Transcriptional plasticity promotes primary and acquired resistance to BET inhibition (original) (raw)
. Author manuscript; available in PMC: 2016 Jun 25.
Published in final edited form as: Nature. 2015 Sep 14;525(7570):543–547. doi: 10.1038/nature14898
Summary
Following the discovery of BRD4 as a non-oncogene addiction target in acute myeloid leukemia (AML)1,2, BET inhibitors are being explored as promising therapeutic avenue in numerous cancers3–5. While clinical trials have reported single-agent activity in advanced hematologic malignancies6, mechanisms determining the response to BET inhibition remain poorly understood. To identify factors involved in primary and acquired BET resistance in leukemia, we performed a chromatin-focused RNAi screen in a sensitive MLL/AF9; NrasG12D -driven AML model, and investigated dynamic transcriptional profiles in sensitive and resistant murine and human leukemias. Our screen reveals that suppression of the PRC2 complex, contrary to effects in other contexts, promotes BET inhibitor resistance in AML. PRC2 suppression does not directly affect the regulation of Brd4-dependent transcripts, but facilitates the remodeling of regulatory pathways that restore the transcription of key targets such as Myc. Similarly, while BET inhibition triggers acute MYC repression in human leukemias regardless of their sensitivity, resistant leukemias are uniformly characterized by their ability to rapidly restore MYC transcription. This process involves the activation and recruitment of WNT signaling components, which compensate for the loss of BRD4 and drive resistance in various cancer models. Dynamic ChIP- and STARR-seq enhancer profiles reveal that BET-resistant states are characterized by remodeled regulatory landscapes, involving the activation of a focal MYC enhancer that recruits WNT machinery in response to BET inhibition. Together, our results identify and validate WNT signaling as a driver and candidate biomarker of primary and acquired BET resistance in leukemia, and implicate the rewiring of transcriptional programs as an important mechanism promoting resistance to BET inhibitors and, potentially, other chromatin-targeted therapies.
BRD4 is a chromatin reader that regulates transcription through linking histone acetylation and core components of the transcriptional apparatus7. Recent studies suggest that BRD4 regulates distinct gene sets through interacting with context-specific enhancers and transcription factors8,9. However, the mechanisms underlying the wide range of sensitivity to BET inhibition remain elusive, and so far no predictive biomarker could be identified. Towards understanding these mechanisms, we sought to functionally identify chromatin factors that are required for rendering AML cells sensitive to JQ1, a well-known BET inhibitor10. To this end, we constructed a microRNA-embedded short hairpin RNA (shRNAmir) library covering 626 chromatin regulators and screened it in the same MLL/AF9;NrasG12D-driven model that led to the identification of BRD4 as a candidate target1 (Fig. 1a). Deep-sequencing following transduction (T0) and seven days of selection (T1) identified chromatin-associated dependencies, including Smarca4 and Brd4 as top hits (Extended Data Fig. 1a-c). To control for unspecific events, we mixed the GFP+ library population with mCherry+ control cells, and subsequently treated with DMSO or JQ1. While mCherry+ cells disappeared over time (Fig. 1b), GFP+ cells survived and eventually grew in the presence of 50 nM JQ1 (corresponding to an IC70 dose; Extended Data Fig. 1d), indicating that resistance emerged from shRNA-mediated effects. Four shRNAs showed an outstanding enrichment that was consistent between replicates, despite almost five weeks of independent culture (Fig. 1c, Extended Data Fig. 1a,e). These included two shRNAs targeting Suz12, one targeting Psip1, and one previously characterized potent Dnmt3a shRNA11. All four shRNAs strongly suppressed their target mRNA (Extended Data Fig.1f), and validated to promote JQ1 resistance in single assays (Fig. 1d, Extended Data Fig. 2a).
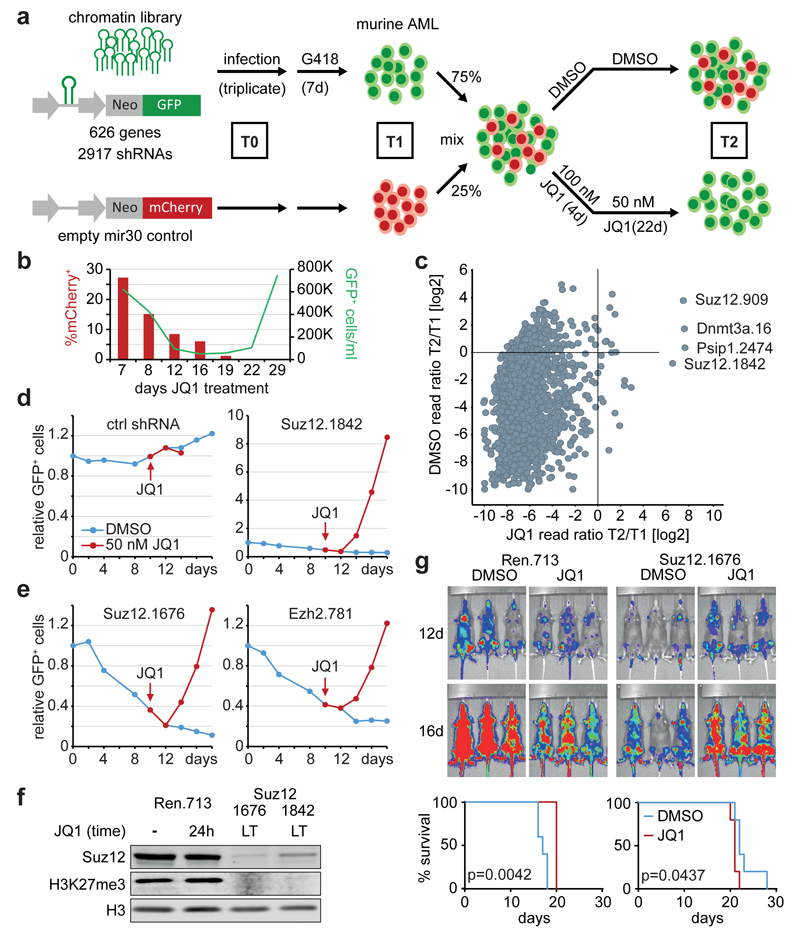
Figure 1. Multiplexed shRNAmir screening identifies chromatin factors that prevent resistance to BET inhibition.
a, Schematic of the multiplexed screening strategy. Mouse MLL/AF9;NrasG12D AML cells were infected with a library targeting 626 chromatin-associated genes (GFP+) or empty control (mCherry+). G418-selected cell populations were mixed and treated with DMSO or 100 nM JQ1 for 4 days followed by 50 nM JQ1 for 22 days. Genomic DNA isolated from T0, T1 and T2 was used to amplify and deep-sequence shRNA guides. b, Relative abundance of mCherry+ control cells and absolute number of GFP+/shRNA-expressing in the screen population cells over time. c, Scatter plot showing the average ratio of normalized reads before (T1) and after 26 days of DMSO- or JQ1 treatment (T2) for all 2917 shRNAs. d, e, Competitive proliferation assays of MLL/AF9;NrasG12D leukemia cells expressing the indicated shRNAs. Shown is the relative fraction of GFP+/shRNA+ cells relative to the initial measurement. After 10 days, each sample was split in half, treated with DMSO or 50 nM JQ1 and analyzed over 8 days. f, Immunoblotting of Suz12 and H3K27me3 in resistant AML cells expressing the indicated Suz12 shRNAs (LT, long-term culture in 50 nM JQ1 over 6 weeks) and Ren.713 control cells (+/- treatment with 200 nM JQ1 for 24h). g) Top, bioluminescent imaging of mice transplanted with 105 MLL/AF9;NrasG12D leukemia cells expressing the indicated shRNAs. Treatment with JQ1 (50 mg/kg/d) or DMSO carrier was initiated at day 3 after transplantation. Bottom, Kaplan–Meier survival curves of control and JQ1-treated mice (n=5). Statistical significance was calculated using a log-rank test.
The finding that suppression of Suz12, a component of the PRC2 complex, promotes resistance to JQ1 was surprising in two ways. First, a recent report, based on studies in nerve sheath tumors, has implicated Suz12 deficiency as a condition that sensitizes to BET inhibition12. Second, several studies (including work from our group) have characterized PRC2 as a requirement in MLL/AF9-driven AML13,14. Interestingly, the most potent Suz12 shRNA in previous studies (Suz12.1676) did not score in the pooled screen and validated to strongly inhibit proliferation in our model (Fig. 1e). However, when we added JQ1, Suz12.1676-expressing cells rapidly enriched, indicating that Suz12 deficiency turns from a detrimental into a favorable condition. Similar effects were observed using potent shRNAs targeting Ezh2 and Eed, two other PRC2 components (Fig. 1e, Extended Data Fig. 2b). We also validated this phenomenon using Tet-regulatable RNAi, where we included a validated Myc shRNA15 to rule out that resistance is merely a consequence of reduced proliferation (Extended Data Fig. 2c). Resistant cells generated through Suz12 suppression showed a global loss of H3K27me3 (Fig. 1f), and were also refractory to the effects of JQ1 in methyl-cellulose assays (Extended Data Fig. 2d) and in vivo (Fig. 1g). While recipient mice of Suz12-deficient cells, consistent with previous observations14, showed a delayed disease progression, JQ1 had no anti-leukemic effects in this context (Fig. 1g). Together, these data demonstrate that loss of Suz12 impairs rather than promotes BET sensitivity in AML, revealing another scenario where PRC2 plays opposing roles in different cancers.
To search for underlying mechanisms, we investigated whether our hits affected the regulation of BRD4-dependent transcripts, and found that the acute response to JQ1 is largely unchanged in the absence of Suz12, Dnmt3a or Psip1 (Fig. 2a). However, under long-term treatment 60-80% of JQ1-induced changes reverted and transcription of the key target Myc1,3 was restored (Fig. 2a-c; Extended Data Fig. 2e). To investigate this rebound phenomenon, we focused subsequent analyses on PRC2, a writer of repressive H3K27me3 marks16. Based on recent reports showing that BRD4 interacts with WHSC1L117, a writer of H3K36 methylation marks that can recruit PRC218, we wondered whether PRC2 is involved in repressing BRD4 targets following BET inhibition. However, ChIPseq analysis before and after JQ1 treatment showed no increase in H3K27me3 at Brd4 occupancy sites (Extended Data Fig. 2f,g), which together with RNAseq studies suggests that PRC2 does not regulate these genes directly.
