The ELF4-ELF3-LUX Complex Links the Circadian Clock to Diurnal Control of Hypocotyl Growth (original) (raw)
. Author manuscript; available in PMC: 2012 Jan 21.
Published in final edited form as: Nature. 2011 Jul 13;475(7356):398–402. doi: 10.1038/nature10182
Abstract
The circadian clock is required for adaptive responses to daily and seasonal changes in environmental conditions1-3. Light and the circadian clock interact to consolidate the phase of hypocotyl cell elongation to dawn under diurnal cycles in Arabidopsis thaliana4-7. Here we identify a protein complex (Evening Complex) composed of EARLY FLOWERING 3 (ELF3), EARLY FLOWERING 4 (ELF4) and the transcription factor LUX ARRHYTHMO (LUX) that directly regulates plant growth8-12. ELF3 is both necessary and sufficient to form a complex between ELF4 and LUX, and the complex is diurnally regulated, peaking at dusk. ELF3, ELF4 and LUX are required for the proper expression of the growth-promoting transcription factors PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) and PIF5 under diurnal conditions4,6,13. LUX targets the complex to the promoters of PIF4 and PIF5 in vivo. Mutations in PIF4 and/or PIF5 are epistatic to the loss of the ELF4-ELF3-LUX complex, suggesting that regulation of PIF4 and PIF5 is a critical function of the complex. Therefore, the Evening Complex underlies the molecular basis for circadian gating of hypocotyl growth in the early evening.
The circadian clock is an endogenous molecular oscillator with a period of ∼24 hours that is present nearly ubiquitously1. In plants, multiple interlocking transcriptional feedback loops contribute to the robust architecture of this oscillator network3. The clock functions to enable anticipation of diurnal rhythmic environmental changes, allowing for the optimal phasing of molecular, physiological and behavioral responses to specific times of day2. Plant growth is a physiological response that is controlled by both the clock and changes in light conditions, and under diurnal growth conditions, maximal plant growth occurs at the end of night4-7.
EARLY FLOWERING 3 and EARLY FLOWERING 4 were first identified in genetic screens for photoperiodism mutants and were found to regulate circadian rhythms8-10,14. ELF3 and ELF4 encode nuclear, plant-specific proteins with no known functional domains9,10,15,16. LUX ARRHYTHMO (also PHYTOCLOCK 1) is a single-MYB domain SHAQYF-type GARP transcription factor identified in a genetic screen for long hypocotyl mutants and aberrant circadian-regulated gene expression11,12. elf3, elf4 and lux share multiple phenotypes, including an arrhythmic circadian oscillator, abnormal hypocotyl growth under diurnal cycles and early flowering4,8-12,14,15,17. ELF3, ELF4 and LUX showed similar expression profiles in microarray experiments (Figure S1; http://diurnal.cgrb.oregonstate.edu/18,19), and these expression profiles were confirmed by quantitative RT-PCR analysis in diurnal and circadian conditions (Figure 1a).
Figure 1. ELF3, ELF4 and LUX are co-expressed and ELF3 directly interacts with both ELF4 and LUX in yeast.

a) Expression of ELF3, ELF4 and LUX under diurnal or circadian conditions. Normalization is relative to maximum. Bars above the graphs represent light conditions during harvesting; black=lights off, white=lights on, grey=lights on during subjective night. Error bars represent the S.E.M., n=3. b) Yeast two-hybrid assay between ELF4 and ELF3, ELF4, LUX, LUX-N or LUX-C. C) Yeast two-hybrid assay between LUX and ELF3, ELF4 or LUX. These experiments were repeated twice (b and c). d) Yeast three-hyrbid containing combinations of ELF4-GAL4-DBD, LUX-GAL4-AD and ELF3. Error bars are S.E.M., n=4, presented as fold of induction over control vectors.
