α-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1 (original) (raw)
. Author manuscript; available in PMC: 2012 Jun 3.
Published in final edited form as: Sci Signal. 2011 May 24;4(174):ra33. doi: 10.1126/scisignal.2001823
Abstract
The Hippo pathway regulates contact inhibition of cell proliferation and, ultimately, organ size in diverse multicellular organisms. Inactivation of the Hippo pathway promotes nuclear localization of the transcriptional coactivator Yap1, a Hippo pathway effector, and can cause cancer. Here, we show that deletion of αE-catenin in the hair follicle stem cell compartment resulted in the development of skin squamous cell carcinoma in mice. Tumor formation was accelerated by simultaneous deletion of αE-catenin and the tumor suppressor-encoding gene p53. An siRNA screen revealed a functional connection between αE-catenin and Yap1. By interacting with Yap1, αE-catenin promoted its cytoplasmic localization, and Yap1 showed constitutive nuclear localization in _αE-catenin-nul_l cells. We also found an inverse correlation between αE-catenin abundance and Yap1 activation in human squamous cell carcinoma tumors. These findings identify αE-catenin as a tumor suppressor that inhibits Yap1 activity and sequesters it in the cytoplasm.
Introduction
The proliferation and differentiation of adult stem and progenitor cells is tightly controlled to generate and then maintain an appropriate number of cells, which is necessary for proper organ function while preventing tumor formation. How stem and progenitor cells measure and accordingly control their rates of accumulation and differentiation is a key question in stem cell biology. It has been proposed that cell-cell adhesion structures called the adherens junctions can be utilized by cells to measure and regulate the local cell “crowdedness” (1, 2). At the core of the adherens junctions are the transmembrane cadherins, which form intercellular bridges that are bound intracellularly to β-catenin and functionally linked to the actin cytoskeleton through α-catenin (3). β-catenin, a structural component of adherens junctions and a transcriptional coactivator in the canonical Wnt signaling pathway, represents one putative link between adherens junctions and the regulation of cell density (4). The constitutive activation of the Wnt pathway is oncogenic and causes tumor development in various organs (5); therefore, sequestering β-catenin to adherens junctions during conditions of high cell density may normally function to decrease rates of cell proliferation by inhibiting Wnt pathway signaling. In this study, we propose a mechanism that couples a different adherens junction protein, α epithelial (αE)-catenin, to the regulation of cell accumulation and cancer.
αE-catenin (which is encoded by Ctnna1) is required for adherens junction formation in multiple epithelial cell types (6–8), where it functionally links the cell membrane-localized cadherin-catenin adhesion complexes to the actin cytoskeleton (9). Intriguingly, αE-catenin expression is often absent or decreased in various primary human tumors and tumor-derived cell lines through deletion, inactivating mutations, or promoter methylation (10, 11). Skin keratinocytes lacking αE-catenin display loss of contact-mediated inhibition of cell proliferation (12), and xenografts containing these keratinocytes results in the formation of skin lesions in nude mice that resemble squamous cell carcinoma (13). Moreover, re-expression of αE-catenin in αE-catenin-negative cell lines attenuates their growth (11, 14). Tumor development in animals lacking a particular protein suggests a role for that protein as a tumor suppressor; however, due to the crucial role of αE-catenin in tissue integrity, previously employed gene knockout strategies that targeted αE-catenin for deletion in embryonic progenitor cells resulted in embryonic or neonatal lethality (1, 8, 12). Therefore, the question of tumor suppressor activity of αE-catenin has remained unresolved.
The Hippo signaling pathway was discovered in Drosophila, where it plays a crucial role in regulating the size of the organs in developing embryos (15). Many genes involved in the Hippo pathway are evolutionarily conserved and analysis of Hippo signaling in mammalian organisms has revealed its role in regulation of stem and progenitor cell accumulation and development of cancer (16–22). The Hippo pathway senses local cell densities and controls tissue growth through a kinase cascade that culminates in phosphorylation of the transcriptional coactivator, Yap1, a posttranslational modification that prevents its nuclear localization, thus blocking its activity (23). However, the molecular mechanisms that link the Hippo pathway to focal cell accumulation, particularly in the mammalian system, are not well understood.
Here, we show that genetic deletion of αE-catenin in the hair follicle stem cell compartment leads to development of squamous cell carcinoma. We also demonstrate that αE-catenin interacts with Yap1 and regulates its nuclear localization. Finally, our examination of human keratoacanthoma tumors revealed frequent loss of αE-catenin abundance, which correlated significantly with nuclear Yap1 localization.
Results
Conditional deletion of α E-catenin in the hair follicle stem cell compartment results in the formation of inflammatory skin lesions and squamous cell skin tumors
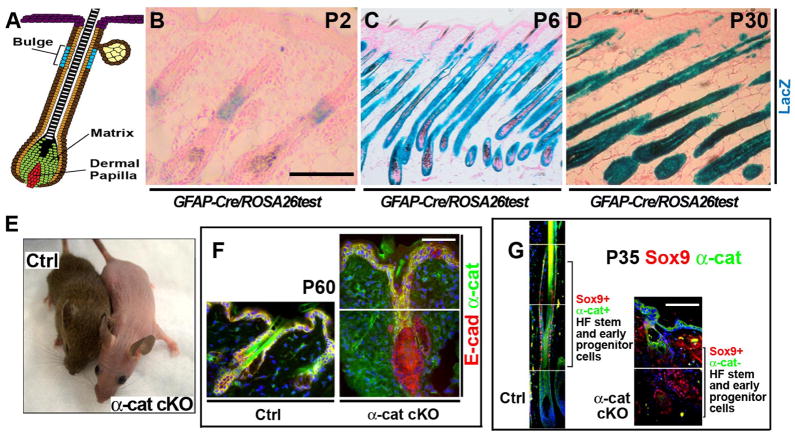
We generated and analyzed mice with a conditional deletion of α-catenin in the hair follicle stem and progenitor cell niche (Fig. 1 and fig. S1). In the GFAP-Cre mouse line (24), Cre was activated in the bulge region of the hair follicle at postnatal day 2 (P2) (Fig. 1, A and B). Because the progeny of P2 bulge stem cells expand throughout the hair follicle, the gene-targeting event occurred in most of the epithelial cells lining the hair follicle at P6, except for hair matrix cells (Fig. 1C). In adult GFAP-Cre animals, all epithelial cells of the hair follicle that originate from the bulge stem cell niche were targeted for Cre-mediated recombination (Fig. 1D). GFAP-Cre/αE-cateninfl/fl mice were viable and fertile; however, they were either completely bald or displayed only patchy hair growth on their backs, due to partially penetrant Cre expression in some of the animals (Fig. 1E). Sox9 is necessary for stem cell specification and maintenance (25) and thus is a specific marker of hair follicle stem and early progenitor cells. In GFAP-Cre/αE-cateninfl/fl mice, Sox9-positive hair follicle stem and early progenitor cells displayed loss of α-catenin (Fig. 1, F and G). Wild-type hair follicles undergo cycles of growth (anagen), degeneration (catagen), and rest (telogen), which together are known as the hair cycle (26). Histological examination of skin from wild-type and GFAP-Cre/αE-cateninfl/fl mice at different time points after birth revealed defective hair follicle morphogenesis and formation of disorganized hair follicles that were unable to produce hair (fig. S2, A to H). Sox9-positive hair stem and progenitor cells persisted in disorganized hair follicles of GFAP-Cre/αE-cateninfl/fl mice, and significantly more Sox9-positive cells incorporated BrdU, indicating that αE-catenin −/− stem and early progenitor cells cycle more actively than their αE-catenin+/+ counterparts (fig. S3). We concluded that deletion of αE-catenin in the hair follicle stem cell compartment resulted in abnormal hair follicle maintenance but did not affect viability, thus enabling the analysis of the long-term consequences of αE-catenin ablation.