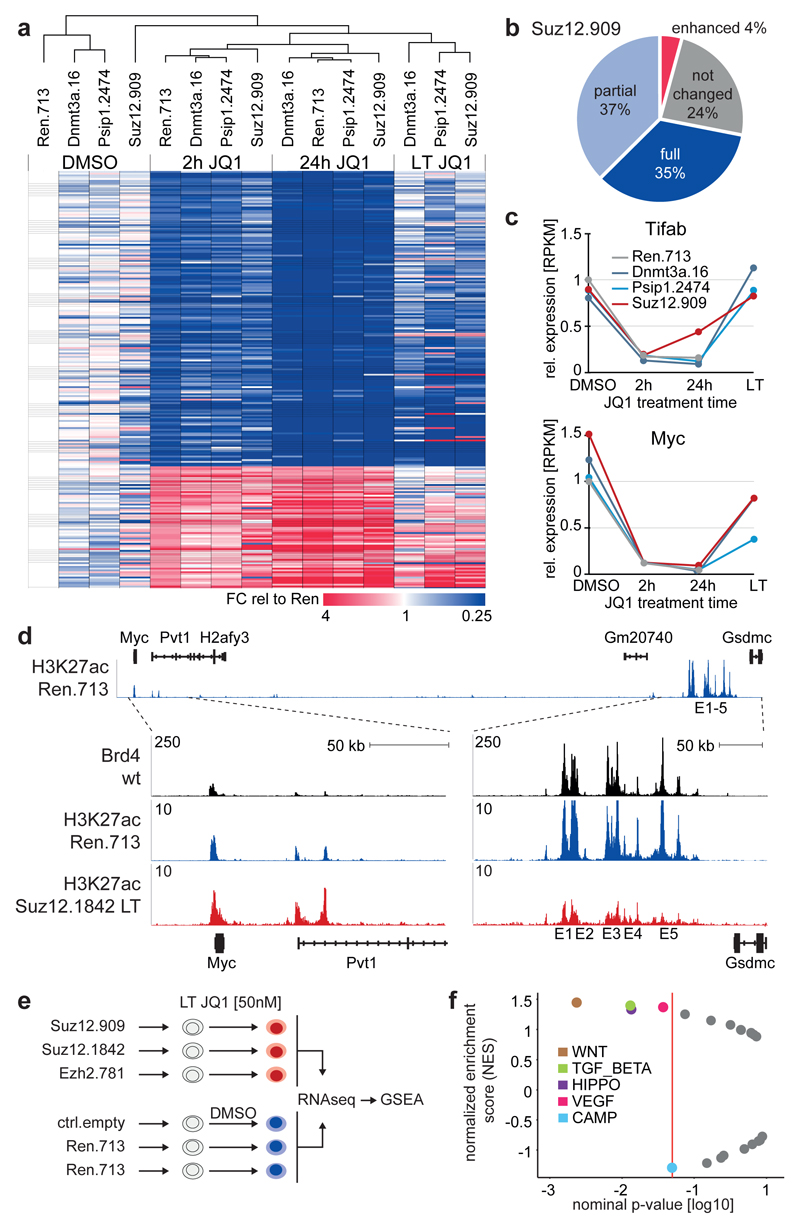
Figure 2. BET resistant AML cells restore the transcription of key Brd4 target genes through remodeling of regulatory landscapes and pathways.
a, Heat map showing RPKM fold-change (FC) of 235 response genes in MLL/AF9;NrasG12D leukemia cells expressing the indicated shRNAs after treatment with DMSO or JQ1 (200 nM) for 2h or 24h, and after long-term (LT) JQ1 treatment (50 nM) for 6 weeks. Response genes were defined based on JQ1-induced changes in Ren.713 expressing leukemia cells (FC>2/<0.5 after 2h and FC>2/<0.33 after 24h). Samples are presented according to non-hierarchical clustering; genes are grouped in up- or down-regulated targets in Ren.713 control cells, and sorted by their average re-expression in resistant leukemia. b, Response genes in (a) were grouped into four categories based on the divergence of their expression in resistant AML compared to AML expressing Ren.713 (see legend of Extended Data Fig. 2e for details). c, Time course of Tifab and Myc transcript levels in MLL/AF9;NrasG12D leukemia expressing the indicated shRNAs, relative to untreated cells expressing Ren.713. d, ChIPseq occupancy profiles of Brd4 and H3K27ac in Myc regulatory regions in sensitive AML cells (wt and Ren.713) and Suz12.1842-expressing resistant AML cells under long-term (LT) treatment with JQ1 (50 nM). The H3K27ac profile in Ren.713 control AML cells is shown on top for the entire region (~2 MB), together with validated transcript models from the mm10 genome assembly. Additional tracks are zoomed in on the proximal region and a distal region containing an established cluster of enhancers (E1-E5)19, as indicated (dotted lines). The y-axis reflects the number of normalized cumulative tag counts in each region. e, Schematic of RNA-seq profiling in resistant AML cells generated through expression of three independent shRNAs targeting Suz12 or Ezh2, which are treated as biological triplicates and compared to triplicate control cells. f, Gene set enrichment analysis (GSEA) of expression changes in 22 KEGG signaling pathways in resistant AML cells generated as described in (e). Plotted are normalized enrichment scores (NES) against nominal p-values; significantly altered gene sets (p<0.05) are indicated in the graph.
Next, we investigated whether the rebound of BRD4 targets is associated with changes in the enhancer landscape. Under long-term JQ1 treatment, resistant cells showed a global loss of enhancer-associated H3K27ac marks, which was reversible upon drug withdrawal (Extended Data Fig. 3a). At the level of Myc, the loss of H3K27ac was most prominent at a 3’-cluster of lineage-specific enhancers known to be required for Myc transcription19 (Fig. 2d). In parallel, we observed a strong focal gain of H3K27ac in the first intron of Pvt1 (a lncRNA 3’ of Myc), which was the tenth most prominent of only 119 genomic regions that gained H3K27ac in resistant cells (Fig. 2d, Extended Data Fig. 3b,c). Together, these findings reveal that BET resistance is associated with profound changes in the enhancer landscape.
To probe regulatory pathways involved in the re-expression of Brd4 targets, we analyzed the transcriptomes of sensitive and resistant cells generated using three independent PRC2 shRNAs (Fig. 2e). Gene set enrichment analysis20 identified alterations in five major signaling pathways (Fig. 2f). Almost half of the genes associated with upregulated pathways harbor H3K27me3 marks in sensitive cells (Extended Data Fig. 3d-f), indicating that loss of PRC2 can facilitate their transcriptional activation. The most significant alteration was an upregulation of genes associated with Wnt signaling, which can drive Myc transcription21 and has important functions in normal and leukemic stem cells (LSC)22,23. Of note, resistant cells in our model were not enriched for LSC-associated surface markers and expression signatures (Extended Data Fig. 4a-c), which does not exclude a role of LSC in resistance. Collectively, our findings suggest that loss of PRC2 promotes resistance through facilitating the derepression of compensatory pathways that restore the transcription of critical BRD4 target genes.
Given this complexity, we sought to explore whether any phenomena in our model of acquired resistance are also associated with primary BET resistance. To this end, we determined the JQ1 sensitivity in 246 human cell lines (Extended Data Fig. 5a), and selected 3 sensitive and 3 resistant lines from 3 cancer subtypes for dynamic transcriptional profiling. While RNAseq profiles distinguished cancer subtypes as expected, sensitive and resistant contexts could not be differentiated, neither through their steady-state transcriptomes, nor through comparing JQ1-induced changes (Extended Data Fig. 5b,c). In fact, BET inhibition triggered highly distinct responses regardless of whether MYC is affected, and even cancers of similar tissue and sensitivity profile do not share greater overlaps (Fig. 3a, Extended Data Fig. 5d-f). To test whether this heterogeneity is associated with differential functions of JQ1 targets, we used Tet-regulated shRNAmirs to profile BRD2/3/4-dependent transcripts in sensitive and resistant leukemia cells. In both cases, JQ1-induced changes showed the largest overlap with BRD4-knockdown profiles (Extended Data Fig. 6a-d), indicating that suppression of BRD4 dominates the effects of BET inhibition in leukemia.
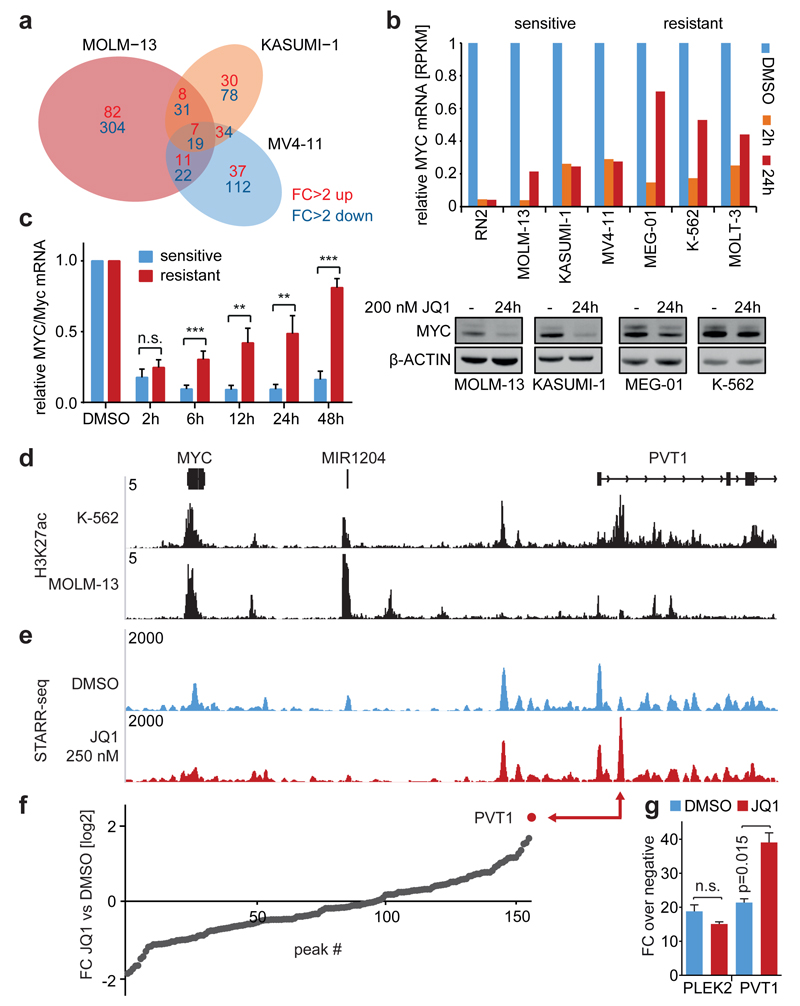
Figure 3. Dynamic transcriptional and enhancer profiling of sensitive and resistant cancer cell lines.
a, Venn diagram depicting the overlap of expression changes in 3 sensitive AML lines after 2h of JQ1 (200 nM). b, Top, MYC mRNA levels in indicated cell lines after 2h or 24h of JQ1 (200 nM), relative to DMSO-treated cells. Bottom, MYC immunoblotting in indicated cell lines before and after 24h of JQ1. c, MYC mRNA levels across 6 sensitive and 6 resistant cell lines at indicated time points following JQ1 (200 nM), normalized to DMSO (mean±SEM; Student’s _t_-test; **, p=0.01; ***, p<0.01). d, ChIPseq occupancy profiles of H3K27ac at the MYC locus in K-562 and MOLM-13 cells. Validated transcript models from the hg19 genome assembly are shown above; y-axes reflect normalized cumulative tag counts in each region. e, STARR-seq fragment densities in K-562 cells treated with DMSO or 250 nM JQ1. Read densities are shown as unique reads per million (rpm). f, Sorted ratios of STARR-seq signals in DMSO versus 250 nM JQ1-treated cells for all 156 identified STARR-seq peaks. g, Luciferase reporter assay measuring the enhancer activity of the PVT1 element and an unrelated enhancer (PLEK2) on a minimal MYC promoter. Shown are fold changes of normalized luciferase signal over background ± 250 nM JQ1 (n=3; mean±SEM, Student’s _t_-test with Welch’s correction).
