Towards an encyclopaedia of mammalian gene function: the International Mouse Phenotyping Consortium (original) (raw)
Introduction
Nearly 10 years after the completion of the human genome project, and the report of a complete sequence of the mouse genome, it is salutary to reflect that we remain remarkably ignorant of the function of most genes in the mammalian genome. This is clearly illustrated from the outputs of genome-wide association studies (GWAS), in which the detection of loci that are associated with genetic diseases is often still just a starting point for more detailed studies that investigate the function of genes in the vicinity of the identified locus. Most importantly, we are far from uncovering the multiple, pleiotropic functions of genes. Determining pleiotropy will be key to deciphering the genetic networks and systems that govern developmental and physiological mechanisms. The mouse has played an important role in deciphering gene function through the analysis of mutations generated by a variety of routes (Justice et al., 2011). The mouse toolkit is exceptionally rich and sophisticated, with the ability to engineer mutations into the genome, more or less at will. However, to date, we have only analysed knockout mutations for around 30% of mouse genes (Eppig et al., 2012). Moreover, the analysis of these mutations has inevitably focused on the phenotypic domains and systems that reflect the interests and expertise of the investigator and, as a consequence, key functions and pleiotropic effects certainly have been missed. We are also remarkably poor at predicting the functions of genes and their pleiotropic effects, so it is imperative to assess gene function without making any prior assumptions.
The mouse genetics community has recognised for a number of years that we need a new approach, involving an unbiased, comprehensive and systematic effort to phenotype mutations for every gene in the mouse genome (Brown et al., 2006). A comprehensive encyclopaedia of mammalian gene function would be a remarkable resource that would underpin new developments in understanding biological systems and disease mechanisms. The International Mouse Phenotyping Consortium (IMPC; www.mousephenotype.org) has accepted the challenge and is embarking on a 10-year project to generate a null mutation for every gene in the mouse genome, to acquire broad-based phenotype data for each mutation, and to disseminate the mutant resource and phenotype data to the scientific community.
The International Knockout Mouse Consortium
A crucial step towards a global assessment of mammalian gene function was the development of an embryonic stem (ES) cell resource of knockout mutations for all protein-coding mouse genes. The International Knockout Mouse Consortium (IKMC) comprises several programmes from North America and Europe – including KOMP, EUCOMM, TIGM and NorCOMM – that have used numerous mutagenesis approaches to develop mutant ES cells for protein-coding genes (see www.knockoutmouse.org). All clones are generated using a C57BL/6N ES cell line. To date, 15,000 targeted ES cell lines are available, and we envisage complete genome coverage for protein-coding genes in a few years. Importantly, over 10,000 genes are available as targeted conditional clones. These ES cells have been targeted using a ‘knockout first, conditional ready’ approach (employed by EUCOMM and KOMP), which enables the derivation of mice carrying a null allele or, alternatively, a ‘conditional ready’ allele (see below) (Skarnes et al., 2011). These ES cell lines are currently the allele of choice for the generation and phenotyping of mice in the IMPC. The ES cells are available from the KOMP repository (www.knockoutmouse.org) or the European Mouse Mutant Cell Repository (EuMMCR).
Mouse clinics and pilot large-scale phenotyping efforts
In parallel with the development of the mouse mutant resources, several mouse genetics centres have established large-scale mouse phenotyping centres, so-called ‘mouse clinics’, that can carry out broad-based primary phenotyping across a number of organ and disease systems. Several programmes have undertaken pilot studies for large-scale mouse phenotyping; these include the European EUMODIC programme (www.eumodic.org), the Mouse Genetics Project (MGP) at the Sanger Institute (www.sanger.ac.uk/resources/mouse), KOMP312 at UC Davis (www.kompphenotype.org) and the German Mouse Clinic (GMC; www.mouseclinic.de). For example, the EUMODIC programme has generated 500 mouse mutants from EUCOMM ES cell lines, and carried out primary phenotyping using the EMPReSS phenotyping protocols (www.empress.har.mrc.ac.uk) that were established and validated under a previous project, EUMORPHIA (www.eumorphia.org). Such programmes provide important insights into the logistics of mouse production and phenotyping, the use of the assays in the phenotyping pipeline for disease model identification, the sensitivity of the assays, and the bioinformatics issues of data capture, analysis and dissemination. The EuroPhenome database (www.europhenome.org) was established to hold and disseminate both raw and annotated data from EUMODIC. As of January 2012, EuroPhenome contained phenotype data on 419 mutant lines, encompassing 26,258 mice, 8.22 million data points and 3416 phenotype annotations. Importantly, excluding viability and fertility assessments, 78% of the mutant lines analysed showed at least one phenotype annotation, and the majority (67%) showed more than one phenotype annotation. Thus, encouragingly, pleiotropy was frequently revealed. Nevertheless, programmes such as these emphasise the need to raise hit rates. This will demand improvements to existing tests that increase sensitivity, but is also likely to involve the introduction of additional phenotyping modalities, such as imaging, which will enrich phenotype discovery. Additionally, 34% of the lines showed embryonic lethality, indicating that a full embryonic assessment of these mutant mice will reveal further interesting phenotypes.