Fig. 1. Conditional deletion of α E-catenin in hair follicles.
(A) Model of growing hair follicle. Hair follicle stem cells localize to the bulge region. Matrix contains committed progenitors that differentiate and give rise to hair and inner root sheath. (B to D) Staining for LacZ activity in frozen sections from newborn P2, P6, and P30 GFAP-Cre/ROSA26Cre test mice. Red is nuclear fast red counterstain. (E) General appearance of 6 month-old αE-cateninfl/fl (Ctrl) and GFAP-Cre/αE-cateninfl/fl (α-cat cKO) mice. (F) Immunofluorescent staining of skin sections from P60 αE-cateninfl/fl (Ctrl) and GFAP-Cre/αE-cateninfl/fl (α-cat cKO) mice with anti-E-cadherin (red) and anti-α-catenin (green) antibodies. Note loss of α-catenin in hair follicles of α-cat cKO skin. (G) Immunofluorescent staining of skin sections from P35 αE-cateninfl/fl (Ctrl) and GFAP-Cre/αE-cateninfl/fl (α-cat cKO) mice with antibodies against Sox9, a stem and early progenitor marker (red) and α-catenin (green). α-catenin is absent in Sox9+ cells in α-cat cKO skin. Blue in F and G is nuclear DAPI stain. Scale bars: 70 μm in (B); 190 μm in (C) and (D); 47 μm in (F) and (G).
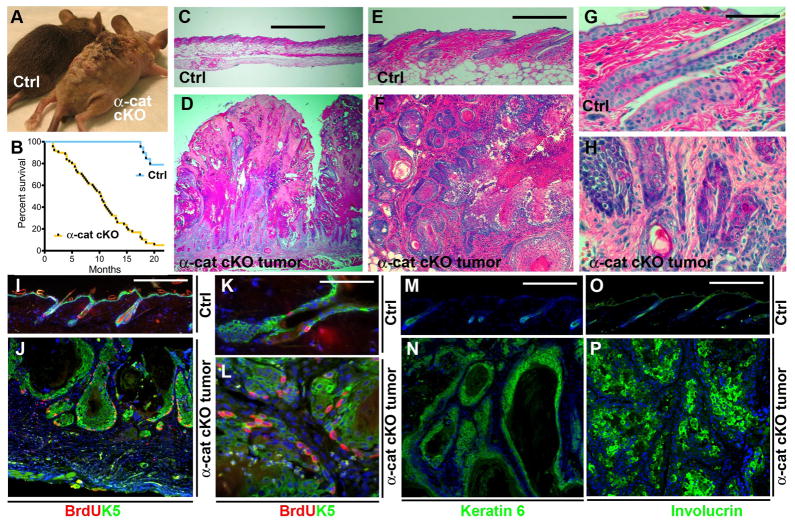
GFAP-Cre/αE-cateninfl/fl mice developed extensive skin lesions over time and needed to be euthanized with a half-survival time of ~10 months (Fig. 2, A and B). Histological analyses of the skin lesions showed prominent skin inflammation and tumors with squamous cell differentiation that resembled human squamous cell carcinoma of the keratoacanthoma type (Fig. 2, C to P, and fig. S4, A to C and G to I). Tumors displayed a massive expansion of the keratinocyte population with prominent signs of cellular atypia, intercellular bridges, and extensive extracellular keratinization. In addition, pearls of keratin that localized to the middle of concentric layers of squamous cells in tumor cell masses were prevalent (Fig. 2, F and H). The outer edges of tumor cell masses contained nondifferentiated proliferating cells, which prominently stained for keratins 5 and 6 (Fig. 2, I to N). The inner layers of tumor cell masses were positive for the differentiated keratinocyte marker, involucrin (Fig. 2P). Overall, the results obtained from immunostaining of tumor sections with cell type-specific markers were consistent with the histological diagnosis of keratoacanthoma squamous cell carcinoma. Tumors in GFAP-Cre/αE-cateninfl/fl animals formed in skin areas that displayed prominent inflammation; however, the onset of tumor formation varied among animals (fig. S4). Moreover, both the tumor and surrounding areas of inflammation were negative for αE-catenin, indicating that they were derived from the αE-catenin−/− keratinocytes of the hair follicles (fig. S5).
Fig. 2. αE-catenin is a tumor suppressor in skin keratoacanthoma.
(A) General appearance of 2 month-old αE-cateninfl/fl (Ctrl) and tumor-bearing GFAP-Cre/αE-cateninfl/fl (α-cat cKO) mice. (B) Kaplan-Meier survival curves of αE-cateninfl/fl (Ctrl) (N=19) and GFAP-Cre/αE-cateninfl/fl (α-cat cKO) (N=59) mice. Statistical significance was determined by the logrank test (P=<0.0001). (C to H) Hematoxylin and eosin staining of skin sections from 2 month-old αE-cateninfl/fl (Ctrl) and tumor-bearing GFAP-Cre/αE-cateninfl/fl (α-cat cKO) mice. Images in E to H show higher magnifications of the sections shown in C and D. (I to P) Immunofluorescent staining of skin sections from 2 month-old αE-cateninfl/fl (Ctrl) and tumor-bearing GFAP-Cre/αE-cateninfl/fl (α-cat cKO) mice with anti-BrdU (red), anti-keratin 5 [green in (I) to (L)], anti-keratin 6 [green in (M) and (N)], anti-involucrin [green in (O) and (P)] antibodies. Blue in I to P is nuclear DAPI stain. Scale bars: 500 μm in (C) and (D); 214 μm in (E) and (F); 53 μm in (G) and (H); 75 μm in (I) and (J); 19 μm in (K) and (L); 75 μm in (M) to (P).