Despite this heterogeneity, a closer look at the few commonly regulated genes (Extended Data Fig. 6e) revealed two interesting findings: (1) One of two transcripts commonly induced by JQ1 was HEXIM1, which we found upregulated in all analyzed cancers (Extended Data Fig. 6f), indicating that it could serve as a pharmacodynamic biomarker. (2) Surprisingly, one of three transcripts repressed after 2h of JQ1 in both sensitive and resistant leukemias was MYC. In sensitive leukemias, this repression was durable and led to strong suppression of the MYC protein (Fig. 3b), while all resistant leukemias showed a rapid rebound of MYC transcription and, consequently, only minimal protein suppression. Similar effects were observed in a larger panel of hematopoietic cell lines (Fig. 3c, Extended Data Fig. 6g), indicating that primary BET resistance is not due to BRD4-independent regulation of critical targets, but driven by compensatory mechanisms that rapidly restore their transcription.
To investigate how this rebound phenomenon is encoded in the enhancer landscape, we performed H3K27ac ChIPseq, which revealed that resistant K-562 cells harbor several H3K27ac peaks around MYC that are missing in sensitive MOLM-13 cells. These include a strong occupancy in the first intron of PVT1 (Fig. 3d), the same region that gained H3K27ac in our mouse model (Fig. 2d). To systematically analyze how these putative MYC enhancers change their activity upon BET inhibition, we performed STARR-seq (a new method for high-throughput functional enhancer analysis24) using a library covering 3.1 MB surrounding MYC (Extended Data Fig. 7a). While the PVT1 element had little activity in DMSO-treated K-562 cells, its enhancer activity strongly increased after 24h of JQ1 treatment (Fig. 3e) and defined the single strongest gain in the entire region (Fig. 3f). We confirmed this effect in conventional reporter assays using a minimal MYC promoter (Fig. 3g) and found that it does not correlate with PVT1 transcription (Extended Data Fig. 7b), indicating that this element primarily acts as a MYC enhancer. Collectively, our results suggest that the rebound of MYC is driven by a focal enhancer that is formed during acquired resistance and pre-established in primary resistant cells.
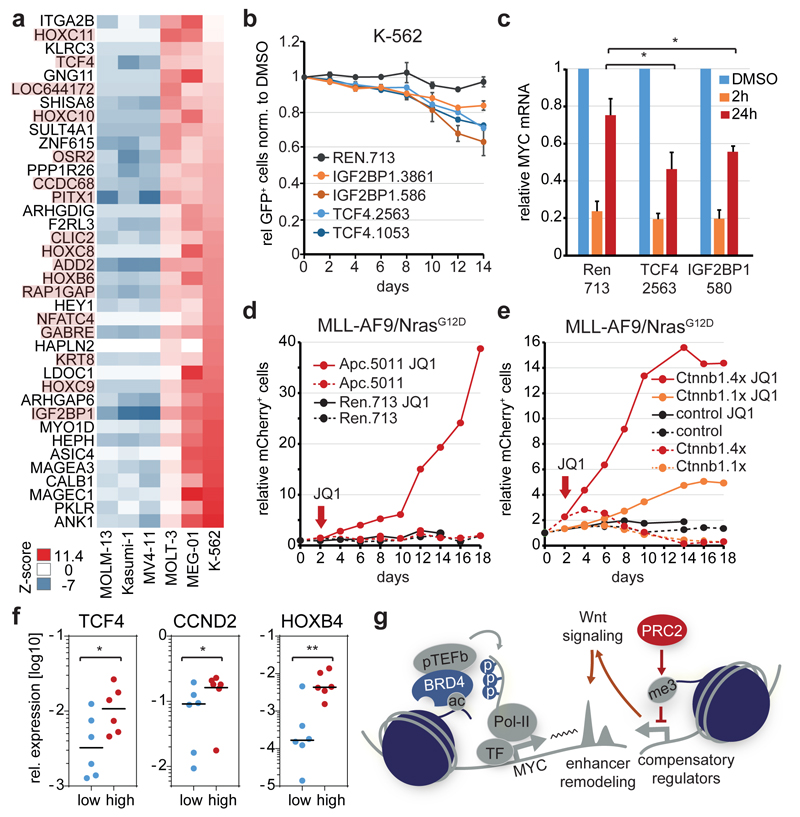
To search for regulatory pathways driving primary resistance, we compared steady-state transcriptomes of sensitive and resistant leukemias. Among only 38 genes consistently upregulated in resistant leukemias, we identified 17 genes previously implicated as Wnt signaling targets (Fig. 4a), which together with findings in our mouse model (Fig. 2f) pointed at Wnt signaling as a candidate driver of BET resistance. From these 17 genes, we selected two well-established Wnt targets, IGF2BP125 and TCF426, to test whether they contribute to the resistant state of K-562 cells. Suppression of TCF4 or IGF2BP1 increased the JQ1 sensitivity and diminished transcriptional rebound phenomena in K-562 cells (Fig. 4b,c; Extended Data Fig. 8a), while overexpression of TCF4 in MOLM-13 cells reduced their sensitivity (Extended Data Fig. 8b,c).
Figure 4. Wnt-signaling promotes primary and acquired BET resistance in leukemia.
a, Heat map of genes differentially expressed between indicated sensitive and resistant leukemia cell lines. Genes previously implicated as Wnt target genes are highlighted in red. b, Competitive proliferation assay of K-562 cells expressing the indicated shRNAs. Shown is the fraction of GFP+/shRNA+ under JQ1 treatment (100 nM), normalized to the fraction in DMSO-treated control cells (n=3, mean±SEM). c, qRT-PCR analysis of MYC mRNA levels in K-562 cells expressing the indicated shRNAs at different time points after JQ1 treatment (200 nM) (n=3; mean±SEM; Student’s _t_-test). d, Competitive proliferation assays of MLL/AF9;NrasG12D leukemia cells expressing the indicated shRNAs. Shown is the fraction of mCherry+/shRNA+ cells relative to the initial measurement. After 2 days, each sample was split in half, treated with DMSO or 50 nM JQ1 and followed up for 16 days. e, Competitive proliferation assays of MLL/AF9;NrasG12D leukemia cells co-expressing mCherry and Ctnnb1 harboring 1 (Ctnnb1.1x) or 4 (Ctnnb1.4x) activating mutations. After 2 days cells were treated with 50 nM JQ1 or DMSO, and the relative fraction of mCherry+/shRNA+ cells was followed over time. f, qRT-PCR analysis of TCF4, CCND2 and HOXB4 mRNA levels (relative to GAPDH) in sensitive (n=6; IC50<200 nM) and resistant (n=6; IC50>500 nM) primary human leukemia samples (Student’s _t_-test; *, p<0.05; **, p<0.01). g, Model of transcriptional plasticity as a driver of primary and acquired BET resistance.
To investigate whether resistant cells engage Wnt transcriptional machinery following BET inhibition, we performed ChIP assays for TCF7L2, a Wnt-dependent transcription factor known to drive MYC and other Wnt targets in complex with CTNNB121. Following JQ1 treatment, established Wnt target genes showed a marked increase in TCF7L2 binding, which was particular prominent at the PVT1 element 3’ of MYC (Extended Data Fig. 8d), suggesting that this site acts as a Wnt-dependent MYC enhancer. To test whether Wnt activation drives denovo resistance, we transduced cells of our sensitive AML mouse model with validated shRNAs targeting Apc (Extended Data Fig. 8e,f), a negative regulator of Wnt signaling, or active mutants of Ctnnb1. While Wnt activation had no or slightly detrimental effects in untreated cells, JQ1 treatment led to a rapid outgrowth of cells expressing two independent Apc shRNAs or Ctnnb1 harboring 4 activating mutations (Ctnnb1.4x) (Fig. 4d,e; Extended Data Fig. 8g). Single-mutant Ctnnb1 had milder effects, indicating that the degree of BET resistance depends on Wnt activation levels. Ctnnb1.4x also completely blunted the response to JQ1 in vivo, and promoted resistance in two independent leukemia models (Extended Data Fig. 8h,i).
After demonstrating that Wnt activation drives resistance in leukemia, we wondered whether our Wnt expression signature would be more generally associated with BET resistance. Through integrating sensitivity profiles of 246 cell lines with available transcriptome data27,28, we found that expression of this gene set is significantly increased in JQ1-resistant contexts (Extended Data Fig. 9a,b). As a first step towards probing Wnt as BET resistance driver in other cancers, we found that suppression of Apc promotes resistance in a mouse model of pancreatic cancer, while treatment with the Wnt inhibitor Pyrvinium29 synergizes with JQ1 in this model and highly resistant ASPC-1 cells (Extended Data Fig. 9c-e). To probe whether Wnt-signaling is associated with BET resistance in primary human leukemia, we quantified 9 Wnt-associated transcripts in sensitive and resistant patient-derived samples (Extended Data Fig. 10a). Remarkably, three of these transcripts (i.e. TCF4, CCND2 and HOXB4) were significantly overexpressed in resistant samples (Fig. 4f), while others showed a similar trend (Extended Data Fig. 10b). To reduce context-specific biases of single markers, we used the three significant transcripts to establish a simple “resistance index”, which strongly correlated with IC50 values (Extended Data Fig. 10c) and may provide a first step towards developing a predictive biomarker.
Through integrative profiling and functional genetics in mouse and human leukemia, our study reveals that BRD4 regulates a remarkably specific and diverse set of target genes. Leukemia cells, through an adaptation process that is facilitated by inactivation of PRC2, can become resistant to BET inhibition by rewiring the transcriptional regulation of key BRD4 targets such as MYC (Fig. 4g). Interestingly, a recent study found that in T-lymphoblastic leukemia MYC transcription can switch from a NOTCH1-dependent to a BRD4-dependent mode under treatment with γ-secretase inhibitors30. Our study reveals that the BRD4-dependency of MYC can be overcome through engaging a focal enhancer and, together with another report31, establishes WNT signalling as a major driver and candidate biomarker of BET resistance in leukaemia. These findings highlight that the heterogeneity and plasticity of transcriptional machinery plays a major role in promoting resistance to chromatin-targeted therapies.