The similarities in expression patterns and phenotypes prompted us to test whether these proteins could interact. Using a yeast two-hybrid assay, we found that ELF4 interacted with ELF3 (Figure 1b). Also, when LUX-fragments were used as baits (full-length LUX showed auto-activation, data not shown), ELF3 showed an interaction with LUX-C (a.a. 144-324), containing the DNA-binding domain of LUX11,12,20, but not with LUX-N (a.a. 1-143) (Figure 1c). ELF4 and LUX did not interact in any combination (Figure 1b and c). As ELF3 could interact independently with either ELF4 or LUX, we hypothesized that ELF3 might form a complex between these two proteins. To test this, ELF3 was used in a yeast three-hybrid system with ELF4-GAL4-DBD and/or LUX-GAL4-AD. Activation of the reporter was observed only when all three proteins were present, suggesting that ELF3 was sufficient to bridge an interaction between ELF4 and LUX (Figure 1d).
Next, we tested whether ELF4, ELF3 and LUX interact in vivo. Antibodies were developed against ELF3 and LUX and an ELF4∷ELF4-HA construct was introduced into the elf4-2 mutant21. The ELF4-HA protein is likely functional, as we identified transformants that rescued the hypocotyl length (Figure S2a) and circadian CHLOROPHYLL A/B BINDING PROTEIN∷LUCIFERASE (CAB2∷LUC) rhythmicity, albeit with a shorter period (Figure S2b-d). We then asked whether ELF4-HA could co-immunoprecipitate endogenous ELF3 and/or LUX at Zeitgeber time (ZT) 12 (Figure 2a). We found that ELF4-HA could co-immunoprecipitate both ELF3 and LUX (Figure 2a and S2f). The experiments in yeast suggested that ELF3 bridges an interaction between ELF4 and LUX, and that ELF3 would be necessary for the co-immunoprecipitation of LUX by ELF4-HA. To test this, we introduced elf3-1 into the ELF4∷ELF4-HA elf4-2 and immunoprecipitated ELF4-HA. Although similar amounts of ELF4 and LUX were present in the extracts, LUX did not co-immunoprecipitate with ELF4-HA (Figure 2a). These results show that ELF3 is necessary for tripartite complex formation with ELF4 and LUX in vivo. Furthermore, hypocotyl length in elf3-1 elf4-3 and elf3-1 lux-4 double mutants grown under 12 h light: 12 h dark (12L:12D) did not exhibit additive effects over elf3 (Figure S3). These results are consistent with the hypothesis that ELF3, ELF4 and LUX function together as a complex to regulate common pathways.
Figure 2. ELF3 bridges a diurnally regulated complex containing ELF4 and LUX in vivo.

a) ELF3 is necessary for ELF4 and LUX to co-precipitate in vivo. Immunoprecipitations (IP) were performed on day 12 at ZT12, 12L:12D. b) ELF3, ELF4 and LUX oscillate and form a complex. (*): 15% gel, (·): bead background band, (–): LUX isoforms. c and d) Evening Complex formation in short and long days. Seedlings were grown under short– or long–day photoperiods (8L:16D or 16L:8D, respectively) and harvested beginning at ZT0 on day 12. Experiments were performed three times with similar results. Bars represent light conditions during harvesting as denoted in Figure 1a.
Since ELF4, ELF3 and LUX mRNA levels oscillate and peak with a similar phase, we analyzed the dynamics of the protein levels under diurnal cycles. _ELF4_∷ELF4-HA tissue was harvested every 4 hours, starting at ZT12 under 12L:12D cycles and then after transferring to constant light at ZT0 the following day. ELF3, LUX and ELF4-HA peaked at ZT12, declined during the night, reached a trough between ZT0-ZT4, and then increased again (Figure 2b,S4). The levels of all three proteins remained elevated into the subjective dark period relative to their respective time points in the dark and the protein peak was now shifted to the middle of the subjective night (Figure 2b). Comparable results were observed for ELF3 and LUX levels in wild-type seedlings (Figure S5). To assay for time-dependent formation of the ELF4-ELF3-LUX “Evening Complex” (EC), ELF4-HA was immunoprecipitated from the diurnal samples. Formation of the EC followed the same pattern as that of its composite parts, suggesting that they would associate when present (Figure 2b).