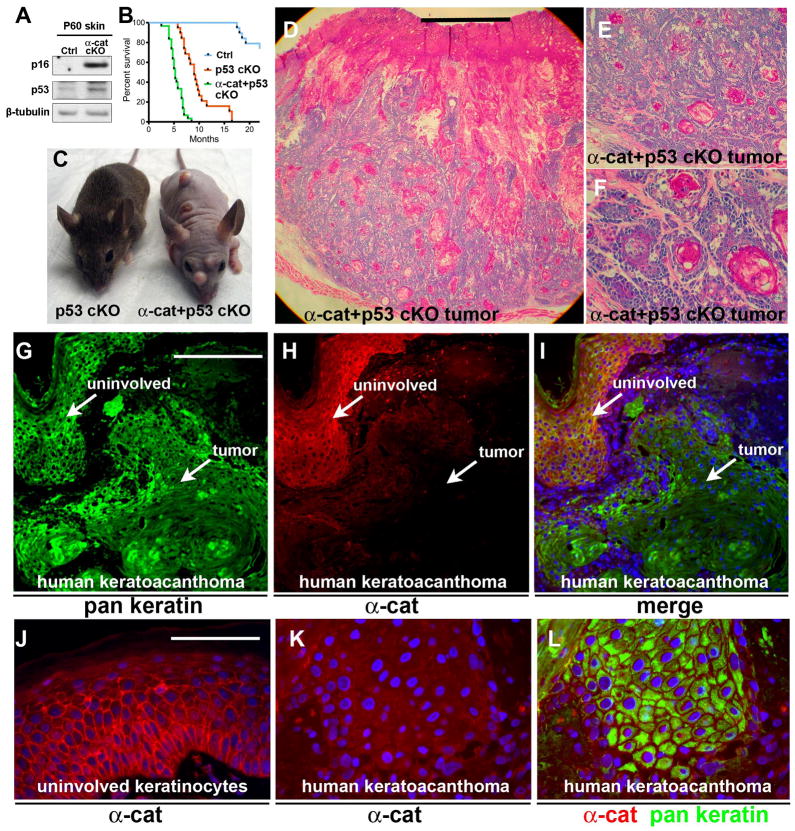
GFAP-Cre/αE-cateninfl/fl/p53fl/fl mice display development of an early onset multifocal keratoacanthoma
Because the onset of skin lesions varied considerably, we hypothesized that in addition to loss of αE-catenin, other genetic or epigenetic events had to take place before tumor development occurred in these animals. The inactivation of the gene encoding the tumor suppressor p53 occurs frequently in human keratoacanthoma (27, 28). We found that the abundance of p16Ink4A and p53 was increased in the skin of GFAP-Cre/αE-cateninflfl mice before they develop skin lesions (Fig. 3A). p53 elicits its tumor-suppressing function by activating apoptotic cell death or cell senescence (29, 30). Indeed, we found that a fraction of αE-catenin−/− cells was positive for apoptotic TUNEL staining (fig. S6A). To determine whether increased p53 signaling attenuates tumor development in GFAP-Cre/αE-cateninflfl mice, we generated and analyzed GFAP-Cre/αE-cateninfl/fl/p53fl/fl mice. Apoptotic cell death was significantly decreased in double mutant GFAP-Cre/αE-cateninfl/fl/p53fl/fl animals (fig. S6B). Furthermore, these mice developed completely penetrant, early onset, multifocal keratoacanthomas (Fig. 3, B to F). Unlike GFAP-Cre/αE-cateninflfl mice, skin tumors in GFAP-Cre/αE-cateninfl/fl/p53fl/fl mice appeared at multiple sites, and displayed no skin inflammation preceding tumor initiation (Fig. 3C and fig. S4). Therefore, it is likely that the deletion of p53 is completely sufficient to bypass the inflammatory response required for tumor development in GFAP-Cre/αE-cateninfl/fl mice. Although the exact cellular origin of human keratoacanthoma is unknown, this cancer is thought to originate in the upper portion of the hair follicle (31), which is the site of the hair follicle stem cell compartment targeted in our GFAP-Cre mice.
Fig. 3. Early onset keratoacanthoma tumors in GFAP-Cre/αE-cateninfl/fl/p53fl/fl mice and loss of αE-catenin abundance in human keratoacanthomas.
(A) Western blot analysis of total protein extracts from skins of αE-cateninfl/fl (Ctrl) and GFAP-Cre/αE-cateninfl/fl mice with anti-p53, anti-Arfp16, and anti-β-tubulin antibodies. N=3. (B) Kaplan-Meier survival curves for GFAP-Cre/αE-cateninfl/fl/p53fl/fl (N=30), GFAP-Cre/p53fl/fl (N=19), and wild-type (Ctrl) (N=19) mice. Curves for GFAP-Cre/αE-cateninfl/f/p53fl/fl and GFAP-Cre/p53fl/fl mice are different P<0.0001 (logrank test). Note: Due GFAP-Cre activity in brain, GFAP-Cre/p53fl/fl mice die from brain tumors; however, they do not have a skin phenotype at the time of euthanasia. (C) General appearance of 4 month-old GFAP-Cre/p53fl/fl (p53 cKO) F) and GFAP-Cre/αE-cateninfl/fl/p53fl/fl (α-cat+p53 cKO) mice. (D to Hematoxylin and eosin staining of skin tumor from 4 month-old GFAP-Cre/αE-cateninfl/fl/p53fl/fl (α-cat+p53 cKO) mouse. Images in E and F show higher magnifications of the section shown in D. (G to L) Immunofluorescent staining of human keratoacanthoma tumor with anti-α-catenin (red) and anti-pan-cytokeratin (green) antibodies. α-catenin is present in normal areas and not detectable in tumor cells. Blue in I to L is nuclear DAPI stain. Scale bars: 1 mm in (D); 0.4 mm in (E); 100 μm in (F); 30 μm in (G) to (I); 7.5 μm in (J) to (L).
αE-catenin is lost or decreased in abundance in human keratoacanthoma
To determine whether the abundance of α-catenin changes in human keratoacanthomas, we performed immunofluorescent staining with anti-αE-catenin antibodies on human tumors. Abundance of α-catenin in the tumors was compared to its abundance in normal areas of the epidermis in the same tissue sections. Of the 29 tumors analyzed, 3 (~10%) had similar α-catenin abundance to that in normal epidermis. Nineteen tumors (~66%) displayed reduced staining and seven tumors (~24%) showed staining for α-catenin comparable to background (Fig. 3, G to L). These data suggest that α-catenin may be frequently decreased in abundance or lost in human keratoacanthomas, a finding consistent with the potential role for αE-catenin as a tumor suppressor in these neoplasms.
An siRNA screen reveals a functional connection between αE-catenin and the transcriptional coactivator Yap1
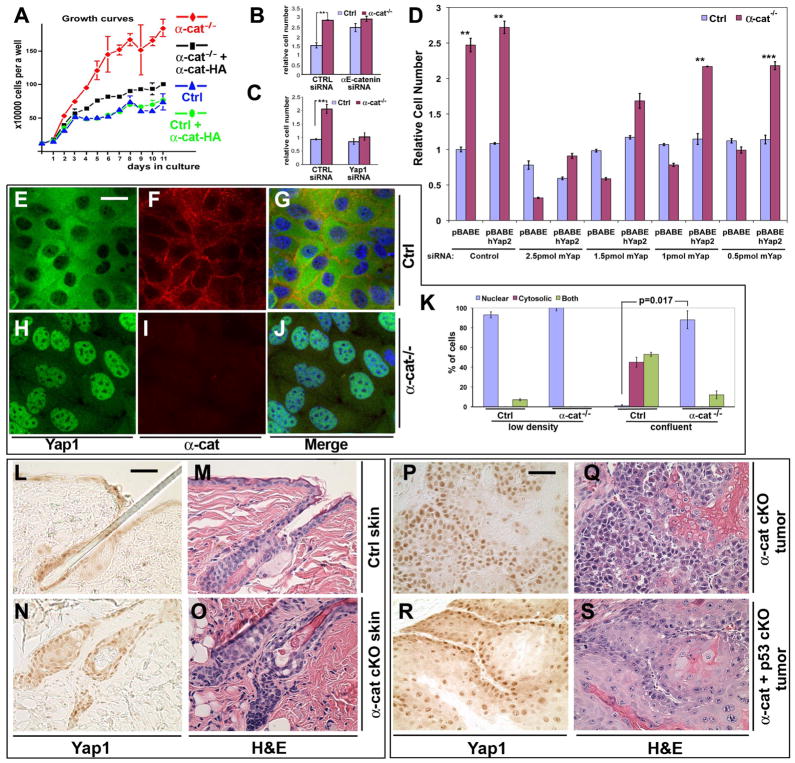
To elucidate the tumor suppressing mechanisms of αE-catenin, we isolated and analyzed mouse αE-catenin−/− keratinocytes. These cells displayed a loss of cell density-dependent inhibition of cell accumulation, a phenotype that was rescued by re-expression of α-catenin (Fig. 4, A to B and figs. S7 and S8).
