Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease (original) (raw)
. Author manuscript; available in PMC: 2015 Aug 28.
SUMMARY
Specific members of the intestinal microbiota dramatically affect inflammatory bowel disease (IBD) in mice. In humans, however, identifying bacteria that preferentially affect disease susceptibility and severity remains a major challenge. Here, we used flow cytometry-based bacterial cell sorting and 16S sequencing to characterize taxa-specific coating of the intestinal microbiota with immunoglobulin A (IgA−SEQ) and show that high IgA−coating uniquely identifies colitogenic intestinal bacteria in a mouse model of microbiota-driven colitis. We then used IgA−SEQ and extensive anaerobic culturing of fecal bacteria from IBD patients to create personalized disease-associated gut microbiota culture collections with pre-defined levels of IgA coating. Using these collections, we found that intestinal bacteria selected on the basis of high coating with IgA conferred dramatic susceptibility to colitis in germ-free mice. Thus, our studies suggest that IgA−coating identifies inflammatory commensals that preferentially drive intestinal disease. Targeted elimination of such bacteria may reduce, reverse, or even prevent disease development.
INTRODUCTION
The composition of the intestinal microbiota varies substantially between individuals and is thought to be a key determinant of host susceptibility to an increasing variety of diseases (Blumberg and Powrie, 2012; Chow et al., 2011; Hooper et al., 2012; Littman and Pamer, 2011; Lozupone et al., 2012). In inflammatory bowel disease (IBD), which includes Crohn's disease and ulcerative colitis, it is believed that the intestinal microbiota plays a key role in driving inflammatory responses during disease development and progression (Abraham and Cho, 2009; Gevers et al., 2014; Knights et al., 2013). This is clearly illustrated in mouse models of IBD, where the effects of the composition of the intestinal microbiota on disease have been examined in detail (Saleh and Elson, 2011). These studies have revealed that particular bacterial taxa within the intestinal microbiota can be uniquely potent drivers of intestinal disease. For example, Prevotellaceae species drive chronic intestinal inflammation in mice with inflammasome-mediated dysbiosis and exacerbate chemically-induced colitis (Elinav et al., 2011; Scher et al., 2013), and Helicobacter species can drive colitis in mice lacking the immunoregulatory cytokine interleukin-10 (Kullberg et al., 1998). Thus, individual members of the intestinal microbiota vary dramatically in their propensity to induce inflammatory responses and, thereby, influence the development and progression of intestinal disease (Saleh and Elson, 2011).
As in mice, specific members of the human intestinal microbiota that impact disease susceptibility and/or severity by stimulating chronic inflammatory responses may also play central roles in the etiology of IBD (Packey and Sartor, 2009; Round and Mazmanian, 2009). However, identifying such potentially disease-driving members of the intestinal microbiota in humans has remained a major challenge (Knights et al., 2013; Round and Mazmanian, 2009).
IgA is the predominant antibody isotype produced at mucosal surfaces and is a critical mediator of intestinal immunity (Pabst, 2012; Slack et al., 2012). Recognition of enteric pathogens by the intestinal immune system results in the production of high-affinity, T cell-dependent, pathogen-specific IgA, which is transcytosed into the intestinal lumen. In the lumen, these antibodies can bind and ‘coat’ offending pathogens, and provide protection against infection through neutralization and exclusion. Indigenous members of the intestinal microbiota also can stimulate IgA production and can become coated with IgA (Pabst, 2012; Slack et al., 2012; van der Waaij et al., 1994). However, as compared to pathogen-induced IgA, commensal-induced IgA is generally believed to be of relatively low-affinity and specificity (Pabst, 2012; Slack et al., 2012). Thus, relative levels of bacterial coating with IgA might be predicted to correlate with the magnitude of the inflammatory response triggered by a specific intestinal bacterial species.
Since indigenous members of the intestinal microbiota that selectively impact disease susceptibility or severity are proposed to share certain features with pathogens that lead them to stimulate potent inflammatory responses (Chow et al., 2011; Hooper et al., 2012; Strober, 2013), we hypothesized that these bacteria would (i) induce high-affinity antigen-specific IgA responses and (ii) become highly coated with IgA relative to the rest of the intestinal microbiota. Thus, IgA coating might distinguish disease-driving bacteria from the remainder of the intestinal microbiota.
To test this hypothesis, we developed a system to determine taxa-specific levels of IgA coating of the intestinal microbiota and used this approach to examine IgA coating patterns in mice and humans in health and disease. We found that high coating with IgA specifically and selectively marked known disease-driving members of the intestinal microbiota in mice with a transmissible colitogenic microbiota. Furthermore, colonization of germ-free mice with members of the intestinal microbiota isolated from human IBD patients and selected based on high IgA coating conferred dramatic susceptibility to colitis in a mouse model of IBD; in contrast, germ-free mice colonized with bacteria selected based on low IgA coating showed minimal disease. Thus, our data show that high IgA coating selectively marks specific members of the mouse and human intestinal microbiota that can drive or exacerbate intestinal inflammation in a mouse model of IBD.
RESULTS
IgA−SEQ identifies highly IgA coated members of the intestinal microbiota
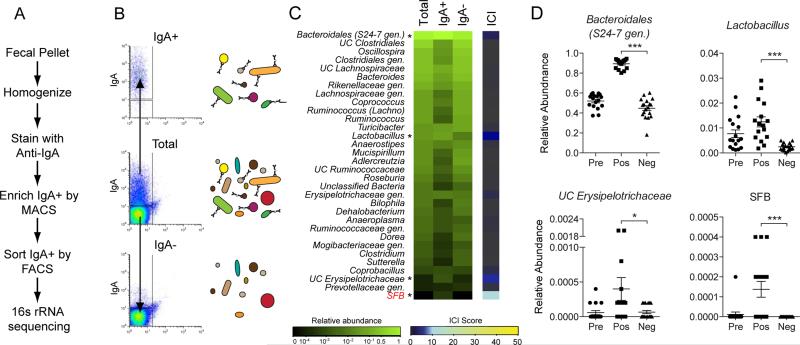
To measure taxa-specific IgA coating in an unbiased and comprehensive manner, we devised an approach that combines antibody-based bacterial cell sorting and 16S ribosomal RNA (rRNA) gene sequencing to isolate and identify IgA coated bacteria from fecal material (IgA−SEQ, Figure 1A). First, we stained fecal bacteria from specific pathogen free (SPF) mice for IgA and confirmed that only a fraction of intestinal bacteria are measurably IgA coated, as determined by flow cytometry (7.4% ± 2.2; Figures S1A-C) (Kawamoto et al., 2012; Tsuruta et al., 2009; van der Waaij et al., 1994); importantly, intestinal bacteria from recombination activating gene 2 (Rag2)-deficient mice, which cannot produce antibodies, showed minimal staining for IgA (0.5% ± 0.3). We subsequently isolated highly IgA coated (IgA+) and non-coated (IgA−) bacteria using a combination of magnetic activated cell sorting (MACS) and fluorescence activated cell sorting (FACS). The specificity and efficacy of our sorting was confirmed by reanalyzing sorted fractions via flow cytometry (Figures 1B and S1E) and ELISA (Figure S1F). After 16S rRNA gene sequencing, microbial compositions were compared and visualized using Principal Coordinates Analysis (PCoA) of weighted UniFrac distances, which revealed that, rather than comprising a random sampling of all intestinal bacteria, IgA+ bacteria represent a distinct sub-community within the intestinal microbiota (P < 0.05, PERMANOVA) (Figures S1G and S1H). Importantly, as was observed in other recent studies using FACS to sort fecal bacteria (Ben-Amor et al., 2005; Maurice et al., 2013; Peris-Bondia et al., 2011), sorting itself did not artificially alter microbial composition (_P_ > 0.05, PERMANOVA). These data demonstrate that IgA coating of the intestinal microbiota is selective across microbial taxa, and show that IgA coated bacteria represent a taxonomically distinct subset of intestinal bacteria in mice.