Methods
Plasmids
The shRNA library used in the screen was cloned into pLMN (pMSCV-miR30-PGK-NeoR-IRES-GFP)15. The same vector was used for validation of primary hits and PRC2 core complex partners. Additional validation studies for Suz12 were performed using pRT3GEN-mir30 (pSIN-TRE3G-turboGFP-miR30-PGK-NeoR) or pLENC (pMSCV-miRE-PGK-NeoR-IRES-mCherry)11. shRNAs targeting Apc were cloned into pLMPC (pMSCV-miRE-PGK-PuroR-IRES-mCherry). A Ctnnb1 cDNA harboring 4 activating mutations (S33A, S37A, T41A, S45A; Ctnnb1.4x) was obtained from Addgene (#24312) and cloned into pMSCV-IRES-mCherry. Ctnnb1 harboring a single mutation (S45P; Ctnnb1.1x) was cloned into pMSCV-IRES-GFP. shRNAs targeting TCF4 and IGF2BP1 were cloned into pRT3GEN (pSIN-TRE3G-turboGFP-miRE-PGK-NeoR)11, and the TCF4 cDNA was cloned into pMSCV-PGK-NeoR-IRES-GFP.
Antibodies
The following antibodies were used: Suz12 (39357, Active Motif), Histone H3K27me3 (39155, Active Motif; 07-449, Millipore), Histone H3K36me3 (Ab9050, Abcam), Histone H3K27ac (ab4729, Abcam), Histone H3 (61277, Active Motif), MYC (56058, Cell Signaling), TCF4 (ab130014, Abcam), IGF2BP1 (RN001M, MBL), TCF7L2 (sc-8631, Santa Cruz), BRD4 (A301-985A1002, Bethyl), BRD3 (A302-583A, Bethyl), BRD2 (A302-368A, Bethyl), APC (MABC202, Millipore) and β‐Actin (A3854, Sigma‐Aldrich and ab49900, Abcam). Secondary antibodies were anti‐mouse (926-32210, LI-COR), anti-rabbit (926-32211, LI-COR) and anti-goat (926-32214, LI-COR). Antibodies used for FACS were: PE anti-mouse CD117/c-Kit (Biolegend, 105808), APC anti-mouse CD11b/Mac-1 (101212, Biolegend), PE-Cy7 anti-mouse Ly-6G/Gr-1 (108416, Biolegend), PE-Cy7 anti-mouse Ly-6A/E/Sca-1 (108114, Biolegend), PerCP/Cy5.5 anti-mouse CD45.2 (109828, Biolegend), PE/Cy7 anti-mouse CD117/c-kit (105814, Biolegend), APC anti-mouse CD150 (115925, Biolegend), PE-Cy7 anti-human CD11B/MAC-1 (101215, Biolegend) and APC anti-human CD36 (336207, Biolegend).
Pooled shRNAmir screening
An shRNA library targeting 626 chromatin-associated mouse genes (Supplementary Information; shRNAs prim. screen performance) was assembled in pLMN15 by combining an existing chromatin-focused library (243 genes, 1094 shRNAs)1 and 1793 additional shRNAs that were designed based on optimized Sensor rules32,33, and cloned, sequence-verified and pooled as previously described1. After spiking in several control shRNAs at equimolar ratios, the final pool of 2917 pLMN-shRNAs was transduced in triplicate into the same MLL-AF9/NrasG12D driven model that we had previously used to identify BRD4 as a target in AML2. To ensure library representation a total of 60 million cells were infected with 5% transduction efficiency using conditions that predominantly lead to a single retroviral integration and represent each shRNA in a calculated number of >800 cells. Throughout G418 drug selection (1 mg ml−1) more than 50 million cells were maintained at each passage to preserve library representation. Two days after infection T0 samples were acquired (6 million GFP+ cells per replicate) using a FACS AriaII (BD Biosciences) and deep sequencing analysis confirmed that the library was well represented in all replicates. After G418 drug selection for 7 days, T1 samples were obtained using FACS (6 million GFP+ cells per replicate), and cells were subsequently cultured in the presence of 0.5 mg ml−1 G418 (15 million cells per replicate were maintained at each passage). To control for unspecific clonal events during the JQ1 resistance screen, stably selected cells were mixed in 4:1 ratio with G418-selected cells expressing an empty miR-30 cassette, mCherry and NeoR (pMSCV-miR30.empty-PGK-NeoR-IRES-mCherry). Replicates were subsequently treated for 4 days with 100 nM JQ1 followed by 50 nM JQ1 for 22 days. Culture medium was exchanged every two days until cells proliferated comparable to wild type cells and afterwards passaged 7 times. Three replica cultured in the presence of vehicle (0.033% DMSO) were maintained in parallel. After 22 days (T2), about 6 million shRNA-expressing (GFP+) cells were sorted for each replicate by FACS. While mCherry+ cells completely disappeared over time, GFP+ cells survived and eventually grew in the presence of 50 nM JQ1 (corresponding to an IC70 dose in this model), indicating that resistance did not emerge from random clonal events, but from shRNA-mediated effects. Genomic DNA from T1 and T2 samples was isolated by two rounds of phenol extraction using PhaseLock tubes (5PRIME), followed by isopropanol precipitation. Deep sequencing libraries were generated by PCR amplification of shRNA guide strands using primers that tag the product with standard Illumina adapters (p7+Loop: CAAGCAGAAGACGGCATACGATAGTGAAGCCACAGATGT; p5+PGK: AATGATACGGCGACCACCGATGGATGTGGAATGTGTGCGAGG). For each sample, DNA from at least 5 × 106 cells was used as template in multiple parallel 50-μl PCR reactions, each containing 0.5 μg template, 1× AmpliTaq Gold buffer, 0.2 mM of each dNTP, 2 mM MgCl2, 0.3 μM of each primer and 2.5 U AmpliTaq Gold (Life Technologies), which were run using the following cycling parameters: 95°C for 10 min; 35 cycles of 95°C for 30 s, 52°C for 45 s and 72°C for 60 s; 72°C for 7 min. PCR products (340 bp) were combined for each sample, column purified using the QIAquick PCR purification kit (Qiagen) and further purified on a 1% agarose gel (QIAquick gel extraction kit, Qiagen). Libraries were analyzed on an Illumina HiSeq 2000 deep sequencer; 22 nucleotides of the guide strand were sequenced using a custom primer (miR30_Eco_RISeq, TAGCCCCTTGAATTCCGAGGCAGTAGGCA). To provide a sufficient baseline for detecting shRNA depletion in experimental samples, we aimed to acquire >500 reads per shRNA in the sequenced shRNA pool to compensate for variation in shRNA representation inherent in the pooled plasmid preparation or introduced by PCR biases. With these conditions, we acquired baselines of >500 reads for all 2917 shRNAs. Sequence processing was performed using a customized Galaxy platform34. For each shRNA and condition, the number of matching reads was normalized to the total number of library-specific reads per lane and imported into a database for further analysis (Access 2013, Microsoft). All primary screen data is provided under Supplementary Information; shRNAs prim. screen performance.
Cell culture, retroviral gene transfer and RNAi studies
Mouse MLL-AF9/NrasG12D AML cells (RN2)1 and other murine leukemia cell lines were derived from bone marrow of terminally diseased mice transplanted with fetal-liver cells engineered to express the indicated oncogenes, as previously described35. Murine leukemia cells were cultured in RPMI 1640 (Gibco-Invitrogen) supplemented with 10% FBS, 20 mM glutamate, 10 mM sodium pyruvate, 10 mM HEPES (pH 7.3), 100 U ml−1 penicillin and 100 μg ml−1 streptomycin. MV4-11, MEG-01 and K-562 cells were grown in IMDM with 10% FBS, 20 mM glutamate, 10 mM sodium pyruvate, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin. All human AML cell lines were cultured in RPMI 1640 (Gibco-Invitrogen) supplemented with 10% FBS, 20 mM glutamate, 10 mM sodium pyruvate, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin, except KASUMI-1 cells, which were cultured in 20% FBS. Human PDAC cell lines CAPAN-2 and MIAPACA-2 were cultured in DMEM (Gibco-Invitrogen) supplemented with 10% FBS, 20 mM Glutamate, 10 mM sodium pyruvate 100 U ml−1 penicillin and 100 μg ml−1 streptomycin. SU-8686 and ASPC-1 were cultured in RPMI 1640 (Gibco-Invitrogen) supplemented with 10% FBS, 20 mM Glutamate, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin. HUPT-4 and HPAF-2 were cultured in EMEM (Sigma-Aldrich) supplemented with 10% FBS, 20 mM glutamate, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin. Human SCLC cell lines DMS-273, SHP-77, NCI-H82 and NCI-H1048 were cultured in RPMI 1640 (Gibco-Invitrogen) supplemented with 10% FBS, 20 mM glutamate, 10% GlutaMAX, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin. Cell lines were obtained from ATCC (http://www.lgcstandards-atcc.org/en.aspx) or DSMZ (http://www.dsmz.de/) and tested for Mycoplasma infection on a regular basis using a commercial biochemical test (Lonza).
Tet-on competent murine pancreatic cancer cells (KRPC2) were generated and characterized as previously described36. In brief, pancreatic progenitor cells isolated from a murine fetus (ED17.5-18.5) harboring a conditional endogenous KrasG12D allele (lox-STOP-lox-KrasG12D)37 and a conditional Trp53 deletion (Trp53fl/fl)38 were transduced with retroviral constructs expressing Myc, 4-OHT-inducible CreERT2, codon-optimized Firefly Luciferase (Luc2), and rtTA3, and orthotopically injected into the pancreas of syngeneic recipient mice (detailed protocols available upon request). Emerging tumors were characterized to harbor histological features of human pancreatic adenocarcinoma and used to derive a cell line (KRPC2), which was cultured in DMEM (Gibco-Invitrogen) with 10% FBS, 20 mM Glutamate, 10 mM sodium pyruvate 100 U ml−1 penicillin and 100 μg ml−1 streptomycin.
Retroviral packaging was performed using Platinium-E cells (Cell Biolabs) according to established protocols11. In brief, for each calcium phosphate transfection, 10‐20 μg plasmid DNA and 5 μg helper plasmid (pCMV-Gag-Pol, Cell Biolabs) were used. Transduction efficiencies of retroviral constructs were measured 48h post infection by flow cytometry (Guava easyCyte, Millipore). Transduced cell populations were usually selected 48h post infection using 500 μg/ml G418 (Gibco, Life Technologies). To generate resistant MLL-AF9/NrasG12D murine leukemia, cells were transduced with retrovirally delivered shRNAs or cDNAs at a transduction efficacy between 5-15% (predominantly yielding single viral integrations per cell). JQ1 treatment was started post infection and medium was carefully exchanged every two days to maintain a 50 nM JQ1 concentration until cells were proliferating normally and daily passaging was required. The starting point of JQ1 treatment is critical and can vary between different targets and shRNAs since the effect is dependent on shRNA potency, protein half-life of the respective target and the half-life of the associated chromatin mark. In case of strong detrimental effects the optimal start of JQ1 treatment needs to be determined for every target.
MOLM-13 and K-562 cells were modified to express the ecotropic receptor and rtTA3 using retroviral transduction of pMSCV-RIEP (pMSCV-rtTA3-IRES-EcoR-PGK-Puro) or lentiviral transduction of pWPXLd-RIEP (pWPXLd-rtTA3-IRES-EcoR-PGK-Puro) followed by puromycin selection (0.5 and 1 ug ml−1 for 1 week, respectively). Derived cell lines were subsequently transduced with ecotropically packaged retroviruses.