Fig. 4. Yap1 is necessary for α-catenin-mediated loss of contact inhibition and hyperplasia.
(A) Growth curves of αE-cateninfl/fl (Ctrl) and αE-catenin−/− (α-cat−/−) cells expressing vector or full-length αE-catenin (Ctrl+α-cat, α-cat−/−+α-cat). Note prominent inhibition of cell accumulation in confluent Ctrl keratinocytes and continued accumulation of α-cat−/− keratinocytes. The phenotype is rescued by re-expression of full-length αE-catenin. (B) αE-cateninfl/fl or αE-catenin−/− keratinocytes were plated at high density in siRNA-Lipofectamine mixture in triplicate and cultured for 5 days. Cell numbers at the end of culture were determined by MTT assay. Under these conditions, αE-catenin−/− cells were not contact inhibited and show increased cell accumulation. This phenotype was replicated in αE-cateninfl/fl cells by transfection of siRNAs targeting αE-catenin. Bar graph shows mean values ± SD. N=3. **, P<0.01 by t-test. **(C)** Yap1 is necessary for the loss of contact inhibition in αE-catenin-null cells. Validation of the functional connection of Yap1 to α-catenin previously identified by siRNA screen (fig. S10), using siRNA oligos targeting disparate regions of Yap1. Western control is shown in fig. S11. Bar graph shows mean values ± SD. N=3. **(D)** Validation of the specificity of Yap1 siRNA hit using re-expression of human Yap2, which is not targeted by anti-mouse Yap1 siRNAs. **(E** to **J)** Representative images of confluent _αE-cateninfl/fl_ (Ctrl) and _αE-catenin−/−_ (_α-cat−/−_) keratinocytes stained with anti-Yap1 and anti-α-catenin antibodies. **(K)** Quantification of Yap1 localization illustrated in (E) to (J) and fig. S12. Bar graph shows mean values ± SD. Number of cells counted was >50 per condition. P value was determined by Mann-Whitney test. (L to O) Hematoxylin and eosin (H&E) and anti-Yap1 immunohistochemical staining of skin sections from 8 month-old αE-cateninfl/fl (Ctrl) and GFAP-Cre/αE-cateninfl/fl (α-cat cKO) mice. (P to S) H&E and anti-Yap1 immunohistochemical staining of keratoacanthoma tumors from GFAP-Cre/αE-cateninfl/fl (α-cat cKO), GFAP-Cre/αE-cateninfl/fl/p53fl/fl (α-cat+p53 cKO) mice. Blue in G and J is nuclear DAPI stain. Scale bars: 21 μm in (E) to (J); 35 μm in (L) to (O); 50 μm in (P) to (S).
To identify signaling pathways involved in αE-catenin-dependent inhibition of cell proliferation, we performed a targeted siRNA-mediated gene knockdown screen for 70 genes previously identified as encoding αE-catenin-interacting proteins or otherwise implicated in contact inhibition or cancer (figs. S8 to S10). siRNA targeting of most of genes, including β-catenin (β-cat), resulted in minimal phenotypes; however, knockdown of Yap1 inhibited the proliferation of αE-catenin−/− cells (fig. S10). The specificity of the Yap1 siRNA was validated by using siRNAs from a different supplier (Dharmacon) and that targeted against a different portion of the Yap1 gene (Fig. 4C and fig. S11), and by re-expression of human Yap2, which was not targeted by mouse Yap1-directed siRNAs (Fig. 4D). We conclude that Yap1 is necessary for the contact inhibition-defective phenotype of αE-catenin−/− keratinocytes.
αE-catenin negatively regulates Yap1 nuclear localization
The Hippo signaling pathway, which has a central role in the regulation of contact inhibition, organ size determination, and tumor suppression (22, 32), impinges on the nuclear localization of the transcriptional co-factor Yap1 (15). Therefore, we examined whether the activity of Yap1 was increased in αE-catenin−/− keratinocytes. In non-confluent cells, Yap1 localized primarily to the nucleus in both αE-catenin+/+ and αE-catenin−/− cells (fig. S12). In contrast, confluent αE-catenin+/+ cells displayed a diffuse cytoplasmic and partially junctional localization of Yap1, whereas confluent αE-catenin−/− keratinocytes showed predominantly nuclear Yap1 staining (Fig. 4, E to K). Similarly, cellular fractionation experiments demonstrated a significant increase in the amount of nuclear Yap1 in αE-catenin−/− keratinocytes (fig. S13). Re-expression of αE-catenin in αE-catenin−/− cells rescued the cytoplasmic localization of Yap1 (fig. S14). In epithelial cells, αE-catenin localizes to both cell-cell junctions and cytoplasm (33). Confocal microscopy analysis revealed partial colocalization between cytoplasmic αE-catenin and Yap1 in confluent keratinocytes (fig. S15).
If Yap1 constitutively localizes to the nucleus in proliferating cells and is cytoplasmic in non-proliferating cells, the differences in nuclear Yap1 localization between αE-catenin+/+ and αE-catenin−/− cells could be due to the differences in their proliferation status. To address this possibility, we analyzed the proliferation status and Yap1 localization in serum starved sub-confluent αE-catenin+/+ and αE-catenin−/− cells by immunostaining with anti-Ki67, which is a marker of proliferative cells, and anti-Yap1 antibodies, respectively. We found that Ki67-negative keratinocytes maintained nuclear Yap1 localization (fig. S16). Therefore, nuclear Yap1 was not a marker of cellular proliferation and differences in nuclear Yap1 between αE-catenin+/+ and αE-catenin−/− cells were not due to the differences in their proliferation status.