Figure 1. IgA−based sorting and 16S sequencing of fecal bacteria from specific pathogen free (SPF) mice.
(A) Overview of IgA−based cell sorting of fecal bacteria combined with 16S rRNA gene sequencing (IgA−SEQ).
(B) Representative results and a cartoon of cell sorting of IgA+ and IgA− fecal bacteria from mice.
(C) Heatmap depicting IgA Coating Index (ICI) scores and average relative abundance of bacterial genera in Total (Presort), IgA+ and IgA− fractions of fecal bacteria from C57Bl/6 SPF mice (n=17 samples). Relative abundance heatmaps are depicted on a logarithmic scale. Genera that are highly coated with IgA (significantly higher relative abundance in the IgA+ fraction as compared to the IgA− fraction by LEfSe; P < 0.05) are marked with an asterisk. Genera with ICI > 10 are labeled in red. UC, unclassified in the Greengenes reference database. Gen., classified as a distinct but unnamed genus in the Greengenes reference database.
(D) Relative abundance of significantly coated bacterial genera in Presort, IgA+ and IgA− fractions. * P < 0.05; *** P < 0.001 (Wilcoxon rank-sum). Indicated are mean ± standard error of the mean.
See also Figure S1 and Table S1.
To identify which specific bacterial taxa were highly coated with IgA, we examined the relative abundance of bacterial genera in total, IgA+ and IgA− bacterial fractions isolated from the feces of SPF mice (Figures 1C, 1D, S1I and Table S1). To quantify and compare relative levels of IgA coating between taxa, we calculated an IgA Coating Index (ICI) for each individual bacterial taxon as follows: ICI = relative abundance (IgA+) / relative abundance (IgA−). We then compared taxonomic abundance using the Wilcoxon rank-sum test and Linear Discriminant Analysis Effect Size (LEfSe; (Segata et al., 2011)) to determine which taxa were enriched in either the IgA+ or IgA− fractions (Figure S1J and Table S1). These analyses revealed that only four genera were significantly enriched in the IgA+ fraction in SPF mice (‘significantly coated’; P < 0.05): an unclassified genus of the family _S24-7_ from the order _Bacteroidales, Lactobacillus_, SFB, and an unclassified _Erysipelotrichaceae_ (Figures 1C and 1D). Among these bacteria, only SFB was significantly enriched in the IgA+ fraction and showed an ICI score greater than 10, which we will define as ‘highly coated’ (_P_ < 0.05; ICI > 10). In addition, 22 taxa were significantly enriched in the IgA− fraction (‘low- or non-coated’; P < 0.05), while the remaining taxa were neither enriched nor depleted by IgA−based separation.
Colitogenic members of the intestinal microbiota are highly coated with IgA in mice with inflammasome-mediated intestinal dysbiosis
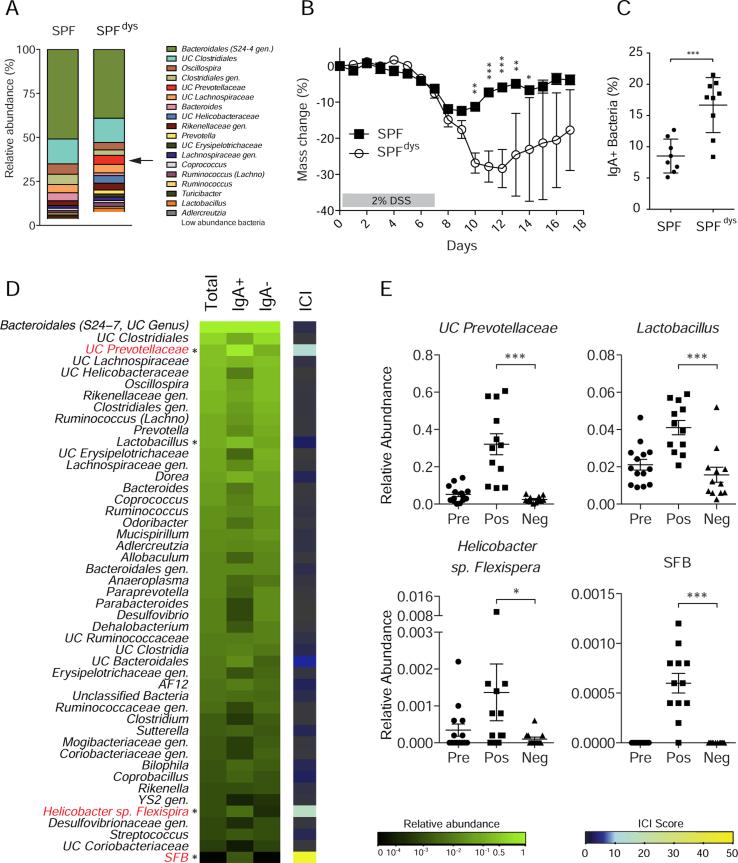
We next tested whether IgA coating would identify disease-driving members of the intestinal microbiota in the context of a colitogenic intestinal dysbiosis. We recently found that mice lacking components of the inflammasome, which is a critical mediator of innate immunity, harbor a colitogenic intestinal microbiota that can be transmitted to wild type SPF mice through co-housing. In this model, susceptibility to colitis is driven by Prevotellaceae species (Elinav et al., 2011). We therefore performed IgA−SEQ on SPF mice that had acquired inflammasome-mediated intestinal dysbiosis through co-housing with _Asc_−/− mice (SPFdys). As previously reported, co-housing with _Asc_−/− mice altered the composition of the SPF intestinal microbiota and strongly increased susceptibility to chemically-induced colitis (Figures 2A, 2B and S2A). Flow cytometric analysis of IgA coating of the intestinal microbiota of SPFdys mice at the steady state revealed an increase in the percentage of intestinal bacteria coated with IgA as compared to SPF mice, suggesting that acquisition of the colitogenic microbiota altered the pattern and/or extent of IgA coating (Figure 2C). Indeed, IgA+ bacteria in SPFdys mice were distinct from IgA−bacteria and from IgA+ bacteria in control SPF mice sampled under identical conditions (Figure S2A; P < 0.05, PERMANOVA). Although 23 taxa in SPFdys mice showed significant expansion as a result of co-housing, only two of these taxa were highly coated with IgA (Table S2, Figures 2D, 2E and S2B; P < 0.05 LEfSe); remarkably, the most abundant highly IgA coated taxon was an unclassified genus from the Prevotellaceae family, which is the defining taxon in inflammasome-mediated intestinal dysbiosis and the main driver of colitis in this model (Elinav et al., 2011). Furthermore, Helicobacter sp. flexispira, which is also acquired during co-housing, was highly coated with IgA in SPFdys mice. As in SPF mice, Lactobacillus remained coated and SFB remained highly coated in SPFdys mice.
