiang M., Zhang J., Tan M., Hu X. (2006). Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol. 141: 475–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Jennings A., Barlow P.W., Forde B.G. (1999). Dual pathways for regulation of root branching by nitrate. Proc. Natl. Acad. Sci. USA 96: 6529–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Rong H., Pilbeam D. (2007b). Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. J. Exp. Bot. 58: 2329–2338 [DOI] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. (1997). Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9: 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6: 520–527 [DOI] [PubMed] [Google Scholar]