Yap1 phosphorylation on Ser127 by Mst1-Lats kinase cascade promotes Yap1 cytoplasmic retention and decreases its transcriptional activity (22). Analyses of total protein extracts with phospho-specific antibodies revealed that phosphorylation of Ser127 in Yap1 was decreased in αE-catenin−/− keratinocytes (fig. S17). However, when this was normalized for total Yap1 protein levels, we found that the changes in specific phosphorylation were not significant (fig. S18, N=5). Western blot and qRT-PCR analyses revealed the abundance of total Yap1 protein and mRNA in αE-catenin−/− keratinocytes was decreased compared to αE-catenin+/+ cells (figs. S17 to S20), which may indicate activation of the Yap1-mediated negative feedback loop (34). In addition, Western blot analysis of αE-catenin−/− keratinocytes did not reveal a decrease in the phosphorylation and thus activation of LATS, the kinase that phosphorylates Yap1 to prevent its nuclear localization (fig. S17). Together, these data suggest that αE-catenin does not regulate nuclear localization of Yap1 by impacting the canonical Hippo kinase cascade, which converges on phosphorylation of Ser127 in Yap1.
We next analyzed the localization of Yap1 in vivo. Yap1 was predominantly localized in nuclei in hair follicle cysts and tumors in GFAP-Cre/αE-cateninflfl and GFAP-Cre/αE-cateninfl/fl/p53fl/fl mice (Fig. 4, L to S). We also found that Yap1 was prominently nuclear in αE-catenin−/− neural progenitor cells in Nestin-Cre/αE-cateninflfl mice (1), suggesting that the connection between α-catenin and nuclear Yap1 is not restricted to keratinocytes (fig. S21).
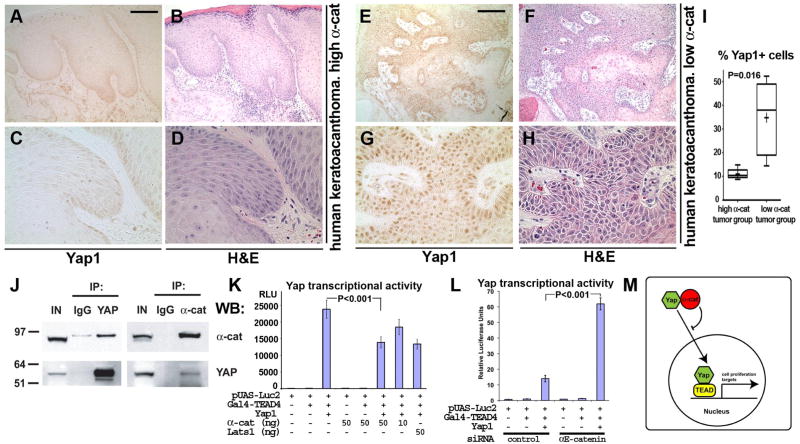
We reasoned that if αE-catenin inhibits nuclear localization of Yap1, human keratoacanthoma tumors with low abundance of αE-catenin should show increased nuclear Yap1. Indeed, immunostaining of sections of human keratoacanthoma tumors with anti-Yap1 antibodies revealed a significant correlation between low αE-catenin abundance and nuclear Yap1 localization (Fig. 5, A to I).
Fig. 5. αE-catenin interacts with Yap1 and inhibits its nuclear localization and transcriptional activity.
(A to H) H&E and anti-Yap1 immunohistochemical staining of human keratoacanthoma tumors with high and low abundance of α-catenin. (I) Box and whisker plot showing quantification of differences in Yap1 abundance in keratoacanthoma tumors with low (N=5) and high (N=5) α-catenin abundance. Cells were counted in 3 randomly selected images per tumor. Number of cells counted per each image was >170. The plots show percentage of tumor cells displaying nuclear staining for Yap1. Lines within boxes, median values. Upper and lower borders of the boxes, 25th and 75th percentiles. Upper and lower bars, maximum and minimum values. Statistical significance was assessed by the Mann-Whitney test. Scale bars: 155 μm in (A) and (B), (E) and (F); 50 μm in (C) and (D), (G) and (H). (J) Coimmunoprecipitation of endogenous α-catenin and Yap1. Proteins were extracted from cultured keratinocytes (IN) and immunoprecipitated (IP) with IgG controls, anti-Yap1, or anti-α-catenin antibodies and analyzed by Western blotting (WB) with anti-α-catenin or anti-Yap1 antibodies. (K and L) αE-catenin inhibits Yap1 transcriptional activity. Indicated constructs with or without siRNAs were cotransfected with a Gal4-TEAD firefly luciferase reporter and a Renilla luciferase control plasmid into HEK 293FT cells for gain-of-function experiments (K) or wild-type mouse keratinocytes for loss-of-function experiments (L). Reporter luciferase activity was normalized to Renilla luciferase activity. Bar graph shows mean values ± SE. Statistical significance was determined by ANOVA test. (M) Model showing the role of α-catenin in regulating Yap1 localization and transcriptional activity.
αE-catenin interacts with Yap1 and inhibits its transcriptional activity
To analyze the potential mechanisms responsible for αE-catenin-mediated regulation of Yap1 localization, we performed an additional siRNA screen to target 34 known members of the Hippo pathway. However, except for Yap1, none of these genes attenuated cell accumulation (fig. S22). Therefore, we hypothesized that αE-catenin-mediated regulation of Yap1 nuclear localization may occur downstream, perhaps at the level of Yap1 itself. Because αE-catenin is normally localized to the cytoplasm and is required for the cytoplasmic distribution of Yap1, we asked whether αE-catenin could regulate its intracellular localization through direct binding. Coimmunoprecipitation experiments revealed an interaction between endogenous αE-catenin and Yap1 proteins in keratinocytes (Fig. 5J). Quantification of Western blots indicated that 11.6±1.78% (N=3) of endogenous Yap1 was associated with αE-catenin.
Yap1 interacts with the transcription factors TEAD, RUNX2, p73, and ErbB4 and activates transcription of various genes involved in regulating cell proliferation and apoptosis (35). To determine whether the interaction between Yap1 and TEAD is required for increased proliferation in αE-catenin−/− keratinocytes, we performed cell density-dependent inhibition of cell accumulation experiments with cells expressing wild-type or Ser94→Ala (S94A) forms of human Yap2. The S94A mutation prevents Yap2 from interacting with the TEAD family of transcription factors, but not other transcription factors (35). We found that wild-type, but not the S94A mutant, human Yap2 supported increased proliferation in αE-catenin−/− keratinocytes (fig. S23). Therefore, TEAD transcription factors appear to be critical partners of Yap1 in αE-catenin−/− keratinocytes.
To analyze whether αE-catenin inhibits Yap1 transcriptional activity, we performed Yap1 transcription coactivation assays with the TEAD family of transcription factors. In gain-of-function experiments in HEK293 cells, overexpression of αE-catenin significantly decreased Yap1-mediated transcriptional activity in a dose-dependent manner (Fig. 5K). The extent of Yap1 signaling inactivation by α-catenin was similar to inactivation caused by LATS1, a pivotal inhibitor of Yap1 signaling. In complementary loss-of-function experiments, knockdown of αE-catenin in wild-type keratinocytes resulted in an increase of Yap1-mediated transcriptional activity (Fig. 5L). Endogenous transcriptional targets of Yap1 are tissue specific. Recently, Cyr61 was identified as a Yap1 transcriptional target in mouse skin keratinocytes (36). We found with qRT-PCR analysis that the abundance of Cyr61 was significantly increased in skins and tumors of GFAP-Cre/αE-cateninflfl mice (fig. S24). Therefore, we conclude that αE-catenin inhibits Yap1-mediated transcription by interacting with Yap1 and promoting its cytoplasmic localization (Fig. 5M).