For flow cytometry immunophenotyping, cultured cells were collected and stained using the indicated antibodies (two million cells in 100 μl FACS buffer: PBS, 5% FCS, 0.005% sodium acetate, and human or mouse TruStain fcX Fc-receptor blocking agent (Biolegend) for 20 min at 4°C. Antibodies were diluted 1:400. Stained samples were analyzed on an LSR Fortessa (BD) flow cytometer. Data analysis was performed using FlowJo software (Treestar). Cell viability assays for drug IC50 and GI50 determination were performed using Alamar blue staining according to manufacturer’s guidelines, or by counting the increase in viable cell numbers over 72h in the presence of different JQ1 concentrations. Dead cells were excluded using propidium iodide (PI) staining. Measurements of cell concentration were performed on a Guava easyCyte (Millipore), gating only viable cells (FSC/SSC/PI−). Synergy was evaluated using Chou-Talalay Combination Index (CI), calculated with the CompuSyn program (ComboSyn, Inc.)39. GI50 values for all tested cell lines are provided in Supplementary Information; JQ1 profiled cell lines.
Competitive proliferation assays using shRNAs in pLMN, pLENC, pLMP or pRT3GEN-mir30/RT3GEN vectors were performed as described previously1,40. Briefly, for assays involving Tet-regulated shRNA expression vectors, Tet-on competent MLL-AF9/NrasG12D mouse AML cells40 or Tet‐on competent derivatives of human leukemia cell lines were transduced with the respective plasmid at infection rates of <5% to ensure single‐copy integration. For constitutive shRNA expression (pLMN) cells were analyzed for GFP+ expression 1 day post infection. pRT3GEN transduced cells were selected for 7 days using G418 (0.5 mg/ml), mixed with 10% uninfected cells, and shRNA expression was induced through addition of doxycycline (DOX) to a final concentration of 1 μg ml-1. The percentage of shRNA‐expressing cells (turboGFP+ or GFP+) was measured daily using flow cytometry. All values were normalized to day 1. Once the percentage of viable cells was below 20%, measurements were discontinued (indicated in the graph by the discontinuation of the respective sample). Sequences for all shRNAs used in single assays are provided in Supplementary Information; shRNAs single assays.
Colony forming assays were performed in Methocult (Stemcell Technologies, Cat. No. M3231). 1000 AML cells (in 100 µl standard medium) were added to 900 µl Methocult supplemented with 1 µl of DMSO or JQ1 (to a final concentration of 200 nM). After 7 days the types of colonies were enumerated and normalised to input.
Gene expression and protein level analysis
RNA was prepared using the RNeasy Plus Mini Kit (Qiagen). Synthesis of cDNA was performed using SuperScript III Reverse Transcriptase (Invitrogen) or Taqman reverse transcription kit (Applied Biosystems) with random hexamers. Quantitative PCR analysis was performed on an ABI 7900HT with SYBR green (ABI). All signals were quantified using the ΔCt method and were normalized to the levels of β-Actin or GAPDH. All primer used in this study are presented in Supplementary Information; Primer.
For immunoblotting of Suz12, Histone H3K27me3, BRD4, TCF4, IGF2BP1 and MYC, 20 μg of whole-cell lysate (lysis buffer: 50 mM TRIS pH 8, 250 mM NaCl, 0.5% NP-40, 5 mM EDTA) were loaded onto each lane. Protein extracts were resolved by SDS polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to nitrocellulose for blotting.
Preparation of RNA-Seq libraries
RNA from human cell lines or mouse AML cells was isolated using the RNeasy Plus Mini Kit (Qiagen). mRNA was obtained using two rounds of poly(A) selection using the Dynabeads mRNA purification kit (Invitrogen) and subsequently fragmented by incubation at 94°C for 3 min (fragmentation buffer: 40 mM Tris base adjusted to pH 8.2 with glacial acetic acid, 100 mM potassium acetate, 30 mM magnesium acetate in DEPC-treated H2O). Cell lines infected with shRNA expressing vectors were FAC-sorted for shRNA expression (GFP+) prior to RNA extraction. The fragmented mRNA was used as template for first-strand cDNA synthesis with random hexamers and a NEBNext DNA Library Prep Master Mix Set for Illumina Synthesis kit (Invitrogen). The second-strand cDNA was synthesized with 100 mM dATP, dCTP, dGTP and dUTP in the presence of RNase H, E. coli DNA polymerase I and DNA ligase (Invitrogen). The incorporation of dUTP allowed elimination of the second strand during library preparation (described below) and thus preservation of strand specificity.
Chromatin immunoprecipitation and ChIP-Seq library construction
ChIP assays were performed exactly as described14 in two to three independent biological replicates. Crosslinking was performed with sequential EGS and formaldehyde or with formaldehyde alone. All samples were quantified by quantitative PCR performed using SYBR green (ABI) on an ABI 7900HT after crosslik reversal. Each immunoprecipitate signal was referenced to an input standard-curve dilution series (immunoprecipitate/input) to normalize for differences in starting cell number and for primer amplification efficiency and normalized to a control region. Two independent biological replicates were performed for each ChIP-seq experiment.
For ChIP-Seq library construction, 1 x 107 leukemia cells were crosslinked using 1% formaldehyde for 20 minutes at room temperature. After purifying immunoprecipitated DNA using QIAquick Gel Extraction Kit (QIAGEN), ChIP-Seq libraries were constructed using the TruSeq ChIP Sample Prep Kit (illumina) following manufacturer’s instructions with the following exceptions: Following adapter ligation, libraries were amplified for 15 cycles. Briefly, purified ChIP DNA was repaired to blunt ends followed by dA-tailing process to ligate adapters for multiplex-based sequencing. Size-selection of adapter-ligated ChIP-DNA was performed with agarose-gel based selection of DNA size ranged from 200 to 350 bp. The final libraries were amplified for 15 cycles, and size-selected with SPRI clean-up by using AMPure XP beads (Beckman Coulter). Library quality was determined using a Bioanalyzer (Agilent). Libraries sizes ranged from 250 to 300 bp. ChIP-Seq libraries were sequenced using an Illumina HiSeq 2000 platform. Barcoded libraries were sequenced in a multiplexed fashion with two to six libraries at equal molar ratio, with single-end reads of 50 bases.
Illumina deep sequencing
2–5 ng of cDNA or DNA precipitated by ChIP was used as starting material for the generation of single-end sequencing libraries as described in Illumina’s ChIP Sequencing sample preparation protocol. DNA fragments of the following sizes were selected for these experiments: 200–350 bp for ChIP-Seq, 150–700 bp for RNA-Seq. For strand-specific RNA-Seq, the uridine residues present in one cDNA strand were digested with uracil-N-glycosylase (New England Biolabs), followed by PCR amplification. Completed libraries were quantified on a Bioanalyzer using the dsDNA 1000 assay kit (Agilent) and the qPCR NGS library quantification kit (Agilent). Cluster generation and sequencing was performed using a HiSeq 2000 system with a read length of 50 or 100 nucleotides according to the manufacturer’s guidelines (Illumina).
Analysis of RNA-seq data
The calculation of RNA expression values was based on the RefSeq database, which was downloaded from UCSC on January 10th, 2014. Genes with overlapping exons were flagged and double entries (i.e. exactly the same gene at two different genomic locations) were renamed. Identical genes with more than one assigned gene symbol were flagged. Genes with several transcripts were merged to consensus-genes consisting of a union of all underlying exons using the fuge software (I. Tamir, unpublished), which resulted in 25,098 gene models. Gene names and accession numbers of identical genes are presented in Supplementary Information; refseq.hg19.2014_0110.imp.map.
Paired-end and single-read fragments were trimmed on their 5’ end (6 cycles PE100, 2 cycles SR50 NEB RNA sample prep protocol), adaptors were removed using cutadapt v1.4.241 and reads with a length of less than 18 bp in any read of the pair were discarded. The trimmed and adaptor-free reads were aligned against rDNA of the respective organism using bowtie2 v2.0.242 for paired-end and bowtie v0.12.543 for single-end reads. The rRNA cleaned paired-end reads were aligned against the transcriptome to estimate the fragment size and the standard deviation of the fragment size using bwa v0.6.244. The rRNA cleaned reads were aligned to the genome with the TopHat splice junction mapper for RNA-Seq reads45. The uniquely aligning reads were used for counting per gene with htseq-count v 0.6.1p46 with the overlap-resolution mode option set to ‘union’ using the processed RefSeq-annotated gene database. Reads per kilobase per million mapped reads (RPKM) values for all samples were calculated following Mortazavi A., et al. 200847. Read counts for all samples were normalized using the median-of-ratios method implemented in DESeq2, version 1.2.1048. Counts were transformed for PCA using DESeq2’s variance stabilizing transformation. For both the resistant and sensitive groups, three independent RNA-Seq experiments on different leukemia cell lines were available. The edgeR R package version 3.4.249 was utilized to calculate the significance of difference in expression of resistant and sensitive leukemia cell lines (DMSO treated) in Figure 4a. For this, raw counts were normalized using the “relative log expression” method analogous to DESeq2. Dispersions were sequentially estimated with the default settings by first estimating a common dispersion for the gene set and then ‘shrinking’ tagwise dispersion values towards that dispersion trend. Statistical modeling and analysis was done by fitting a model taking the separation into sensitive and resistant cell lines into account. Differential expression of genes between those two groups was subjected to statistical analysis and resulting p-values were adjusted for multiple testing with the Benjamini-Hochberg correction50. The resulting list was further filtered by only considering genes which all have higher normalized counts in the sensitive cell lines vs the resistant cell lines or vice versa.
GSEA analysis
Gene set enrichment analysis20 was performed using GSEA v2.07 software with 1,000 gene set permutations. KEGG signaling gene sets and Wnt signaling associated gene sets were obtained from the Molecular Signatures Database v4.0 (MSigDB, http://www.broadinstitute.org/gsea/msigdb/index.jsp). To perform GSEA with human gene signatures, human-mouse homologs from the mouse gene sets were identified and converted into human gene names using the NCBI homologene database (build68). Ambiguous homolog mapping i.e. different mouse gene names pointing to the same human homolog was excluded. A detailed description of GSEA methodology and interpretation is provided at http://www.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html. In brief, the normalized enrichment score (NES) provides ‘the degree to which a gene set is overrepresented at the top or bottom of a ranked list of genes’. The nominal p value (Nom p-val) describes ‘the statistical significance of the enrichment score’. The false discovery rate _q_-value (FDR _q_-val) is ‘the estimated probability that a gene set with a given NES represents a false positive finding’. ‘In general, given the lack of coherence in most expression datasets and the relatively small number of gene sets being analyzed, an FDR cutoff of 25% is appropriate.’ All gene sets used in this study are provided in Supplementary Information; GSEA gene sets.