Figure 2. IgA coating identifies colitogenic bacteria in mice with inflammasome-mediated intestinal dysbiosis (SPFdys).
(A) Average relative abundance of bacterial genera of greater than 1% abundance in the intestinal microbiota of SPF and SPFdys mice. UC Prevotellaceae is marked with an arrow. SPFdys mice were co-housed with _Asc_−/− mice to allow for the acquisition of dysbiosis.
(B) Dextran Sodium Sulfate (DSS)-induced colitis in SPF (n=5) and SPFdys (n=4) mice. * P < 0.05; ** P < 0.01; *** P < 0.001 (one-way ANOVA).
(C) IgA coating of fecal bacteria from 10-16 week old SPF (n=8) and SPFdys (n=9) mice at the steady state. *** P < 0.001 (unpaired Student's _t_-test).
(D) Heatmap depicting IgA Coating Index (ICI) scores and average relative abundance of bacterial genera in Total (Presort), IgA+ and IgA− fractions of fecal bacteria from 10-16 week old SPFdys mice (n=14 samples) sampled under steady state conditions. Relative abundance heatmaps are depicted on a logarithmic scale. Genera that are highly coated with IgA (significantly higher relative abundance in the IgA+ fraction as compared to the IgA− fraction by LEfSe; P < 0.05) are marked with an asterisk. Genera with ICI > 10 are labeled in red.
(E) Relative abundance of significantly coated genera in Presort, IgA+ and IgA− fractions. * P < 0.05; *** P < 0.001 (Wilcoxon rank-sum).
Indicated are mean ± standard error of the mean. UC, unclassified in the Greengenes reference database. Gen., classified as a distinct but unnamed genus in the Greengenes reference database. See also Figure S2 and Table S2.
Strikingly, all of the bacteria that we find are highly coated in SPFdys mice (Prevotellaceae,Helicobacter and SFB) are known to drive intestinal inflammation and disease development in mouse models of colitis (Elinav et al., 2011; Kullberg et al., 1998; Stepankova et al., 2007). In addition, SFB is a potent driver of intestinal T helper 17 (Th17) cell responses in mice and has been shown to exacerbate development of arthritis (Ivanov et al., 2009; Wu et al., 2010).
Antigen-specific binding of IgA to the intestinal microbiota can result from both high affinity, T cell-dependent responses and lower affinity, T cell-independent responses (Bemark et al., 2012). In addition, IgA can bind members of the intestinal microbiota non-specifically, for example through glycan-dependent binding to certain Gram-positive bacteria (Mathias and Corthesy, 2011). Importantly, we found that IgA coating of SFB, UC Prevotellaceae, and Helicobacter sp. flexispira was significantly reduced in SPFdys mice lacking T cells, which shows that high IgA coating detected via IgA−SEQ is largely the result of high-affinity, antigen-specific, T cell-dependent antibody responses rather than low-affinity T cell-independent responses (Figure S2C). In contrast, coating of Lactobacillus, which was significantly but not highly coated (ICI < 10), was increased in T cell-deficient mice, suggesting that these IgA antibodies resulted from either T cell-independent immune responses or non-specific binding.
IgA−SEQ identifies highly coated members of the intestinal microbiota in IBD patients
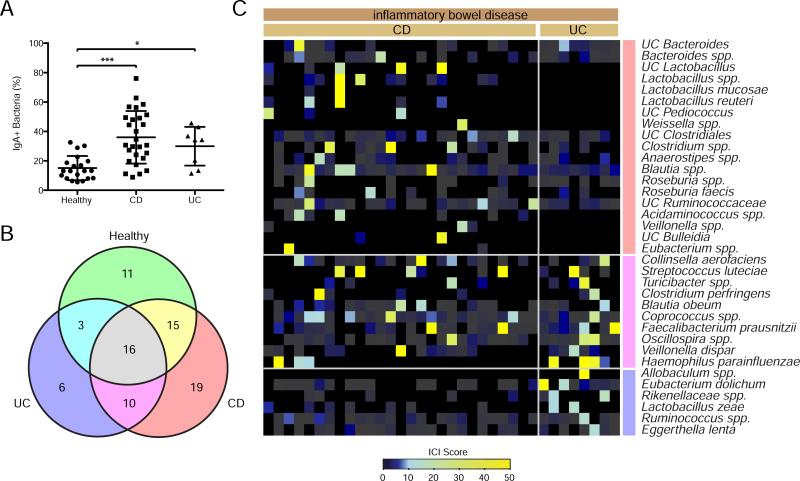
Interactions between the intestinal microbiota and the immune system play a critical role in IBD development and progression in humans; however, the specific bacteria responsible for these effects have remained elusive (Abraham and Cho, 2009; Knights et al., 2013; Round and Mazmanian, 2009). Since our data show that IgA coating can identify colitogenic members of the intestinal microbiota in mice, we next examined coating of fecal bacteria from 27 patients with Crohn's disease (CD), 8 patients with ulcerative colitis (UC) and 20 healthy controls to identify such organisms in human disease. Similar to mice with intestinal dysbiosis, and as previously reported (van der Waaij et al., 2004), the proportion of intestinal bacteria that are coated with IgA was significantly increased in CD and UC patients as compared to healthy controls (Figures 3A and S1D). As expected, both healthy control subjects and IBD patients exhibited considerable diversity in their gut microbiota compositions and patterns of IgA coating (Table S3 and Figure S3). While many species were highly coated (ICI > 10) in both IBD and control groups, 35 species were uniquely highly coated in patients with IBD (Figures 3B and 3C). For example, Streptococcus luteciae, Haemophilus parainfluenzae, and Collinsella aerofaciens were detected in both IBD patients and some healthy controls, but were only highly coated in IBD. In addition, multiple species that were uniquely present in IBD were highly coated in at least one patient (e.g., unclassified Bulleidia, Allobaculum spp., Lactobacillus mucosae, unclassified Pediococcus, and Weissella spp.). Finally, both CD- and UC-specific highly IgA coated bacterial species could be observed; for instance, unclassified Clostridiales, unclassified Ruminococcaceae, and Blautia spp. were uniquely coated in CD, and Eubacterium dolichum and Eggerthella lenta were uniquely coated in UC.