Discussion
αE-catenin is a tumor suppressor protein
We demonstrate that conditional deletion of αE-catenin in the skin hair follicle stem cell compartment results in the development of keratoacanthoma and correspondingly that αE-catenin is frequently absent in human keratoacanthoma tumors. These data provide genetic evidence of the tumor suppressing function of αE-catenin. Although αE-catenin is frequently missing in various human epithelial and hematopoietic system tumors (10, 11, 37), it has been difficult to provide genetic, causal evidence of its tumor suppression in mice. Conditional deletion of αE-catenin during development in embryonic progenitor cells results in hyperplasia of the _αE-catenin_-null epidermis and brain, but also neonatal or perinatal lethality (1, 12, 38). In contrast, conditional deletion of αE-catenin in differentiated cells results in mild phenotypes with no reported tumor development (39, 40). Precisely targeting the deletion of αE-catenin to the bulge stem cell population after the major developmental events were already completed enabled us to establish direct genetic proof of the tumor suppressing function of αE-catenin.
Similar to other known tumor suppressors, deletion of αE-catenin in stem and progenitor cells is not sufficient for immediate tumor development. We found that αE-catenin−/− hair follicle stem and progenitor cells could persist many months without signs of tumor initiation. Because we detected skin tumors in GFAP-Cre/αE-cateninfl/fl mice only in the areas displaying a prominent inflammatory response, inflammation may be a critical factor that cooperates with the loss of αE-catenin to allow direct tumor development. Indeed, inflammation plays a central role in human cancer development and progression (41).
Unlike the mice lacking only αE-catenin, the double mutant mice lacking both αE-catenin and p53 displayed rapid and multifocal tumor development that initiated directly in the hair follicle, without signs of skin inflammation before tumor initiation. Therefore, it is likely that the deletion of p53 is sufficient to bypass the inflammatory response, which may be required for tumor development in GFAP-Cre/αE-cateninfl/fl mice. p53 attenuates tumor development by inducing cell cycle withdrawal and senescence or by activating programmed, apoptotic cell death (29). Although skin cells from GFAP-Cre/αE-cateninfl/fl and Cre/αE-cateninfl/fl/p53fl/fl mice did not show differences in cellular senescence, those from Cre/αE-cateninfl/fl/p53fl/fl mice showed a significant decrease in apoptotic cell death (fig. S6). Hence, the canonical role of p53 in regulation of apoptotic cell death is the most likely explanation for the tumor-promoting function of p53 in GFAP-Cre/αE-cateninfl/fl/p53fl/fl mice.
Molecular mechanisms of αE-catenin function in regulation of cell accumulation
αE-catenin is required for contact-mediated inhibition of cell proliferation. Intriguing correlations between αE-catenin membrane localization, cellular crowdedness, and contact-mediated control of cell proliferation have resulted in the formulation of a “cell crowd control” hypothesis, which postulates that in progenitor cells, the adherens junctions link information about the local progenitor cell density to signaling pathways that influence proliferation or cell cycle withdrawal (1, 2). This could be a mechanism underlying tissue homeostasis, a process that is necessary to ensure that an appropriate number of cells are generated for adult tissue maintenance. If αE-catenin is involved in connecting information about stem cell niche crowdedness with cell cycle regulation, it is important to determine the molecular mechanisms responsible for this function, because they are likely to be linked to the tumor-suppressing function of this protein. To reveal these mechanisms, we used an unbiased approach and performed an siRNA screen for genes that may be involved in αE-catenin function in contact inhibition. This screen identified Yap1 as a critical component necessary for increased proliferation in αE-catenin−/− cells. Further analyses revealed that Yap is constitutively nuclear in αE-catenin−/− cells. In addition, we showed that αE-catenin bound to Yap1 and determined its subcellular (cytoplasmic or nuclear) distribution. Thus, we conclude that αE-catenin is a critical regulator of Yap activity and abnormal activation of Yap1 signaling may be responsible for increased proliferation of αE-catenin−/− cells.
The Hippo pathway determines organ size in Drosophila and in mammalian organisms, it is implicated in the regulation of stem cell self-renewal and differentiation, as well as in cancer development (16, 20, 32, 35, 42). Similarly to αE-catenin, the Hippo pathway was previously implicated in the regulation of contact inhibition; however, the mechanisms connecting extracellular signals to Hippo signaling are not well understood (22, 43). Our study functionally and physically connects Yap1 with αE-catenin and suggests that Hippo signaling is regulated downstream of the canonical Hippo kinases by the binding of αE-catenin to Yap1 and its cytoplasmic retention. The tight junction protein Amot has also been implicated in regulating contact-mediated proliferation and nuclear Yap1 localization by tethering Yap1 outside of the nucleus (44, 45). This function of Amot is similar to the function of αE-catenin described in this study. We found that siRNA-mediated targeting of Amot or Amot-like genes in keratinocytes did not affect contact-mediated inhibition of cell accumulation (fig. S22). Therefore, it appears that the function of Amot and αE-catenin in keratinocytes may be different.
To summarize, we show here that αE-catenin suppresses tumor development in murine skin epidermis and functions to regulate Yap activity. Because αE-catenin is an adherens junction protein and adherens junctions are regulated by cell crowdedness, the connection between αE-catenin and Yap may potentially provide progenitor cells with a mechanism that can measure the local cell accumulation and adjust their rates of proliferation accordingly to ensure normal tissue homeostasis and to protect from tumor development.
Materials and Methods
Animals and labeling experiments
Homozygous αE-cateninfl/fl mice (12) were crossed with GFAP-Cre mice (Jackson Laboratory, FVB-Tg(GFAP-cre)25Mes/J) (24). To monitor Cre-mediated excision, mice were crossed with the reporter strain ROSA26Cre test (B6.129S4-Gt(ROSA)26Sortm1Sor/J; Jackson Laboratories) (46). To generate GFAP-Cre/αE-cateninfl/fl/p53fl/fl mice, GFAP-Cre/αE-cateninfl/fl animals were crossed with p53fl/fl mice (NCI Mouse Repository, 01XC2). To identify cycling Sox9+ stem and progenitor cells, adult mice were intraperitoneally injected with 50 μg/g of BrdU and euthanized after 1 hour. BrdU+ cells were detected in skin sections by immunofluorescent staining with anti-BrdU and anti-Sox9 antibodies.
Histology, immunofluorescent staining, and immunohistochemistry
Tissues for histology were fixed in 4% paraformaldehyde in PBS, processed, and embedded in paraffin. Sections (5 μM) were stained with hematoxylin and eosin and photographed using Olympus BX41 microscope and Microfire camera (Optronics) or Zeiss LSM 510 confocal microscope. Sections were deparaffinized and processed for immunofluorescent staining and immunohistochemistry as previously described (47).