The IMPC
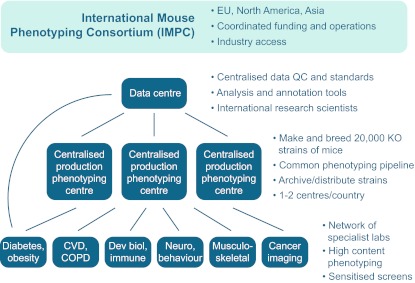
The outcomes of programmes such as EUMODIC, the MGP, KOMP 312 and the GMC demonstrated the feasibility and potential of large-scale mouse phenotyping. In addition, these projects represented an important advance in developing a plan for an international initiative that would expand the ongoing phenotyping effort from hundreds of mouse lines to thousands, with the ultimate aim of tackling the whole genome. This is the goal of the IMPC, which was launched in September 2011. The IMPC envisages a 10-year project with the aim of completing the generation and phenotyping of 20,000 mouse mutant lines by 2021. The programme encompasses a broad international consortium of mouse genetics centres, supported by diverse funding bodies (Table 1). Each mouse genetics centre will participate in the production and phenotyping of mouse mutant lines, and will deposit the data into a Data Centre for analysis and dissemination (Fig. 1). IMPC mouse genetics centres also engage with other biomedical research centres that will receive mutant mouse lines for more detailed secondary phenotype analysis. These networks will provide expertise and input that impact the design and evolution of the IMPC’s phenotyping pipelines, as well as data analysis and annotation approaches.
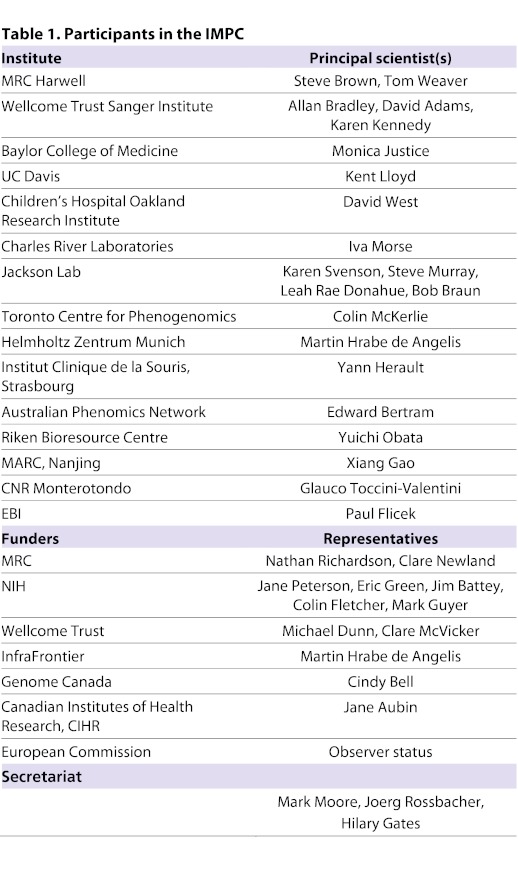
Table 1.
Participants in the IMPC
Fig. 1.
Organisation of the IMPC. A core of mouse production and phenotyping centres is allied to the data centre supporting the IMPC. The core mouse production and phenotyping centres also network with a variety of other biomedical research centres that will undertake more specialised phenotyping on selected lines, as well as provide additional expertise and input into the IMPC phenotyping programme. COPD, chronic obstructive pulmonary disease; CVD, cardiovascular disease; Dev biol, developmental biology; KO, knockout; Neuro, neurological; QC, quality control.