Figure 3. IgA coating of fecal bacteria from healthy humans and inflammatory bowel disease patients.
(A) IgA coating of fecal bacteria from 20 healthy subjects, 27 Crohn's disease patients (CD) and 8 Ulcerative colitis patients (UC). *P < 0.05; ***P < 0.001 (one-way ANOVA).
(B) Venn-diagram depicting the distribution of highly coated bacterial species in healthy, UC and CD patients. Bacterial taxa that showed an ICI score greater than 10 in at least one subject were classified as highly coated within that group.
(C) Heatmap depicting IgA coating index (ICI) scores for bacterial species that are uniquely highly coated (ICI > 10) in IBD and never highly coated or never present in healthy controls. Bars to the right of the heatmap correspond with the color-coding of the Venn-diagram in panel (B). Each column represents an individual human subject.
UC, unclassified in the Greengenes reference database. Spp., classified as a distinct but unnamed species in the Greengenes reference database.
See also Figure S3 and Table S3.
Establishment of a gnotobiotic mouse model to evaluate the effects of IgA+ and IgA− members of the human microbiota on intestinal inflammation
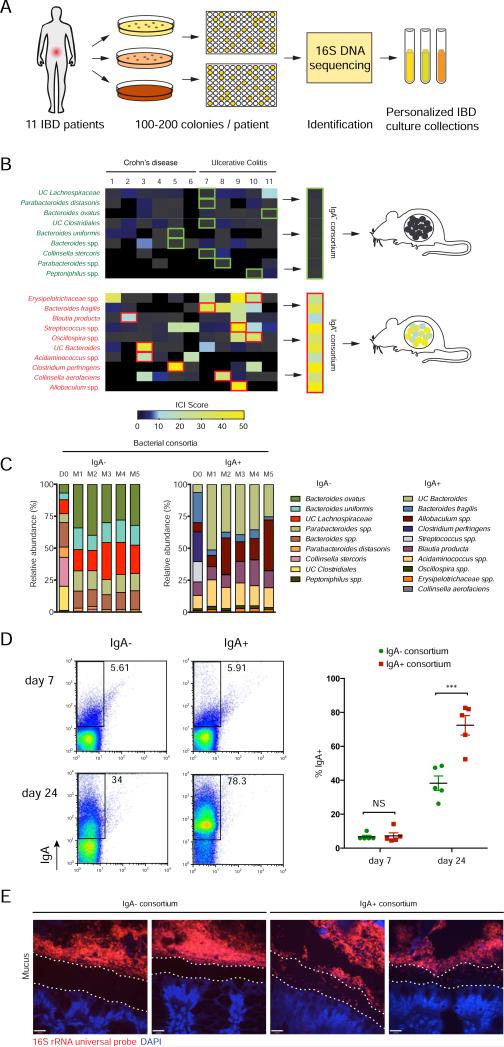
In order to directly test whether IgA coating marks human intestinal bacteria that preferentially drive intestinal inflammation, we next attempted to isolate representative IgA coated and non-coated bacteria from human IBD patients. We assembled personalized gut microbiota culture collections from eleven IBD patients using standard anaerobic culture media and a custom rich medium designed to recover intestinal bacterial species from humans (Goodman et al., 2011). First, we selected and cultured 100-200 single colonies per patient sample, and taxonomically classified these isolates through high-throughput 16S sequencing (Figure 4A). We then cross-referenced the individual bacterial isolates in each of the personalized gut microbiota culture collections with the matching data from our IgA−SEQ studies to classify all isolates based on their level of IgA coating. Finally, we rationally selected and combined individual isolates from these culture collections to assemble representative consortia consisting of either isolates that were classified as highly coated (IgA+ consortium) or isolates that were classified as low coated (IgA− consortium) (Figure 4B; see methods for criteria used to select the IgA+ and IgA− consortia). Importantly, the taxa comprising the IgA+ and IgA−consortia would not have been chosen simply based on traditional evaluations of the relative abundance of these taxa in healthy versus sick individuals (Fig. S4).
Figure 4. Isolation of personalized IBD-associated gut microbiota culture collections, assembly of IgA+ and IgA− consortia and colonization of germ-free mice.
(A) Strategy for isolation of personalized IBD-associated gut microbiota culture collections.
(B) Selection of individual bacterial isolates comprising IgA+ and IgA− consortia and colonization of germ-free mice. Specific isolates that were included in the consortia are boxed in green (IgA−) or red (IgA+).
(C) Barplots depicting relative abundance of bacterial taxa in IgA+ and IgA− consortia pre-gavage (D0) and in the feces of IgA+ and IgA− colonized mice 2 weeks post-colonization. All members of the pre-gavage consortia were detectable in colonized mice except Peptinophilus spp. in the IgA− consortium and Streptococcus spp. in the IgA+ consortium. UC, unclassified in the Greengenes reference database. Spp., classified as a distinct but unnamed species in the Greengenes reference database.
(D) IgA coating of fecal bacteria from germ-free mice colonized with IgA+ (n=5) or IgA−consortia (n=5) on days 7 and 24 post-colonization. Representative plots are shown. *** P < 0.005 (unpaired Student's t-test). Indicated are mean ± standard error of the mean.
(E) Microbiota localization as visualized by 16S rRNA FISH (red) and DAPI (blue) staining. The mucus layer is demarked by two dotted lines.
See also Figure S4.
To directly test the effects of IgA+ versus IgA− bacteria from IBD patients on intestinal inflammation, we next colonized germ-free mice with the assembled IgA+ or IgA− consortia. As a first test of the feasibility of this system, we examined the composition of the intestinal microbiota in these mice two weeks post-colonization and found that all but two bacteria, one from each consortium, were able to successfully colonize germ-free mice (Figure 4C).
We next wished to test whether the human IgA+ consortium would also preferentially induce the production of IgA when transplanted into germ-free mice. At seven days post-colonization, before the induction of a specific IgA response, we found that fecal bacteria from mice colonized with the IgA+ or IgA− consortia showed equivalent low levels of IgA coating by flow cytometry (Figure 4D). However, as compared to the IgA− consortium, the IgA+ consortium showed dramatically higher levels of IgA coating by day 24 post-colonization, which suggests that the IgA+ consortium had induced a strong and specific IgA response. These data show that bacteria that preferentially drive IgA responses in human IBD patients can also drive strong IgA responses in gnotobiotic mice.