Photoperiodic control of flowering and growth is compromised in elf3, elf4 and lux mutants4,8-12,14,15,17,22. To determine how ELF4, ELF3 and LUX respond to altered photoperiods, we analyzed the levels and formation of the EC in plants grown under short (8L:16D) and long days (16L:8D). Peak levels of ELF4, ELF3 and LUX followed their respective RNA profiles under different photoperiods (Figure 2c and d, S4), similar to previous reports9,10,15,22. Complex formation was also sensitive to photoperiod, peaking earlier in short days compared to long days (Figure 2c and d).
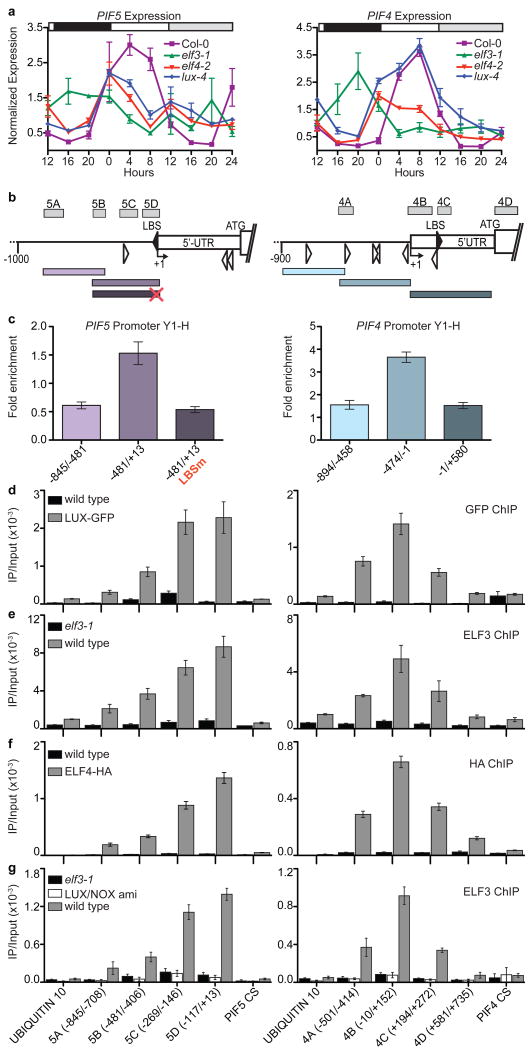
To investigate the molecular role of the EC, we focused on the diurnal hypocotyl growth phenotype shared by all mutants5,8-11,20. Previous work demonstrated that the bHLH transcription factors PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) and PIF5 (also known as PHYTOCHROME-INTERACTING FACTOR LIKE 6) are critical components that determine the hypocotyl elongation rate in seedlings, and that both genes act downstream of light- and clock-signaling pathways4,6,7,13,23. Expression of PIF4 and PIF5 was nearly anti-phasic to the EC under different photocycles (Figure S6). This raised the possibility that the EC may be repressing the transcription of PIF4 and PIF5, consistent with recent reports that ELF3 and LUX act as transcriptional repressors in the circadian clock20,24. The levels of PIF4 and PIF5 are elevated in elf3-1, elf4-2 and lux-4, particularly during the early evening (Figure 3a). Recent work demonstrated that the addition of an activation domain to LUX (LUX-VP64) induced a neomorphic hypocotyl elongation phenotype20 and we found that PIF4/5 expression levels were increased in this background (Figure S7). These results, as well as the presence of full consensus LUX binding sites (LBS)20 in the 5′-UTR of both PIF4 and PIF5 (Figure 3b), suggested that LUX may participate directly in the modulation of PIF4/5 expression. Indeed, LUX could directly bind the PIF4 and PIF5 promoters in yeast, and LUX binding to the PIF5 promoter (-481/+13 bp) was lost when the consensus LBS was mutated (Figure 3c).