Antibodies, LacZ, and apoptosis staining
The antibodies used were anti-GFAP (Sigma, G3893), anti-β-catenin (Sigma, C2206), anti-α-catenin (Epitomics, 2028-1, or Sigma, C2081), anti-E-cadherin (Invitrogen, 13-1900, or rabbit polyclonal IgGs developed against cytoplasmic domain of dog E-cadherin), anti-GFAP (Advanced ImmunoChemical, 031223), anti-Sox9 (Santa Cruz Biotech., sc-20095), anti-Ki67 (Novocastra, NCL-Ki67p), anti-BrdU (Developmental Studies Hybridoma Bank), anti-α6-integrin (BD Pharmingen, CD49f, clone GoH3), anti-keratin5, anti-keratin6, anti-involucrin (gift from Dr. Julia Segre, NIH/NHGRI), anti-pan cytokeratin (Sigma, C1801), anti-p120 catenin (BD Transduction Labs, 610133), anti-β-actin (Sigma, A5441), anti-Yap1, anti-phospho-Yap1 (Santa Cruz Biotech., sc-101199; Cell Signaling, 4912, 4911), anti-LATS1 (Cell Signaling, 3477), anti-LATS (S909) (Cell Signaling, 9157), anti-MST1 (T183)/MST2 (T180) (Cell Signaling, 3681), and anti-p21 (Calbiochem, OP76). LacZ staining was performed on 7 μm frozen skin sections as described (48). Apoptosis was detected using ApopTag® Plus Peroxidase In Situ kit (Millipore). To determine senescence, frozen skin sections were stained for SA-β-Gal activity using a senescence detection kit (Calbiochem) as previously described (49).
Primary keratinocyte culture, 96-well siRNA screen for contact inhibition, and cell fractionation
Keratinocytes were isolated as described (50). αE-cateninflox/flox keratinocytes were first established in low calcium media then later adapted and maintained in normal calcium E media (51). αE-catenin−/− cells were generated with AD5-CMV-Cre or Ad5-CMV-GFP adenoviruses (Vector Development Laboratory, Baylor College of Medicine). For αE-catenin rescue experiments, keratinocytes were infected with retrovirus containing full-length mouse αE-catenin and selected with G418. For the siRNA screen, 104 keratinocytes were plated directly into Lipofectamine 2000-siRNA mixture on 96 well plates. Each gene was targeted with a mixture of 4 independent siRNA oligos (Qiagen, 5pmol per well, each). Cells were allowed to grow for 5 days after seeding; media was changed daily; and final cell numbers were determined by manual cell counting or by MTT assay (Promega). For validation, SMARTpool siRNA oligos targeting Yap1 were purchased from Dharmacon. Cytoplasmic and nuclear fractions were isolated using the NE-PER Nuclear and Cytoplasmic Extraction reagents (Pierce).
DNA constructs
Retroviral αE-catenin expression vector was generated by cloning full-length mouse αE-catenin into the MCS of pLNCX2-GFP vector using BglII–EcoRI. pLNCX2-GFP vector was constructed by inserting the MCS-IRES-eGFP fragment from pBMN-I-GFP vector (G. Nolan, Addgene) into the BamHI-SalI sites of pLNCX2 vector. Retroviruses were produced using the Phoenix system (provided by G. Nolan, Stanford University, Stanford, CA (52). Plasmids carrying flag-tagged human Yap1 and Lats1 were generated by Dr. Marius Sudol and obtained from Addgene (53). Retroviral human Yap2 expression construct was generated by Dr. Joan Brugge and obtained from Addgene (54). GAL4-TEAD4 and GST-Yap2 plasmids were generated by Dr. Kun-Liang Guan and obtained from Addgene (35). pUAS-Luc2 was generated by Dr. Liqun Luo and obtained from Addgene (55). Retroviral expression construct encoding S94A mutant human Yap2 was generated using a site-directed mutagenesis kit (Stratagene) and the following oligos: 5′-ggaagctgcccgacgccttcttcaagccg-3′ and 5′-cggcttgaagaaggcgtcgggcagcttcc-3′.
Luciferase assay
HEK293FT cells were seeded in 96 well plates. Firefly luciferase reporter, CMV-Renilla luciferase, and the indicated plasmids were cotransfected. 48 hours after transfection, cells were lysed and luciferase activity was assayed using the Dual Glo Luciferase assay system (Promega) following the manufacturer’s instructions. Firefly luciferase activity was normalized to Renilla Luciferase activity. All assays were done in quadruplicate.
RNA extraction and qRT-PCR
Total RNA was extracted using TRIZOL (Invitrogen) and cDNA was prepared using Superscript III First Strand Synthesis kit (Invitrogen). qPCR was performed using Prism 7900HT (Applied Biosystems), platinum qPCR mix (Invitrogen), and Universal ProbeLibrary kit utilizing the primers, probes and PCR conditions recommended by the Universal ProbeLibrary assay center (www.roche-applied-science.com/sis/rtpcr/upl/adc.jsp). qPCR data was normalized to ribosomal protein Rps16.
Data analysis
Statistical significance was determined by the unpaired Student’s t test, ANOVA test, or the Mann-Whitney test. P value is indicated by asterisk or asterisks in the figures. * denotes P<0.05; **, P<0.01; ***, P<0.001. Differences at P<0.05 and lower were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Elaine Fuchs for encouragement and helpful advice; all members of our laboratory for suggestions and comments; Dr. H. Denny Liggitt for help with skin pathology analysis. Elise Herman, Liem Nguyen, Nicholas Ramirez, and Dr. Ewa Stepniak for help with the maintenance of mutant mice. Drs. Julia Segre, Stephen J. Kaufman and the Developmental Studies Hybridoma Bank for gifts of antibodies; This work was supported by NCI grants R01 CA098161 and R01 CA131047 to VV. Dr. Mark Silvis was partially supported by the Chromosome Metabolism and Cancer Training Grant NIH T32 CA09657.