The bacteria that we identified as highly coated in SPFdys mice are known to colonize normally sterile mucosal environments, such as the inner mucus layer and intestinal crypts (Elinav et al., 2011; Ivanov et al., 2009). To test whether the bacteria in our human IgA+ consortium also exhibit such characteristics, we performed bacterial 16S rRNA fluorescence in situ hybridization (FISH) on colons from IgA+ and IgA− consortia colonized mice. Remarkably, we observed the presence of many bacteria in the mucus layer of mice colonized with the IgA+ consortium (Figure 4E). In contrast, in mice colonized with the IgA− consortium, the inner mucus layer remained devoid of any detectable bacteria. Thus, one mechanism by which the bacteria comprising the IgA+ consortium may selectively induce IgA responses is through the invasion or colonization of normally sterile mucosal environments.
IgA inducing members of the intestinal microbiota cultured from IBD patients exacerbate DSS colitis in gnotobiotic mice
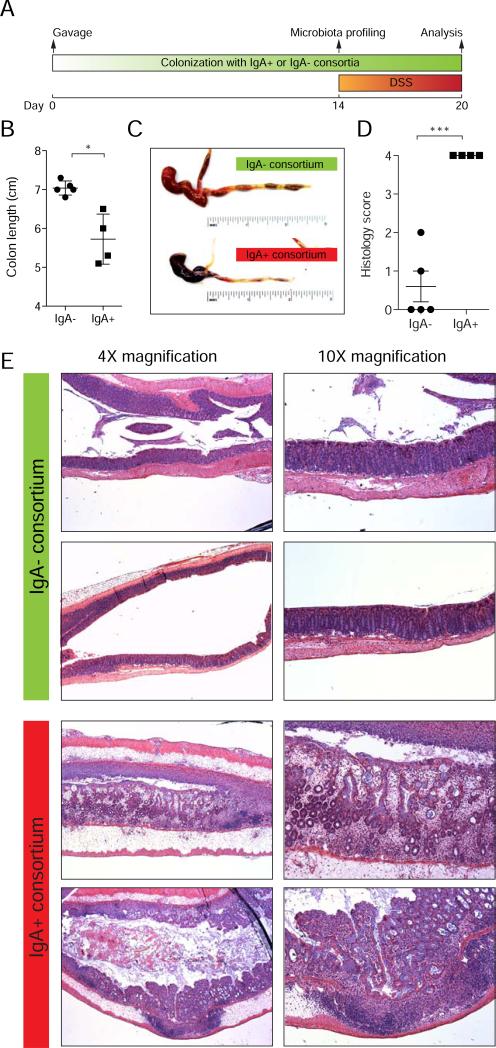
Germ-free mice colonized with the IgA− and IgA+ consortia for two weeks showed no signs of spontaneous intestinal pathology (Figure S9), suggesting that these bacterial strains are not immediately pathogenic in wild-type mice under healthy conditions. However, after the induction of colitis with DSS, IgA+ mice exhibited obvious and severe intestinal inflammation with extensive bleeding throughout the intestine, as well as significant shortening of the colon, while the intestines of IgA− mice appeared normal (Figures 5A-C). Histological examination revealed that IgA+ mice showed significant cellular infiltration and extensive loss of tissue integrity in the colon, while IgA− mice showed minimal visible inflammation (Figures 5D and 5E). These data show that bacteria isolated from IBD patients and chosen based on high IgA coating selectively drive severe intestinal inflammation in a mouse model of IBD.
Figure 5. IBD-associated IgA+ bacteria exacerbate DSS-induced colitis in gnotobiotic mice.
(A) Timeline of colonization and DSS treatment in germ-free mice colonized with IgA+ and IgA− consortia.
(B) Colon length after DSS. * P < 0.05 (unpaired Student's t-test).
(C) Gross pathology of large bowels after DSS. Note the extensive bleeding and diarrhea in the IgA+ colonized mice.
(D) Colon histopathology scores after DSS. Scores were assigned as follows: 0, Intact colonic architecture. No acute inflammation or epithelial injury; 1, Focal minimal acute inflammation; 2, Focal mild acute inflammation; 3, Severe acute inflammation with multiple crypt abscesses and/or focal ulceration; 4, Severe acute inflammation, multiple crypt abscesses, epithelial loss and extensive ulceration. *** P < 0.0001 (unpaired Student's t-test).
(E) Representative histology pictures from hematoxylin and eosin stained colons after DSS. Note that IgA+ colonized mice exhibit extensive inflammation, crypt abscesses, epithelial loss, and ulceration, while all IgA− colonized mice showed either no inflammation or minimal/mild focal inflammation. Data are representative of 3 independent experiments.
See also Figure S5.
Finally, we observed that bacterial isolates from different patients that were taxonomically assigned as the same species via 16S sequencing often showed differential IgA coating. While many factors may contribute to this phenomenon, one possibility is that these isolates represent distinct bacterial strains that display divergent behaviors that lead to differential IgA induction. As a proof-of-principle test of this scenario, we identified and characterized two isolates of B. fragilis from our gut microbiota culture collections that showed either high (ICI = 37.8) or low (ICI = 0.68) IgA coating (Figure S5F). Whole genome sequencing demonstrated that these isolates represent genetically distinct strains (Figure S5G). Finally, we monocolonized germ-free mice with these two strains and examined their effects on the development of DSS-induced colitis. We found that mice colonized with the IgA+ strain exhibited more severe colitis than IgA− colonized mice, as measured by colon length and histopathology (Figure S5H-J). These data suggest that strains of the same bacterial species that exhibit differential effects on the intestinal immune system and inflammatory disease can be distinguished based on IgA coating.
DISCUSSION
We examined taxa-specific coating of the intestinal microbiota with the secreted immunoglobulin IgA based on the hypothesis that levels of IgA coating might distinguish between members of the microbiota that impact disease susceptibility and/or severity by stimulating inflammatory responses and the remaining members of the microbiota. We found that high coating with IgA specifically marked a select group of known inflammation- and disease-driving intestinal bacteria in mice with inflammasome-mediated colitogenic dysbiosis. We also found that bacteria isolated from patients with IBD and selected based on high IgA coating induced potent IgA responses and dramatically exacerbated development of DSS-induced colitis in gnotobiotic mice. Thus, our data suggest that high coating with IgA selectively marks inflammatory, and therefore, potentially disease-driving commensals in mice and humans.
We combined bacterial cell-sorting based on IgA coating and 16S rRNA gene sequencing in order to examine the intestinal immune response to the intestinal microbiota in an unbiased and comprehensive manner. Recently, FACS-based bacterial cell sorting has been combined with next-generation sequencing by others as a way to examine: the active human gut microbiota (Peris-Bondia et al., 2011); responses of the intestinal microbiota to xenobiotics (Maurice et al., 2013); the effect of IgA coating on bacterial gene expression (Cullender et al., 2013); and IgA coating of the healthy intestinal microbiota (D'Auria et al., 2013). This type of approach, which combines taxonomic information with functional information regarding bacterial viability, behavior, or other bacterial features, will likely become increasingly common in future studies of the microbiota and its interactions with the host. Our data clearly illustrate the utility of this approach as a way to functionally classify intestinal bacteria based on their interactions with and recognition by the host immune system.