Figure 3. The Evening Complex regulates PIF5 and PIF4 expression through recruitment by LUX.
a) PIF5 and PIF4 expression in elf3, elf4, lux and wild type (Col-0). Bars indicate light conditions as denoted in Figure 1a. Error bars represent the S.E.M., n=3. b) PIF5 (left) and PIF4 (right) promoters denoting degenerate (GATWCK or GATWYG) or consensus (GATWCG) LBS, unfilled or filled arrowheads, respectively. Numbers are relative to transcriptional start (+1). Bars are ChIP amplicons (above) and fragments for yeast one-hybrid (below), red × denotes mutated LBS. c)Yeast one-hybrid (Y 1-H) with LUX-AD and PIF5 and PIF4 promoter fragments. Fold enrichment is relative to controls. d) LUX, e) and g) ELF3 and f) ELF4 ChIP on PIF5 and PIF4 at ZT14, extended light. CS=coding sequence (d-g). Error bars represents the S.E.M. n=3 (d and e). g) Endogenous ELF3 ChIP from elf3-1, LUX/NOX ami and wild type. Error bars represent the S.D. of the average of two technical replicates measured twice. Experiments were repeated with similar results (f and g).
To determine if members of the EC were bound to the PIF4 and PIF5 promoters in vivo, chromatin immunoprecipitations (ChIP) was performed in LUX_∷LUX-GFP transgenic lines followed by amplification of PIF4 and PIF5 promoter sequences. These experiments revealed in vivo binding to the LBS in the PIF4 and PIF5 promoters, but not to control sequences in their coding regions or in the UBIQUITIN10 (UBQ10) promote_r (Figure 3d). The formation of the EC (Figure 2) suggested that all members might participate in the regulation of PIF4/5 expression; therefore, we performed similar ChIP experiments for ELF3 and ELF4-HA. We found that ELF3 and ELF4-HA showed specific enrichment at PIF4 and PIF5 promoter sequences that were also bound by LUX (Figure 3e-f). Additionally, ELF3 ChIP experiments performed at the trough of EC levels (ZT2) showed a lower specific enrichment relative to ZT14 (Figure S8).
The localization pattern of the EC members on the PIF4/5 promoters suggested that the transcription factor LUX might be responsible for recruitment. ELF3 ChIP experiments in lux-4 seedlings demonstrated a lower, but incomplete loss of recruitment of ELF3 to the PIF4/5 promoters (Figure S9). Previous work identified a MYB transcription factor highly similar to LUX, named NOX11,12,20. NOX binds to similar sequences as LUX in yeast20 and could also form a complex with ELF4 and ELF3 (Figure S10a). We generated an artificial microRNA transgenic line that would simultaneously reduce the levels of both NOX and LUX (LUX/NOX ami), designed using a web-based amiRNA algorithm (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi)25,26. LUX protein and NOX expression levels are reduced in this line (Figure S10b and c), which showed similar defects in circadian rhythms to lux-4 (Figure S10e and f); however, we observed an increase in hypocotyl length and PIF4/5 expression level compared to lux-4 (Figure S10d and g). When ELF3 ChIP was performed in the LUX/NOX ami line, we observed loss of ELF3 signal at the PIF4 and PIF5 promoters (Figure 3g). ELF3 was still present in extracts from these plants (Figure S10c), suggesting that recruitment of ELF3 (and therefore the EC) is mediated by both LUX and NOX.