ChIP-Seq data analysis
Adaptors of the single-read fragments were removed using cutadapt v1.4.241 and reads with a length of less than 18bp in any read of the pair were discarded. The remaining reads were aligned to the genome (mouse: mm10, human: hg19) using Bowtie v1.0.043. Reads with same start and end position on the same strand were removed from the alignment. To identify ChIP-Seq peaks, we used the MACS2 v2.1.0.20141030 (https://github.com/taoliu/MACS/) peak finding algorithm. A 3-fold enrichment relative to input control samples was used for peak calling as well as the option to call broad peaks. Building a shifting model was disabled and the small nearby and large nearby region parameters were set to 5,000 and 20,000 respectively. The extension size was set to the respective median insert size of the ChIP-Seq treatment sample for paired-end data and the estimated fragment size for single-end data. Read numbers for the resulting peaks were quantified using the BEDTools suite51 and normalized to total mapped reads. The Brd4 ChIP-Seq track was obtained from published data sets19 (GEO sample GSM1262345). Peaks were assigned to modified versions of the RefSeq database (GRCh37/hg19 downloaded on January 10, 2014 from UCSC Table Browser and GRCm38/mm10 downloaded on June 30, 2014 from UCSC Table Browser) by assigning peaks to the closest upstream/downstream TSS using BEDTools. For these versions, overlapping genes were flagged and double entries (i.e. exactly the same gene at two different genomic locations) were renamed. Identical genes with more than one assigned gene symbol were flagged. For the density heatmaps, ChIP-Seq peaks were identified using the MACS version 1.4.0beta (Model based Analysis of ChIP-Seq)52 peak finding algorithm. A p value threshold of enrichment of 1 x 10-5, a false discovery rate (FDR) of less than 1%, and a 5-fold enrichment relative to input control samples were used for all peak calling. The Brd4 peaks were considered as promoter if the peak showed at least 1bp overlap within +/- 200bp of RefSeq gene TSSs. If Brd4-called peaks showed no overlap within +/- 200bp of RefSeq gene TSSs, they were considered as enhancer-bound peaks. Heatmap matrices were created by counting tags using 10kb window (+/- 5kb of the peak summit) and 20bp bin size. Further visualization of heatmap matrices were done using Java TreeView 1.1.6r4 (http://jtreeview.sourceforge.net). For comparing DMSO and JQ1 treated ChIP-Seq datasets, heatmap matrices were normalized to total mapped reads counts before visualization with Java TreeView.
STARR-seq screens
STARR-seq screens were done as described previously24, with the following exceptions. DNA from 21 bacterial artificial chromosomes (BACs), which were available for the extended MYC locus covering approximately 91% of the surrounding 3Mb of genomic region, plus 25 genic control BACs, were used for library generation. The fragmented BAC DNA was cloned into the human STARR-seq screening vector24 containing a minimal MYC promoter. We transfected 500µg of this library into 1×108 K-562 cells per condition, using a BTX Agile Pulse MAX system (Harvard Apparatus) following the manufacturer’s protocol. Electroporation conditions: two pulses at 750V for 0.5 ms (100 ms interval), followed by 5 pulses of 250V for 2 ms (100 ms interval) in BTXpress cytoporation medium T. After electroporation, the cells were grown at 37°C in RPMI-1640 containing either DMSO or 250nM JQ1 and were harvested after 24h. For each screen we isolated poly-A RNA and processed it as described before24, with a different extension time of 45s in the final amplification of the cDNA and modified primers for the first cDNA amplification. All BACs used for the STARR-seq screen are provided in Supplementary Information; STARR-seq BACs.
STARR-seq deep sequencing and analysis
All sequencing was performed as a 50-cycle paired-end run on an Illumina HiSeq2000 machine. STARR-seq and input read processing was performed as described24, with the following exceptions. Libraries from different conditions were normalized to each other by subsampling to 105 reads per condition using reservoir sampling. Peaks were called using macs2 (v 2.1.0)52 with the input library as background. The peaks were then filtered for a minimal fold enrichment of 3 and only peaks with at least 5 reads per kb were considered for further analysis. Peak calling was performed for STARR-seq reads from both conditions (DMSO and JQ1) and the resulting peaks were merged using bedtools merge51. STARR-seq enrichment was calculated using bedtools coverage51. Differential fold enrichment was calculated as the ratio between enrichments in JQ1 over DMSO and expressed in log2.
Luciferase assays and data analysis
Luciferase assays were done as described previously24, with the following exceptions. Enhancer candidate regions were amplified from genomic DNA and shuttled into a modified pGL4 luciferase reporter vector containing a minimal MYC promoter (chr8:128,748,440-128,748,550). Each construct was tested by co-transfecting 106 K-562 cells with 45µg of the firefly luciferase construct and 5µg of a renilla luciferase control plasmid (pGL4.75, Promega). After electroporation, the cells were grown at 37°C in RPMI-1640 containing either DMSO or 250nM JQ1 and were harvested after 46h. Relative luciferase signals (firefly/renilla) were normalized to signal from two negative background sequences chosen based on their absence of STARR-seq signal and expressed as fold change over background. We analyzed the statistical significance of differential luciferase signals with or without JQ1 treatment using an unpaired t test with Welch’s correction for three biological replicates. Primer sequences which were used to amplify the particular enhancer regions are deposited in Supplementary Information; Primer.
Animal experiments
Mouse MLL-AF9/NrasG12D leukemia cells (RN2) were transplanted by tail-vein injection of 1 × 105 cells into sub-lethally (5.5 Gy) irradiated female B6/SJL (CD45.1) recipient mice at the age of 6-10 weeks (n=5, for each individual experiment). For whole-body bioluminescent imaging, mice were intraperitoneally injected with 50 mg kg−1 D-Luciferin (Goldbio), and after 5 min analyzed using an IVIS Spectrum system (Caliper LifeSciences). For JQ1 treatment trials, a stock of 100 mg ml−1 JQ1 in DMSO was 20-fold diluted by drop wise addition of a 10% 2-hydroxypropyl-β-cyclodextrin carrier (Sigma) under vortexing, yielding a 5 mg ml−1 final solution. Mice were intraperitoneally injected daily with freshly diluted JQ1 (50 mg kg−1) or a similar volume of carrier containing 5% DMSO. JQ1 treatment was started 3 days after transplantation of RN2 cells stably transduced with pLMN shRen.713 and shSuz12.1676. Mice transplanted with RN2 cells expressing beta-catenin cDNA or empty vector were treated with JQ1 daily starting 1 day after transplantation. All animals were maintained in the pathogen-free animal facility of the Research Institute of Molecular Pathology, and all procedures were carried out according to an ethical animal license, which is approved and regularly controlled by the Austrian Veterinary Authorities. No statistical methods were used to predetermine sample size. The investigators were not blinded to allocation during experiments and outcome assessment. Randomization was not applied because all animals used in this study were similar for age, sex and strain background.
Patient sample analysis
Ten patients with AML (females, n=4; males, n=6) and two with CML blast phase (BP) (males, n=2) were examined. Diagnoses were established according to the proposal of the French-American-British (FAB) cooperative study group53,54 and the classification of the World Health Organization (WHO)55. Informed consent was obtained prior to BM puncture in each case. The study was approved by the Institutional Review Board (Ethics Committee) of the Medical University of Vienna. Mononuclear cells (MNC) were isolated using Ficoll and stored in liquid nitrogen until used. After thawing the viability of cells ranged from 75% to 85% as assessed by trypan blue exclusion.RNA was isolated from BM or peripheral blood MNC using the RNeasy MinElute CleanupKit (Qiagen). cDNA was synthesized using RQ1 DNase buffer, RQ1 DNase and RQ1 DNase Stop Solution (Promega), Moloney murine leukemia virus reverse transcriptase, random primers, First Strand buffer, dNTPs (100 mM), and RNasin (Invitrogen) according to the manufacturer’s instructions. Details of the patient material used is deposited in Supplementary Information; Patient information).
MNC were cultured in 96-well plates (TPP) in RPMI-1640 medium (Lonza) with 10% fetal calf serum in the absence or presence of JQ1 (50–5,000 nM) at 37°C for 48 hours. AML MNC were incubated with JQ1 in the presence of 100 ng/ml recombinant human (rh) G-CSF (Amgen), 100 ng/ml rhSCF (Peprotech) and 100 ng/ml rhIL-3 (Novartis Pharma AG). CML BP cells were incubated with JQ1 without cytokines. After incubation, 0.5 µCi 3H-thymidine (Perkin Elmer) was added for 16 hours. Cells were then harvested on filter membranes in a Filtermate 196 harvester (Packard Bioscience). Filters were air-dried and the bound radioactivity was measured in a Top-Count NXT β-counter (Packard Bioscience). All experiments were performed in triplicates. Proliferation was calculated as percent of medium control, and the inhibitory effects of JQ1 were expressed as IC50 values.
Statistical analyses
Results are expressed as means ± standard error of the mean (SEM). If not stated otherwise statistical significance was calculated by two-tailed unpaired _t_-test on two experimental conditions with P < 0.05 considered statistically significant. Statistical significance level was set as follows: ****, p<0.0001 ***, p<0.001, **, p<0.01, *, p<0.05. No statistical methods were used to predetermine sample size.
Extended Data
Extended Data Figure 1. Multiplexed shRNAmir screening for chromatin-associated dependencies in AML maintenance and BET resistance.
a, Scatter plot illustrating the correlation of normalized reads per shRNA at all three time points (T0, T1 and T2; top to bottom) compared to the plasmid pool in all three independent replicates (left to right). The schematic to the right illustrates the different sampling time points subjected to deep sequencing. Top hits enriched under JQ1 treatment and positive controls from the initial negative-selection screen (T1) are marked with colored dots according to the legend on the right. b, Pooled negative-selection screening depicting changes in representation of 2917 shRNAs during 7 days of culture. shRNA fold depletion values were calculated by dividing the number of reads after 7 days of culture (T1) by the number of reads obtained from plasmid pool, and are plotted as the mean of three replicates in ascending order. Completely depleted shRNAs (0 reads at T1) obtained a fold depletion value of 10-3. Positive control shRNAs targeting essential genes are marked in red; negative-control shRNAs are depicted in green. c, Scatter plot depicting all genes ranked by the sum of their average depletion score of all shRNAs across all three replicates. Top scoring hits were defined as genes for which at least two shRNAs showed an average depletion of 8-fold after 7 days of shRNA expression and are marked in red (45 genes) d, IC50 determination for JQ1 in murine MLL/AF9;NrasG12D AML cells (RN2). Obtained numbers of viable cells ml-1 were normalized to DMSO (n=3, mean±SEM). e, Table showing the top 10 enriched shRNAs at T2. shRNAs targeting Suz12, Dnmt3a and Psip1 are strongly enriched in all three independent replicates. f, Relative mRNA abundance (RPKM) of shRNA target genes in JQ1-resistant leukemia cells expressing the indicated shRNAs, plotted relative to leukemia cells expressing Ren.713.