To maintain intestinal homeostasis, the mucosal immune system must selectively recognize and respond to pathogenic species while simultaneously maintaining tolerance to harmless and symbiotic members of the intestinal microbiota (Belkaid and Hand, 2014). Because most innate immune receptors involved in the detection of bacterial pathogens sense microbial components present in both pathogens and commensals (e.g., lipopolysaccharide), the mechanisms by which the immune system distinguishes between pathogens and commensals remain largely unknown. However, one way the immune system is thought to distinguish between pathogens and commensals is by sensing pathogen-associated activities or behaviors, such as adherence to the intestinal epithelium, tissue invasion or destruction, or the ability to colonize normally sterile mucosal environments, such as intestinal crypts (Sansonetti, 2011). The inflammatory commensals that we identify via IgA−SEQ appear to exhibit similar activities or behaviors. For example, Prevotellaceae species invade the mucus layer in the large intestine and colonize colonic crypts (Elinav et al., 2011); furthermore, SFB firmly adhere to the epithelium in the small intestine (Ivanov et al., 2009). Finally, we found that members of the human IgA+ consortium could be observed in the colonic mucus layer. Since the invasion of normally sterile sites proximal to the epithelium would naturally lead to increased stimulation of the innate immune system and increased availability of antigen for the induction of specific T cell and antibody responses, these behaviors may at least partially explain the propensity of IgA coated bacteria to preferentially induce both IgA responses and the inflammatory responses that lead to exacerbated disease susceptibility.
A variety of specific innate and adaptive immune mechanisms are known to influence IgA responses to the intestinal microbiota. For example, the Toll-like receptors have been implicated both T-dependent and T-independent IgA responses to the gut microbiota (Tezuka et al., 2007). Furthermore, specific T cell subsets, including T helper type 17 cells and regulatory T cells, as well as gamma-delta T cells have been implicated in coordinating IgA responses (Cong et al., 2009; Fujihashi et al., 1996; Hirota et al., 2013; Kawamoto et al., 2012). The specific responses we observed by IgA−SEQ were largely T-dependent; however, T-independent responses also clearly contributed to IgA responses to specific taxa. It will be interesting to determine the roles of specific T cell subsets, as well as specific innate immune receptors, in the induction of IgA responses to the colitogenic bacteria that we identify here.
Since highly IgA coated bacteria are constitutive inhabitants of the intestine and can drive disease, it appears that the host's IgA response to these bacteria is insufficient to lead to bacterial clearance or complete neutralization. Nonetheless, the IgA response may still reduce the level of inflammation caused by such bacteria. Indeed, SFB expand in the absence of an effective IgA response (Kato et al., 2014; Shinkura et al., 2004). Furthermore, bacterial-specific IgA has been shown to minimize intestinal inflammation through bacterial exclusion (Peterson et al., 2007). It will be interesting to examine whether IgA inducing bacteria drive even stronger pathological inflammatory responses in mice that are unable to mount an efficient IgA response.
Unlike classical pathogens, the high IgA inducing bacteria that we identified in mice are insufficient to cause disease on their own and instead drive or exacerbate disease only under specific predisposing genetic or environmental conditions: Prevotellaceae species exacerbate chemically-induced colitis (Elinav et al., 2011), and Helicobacter species drive colitis in the absence of the immunoregulatory cytokine IL-10 (Kullberg et al., 1998). A similar paradigm may apply to the development of IBD in humans where the coincidence of potent inflammatory bacteria with matching predisposing genetic or environmental lesions (e.g., mutations in Nod2) may be necessary for disease development (Knights et al., 2013). In support of this model, we found that bacteria selected from IBD patients based on high IgA coating did not cause overt disease at the steady state, showing that these bacteria are insufficient to cause disease on their own. Also, we found that healthy control subjects harbored multiple highly IgA coated species, but these subjects obviously did not show symptoms of pathological intestinal inflammation. Like Helicobacter and Prevotellaceae species in mice, high IgA inducing bacteria from healthy humans might also have the capacity to drive pathogenic inflammatory responses in patients with predisposing genetic lesions or environmental factors. Alternatively, IBD patients might harbor high IgA inducing bacteria that are uniquely potent at driving inflammatory responses in the intestine. Finally, a combination of these two scenarios may determine disease susceptibility and progression in humans.
We found that each individual human displayed a distinct pattern of IgA coating. This high inter-individual diversity in IgA coating patterns likely results from multiple factors, many of which illustrate the potential advantages of IgA−SEQ over traditional 16S and metagenomic sequencing. First, the collective human intestinal microbiota is enormously diverse and likely contains many inflammatory species that remain to be discovered and characterized (Chow et al., 2011; Huttenhower and Consortium, 2012; Lozupone et al., 2012; Saleh and Elson, 2011). Our data suggest that each individual harbors a unique combination of IgA inducing inflammatory commensals and provides the first insights into the diversity of such bacteria within the collective human intestinal microbiota. Second, 16S-based classifications cannot distinguish between strains of the same bacterial species, which may differ dramatically in their behaviors and effects on the host. Indeed, we found that two isolates of B. fragilis that displayed differential IgA coating were genetically distinct, and that gnotobiotic mice monocolonized with these two strains exhibited differential susceptibility to DSS colitis. Therefore, unlike traditional 16S- and metagenomic-based microbiota profiling, IgA−SEQ, particularly in combination with gnotobiotic mouse models, may provide important insights into strain-specific features that influence host-bacterial interactions. Third, host genetics may influence the behavior, detection, or handling of genetically identical bacterial species. For example, a particular bacterial species may only become IgA coated in hosts with mutations in Nod2, which are common in IBD (Abraham and Cho, 2009; Knights et al., 2013). Thus, examination of IgA coating may allow for the identification of bacteria that drive inflammation and cause disease specifically in individuals with particular predisposing genetic lesions. Fourth, environmental factors (e.g., diet) may have conceptually similar effects to genetic factors, where bacteria may induce inflammatory and IgA responses only under specific environmental conditions. Finally, two or more of these factors may affect IgA coating patterns and the role of specific bacterial species in disease. In the future, examination of IgA coating may allow us to decode some of these complex host-microbiota-environment relationships and begin to directly connect specific disease-driving members of the intestinal microbiota with the corresponding genetic and/or environmental factors that, together, lead to the development of disease.