Previous reports showed that ectopic overexpression of the MYB transcription factors CIRCADIAN CLOCK ASSOCIATED 1 (in CCA1-OX) or LATE ELONGATED HYPOCOTYL (in lhy-1) resulted in phenotypes similar to elf3, elf4 or lux (Figure 3a and S11)4-6. As CCA1 and LHY forms a complex that controls the expression of evening element containing genes27, such as ELF4 and LUX11, 28, the misexpression of PIF4 and PIF5 seen in CCA1-OX or lhy-1 could be a result of EC misregulation. Therefore, we analyzed the expression of ELF4, ELF3 and LUX in the lhy-1 background using the DIURNAL database18, 19. We found that ELF4 is clamped low, while ELF3 and LUX are shifted 4 and 12 hours later, respectively (Figure S11). These results are consistent with the circadian clock playing a critical role in the proper expression and phasing of the EC proteins.
If improper regulation of PIF4 and PIF5 underlies the hypocotyl growth defects observed in the EC mutants, then loss of PIF4 and PIF5 should be epistatic to loss of the EC. To test this, we introduced pif4 and pif5 mutant alleles into the elf3-2 mutant background, since mutating ELF3 caused dissolution of the EC (Figure 2a). Loss of PIF4 or PIF5 additively mitigated the hypocotyl length defect elf3-2 (Figure 4a and b), indicating that the hypocotyl phenotypes of EC mutants are mainly caused by misexpression of PIF4 and PIF5. In addition, loss of PIF4 or/and PIF5 did not restore circadian rhythms in an elf3 background (Figure S12), consistent with PIF4 and PIF5 being clock outputs that do not feedback into the oscillator4.
Figure 4. Hypocotyl growth defects are rescued by loss of PIF5 and PIF4 in EC member mutant backgrounds.

a) Growth defects in elf3-2 background require PIF4/5. Scale bar = 5mm. b) Scatter plot of hypocotyl measurements from wild type, elf3-2, pif4-101 and pif5-1 single and compound mutants. This experiment was repeated with similar results. c) Model represents EC action on PIF4/5 expression during early evening to gate hypocotyl growth in Arabidopsis seedlings. The circadian-regulated EC represses P_IF4/5_ expression in the evening. Throughout the day, post-transcriptional light-mediated degradation of PIF4/5 proteins inhibit growth. Near dawn, concomitant rise of PIF4/5 RNA and PIF4/5 protein levels promote growth (white arrow).
To summarize, we have identified a novel multi-protein complex that directly links the circadian clock to diurnal regulation of hypocotyl growth. The ELF4-ELF3-LUX complex is regulated by the clock and light (Figure 1a, 2b-d) and represses the expression of PIF4 and PIF5 in the early evening (Figure 4c). This is combined with light-regulated turnover of PIF4/5 to permit maximal hypocotyl growth at dawn in diurnal conditions4,13 (Figure 4c). ELF3 is necessary and sufficient to bring together ELF4 and LUX to form a complex (Figure 1d and 2a), providing a mechanistic framework to understand their shared phenotypes in regulating circadian rhythms, growth and flowering. The role of ELF3 as an adaptor protein is similar to its previously described capacity to modulate GIGANTEA levels through association with CONSTITUTIVELY PHOTOMORPHOGENIC 1 to regulate flowering and circadian rhythms22. The EC is composed of multiple proteins known to regulate signaling from the environment5,9-12,14-17,20,22,24,28; therefore, elucidating Evening Complex function will ultimately contribute to understanding how the clock gates biochemical, physiological and developmental outputs.