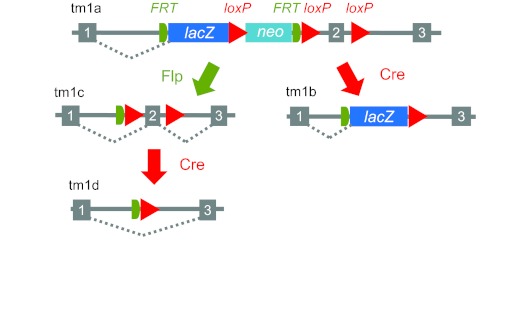
The IMPC will accomplish its goals in two phases. Phase 1, which is already funded and will run from 2011 to 2016, will generate and phenotype 5000 mouse mutants. Phase 2, which will run from 2016 to 2021, is planned to generate and phenotype 15,000 mouse mutants. The consortium will analyse null homozygous mutants generated from the IKMC resource and, in cases in which homozygotes are lethal, undertake analysis of embryos and heterozygous adults. All mice produced will be isogenic on the C57BL/6N background. Initially, mice carrying the knockout first ‘tm1a’ allele are generated and, following breeding to an appropriate Cre driver line, a key early exon within the gene is eliminated along with the selection cassette, producing the ‘tm1b’ null allele (Fig. 2). Alternatively, Flp engineering can produce the ‘tm1c’ or conditional ready allele.
Fig. 2.
Design of the knockout first, conditional ready allele employed by the IMPC. The tm1a allele is generated in ES cells (which itself provides a knockout through splicing into the lacZ reporter). Mice generated with the tm1a allele are crossed to an appropriate Cre driver line to both eliminate a key exon within the gene and remove the selection cassette, generating the tm1b null allele. The tm1b allele will be phenotyped by the IMPC. Alternatively, Flp action on the tm1a allele can produce the tm1c ‘conditional ready’ allele. Grey boxes represent exons, and grey lines show introns. Figure reproduced from Skarnes et al. (Skarnes et al., 2011) with permission.
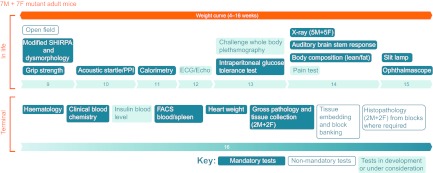
The IMPC have agreed to a consensus core adult phenotyping pipeline that will reveal phenotypes relevant to key disease areas (Fig. 3). This pipeline involves the analysis of cohorts of seven male and seven female mice with a broad spectrum of in-life tests between the ages of 9 and 15 weeks, followed by various terminal tests at 16 weeks. Additional important phenotype platforms are under development and assessment, with the aim of implementing them early in the life of the IMPC. When including the tests that are still under development, the IMPC pipeline incorporates 20 phenotyping platforms, encompassing diverse biological systems such as neurological and neuromuscular, sensory, behavioural, cardiovascular, metabolic, respiratory, bone, haematological, and clinical chemistry, among others. Moreover, it is envisaged that individual centres will incorporate additional tests of local interest into the adult phenotyping pipeline. The embryonic phenotyping pipeline will be finalised by April 2012. Importantly, the mutant allele being used by the IMPC allows tracking of gene expression using a lacZ (β-galactosidase) reporter, allowing the capture of both adult and embryonic expression patterns.
Fig. 3.
The IMPC adult phenotyping pipeline. The consensus IMPC pipeline for phenotyping in the adult is shown, highlighting mandatory tests, non-mandatory tests (which a number of centres will employ, but not all) and tests under development. In-life phenotyping is carried out from 9 to 15 weeks, after which a variety of terminal tests are employed. ECG, electrocardiogram; Echo, echocardiogram; F, female; M, male; PPI, prepulse inhibition. Schematic reproduced from www.mousephenotype.org, with permission.