The etiology of IBD involves a combination of genetic, environmental and microbial factors (Abraham and Cho, 2009; Knights et al., 2013). Here, we focused on the microbial contribution to IBD and attempted to identify members of the human gut microbiota that may preferentially impact IBD susceptibility and/or severity. One reason why it has been difficult to identify disease-driving bacteria in humans is that the strategies traditionally used to identify these bacteria in mice, including co-housing and gut microbiota transfer cannot be applied to humans (Dantas et al., 2013). In addition, due to the diversity of the intestinal microbiota in humans, the specific bacteria that drive disease may differ dramatically from patient to patient and, therefore, identifying these bacteria may require an individualized, rather than population-based, approach (Huttenhower and Consortium, 2012; Lozupone et al., 2012). We found that high IgA coating can identify colitogenic bacteria from patients with IBD by combining: (i) a functional classification of the intestinal microbiota based on the host's individual immune response; (ii) anaerobic culturing of members of the intestinal microbiota from diseased patients; and (iii) colonization of germ-free mice with human microbial consortia selected rationally based on their propensity to induce inflammation and, therefore, become highly coated with IgA. Using this approach, we showed that a subset of the intestinal microbiota from IBD patients that is characterized by high coating with IgA selectively confers susceptibility to colitis in a mouse model of IBD. Thus, our data suggest that the host's individual IgA response to the intestinal microbiota can be used as a guide to identify members of the microbiota that preferentially impact disease susceptibility and/or severity. The ability to identify these important bacterial taxa in humans in an individualized manner represents a first step towards the development of personalized, microbiota-focused therapies that may reduce, reverse, or even prevent disease development through targeted elimination or replacement of disease-driving members of the intestinal microbiota.
EXPERIMENTAL PROCEDURES
Animals
_Asc_−/−, _Rag2_−/−, and _Tcrb_−/−;_Tcrd_−/− mice were bred and maintained at the Yale School of Medicine and all treatments were in accordance with Yale Animal Care and Use Committee guidelines. Mice were strictly maintained under SPF conditions with consistent monitoring for and exclusion of viral, fungal and bacterial pathogens, as well as helminths and ectoparasites. Wild type C57Bl/6 mice were from the National Cancer Institute (NCI), and germ-free C57Bl/6 mice were purchased from the University of Michigan and the University of North Carolina germ-free facilities. Germ-free mice were singly housed, and age and sex matched mice were used for all studies.
Inflammasome-mediated intestinal dysbiosis
Intestinal dysbiosis was induced by co-housing two wild type C57Bl/6 mice from NCI with two Asc−/− mice for at least 6 weeks.
DSS colitis
SPF and SPFdys mice were treated with 2% Dextran Sodium Sulfate (MP Biomedicals) in the drinking water ad libitum for 7 days to induce colitis. Weight was measured daily for 14 days. Gnotobiotic mice were treated with filter sterilized 2.5% DSS in the drinking water ad libitum for 6 days before end-point euthanasia.
ELISA
Pre-sort, IgA+ and IgA− fractions (after MACS sorting) were probed for IgA by ELISA (Coating: MP Biomedicals 55478, Detection: Sigma B2766).
Fecal IgA flow cytometry and sorting of IgA+ and IgA− bacteria
Fecal homogenates were stained with PE-conjugated Anti-Mouse IgA (eBioscience clone mA-6E1) or PE-conjugated Anti-Human IgA (Miltenyi Biotec clone IS11-8E10) prior to flow cytometric analysis or MACS and FACS sorting. See the Extended Experimental Procedures for further information.
16S rRNA gene sequencing, bacterial genome sequencing and statistical analyses
16S rRNA sequencing of the V4 region and bacterial genome sequencing were performed on an Illumina miSeq using barcoded primers. Microbial diversity and statistical analyses were performed with QIIME, the Vegan package for R and LEfSe (Caporaso et al., 2010; Segata et al., 2011). See the Extended Experimental Procedures for further information.
Human fecal samples
The human study protocol was approved by the Institutional Review Board (Protocol No.10-1047) of the Icahn Medical School at Mount Sinai, New York. The healthy subjects were recruited through the Mount Sinai Biobank or an advertisement. Fresh fecal samples were collected at home, stored at −20°C in an insulated foam shipper, mailed to Mount Sinai overnight and then stored at −80°C for further analysis. A short questionnaire was also administrated to collect participants’ health information. Informed consent was obtained from all subjects.
Culturing of human fecal bacteria and generation of Personalized Microbiota Culture Collections
Culture methods were essentially as in Goodman et al. (Goodman et al., 2011) with minor modifications. Briefly, serial dilutions of fecal material from 11 IBD patients were plated on three types of media: CDC Anaerobe 5% Sheep Blood Agar with or without Kanamycin and Vancomycin (BD Bioscience), and Gut Microbiota Medium (GMM) Agar. 100 to 200 colonies per patient were picked and cultured individually in GMM for 5 days to establish Culture Collections.
Assembly of IgA+ and IgA− consortia and colonization of germ-free mice
Members of the IgA+ and IgA− consortia were selected from our IBD microbiota culture collections based on ICI scores determined by IgA−SEQ. The criteria for selecting the members of these consortia were as follows: Strains comprising the IgA+ consortium were selected based on high coating in the patient from whom they were isolated (ICI >10), and were rarely or never highly coated in healthy controls; in other words, they were selected to represent bacteria that are uniquely or preferentially highly coated in IBD. Strains comprising the IgA− consortium were selected based on low coating (ICI < 1) in the patient from whom they were isolated and were rarely or never highly coated in IBD patients or controls. Bacterial strains that met these criteria were cultured in GMM for 4 days before mixing to form consortia. Singly housed germ-free C57Bl/6 mice were colonized with 100 μl of the appropriate consortium by oral gavage.
Fluorescence in situ hybridization
16S rRNA FISH was performed with a universal bacterial probe (EUB388; 5’G*CTGCCTCCCGTAGGAGT-3’[Cy5]) on sections fixed with Carnoy's solution to preserve the mucus layer as described previously (Canny et al., 2006).
Histology
Colons were fixed in Bouin's solution and embedded in paraffin. Sections were stained with hematoxylin and eosin and scored in a blinded manner by a trained pathologist.
Monocolonization with B. fragilis isolates
Germ-free C57Bl/6 mice were monocolonized with B. fragilis strains by oral gavage. Mice were treated with 2.5% DSS in the drinking water starting at 5 days after colonization. Monocolonization was confirmed by 16S sequencing of feces.
Supplementary Material
1
9
10
2
3
4
5
6
7
8
HIGHLIGHTS.
- Bacterial members of the intestinal microbiota are differentially coated with IgA
- A limited number of intestinal bacterial species are highly coated with IgA
- IgA coating defines a subset of bacteria that selectively stimulate intestinal immunity
- High IgA coating marks colitogenic bacteria in inflammatory bowel disease
ACKNOWLEDGEMENTS
The authors thank members of the Flavell laboratory and A.M. Rhebergen for useful advice, discussions and review of the manuscript. This work was supported by a Rubicon Fellowship from the Netherlands Organization of Scientific Research (MRdZ), the Cancer Research Institute Irvington Fellowship Program and NIH T32 AR 7107-37 (NWP), the Blavatnik Family Foundation and the Department of Defense (W81XWH-11-1-0745) (RAF), and the Howard Hughes Medical Institute (RAF and MRdZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
NWP and MRdZ designed and performed all experiments, analyzed all data and wrote the manuscript and RAF supervised all research, assisted with experimental design and co-wrote the manuscript. ALG, TWC and NAB assisted with and offered guidance on the anaerobic culturing and gnotobiotic experiments. PHD wrote the paired-end assembly pipeline. JS optimized the FISH experiments. LH performed histology scoring. JH, IP, WZ, ER and JHC provided fecal samples.