Extended Data Figure 2. PRC2 suppression confers resistance to JQ1.
a, Competitive proliferation assays of MLL/AF9;NrasG12D leukemia cells transduced with pLMN constructs expressing the indicated shRNAs. Shown is the fraction of GFP+/shRNA+ cells (relative to the initial ratio) under treatment with JQ1 (50 nM) or DMSO over time (continuation from Fig. 1d). JQ1 treatment was initiated at the indicated time points (red arrow). b, Competitive proliferation assays evaluating one validated shRNA targeting Eed as an additional core component of the PRC2 complex (as in a). c, Competitive proliferation assays of Tet-on competent MLL/AF9;NrasG12D leukemia cells expressing the indicated shRNAs from a Tet‐inducible vector (pRT3GEN-miR30). Transduced cells were selected with G418 and subsequently mixed with wild-type (wt) cells in a ratio of 95% to 5%. shRNA-expression was induced using dox treatment (1 µg/ml; from day 0), and the fraction of GFP+ cells was measured over time and plotted relative to day two. JQ1 treatment (50 nM) was initiated in one of the duplicate samples at the indicated time point (red arrow). Once the percentage of viable cells was below 10%, measurements were discontinued (indicated in the graph by the discontinuation of the respective sample). The negative effect induced by Suz12 suppression is reverted upon treatment with JQ1, which is not the case when Myc is suppressed. d, Bar chart showing colony forming cell frequencies of MLL/AF9;NrasG12D leukemia cells expressing Ren.713 or Suz12.1676 shRNAs in the presence of DMSO or 200nM JQ1; type 1: myeloblasts; type 2: maturing myeloblasts, type 3: terminally differentiated myeloid cells (n=3, mean±SEM). e, Pie charts depicting the fraction of JQ1 response genes which are re-expressed to the indicated extent in mouse resistant AML cells expressing shRNAs targeting Dnmt3a and Psip1 (continuation of Fig. 2b). JQ1-response genes (Fig. 2a) were grouped into four categories based on the divergence of their expression in resistant AML compared to AML expressing Ren.713 (not changed compared to expression after 24h JQ1, less than 1.5 fold; restoration relative to DMSO control: full restoration, less than 1.5 fold; enhanced, above 1.5 fold; partial restoration, restored but less than 1.5 fold) f, Heat map showing the distribution of H3K27me3, H3K36me3 and H3K4me3 ChIPseq peaks (fold enrichment >5 over input, FDR<1%) relative to Brd4 enhancer and promoter binding sites in MLL/AF9;NrasG12D AML cells without and with JQ1 treatment. g, ChIPseq occupancy profiles of Brd4 and H3K27me3 and H3K36me3 chromatin marks at enhancer regions downstream of Myc and upstream of Tifab following 3 days of treatment with vehicle or JQ1 (25 nM). Y-axis reflects the number of normalized cumulative tag counts in each region.
Extended Data Figure 3. Changes in the regulatory landscape of BET inhibitor resistant mouse AML cells.
a, Global H3K27 acetylation density of Suz12.1842-expressing resistant MLL/AF9;NrasG12D leukemia cells under long term (LT) treatment with 50 nM JQ1 (red bar), after 4 days of drug withdrawal (orange bar) and in Ren.713 controls (blue bar; statistical significance determined using Student’s _t_-test). b, Left panel, sorted fold change (FC) ratios of H3K27ac peaks in long-term JQ1-treated MLL–AF9;NrasG12D leukemia cells expressing Suz12.1842 compared to cells expressing Ren.713 control shRNA. Included were all peaks showing >10 reads per million in at least one condition. Right panel, top 15 gained peaks and their associated genes (defined using the closest transcription start site, TSS) . The Myc proximal enhancer in the first intron of Pvt1 is highlighted in red as one of the most differentially enriched peaks (FC=4.18). c, QRT-PCR validation of presented H3K27ac ChIPseq at the indicated regions downstream of the Myc locus (n=3, mean±SEM, statistical significance determined using Student’s t-test). d, GSEA plots of three publicly available gene sets associated with signaling pathways comparing expression changes in resistant MLL/AF9;NrasG12D AML cells induced by suppression of PRC2 complex members, compared to control cells expressing Ren.713 shRNA (n=2) or empty vector (continuation of Fig. 2e and f). e, Core signature genes of KEGG-curated Wnt and TGF-beta gene sets with increased expression in resistant murine MLL-AF9/NrasG12D, compared to sensitive cells. Red colored bars indicate association with H3K27 methylation in JQ1-sensitive MLL/AF9;NrasG12D AML cells. f, H3K27 methylation density at three exemplified genes with high expression changes in JQ1-resistant murine AML.
Extended Data Figure 4. Resistant MLL/AF9;NrasG12D AMLs generated through Suz12 suppression are not enriched for LSC-associated surface markers or expression signatures.
a, Immunophenotyping of JQ1-resistant mouse AML cells expressing two independent Suz12 shRNAs stably cultured in 50 nM JQ1 for >4 weeks (LT) compared to cells expressing Ren.713 control shRNA. Data are representative of two independent biological replicates. b, Percentage of c-kit+ cells in CD45.2+ bone marrow cells isolated from terminally diseased CD45.1+ mice following transplantation with MLL/AF9;NrasG12D cells expressing Ren.713 or Suz12.1676 and in-vivo treatment with DMSO carrier or JQ1 (50 mg/kg/d) (n=5, mean±SEM). c, GSEA analysis evaluating changes in macrophage and LSC gene signatures in resistant MLL/AF9;NrasG12D AML expressing three PRC2 shRNAs compared to cells expressing Ren.713 shRNA (n=2) or empty vector (see also Fig. 2e,f).
Extended Data Figure 5. Comparison of JQ1 response genes in sensitive and resistant cancer cell lines.
a, JQ1 sensitivity profiling in 246 human cancer cell lines of different tissue contexts. Shown are GI50 values determined using Alamar blue staining after 72h. b, PCA analysis of steady-state transcriptomes (based on RPKM) and c, Transcription changes (based on fold change, FC) following 2h of JQ1 treatment (200 nM) in indicated sensitive and resistant cancer cell lines of different tissue context. Steady-state profiles cluster based on tissue context, while neither baseline nor dynamic expression analysis can accurately distinguish sensitive and resistant contexts. d, MYC mRNA levels (RPKM) at different time points after JQ1 treatment (200 nM) in indicated cell lines, relative to levels in DMSO-treated cells. Individual cell line pairs are grouped for their tissue context and colored according to their sensitivities (green, sensitive; red, resistant). e, Number of genes 2-fold up- or downregulated upon JQ1 treatment (200 nM) after 2h and 24h (minimum expression >3 RPKM). f, Pairwise overlap of commonly up- or downregulated genes after 2h of JQ1 treatment (200 nM) relative to DMSO control. Each cell color corresponds to the relative number of commonly up- or downregulated genes in the cell lines listed in the respective row and column. The total number of genes regulated per cell line is indicated in black next to each cell line. Only little overlap is observed between cell lines of the same tissue context as well as between JQ1 sensitive or resistant cell lines.
Extended Data Figure 6. shRNA-based analysis of BRD2/3/4-dependent target genes and detailed analysis of effects on HEXIM1 and MYC transcription in different cell lines.
a, Determination of knockdown levels of a set of BRD2, BRD3 and BRD4 shRNAs determined using an established fluorescence-based shRNAmir reporter assay11. Knockdown levels were quantified relative to Ren.713 control shRNA. b, Competitive proliferation assays in MOLM-13 cells functionally evaluating the potency of BRD2, BRD3 and BRD4 shRNAs over time using a Tet‐regulated vector (pRT3GEN) in presence of doxycycline. Only shRNAs targeting BRD4 induce a proliferative disadvantage and lead to rapid depletion of GFP+ cells over time. Red labels indicate most potent shRNAs based on results obtained from reporter and competitive proliferation assay. c, Determination of BRD2, BRD3 and BRD4 mRNA levels in K-562 and MOLM-13 cells expressing the indicated shRNA. d, Number of genes commonly up- or down-regulated with a fold change (FC) >3 in MOLM-13 or K-562 cells following expression of indicated validated shRNAs or treatment with 200 nM JQ1 for 24h. JQ1-induced expression changes show the largest overlap with cells expressing a validated BRD4 shRNA, suggesting that suppression of BRD4 is the key effector mechanism of JQ1 in leukemia. e, Venn diagrams showing the overlap of genes commonly up- or down-regulated following 2h and 24h of JQ1 either in all contexts, or specifically in sensitive or resistant leukemias. f, HEXIM1/Hexim1 expression (RPKM) in all 18 analyzed human cell lines and murine MLL/AF9;NrasG12D AML after 2h and 24h treatment with 200 nM JQ1, compared to DMSO control (statistical significance was determined using a paired Student’s _t_-test). g, Relative MYC mRNA levels determined by qRT-PCR quantification in the indicated cell lines after incubation with 200 nM JQ1 measured at different treatment time points. Cell lines are grouped according to their sensitivity and the respective IC50 is presented below. Resistant cell lines rapidly restore MYC transcription (n=3, mean±SEM).
Extended Data Figure 7. Dynamic STARR-seq analysis of enhancer activity in K-562 cells.
a, Schematic representation of the STARR-seq cloning and screening strategy in K-562 cells. BACs available for the extended MYC locus (covering approximately 91% of a 3.1 MB region at the MYC locus) and 25 genic control BACs were fragmented and cloned into a modified STARR-seq vector containing a minimal MYC promoter. This library was then screened for enhancer activity using STARR-seq in K-562 cells with or without 250 nM JQ1. The schematic shows the underlying principle of STARR-seq: a bona-fide enhancer can activate its own transcription from a minimal MYC promoter. mRNA corresponding to active enhancer elements will therefore become more abundant among the cellular RNA compared to inactive fragments. b, PVT1 mRNA levels (RPKM) at different time points after JQ1 treatment (200 nM) in indicated cell lines, relative to levels in DMSO-treated cells. PVT1 expression is generally reduced upon JQ1 treatment indicating no association with enhancer activation.
Extended Data Figure 8. Analysis and functional validation of Wnt signaling as a key driver of BET resistance.
a, Protein levels of TCF4 and IGF2BP1 in K-562 cells transduced with pRT3GEN expressing indicated shRNAs after 7 days of doxycycline treatment, compared to Ren.713 and wt control samples. b, Competitive proliferation assay of JQ1-sensitive MOLM-13 cells expressing GFP-linked TCF4 cDNA or empty vector. Plotted is the relative fraction of GFP+ cells 72h after JQ1 treatment using the indicated doses (n=3, means±SEM, ***, p<0.001, **, p<0.01, *, p<0.05 as determined by Student’s _t_-test). Cells over-expressing TCF4 exhibit a dose-dependent competitive advantage under JQ1 treatment. c, Protein levels of TCF4 after overexpression of TCF4 cDNA subcloned into pMSCV-PGK-NeoR-IRES-GFP in MOLM-13 after 4 weeks of G418 (0.5 mg/ml) selection, compared to MOLM-13 transduced with empty control vector and to TCF4 protein levels in K-562 cells. d, ChIP qRT-PCR analysis of TCF7L2 binding to AXIN2, SP5, MYC promoter and the PVT1 enhancer element in K-562 cells at indicated time points after treatment with 200 nM JQ1 (n=2 biological replicates, mean±SEM). TCF7L2 binding increases gradually over time at promoters of Wnt target genes and the PVT1 enhancer at the MYC locus. e, Protein levels of Apc in 3T3 murine fibroblast cells 7 days after infection with the indicated shRNAs cloned into pLMP compared to Ren.713 and wt control samples. f, Relative Axin2 mRNA expression levels, determined by qRT-PCR normalized to B2m, after expression of the indicated shRNAs targeting Apc. g, Competitive proliferation assays of MLL/AF9;NrasG12D leukemia cells transduced with pLMP constructs expressing shRNAs targeting Apc, in combination with JQ1 (50 nM) or DMSO over time (continuation from Fig. 4d). h, Top: bioluminescent imaging of mice transplanted with 105 MLL/AF9;NrasG12D leukemia cells expressing constitutively active Ctnnb1 (Ctnnb1.4x). Treatment with JQ1 (50 mg−1 kg−1 d−1) or DMSO carrier started at d1 after injection. Bottom: Kaplan–Meier survival curves of control and JQ1-treated mice demonstrate decreased survival rates in mice treated with JQ1 (n=5). Statistical significance was calculated using the log-rank test. i, Competitive proliferation assays of BCR/ABLp210;p53-/- and MLL/ENL;NrasG12D leukemia cells transduced with constitutively active Ctnnb1 (Ctnnb1.4x) or empty vector control in the absence or presence of JQ1. Measurements started 4 days after transduction together with JQ1 treatment (50 nM). Shown is the fraction of GFP+ cells (relative to initial) over time.