Mouse mutant production has already begun, and we expect significant phenotyping data to be generated this year. Importantly, a gene selection system known as the international microinjection tracking system (iMits) has already been developed and is in use to handle the selection and assignment of genes to individual centres of the consortium, as well as to track the progress of ES cell clones from quality control to microinjection (and ultimately through mutant production and phenotyping). All of the mice being generated (both tm1a and tm1b alleles) are being frozen as sperm, providing an unparalleled resource of mouse mutants to the wider biomedical sciences community.
The underlying bioinformatics support for the IMPC is crucial to the success of the project and represents a significant challenge. Data are collected in each of the production and phenotyping centres using local laboratory information management systems, and, following local quality control, are exported to the IMPC data coordination centre (DCC). This data-staging centre will allow validation and quality control of the data before they are deposited into the IMPC central data archive. The DCC has already developed the IMPReSS database (see www.mousephenotype.org/impress), which holds and updates information on the standard phenotyping protocols, including phenotype and metadata parameters, and which is crucial for delivering robust and valid data outputs across the consortium. A variety of analytical pipelines will run on the central data archive, and the IMPC informatics programme will enhance existing tools for data analysis, including the development of automated annotation pipelines, which will assign phenotype ontology terms to the significant phenotypic outliers identified. Ultimately, it will be essential to integrate IMPC data with other data repositories, particularly those related to human mutations and disease, to ensure that the rich value of the gene-phenotype data is maximally exploited.
Future challenges
There remain several important challenges for the IMPC as we enter Phase 1, and as we ramp up for Phase 2. First, we must improve the phenotyping pipeline, continually assessing the productivity and sensitivity of phenotyping platforms and introducing alternative tests where necessary. Already we have learnt a great deal from the existing phenotyping programmes, and the scale of the IMPC will allow us to rapidly acquire and assess substantive data on existing or new phenotype tests. New phenotyping modalities, particularly using improved imaging platforms, will be needed to enhance phenotype hit rates so that we maximise the chance of acquiring phenotypic information for every mutant analysed. Second, it is imperative that we improve the sophistication and precision of phenotype descriptors. This will require continuing improvements in phenotype ontologies and also their application in annotation pipelines, which will have an enormous bearing on the utility of the resource and data to the wider biomedical sciences community. We recognise that providing intelligent tools by which, for example, a clinician can search with disease terms to identify potential mouse models of interest, will be essential. Moreover, the related but wider issue of relating disease states in mouse to man and vice versa will require more attention as the encyclopaedia develops. Third, we need to consider how to uncover phenotypes that will probably only be revealed by adopting platforms and approaches that are beyond the current resources and scope of the IMPC. For example, the acquisition of pathology data on each mutant would add considerable richness to the phenotype assessment (Schofield et al., 2012); although tissues will be banked from every mutant mouse, there is not the resource to undertake this type of analysis on a global scale. Furthermore, some phenotypes (such as immunological phenotypes) will only be revealed under challenge; although individual centres might carry out challenge phenotyping on subsets of mutants, it will be impossible to undertake these analyses comprehensively. Phenotypes that develop with aging will also not be uncovered in the IMPC pipeline because the costs of aging and re-phenotyping each mutant line would be prohibitive. In all of these areas, we need to develop approaches that bring these additional phenotypes within our grasp.
The IMPC has begun a project that has the potential to transform mammalian genetics and biology, markedly improving our understanding of gene function, disease networks, and developmental and physiological systems. The first 5 years of the IMPC represent an unparalleled opportunity to gather novel data on a substantial fraction of mouse genes, as well as to develop and improve our approaches to mammalian phenotyping, identify new disease models and expand their use in the wider biomedical sciences community.
REFERENCES
- Brown S. D. M., Hanckcok J. M., Gates H. (2006). Understanding mammalian genetic systems: The challenge of phenotyping in the mouse. PLoS Genet. 2, e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig J. T., Blake J. A., Bult C. J., Kadin J. A., Richardson J. E. (2012). The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 40, D881–D886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice M. J., Siracusa L. D., Stewart A. F. (2011). Technical approaches for mouse models of human disease. Dis. Model. Mech. 4, 305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield P. N., Vogel P., Gkoutos G. V., Sundberg J. P. (2012). Exploring the elephant: histopathology in the high-throughput phenotyping of mutant mice. Dis. Model. Mech. 5, 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W., Iyer V., Mujica A. O., Thomas M., Harrow J., Cox T., et al. (2011). A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]