REFERENCES
- Abraham C, Cho JH. Inflammatory bowel disease. The New England journal of medicine. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of computational biology : a journal of computational molecular cell biology. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemark M, Boysen P, Lycke NY. Induction of gut IgA production through T cell-dependent and T cell-independent pathways. Annals of the New York Academy of Sciences. 2012;1247:97–116. doi: 10.1111/j.1749-6632.2011.06378.x. [DOI] [PubMed] [Google Scholar]
- Ben-Amor K, Heilig H, Smidt H, Vaughan EE, Abee T, de Vos WM. Genetic diversity of viable, injured, and dead fecal bacteria assessed by fluorescence-activated cell sorting and 16S rRNA gene analysis. Applied and environmental microbiology. 2005;71:4679–4689. doi: 10.1128/AEM.71.8.4679-4689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Science translational medicine. 2012;4:137rv137. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canny G, Swidsinski A, McCormick BA. Interactions of intestinal epithelial cells with bacteria and immune cells: methods to characterize microflora and functional consequences. Methods in molecular biology. 2006;341:17–35. doi: 10.1385/1-59745-113-4:17. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Current opinion in immunology. 2011;23:473–480. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, Angenent LT, Bell ME, Hay AG, Peterson DA, et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell host & microbe. 2013;14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Auria G, Peris-Bondia F, Dzunkova M, Mira A, Collado MC, Latorre A, Moya A. Active and secreted IgA-coated bacterial fractions from the human gut reveal an under-represented microbiota core. Scientific reports. 2013;3:3515. doi: 10.1038/srep03515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantas G, Sommer MO, Degnan PH, Goodman AL. Experimental approaches for defining functional roles of microbes in the human gut. Annual review of microbiology. 2013;67:459–475. doi: 10.1146/annurev-micro-092412-155642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome research. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihashi K, McGhee JR, Kweon MN, Cooper MD, Tonegawa S, Takahashi I, Hiroi T, Mestecky J, Kiyono H. gamma/delta T cell-deficient mice have impaired mucosal immunoglobulin A responses. The Journal of experimental medicine. 1996;183:1929–1935. doi: 10.1084/jem.183.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell host & microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Turner JE, Villa M, Duarte JH, Demengeot J, Steinmetz OM, Stockinger B. Plasticity of Th17 cells in Peyer's patches is responsible for the induction of T cell-dependent IgA responses. Nature immunology. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Consortium HMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato LM, Kawamoto S, Maruya M, Fagarasan S. Gut TFH and IgA: key players for regulation of bacterial communities and immune homeostasis. Immunology and cell biology. 2014;92:49–56. doi: 10.1038/icb.2013.54. [DOI] [PubMed] [Google Scholar]
- Kawamoto S, Tran TH, Maruya M, Suzuki K, Doi Y, Tsutsui Y, Kato LM, Fagarasan S. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336:485–489. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut. 2013;62:1505–1510. doi: 10.1136/gutjnl-2012-303954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg MC, Ward JM, Gorelick PL, Caspar P, Hieny S, Cheever A, Jankovic D, Sher A. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infection and immunity. 1998;66:5157–5166. doi: 10.1128/iai.66.11.5157-5166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell host & microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias A, Corthesy B. N-Glycans on secretory component: mediators of the interaction between secretory IgA and gram-positive commensals sustaining intestinal homeostasis. Gut microbes. 2011;2:287–293. doi: 10.4161/gmic.2.5.18269. [DOI] [PubMed] [Google Scholar]
- Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst O. New concepts in the generation and functions of IgA. Nature reviews Immunology. 2012;12:821–832. doi: 10.1038/nri3322. [DOI] [PubMed] [Google Scholar]
- Packey CD, Sartor RB. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Current opinion in infectious diseases. 2009;22:292–301. doi: 10.1097/QCO.0b013e32832a8a5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris-Bondia F, Latorre A, Artacho A, Moya A, D'Auria G. The active human gut microbiota differs from the total microbiota. PloS one. 2011;6:e22448. doi: 10.1371/journal.pone.0022448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell host & microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nature reviews Immunology. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh M, Elson CO. Experimental inflammatory bowel disease: insights into the host-microbiota dialog. Immunity. 2011;34:293–302. doi: 10.1016/j.immuni.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti PJ. To be or not to be a pathogen: that is the mucosally relevant question. Mucosal immunology. 2011;4:8–14. doi: 10.1038/mi.2010.77. [DOI] [PubMed] [Google Scholar]
- Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome biology. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkura R, Ito S, Begum NA, Nagaoka H, Muramatsu M, Kinoshita K, Sakakibara Y, Hijikata H, Honjo T. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nature immunology. 2004;5:707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- Slack E, Balmer ML, Fritz JH, Hapfelmeier S. Functional flexibility of intestinal IgA - broadening the fine line. Frontiers in immunology. 2012;3:100. doi: 10.3389/fimmu.2012.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepankova R, Powrie F, Kofronova O, Kozakova H, Hudcovic T, Hrncir T, Uhlig H, Read S, Rehakova Z, Benada O. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflammatory bowel diseases. 2007;13:1202–1211. doi: 10.1002/ibd.20221. [DOI] [PubMed] [Google Scholar]
- Strober W. Impact of the gut microbiome on mucosal inflammation. Trends in immunology. 2013;34:423–430. doi: 10.1016/j.it.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezuka H, Abe Y, Iwata M, Takeuchi H, Ishikawa H, Matsushita M, Shiohara T, Akira S, Ohteki T. Regulation of IgA production by naturally occurring TNF/iNOS-producing dendritic cells. Nature. 2007;448:929–933. doi: 10.1038/nature06033. [DOI] [PubMed] [Google Scholar]
- Tsuruta T, Inoue R, Nojima I, Tsukahara T, Hara H, Yajima T. The amount of secreted IgA may not determine the secretory IgA coating ratio of gastrointestinal bacteria. FEMS immunology and medical microbiology. 2009;56:185–189. doi: 10.1111/j.1574-695X.2009.00568.x. [DOI] [PubMed] [Google Scholar]
- van der Waaij LA, Kroese FG, Visser A, Nelis GF, Westerveld BD, Jansen PL, Hunter JO. Immunoglobulin coating of faecal bacteria in inflammatory bowel disease. European journal of gastroenterology & hepatology. 2004;16:669–674. doi: 10.1097/01.meg.0000108346.41221.19. [DOI] [PubMed] [Google Scholar]
- van der Waaij LA, Mesander G, Limburg PC, van der Waaij D. Direct flow cytometry of anaerobic bacteria in human feces. Cytometry. 1994;16:270–279. doi: 10.1002/cyto.990160312. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
9
10
2
3
4
5
6
7
8