References and Notes
- 1.Lien WH, Klezovitch O, Fernandez TE, Delrow J, Vasioukhin V. alphaE-catenin controls cerebral cortical size by regulating the hedgehog signaling pathway. Science. 2006;311:1609–1612. doi: 10.1126/science.1121449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lien WH, Klezovitch O, Vasioukhin V. Cadherin-catenin proteins in vertebrate development. CurrOpin Cell Biol. 2006;18:499–506. doi: 10.1016/j.ceb.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Meng W, Takeichi M. Adherens junction: molecular architecture and regulation. Cold Spring Harb Perspect Biol. 2009;1:a002899. doi: 10.1101/cshperspect.a002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol. 2010;2:a002915. doi: 10.1101/cshperspect.a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 6.Hirano S, Kimoto N, Shimoyama Y, Hirohashi S, Takeichi M. Identification of a neural alpha-catenin as a key regulator of cadherin function and multicellular organization. Cell. 1992;70:293–301. doi: 10.1016/0092-8674(92)90103-j. [DOI] [PubMed] [Google Scholar]
- 7.Herrenknecht K, Ozawa M, Eckerskorn C, Lottspeich F, Lenter M, Kemler R. The uvomorulin-anchorage protein alpha catenin is a vinculin homologue. Proc Natl Acad Sci USA. 1991;88:9156–9160. doi: 10.1073/pnas.88.20.9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torres M, Stoykova A, Huber O, Chowdhury K, Bonaldo P, Mansouri A, Butz S, Kemler R, Gruss P. An alpha-E-catenin gene trap mutation defines its function in preimplantation development. Proc Natl Acad Sci USA. 1997;94:901–906. doi: 10.1073/pnas.94.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benjamin JM, Nelson WJ. Bench to bedside and back again: molecular mechanisms of alpha-catenin function and roles in tumorigenesis. Semin Cancer Biol. 2008;18:53–64. doi: 10.1016/j.semcancer.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lien W-H, Vasioukhin V. In: Loss of Cadherin-Catenin Adhesion System in Invasive Cancer Cells. Thomas-Tikhonenko A, editor. Cancer Genome and Tumor MicroenvironmentSpringer; New York: 2010. pp. 33–66. [Google Scholar]
- 11.Liu TX, Becker MW, Jelinek J, Wu WS, Deng M, Mikhalkevich N, Hsu K, Bloomfield CD, Stone RM, DeAngelo DJ, Galinsky IA, Issa JP, Clarke MF, Look AT. Chromosome 5q deletion and epigenetic suppression of the gene encoding alpha-catenin (CTNNA1) in myeloid cell transformation. Nature medicine. 2007;13:78–83. doi: 10.1038/nm1512. [DOI] [PubMed] [Google Scholar]
- 12.Vasioukhin V, Bauer C, Degenstein L, Wise B, Fuchs E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell. 2001;104:605–617. doi: 10.1016/s0092-8674(01)00246-x. [DOI] [PubMed] [Google Scholar]
- 13.Kobielak A, Fuchs E. Links between alpha-catenin, NF-kappaB, and squamous cell carcinoma in skin. Proc Natl Acad Sci USA. 2006;103:2322–2327. doi: 10.1073/pnas.0510422103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bullions LC, Notterman DA, Chung LS, Levine AJ. Expression of wild-type alpha-catenin protein in cells with a mutant alpha-catenin gene restores both growth regulation and tumor suppressor activities. Mol Cell Biol. 1997;17:4501–4508. doi: 10.1128/mcb.17.8.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LS, Abujarour R, Ding S, Guan KL. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staley BK, Irvine KD. Warts and yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr Biol. 2010;20:1580–1587. doi: 10.1016/j.cub.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee JS, Johnson RL. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, Park O, Chang J, Simpson RM, Wang CY, Gao B, Jiang J, Yang Y. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci USA. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 21.Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, Bardeesy N. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh H, Irvine KD. Yorkie: the final destination of Hippo signaling. Trends Cell Biol. 2010;20:410–417. doi: 10.1016/j.tcb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuo L, Theis M, Alvarez-Maya I, Brenner M, Willecke K, Messing A. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
- 25.Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchs E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell. 2009;137:811–819. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burnworth B, Arendt S, Muffler S, Steinkraus V, Brocker EB, Birek C, Hartschuh W, Jauch A, Boukamp P. The multi-step process of human skin carcinogenesis: A role for p53, cyclin D1, hTERT, p16, and TSP-1. European journal of cell biology. 2006 doi: 10.1016/j.ejcb.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 29.Donehower LA. Using mice to examine p53 functions in cancer, aging, and longevity. Cold Spring Harb Perspect Biol. 2009;1:a001081. doi: 10.1101/cshperspect.a001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cichowski K, Hahn WC. Unexpected pieces to the senescence puzzle. Cell. 2008;133:958–961. doi: 10.1016/j.cell.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roenigk RK, Roenigk HH. Roenigk & Roenigk’s dermatologic surgery: principles and practice. M. Dekker; New York: 1996. [Google Scholar]
- 32.Zhao B, Lei QY, Guan KL. The Hippo-YAP pathway: new connections between regulation of organ size and cancer. Curr Opin Cell Biol. 2008;20:638–646. doi: 10.1016/j.ceb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benjamin JM, Kwiatkowski AV, Yang C, Korobova F, Pokutta S, Svitkina T, Weis WI, Nelson WJ. AlphaE-catenin regulates actin dynamics independently of cadherin-mediated cell-cell adhesion. J Cell Biol. 2010;189:339–352. doi: 10.1083/jcb.200910041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neto-Silva RM, de Beco S, Johnston LA. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell. 2010;19:507–520. doi: 10.1016/j.devcel.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H, Pasolli HA, Fuchs E. Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proc Natl Acad Sci U S A. 2011;108:2270–2275. doi: 10.1073/pnas.1019603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, Appelbaum E, Deschryver K, Davies S, Guintoli T, Crowder R, Tao Y, Snider JE, Smith SM, Dukes AF, Sanderson GE, Pohl CS, Delehaunty KD, Fronick CC, Pape KA, Reed JS, Robinson JS, Hodges JS, Schierding W, Dees ND, Shen D, Locke DP, Wiechert ME, Eldred JM, Peck JB, Oberkfell BJ, Lolofie JT, Du F, Hawkins AE, O’Laughlin MD, Bernard KE, Cunningham M, Elliott G, Mason MD, Thompson DM, Jr, Ivanovich JL, Goodfellow PJ, Perou CM, Weinstock GM, Aft R, Watson M, Ley TJ, Wilson RK, Mardis ER. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beronja S, Livshits G, Williams S, Fuchs E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nature medicine. 2010;16:821–827. doi: 10.1038/nm.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheikh F, Chen Y, Liang X, Hirschy A, Stenbit AE, Gu Y, Dalton ND, Yajima T, Lu Y, Knowlton KU, Peterson KL, Perriard JC, Chen J. alpha-E-catenin inactivation disrupts the cardiomyocyte adherens junction, resulting in cardiomyopathy and susceptibility to wall rupture. Circulation. 2006;114:1046–1055. doi: 10.1161/CIRCULATIONAHA.106.634469. [DOI] [PubMed] [Google Scholar]
- 40.Nemade RV, Bierie B, Nozawa M, Bry C, Smith GH, Vasioukhin V, Fuchs E, Hennighausen L. Biogenesis and function of mouse mammary epithelium depends on the presence of functional alpha-catenin. Mech Dev. 2004;121:91–99. doi: 10.1016/j.mod.2003.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badouel C, Garg A, McNeill H. Herding Hippos: regulating growth in flies and man. Curr Opin Cell Biol. 2009;21:837–843. doi: 10.1016/j.ceb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 43.Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 44.Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Dev Cell. 2010;19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 45.Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 47.Klezovitch O, Fernandez TE, Tapscott SJ, Vasioukhin V. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 2004;18:559–571. doi: 10.1101/gad.1178004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci U S A. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Zinkel S, Polonsky K, Fuchs E. Transgenic studies with a keratin promoter-driven growth hormone transgene: prospects for gene therapy. Proc Natl Acad SciU S A. 1997;94:219–226. doi: 10.1073/pnas.94.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6:331–343. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- 52.Swift S, Lorens J, Achacoso P, Nolan GP. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. In: Coligan John E, et al., editors. Current protocols in immunology. Unit 10. Chapter 10. 2001. p. 17C. [DOI] [PubMed] [Google Scholar]
- 53.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 54.Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lien WH, V, Gelfand I, Vasioukhin V. Alpha-E-catenin binds to dynamitin and regulates dynactin-mediated intracellular traffic. J Cell Biol. 2008;183:989–997. doi: 10.1083/jcb.200805041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.