Extended Data Figure 9. Wnt signatures are generally associated with BET resistance and Wnt activation drives resistance of pancreatic cancer models.
The BETi_resistance_Rathert signature was defined by combining the core enriched genes from the two significant Wnt gene sets (KEGG_WNT_SIGNALING and ST_WNT_SIGNALING) in murine resistant AML, filtered for significant upregulation (DESeq padj<0.1). These were combined with the Wnt-associated genes found differentially expressed in resistant human leukemia cell lines and primary patient samples, resulting in a total of 26 genes. **a**, Left: Microarray expression data of all 26 signature genes was curated from the Cancer Cell Line Encyclopedia (CCLE)28 and normalized to the geometric mean of each individual gene throughout all samples (relative expression). The sum of the relative expression of all genes (resistance index) was plotted for all CCLE cell lines showing a GI50>450 nM (resistant, 54 total) or a GI50<150nM (sensitive, 55 total) based on our sensitivity profiling (statistical significance was determined using Student’s _t_-test). Right: GSEA plot comparing the expression of 26 signature genes associated with JQ1 resistance across 54 resistant (GI50>450 nM) and 55 sensitive cell lines (GI50<150nM) available from CCLE. **b**, Left: as in (a) the resistance index was calculated as the sum of relative expression values of all 26 signature genes, which were based on RPKM extracted from an independent RNA-seq dataset27. Plotted are all 49 resistant (GI50>450 nM) and all 50 sensitive (GI50<150nM) cell lines analyzed in both sensitivity profiling and RNA-seq27 (statistical significance determined using Student’s _t_-test). Right: GSEA plot comparing the expression of 26 signature genes associated with JQ1 resistance across 49 resistant (GI50>450 nM) and 50 sensitive cell lines (GI50<150nM) available from27. c, Competitive proliferation assays of murine pancreatic adenocarcinoma (KRPC2) cells transduced with pLEPC constructs expressing potent validated shRNAs targeting Apc, cultured in the presence of JQ1 (800 nM) or DMSO. Shown is the relative number of mCherry+ cells over time, relative to initial. d, Cell viability of murine KRPC2 was determined following 5 days of treatment with Pyrvinium and/or JQ1 as indicated (n=3, mean±SEM, statistical significance determined using Student’s _t_-test). The combination index (CIs) for drug combinations was calculated using the CompuSyn software and percent inhibition (fraction affected, Fa) resulting from combined action of the two drugs versus effects of either drug alone. CI values <1.0 indicate synergism of the two agents. e, As in (d) cell viability was determined for human ASPC-1 pancreatic cancer cells following 5 days of treatment with Pyrvinium and/or JQ1 as indicated (n=3, mean±SEM, statistical significance determined using Student’s _t_-test). The combination index (CIs) for drug combinations was obtained using percent inhibition (fraction affected, Fa) resulting from combined action of the two drugs versus effects of either drug alone.
Extended Data Figure 10. Expression analysis of Wnt-associated genes in primary AML patient samples.
a, Determination of JQ1 response profiles in 12 primary AML patient samples. Sensitivity was determined using 3H-thymidine uptake across different JQ1 concentrations. b, qRT-PCR analysis of mRNA levels of additional Wnt-associated genes in primary human leukemia samples relative to GAPDH (continuation of Fig. 4g). Patient groups with low JQ1 IC50 (<200nM, blue dots) were compared to patients with high IC50 (>500nM, red dots). Statistical significance was determined using a Student’s _t_-test. c, Definition of a JQ1 resistance index. Expression of HOXB4, TCF4 and CCND2 in each primary AML patient sample was normalized to the geometric mean of all samples. The sum of these relative expression values of all three genes were added up to a resistance index, which was plotted in comparison to the JQ1 IC50 of each sensitive (IC50<200 nM, blue dots) and resistant (IC50>500 nM, red dots) AML patient sample. Two tailed Pearson correlation coefficient R and p-value are shown.
Supplementary Material
ED Figure 1
ED Figure 2
ED Figure 3
ED Figure 4
ED Figure 5
ED Figure 6
ED Figure 7
ED Figure 8
ED Figure 9
ED Figure 10
Supplementary Information
Acknowledgements
We thank M. Weißenböck, B. Hopfgartner and M. Fellner for excellent technical support, S.-M. Kula and the IMP/IMBA Molecular Biology Service for help with library construction, G. Schmauß, T. Lendl, M. Weninger and G. Stengl and the Biooptics Service Facility for FACS; A. Sommer and his team at Campus Science Support Facilities (www.csf.ac.at) for Illumina sequencing, S.W. Lowe for reagents and discussions, and all members of the Zuberlab for reagents, protocols and fruitful discussions. This research was funded by a Starting Grant of the European Research Council (ERC n° 336860; to J.Z.), SFB grants F4704 and F4710 of the Austrian Science Fund (FWF), a Fellowship of the People Programme (Marie Curie Actions) of the European Union (to P.R.). F.M. is an EMBO long-term fellow (EMBO ALTF 491-2014) and research in the Starklab is supported by an ERC Starting grant (n° 242922; to A.S.). The Zuberlab and research at the IMP is generously supported by Boehringer Ingelheim.
Footnotes
Author contributions
P.R. designed the shRNA library and performed the RNAi screen, P.R. and M.R. planned and performed most experiments and analyzed data. T.N. and E.A. performed bioinformatics analysis of RNA- and ChIP-seq data. F.M., Ł.M.B. and A.S. performed and analyzed STARR-seq experiments. M.M. generated validated BRD2/3/4 shRNAs and performed experiments. J.R-S. and C.V. contributed ChIP-seq data and provided advice. B.P., S.C-R. and P.V. performed analyses in primary patient samples. S.D. performed experiments in PDAC. T.H. improved protocols for shRNA deep-sequencing. J.J. contributed critical reagents and performed CFU assays. L.E.D. provided critical reagents. N.S., U.T-G. and D.G. profiled JQ1 sensitivity and performed RNAseq in human cell lines. M.P. and N.K. provided critical advice and support. J.Z. designed experiments, analyzed data, and supervised this research. P.R., M.R. and J.Z. co-wrote the paper.
Author Information
All RNA-seq, ChIPseq and STARR-seq data are deposited at Gene Expression Omnibus (Accession GSE63782). Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of the paper.
The authors declare no competing financial interests.
References
- 1.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asangani IA, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–82. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puissant A, et al. Targeting MYCN in Neuroblastoma by BET Bromodomain Inhibition. Cancer Discov. 2013;3:308–23. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herait PE, et al. BET-bromodomain inhibitor OTX015 shows clinically meaningful activity at nontoxic doses: interim results of an ongoing phase I trial in hematologic malignancies. Proc 105th Annu Meet Am Assoc Cancer Res. 2014 Apr 5-9; Abstract CT231. [Google Scholar]
- 7.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 8.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roe J-S, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Mol Cell. 2015:1–12. doi: 10.1016/j.molcel.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 12.De Raedt T, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature. 2014;514:247–51. doi: 10.1038/nature13561. [DOI] [PubMed] [Google Scholar]
- 13.Neff T, et al. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc Natl Acad Sci U S A. 2012;109:5028–33. doi: 10.1073/pnas.1202258109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J, et al. The Polycomb complex PRC2 supports aberrant self-renewal in a mouse model of MLL-AF9;Nras(G12D) acute myeloid leukemia. Oncogene. 2013;32:930–8. doi: 10.1038/onc.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuber J, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011;25:1628–40. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–55. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 17.Rahman S, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–52. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ballaré C, et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat Struct Mol Biol. 2012;19:1257–65. doi: 10.1038/nsmb.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi J, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27:2648–62. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He TC, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–12. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 22.Reya T, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–14. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–3. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnold CD, et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–7. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 25.Noubissi FK, et al. CRD-BP mediates stabilization of betaTrCP1 and c-myc mRNA in response to beta-catenin signalling. Nature. 2006;441:898–901. doi: 10.1038/nature04839. [DOI] [PubMed] [Google Scholar]
- 26.Kolligs FT, et al. ITF-2, a downstream target of the Wnt/TCF pathway, is activated in human cancers with beta-catenin defects and promotes neoplastic transformation. Cancer Cell. 2002;1:145–155. doi: 10.1016/s1535-6108(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 27.Klijn C, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2014;33:306–312. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 28.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–307. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorne Ca, et al. Small-molecule inhibition of Wnt signaling through activation of casein kinase 1α. Nat Chem Biol. 2010;6:829–836. doi: 10.1038/nchembio.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knoechel B, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46:364–70. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fong CY, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015 doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fellmann C, et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol Cell. 2011;41:733–46. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dow LE, et al. A pipeline for the generation of shRNA transgenic mice. Nat Protoc. 2012;7:374–93. doi: 10.1038/nprot.2011.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuber J, et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009;23:877–89. doi: 10.1101/gad.1771409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lito P, et al. Disruption of CRAF-Mediated MEK activation is required for effective mek inhibition in KRAS mutant tumors. Cancer Cell. 2014;25:697–710. doi: 10.1016/j.ccr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. 2001:3243–3248. doi: 10.1101/gad.943001.GENES. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 39.Chou T. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 40.Zuber J, et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2011;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:10. [Google Scholar]
- 42.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mortazavi A, Williams Ba, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 48.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 51.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett JM, et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann Intern Med. 1985;103:620–625. doi: 10.7326/0003-4819-103-4-620. [DOI] [PubMed] [Google Scholar]
- 54.Bennett JM, et al. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol. 1976;33:451–458. doi: 10.1111/j.1365-2141.1976.tb03563.x. [DOI] [PubMed] [Google Scholar]
- 55.Vardiman JW, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–51. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ED Figure 1
ED Figure 2
ED Figure 3
ED Figure 4
ED Figure 5
ED Figure 6
ED Figure 7
ED Figure 8
ED Figure 9
ED Figure 10
Supplementary Information