Methods Summary
All wild-type, mutant and transgenic lines were in the Arabidopsis thaliana ecotype Columbia-0 (Col-0). All transgenic and mutant lines were brought to homozygosity before use. The procedures for Arabidopsis husbandry; yeast one-hybrid, two-hybrid, and three-hybrid analyses; biolumenscent imaging; immunoprecipitations; chromatin immunoprecipitations; and hypocotyl measurements were as described previously20,29,30, with modifications detailed in Methods. In all growth chambers, light was supplied at 80 μmol m-2 sec-1 by cool-white fluorescent bulbs at 22 °C. For yeast two-hybrid analyses, SD-WL medium selects for the presence of both bait and prey vectors, and SD-WLHA medium selects for an interaction between bait and prey proteins. IPP2, APX3 and At1g11910 were used to normalize real-time PCR expression analyses, and all primers for quantitative PCR are listed in Supplemental Table 1. ELF4∷ELF4-HA construct includes 580 bp of promoter sequence cloned from Col-0 DNA amplified using primers listed in Supplemental Table 1. The sequence TATGATATCCTTGCGTACCCA is the target of the LUX/NOX amiRNA. Antibodies were generated in rabbits (Sigma Genosys) against either an ELF3 specific peptide (CSIQEERKRYDSSKP), or a full-length LUX protein fused to glutathione-S-transferase (GST). Antibodies were affinity purified against the same ELF3 peptide using a SulfoLink Immobilization kit (Thermo Scientific) or using a GST-LUX affinity column. All immunoprecipitations were performed with Protein G Dynabeads (Invitrogen). For westerns, ACTIN served as a loading control. Blots for ELF4 represent 20% of the total IP sample, as ELF4 must be run on a separate 15% gel, identified by (*), due to its low molecular weight. The dot (·) denotes a background signal arising from the cross-linked HA beads (data not shown). LUX runs as high and low molecular weight isoforms, denoted by (–). Hypocotyl measurements were performed on evenly spaced seedlings grown under 12L:12D, measured on day 10.
Supplementary Material
1
Acknowledgments
Pagkapol Y. Pongsawakul illustrated the model in Figure 4C. We thank Ghislain Breton, Tsuyoshi Hirota, Jose Pruneda-Paz, Dawn Nagel, Elsebeth Kolmos and Benjamin J. Cole for critical reading of the manuscript. We also thank Justina Halverson, Ana Lily Quiroz and Cenobio Valdivia for excellent technical assistance. pif4-101 pif5-1 seedlings were a generous gift from Séverine Lorrain and Christian Fankhauser. Frank Harmon originally identified the nature of the elf4-3 mutation and designed the dCAPS strategy. This work was supported by a University of California, San Diego Chancellor's Undergraduate Research Scholarship to J.J.K., grants from the European Molecular Biology Organization (ALTF 236-2005 to A.H.), the US National Science Foundation (IBN-0416762 to T.F.S.), and US National Institutes of Health (NRSA GM083585 to D.A.N., NRSA GM080930 to E.E.H., R01 GM79712 to T.I., R01 GM50006 and GM67837 to S.A.K.).
Footnotes
Accession Numbers: The following database accession numbers for the genes analyzed are At4g35000 (APX3), At1g11910 (ASPARTYL PROTEASE FAMILY PROTEIN), At2g25930 (ELF3), At2g40080 (ELF4), At3g02780 (IPP2), At3g46640 (LUX), At5g59570 (NOX), At2g43010 (PIF4), and At3g59060 (PIF5).
Author contribution: D.A.N., A.H., E.E.H., T.I., T.F.S., E.M.F and S.A.K. designed the experiments. D.A.N. performed and analyzed all immunoprecipitations and ChIPs, generated and characterized the ELF3 antibody and the transgenic lines and generated the plasmids for yeast three-hybrid analysis, he also co-performed the gene expression analysis with A.H. A.H. performed the yeast two- and one-hybrid assays and generated and characterized the LUX and LUX/NOX ami transgenic lines. E.E.H. generated the LUX antibody, characterized transgenic lines and co-performed western blot analysis with D.A.N. J.J.K. measured hypocotyls, performed and analyzed the yeast three-hybrid assay and assisted with the yeast one-hybrid analysis and generation of the elf3-2 pif4-101 pif5-1 seedlings. T.I. performed the original yeast two-hybrid assays. T.F.S. generated the _ELF4_∷ELF4-HA elf4-2 line. E.M.F characterized the ELF4 transgenic lines. D.A.N. and S.A.K. wrote the manuscript.
References
- 1.Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–48. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 2.Yakir E, Hilman D, Harir Y, Green RM. Regulation of output from the plant circadian clock. FEBS J. 2007;274:335–45. doi: 10.1111/j.1742-4658.2006.05616.x. [DOI] [PubMed] [Google Scholar]
- 3.Harmer SL. The circadian system in higher plants. Annual review of plant biology. 2009;60:357–77. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 4.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–61. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 5.Michael TP, et al. A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 2008;6:e225. doi: 10.1371/journal.pbio.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niwa Y, Yamashino T, Mizuno T. Circadian Clock Regulates Photoperiodic Response of Hypocotyl Elongation through a Coincidence Mechanism in Arabidopsis thaliana. Plant Cell Physiol. 2009 doi: 10.1093/pcp/pcp028. [DOI] [PubMed] [Google Scholar]
- 7.de Montaigu A, Tóth R, Coupland G. Plant development goes like clockwork. Trends Genet. 2010;26:296–306. doi: 10.1016/j.tig.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Zagotta MT, et al. The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 1996;10:691–702. doi: 10.1046/j.1365-313x.1996.10040691.x. [DOI] [PubMed] [Google Scholar]
- 9.Hicks KA, Albertson TM, Wagner DR. EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell. 2001;13:1281–92. doi: 10.1105/tpc.13.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doyle MR, et al. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature. 2002;419:74–77. doi: 10.1038/nature00954. [DOI] [PubMed] [Google Scholar]
- 11.Hazen SP, et al. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci U S A. 2005;102:10387–92. doi: 10.1073/pnas.0503029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Onai K, Ishiura M. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells. 2005;10:963–72. doi: 10.1111/j.1365-2443.2005.00892.x. [DOI] [PubMed] [Google Scholar]
- 13.Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2007 doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- 14.Hicks KA, et al. Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science. 1996;274:790–2. doi: 10.1126/science.274.5288.790. [DOI] [PubMed] [Google Scholar]
- 15.Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell. 2001;13:1293–304. doi: 10.1105/tpc.13.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khanna R, Kikis EA, Quail PH. EARLY FLOWERING 4 functions in phytochrome B-regulated seedling de-etiolation. Plant Physiol. 2003;133:1530–8. doi: 10.1104/pp.103.030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thines B, Harmon FG. Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc Natl Acad Sci USA. 2010;107:3257–62. doi: 10.1073/pnas.0911006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mockler TC, et al. THE DIURNAL PROJECT: Diurnal and Circadian Expression Profiling, Model-Based Pattern Matching and Promoter Analysis. Cold Sping Harbor Symposium on Quantitative Biology. 2007;1:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 19.Michael TP, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008;4:e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helfer A, et al. LUX ARRHYTHMO Encodes a Nighttime Repressor of Circadian Gene Expression in the Arabidopsis Core Clock. Curr Biol. 2011;21:126–33. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazen SP, et al. Rapid array mapping of circadian clock and developmental mutations in Arabidopsis. Plant Physiol. 2005;138:990–7. doi: 10.1104/pp.105.061408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu JW, et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell. 2008;32:617–30. doi: 10.1016/j.molcel.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–4. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 24.Dixon LE, et al. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol. 2011;21:120–5. doi: 10.1016/j.cub.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwab R, Ossowski S, Riester… M. Highly specific gene silencing by artificial microRNAs in Arabidopsis. The Plant Cell. 2006 doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ossowski S, Schwab R. Gene silencing in plants using artificial microRNAs and other small RNAs. The Plant Journal. 2008 doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- 27.Lu SX, Knowles SM, Andronis C, Ong MS, Tobin EM. CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009;150:834–43. doi: 10.1104/pp.108.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kikis EA, Khanna R, Quail PH. ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 2005;44:300–13. doi: 10.1111/j.1365-313X.2005.02531.x. [DOI] [PubMed] [Google Scholar]
- 29.Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–5. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Para A, et al. PRR3 Is a Vascular Regulator of TOC1 Stability in the Arabidopsis Circadian Clock. Plant Cell. 2007;19:3462–73. doi: 10.1105/tpc.107.054775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1